Abstract
We recently reported the inflammation of the cystic fibrosis (CF) mouse small intestine, and we hypothesized bacterial overgrowth as a possible cause. Quantitative PCR of bacterial 16S genomic DNA in the CF mouse small intestine revealed an increase of greater than 40-fold compared to controls. Sequencing of 16S PCR products and Gram staining showed that the majority of bacteria in the CF mouse intestine were gram negative. Bacteria were observed to colonize the mucus that accumulates in the intestinal lumen of mice with CF. Impaired Paneth cell defenses were suggested by observation of partially dispersed Paneth granules in the mucus plugs of CF mouse intestinal crypts, and this mucus was strongly immunoreactive for Paneth cell bactericidal products. The role of bacterial overgrowth in intestinal inflammation in CF was tested by treating mice with oral antibiotics (ciprofloxacin and metronidazole) for 3 weeks, which reduced bacterial load in the CF mouse small intestine over 400-fold. Antibiotic treatment decreased the expression of the inflammation-related genes mast cell protease 2, leucine-rich α2 glycoprotein/leucine-rich high endothelial venule glycoprotein, suppressor of cytokine signaling 3, hematopoietic cell transcript 1, and resistin-like molecule β/found in inflammatory zone 2, all of which were no longer expressed at levels significantly different from control levels. The reduction of intestinal bacteria also significantly improved the growth of CF mice but had no effect on the growth of wild-type mice. These data suggest that bacterial overgrowth in the CF mouse small intestine has a role in inflammation and contributes to the failure to thrive in this mouse model of CF.
Cystic fibrosis (CF) is caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene (35). CFTR is a cAMP-activated Cl− channel expressed in epithelial cells, and it also regulates the activity of other apically located electrolyte transport proteins (12). Loss of CFTR function results in the secretion of fluid at a lower volume than normal and with abnormal electrolyte composition. In the intestine, this results in a dehydrated state of the lumen, which is believed to contribute to the insolubility of secreted mucus and glycoproteins. As a consequence, intestinal obstruction occurs in the form of meconium ileus in newborns and distal intestinal obstructive syndrome in older patients (8).
Although the primary CF defect results in altered electrolyte transport, inflammation is a hallmark of the disease. Inflammation in CF occurs in the airways (2) and probably also in the gastrointestinal tract (34, 40). Studies of the small intestine in humans with CF reported an increase in mononuclear cells in the duodenum (34) and increases in luminal albumin, immunoglobulins, eosinophil cationic protein, neutrophil elastase, interleukin-1β, and interleukin-8, which suggest increased epithelial permeability and inflammation (40). The cause of intestinal inflammation in humans with CF is not known, but 30 to 40% of CF patients have been reported to have microbial overgrowth in the small intestine (23, 30).
The CFTR-deficient mouse serves as a model of the gastrointestinal complications of CF, and these mice exhibit pronounced intestinal obstruction due to decreased fluid secretion and the accumulation of poorly cleared mucus and glycoprotein secretions (13). It has recently been reported that these mice have inflammation of the small intestine with infiltration of mast cells and neutrophils, as well as upregulation of several inflammation-associated genes (29). Thus, the CFTR null mouse will be useful to explore intestinal inflammation in CF.
While there is a strong association of microbial infection and inflammation in CF, accumulating evidence indicates that susceptibility to inappropriate inflammation, at least in the airways, may be inherent to the diseased tissues even in the absence of specific pathogenic microbial colonization (26). A possible mechanism for an inherent inflammation in CF has been suggested to result from misfolding of ΔF508 CFTR, the protein resulting from the most common CFTR mutation. Misfolded CFTR has been shown in cultured cells to induce endoplasmic reticulum stress responses leading to proinflammatory NF-κB activation (20, 47). The CFTR null mouse does not express CFTR and, therefore, is not expected to have an inherent inflammation caused by protein misfolding. Instead, the observed inflammation in the CF mouse intestine likely occurs as a result of the altered luminal environment caused by the decrease in fluid secretion and the accumulation of secreted mucus and glycoproteins. We propose that the altered environment allows bacterial overgrowth, which leads to inflammation.
In the present study we used the CF mouse to test for a role of bacterial overgrowth in inflammation of the small intestine. The data demonstrate that bacterial overgrowth occurs in the CF mouse intestine and that a reduction of bacterial load reduces immune cell infiltration and the expression of inflammatory genes and improves the body weight of these mice.
MATERIALS AND METHODS
Animals.
CFTR+/− mice (cftrtm1UNC) were obtained from Jackson Laboratories and were backcrossed on the C57BL/6 background until congenic (5). CFTR+/− mice were intercrossed to obtain wild-type (CFTR+/+) and CF (CFTR−/−) mice. Mice of both sexes were used between 6 to 9 weeks of age and, with the exception of body weights, no gender-related differences in the measured parameters were observed. To prevent lethal intestinal obstruction, all mice were maintained on a complete elemental liquid diet (Peptamen; Nestlé, Deerfield, Ill.) from the age of 10 days (7). Endogenous intestinal bacteria were reduced by the use of the broad-spectrum antibiotics ciprofloxacin (50 mg/kg of body weight/day) and metronidazole (100 mg/kg of body weight/day) (Sigma-Aldrich, St. Louis, Mo.) for 3 weeks beginning at weaning (postnatal day 20). The antibiotics were administered orally by addition to the liquid diet of experimental groups of mice. Some wild-type mice were placed on a restricted caloric intake (pair fed) to match Peptamen consumption by CF mice. All procedures were approved by the University of Kansas Medical Center Institutional Animal Care and Use Committee.
Analysis of gene expression.
Total RNA was prepared from the entire small intestine by the TriZol method as previously described (29). Real-time quantitative reverse transcription-PCR (QRT-PCR) was performed by using a LightCycler (Roche, Indianapolis, Ind.) instrument. Expression of the following inflammatory genes was measured by using the previously described gene-specific primer pairs (29): resistin-like molecule β/found in inflammatory zone 2 (RELMβ/FIZZ2), leucine-rich α2 glycoprotein/leucine-rich high endothelial venule glycoprotein (Lrg/Lrhg), hematopoietic cell transcript 1 (HemT1), mast cell protease 2 (Mcpt2), and suppressor of cytokine signaling 3. Data are expressed relative to GAPDH (glutaraldehyde-3-phosphate dehydrogenase) mRNA levels which are not changed in the CF mouse intestine (29).
Measurement of bacterial load and classification of bacteria.
Mice were sacrificed with CO2 gas; the small intestines were ligated at the stomach and cecum and removed in their entirety. The ligatures were removed, and 30 ml of room temperature phosphate-buffered saline (PBS) containing a mucolytic agent (10 mM dithiothreitol [36, 38]) was flushed through the lumens. The flushed material was centrifuged at 20,000 × g for 30 min to pellet bacteria. The pellet was processed to extract microbial DNA by using a stool DNA kit, following the supplier's instructions, and included the option of heating the bacterial suspension in the supplied ASL buffer (proprietary composition) for 5 min in a boiling water bath at step 3 of the protocol (QIAGEN, Valencia, Calif.). This procedure was equally effective at extracting genomic DNA from laboratory strains of gram-negative (Escherichia coli) and gram-positive (Enterococcus faecalis) bacteria (data not shown), and this kit was recently reported to be as effective as the bead-beating technique (24). Microbial load was measured by real-time quantitative PCR (QPCR) by using microbial 16S (small ribosomal subunit gene)-specific primers: plus primer, 5′-TCC TAC GGG AGG CAG CAG T A-3′; minus primer, 5′-GGA CTA CCA GGG TAT CTA ATC CTG TT-3′ (28). The 16S PCR product from a lab strain of E. coli (XL1-Blue; Stratagene) was cloned into the pDRIVE plasmid (QIAGEN), and linearized plasmid was used to generate a standard curve for copy number determinations in the real-time QPCR assays as described (29). The minimum detectable copy number was 27 copies per reaction. When a sample had undetectable 16S DNA, the minimum copy number value was used for analysis.
To determine which bacterial species were present in the wild-type and CF mouse small intestine, pools of equal amounts of bacterial DNA were made from four wild-type and four CF samples. The pooled DNA samples were amplified by PCR by using the 16S primers, cloned, transformed into XL1-Blue cells, and cultured on agar plates overnight at 37°C. About 100 colonies each from the wild-type and CF samples were picked at random to prepare plasmid DNA samples, which were submitted for DNA sequencing by using the T7 primer site in the plasmid. A BLAST search of the GenBank database for the 16S sequences was performed to identify the bacteria present.
Histology.
Mice (three to five per genotype and treatment group) were sacrificed with CO2 gas, the small intestines were removed in their entirety, and the lumens were flushed with ice-cold PBS. Tissue samples were fixed in 4% paraformaldehyde for paraffin sections or 4% paraformaldehyde and 1.6% glutaraldehyde for electron microscopic analysis. Paraffin sections (5-μm) were stained with hematoxylin and eosin for general histological evaluation, with Gram stain to determine gram-negative and gram-positive microbes, with an antibody to lysozyme (DakoCytomation, Carpinteria, Calif.), and with an antineutrophil antibody for neutrophils (29). Plastic sections (1-μm) were stained with the cationic dye toluidine blue to reveal bacteria (1). Thin sections (100-nm) were stained with osmium tetroxide, lead citrate, and uranyl acetate and were examined by electron microscope to assess the ultrastructure of the intestinal tissue and to verify the presence of bacteria.
Statistics.
Analysis of variance (ANOVA) with a post hoc Tukey's test was used (Systat software, Chicago, Ill.). P values of less than 0.05 were considered significant.
RESULTS
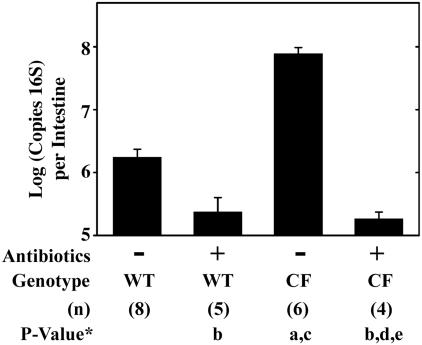
The CF mouse small intestine exhibits an innate type of inflammatory response (29), which led us to propose there might be bacterial overgrowth. To measure bacterial load in CF mice compared to the load in wild-type mice, the small intestines of wild-type and CF mice were isolated, and DNA was prepared from the flushed luminal contents. Bacterial DNA was quantified by using generic bacterial 16S PCR primers and a QPCR technique. The CF mouse had more than 40-fold more copies of 16S per intestine than the wild-type mouse (Fig. 1). The 16S PCR products were cloned, and we sequenced about 100 clones each from wild-type and CF mice. In wild-type mice, there was a diverse range of bacteria present (Table 1). The fact that about one-third of the 16S sequences matched uncultured bacteria and could not be assigned to known species demonstrates the utility of the PCR approach used, as these sequences would have been missed by culture techniques. In contrast to the diverse microbial ecology of the wild-type mouse small intestine, the CF mouse small intestine had bacteria that were predominantly in the family Enterobacteriaceae which constituted about 95% of the clones (Table 1). Quantitation of bacterial load by 16S analysis has a degree of uncertainty because different bacterial species have different numbers of 16S operons in their genomes (19; http://rrndb.cme.msu.edu). Based on the bacteria for which the number of 16S operons is known, we estimate that the error in quantitation of bacterial load could be as high as a 1.7-fold overestimation of bacteria in the CF mouse small intestine. Even if all the unknown species in the wild-type samples had one copy of the 16S gene, the error would be threefold. The diversity of species present in the CF mouse small intestine could also be underestimated to some extent because Enterobacteriaceae have seven copies of the 16S gene and may be overrepresented in the DNA amplified by PCR of the bacterial genomic extracts.
FIG. 1.
Bacterial load in the small intestine of wild-type and CF mice and effect of antibiotic treatment. Material was flushed from the small intestine lumen by using a mucolytic agent (10 mM dithiothreitol) in PBS. DNA was extracted from the pellet and analyzed by QRT-PCR by using bacterial 16S gene-specific primers. Data are presented as the means ± SEM. The number of mice (n) used for each group is given in parentheses below the graph. P values were determined by ANOVA with a post hoc Tukey test as follows: P < 0.05 versus untreated wild-type mice (a); P > 0.05 versus untreated wild-type mice (b); P < 0.05 versus antibiotic-treated wild-type mice (c); P > 0.05 versus antibiotic-treated wild-type mice (d); P < 0.05 versus untreated CF mice (e).
TABLE 1.
Classification of bacteria in wild-type and CF mouse small intestines by sequencing of cloned 16S PCR products
| Classificationa | No. of clones
|
|
|---|---|---|
| Wild-type mice | CF mice | |
| Uncultured bacteriab | 37 | 2 |
| Klebsiella oxytoca, Klebsiella pneumoniae, and Klebsiella granulomatis | 20 | 0 |
| Enterobacter aerogenes or Enterobacter asburiae | 13 | 0 |
| Other Enterobacteriacaec | 5 | 95 |
| Lactococcus lactis | 9 | 0 |
| Acinetobacter sp. | 5 | 0 |
| Pantoea sp. and Pantoea agglomerans | 4 | 0 |
| Unclassified β-Proteobacter | 2 | 0 |
| Helicobacter typhlonicus | 1 | 1 |
| Bacteroides acidofaciens | 1 | 0 |
| Streptococcus gordonii or Streptococcus parasanguinis | 1 | 0 |
| Clostridium perfringens | 0 | 6 |
When the 16S sequence was definitive, both genus and species are listed. When more than one species matched the sequence data or more than one species was unequivocally identified, all species are listed.
The wild-type group includes five clones that did not match anything in the GenBank database even though the cloned PCR product inserts were of the appropriate size for the 16S primers used (460 to 475 bp).
The sequences of the 16S PCR products in this group are identical between various family members (E. coli, Escherichia fergusonii, Escherichia vulneris, Photorhabdus liminescens, and Shigella flexneri).
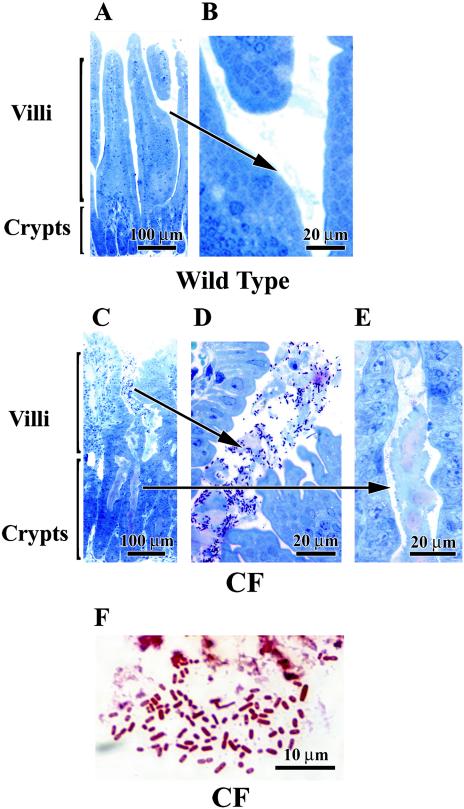
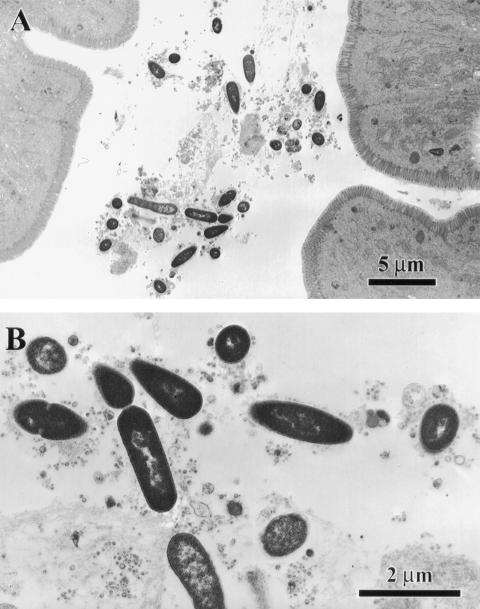
To visualize the relationship between mucus and bacteria in the intestine, tissue from the middle region of the small intestine (jejunum and ileum) was processed for 1-μm plastic sections and stained with toluidine blue. Histologically, the wild-type mouse intestine had little mucus in the crypt or in the lumen between villi (Fig. 2A), and bacteria were not apparent in the small amounts of mucus present (Fig. 2B). In the CF mouse intestine the presence of bacteria was confirmed at the level of light microscopy as well as ultrastructurally. As shown in Fig. 2C to E, the CF mouse small intestine has abundant mucus in the crypts (Fig. 2E) as well as in the intestinal lumen around villi (Fig. 2D), as previously reported in these mice (41). Numerous bacteria were observed in the mucus in the lumens (Fig. 2D) but not in the mucus in the crypts (Fig. 2E). Gram staining of the CF intestine showed that the majority of bacteria in the CF intestine were gram-negative bacilli (Fig. 2F), confirming the 16S gene analysis. By ultrastructural examination of the CF mouse small intestine, rod-shaped bacteria were observed embedded in the mucus along villus cells (Fig. 3). Bacteria were only seen in the lumens, and there was no apparent adherence to the epithelial cell surfaces or invasion of the tissue by bacteria.
FIG. 2.
Histological demonstration of bacterial overgrowth of the CF mouse small intestine. Intestinal tissue was prepared for 1-μm thick plastic sections and stained with toluidine blue. (A and B) Wild-type mouse small intestine. The crypt lumens are very small and there is only occasional mucus surrounding the villi (arrow). (C to E) CF mouse small intestine. In panels C and E, the crypts are dilated and filled with mucus but devoid of bacteria. In panel D, the villi are surrounded by mucus that is heavily colonized by bacteria (dark spots in the lumen). (F) Gram staining of paraffin section of CF mouse intestine. The bacteria present in mucus in the intestinal lumens are largely gram-negative microorganisms, as shown by the red staining. Panels of tissues with either confirmed gram-positive or confirmed gram-negative infections were processed in parallel for comparison of staining patterns (blue and red, respectively; data not shown).
FIG. 3.
Ultrastructural appearance of bacteria in the CF mouse small intestine lumen. The plastic-embedded samples shown in Fig. 2 were further processed for electron microscopy. (A) Embedded in the mucus along the villus epithelium are rod-shaped bacteria. (B) Bacteria shown at higher magnification.
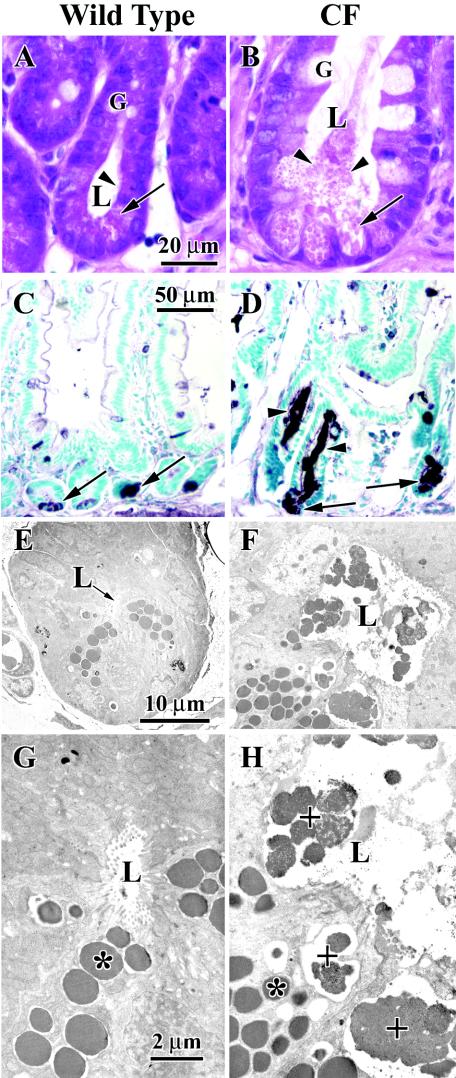
It was recently reported that CF mice have evidence of impaired Paneth cell defenses with decreased clearance of introduced bacteria (3). Therefore, we examined the morphology of crypts in the CF mouse intestine, paying special attention to the Paneth cells which contain eosinophilic secretory granules. Wild-type mouse tissue had narrow crypts without apparent material in the lumens, and Paneth cells at the base of the crypts had typical eosinophilic granules (Fig. 4A). In contrast, the crypts of CF mice were greatly dilated and filled with eosinophilic material which appeared to emanate from dilated Paneth cells at the base of the crypts (Fig. 4B). By using lysozyme immunohistochemistry as a Paneth cell granule marker, Paneth cells at the base of the crypts were immunostained in both wild-type (Fig. 4C) and CF (Fig. 4D) mouse intestines. Strikingly, the mucus in the crypts of mice with CF was strongly stained for lysozyme (Fig. 4D), indicating that Paneth cell-secreted proteins are trapped in the mucus-obstructed crypt lumen. At the ultrastructural level, the crypts of CF mice had morphologically identifiable Paneth granules in the crypt lumens that are only partially dissolved (Fig. 4D and F). In contrast, crypts in wild-type mice had identifiable granules only within the Paneth cells and not in the crypt lumens (Fig. 4C and E). These observations confirm those of Clarke et al. (3) and are consistent with their conclusion that Paneth cell innate defenses are impaired.
FIG. 4.
Histology and ultrastructure of small intestinal crypts in wild-type and CF mice. (A and B) Conventional hematoxylin and eosin staining. In the wild-type mouse tissue shown in panel A, the crypt lumen is small and clear. At the base of the crypts Paneth cells with characteristic eosinophilic secretory granules are observed (arrow), and there are traces of eosinophilic material in the lumen (arrowhead). Goblet cells are compact in appearance. In the CF mouse tissue shown in panel B, the crypt lumen is greatly dilated and filled with eosinophilic material (arrowheads) that appears to emanate from distended Paneth cells (arrow) at the base of the crypt. Goblet cells also appear distended. (C and D) Immunohistochemistry for the Paneth cell protein lysozyme. In the wild-type mouse tissue shown in panel C, ly-sozyme is primarily localized to the Paneth cells at the base of the crypt (arrows). In the CF mouse tissue shown in panel D, lysozyme immunoreactivity is greater and both the Paneth cells (arrows) and the crypt lumen (arrowheads) are strongly labeled. (E to H) Transmission electron microscopy. In the wild-type mouse tissue shown in panels E and G, Paneth cell granules (*) are observed inside Paneth cells, and there is little material in the small lumen (indicated by an arrow in panel E and by L in panel G). (F and H) In the CF mouse tissue Paneth cell granules are observed inside cells (*), and there are almost intact granules in the lumen (+) of the crypt. L, lumen; G, goblet.
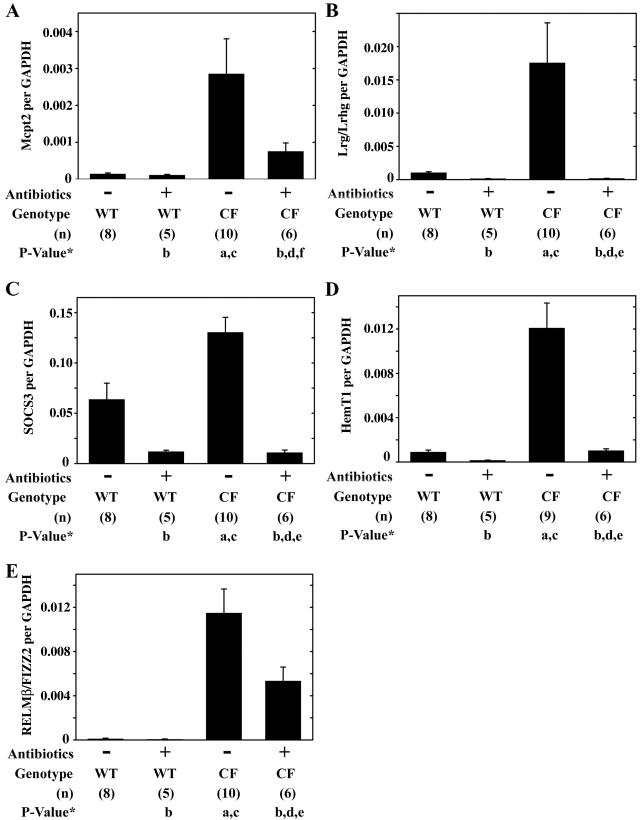
To explore the relationship between bacterial overgrowth and the intestinal phenotype of mice with CF, mice were treated with a combination of broad-spectrum antibiotics (ciprofloxacin and metronidazole). Antibiotic treatment significantly reduced the bacterial load in CF mice by over 400-fold, and after treatment the bacterial load was not significantly different from that in wild-type mice (Fig. 1). The CF phenotype of these mice includes increased expression of various inflammatory markers, infiltration of mast cells and neutrophils in the small intestine (29), and decreased body weight compared to wild-type mice (5, 29). We used QRT-PCR to measure gene expression in wild-type and CF mouse small intestine, without and with antibiotic treatment. The mast cell marker Mcpt2 was significantly increased in CF mice as previously reported (29), and after antibiotic treatment expression levels were reduced and were not significantly different from those of wild-type mice (Fig. 5A). The acute phase serum protein Lrg/Lrhg (45), which is a putative neutrophil marker (31) and is also expressed on mesenteric lymph node high endothelial venules (37), was significantly elevated in the CF mouse intestine and was significantly reduced after antibiotic treatment (Fig. 5B). Expression of the anti-inflammatory gene suppressor of cytokine signaling 3 (44) was significantly increased in CF mice compared to levels in wild-type mice and was significantly downregulated in the antibiotic-treated CF mouse intestine (Fig. 5C). The marker of blood cell proliferation, HemT1 (48), was significantly increased in the CF mouse intestine and was reduced to the levels of wild-type mice after antibiotic treatment (Fig. 5D). The final gene examined was RELMβ/FIZZ2 which is associated with bacterial colonization and inflammation (14, 16, 43) as well as cell growth (42). RELMβ/FIZZ2 expression was highly increased in the CF mouse small intestine, as previously reported (29); after antibiotic treatment RELMβ/FIZZ2 expression was reduced to a level that was intermediate between the levels of the wild-type and CF mouse intestine and was not statistically different from either (Fig. 5E). Antibiotic-treated wild-type mice had small decreases in expression of the genes measured, but the changes were not statistically significant by ANOVA versus untreated wild-type mice (Fig. 5).
FIG. 5.
Effects of antibiotic treatment on the expression of inflammatory genes in the small intestine of wild-type and CF mice. Wild-type (WT) and CF mice were untreated (−) or given oral antibiotics (+) for 3 weeks, and total RNA was prepared from the entire small intestine. Gene expression was determined by using QRT-PCR for the indicated genes, and data are expressed as copy number per copy of GAPDH. Data are presented as the means ± SEM. The number of mice (n) used for each group is given in parentheses below the graph. P values were determined by ANOVA with a post hoc Tukey’s test as follows: (a) P < 0.05 versus untreated wild-type mice; (b) P > 0.05 versus untreated wild-type mice; (c) P < 0.05 versus antibiotic-treated wild-type as follows; (d) P > 0.05 versus antibiotic-treated wild-type mice; (e) P < 0.05 versus untreated CF mice; (f) P > 0.05 versus untreated CF mice.
When fed ad libitum, CF mice are significantly smaller than wild-type mice (5). To evaluate the possibility that changes in gene expression in CF mice are due to reduced calorie intake, we measured the Peptamen consumption of CF mice and pair fed a group of wild-type mice; QRT-PCR was then used for three of the inflammation-associated genes analyzed above. CF mice consumed on average 20% ± 8% (females; seven wild-type and eight CF mice) and 24% ± 7% (males; nine wild-type and seven CF mice) less Peptamen than wild-type mice. After 3 weeks of caloric restriction, wild-type mice were smaller by 20% ± 3% (females; n = 3) and 24% ± 2% (males; n = 3) than ad libitum fed wild-type mice. Caloric restriction for 3 weeks did not affect gene expression. Pair-fed wild-type mice had 4.1 ± 1.2 copies of Mcpt2 per 104 copies of GAPDH (mean ± standard error of the mean [SEM]; n = 6), which was not significantly different than the number of copies in ad libitum fed wild-type mice (P > 0.9 by ANOVA) and was significantly less than the number of copies in CF mice (P < 0.02) (Fig. 5A). Calorie-restricted mice had 17 ± 3.3 copies of Lrg per 104 copies of GAPDH, which was not significantly different than the number of copies in ad libitum fed wild-type mice (P > 0.9) and was significantly less than the number of copies in CF mice (P < 0.05) (Fig. 5B). And calorie-restricted mice had 3.2 ± 0.62 copies of RELMβ/FIZZ2 per 104 copies of GAPDH, which was not significantly different than the value for ad libitum fed wild-type mice (P > 0.9) and was significantly less than the value for CF mice (P < 0.02) (Fig. 5E). Thus, reduced food consumption by wild-type mice did not affect expression of the inflammatory genes studied.
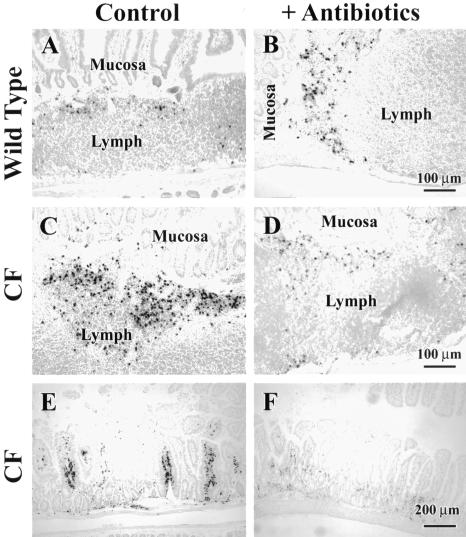
To histologically evaluate the effect of antibiotics on CF mouse intestinal inflammation, tissue was immunostained for neutrophils. Wild-type mice have some neutrophils in their small intestines, primarily near lymph nodules (Fig. 6A). After antibiotic treatment there was no appreciable effect on the numbers of neutrophils (Fig. 6B). As previously reported, CF mice have increased numbers of neutrophils both near lymph nodules (Fig. 6C) and frequently in the lamina propria in areas away from lymph nodules (Fig. 6E). After antibiotic treatment of CF mice, there was a decrease in neutrophil numbers near lymph nodules (Fig. 6D) and in the lamina propria (Fig. 6F), consistent with the decrease in Lrg/Lrhg gene expression (Fig. 5B).
FIG. 6.
Effects of antibiotic treatment on neutrophil infiltration in wild-type and CF mouse small intestine. Mice were untreated (Control) or given oral antibiotics (+Antibiotics) for 3 weeks, and paraffin sections of the central portion of the intestine (jejunum and ileum region) were prepared for neutrophil immunohistochemistry. (A) Untreated wild-type mouse small intestine has some neutrophils, mostly near lymph nodules (dark-staining structures). (B) Antibiotic-treated wild-type mouse intestine does not have appreciably different numbers of neutrophils compared to untreated wild-type mice. (C and E) Untreated CF mouse small intestine has increased neutrophils near lymph nodules, shown in panel C, and in the lamina propria, shown in panel E, compared to untreated wild-type mice. (D and F) Antibiotic-treated CF mouse intestine has fewer neutrophils near lymph nodules, shown in panel D, and in the lamina propria, shown in panel F, compared to untreated CF mice. Representative images from three to five mice per group are shown.
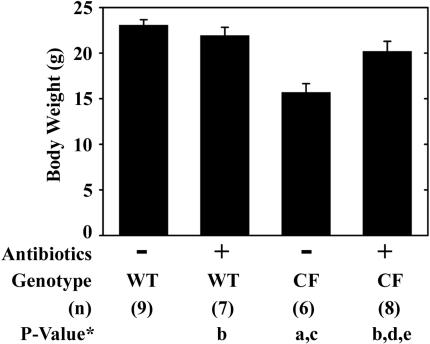
Finally, we assessed whether antibiotic treatment of CF mice would affect body weight gain. CF mice are about 30% smaller than wild-type mice on the same Peptamen liquid diet, as previously reported (5, 18, 46) (Fig. 7). The body weight of CF male mice at the end of the 3-week antibiotic treatment was significantly increased compared to untreated CF mice and was not significantly different than that of wild-type mice (Fig. 7). Importantly, antibiotic treatment had no effect on the body weight of wild-type mice (Fig. 7).
FIG. 7.
Effect of antibiotic treatment on body weights of wild-type and CF mice. Body weights of male mice at 6 weeks of age, either untreated or treated with oral antibiotic administration for 3 weeks, are shown. Data are presented as the means ± SEM. The number of mice (n) used for each group is given in parentheses below the graph. P values were determined by ANOVA with post hoc Tukey’s test as follows: (a) P < 0.05 versus untreated wild-type mice; (b) P > 0.05 versus untreated wild-type mice; (c) P < 0.05 versus antibiotic-treated wild-type mice; (d) P > 0.05 versus antibiotic-treated wild-type mice; (e) P < 0.05 versus untreated CF mice.
DISCUSSION
In this work we demonstrated that the CF mouse has small intestinal bacterial overgrowth. Classification of the bacteria in the CF mouse small intestine by 16S gene analysis revealed that there is a loss of species diversity compared to wild-type mice and that the microbes present are overwhelmingly gram-negative organisms of the Enterobacteriaceae family. When the bacterial load was reduced by broad-spectrum antibiotics, there were positive effects on the CF phenotype. Antibiotic-treated CF mice had significant reductions in inflammation as assessed by the measurement of gene expression and infiltration of neutrophils and also had improved body weight gain.
Small intestine bacterial overgrowth has been found in 30 to 40% of CF patients by breath testing techniques (23, 30), but the cause is unknown. The normal small intestine has low numbers of bacteria, and there are several mechanisms that play roles in minimizing microbial colonization (for a review, see reference 39).
First is the defense mechanism whereby most ingested microbes are killed by gastric acid in the stomach (22). However, stomach function is not believed to be compromised in CF patients (8), and so it is unlikely that impairment of this defense accounts for the intestinal bacterial overgrowth. Second is the mechanism whereby microbes that survive the stomach to enter the small intestine are continually swept along to the large intestine by peristalsis. The rate of transit through the small intestine normally outpaces the replication rate of microbes, limiting the accumulation of bacteria (17). Slower transit rates in the small intestine may play a role in bacterial overgrowth of the CF human small intestine, and there is evidence from several studies that orocecal transit time is increased in CF patients (4, 9, 23, 27). It is not currently known if orocecal transit time is altered in the CF mouse, and this remains an interesting possibility.
A third factor in epithelial innate defense is mucus secreted by goblet cells. Mucus glycoproteins contain carbohydrate structures which bind bacteria and reduce adherence of the microbes to the epithelial surfaces (25). Mucus-bound bacteria are then swept away down the intestinal tract, which limits their numbers in the small intestine (6). In the CF mouse intestine, mucus accumulates even though there is actually a small decrease in mucin gene expression (33) (S. Kaur and R. C. De Lisle, unpublished data). We observed that bacteria in the CF mouse intestine had colonized the abundant mucus that accumulates along the villus surfaces. This finding is consistent with the idea that the slow clearance of secreted mucus in the CF human intestine provides an anchorage for bacterial adherence and abnormal colonization.
The innate immune defenses of the Paneth cells constitute a fourth mechanism. The Paneth cells are at the base of intestinal crypts, and they secrete a family of α-defensins (cryptdins) and lysozyme that have bacteriostatic activities (32). The high levels of mucus that accumulate in the CF intestine may impair functions of the Paneth cell. Recently, it was demonstrated that partially dissolved Paneth cell granules can be observed in mucus-occluded small intestinal crypts in CF mice (3). In the study, intestinal obstruction was prevented by use of a laxative solution instead of the Peptamen liquid diet we used. It was further demonstrated that the extent of mucus accumulation and nearly intact granules in the lumen were increased when the laxative was removed from the drinking water. The investigators also presented evidence for impaired clearance of an introduced cryptdin-sensitive enteric pathogen (Salmonella enterica serovar Typhimurium). We confirmed in Peptamen-fed CF mice that Paneth cell granules released into the crypt lumens appear nearly intact. We extended these studies by showing high levels of Paneth cell-secreted bactericidal protein (lysozyme) (32) within the crypt lumens of CF mice. Interestingly, there were no bacteria in the crypts which have high levels of secreted Paneth cell bactericidal proteins, suggesting that these defenses are active in the crypt but not in the intestinal lumen in CF mice.
It appears that the loss of Paneth cell innate defenses alone is not sufficient for bacterial overgrowth and inflammation. A transgenic mouse that expresses a toxin under a Paneth cell-specific promoter lacks Paneth cells but does not exhibit bacterial overgrowth of the small intestine (10). We propose that two factors are needed for bacterial overgrowth in the CF intestine: (i) accumulated mucus along the villus surface which provides an anchorage for bacteria and (ii) impairment of Paneth cell defenses by mucus occlusion of the crypt which permits bacterial overgrowth of the small intestine.
A reduction of the bacterial load with broad-spectrum antibiotics reduced expression of inflammatory genes, indicating that bacterial overgrowth may be responsible for inflammation of the CF mouse small intestine. Since we did not observe invasion of the epithelium by bacteria, inflammation may likely be due to bacterial products released in the intestinal lumen (e.g., lipopolysaccharide) and not to an invasive pathogen. This needs to be confirmed by more direct means to measure levels of bacterial translocation to mesenteric lymph nodes and spleen in CF mice.
An important effect of the reduction in the bacterial load was an improvement in the growth of CF mice. The mechanism of the effect of antibiotics on body weight is not known but may be due to reduced competition for nutrients by intestinal bacteria (for a review, see reference 11), as well as to improved lipid digestion and absorption, which can be impaired by bile salt precipitation after bacterial deconjugation (15). While the data are consistent with a central role of bacterial overgrowth in CF pathologies in the small intestine, it is also possible that the positive effects of antibiotic therapy are due to the anti-inflammatory activities that some antibiotics exhibit (21). Whether the effects of antibiotics are due to anti-inflammatory activities and if such nonantibiotic effects are sufficient to improve the health of the CF mouse will require further study. Nevertheless, these studies show a dramatic reduction in intestinal inflammation and an improvement in the health of these animals when treated with broad-spectrum antibiotics.
The results suggest that inflammation in the CF intestine is associated with abnormal microbial growth rather than an inherent alteration of the immune system in the absence of functional CFTR. It remains possible that if the ΔF508 CFTR mutation were present, there would be an inherent inflammatory state in addition to the one we observed in the CFTR null mouse. It might be predicted that intestinal inflammation would be greater in ΔF508 CFTR mice than in CFTR null mice, and this needs to be determined.
In summary, there is bacterial overgrowth in the CF mouse small intestine which is associated with mucus accumulation and impaired Paneth cell innate defense functions. Accompanying bacterial overgrowth is inflammation. A reduction of the bacterial load significantly improves several parameters of CF pathology, including body weight gain. Given that small intestinal bacterial overgrowth is common in CF patients, it will be important to investigate the role of microbes in this disease in inflammation and failure to thrive.
Acknowledgments
We thank Larysa Stroganova for diligent maintenance of the CF mouse colony and accurate genotyping and Rosetta Barkley for her expert assistance with histological preparations.
This work was supported by NIH grant DK56791 to R.C.D.
Editor: J. D. Clements
REFERENCES
- 1.Caselli, M., M. Gaudio, C. M. Chiamenti, L. Trevisani, S. Sartori, L. Saragoni, P. Boldrini, A. Dentale, M. Ruina, and V. Alvisi. 1998. Histologic findings and Helicobacter pylori in duodenal biopsies. J. Clin. Gastroenterol. 26:74-80. [DOI] [PubMed] [Google Scholar]
- 2.Chmiel, J. F., M. Berger, and M. W. Konstan. 2002. The role of inflammation in the pathophysiology of CF lung disease. Clin. Rev. Allergy Immunol. 23:5-27. [DOI] [PubMed] [Google Scholar]
- 3.Clarke, L. L., L. R. Gawenis, E. M. Bradford, L. M. Judd, K. T. Boyle, J. E. Simpson, G. E. Shull, H. Tanabe, A. J. Ouellette, C. L. Franklin, and N. M. Walker. 2004. Abnormal Paneth cell granule dissolution and compromised resistance to bacterial colonization in the intestine of CF mice. Am. J. Physiol. Gastrointest. Liver Physiol. 286:G1050-G1058. [DOI] [PubMed] [Google Scholar]
- 4.Dalzell, A. M., N. S. Freestone, D. Billington, and D. P. Heaf. 1990. Small intestinal permeability and orocaecal transit time in cystic fibrosis. Arch. Dis. Child. 65:585-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Lisle, R. C., K. S. Isom, D. Ziemer, and C. U. Cotton. 2001. Changes in the exocrine pancreas secondary to altered small intestinal function in the CF mouse. Am. J. Physiol. Gastrointest. Liver Physiol. 281:G899-G906. [DOI] [PubMed] [Google Scholar]
- 6.Deplancke, B., and H. R. Gaskins. 2001. Microbial modulation of innate defense: goblet cells and the intestinal mucus layer. Am. J. Clin. Nutr. 73:1131S-1141S. [DOI] [PubMed] [Google Scholar]
- 7.Eckman, E. A., C. U. Cotton, D. M. Kube, and P. B. Davis. 1995. Dietary changes improve survival of CFTR S489X homozygous mutant mouse. Am. J. Physiol. Lung Cell. Mol. Physiol. 269:L625-L630. [DOI] [PubMed] [Google Scholar]
- 8.Eggermont, E. 1996. Gastrointestinal manifestations in cystic fibrosis. Eur. J. Gastroenterol. Hepatol. 8:731-738. [PubMed] [Google Scholar]
- 9.Escobar, H., M. Perdomo, F. Vasconez, C. Camarero, M. T. del Olmo, and L. Suarez. 1992. Intestinal permeability to 51Cr-EDTA and orocecal transit time in cystic fibrosis. J. Pediatr. Gastroenterol. Nutr. 14:204-207. [DOI] [PubMed] [Google Scholar]
- 10.Garabedian, E. M., L. J. Roberts, M. S. McNevin, and J. I. Gordon. 1997. Examining the role of Paneth cells in the small intestine by lineage ablation in transgenic mice. J. Biol. Chem. 272:23729-23740. [DOI] [PubMed] [Google Scholar]
- 11.Gaskins, H. R., C. T. Collier, and D. B. Anderson. 2002. Antibiotics as growth promotants: mode of action. Anim. Biotechnol. 13:29-42. [DOI] [PubMed] [Google Scholar]
- 12.Greger, R., R. Schreiber, M. Mall, A. Wissner, A. Hopf, M. Briel, M. Bleich, R. Warth, and K. Kunzelmann. 2001. Cystic fibrosis and CFTR. Pflugers Arch. 443(Suppl. 1):S3-S7. [DOI] [PubMed] [Google Scholar]
- 13.Grubb, B. R., and S. E. Gabriel. 1997. Intestinal physiology and pathology in gene-targeted mouse models of cystic fibrosis. Am. J. Physiol. Gastrointest. Liver Physiol. 273:G258-G266. [DOI] [PubMed] [Google Scholar]
- 14.He, W., M. L. Wang, H. Q. Jiang, C. M. Steppan, M. E. Shin, M. C. Thurnheer, J. J. Cebra, M. A. Lazar, and G. D. Wu. 2003. Bacterial colonization leads to the colonic secretion of RELMbeta/FIZZ2, a novel goblet cell-specific protein. Gastroenterology 125:1388-1397. [DOI] [PubMed] [Google Scholar]
- 15.Hofmann, A. F., and K. J. Mysels. 1992. Bile acid solubility and precipitation in vitro and in vivo: the role of conjugation, pH, and Ca2+ ions. J. Lipid Res. 33:617-626. [PubMed] [Google Scholar]
- 16.Holcomb, I. N., R. C Kabakoff, B. Chan, T. W. Baker, A. Gurney, W. Henzel, C. Nelson, H. B. Lowman, B. D. Wright, N. J. Skelton, G. D. Frantz, D. B. Tumas, F. V. Peale, Jr., D. L. Shelton, and C. C. Hebert. 2000. FIZZ1, a novel cysteine-rich secreted protein associated with pulmonary inflammation, defines a new gene family. EMBO J. 19:4046-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Husebye, E. 1999. The patterns of small bowel motility: physiology and implications in organic disease and functional disorders. Neurogastroenterol. Motil. 11:141-161. [DOI] [PubMed] [Google Scholar]
- 18.Ip, W. F., I. Bronsveld, G. Kent, M. Corey, and P. R. Durie. 1996. Exocrine pancreatic alterations in long-lived surviving cystic fibrosis mice. Pediatr. Res. 40:242-249. [DOI] [PubMed] [Google Scholar]
- 19.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knorre, A., M. Wagner, H. E. Schaefer, W. H. Colledge, and H. L. Pahl. 2002. DeltaF508-CFTR causes constitutive NF-kappaB activation through an ER-overload response in cystic fibrosis lungs. Biol. Chem. 383:271-282. [DOI] [PubMed] [Google Scholar]
- 21.Konstan, M. W., and P. B. Davis. 2002. Pharmacological approaches for the discovery and development of new anti-inflammatory agents for the treatment of cystic fibrosis. Adv. Drug Delivery Rev. 54:1409-1423. [DOI] [PubMed] [Google Scholar]
- 22.Laine, L., D. Ahnen, C. McClain, E. Solcia, and J. H. Walsh. 2000. Review article: potential gastrointestinal effects of long-term acid suppression with proton pump inhibitors. Aliment. Pharmacol. Ther. 14:651-668. [DOI] [PubMed] [Google Scholar]
- 23.Lewindon, P. J., T. A. Robb, D. J. Moore, G. P. Davidson, and A. J. Martin. 1998. Bowel dysfunction in cystic fibrosis: importance of breath testing. J. Paediatr. Child Health 34:79-82. [DOI] [PubMed] [Google Scholar]
- 24.Li, M., J. Gong, M. Cottrill, H. Yu, C. de Lange, J. Burton, and E. Topp. 2003. Evaluation of QIAamp DNA Stool Mini Kit for ecological studies of gut microbiota. J. Microbiol. Methods 54:13-20. [DOI] [PubMed] [Google Scholar]
- 25.Moncada, D. M., S. J. Kammanadiminti, and K. Chadee. 2003. Mucin and Toll-like receptors in host defense against intestinal parasites. Trends Parasitol. 19:305-311. [DOI] [PubMed] [Google Scholar]
- 26.Muhlebach, M. S., P. W. Stewart, M. W. Leigh, and T. L. Noah. 1999. Quantitation of inflammatory responses to bacteria in young cystic fibrosis and control patients. Am. J. Respir. Crit. Care Med. 160:186-191. [DOI] [PubMed] [Google Scholar]
- 27.Murphy, M. S., A. L. Brunetto, A. D. Pearson, M. A. Ghatei, R. Nelson, E. J. Eastham, S. R. Bloom, and A. A. Green. 1992. Gut hormones and gastrointestinal motility in children with cystic fibrosis. Dig. Dis. Sci. 37:187-192. [DOI] [PubMed] [Google Scholar]
- 28.Nadkarni, M. A., F. E. Martin, N. A. Jacques, and N. Hunter. 2002. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148:257-266. [DOI] [PubMed] [Google Scholar]
- 29.Norkina, O., S. Kaur, D. Ziemer, and R. C. De Lisle. 2004. Inflammation of the cystic fibrosis mouse small intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 286:G1032-G1041. [DOI] [PubMed] [Google Scholar]
- 30.O'Brien, S., H. Mulcahy, H. Fenlon, A. O'Broin, M. Casey, A. Burke, M. X. FitzGerald, and J. E. Hegarty. 1993. Intestinal bile acid malabsorption in cystic fibrosis. Gut 34:1137-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Donnell, L. C., L. J. Druhan, and B. R. Avalos. 2002. Molecular characterization and expression analysis of leucine-rich alpha2-glycoprotein, a novel marker of granulocytic differentiation. J. Leukoc. Biol. 72:478-485. [PubMed] [Google Scholar]
- 32.Ouellette, A. J. 1997. Paneth cells and innate immunity in the crypt microenvironment. Gastroenterology 113:1779-1784. [DOI] [PubMed] [Google Scholar]
- 33.Parmley, R. R., and S. J. Gendler. 1998. Cystic fibrosis mice lacking Muc1 have reduced amounts of intestinal mucus. J. Clin. Investig. 102:1798-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raia, V., L. Maiuri, G. De Ritis, B. De Vizia, L. Vacca, R. Conte, S. Auricchio, and M. Londei. 2000. Evidence of chronic inflammation in morphologically normal small intestine of cystic fibrosis patients. Pediatr. Res. 47:344-350. [DOI] [PubMed] [Google Scholar]
- 35.Riordan, J. R., J. M. Rommens, B.-S. Kerem, N. Alon, R. Rozmahel, Z. Grzelczak, J. Zielenski, S. Lok, N. Plavsic, J.-L. Chou, M. L. Drumm, M. C. Iannuzzi, F. S. Collins, and L.-C. Tsui. 1989. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science 245:1066-1073. [DOI] [PubMed] [Google Scholar]
- 36.Rose, M. C., C. F. Brown, J. Z. Jacoby III, W. S. Lynn, and B. Kaufman. 1987. Biochemical properties of tracheobronchial mucins from cystic fibrosis and non-cystic fibrosis individuals. Pediatr. Res. 22:545-551. [DOI] [PubMed] [Google Scholar]
- 37.Saito, K., T. Tanaka, H. Kanda, Y. Ebisuno, D. Izawa, S. Kawamoto, K. Okubo, and M. Miyasaka. 2002. Gene expression profiling of mucosal addressin cell adhesion molecule-1+ high endothelial venule cells (HEV) and identification of a leucine-rich HEV glycoprotein as a HEV marker. J. Immunol. 168:1050-1059. [DOI] [PubMed] [Google Scholar]
- 38.Sandberg, J. W., C. Lau, M. Jacomino, M. Finegold, and S. J. Henning. 1994. Improving access to intestinal stem cells as a step toward intestinal gene transfer. Hum. Gene Ther. 5:323-329. [DOI] [PubMed] [Google Scholar]
- 39.Singh, V. V., and P. P. Toskes. 2003. Small bowel bacterial overgrowth: presentation, diagnosis, and treatment. Curr. Gastroenterol. Rep. 5:365-372. [DOI] [PubMed] [Google Scholar]
- 40.Smyth, R. L., N. M. Croft, U. O'Hea, T. G. Marshall, and A. Ferguson. 2000. Intestinal inflammation in cystic fibrosis. Arch. Dis. Child. 82:394-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snouwaert, J. N., K. K. Brigman, A. M. Latour, N. N. Malouf, R. C. Boucher, O. Smithies, and B. H. Koller. 1992. An animal model for cystic fibrosis made by gene targeting. Science 257:1083-1088. [DOI] [PubMed] [Google Scholar]
- 42.Steppan, C. M., E. J. Brown, C. M. Wright, S. Bhat, R. R. Banerjee, C. Y. Dai, G. H. Enders, D. G. Silberg, X. Wen, G. D. Wu, and M. A. Lazar. 2001. A family of tissue-specific resistin-like molecules. Proc. Natl. Acad. Sci. USA 98:502-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stutz, A. M., L. A. Pickart, A. Trifilieff, T. Baumruker, E. Prieschl-Strassmayr, and M. Woisetschlager. 2003. The Th2 cell cytokines IL-4 and IL-13 regulate found in inflammatory zone 1/resistin-like molecule alpha gene expression by a STAT6 and CCAAT/enhancer-binding protein-dependent mechanism. J. Immunol. 170:1789-1796. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki, A., T. Hanada, K. Mitsuyama, T. Yoshida, S. Kamizono, T. Hoshino, M. Kubo, A. Yamashita, M. Okabe, K. Takeda, S. Akira, S. Matsumoto, A. Toyonaga, M. Sata, and A. Yoshimura. 2001. CIS3/SOCS3/SSI3 plays a negative regulatory role in STAT3 activation and intestinal inflammation. J. Exp. Med. 193:471-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takahashi, N., Y. Takahashi, and F. W. Putnam. 1985. Periodicity of leucine and tandem repetition of a 24-amino acid segment in the primary structure of leucine-rich alpha 2-glycoprotein of human serum. Proc. Natl. Acad. Sci. USA 82:1906-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Heeckeren, A. M., M. Schluchter, L. Xue, J. Alvarez, S. Freedman, J. St. George, and P. B. Davis. 2004. Nutritional effects on host response to lung infections with mucoid Pseudomonas aeruginosa in mice. Infect. Immun. 72:1479-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weber, A. J., G. Soong, R. Bryan, S. Saba, and A. Prince. 2001. Activation of NF-kappaB in airway epithelial cells is dependent on CFTR trafficking and Cl− channel function. Am. J. Physiol. Lung Cell Mol. Physiol. 281:L71-L78. [DOI] [PubMed] [Google Scholar]
- 48.Xue, H., D. O'Neill, J. Morrow, and A. Bank. 1999. A novel mouse gene, HemT, encoding an hematopoietic cell-specific transcript. Gene 231:49-58. [DOI] [PubMed] [Google Scholar]