Abstract
Vibrio cholerae is a gram-negative bacterium that has been associated with cholera pandemics since the early 1800s. Whole-cell, killed, and live-attenuated oral cholera vaccines are in use. We and others have focused on the development of a subunit cholera vaccine that features standardized epitopes from various V. cholerae macromolecules that are known to induce protective antibody responses. TcpA protein is assembled into toxin-coregulated pilus (TCP), a type IVb pilus required for V. cholerae colonization, and thus is a strong candidate for a cholera subunit vaccine. Polypeptides (24 to 26 amino acids) in TcpA that can induce protective antibody responses have been reported, but further characterization of their amino acid targets relative to tertiary or quaternary TCP structures has not been done. We report a refinement of the TcpA sequences that can induce protective antibody. One sequence, TcpA 15 (residues 170 to 183), induces antibodies that bind linear TcpA in a Western blot as well as weakly bind soluble TcpA in solution. These antibodies bind assembled pili at high density and provide 80 to 100% protection in the infant mouse protection assay. This is in sharp contrast to other anti-TcpA peptide sera (TcpA 11, TcpA 13, and TcpA 17) that bind very strongly in Western blot and solution assays yet do not provide protection or effectively bind TCP, as evidenced by immunoelectron microscopy. The sequences of TcpA 15 that induce protective antibody were localized on a model of assembled TCP. These sequences are centered on a site that is predicted to be important for TCP structure.
Vibrio cholerae is a gram-negative bacterium that causes pandemics of diarrheal disease that can be fatal in untreated individuals. In some parts of the world, economic, political, and social conditions are consistent with the potential for future cholera epidemics. V. cholerae must colonize the human intestinal tract following ingestion of contaminated water or food before virulence genes are expressed (9). The toxin-coregulated pilus (TCP) is a homopolymer of a 20.5-kDa TcpA pilin which is expressed upon infection and is required for colonization (16). Two biotypes of V. cholerae are associated with pandemic disease, classical and El Tor. The TcpAs of the two biotypes are 82% identical, with variability being found in the C terminus of the protein (11). Other TcpA alleles are found in non-O1/non-O139 strains, where the sequence of the TcpA subunit exhibits several additional amino acid substitutions, especially in the area of the last 60 amino acids, which comprise a disulfide-bound structure that is required for a functional TCP (1).
Antibodies specific for TcpA are protective (12-15). TCP is not considered a dominant immunogen because a single-dose, experimental infection of North American volunteers with V. cholerae O1 strain O395 induced only a low-titer antibody response to TcpA (5). However, 50% of a limited number of sera (three of six) from a highly cholera-endemic area of Indonesia had TcpA-specific immunoglobulin G (IgG) and IgA antibodies (5).
Recent data from Calderwood's group suggest that antibodies to TCP are more prevalent in individuals recovering from cholera than was previously thought (4). Rabbit anti-TCP antibodies made to purified pilus, when mixed with virulent V. cholerae (500 50% lethal doses) and administered to infant mice, provided complete immunity to cholera (13). Protection was shown to be associated with anti-TCP antibody based on adsorption experiments. Similar results were obtained with monoclonal antibodies specific for TcpA as well as with polyclonal antibody raised against synthetic peptides corresponding to a region of TcpA adjacent to the domain recognized by the monoclonal antibody (12, 15). We reported that the previously identified TcpA peptides TcpA 4 (amino acids 145 to 168), 5 (amino acids 157 to 181), and 6 (amino acids 174 to 199) can be variably immunogenic in mice of different major histocompatibility complex haplotypes (10, 12). The mechanism of the anti-TcpA antibody's protective effect is not known but is likely related to reduced colonization of V. cholerae in the face of anti-pilin antibody. The means by which colonization is affected are perhaps not related to inhibition of bacterial adhesion to host cells but rather to effects on microcolony formation and thus the association of the bacteria in the intestine (8).
The crystal structure of classical TcpA was recently published (3). The TcpA monomer consists of a long N-terminal alpha-helical tail and a globular domain that results from a disulfide bond between Cys residues 120 and 186. The N terminus has two alpha-helical (α1, α2) stretches that are on one side of a packed hydrophobic core (β-sheets). The α/β loop (residues connecting α1 to the β sheet) is predicted to be involved in pilus structure. A third (α3) and fourth (α4) α helix are on the other side of the hydrophobic core. The last two β strands, β4 and β3, along with the last two alpha-helices form the D region, a discrete face containing residues important for TCP function and residues able to induce protective antibodies (8, 13-15).
The group that crystallized V. cholerae TcpA generated a computer TCP model based on energy-minimized structures that recapitulated physical measurements from cryoelectron micrographs and x-ray defraction images (3). TCP is about 80 Å in diameter and can be several micrometers long. TCP pili are left-handed, three-start alpha-helical structures with a 22.5-Å rise and a pitch of 45 Å. Interestingly, the model predicts that there is an area of interaction between TcpA monomers that might be important for TCP structure. The alpha-helical N termini of the monomers are predicted to be buried in the center of the pilus and impart a considerable amount of binding energy to maintain the structure. There is also extensive contact between the D region and the α/β loop of neighboring monomers. Amino acids 121 to 186 in the D region and 50 to 77 in the loop are in close proximity, such that L176 and T125 form hydrogen bounds with Y51 and L76, respectively.
We report here data that refine the location of a B-cell epitope of TcpA that is able to induce protective antibody responses. Rabbit serum resulting from immunization with keyhole limpet hemocyanin-conjugated TcpA 15 peptide (TcpA amino acids 170 to 183) contained protective antibody that bound soluble TcpA as well as assembled pilus. TcpA 15 residue D175 was required for Western blot and immunoelectron reactivity with anti-TcpA 15 serum. TcpA 15 is predicted to be involved in TcpA subunit interactions required for TCP structure.
MATERIALS AND METHODS
Animals.
Six-month-old New Zealand White female rabbits were used for the peptide immunization studies under contract with the Pocono Rabbit Farm and Laboratory (Canadensis, Pa.). Pregnant female CD-1 mice were purchased from Charles River (Raleigh, N.C.) for use in the infant mouse protection studies. All mice were housed under standard conditions in the Animal Resources Center located at the Dartmouth Medical School, Hanover, N.H.
Bacterial stocks and growth conditions.
The bacterial stocks used in these studies are listed in Table 1. Strains were maintained as frozen glycerol stocks stored at −80°C. Bacteria were streaked onto Luria-Bertani (LB) agar plates and incubated overnight at 37°C. Bacteria from single colonies were inoculated into LB medium at pH 6.5 and incubated on a rotary wheel at 30°C for 16 to 18 h to induce maximal TCP surface expression unless otherwise noted.
TABLE 1.
Strains used for Western blot, immunoprecipitation, immunoelectron microscopy, and infant mouse protection assays
| Strain | Species | TCP |
|---|---|---|
| O395 | V. cholerae O1 classical | Wild-type TCP |
| RT4031 | V. cholerae O1 classical | TCP negative |
| RT4018 | V. cholerae O1 classical | P169→A |
| RT4056 | V. cholerae O1 classical | K172→A |
| RT4054 | V. cholerae O1 classical | D175→A |
| RT4506 | V. cholerae O1 classical | T180→A |
| RT4058 | V. cholerae O1 classical | H181→A |
| RT4060 | V. cholerae O1 classical | E183→A |
| RT4055 | V. cholerae O1 classical | K184→A |
| RT4062 | V. cholerae O1 classical | K187→A |
| RT4469 | E. coli Origami/pET15b- cl-TcpA clone | Wild-type TCP NH2 terminal His tag |
TcpA peptides.
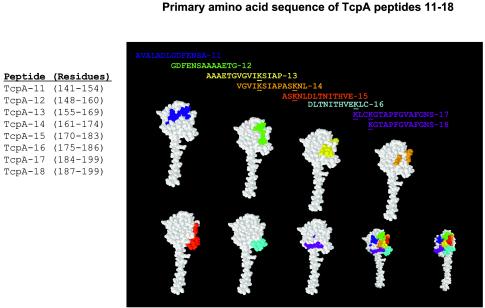
TcpA peptides 11 through 18, derived from the sequence of the classical biotype of V. cholerae O1, were prepared and conjugated to keyhole lymphocyte hemocyanin or bovine serum albumin or left unconjugated by Macromolecular Resources (Colorado State University, Fort Collins, Colo.). The peptides used for these studies were TcpA 11 (amino acids 141 to 154), AVALADLGDFENSA; TcpA 12 (amino acids 148 to 160), GDFENSAAAAETG; TcpA 13 (amino acids 155 to 169), AAAETGVGVIKSIAP; TcpA 14 (amino acids 161 to 174), VGVIKSIAPASKNL; TcpA 15 (amino acids 170 to 183), ASKNLDLTNITHVE; TcpA 16 (amino acids 175 to 186), DLTNITHVEKLC; TcpA 17 (amino acids 184 to 199), KLCKGTAPFGVAFGNS; and TcpA 18 (amino acids 187 to 199), KGTAPFGVAFGNS. The lyophilized peptides were resuspended in 5.0% dimethyl sulfoxide-0.25× phosphate-buffered saline (PBS) to a final concentration of 0.5 mg/ml and stored at −80°C in 1.0-ml aliquots until used. The relative positions of these peptides within TcpA are shown in Fig. 1. The locations of the individual TcpA peptides as well as all the TcpA peptides on the space-filling TcpA monomer model (3) are shown in Fig. 1.
FIG. 1.
Classical, toxin-coregulated pilus protein A (TcpA) peptides that were synthesized for this study and their locations on the classical TcpA monomer. TcpA peptides 11 to 18 were synthesized with a C residue at the C termini and conjugated by a disulfide bond to keyhole limpet hemocyanin (KLH) for immunization of rabbits as described in Materials and Methods. Colored peptides are correlated with the colored amino acids in the space-filling model of TcpA and define the locations of the primary amino acid sequences of the TcpA peptides on the folded TcpA monomer. RasMol version 2.7.2.1 molecular visualization software was used to display and manipulate the Brookhaven Protein Data Bank (available through the National Center for Biotechnology Information [http://www.ncbi.nlm.nih.gov]) TcpA file 1OQV (3).
Immunization and serum collection.
Synthetic TcpA peptides, conjugated to keyhole limpet hemocyanin and emulsified in Freund's adjuvant were injected into rabbits in decreasing doses employing multiple routes of injection. We administered 200 μg of immunogen for the first three injections in the popliteal lymph nodes (day 0), subcutaneous and intradermal (day 14), or subcutaneous inoculation and popliteal lymph nodes (day 28). On day 56, 100 μg of immunogen was injected subcutaneously. Rabbits were bled on days 0, 28, 42, 70, and 87, which represent preimmune, primary, secondary, tertiary, and quaternary sera, respectively.
Serology.
The presence of anti-TcpA-specific antibodies in individual sera was measured by enzyme-linked immunosorbent assay; 96-well, high-binding, flat-bottomed microtiter plates (Corning Life Sciences; Acton, Mass.) were coated with 100 μl per well of 0.5 μg of TcpA peptides conjugated to bovine serum albumin in 0.1 M Na2HPO4 binding buffer, pH 9.0, and incubated overnight at 4°C. The plates were washed four times with a Molecular Devices Skan Washer 400 microtiter plate washer with 250 μl per well of 1× PBS-0.05% Tween 20 (Fisher Scientific; Pittsburgh, Pa.). Nonspecific binding was blocked with 200 μl per well of buffer consisting of 1× PBS-1.0% bovine serum albumin-0.5% Tween 20 for 2 h at room temperature. The plates were washed four more times, after which 50 μl of antiserum (serially diluted twofold) was added to each well and incubated overnight at 4°C. The initial dilution of 1:1,000 was used for all serum samples. The plates were then brought to room temperature and washed four times, and 50 μl of horseradish peroxidase-labeled goat anti-rabbit IgG (heavy and light chain specific) (Jackson ImmunoResearch Laboratories, West Grove, Pa.) detector antibodies (diluted 1:4,000) were added to each well and incubated at room temperature for 2 h in the dark. The plates were washed six times and then developed with 100 μl per well of 3,3′,5,5′-tetramethylbenzidine (TMB) peroxidase substrate (Kirkegaard & Perry Laboratories, Gaithersburg, Md.) for 30 min at room temperature. The reaction was stopped with an equal volume of 0.18 M H2SO4. Optical densities (ODs) were read with a Dynex Technologies MRX microtiter plate reader (Thermo Labsystems, Helsinki, Finland) with Dynex Revelation version 3.04 software at λ = 450 nm with 630 nm as the reference wavelength.
Immunoelectron microscopy.
We centrifuged 250 μl of an overnight culture of V. cholerae O1 and resuspended it in 1 ml of LB at pH 6.5. A humidified chamber was prepared by lining the bottom of a glass petri dish with moistened filter paper, and 4% Formvar-coated, 300-mesh nickel support grids were placed in the dish on top of the parafilm. Each grid was covered with a 15-μl drop of bacterial suspension and sealed in the petri dish with parafilm. The grids were incubated at 30°C for 3 h in a dry-heat incubator. Excess medium was wicked from the grids by carefully touching their edges to Whatman filter paper. For all subsequent manipulations, a 10-in.-square sheet of parafilm was fixed to a flat surface on the bench and the grids were transferred from the petri dish to the parafilm. The grids were covered with an inverted glass Pyrex baking dish to prevent evaporation during incubation periods. Grids were then fixed for 1 h at room temperature by placing a 15-μl drop of fixative buffer (2.0% paraformaldehyde, 0.5% glutaraldehyde, 50 mM sodium cacodylate, pH 7.4) over each grid. Following incubation, the grids were drained of excess fixative and washed four times in Tris-buffered saline with 0.05% Tween-20 (TBST).
Nonspecific antibody binding was blocked with TBST with 1% bovine serum albumin for 30 min at room temperature. Anti-TcpA serum was diluted 1:500 in TBST-1% bovine serum albumin and allowed to incubate on the grids for 1 h at room temperature. The grids were washed four times in TBST before adding 10-nm colloidal gold-conjugated goat anti-rabbit IgG detector antibody (ICN Biomedicals, Irvine, Calif.) diluted 1:100 in TBST and incubated for 30 min. Unbound antibody was removed with four more washes, and then the grids were negatively stained for 2 min with 0.5% phosphotungstic acid, pH 6.5 (Electron Microscopy Sciences, Hatfield, Pa.). The grids were then removed to a storage box and placed in a desiccating environment at room temperature until viewed. The grids were viewed with a Jeol, JEM-1010 40 kV to 100 kV transmission electron microscope housed at the Julius A. Rippel Electron Microscopy Facility (Dartmouth College, Hanover, N.H.). All images were photographed at 25,000× magnification. The resulting micrographs were scanned into Adobe Photoshop 7.0 with a UMax PowerLook (Dallas, Tex.) 1120 overhead transmission flatbed scanner at 1,200 dots per inch and then converted into jpg format.
Infant mouse challenge.
The suckling mouse challenge model for cholera was used for assessing the protective quality of anti-TcpA-specific antibodies in vivo (2, 13). Cultures of V. cholerae (classical Ogawa strain O395) were grown for 16 h under toxin-coregulated pilus-expressing conditions (LB at pH 6.5 and 30°C); 25 μl of bacterial suspension, diluted to 4.4 50% lethal doses, was combined with 25 μl of either preimmune serum (negative control) or tertiary anti-TcpA-specific serum immediately before intragastric administration to 4- to 5-day-old CD-1 mice. Challenged mice were kept at 30°C and monitored every 4 h starting 24 h postchallenge until the termination of the experiment at 48 h. GraphPad Prism 4.0 was used to generate survival curves. The assay was repeated six times for the TcpA 15 tertiary bleed sample with a range of concentrations (4.4 to 70.0 50% lethal doses) of challenge bacteria.
SDS-PAGE and Western blot.
Bacteria from overnight cultures were transferred to microcentrifuge tubes and centrifuged at 13,000 × g for 1 min. The resulting pellet was resuspended in 0.20 volume of sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) sample buffer containing 2-mercaptoethanol. Each sample was boiled for 10 min and then centrifuged for 10 min at 13,000 × g. Samples were loaded onto a 10% polyacrylamide gel and electrophoresed for 1 h at 150 V. The gel was allowed to come to equilibrium in Western blot transfer buffer for 10 min, and then proteins were electrophoretically transferred to a nitrocellulose membrane (Schleicher & Schuell Bioscience Inc., Keene, N.H.) with a Bio-Rad semidry transfer system at 23 V for 35 min. The nitrocellulose membrane was blocked for 2 h at room temperature or overnight at 4°C with 5% nonfat dry-milk in PBS-0.05% Tween 20 (PBST).
Antiserum from rabbits injected with TcpA peptides was diluted 1:10,000 to 1:50,000 in blocking buffer, and the nitrocellulose was incubated for 2 h at room temperature and then washed six times for five min in PBST. Goat anti-rabbit horseradish peroxidase IgG detector antibody (Jackson ImmunoResearch Laboratories) was diluted 1:8,000 in PBST and incubated with the nitrocellulose for 30 min at room temperature. The nitrocellulose was then washed six times for 5 min. For Western blotting with the TcpA-specific monoclonal antibody, purified 169.1 was diluted 1:500 and incubated for 30 min before being washed as above and the addition of goat anti-mouse IgG2b horseradish peroxidase-conjugated antibody (1:8,000) followed by subsequent washes. Enhanced chemiluminescent reagent (Amersham Biosciences, Piscataway, N.J.) was applied to the nitrocellulose, and the membrane was exposed to Kodak BioMax film for various periods of time (15 s up to 1 h). Developed film was scanned into a computer file with a Umax PowerLook 1120 overhead transmission flatbed scanner and then converted into jpg format with Adobe Photoshop 7.0.
TcpA purification.
Overnight cultures of Escherichia coli Origami DE cells containing the plasmid pET15b-cl-TcpA clone (expressing the polyhistidine-tagged wild-type TCP protein) were cultured in a shaker incubator at 37°C, 250 rpm (3); 10 ml of the overnight culture was inoculated into 1 liter of LB broth and grown to an OD600 of 0.6. Isopropyl-β-d-thiogalactoside was added to a final concentration of 0.01 M, and the cultures were grown overnight at 30°C, 250 rpm. Cells were centrifuged in a Sorvall GSA rotor at 6,000 rpm (5,858 × g) for 15 min at 4°C. The pellet was resuspended in 100 ml of cold equilibration-wash buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 1 mM phenylmethylsulfonyl fluoride, pH 7.0), sonicated, and centrifuged in a Sorvall SS-34 rotor at 14,000 rpm (23,420 × g) for 20 min at 4°C. The supernatant was passed over a Talon Metal affinity resin column (BD Clontech, Palo Alto, Calif.), and bound protein was eluted with 50 mM NaH2PO4-300 mM NaCl-250 mM imidazole, pH 7.0. Fractions containing TcpA were pooled and concentrated with a Sephacryl S-100HR column (Amersham Biosciences, Piscataway, N.J.).
Immunoprecipitation of biotinylated TcpA.
We biotinylated 1 mg of soluble polyhistidine-tagged classical TcpA with Pierce EZ-Link sulfo-NHS-LC-biotin, following the manufacturer's protocol (Pierce Biotechnology Inc., Rockford, Ill.); 5 μl of rabbit TcpA polypeptide tertiary antiserum or preimmune antiserum was incubated with approximately 2 μg of biotinylated soluble TcpA overnight at 4°C; 20 μl of streptavidin-Sepharose (Amersham Biosciences, Piscataway, N.J.) was added to each sample and incubated at room temperature for 2 h. Each sample was washed three times with PBS-1% nonfat dry milk. The pellet was resuspended in 20 μl of SDS-PAGE sample buffer containing 2-mercaptoethanol. A 1:20 dilution in sample buffer with 2-mercaptoethanol was made, boiled for 10 min, and centrifuged for 10 min, and 10 μl of the supernatant was loaded onto a 10% polyacrylamide gel. The proteins were transferred to a nitrocellulose membrane and blocked with PBST containing 3% nonfat dry milk for 2 h at room temperature. The membrane was then incubated for 2 h at room temperature with goat anti-rabbit IgG-horseradish peroxidase antibody diluted 1:5,000 in blocking buffer. Unbound antibody was removed by washing before the detection of bound antibody.
Immunoprecipitation of unbiotinylated TcpA.
HIS-Select HC nickel affinity beads (Sigma, St. Louis, Mo.) were washed with 2 volumes of deionized water and then 1 volume of wash buffer (50 mM sodium phosphate, pH 8.0, 0.3 M sodium chloride) to remove ethanol; 15 μl of washed beads was incubated overnight at 4°C with 100 μg of soluble classical TcpA lysate. The samples were washed five times with 1 ml of wash buffer, resuspended in 25 μl of wash buffer, and 10 μl of TcpA 14-mer tertiary or preimmune antiserum (diluted 1:25 in wash buffer) was added and incubated overnight at 4°C. The samples were washed three times with 1 ml of wash buffer, resuspended in 20 μl of 1x SDS-PAGE sample buffer with 2-mercaptoethanol, boiled for 10 min, and centrifuged 10 min, and the supernatant was loaded onto a 10% polyacrylamide gel. Proteins were transferred to nitrocellulose and blocked 2 h with PBST containing 5% nonfat dry milk-horseradish peroxidase-labeled detector antibody diluted 1:5,000 in PBST and incubated for 2 h, then the nitrocellulose was washed and the ECL reagent was added, and the blots were exposed to film.
TcpA and TCP structure modeling.
RasMol version 2.7.2.1 molecular visualization software (Herbert J. Bernstein, Bernstein and Sons, P.O. Box 177, Bellport, N.Y.) was used to manipulate the files 1OQV (TcpA) and 1OR9 (TCP model) which were downloaded from the Protein Data Base (available through the National Center for Biotechnology Information [http://www.ncbi.nlm.nih.gov]).
RESULTS
Immunogenicity of TcpA peptides as determined by enzyme-linked immunosorbent assay.
Previous work showed that TcpA is required for colonization of V. cholerae in humans and mice (5, 16). The nucleotide and amino acid sequences for both biotypes of TcpA have been reported previously (11). Peptides from the carboxyl end (amino acids 144 to 199) of classical TcpA, TcpA 4 (amino acids 145 to 168), TcpA 5 (amino acids 157 to 181), and TcpA 6 (amino acids 174 to 199) were shown to induce protective antibody responses in rabbits (12-15). Since the antibody combining site can accommodate some 7 to 10 amino acids depending on the orientation and conformation of the B-cell epitope, the precise localization of potential protective epitopes within the larger 24 to 26 TcpA polypeptides was unknown.
We made a series of smaller overlapping TcpA polypeptides of between 12 and 16 amino acids that spanned the region covered by the larger TcpA 4, 5, and 6 peptides. These smaller TcpA peptides were coupled to keyhole limpet hemocyanin and assessed for their immunogenicity in rabbits (Fig. 1). The N termini of the sequential peptides (TcpA 11 to 18) was moved, depending on the peptide, progressively 3 to 10 amino acids towards the carboxyl end to produce immunogens with variable N-terminal ends that, given the linkage to keyhole limpet hemocyanin, were the most likely structures available to induce B-cell responses. The locations of the individual TcpA peptides (colored space filling residues) are shown on the TcpA crystal structure that was recently reported by Craig and colleagues (3) (Fig. 1). If the complete set of TcpA peptides we generated are highlighted on the TcpA crystal structure, it can be seen that the surface area of the globular domain of TcpA that is potentially immunogenic can be extensively examined with these peptides.
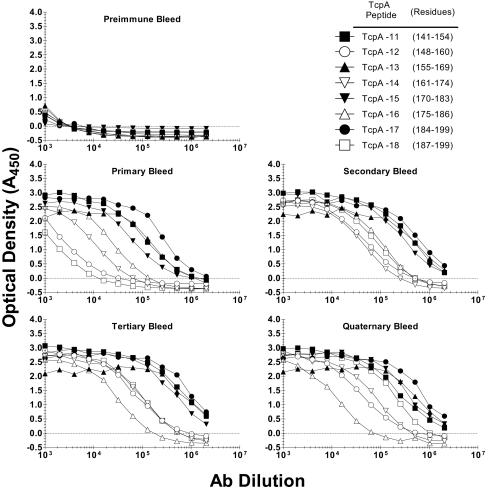
Rabbits were universally able to make anti-TcpA polypeptide antibodies, the titers of which increased with time and booster immunizations. The endpoint titers for all antipeptide sera at the tertiary bleed (day 70) were between 1:10,000 and 1:1,000,000 (Fig. 2). Tertiary bleed sera were used as the standard sera for subsequent analyses. As a group, the even-numbered peptides were less immunogenic than the odd-numbered peptides, with the average endpoint titers being 2.0 × 106 for the odd-numbered antipeptide sera and 0.4 × 106 for the even-numbered peptide sera.
FIG. 2.
Rabbit anti-TcpA 11 to 18 sera specific for sequences from classical V. cholerae O1 toxin-coregulated pilus protein A (TcpA). Rabbits were immunized with synthetic TcpA polypeptides coupled to keyhole limpet hemocyanin (KLH). The immunization protocol employed multiple routes and multiple injections. Serum samples were collected on days 0 (preimmune), 28 (primary), 42 (secondary), 70 (tertiary), and 87 (quaternary). Primary rabbit serum was serially diluted in twofold increments, starting at 1:1,000. Enzyme-linked immunosorbent assay results are reported as optical densities measured at 450 nm.
Recognition of TcpA sequence in the context of full-length linear TcpA.
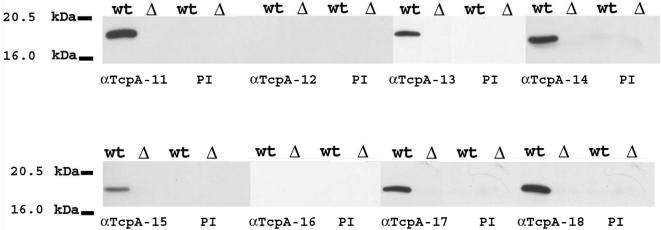
As all TcpA peptides were immunogenic as assessed by enzyme-linked immunosorbent assay analysis, we performed Western blotting analyses to assess the capacity of the antipeptide sera to bind TcpA sequences in the context of full-length TcpA. Negative controls included prebleed sera from all rabbits and assessment of tertiary sera against bacteria deleted for tcpA (Fig. 3). The capacity of the individual rabbit sera to recognize TcpA in Western blotting with the immunizing sequences now being contextually associated with other TcpA sequences revealed differences in the ability of the antisera to recognize linear TcpA (Fig. 3). Several anti-TcpA peptide antibodies, TcpA 11, TcpA 14, and TcpA 18, were highly reactive in Western blotting, but others were less so (TcpA 13, TcpA 15, and TcpA 17) or negative (TcpA 12 and TcpA 16). The dilutions of the various antisera, while comparable, were not identical. It is possible that the difference in the signals was related to a particular serum's antibody concentration or the affinity of those antibodies.
FIG. 3.
Western blot (WB) analysis of toxin-coregulated pilus protein A (TcpA). Whole-cell lysates of classical V. cholerae O395 (wild-type [wt] TcpA and TcpA deleted [ΔtcpA]) were electrophoresed through 10% polyacrylamide gel and detected by anti-classical TcpA 11 to 18 peptide tertiary sera. The corresponding preimmune (PI) serum was used as the negative control. Some anti-TcpA peptide sera were more reactive in Western blotting (TcpA 11, TcpA 14, and TcpA 18), while others were less so (TcpA 13, TcpA 15, and TcpA 17), and two were negative (TcpA 12 and TcpA 16).
Alternatively, the diminution in signal could be due to the availability of the epitope. The degree of reactivity in Western blotting did not correlated with the enzyme-linked immunosorbent assay titers of the anti-TcpA peptide sera. Anti-TcpA 17 and anti-TcpA 18 had similar enzyme-linked immunosorbent assay titers but reacted differently in Western blotting against full-length TcpA.
Assessment of the ability of anti-TcpA polypeptide sera to recognize soluble, recombinant TcpA.
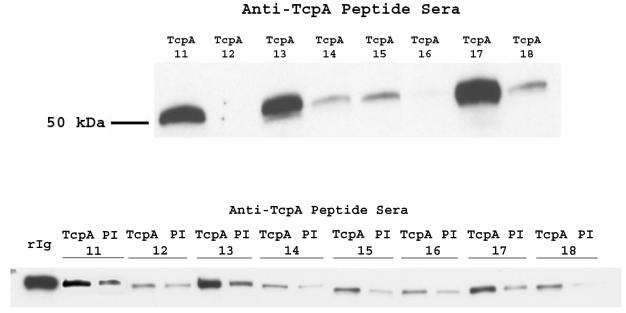
Classical TcpA has recently been crystallized, and the structure was solved at the level of 1.3 Å (3). The form of classical TcpA that was used for the crystal structure was genetically substituted at the first 28 amino acids with codons for 21 N-terminal histidines and grown in the E. coli stain Origami to enhance formation of the TcpA disulfide bond. We wanted to determine if the various anti-TcpA polypeptide sera could react with folded TcpA subunits. In order to maximize the signal from the pulldown assay, biotinylated TcpA derived from nickel column-purified TcpA was reacted with rabbit antiserum, and the complex was pulled down by streptavidin-Sepharose and probed for bound rabbit antibody (Fig. 4, top panel, 50 kDa, heavy chain of rabbit IgG). Additionally, to confirm the results and to avoid the potential confounding influence of biotinylation on epitope availability, we used nickel-Sepharose beads to pull down antigen-antibody complexes (Fig. 4, bottom panel).
FIG. 4.
Soluble toxin-coregulated pilus protein A (TcpA) can interact with some anti-TcpA peptide antisera. (Top) A pulldown analysis which assessed the ability of anti-classical TcpA peptide serum to interact with TcpA monomers was performed. Purified, biotin-labeled TcpA was incubated overnight at 4°C with anti-TcpA peptide prebleed or tertiary serum, and then 20 μl of streptavidin-Sepharose (in PBS-1% nonfat milk) was added for 2 h at room temperature, followed by washing, electrophoresis, and subsequent Western blot analysis with horseradish peroxidase-labeled goat anti-rabbit IgG, antibody. (Bottom) Purified TcpA lysate was incubated overnight at 4°C with His-Select nickel affinity beads and washed, and anti-TcpA peptide preimmune (PI) or tertiary serum was added and incubated overnight at 4°C. Samples were washed before pellets were resuspended and analyzed in a Western blot as described for the top panel. rIg, rabbit IgG control.
The enzyme-linked immunosorbent assay titers of the various anti-TcpA sera were not predicative of the ability of the antisera to interact with soluble TcpA. Both low-titer (anti-TcpA 14) and high-titer (anti-TcpA 18) sera failed to interact strongly with soluble TcpA. Similarly, the degree of binding in Western blotting was not predicative of binding soluble TcpA. The most efficient sera at binding soluble TcpA, anti-TcpA 13 and anti-TcpA 17, were in the second tier for Western blotting binding signals. The lack of interaction of some sera in the pulldown assay could be due to the fact that the linear immunogen sequence is not exposed on the TcpA protein or that the labeling of TcpA with biotin affected the B-cell epitope. All of the TcpA polypeptides (except TcpA 11 and TcpA 12) contain a Lys, and thus biotin bound to Lys could be completely or partially obscuring the B-cell epitope. We repeated the assay with unbiotinylated TcpA. The IgG-TcpA complex was isolated with nickel-Sepharose beads and then probed with anti-rabbit IgG-horseradish peroxidase. The degree of association of both forms of TcpA with TcpA-specific rabbit IgG was compared by quantitating the pixel intensity of the Western blotting results and then grouping the responses in rank order (data not shown). The results were essentially the same for both forms of TcpA.
Efficacy of anti-TcpA polypeptide rabbit sera to protect infant mice from challenge with virulent V. cholerae.
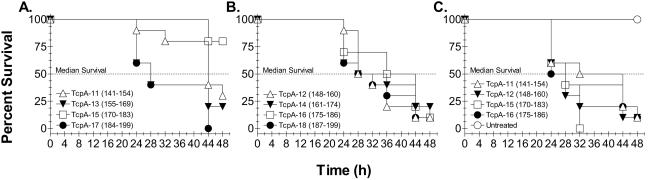
There are two means to test the efficacy of potential cholera vaccine formulations. The ultimate test is in human volunteers, but the more proximal means to assess vaccine efficacy is the infant mouse protection assay. Antiserum is combined with a known dose of virulent V. cholerae and introduced via gavage into the gut of 4- to 5-day-old infant CD-1 mice. We used the infant mouse protection assay to assess selected prebleed rabbit sera and all of the anti-TcpA polypeptide antisera for their ability to protect against lethal oral challenge with 4 to 70 50% lethal doses of virulent V. cholerae (Fig. 5). The only antiserum that was protective (80%) was anti-TcpA 15. Importantly, prebleed sera from the rabbit immunized with TcpA 15 were completely ineffective at protecting infant mice from oral challenge (Fig. 5C). The only other peptide antiserum that appeared to offer some protection in the infant mouse model was anti-TcpA 11. This serum did not offer long-term protection compared to the TcpA 15 antiserum, but the immunized mice clearly took longer to die than the mice from groups challenged with virulent V. cholerae combined with the other anti-TcpA peptide sera. Multiple protection experiments revealed that the anti-TcpA 15 serum could provide between 70 and 100% protection against 4.4 to 70 50% lethal doses.
FIG. 5.
Survival of infant mice following oral challenge with virulent classical V. cholerae O1 bacteria treated with anti-TcpA (toxin-coregulated pilus protein A) peptide serum. Groups of 10 4- to 5-day-old CD-1 mice were orally gavaged with 4.4 × 106 CFU of classical V. cholerae O1 Ogawa bacteria (representing 4.4 50% lethal doses) which was mixed with tertiary antisera (A and B) or preimmune antisera (C); some pups were not infected (C). Mice were kept at 30°C and monitored for death every 4 h starting 24 h after oral challenge. The protection data for anti-TcpA 15 serum are representative of seven independent challenge assays. In six independent experiments, protective efficacy ranged between 70 and 100% (mean, 91.4%) for anti-TcpA 15 serum.
Efficacy of anti-TcpA polypeptide rabbit sera to bind assembled TcpA subunits of virulent V. cholerae.
The ultimate biochemical assessment of anti-TcpA peptide sera is to determine the ability of antisera generated to isolated linear amino acid sequences to bind assembled TCP. The association of TcpA monomers and then the association of three alpha-helical strands that is predicted from the TCP model (3) suggests that certain TcpA residues will be buried in monomer-monomer interaction or in individual fiber interactions.
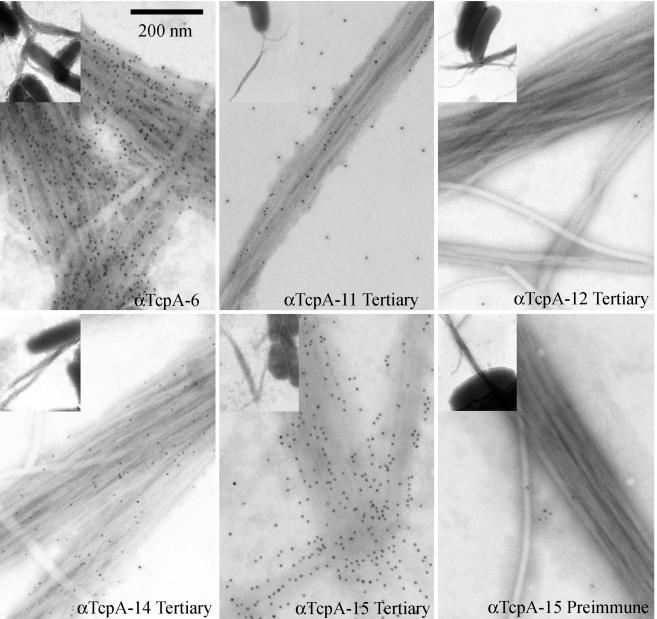
We used immunoelectron microscopy to determine the ability of anti-TcpA peptide sera to bind assembled TcpA (Fig. 6). The electron micrographs show that the negative control serum (preimmune TcpA 15) does not interact with assembled TCP. Similarly, hyperimmunization of a rabbit does not produce serum that nonspecifically binds TCP due to high levels of IgG (TcpA 12 panel). As would be consistent with its protective capacity, the positive control serum (anti-TcpA 6) sera bound TCP with high density. Representative micrographs of anti-TcpA sera that bound TCP with high or moderate to low density are shown. Of the various TcpA 11 to 18 antisera, only anti-TcpA 15 serum, which was protective, bound TCP with high density. The other anti-TcpA peptide sera bound less effectively than anti-TcpA 15 serum. Analysis of multiple images from independent grids suggests that the other sera bind either moderately (TcpA 11 and TcpA 13) or poorly (TcpA 14, TcpA 17, and TcpA 18) to TCP (data not shown).
FIG. 6.
Transmission immunoelectron microscopic analysis of anti-TcpA (toxin-coregulated pilus protein A)-specific antibodies binding to classical V. cholerae O1 TCP. Bacteria were incubated with various TcpA peptide-specific sera on 4% Formvar-coated, 300-mesh nickel support grids, washed to remove unbound antibody, and fixed in 2.0% paraformaldehyde-0.5% glutaraldehyde buffer. Bound antibody was detected with 10.0-nm gold particle-conjugated goat anti-rabbit IgG. Grids were negatively stained with 0.5% phosphotungstic acid (pH 6.5) and viewed with a transmission electron microscope at 25,000× magnification (inset). The inset shows the relationship of TCP and bacteria. The pilus shown in the individual panels corresponds to a magnified view of the pilus in the inset.
Assessment of TcpA amino acids required for TcpA 15 antiserum reactivity.
Previously, Kirn and colleagues reported the effect of engineered missense mutations within tcpA on the structure and functional activity of TCP (8). They found that six residues in classical TcpA within a 16-amino-acid run (K172 to K187) could be mutated to Ala and not affect expression of TCP but could affect TCP functions: autoagglutination capacity, colonization efficiency, phage transduction efficiency, or serum resistance. These residues (K172, D175, H181, E183, K184, and K187) were termed interactive residues. TcpA 15 peptide contains four of the six interactive residues.
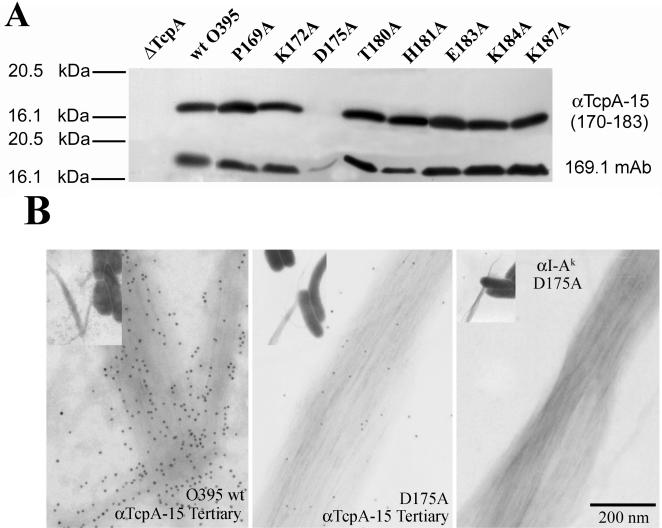
We examined pilin containing altered interactive residues by Western blotting analysis to determine if any of the amino acid changes affected the ability of anti-TcpA 15 serum to bind linear TcpA (Fig. 7A). Pilins with substitution of Ala residues in TcpA residues upstream or downstream of amino acids 170 to 183 were used as controls. The anti-TcpA monoclonal antibody 169.1 served as a positive control (15). This monoclonal antibody reacts with TcpA 5 (157 to 181), binds TCP with high density, and provides protection in the infant mouse assay (15; unpublished observations). Anti-TcpA 15 serum was able to react with TcpA pilin that had amino acid changes within the TcpA 15 sequence (170 to 183) and outside of the TcpA 15 sequence. The exception was that anti-TcpA 15 serum was unable to bind TcpA with an Ala substitution at 175, which is an aspartate in classical TcpA. The alteration (diminution) of binding was also seen when monoclonal antibody 169.1 was used to probe the D175A derivative. We used immunoelectron microscopy to assess the ability of anti-TcpA15 serum to bind the D175A mutant within TcpA that was folded and assembled into TCP. The Ala substitution of D175 severely reduced anti-TcpA 15 binding to assembled, morphologically normal TCP (Fig. 7B). The isotype-matched monoclonal antibody control did not bind to the D175A pilus.
FIG. 7.
Western blot and immunoelectron microscopic analyses of V. cholerae expressing wild-type TcpA (toxin-coregulated pilus protein A), TcpA-deleted strains, and various pilin mutant strains that express TcpA with an Ala substitution in a specific interactive residue (8) (A) Anti-TcpA 15 tertiary serum (upper panel) diluted 1:12,500 was used to probe the membranes and compared to mouse monoclonal antibody 169.1 (lower panel), which is specific for TcpA 5 and known to provide protection in the infant mouse assay (15). (B) The effect of the D175A alteration on binding of anti-TcpA 15 serum to TCP was tested. The D175A change resulted in the complete loss of binding of TcpA 15 serum. An isotype control monoclonal antibody was negative for binding to TCP.
DISCUSSION
Colonization of bacteria at specific host sites is often a rate-limiting step in the pathogenesis of the particular pathogen. In cholera, for disease to occur, a large inoculum of V. cholerae is required for a sufficient number of live bacteria to transit the acidic pH of the stomach and the bile-rich regions of the intestine. Responding to environmental cues, surviving bacteria upregulate a set of genes required for colonization (9). One of these genes is tcpA, which is contained on the vibrio pathogenicity island (6). TcpA and a number of coregulated proteins (TcpB, -C, -D, -E, -F, -Q, -R, -S, and -T) are required for assembly, export, and function of TCP, a type IVb pilus (3, 7, 16). These pili are thought to be important for “self-adherence” so that V. cholerae can form microcolonies in the area of crypt epithelium (8). An expressed TcpA product and assembled pilus is not sufficient for colonization, as ΔtcpF strains produce morphologically normal TCP but do not colonize infant mice (7).
The mechanism by which anti-TcpA or anti-TCP antibodies are protective is not known. Our results presented here and the recently published model of the type IVb pilus of V. cholerae suggest a potential protective mechanism for anti-TCP antibodies, that is, the ability of TcpA-specific antibodies to bind pilus in a specific area associated with the quaternary structure generated by the assembly of TcpA monomers. The residues in TcpA 15, amino acids 170 to 183, are contained in TcpA 5 (amino acids 157 to 181) and TcpA 6 (amino acids 174 to 199). Both TcpA 5 and TcpA 6 induced protective antibody responses in rabbits (12, 14). If the B-cell epitope(s) in TcpA 5 and TcpA 6 is the same as those contained within TcpA 15, this would localize the protective B-cell epitope to amino acids 174 to 181, the common sequence between TcpA 5, TcpA 6, and TcpA 15. Because of the relative position (very close to the N terminus) in TcpA 6 of D175, a critical amino acid recognized by anti-TcpA 15 serum, it is more likely that the sequence from 174 to 181 represents an area that is sensitive to antibody binding that disrupts TCP function rather than the antibodies being specific for exactly the same B-cell epitope.
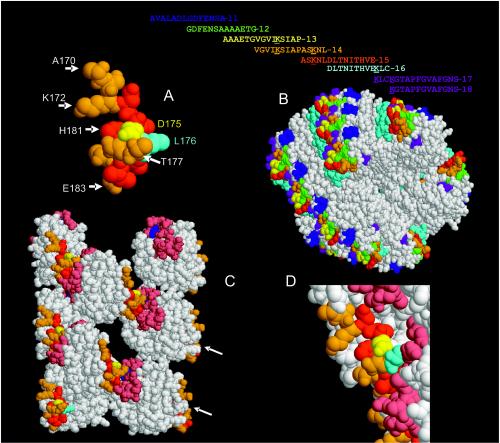
If the sequence of TcpA15 is examined for conserved residues between V. cholerae biotypes and isolates that can colonize infant mice (1, 11) an interesting picture emerges. TcpA 15 contains four (K172, D175, H181, and E183) of the six interactive amino acids (K172, D175, H181, E183, K184, and K187) previously identified as being important for TCP function (Fig. 8A). Substitution of Ala for the various wild-type interactive amino acids selectively alters some functions of TCP. These functions include autoagglutination capacity (K172, K184, and K187) efficient host colonization (D175, H181, E183, K184, and K187), serum resistance (K172, K184, and K187), and phage transduction efficiency (8). In particular, substitution of Ala at residue D175 still allows V. cholerae to maintain its autoagglutination phenotype, but this strain has a 100-fold defect in its colonization capacity. Aspartate 175 is important for anti-15 TcpA binding to SDS-linearized TcpA in a Western blotting assay and for binding of assembled pilus. TcpA 15 is nested within several of the TcpA peptides (TcpA 13, TcpA 14, and TcpA 17) that were not able to induce protective serologic responses (Fig. 8B).
FIG. 8.
Computer-generated space-filling model of TcpA (toxin-coregulated pilus protein A) 15 and its location on TCP. The RasMol version 2.7.2.1 software was used to display and manipulate the Brookhaven Protein Data Bank files 1OQV (TcpA, panel A) and 1OR9 (TCP model, panels B to D) (3). (A) TcpA 15 comprises amino acids 170 to 183. TcpA 15 contains four (K172, D175, H181, and E183) of the six interactive amino acids (K172, D175, H181, E183, K184, and K187) previously identified as being important for TCP function. The TcpA residues (N173, L174, L176 [cyan], I179, H181, and V182) that do not vary between biotypes or colonizing-competent environmental isolates of V. cholerae are colored red. Eight of the 14 residues that comprised TcpA 15, A170, S171, K172, D175 (yellow amino acid), T177, N178, T180, and E183 (orange amino acids), vary between naturally occurring, functional TcpA alleles. Residue L176 is involved with Y51 in a hydrogen bond interaction. (B) The primary amino acid sequence of TcpA peptides (11 to 18) used to immunize rabbits are shown in the one-letter code and color coded for the individual TcpA peptides which are displayed on the TCP fiber model. Lysine residues are underlined. A TCP fiber model has been proposed (3). TcpA 15 is red and is partially exposed, nested between other TcpA peptides, all of which are displayed in a repeating pattern on the surface of the TCP fiber. (C) Residues 30 to 199 of selected chains (C, B, E, H, G. J, M, and L) of the TCP model were restricted (displayed without N termini) with the RasMol program and displayed. This TCP representation shows that D175 (yellow) is exposed and available for antibody binding. There is a recessed “pocket,” 30 by 20 Å, where TcpA 15 is located within the quaternary structure of TCP. Orange residues represent amino acids that can vary in TcpA 15, while red residues are conserved. Leucine 176 is cyan, and Y51 is blue. Residues 50 to 70 (pink, except 51) represent the sequence area in the α/β loop that bind the D region on an adjacent monomer. (D) Enlarged area showing the face of TcpA on adjacent monomers that are proposed to interact. The color scheme for the amino acids is as above.
TcpA 15 can be divided into two parts based on the conserved nature of the residues (Fig. 8A). Hydrophilic residues K172, E175, and E183 are on and adjacent to the 4α helix in the D region (3). Histidine 181, which is partially buried, is a conserved residue that if changed to an Ala produces morphologically normal pilus but results in a state of either excess TcpA production or destabilized TCP, as increased amounts of TcpA monomers are found in spent culture supernatants (8). Two of the interacting residues, 172 (A, K, T, or V) and 175 (D, K, or N) are allowed to vary between biotypes yet still maintain a functional TcpA that can form pili that mediate wild-type levels of colonization (1, 11). Residues N173, L174, L176, I179, H181, V182, and E183 are conserved amino acids between the classical, El Tor biotypes, and other colonization-competent vibrios. Residues in the D region of the TcpA subunit are involved in subunit interaction. In particular, L176, which does not vary between classical and El Tor TcpA, is predicted to form a hydrogen bond with Y51 in the α/β loop of an adjacent classical TcpA subunit (Fig. 8C) (3). The maintenance of specific residues among colonization-competent V. cholerae suggests the involvement of residues for a necessary function which could be targets for protective antibodies. However, based on the TCP model, many of the conserved residues appear to be partially buried in the TCP fiber model, while other TcpA 15 residues, specifically K172, D175, and E183, project away from the pilus (Fig. 8D).
While it is clear the anti-TcpA 15 serum is protective, it is not evident why antisera to the TcpA residues defined by TcpA 11, TcpA 13, TcpA 14, and TcpA 17 are not. If the protective effect was from generalized “blocking” of an area related to function provided by amino acids 170 to 183, then the footprint (700 to 1,200 Å2) of anti-TcpA 11, TcpA 13, TcpA 14, and TcpA 17 Fabs (fraction antigen binding; heavy and light chains) might be expected to occlude the sequence defined by TcpA 15. The area on the TCP model defined by the TcpA peptides that surround TcpA 15 is on the order of 10 to 20Å (Fig. 8D). Thus, either these nonprotective antibodies do not have enough affinity or are not in high enough concentration or the epitopes they bind are not as easily accessible as are the sequences bound by TcpA 15. This concept is supported in part by the data presented here: anti-TcpA 11, anti-TcpA 13, and anti-TcpA 17 serum bind soluble TcpA very well but not assembled pilus. Based on the model of TCP (3) and the protective efficacy of anti-TcpA 15 serum, TcpA 15 is positioned to have a major impact on TCP structure (Fig. 8D). The issue that needs to be resolved is how antibody binding in this region provides protection.
Acknowledgments
This work was supported by NIH grants to R.K.T. (AI25096) and W.F.W. (AI 47373).
We thank Lisa Craig, Scripps Institute, for providing the Escherichia coli Origami DE3 competent cells expressing the polyhistidine-tagged wild-type TCP protein.
Editor: J. D. Clements
REFERENCES
- 1.Boyd, E. F., and M. K. Waldor. 2002. Evolutionary and functional analyses of variants of the toxin-coregulated pilus protein TcpA from toxigenic Vibrio cholerae non-O1/non-O139 serogroup isolates. Microbiology 148:1655-1666. [DOI] [PubMed] [Google Scholar]
- 2.Chaicumpa, W., and D. Rowley. 1972. Experimental cholera in infant mice: protective effects of antibody. J. Infect. Dis. 125:480-485. [DOI] [PubMed] [Google Scholar]
- 3.Craig, L., R. K. Taylor, M. E. Pique, B. D. Adair, A. S. Arvai, M. Singh, S. J. Lloyd, D. S. Shin, E. D. Getzoff, M. Yeaker, K. T. Forest, and J. A. Tainer. 2003. Type IV pilin fold and assembly based on X-ray crystallographic and EM analyses of Vibrio cholerae toxin coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol. Cell 11:1139-1150. [DOI] [PubMed] [Google Scholar]
- 4.Hang, L., J. M. Asaduzzaman, E. A. Bridges, C. Vanderspurt, T. J. Kirn, J. D. Hillman, A. Progulske-Fox, M. Handfield, E. T. Ryan, and S. B. Calderwood. 2003. Use of in vivo-induced antigen technology (IVIAT) to identify genes uniquely expressed during human infection with Vibrio cholerae. Proc. Natl. Acad. Sci. USA 100:8508-8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herrington, D. A., R. H. Hall, G. Losonsky, J. J. Mekalanos, R. K. Taylor, and M. M. Levine. 1988. Toxin, toxin-coregulated pili and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J. Exp. Med. 168:1487-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karaolis, D. K., S. Somara, D. R. Maneval, Jr., J. A. Johnson, and J. B. Kaper. 1999. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature 399:375-379. [DOI] [PubMed] [Google Scholar]
- 7.Kirn, T. J., N. Bose, and R. K. Taylor. 2003. Secretion of a soluble colonization factor by the TCP type 4 pilus biogenesis pathway in Vibrio cholerae. Mol. Microbiol. 49:81-92. [DOI] [PubMed] [Google Scholar]
- 8.Kirn, T. J., M. J. Lafferty, C. M. P. Sandoe, and R. K. Taylor. 2000. Delineation of pilin domains required for bacterial association into microcolonies and intestinal colonization by Vibrio cholerae. Mol. Microbiol. 35:896-910. [DOI] [PubMed] [Google Scholar]
- 9.Lee, S. H., D. L. Hava, M. K. Waldor, and A. Camilli. 1999. Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell 99:625-634. [DOI] [PubMed] [Google Scholar]
- 10.Meeks, M. D., T. K. Wade, R. K. Taylor, and W. F. Wade. 2001. Immune response genes modulate serologic responses to Vibrio cholerae TcpA pilin peptides. Infect. Immun. 69:7687-7694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rhine, J. A., and R. K. Taylor. 1994. TcpA pilin sequences and colonization requirements for O1 and O139 Vibrio cholerae. Mol. Microbiol. 13:1013-1020. [DOI] [PubMed] [Google Scholar]
- 12.Sun, D., M. J. Lafferty, J. A. Peek, and R. K. Taylor. 1997. Domains within the Vibrio cholerae toxin coregulated pilin subunit that mediate bacterial colonization. Gene 192:79-85. [DOI] [PubMed] [Google Scholar]
- 13.Sun, D., J. J. Mekalanos, and R. K. Taylor. 1990. Antibodies directed against the toxin-coregulated pilus isolated from Vibrio cholerae provide protection in the infant mouse experimental cholera model. J. Infect. Dis. 161:1231-1236. [DOI] [PubMed] [Google Scholar]
- 14.Sun, D., J. M. Seyer, I. Kovari, R. A. Sumrada, and R. K. Taylor. 1991. Localization of protective epitopes within the pilin subunit of the Vibrio cholerae toxin-coregulated pilus. Infect. Immun. 59:114-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun, D., D. N. Tillman, T. N. Marion, and R. K. Taylor. 1992. Production and characterization of monoclonal antibodies to the toxin coregulated pilus (TCP) of Vibrio cholerae that protect against experimental cholera in infant mice. Serodiagn. Immunother. Infect. Dis. 4:73-81. [Google Scholar]
- 16.Taylor, R. K., V. L. Miller, D. Furlong, and J. J. Mekalanos. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. USA 84:2833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]