Abstract
Mutants of Listeria monocytogenes with deletions in genes of the common branch of the biosynthesis pathway leading to aromatic compounds were constructed as possible virulence-attenuated carrier strains for protein antigens or vaccine DNA. aroA, aroB, and in particular aroE mutants showed strongly reduced growth rates in epithelial cells and even in rich culture media. The metabolism of the aro mutants under these conditions was predominantly anaerobic. Aerobic metabolism and a wild-type growth rate were, however, regained upon the addition of vitamin K2, suggesting that the aro mutants are deficient in oxidative respiration due to the lack of menaquinone. Replication of the aro mutants in the host cell's cytosol and cell-to-cell spread were drastically slowed down, and all aro mutants showed high virulence attenuation in mice, i.e., the 50% lethal dose in BALB/c mice was increased at least 104-fold for the aroA, aroB, and aroA/B mutants and >105-fold for the aroE mutant compared to the parent strain. Nevertheless, mice preimmunized with aro mutant bacteria elicited good T-cell response and full protection against a subsequent challenge with the virulent wild-type strain. A total of 5 × 106 aroA, aroB, and aroA/B mutant bacteria were sufficient to obtain a protective T-cell response, while 5 × 108 aroE or aroA/E mutants were necessary to achieve comparable numbers of antigen-specific T cells. These numbers were well tolerated without causing any signs of disease, indicating that Listeria strains with deletions in genes of the basic branch of the aromatic amino acid pathway could be useful vaccine carriers for inducing T-cell immunity.
Listeria monocytogenes is a gram-positive facultative intracellular pathogen which specifically infects professional antigen-presenting cells, like monocytes, macrophages, and dendritic cells (18, 25, 36, 48). After uptake into these cells, the bacteria escape into the cytosol by disrupting the phagosomal membrane by the action of the cytolysin listeriolysin (LLO) (40). In the cytoplasm, efficient bacterial replication occurs, and listeriae spread into neighboring cells without being released into the extracellular environment. Hence, cytotoxic T cells play a major role in the adaptive immune response against L. monocytogenes infections (27). For this reason, L. monocytogenes has received increasing attention as a carrier strain for the generation of live vaccines against infectious and neoplastic diseases (reviewed in references 17 and 37).
Since these bacteria have the potential to cause severe systemic infections with symptoms such as sepsis and encephalomeningitis, particularly in immunocompromised and pregnant patients (48), L. monocytogenes vaccine strains must be avirulent but still strongly immunogenic. In the past several years, attenuated strains of L. monocytogenes which lacked essential virulence genes were introduced for this purpose (4, 5, 9-11, 16, 19, 24, 28, 33, 38). These strains are basically avirulent and were even used in a safety study with human volunteers (2). Auxotrophic mutations successfully applied for virulence attenuation in other bacterial carrier systems were also used for obtaining attenuated L. monocytogenes strains (1, 29, 45, 47). However, these mutations either led to insufficient virulence attenuation (1, 29, 45) or required coinjection of the impaired metabolite for the induction of an efficient immune response against L. monocytogenes (47).
In a number of bacterial pathogens, mutations in the basic branch of the aromatic amino acid biosynthesis pathway proved to be particularly efficient in attenuating virulence, e.g., in Salmonella enterica serovar Typhimurium (21), Shigella flexneri (26), Shigella dysenteriae (49), Pseudomonas aeruginosa (42), Bordetella pertussis (43), Neisseria gonorrhoeae (7), and Bacillus anthracis (22), but failed in Mycobacterium tuberculosis (35).
Here, we describe mutants with deletions in aroA (encoding the first enzyme in aromatic amino acid biosynthesis, 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase), aroB (encoding the second enzyme, 3-dehydroquinate synthase), and aroE (encoding 5-enolpyruvylshikimate-3-phosphate synthase). All three single mutants, as well as aroA/B and aroA/E double mutants, still contained the entire set of virulence genes but were strongly attenuated and remained highly immunogenic.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this work are listed in Table 1. Escherichia coli DH10b was used as a host for all DNA manipulations.
TABLE 1.
Listerial strains used
| Strain | Genetic property | Reference or source |
|---|---|---|
| L. monocytogenes EGDe | Wild-type strain | 15 |
| Mutants | ||
| aroA | Deletion of aroA | J. Stritzker and W. Goebel, submitted for publication |
| aroB | Deletion of aroB | This work |
| aroE | Deletion of aroE | This work |
| aroA/B | Deletion of aroA and aroB | This work |
| aroA/E | Deletion of aroA and aroE | This work |
| pheA trpEG tyrA | Deletion of pheA, trpEG, and tyrA | This work |
| aroE revertant | aroE integration into the aroE mutant | This work |
Bacteria were cultivated either in brain heart infusion (BHI) or in modified Welshimer's broth minimal medium (41) containing 2% glucose. Additional supplements (from Sigma, Steinheim, Germany) consisted of 0.1 g (each) of l-tryptophan, l-phenylalanine, and l-tyrosine/liter; 0.3 g of shikimate/liter; 0.3 g of p-aminobenzoic acid/liter; 0.1 g of folic acid/liter; and 50 mg of vitamin K2/liter.
For anoxic BHI cultures, the medium was degassed at 60°C for 45 to 60 min under vacuum and overlaid with mineral oil. For infections, Listeria strains were grown to a late-logarithmic growth phase (180 Klett units for L. monocytogenes EGDe, 150 Klett units for aroA, aroB, and aroA/B mutants; and 90 Klett units for aroE and aroA/E mutants) at 37°C in BHI, washed with phosphate-buffered saline (PBS) (pH 7.4), resuspended in 20% (vol/vol) glycerol-PBS, and stored at −80°C.
Construction of Listeria mutant strains.
All deletion mutageneses were constructed using L. monocytogenes EGDe as a parental strain and were performed by a homologous-recombination technique (50) using constructs derived from the mutagenesis vector pLSV101 (derived from pLSV1, kindly provided by T. Fuchs).
For in-frame deletion of aroB, a 424-bp fragment (primers aroB5′-1 [5′-AAAGGTGCTGAATTCGGTGTC-3′] and aroB5′-2 [5′-TGTAATCCCGGGCATGTTTAAAACTCCTTC-3′]) that was localized upstream of the deletion locus and a 269-bp fragment (primers aroB3′-3 [5′-GGGAATCCCGGGTGAGAGCGATTCGAG-3′] and aroB3′-4 [5′-ACATAGGAAGCGAATTCGGCA-3′]) downstream of the deletion locus were amplified. Both fragments were digested with PspAI, ligated, and further amplified with primers aroB5′-1 and aroB3′-4, mentioned above. The recombination fragment was then cloned into pLSV101 using EcoRI and BamHI restriction, resulting in pLSV101 ΔaroB, which was transformed into L. monocytogenes EGDe (for aroB mutant construction) or aroA (for aroA/B mutant construction). After cultivation at 42°C with 5 μg of erythromycin/ml, clones with a chromosomally integrated knockout plasmid could be selected. The resulting clones were further cultivated at 30°C without erythromycin to obtain the aroB chromosomal deletion mutants.
Deletion of aroE in L. monocytogenes EGDe and aroA was performed by the same method described above. The following primers were used (nomenclature as indicated for aroB primers): aroE5′-1 (5′-GCTTGGGATCCTTGAAAGTGAAGATGC-3′), aroE5′-2 (5′-GTAATCCCGGGCATCGTCATTTCGCT-3′), aroE3′-3 (5′-CACTACCCGGGTAACAACTTCTACCAG-3′), and aroE3′-4 (5′-TGATAGAATTCAGCTAACACGAGCAAA-3′).
From the aroE mutant, an aroE reversion mutant was constructed by using the same method. Therefore, aroE was amplified using aroE5′-1 and aroE3′-4 with wild-type chromosomal DNA as a template, and a pLSV101 aroE reversion was obtained after BamHI and EcoRI restriction and ligation.
Construction of the pheA trpEG tyrA mutant was performed by first making an in-frame deletion of trpEG (primers trpEG5′-1 [5′-TAAAATGAATTCATCAAACAAGAATAGTACTTT-3′], trpEG5′-2 [5′-TTTCATCCCGGGCATCTAAATCATCCCTTCTT-3′], trpEG3′-3 [5′-CACATTCCCGGGTGTTTTAGAAGTGTTAGCG-3′], and trpEG3′-4 [5′-ACTATTGGATCCATCTCCGCCTGTCCCGCA-3′]), followed by in-frame deletion of tyrA in trpEG (primers tyrA5′-1 [5′-CAATTAGAATTCAAGCTTTTATTGAGGCATGC-3′], tyrA5′-2 [5′-GCTCCTACCCGGGCATTCAAAAACGCCTCCAAAC -3′], tyrA3′-3 [5′-TTGAATGCCCGGGTAGGAGCGAAATGACGATGA-3′], and tyrA3′-4 [5′-ATGGATGGATCCCCTAAAATAACTGTATC-3′]), and finally by in-frame deletion of pheA in trpEG tyrA (primers pheA5′-1 [5′-GTCACAGAATTCTTGTAGCTAATAAAATGGAT-3′], pheA5′-2 [5′-ATAAGCCCCGGGCATTTTCAACTCTCCTTTTT-3′], pheA3′-3 [5′-TTACAACCCGGGTAAATAAAGAGTTATCGCAATT-3′], and pheA3′-4 [5′-CTAATCGGATCCAAGGGTTTAAACTCGGATC-3′]).
All in-frame deletion mutants were confirmed by PCR analysis and sequencing of the corresponding sections of the chromosome.
Metabolic analysis.
Listerial strains were grown in BHI containing 2% glucose. Ten milliliters of sterile filtered incubation medium was adjusted to pH 2 by adding 5 ml of 6 N HCl. The acidified medium was successively extracted twice with 40 ml of ethyl ether and 40 ml of dichloromethane (all solvents were distilled before use). The combined organic layers were dried over sodium sulfate and filtered. The solvent was removed at 42°C using a 25-cm-long Vigreux distillation column. Finally, the residue was filled up to a standardized volume of 1 ml, and 1 μl was injected for high-resolution gas chromatography-mass spectrometry (HRGC-MS) analysis.
HRGC-MS analysis was performed using an Agilent 5973 mass selective detector coupled to an Agilent 6890 gas chromatograph with split injector (1:20) and an Agilent 7683 automatic liquid sampler. The system was equipped with Chem Station software. A J&W DB Wax fused-silica capillary column (30-m by 0.25-mm inside diameter; 0.25-μm film thickness) was employed. The temperature program was 3 min isothermal at 50°C and then increased at 4°C/min to 220°C, using a flow rate of 1.0 ml of helium/min. The MS operating values were as follows: ionization voltage, 70 eV (electron impact ionization); ion source and interface temperatures, 230°C.
Identifications were carried out by comparison of mass spectral and chromatographic retention data of the target compounds with those of authentic reference substances (lactic acid, acetic acid, and acetoin were obtained from Fluka, Steinheim, Germany).
Cell culture, infection experiments, and plaque assay.
Caco-2 (human colon adenocarcinoma) and 3T3 (mouse fibroblast) cells were cultured in RPMI 1640 medium supplemented with 2 mM l-glutamine (Gibco) and 10% fetal calf serum (Biochrom, Berlin, Germany).
For infection, cells were seeded in 24-well plates 1 day prior to infection. After a wash step with RPMI 1640 containing 2 mM l-glutamine, the cells were infected with 10 bacteria per cell for 1 h. The cells were washed with RPMI 1640 containing 2 mM l-glutamine and cultivated with gentamicin-containing medium (100 μg/ml), which was replaced with medium containing 10 μg of gentamicin/ml after 1 h. Counts of viable intracellular bacteria were determined by plating serial dilutions of mechanically lysed cell suspensions on BHI agar.
Plaque formation by L. monocytogenes in 3T3 cells was performed as described earlier (46). In brief, a confluent layer of 3T3 fibroblasts was infected with 1 bacterium per 10 cells. Instead of replacing medium containing 100 μg of gentamicin/ml with medium containing 10 μg of gentamicin/ml, the cells were overlaid with medium containing 0.5% agarose, as well as 10 μg of gentamicin/ml. After 2 days, plaques were visualized by the addition of an overlay of medium containing 0.5% agarose, 10 μg of gentamicin/ml, and 0.1% neutral red (1% stock solution was filter sterilized). Plaques were visualized after 4 to 8 h, and their diameters were measured (at least 50 plaques/strain).
Animal experiments.
All animal experiments were approved by the government of the district of Unterfranken and conducted according to the German animal protection guidelines. Female BALB/c mice (6 to 8 weeks of age) were purchased from Charles River Laboratories, Sulzfeld, Germany.
Mice were infected via intravenous (i.v.) injection of 100 μl of an endodotxin-free PBS (pH 7.4) bacterial suspension. The mean number of viable bacteria ± standard deviation in liver and spleen homogenates for five mice were determined by plating serial dilutions on BHI agar plates, which were incubated at 37°C for 2 days before CFU were counted.
For protection assays, 7-week-old BALB/c mice were infected with either PBS alone (negative control), 103 CFU of L. monocytognes EGDe (positive control), or 5 × 106 CFU of the corresponding aro mutants. After 4 weeks, the mice were challenged with 103 CFU of L. monocytogenes EGDe bacteria, and the bacterial loads in the spleen and liver were determined 4 days after challenge.
Ex vivo enumeration of peptide-specific CD8 T cells.
The frequency of peptide-specific CD8 T lymphocytes was determined in a gamma interferon (IFN-γ)-specific enzyme-linked immunospot (ELISPOT) assay as described previously (14). Mice were immunized either with 103 CFU of L. monocytogenes EGDe or with graded doses between 5 × 102 and 5 × 108 CFU of the various L. monocytogenes deletion mutants. Unseparated splenocytes (6 × 105/well) were stimulated for 6 h in round-bottom 96-well microtiter plates in the presence of 10−6 M peptide. Subsequently, activated cells (4 × 105 or 4 × 104/well) were transferred to anti-IFN-γ-coated (clone RMMG-1; Biosource, Camarillo, Calif.) membrane-backed 96-well ELISPOT plates (Nunc, Wiesbaden, Germany), which after 18 h of incubation were developed with biotin-labeled rat anti-mouse IFN-γ monoclonal antibody (clone XMG1.2; BD Biosciences), horseradish peroxidase-streptavidin conjugate (Dianova, Hamburg, Germany), and aminoethylcarbazole dye solution (Sigma, Deisenhofen, Germany). The frequency of antigen-specific cells was calculated as the number of spots per splenocyte seeded. In addition, the total number of antigen-specific T cells per spleen was calculated from the T-cell frequency and the total number of cells per spleen. The specificity and sensitivity of the ELISPOT assay were controlled with IFN-γ-secreting CD8 T-cell lines specific for amino acids 217 to 225 and 449 to 457 of p60 and amino acids 91 to 99 of LLO. Recovery of seeded T cells was generally >90%.
RESULTS
Growth of aro mutants in culture media.
Mutants with deletions in different genes of the common biosynthesis pathway (aroA, aroB, aroA/B, aroE, and aroA/E) (Fig. 1) leading to the aromatic amino acids were constructed. As expected, these mutants were unable to grow in minimal medium without aromatic amino acids. Growth of the aroA, aroB, and aroA/B mutants, but not the aroE mutant, was restored after the addition of shikimate. Only partial growth of the aroE and aroA/E mutants was obtained when all aromatic amino acids, including p-amino benzoate, were provided (Table 2). Unexpectedly, the growth rates of all L. monocytogenes aro mutants were markedly lower than that of the wild-type L. monocytogenes EGDe or the aroE revertant strain when the aro mutants were cultured in rich BHI medium (Table 2).
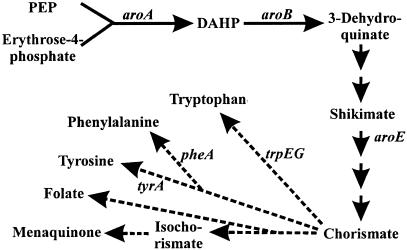
FIG. 1.
Common amino acid biosynthesis pathway in L. monocytogenes. PEP, phosphoenolpyruvate; DAHP, 3-deoxy-arabino-heptulonate 7-phosphate. Each solid arrow represents one enzyme. Dashed arrows represent several enzymes, one of which is indicated in some cases.
TABLE 2.
Generation times of L. monocytogenes strains under different broth culture conditions
| Medium | Generation time (min)a
|
|||||||
|---|---|---|---|---|---|---|---|---|
| EGDe | aroA | aroB | aroA/B | aroE | aroA/E | pheA trpEG tyrA | aroE revertant | |
| BHI | 43 ± 4 | 74 ± 3 | 79 ± 5 | 73 ± 4 | 151 ± 21 | 164 ± 23 | 44 ± 4 | 43 ± 3 |
| BHI folateb | 45 ± 5 | 79 ± 6 | ND | ND | 135 ± 16 | ND | ND | ND |
| BHI anoxicc | 77 ± 5 | 70 ± 7 | ND | ND | 93 ± 2 | ND | ND | ND |
| BHI vitamin K2b | 47 ± 6 | 50 ± 7 | ND | ND | 45 ± 5 | ND | ND | ND |
| MWBd | 105 ± 5 | NG | NG | NG | NG | NG | NG | ND |
| MWB aaab | 102 ± 7 | 367 ± 124 | ND | ND | >1,000 | ND | ND | ND |
| MWB aaa PABAb | 101 ± 6 | 354 ± 5 | ND | ND | >1,000 | ND | ND | ND |
| MWB Shikimateb | 103 ± 10 | 94 ± 3 | ND | ND | NG | ND | ND | ND |
| MWB aaa Vitamin K2 | 94 ± 7 | 98 ± 24 | ND | ND | 118 ± 32 | ND | ND | ND |
Generation time mean values ± standard deviations (three samples). ND, not determined; NG, not grown.
Supplements were added in the following concentrations: folate, 0.1 g/liter; vitamin K2, 50 mg/liter; aaa (aromatic amino acids), 0.1 g/liter (each); PABA (para-aminobenzoic acid), 0.1 g/liter; shikimate, 0.3 g/liter.
Anoxic growth conditions were achieved by degassing of BHI containing 2% glucose and an overlay of mineral oil.
Modified Welshimer's broth (MWB) is a defined minimal medium for L. monocytogenes (41).
To investigate whether the reduced growth rate is due to impaired uptake of the three aromatic amino acids or to the lack of folate and/or ubiquinone-menaquinone biosynthesis, a triple-deletion pheA trpEG tyrA mutant was constructed which is unable to produce phenylalanine, tyrosine, and tryptophan but is not hampered in folate and ubiquinone-menaquinone biosynthesis (Fig. 1). The growth rate of this mutant strain in BHI was similar to that of the wild-type strain (Table 2).
The lack of folate was excluded as a cause for the reduced growth of the aro mutants in BHI, as the addition of trimethoprim or sulfanilamide (both drugs inhibit folate biosynthesis) to BHI led to growth inhibition of the wild-type strain, but also of the aro mutants (data not shown), and supplementation of the BHI medium with additional folate did not increase the growth rate of the aro mutants (Table 2).
Ubiquinone (and/or the prokaryote-specific menaquinone) is an essential component of the respiratory chain, with precursors derived from the common branch of the aromatic amino acid pathway, and a defect in ubiquinone (menaquinone) biosynthesis is expected to strongly impair oxidative phosphorylation even in the presence of oxygen. Indeed, growth of the L. monocytogenes wild-type strain in BHI under anoxic conditions resulted in a replication rate which was similar to that observed for the L. monocytogenes aro mutants under oxic conditions. In contrast, no change in the growth rates of the aro mutants was observed when these mutants were grown in BHI under oxic or anoxic conditions (Table 2). Furthermore, the observed accumulation of lactate and the reduced acetate concentration, as well as the absence of acetoin, a typical overflow substance secreted by L. monocytogenes growing aerobically, in the culture supernatant of the aroE mutant (Table 3) suggest that L. monocytogenes aro mutants carry out a predominantly anaerobic metabolism (44). Finally, addition of vitamin K2 (menaquinone 4) restored the growth rate of the aroA and aroE mutants (Table 2) and led to the disappearance of lactate and reappearance of acetoin in the supernatant of the aroE mutant, which was also observed in the aroE revertant grown in BHI alone (Table 3).
TABLE 3.
Relative amounts of acetoin, acetate, and lactate produced compared to L. monocytogenes EGDe grown in BHI
| Strain | Medium | Amt produced
|
||
|---|---|---|---|---|
| Acetoina | Acetatea | Lactateb | ||
| aroE | BHIc | 0.00 ± 0.00 | 0.03 ± 0.03 | 0.74 ± 0.57 |
| Vitamin K2d | 1.44 ± 0.18 | 1.41 ± 0.95 | 0.07 ± 0.06 | |
| Anoxice | 0.00 ± 0.00 | 0.03 ± 0.01 | 0.71 ± 0.16 | |
| EGDe | BHI | 1.00 | 1.00 | 0.02 ± 0.02 |
| Vitamin K2 | 1.03 ± 0.29 | 1.14 ± 0.50 | 0.05 ± 0.00 | |
| Anoxic | 0.05 ± 0.04 | 0.08 ± 0.05 | 1.00 | |
| aroE revertant | BHI | 1.24 ± 0.29 | 1.53 ± 0.14 | 0.02 ± 0.01 |
Acetoin and acetate values are given as relative amount ± standard deviation (for three independent measurements) compared to the amount produced by the wild-type strain under aerobic growth conditions.
Lactate amounts ± standard deviations (for three independent measurements) compared to the amount produced by the wild-type strain under anoxic growth conditions.
BHI containing 2% glucose.
BHI containing 2% glucose and vitamin K2 at a final concentration of 50 μg/ml.
Degassed BHI containing 2% glucose overlaid with mineral oil.
Reduced cytosolic replication and cell-to-cell spread of listerial aro mutants in vitro.
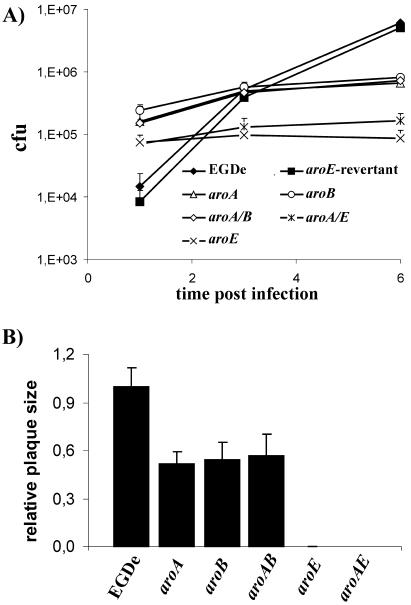
Internalization of all aro mutants by nonphagocytic epithelial (Caco-2) and endothelial (HBMEC) cells is significantly enhanced due to increased internalin A and internalin B production in these mutants compared to the wild-type strain (J. Stritzker, C. Schoen, and W. Goebel, submitted for publication). While release of the aro mutants from the primary phagosome into the host cell cytosol seemed to be comparable to that of the wild-type strain, growth of the aro mutants in the cytosol was significantly reduced. The aroA, aroB, and aroA/B mutants grew in this cellular compartment with a generation time of ∼90 min, which is about twice as long as that of the wild-type strain; the aroE and aroA/E mutant strains replicated even more slowly and stopped intracellular growth ∼3 h postinfection (Fig. 2A). Similar behavior of the aro mutants was observed in plaque assays with 3T3 fibroblasts (Fig. 2B). The plaque size of the aroA, aroB, and aroA/B mutants in these assays was about half that of the wild-type strain, indicating a reduced spreading capacity of these aro mutants in comparison to the wild-type strain. No plaques were observed in this assay after infection with the aroE or aroA/E mutant, although actin tails were formed (data not shown).
FIG. 2.
(A) Mean bacterial counts plus standard deviations for three wells after infection of 2 × 105 Caco-2 cells using 2 × 106 L. monocytogenes EGDe or aroA, aroB, aroA/B, aroE, or aroA/E mutant organisms. (B) For plaque assays, a confluent layer of 3T3 fibroblasts was infected with 1 bacterium per 10 cells, and the diameters of at least 50 plaques were measured; their mean diameters plus standard deviations are compared to those after infection with L. monocytogenes EGDe.
L. monocytogenes with mutations in the aromatic amino acid pathway is strongly attenuated in vivo.
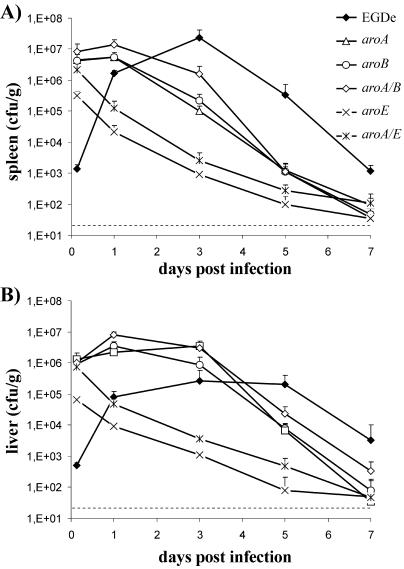
The growth characteristics of the L. monocytogenes aro mutants in BHI and in mammalian cell cultures described above imply that the virulence of these mutant strains will be attenuated in vivo. To further investigate the extent of virulence attenuation, groups of 7-week-old female BALB/c mice (three individuals per group) were infected i.v. with 108 viable bacteria of the corresponding strains (which is 4 orders of magnitude higher than the 50% lethal dose of L. monocytogenes EGDe). No signs of disease were observed after infection. To test whether the attenuated bacteria were still able to replicate in vivo, counts of viable bacteria in the liver and spleen were determined at various time points postinfection (i.v. injection with 5 × 107 aro mutant bacteria per mouse). Since the 50% lethal dose of L. monocytogenes EGDe is 3.5 orders of magnitude lower, no wild-type bacteria were included in this experiment. As indicated by the increasing bacterial load between 4 and 24 h postinfection (Fig. 3), aroA, aroB, and aroA/B mutants replicated in the liver and spleen and were present at rather high levels without causing any obvious signs of disease besides enlargement of the liver and spleen (data not shown). Infection with 103 L. monocytogenes organisms, however, resulted in mild symptoms of disease. In contrast to the other aro mutants, the aroE and aroA/E mutants did not replicate in the liver or spleen, as indicated by the rapid decrease in the numbers of viable bacteria in the infected organs (Fig. 3). However, these strongly attenuated mutants survived at a low level for up to 7 days in the liver and spleen.
FIG. 3.
Colonization of spleens (A) and livers (B) of BALB/c mice following i.v. infection with 5 × 107 CFU of aroA, aroB, aroA/B, aroE, or aroA/E mutant or 103 L. monocytogenes EGDe bacteria. Each point represents the mean number of CFU/g of organ plus standard deviation for five mice. Dashed lines represent the limit of detection.
Attenuated aro mutant L. monocytogenes strains induce a protective CD8 T-cell response.
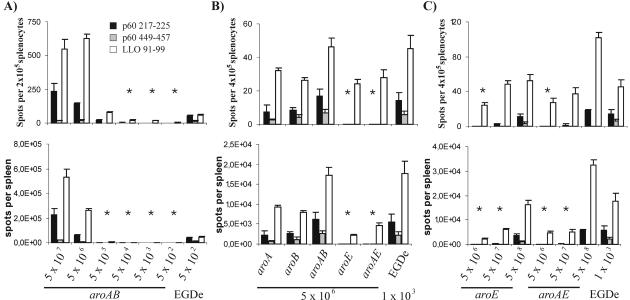
In order to evaluate the immunogenic potential of L. monocytogenes strains with mutations in the aromatic amino acid biosynthetic pathway, we measured the T-cell response against the well-defined H-2d-restricted CD8 T-cell epitopes p60 217 to 225, p60 449 to 457, and LLO 91 to 99 of L. monocytogenes (14). Priming of an L. monocytogenes-specific CD8 T-cell response by the aroA/B mutant, which appears to be best suited to be a possible vaccine carrier strain due to its putative low reversion rate to the wild type (since aroA and aroB are located in different parts of the genome and may not be complemented in one step by recombination) and high attenuation, required at least 5 × 106 CFU (Fig. 4A). After immunization, a significantly stronger CD8+-T-cell response was obtained after i.v. immunization with 5 × 106 CFU of the aroA, aroB, and aroA/B mutants compared to immunization of mice with the aroE or aroA/E mutant (Fig. 4B). While aroA, aroB, and aroA/B mutants primed a significant CD8 T-cell response against all three epitopes tested, the aroE or aroA/E mutant strain selectively primed a T-cell response only against the immunodominant epitope LLO 91 to 99. Thus, the aroA/B mutant combines excellent immunogenicity with high biosafety due to the double deletion of two independent, widely separated genes. In contrast to the aroA, aroB, and aroA/B mutants, the aroE and aroA/E mutants were significantly less immunogenic. The CD8 T-cell response induced by immunization with the aroE and aroA/E mutants was enhanced when more bacteria were inoculated (Fig. 4C); however, CD8 T cells never reached the frequency which was obtained after immunization with the aroA/B mutant (Fig. 4A and B). The Listeria-specific CD8 T-cell response observed after immunization with the aroA/B mutant, expressed both as frequency of antigen-specific CD8 T cells per splenocyte (Fig. 4, top graphs) and as total T-cell response per spleen (Fig. 4, bottom graphs), was superior to those of the other mutants tested.
FIG. 4.
ELISPOT assay of spleen cells after i.v. infection of 7-week-old BALB/c mice with different numbers of CFU of aro mutants and L. monocytogenes EGDe. The results are shown as spots per splenocyte plus standard deviation (top graphs) and mean spots per spleen plus standard deviation (bottom graphs) for three mice. The asterisks indicate P values of <0.05 between wild-type and mutant strains for all three epitopes, determined using a one-sided Student t test. (A) Spots per spleen are shown, depending on the applied infective dose of the aroA/B mutant. (B) CD8 T-cell response when 5 × 106 CFU of the aro mutants and 103 CFU of the wild-type strain were applied. (C) For efficient T-cell responses, at least 5 × 107 CFU of the aroE or aroA/E mutant is necessary.
The protection obtained after immunization of mice with the different aro mutant Listeria strains correlated well with the frequency of peptide-specific CD8 T cells in vivo. Immunization with 5 × 106 CFU of the aroA, aroB, and aroA/B mutants protected mice at least as well as immunization with 103 wild-type bacteria (Fig. 5), and in only some mice, very few bacteria could be detected (detection limits, 25 CFU/g of liver and 100 CFU/g of spleen). In correlation with the lower frequencies of L. monocytogenes-specific CD8 T cells observed after immunization with either the aroE or aroA/E mutant, significantly less protection against challenge with wild-type L. monocytogenes was obtained with these mutants compared to mice infected with the aroA, aroB, or aroA/B mutant. Thus, the L. monocytogenes aroA/B mutant combines high attenuation and biosafety with high immunogenicity, which points to this strain as a possible vaccine carrier strain.
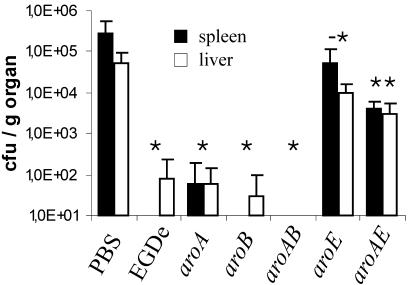
FIG. 5.
Colonization of spleen and liver in immunized mice 4 days after i.v. infection with 103 CFU of L. monocytogenes EGDe bacteria. Immunization was performed with 5 × 106 CFU of aro mutants, 103 CFU of L. monocytogenes EGDe (positive control), or PBS (negative control). Each bar represents the mean number of CFU/g of organ plus standard deviation for five mice. *, P < 0.05 between mice immunized with bacteria and PBS controls; -*, P < 0.05 between aroE mutant-immunized animals and L. monocytogenes EGDe-immunized mice, and P < 0.07 between aroE mutant-immunized animals and PBS-immunized controls; **, P < 0.05 between aroA/E-immunized animals and PBS- and L. monocytogenes EGDe-immunized animals, respectively, determined using a one-sided Student t test.
DISCUSSION
In this study, we investigated the impacts of aroA, aroB, and aroE mutations on replication under extra- and intracellular conditions and on the virulence and immunogenicity of L. monocytogenes.
Although all aro mutants should be unable to produce the aromatic amino acids, including p-amino benzoate, as well as menaquinone, the results obtained showed unexpectedly major differences in the growth rates in culture media and inside mammalian cells and in the degree of virulence attenuation between the aroA, aroB, and aroA/B mutants (with deletions in genes determining the first two enzymes of the common pathway) and the aroA/E and aroE mutants [with a deletion in the gene determining the enzyme for the conversion of shikimate-3-phosphate to 5-O-(1-carboxyvinyl)-3-phosphoshikimate]. The growth phenotypes of all L. monocytogenes aro mutants were also different from those of the intensively studied aroA mutants of S. enterica serovar Typhimurium and other Enterobacteriaceae (note that the aroA mutation in the latter gram-negative bacteria corresponds to the aroE mutation in L. monocytogenes and other gram-positive bacteria). The Salmonella aroA mutant grows like wild-type Salmonella in rich BHI medium, indicating that all essential aromatic compounds which this mutant lacks, can be provided by the culture medium (31). In contrast, the aro mutants of L. monocytogenes, and in particular the aroE mutant, showed strongly reduced growth in these media compared to the wild-type strain. This growth reduction is not due to impaired uptake of the aromatic amino acids by the aro mutants, since a triple mutant with specific mutations in the biosynthesis pathways leading to Phe, Tyr, and Trp grows at the same rate as the wild-type strain in BHI and minimal media supplemented with the aromatic amino acids. Our results rather suggest that the lack of menaquinone biosynthesis is the major cause of the impaired growth of the aro mutants, since L. monocytogenes can apparently synthesize only menaquinone but not ubiquinone (http://genolist.pasteur.fr/ListiList/), whereas in S. enterica serovar Typhimurium, all genes for ubiquinone and menaquinone biosynthesis have been identified in the genome sequence. Ubiquinone and menaquinone are major electron carriers in the aerobic respiration chain, receiving electrons from reduced NAD generated via oxidation of reduced carbon sources by the corresponding dehydrogenases.
Indeed, all L. monocytogenes aro mutants carried out predominantly anaerobic metabolism, as indicated by the low concentration of acetoin and acetate and the increased accumulation of lactate in the culture media (44), as well as the nearly identical growth rates under oxic and anoxic conditions and the restored aerobic growth of the aro mutants if vitamin K2 was added to the culture medium. Preliminary data from comparative transcriptome and proteome patterns of the L. monocytogenes wild-type strain and the aroA mutant also indicate a switch to anaerobic metabolism in the aroA mutant, even under aerobic growth conditions (Stritzker et al., unpublished results).
One would expect that the S. enterica serovar Typhimurium aroA mutant should also be unable to produce these two central electron carriers essential for NADH-driven oxidative respiration unless S. enterica serovar Typhimurium can circumvent this defect by a respiration mechanism that does not require ubiquinone-menaquinone to be absent in L. monocytogenes. Alternatively, ubiquinone (which S. enterica serovar Typhimurium is able to synthesize) or ubiquinone precursors (e.g., 2,3-dihydroxybenzoate or p-hydroxybenzoate) are present in the rich culture media used, like BHI, and complement the ubiquinone defect of the aroA mutant.
The reason for the significant growth difference between the aroA, aroB, and aroA/B and the aroE mutants is less clear. A fortuitous additional mutation in the aroE deletion mutant can be ruled out, since the same growth phenotype was obtained with independently constructed aroE deletion mutants and the aroA/E double mutant, and an aroE revertant showed wild-type replication in extracellular and intracellular growth and also produced wild-type amounts of acetoin, acetate and lactate under aerobic growth conditions. Toxic effects of the possible accumulation of intermediates from the common pathway, e.g., dehydroquinate or shikimate, or the additional ATP consumption in the reaction leading to shikimate-3-phosphate (possible in the aroE mutant but not in the aroA, aroB, and aroA/B mutants) also seem to be unlikely causes of the observed growth difference between the two mutants, since an aroA/E mutant shows the same phenotype as the aroE mutant. Possibly a yet-unknown side reaction leads to low-level generation of a component in the common pathway including menaquinone (downstream of the first intermediate, 2-dehydro-3-deoxy-d-arabino-heptulosonate), which can be converted at low efficiency to the end products of the aromatic amino acid pathway by the aroA, aroB, and aroA/B mutants but not by the aroE and aroA/E mutants. In addition, excess oxygen under oxic growth conditions may harm the aroE and aroA/E mutants but not the aroA, aroB, and aroA/B mutants, since the assumed low-level synthesis of menaquinone in the latter group of mutants may suffice to detoxify the oxygen. This assumption is supported by the enhanced growth rate of the aroE mutant under nonoxic growth conditions, which resembles those of the aroA mutant and the wild-type strain (Table 2).
Similar to the aroA mutants of S. enterica serovar Typhimurium and other Enterobacteriaceae (6, 21, 30, 42, 49), all aro mutants of L. monocytogenes described here are strongly virulence attenuated. However, the physiological basis for the virulence attenuation of the two intracellular bacteria may be different. A shortage of aromatic amino acids in the vacuolar compartment in which S. enterica serovar Typhimurium replicates seems to be a major reason for its virulence attenuation (31). This is most likely not the case for the L. monocytogenes aro mutants. L. monocytogenes replicates in the cytosol of infected host cells, and all aro mutants reach this cellular compartment at least as efficiently as the wild-type strain, since the internalin A (B)-mediated internalization of these mutants by mammalian cells is significantly enhanced and listeriolysin production necessary for escape into the cytosol is as high as that of the wild-type strain (data not shown). For listeriae residing in mammalian-cell cytosol, the supply of aromatic amino acids does not appear to be a limiting factor for the following reasons: (i) L. monocytogenes mutants with single, double, and even triple mutations in phenylalanine, tyrosine, and/or tryptophan biosynthesis are not significantly hampered in intracellular growth (data not shown); (ii) no mutants with defects in any of these pathways were ever isolated in previous studies screening for virulence-defective L. monocytogenes mutants (13, 23; K. Knuth, C. Stuehler, T. Fuchs, and W. Goebel, unpublished data); and (iii) previous studies using L. monocytogenes mutants auxotrophic for aromatic amino acids showed only marginal virulence attenuation and no intracellular growth reduction (1, 29).
We assume that the predominant anaerobic metabolism of the aro mutants is the major cause of the reduced intracellular growth and, hence, the virulence attenuation of these mutants. The stronger attenuation of the aroE and aroA/E mutants than of the aroA, aroB, and aroA/B mutants is in line with this assumption. Efficient replication of L. monocytogenes in the cytosol requires aerobic metabolism, as indicated by the strongly reduced growth rate of a mutant with inactive pyruvate dehydrogenase (32) and of mutants with defects in either the oxidative branch of the citrate cycle or the oxidative respiration chain (Knuth et al., unpublished).
As reported recently, disruption of the basic branch of the aromatic amino acid pathway failed in M. tuberculosis (35). Since M. tuberculosis, which has a strict aerobic metabolism, is also able to synthesize only menaquinone (but, like L. monocytogenes, is unable to produce ubiquinone) (http://www.genome.ad.jp/kegg/), this compound cannot be produced when genes in the basic branch of the aromatic amino acid pathway are deleted. However, since M. tuberculosis is unable to switch to anaerobic metabolism, aro mutants will not survive.
The CD8 T-cell response in BALB/c mice after immunization with different L. monocytogenes aro mutants correlated well with their replication potential in the host cell's cytosol. Compared to the aroE and aroA/E mutants, immunization with the aroA, aroB, or aroA/B mutant primed a significantly stronger p60- and LLO-specific CD8 T-cell response. The CD8 T-cell response and the protection obtained after immunization with 5 × 106 CFU of the aroA/B mutant were at least as strong as those after immunization with a sublethal dose of the L. monocytogenes wild-type strain (103 bacteria). However, in the case of the aroA/B mutant, immunization was accompanied by only weak pathological side effects, such as slight enlargement of the liver and spleen. Thus, the L. monocytogenes aroA/B mutant combines high attenuation and biosafety due to an expected extremely low reversion rate with high immunogenicity, which points to this strain as a good vaccine carrier strain.
Some of the known virulence factors of L. monocytogenes, e.g., LLO, are required for efficient major histocompatibility complex class I-restricted antigen presentation in infected antigen-presenting cells, L. monocytogenes strains attenuated by the knockout of virulence factors, like hly, actA, or a combination of actA and plcB, therefore, have impaired immunogenicity. The LLO-deficient mutant, although highly attenuated in virulence, is strongly hampered in antigen presentation via the major histocompatibility complex class I pathway (4) and in delivery of DNA vaccines (20) and is also missing an important antigen (3, 8, 12, 34). In a previous study, it was shown that an actA mutant shows highly reduced release of vaccine DNA, even in the presence of a phage lysin used as a tool to disrupt the listerial carrier (39), and compared to the wild-type strain, was inferior to a vaccine carrier expressing the complete set of L. monocytogenes-specific virulence factors. In the same study, elevated plasmid DNA transfer and antigen presentation using an aroA mutant compared to wild-type L. monocytogenes were shown.
The aro mutations described were also introduced into a previously developed listerial lethal balanced system stabilizing introduced antigen-encoding plasmid DNA (39). The resulting plasmid-stabilized aro strains may be valuable tools for eliciting cellular immune responses using either DNA or protein delivery strategies.
Acknowledgments
We thank F. Heckel for support with HRGC-MS analysis, T. Williams for assistance with in vivo experiments, and M. Kuhn for discussion and critical reading of the manuscript.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (Go168/27-1) awarded to W.G.
Editor: J. D. Clements
REFERENCES
- 1.Alexander, J. E., P. W. Andrew, D. Jones, I. S. Roberts, H. Marquis, H. G. Bouwer, D. J. Hinrichs, and D. A. Portnoy. 1993. Characterization of an aromatic amino acid-dependent Listeria monocytogenes mutant: attenuation, persistence, and ability to induce protective immunity in mice. Infect. Immun. 61:2245-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angelakopoulos, H., K. Loock, D. M. Sisul, E. R. Jensen, J. F. Miller, and E. L. Hohmann. 2002. Safety and shedding of an attenuated strain of Listeria monocytogenes with a deletion of actA/plcB in adult volunteers: a dose escalation study of oral inoculation. Infect. Immun. 70:3592-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barry, R. A., H. G. A. Bouwer, T. R. Clark, K. A. Cornell, and D. J. Hinrichs. 2003. Protection of interferon-gamma knockout mice against Listeria monocytogenes challenge following intramuscular immunization with DNA vaccines encoding listeriolysin O. Vaccine 21:2122-2132. [DOI] [PubMed] [Google Scholar]
- 4.Barry, R. A., H. G. Bouwer, D. A. Portnoy, and D. J. Hinrichs. 1992. Pathogenicity and immunogenicity of Listeria monocytogenes small-plaque mutants defective for intracellular growth and cell-to-cell spread. Infect. Immun. 60:1625-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boury, N. M., and C. J. Czuprynski. 1995. Listeria monocytogenes infection increases neutrophil adhesion and damage to a murine hepatocyte cell line in vitro. Immunol. Lett. 46:111-116. [DOI] [PubMed] [Google Scholar]
- 6.Bowe, F., P. O'Gaora, D. Maskell, M. Cafferkey, and G. Dougan. 1989. Virulence, persistence, and immunogenicity of Yersinia enterocolitica O:8 aroA mutants. Infect. Immun. 57:3234-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chamberlain, L. M., R. Strugnell, G. Dougan, C. E. Hormaeche, and R. Demarco de Hormaeche. 1993. Neisseria gonorrhoeae strain MS11 harbouring a mutation in gene aroA is attenuated and immunogenic. Microb. Pathog. 15:51-63. [DOI] [PubMed] [Google Scholar]
- 8.Cornell, K. A., H. G. Bouwer, D. J. Hinrichs, and R. A. Barry. 1999. Genetic immunization of mice against Listeria monocytogenes using plasmid DNA encoding listeriolysin O. J. Immunol. 163:322-329. [PubMed] [Google Scholar]
- 9.Darji, A., W. Mohamed, E. Domann, and T. Chakraborty. 2003. Induction of immune responses by attenuated isogenic mutant strains of Listeria monocytogenes. Vaccine 1:S102-S109. [DOI] [PubMed] [Google Scholar]
- 10.Dietrich, G., A. Bubert, I. Gentschev, Z. Sokolovic, A. Simm, A. Catic, S. H. Kaufmann, J. Hess, A. A. Szalay, and W. Goebel. 1998. Delivery of antigen-encoding plasmid DNA into the cytosol of macrophages by attenuated suicide Listeria monocytogenes. Nat. Biotechnol. 16:181-185. [DOI] [PubMed] [Google Scholar]
- 11.Dramsi, S., S. Levi, A. Triller, and P. Cossart. 1998. Entry of Listeria monocytogenes into neurons occurs by cell-to-cell spread: an in vitro study. Infect. Immun. 66:4461-4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fensterle, J., L. Grode, J. Hess, and S. H. Kaufmann. 1999. Effective DNA vaccination against listeriosis by prime/boost inoculation with the gene gun. J. Immunol. 163:4510-4518. [PubMed] [Google Scholar]
- 13.Gahan, C. G., and C. Hill. 2000. The use of listeriolysin to identify in vivo induced genes in the gram-positive intracellular pathogen Listeria monocytogenes. Mol. Microbiol. 36:498-507. [DOI] [PubMed] [Google Scholar]
- 14.Geginat, G., S. Schenk, M. Skoberne, W. Goebel, and H. Hof. 2001. A novel approach of direct ex vivo epitope mapping identifies dominant and subdominant CD4 and CD8 T cell epitopes from Listeria monocytogenes. J. Immunol. 166:1877-1884. [DOI] [PubMed] [Google Scholar]
- 15.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 16.Goossens, P. L., G. Milon, P. Cossart, and M. F. Saron. 1995. Attenuated Listeria monocytogenes as a live vector for induction of CD8+ T cells in vivo: a study with the nucleoprotein of the lymphocytic choriomeningitis virus. Int. Immunol. 7:797-805. [DOI] [PubMed] [Google Scholar]
- 17.Gunn, G. R., A. C. Zubair, and Y. Paterson. 2002. Recombinant intra-cellular bacteria as carriers for tumor antigens, p. 314-348. In G. Dietrich and W. Goebel (ed.), Vaccine delivery strategies, vol. 1. Horizon Scientific Press, Wymondham, United Kingdom. [Google Scholar]
- 18.Guzman, C. A., M. Rohde, T. Chakraborty, E. Domann, M. Hudel, J. Wehland, and K. N. Timmis. 1995. Interaction of Listeria monocytogenes with mouse dendritic cells. Infect. Immun. 63:3665-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harty, J. T., and M. J. Bevan. 1995. Specific immunity to Listeria monocytogenes in the absence of IFN gamma. Immunity 3:109-117. [DOI] [PubMed] [Google Scholar]
- 20.Hense, M., E. Domann, S. Krusch, P. Wachholz, K. E. Dittmar, M. Rohde, J. Wehland, T. Chakraborty, and S. Weiss. 2001. Eukaryotic expression plasmid transfer from the intracellular bacterium Listeria monocytogenes to host cells. Cell Microbiol. 3:599-609. [DOI] [PubMed] [Google Scholar]
- 21.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238-239. [DOI] [PubMed] [Google Scholar]
- 22.Ivins, B. E., S. L. Welkos, G. B. Knudson, and S. F. Little. 1990. Immunization against anthrax with aromatic compound-dependent (Aro-) mutants of Bacillus anthracis and with recombinant strains of Bacillus subtilis that produce anthrax protective antigen. Infect. Immun. 58:303-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klarsfeld, A. D., P. L. Goossens, and P. Cossart. 1994. Five Listeria monocytogenes genes preferentially expressed in infected mammalian cells: plcA, purH, purD, pyrE and an arginine ABC transporter gene, arpJ. Mol Microbiol. 13:585-597. [DOI] [PubMed] [Google Scholar]
- 24.Kocks, C., J. B. Marchand, E. Gouin, H. d'Hauteville, P. J. Sansonetti, M. F. Carlier, and P. Cossart. 1995. The unrelated surface proteins ActA of Listeria monocytogenes and IcsA of Shigella flexneri are sufficient to confer actin-based motility on Listeria innocua and Escherichia coli respectively. Mol. Microbiol. 18:413-423. [DOI] [PubMed] [Google Scholar]
- 25.Kolb-Mäurer, A., S. Pilgrim, E. Kampgen, A. D. McLellan, E. B. Bröcker, W. Goebel, and I. Gentschev. 2001. Antibodies against listerial protein 60 act as an opsonin for phagocytosis of Listeria monocytogenes by human dendritic cells. Infect. Immun. 69:3100-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kotloff, K. L., F. Noriega, G. A. Losonsky, M. B. Sztein, S. S. Wasserman, J. P. Nataro, and M. M. Levine. 1996. Safety, immunogenicity, and transmissibility in humans of CVD 1203, a live oral Shigella flexneri 2a vaccine candidate attenuated by deletions in aroA and virG. Infect. Immun. 64:4542-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lara-Tejero, M., and E. G. Pamer. 2004. T cell responses to Listeria monocytogenes. Curr. Opin. Microbiol. 7:45-50. [DOI] [PubMed] [Google Scholar]
- 28.Manohar, M., D. O. Baumann, N. A. Bos, and J. J. Cebra. 2001. Gut colonization of mice with actA-negative mutant of Listeria monocytogenes can stimulate a humoral mucosal immune response. Infect. Immun. 69:3542-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marquis, H., H. G. Bouwer, D. J. Hinrichs, and D. A. Portnoy. 1993. Intracytoplasmic growth and virulence of Listeria monocytogenes auxotrophic mutants. Infect. Immun. 61:3756-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nnalue, N. A., and B. A. Stocker. 1987. Test of the virulence and live-vaccine efficacy of auxotrophic and galE derivatives of Salmonella choleraesuis. Infect. Immun. 55:955-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Callaghan, D., D. Maskell, F. Y. Liew, C. S. Easmon, and G. Dougan. 1988. Characterization of aromatic- and purine-dependent Salmonella typhimurium: attention, persistence, and ability to induce protective immunity in BALB/c mice. Infect. Immun. 56:419-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Riordan, M., M. A. Moors, and D. A. Portnoy. 2003. Listeria intracellular growth and virulence require host-derived lipoic acid. Science 302:462-464. [DOI] [PubMed] [Google Scholar]
- 33.Paglia, P., I. Arioli, N. Frahm, T. Chakraborty, M. P. Colombo, and C. A. Guzman. 1997. The defined attenuated Listeria monocytogenes delta mp12 mutant is an effective oral vaccine carrier to trigger a long-lasting immune response against a mouse fibrosarcoma. Eur. J. Immunol. 27:1570-1575. [DOI] [PubMed] [Google Scholar]
- 34.Pamer, E. G., J. T. Harty, and M. J. Bevan. 1991. Precise prediction of a dominant class I MHC-restricted epitope of Listeria monocytogenes. Nature 353:852-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parish, T., and N. G. Stoker. 2002. The common aromatic amino acid biosynthesis pathway is essential in Mycobacterium tuberculosis. Microbiology 148:3069-3077. [DOI] [PubMed] [Google Scholar]
- 36.Paschen, A., K. E. Dittmar, R. Grenningloh, M. Rohde, D. Schadendorf, E. Domann, T. Chakraborty, and S. Weiss. 2000. Human dendritic cells infected by Listeria monocytogenes: induction of maturation, requirements for phagolysosomal escape and antigen presentation capacity. Eur. J. Immunol. 30:3447-3456. [DOI] [PubMed] [Google Scholar]
- 37.Paterson, Y. 2003. Rational approaches to immune regulation. Immunol. Res. 27:451-462. [DOI] [PubMed] [Google Scholar]
- 38.Peters, C., E. Domann, A. Darbouche, T. Chakraborty, and M. E. Mielke. 2003. Tailoring host immune responses to Listeria by manipulation of virulence genes—the interface between innate and acquired immunity. FEMS Immunol. Med. Microbiol. 35:243-253. [DOI] [PubMed] [Google Scholar]
- 39.Pilgrim, S., J. Stritzker, C. Schoen, A. Kolb-Mäurer, G. Geginat, M. J. Loessner, I. Gentschev, and W. Goebel. 2003. Bactofection of mammalian cells by Listeria monocytogenes: improvement and mechanism of DNA delivery. Gene Ther. 10:2036-2045. [DOI] [PubMed] [Google Scholar]
- 40.Portnoy, D. A., T. Chakraborty, W. Goebel, and P. Cossart. 1992. Molecular determinants of Listeria monocytogenes pathogenesis. Infect. Immun. 60:1263-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Premaratne, R. J., W. J. Lin, and E. A. Johnson. 1991. Development of an improved chemically defined minimal medium for Listeria monocytogenes. Appl. Environ. Microbiol. 57:3046-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Priebe, G. P., G. J. Meluleni, F. T. Coleman, J. B. Goldberg, and G. B. Pier. 2003. Protection against fatal Pseudomonas aeruginosa pneumonia in mice after nasal immunization with a live, attenuated aroA deletion mutant. Infect. Immun. 71:1453-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roberts, M., D. Maskell, P. Novotny, and G. Dougan. 1990. Construction and characterization in vivo of Bordetella pertussis aroA mutants. Infect. Immun. 58:732-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romick, T. L., H. P. Fleming, and R. F. McFeeters. 1996. Aerobic and anaerobic metabolism of Listeria monocytogenes in defined glucose medium. Appl. Environ. Microbiol. 62:304-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saklani-Jusforgues, H., E. Fontan, and P. L. Goossens. 2001. Characterisation of a Listeria monocytogenes mutant deficient in d-arabitol fermentation. Res. Microbiol. 152:175-177. [DOI] [PubMed] [Google Scholar]
- 46.Sun, A. N., A. Camilli, and D. A. Portnoy. 1990. Isolation of Listeria monocytogenes small-plaque mutants defective for intracellular growth and cell-to-cell spread. Infect. Immun. 58:3770-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson, R. J., H. G. Bouwer, D. A. Portnoy, and F. R. Frankel. 1998. Pathogenicity and immunogenicity of a Listeria monocytogenes strain that requires d-alanine for growth. Infect. Immun. 66:3552-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vazquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Dominguez-Bernal, W. Goebel, B. Gonzalez-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walker, J. C., and N. K. Verma. 1997. Cloning and characterisation of the aroA and aroD genes of Shigella dysenteriae type 1. Microbiol. Immunol. 41:809-813. [DOI] [PubMed] [Google Scholar]
- 50.Wuenscher, M. D., S. Köhler, W. Goebel, and T. Chakraborty. 1991. Gene disruption by plasmid integration in Listeria monocytogenes: insertional inactivation of the listeriolysin determinant lisA. Mol. Gen. Genet. 228:177-182. [DOI] [PubMed] [Google Scholar]