Abstract
Infection with intestinal nematodes induces profound pathological changes to the gut that are associated with eventual parasite expulsion. We have applied expression profiling as an initial screening process with oligonucleotide microarrays (Affymetrix MG-U74AV2 gene chips) and time course kinetics to investigate gene transcription triggered by the intraepithelial nematode Trichinella spiralis in jejunal epithelium from BALB/c mice. Of the 4,114 genes detected, 2,617 were present in all uninfected and T. spiralis-infected replicates, 8% of which were notably upregulated, whereas 12% were downregulated at the time of worm expulsion (day 14 postinfection). Upregulation of goblet cell mucin gene transcripts intestinal mucin gene 3 (MUC3), calcium chloride channel 5 (CLCA5), and goblet cell gene 4 (GOB4) is consistent with enhanced production and alteration of mucus, whereas a 60- to 70-fold upregulation of transcripts for mast cell proteases 1 and 2 (MCPT-1 and -2) is consistent with intraepithelial mucosal mast cell recruitment. Importantly, there was novel expression of sialyltransferase 4C (SIAT4C), small proline-rich protein 2A (SPRR2A), and resistin-like molecule β (RELMβ) on day 14 postinfection. In contrast, DNase I and regenerating protein 3 (REG3) transcripts were substantially downregulated. Time course analyses revealed early (within 48 h of infection) induction of Siat4c, Sprr2A, and Relmβ and later (within 120 h) induction of Mcpt-1 and -2. The findings demonstrate early innate responses and later inflammatory changes within the epithelium. The early epithelial responses may be associated both with repair (Sprr2A) and with the development of innate immunity (Siat4c and Relmβ).
Intestinal infection can induce substantial pathological changes in the epithelial compartment (1, 37, 59). These may be preceded by specific immune responses in the lamina propria, Peyer's patches and mesenteric lymph nodes (66) and may trigger the activation of intraepithelial lymphocytes and the recruitment of inflammatory cells into the epithelial compartment (7, 39). The epithelium can itself respond to pathogens, for example, by the secretion of chloride ions and water by enterocytes (11, 51), by increased polyimmunoglobulin receptor-mediated transport of pathogen-specific immunoglobulin A (IgA) across the enterocytes (10), and by the release of mucus from goblet cells and of antimicrobial peptides from Paneth cells (37, 49). Thus, the resulting elimination of the pathogen and the associated pathology will be a complex product of both innate and pathogen-specific effector mechanisms.
The intestinal nematode Trichinella spiralis dwells within epithelium and actively invades epithelial cells (36, 69). This is associated with major pathological changes within the epithelial compartment such as villus atrophy and crypt hyperplasia, increased epithelial permeability, goblet cell and Paneth cell hyperplasia, and infiltration by mucosal mast cells (20, 25). Some of these profound changes are immunologically mediated and are temporally associated with the rejection of adult worms (25, 33). Infection of BALB/c mice with T. spiralis is a particularly well-characterized model of infection, where parasite expulsion is partially dependent on the recruitment and activity of mucosal mast cells (28, 65)
Although the molecular pathology of intestinal infection is generally studied in the context of specific mechanisms, a global approach such as that afforded by transcriptomics may highlight some of the molecular pathways involved or indeed bring novel mechanisms to light. The aim of the present study was to examine, by using oligonucleotide microarrays (Affymetrix MG-U74AV2), the effect of T. spiralis on the level of gene transcription in small intestinal epithelium during the expulsion phase (day 14 postinfection [p.i.]) when the associated pathological changes are most marked (25, 33). Importantly, these analyses were carried out with preparations of essentially pure epithelium, harvested by the EDTA perfusion technique of Bjerknes and Cheng (6). Thus, we have been able to define the baseline gene expression profiles of mouse small intestinal epithelium and observed changes in these profiles induced by T. spiralis infection.
MATERIALS AND METHODS
Parasite infections and sample preparation.
Maintenance, infection, and recovery of T. spiralis larvae was based on standard methods (67). Age- and sex-matched BALB/c mice (8 to 15 weeks old) were infected by gavage with 200 to 300 muscle larvae/mouse in 0.2 ml of phosphate-buffered saline-0.1% agar freshly isolated from muscle cysts from 30- to 90-day-infected BALB/c mice. To determine whether infections were successful, adult worms were isolated from groups of four to five of the mice 6 to 7 days after infection as described previously (28). For microarray and for reverse transcription-PCR (RT-PCR) analysis, mice were killed on day 14 after infection (worm expulsion phase), and epithelia were purified as described previously (50). Samples were also prepared from four age- and sex-matched uninfected controls. Similar infections were set up in which mice (n = 4) were killed on days 1, 3, 7, 14, 28, and 56 p.i. for epithelium isolation. Stripped epithelium was transferred directly into TriReagent (Sigma, Poole, United Kingdom) for RNA isolation as previously described (50). Subsequent purification and removal of contaminating DNA by using DNA-free DNase (Ambion, Inc.) has been described previously (28). In order to check the cellular composition of stripped epithelium preparations, samples from the epithelial isolates used for microarray analysis were fixed in 4% paraformaldehyde for embedding, sectioning, and hematoxylin-eosin staining (50). Sections were also stained with Alcian Blue for mucin glycoproteins (63). Goblet cells and enterocytes were enumerated from Alcian Blue-stained sections, counting a total of 1,000 to 2,000 cells/sample in four fields under a ×25 objective lens (0.96 mm2). Paneth cells were readily identified histologically from the hematoxylin-eosin-stained sections and were enumerated in the same four areas from each sample. The data are summarized in Table 2.
TABLE 2.
Cellular composition of epithelial isolates
| Day | % Cells (SD)
|
||
|---|---|---|---|
| Enterocytes | Goblet cells | Paneth cells | |
| 0 | 90.0 (0.8) | 8.9 (0.7) | 1.2 (0.1) |
| 14 | 76.4 (1.5)* | 15.5 (0.7)* | 8.2 (1.2)* |
The percentages of enterocytes, goblet cells, and Paneth cells in the epithelial isolates used for microarray analysis are shown for day 0 and day 14 of infection (n = 4). *, P ≤ 0.05 (nonparametric Mann-Whitney test). Note that the day 14 epithelial isolates contained a significantly higher proportion of goblet cells and Paneth cells than the day 0 epithelial isolates.
Processing of RNA for microarray.
For microarray processing, total RNA was prepared from epithelium isolated from uninfected (day 0) and infected (day 14) mice (n = 4 per group) as described above. RNA quality was initially assessed by using formaldehyde gels and by measurement of absorbance at 260/280 nm according to standard procedures. A further assessment of RNA quality was then undertaken with an Agilent Bioanalyzer (Agilent Technologies). First, 25 μg each of four uninfected and four infected samples was subjected to DNase treatment as described above. Then, 1 μg of RNA was reverse transcribed and subjected to RT-PCR to test for the presence or absence of transcripts for the housekeeping gene (mitochondrial creatine kinase [CKMT1]) or mouse mast cell protease 1 (Mcpt-1) as an additional check for RNA quality. Samples (10 μg) were labeled and hybridized to Affymetrix MG-U74AV2 GeneChip arrays according to the standard Affymetrix protocol. This array contains probe sets which represent 12488 transcripts. Scanning and data capture were also undertaken according to standard Affymetrix methods. After datum capture, all arrays were scaled to an overall target intensity of 100 prior to comparison. Filtering and sorting of the data was undertaken in Affymetrix Data Mining Tool software, statistical analyses were carried out in Excel (Microsoft), and GeneSpring (Silicon Genetics) was used for explorative analyses. Briefly, genes of interest were identified by sorting and filtering the data by using the signal value and detection call. For statistical analysis, data were log2 transformed, and a Welch t test (which does not assume equal variance) was then used to identify the most statistically notable changes in gene expression between the day 0 and day-14-infected samples. Prior to explorative hierarchical clustering, all genes defined as “absent” in all eight replicates according to the microarray suite analysis were removed. The coefficient of variance for each remaining gene across all eight replicates was then calculated in Excel, and all genes with a coefficient of variance of <0.4 were discarded. After this second filtering step, which excludes genes not notably changing in expression across all replicates, 1,662 genes remained. This figure represents ca. 13% of the total genes represented on the array. Z-scores were then calculated in Excel for each signal value, and the data were imported into GeneSpring for hierarchical clustering by using Pearson correlation as a distance measure, as outlined by Tavazoie et al. (60). Additional experimental data and links to supplementary data can be obtained from the Scottish Centre for Genomic Technology and Informatics GPX database (www.gti.ed.ac.uk; experiment accession number GXE-00003).
Detection of transcripts by semiquantitative RT-PCR.
In order to further validate the Affymetrix data, 1 μg of RNA was reverse transcribed as previously described (68). Then, a 1/20 volume was amplified by PCR with the gene-specific primers described below, with equivalent quantities of non-reverse-transcribed RNA as negative controls. Reaction conditions were optimized to ensure the number of thermocycles used correlated with the amplification stage of the PCR, and the magnesium concentration was optimized if necessary by using a PCR Optimisation Kit according to the manufacturer's protocol (Roche, Welwyn Garden City, United Kingdom). Primers (68) and PCR conditions (27) for the protease genes Mcpt-1 and Mcpt-2 have already been described. Primers, probes, annealing temperatures, and cycling times for the decay acceleration factor 1 (Daf1), DNase I, resistin-like molecule β (Relmβ), the regenerating proteins 3α, 3β, and 3γ (Reg3α, β and 3γ), the small proline-rich protein 2A (Sprr2A), sialyltransferase 4C (Siat4c), and Ckmt1 as a housekeeping gene are summarized in Table 1. After optimization, all PCRs were carried out in a final magnesium concentration of 1.5 mM (pH 8.3). Amplifications were carried out for 40 s at 94°C, for 40 s at each annealing temperature, and for 120 s at 72°C for the appropriate number of thermocycles. Gene-specific primers and internal complementary probes were designed by using the “RawPrimer” program (http://alces.med.umn.edu/rawprimer.html), checking the specificity by searching the NCBI GenBank nucleotide database (http://www.ncbi.nlm.nih.gov), with the exception of the published primers specific for Sprr2A (56) and Daf1 (35). PCR products were visualized on ethidium bromide-stained 1.6% agarose gels, and images were recorded by using a Kodak Digital Science Image Station 440CF and 1D Image Analysis software. In order to confirm the identity of the PCR products, gels were subjected to Southern blotting according to standard protocols and hybridization with digoxigenin-labeled internal gene-specific probes (see Table 1) as described previously (68).
TABLE 1.
Primers, probes, annealing temperatures, and number of cycles used for RT-PCR analysis for genes of interest
| Gene | Oligonucleotidesa | Product size (bp) | Annealing temp (°C) | No. of cycles |
|---|---|---|---|---|
| Reg3α | Fwd, 5′-TGGGAGTGGAGTAACTCCGAT; Rev, 5′-AAAGCAAGTACAGCCTTGCCG; Probe, 5′-GGGAGACTATTACTGCGATGGGA | 257 | 55 | 28 |
| Reg3β | Fwd, 5′-ATCTACTGCCTTAGACCGTGCTT; Rev, 5′-AGAATTGAGCCCAAACTTATACC; Probe, 5′-AGACAGCAAACATCCCGAATT | 294 | 60 | 32 |
| Reg3γ | Fwd, 5′-TTACATCAACTGGGAGACGAATC; Rev, 5′-TGAGAATTAAGGCCATAGTGCAC; Probe, 5′-AACATTCTCCCCACAATCGAA | 321 | 55 | 28 |
| Sprr2A | Fwd, 5′-CTGAGACTCAAGTACGATGTCTTACTACC; Rev, 5′-TTTTCTGTGAGGAGCCATCATAGGCAC; Probe, 5′-CCTTGTCCTCCTCCAAAGTG | 447 | 60 | 28 |
| DNase I | Fwd, 5′-GATTTCCAAGCAGAATACGGACT; Rev, 5′-GAGATGGTGGAATGCTGTAGTACC; Probe, 5′-TGGCTGAAGCCATCAGTGACCATTA | 175 | 60 | 34 |
| Siat4c | Fwd, 5′-ATAAAGAGCCTGGAGTGTCGTCG; Rev, 5′-TTTCCAGAAGCCTTTTCGCAC; Probe, 5′-GTCATCAACAAGTACGACGTG | 175 | 60 | 34 |
| Daf1 | Fwd, 5′-ATGATCCGTGGGCGGGCGCCT; Rev, 5′-ATCTATGCACCGGGGTGGTGGAC; Probe, 5′-CAAGGAAAGGAGAGGAAGGATGTC | 846 | 60 | 28 |
| Relmβ | Fwd, 5′-AAGGAAGCTCTCAGTCGTCAA; Rev, 5′-GAGTCAGGTTTCCTGGTCGAG; Probe, 5′-CTGTGGATCGTGGGATATCCG | 286 | 55 | 28 |
| Ckmt1 | Fwd, 5′-TGTGATCCAAGAGCGGCATAA; Rev, 5′-GCATCCACCACAACACGTTCTAC; Probe, 5′-GACCTTGATGCCAGTAAAATTCGTT | 209 | 58 | 32 |
Fwd, forward; Rev, reverse.
RESULTS
Cellular composition of the epithelial isolates used for microarray analysis.
Examination of samples of stripped epithelium by light microscopy confirmed that epithelial isolates used for microarray analysis consisted predominantly of epithelium and contained both crypts and villi, as described previously (50). This was further confirmed by examination of hematoxylin-eosin-stained histological sections, and levels of contaminating nonepithelial cells were below 10%. The percentages of enterocytes, goblet cells, and Paneth cells in the day 0 and day 14 paraformaldehyde-fixed epithelial isolates are presented in Table 2. The day 14 samples had a significantly (P ≤ 0.05) higher percentage of goblet cells and Paneth cells and lower percentage of enterocytes than the day 0 samples, reflecting the goblet and Paneth cell hyperplasia associated with T. spiralis infection (20, 25). It was not possible to identify mast cells in the paraformaldehyde-fixed epithelial isolates by staining them with toluidine blue, by using monoclonal antibody RF6.1 to Mcpt-1, or for esterase by conventional techniques (27), probably due to degranulation during fixation. However, intraepithelial mucosal mast cells have repeatedly been shown to be present in T. spiralis-infected mice at the time of worm expulsion (e.g., references 18, 28, and 38; reviewed in reference 20), and Mcpt-1 transcripts are also readily detectable in epithelial isolates (50).
Strategy for analysis of gene transcription data.
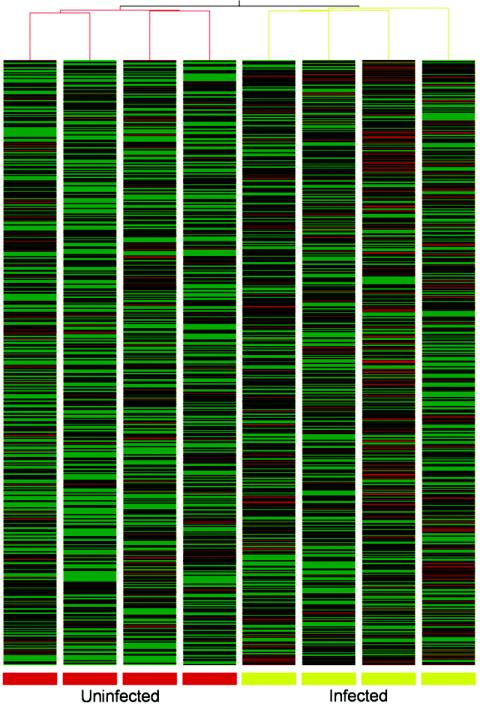
The quantitative data from the microarray analysis were analyzed in several ways. Ranking the transcripts in the uninfected epithelium in order of level of hybridization signal intensities provided information on the baseline expression of genes in this tissue. By comparing the sets of data from uninfected and infected epithelium and filtering the data on the magnitude of change and associated P value (unpaired Welch t test), it was possible to determine which transcripts were notably and consistently affected by infection and to select the most extreme examples for further time course analysis. Hierarchical clustering of a subset of the most variable genes in this experiment was undertaken in GeneSpring. This resulted in a clear segregation of the uninfected and infected samples (Fig. 1). Finally, grouping together transcripts that are known to be specific for particular intestinal epithelial cell types or processes allowed us to observe how these genes change in response to infection.
FIG. 1.
Clustering of differentially expressed genes. To investigate the similarity of samples in this experiment, hierarchical clustering (using Pearson correlation as a distance measure) of a filtered subset of 1,662 variable genes (coefficient of variation > 0.4) was undertaken in GeneSpring (Silicon Genetics). The dendrogram relates samples by their gene expression pattern with short branch length, indicating similarity. Each column represents one sample, and each row represents a gene. Biological replicates (uninfected, red branches; infected, green branches) cluster together. To aid in visualization of the data, gene expression is shown as a color representation of the Z-score (see Materials and Methods for a description of data analysis). Green represents a decrease in expression from the mean value, whereas red represents an increase.
Baseline gene expression in uninfected jejunal epithelium and changes as a consequence of T. spiralis infection.
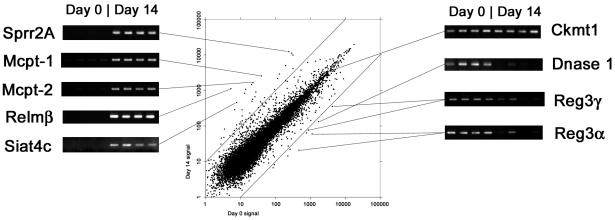
The initial aim of this study was to define the expression profiles of genes in isolated epithelium from the uninfected mice. The 50 transcripts giving the highest hybridization signal values are listed in Table 3. The cellular specificity of expression for each transcript is stated, where known, from literature surveys. From these results, it is clear that the majority of highly expressed transcripts are epithelium specific and, in particular, Paneth cell products are well represented. We then investigated differences between uninfected and infected epithelium. Overall, 4,114 (33%) of the 12,488 genes represented on the MG-U74Av2 gene gave a mean positive detection value across all of the epithelial isolates. Of these, 2,617 genes were present (detection value P) in all uninfected and T. spiralis-infected replicates, 216 (8%) of which increased on infection (P ≤ 0.05 [Welch t test]), whereas 330 (12%) decreased (P ≤ 0.05). Transcription of a further 9 genes became completely undetectable after infection (P in all uninfected samples and A [absent] in all infected samples), whereas 47 genes were apparently induced (A in all uninfected samples and P in all infected samples). Variations from normality were identified by sorting and filtering the data and by using a scatter-graph representation of the gene expression results (Fig. 2). It is possible that genes that show extreme variation in transcription between control and infected epithelium could play an important role in nematode expulsion or in epithelial pathology. We therefore selected candidates for further analysis from the subset of transcripts for which there was an “effective-fold change” increase of ≥8 or decrease of ≥10 on infection. For the purposes of comparison, the effective-fold change includes instances in which the lower value is beneath the threshold level of detection. Increases in transcription level were noted for Sprr2A (37-fold), Mcpt-1 and Mcpt-2 (64- and 71-fold, respectively), Relm-β (206-fold), and Siat4c (76-fold). These findings were confirmed by RT-PCR with gene-specific primers (Fig. 2). Decreased transcription of Islet of Langerhans Reg proteins 3α and 3γ (also known as pancreatitis associated proteins 2 and 3) by 19- and 12-fold, respectively, and of DNase I by 13-fold, was also confirmed by RT-PCR, whereas Ckmt1 did not vary between the day 0 and day 14 samples.
TABLE 3.
The fifty most abundant transcripts detected in uninfected jejunal epithelium
| Probea | GenBank no. | Name | Cell type (fold change)b |
|---|---|---|---|
| 92812_f_at | M33225 | Cryptdin-1 | P |
| 96094_at | U79575 | Apolipoprotein A-I | |
| AFFXActinMur/M12481_3_at | V00478 | Actin cytoplasmic 1 | |
| 100351_f_at | U02997 | Cryptdin-2 | P |
| 99551_f_at | U12560 | Cryptdin-5 | P |
| 100078_at | M13966 | Apolipoprotein A-IV | |
| 95766_f_at | U03066 | Cryptdin-16 | P |
| 93863_f_at | U12561 | Cryptdin-6/12 | P |
| 100213_f_at | U93862 | 60S ribosomal protein L41 | |
| 93879_f_at | U12559 | Cryptdin-3 | P |
| 101255_at | X51703 | Ubiquitin | |
| 96009_s_at | D13509 | Pancreatitis-associated protein 1 | |
| 100106_at | D38410 | Intestinal trefoil factor | G |
| 99926_at | AB001489 | Polyimmunoglobulin receptor | E |
| AFFX-GapdhMur/M32599_3_at | M32599 | Glyceraldehyde-3-phosphate dehydrogenase | |
| 160221_at | AA709879 | RIKEN cDNA 2200008F12 gene (similar to Q9CQW5 [mouse galectin-2]) | |
| 160145_at | AI593999 | RIKEN cDNA 1810010M01 gene | |
| 98092_at | AA790307 | Onzin | |
| 94061_at | M13018 | Cysteine-rich protein 1 | |
| 101794_f_at | U12562 | Cryptdin 1 | P |
| 94805_f_at | M33988 | Histone H2A.1 | |
| 92735_at | X74266 | Phospholipase A2 (membrane associated) | P |
| 95621_at | AA606367 | RIKEN cDNA 9030623C06 gene | |
| 102755_at | AB016496 | Intelectin-1 | P |
| 160099_at | AF026799 | Galectin-6 | |
| 94766_at | M22432 | Elongation factor 1-alpha 1 | |
| 97887_at | Z15090 | Apolipoprotein C-II | |
| 99993_at | U77083 | Aminopeptidase N | (−2.8) |
| 97889_at | M65034 | Fatty acid-binding protein (intestinal) | E |
| 101009_at | X12789 | Keratin type II (cytoskeletal 8) | |
| 99631_f_at | L06465 | Cytochrome c oxidase polypeptide VIa-liver (mitochondrial) | |
| 99660_f_at | X52940 | Cytochrome c oxidase polypeptide VIIc (mitochondrial) | |
| 96573_at | X13055 | Actin cytoplasmic 2 | |
| 101213_at | X15267 | 60S acidic ribosomal protein P0 | |
| 95569_at | U95182 | Uroguanylin | E |
| 98000_at | J04634 | Cell surface antigen 114/A10 (lymphocyte antigen 64) | (−2.4) |
| 101886_f_at | AJ413953 | ARF-related protein 1 | |
| 94738_s_at | M33227 | Cryptdin-related protein 4C-1 | P (−6.5) |
| 102364_at | J04509 | Transcription factor jun-D | |
| 97826_at | AI465965 | Clone IMAGE:4988271 similar to human IgG Fc binding protein [BAA19526] | |
| 102599_at | X06407 | Translationally controlled tumor protein | |
| 94075_at | Y14660 | Fatty acid-binding protein (liver) | |
| 95072_at | AK002815 | Cytochrome c-1 | |
| 94492_at | AB025406 | Destrin (actin-depolymerizing factor) | |
| 94767_at | U93864 | 40S ribosomal protein S11 | |
| 101753_s_at | X51547 | Lysozyme C type P | P |
| 97540_f_at | M69069 | Histocompatibility 2 (D region locus 1) | |
| 160611_at | AK004724 | Family 4 cytochrome P450 | |
| 93798_at | AI839988 | Similar to ATPase (Na+K+ transporting) alpha 1 polypeptide | |
| 93120_f_at | J00400 | H-2 class I histocompatibility antigen (K-B alpha chain) |
Affymetrix probe set identification numbers are shown, ranked in decreasing order of abundance in uninfected epithelium.
The cell specificity, where known: P, Paneth cell; G, goblet cell; E, enterocyte. When any transcript was found to change significantly (P ≤ 0.001 [t test with Welch correction; n = 4]) on infection, the fold change is noted in parentheses).
FIG. 2.
Gene expression in jejunal epithelium at day 0 and day 14 of T. spiralis infection. Preparation of epithelium, extraction of mRNA and microarray procedures are described in Materials and Methods. The mean day 0 signal (n = 4) is plotted against mean day 14 signal (n = 4) for each gene probe set present on the Affymetrix MG-U74AV2 array. Diagonal lines indicate the 10-fold level of up- and downregulation. The altered expression of selected transcripts is confirmed by separate RT-PCR analysis with specific primers (see Materials and Methods). Transcripts for Sprr2A, Mcpt-1 and Mcpt-2, Relmβ, and Siat4c are clearly upregulated. Transcripts for DNase I and Reg3α and -γ are downregulated, whereas mitochondrial creatine kinase (Ckmt1), a highly expressed housekeeping gene, is unchanged.
Analysis of gene transcription by cell type and function.
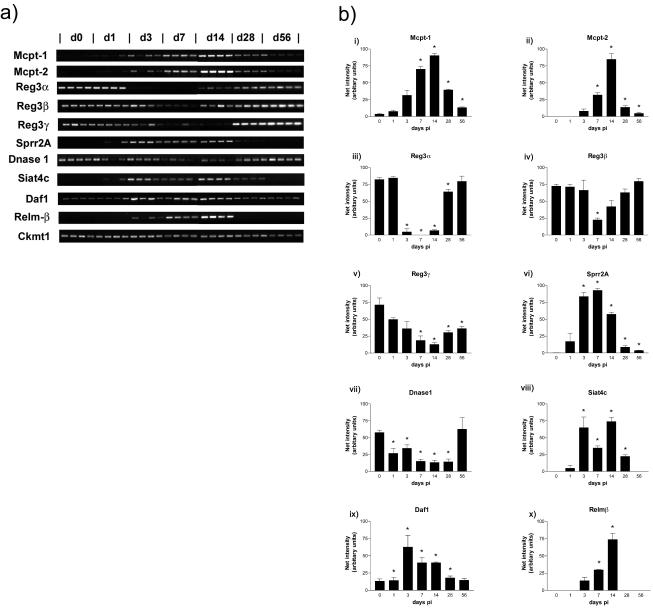
Analyses of gene transcription were carried out with respect to the different cell types known to undergo changes that are maximal during T. spiralis expulsion, such as goblet cell, Paneth cell, and mast cell hyperplasia and enterocyte-associated pathology (20, 25). Transcripts specific for these cell types or sharing a given set of functions are listed in Table 4. To confirm the data obtained and to examine the temporal expression of genes or gene families of interest that changed 8- to 10-fold or more during infection, we performed semiquantitative RT-PCR analysis with epithelial RNA from uninfected mice (day 0) and infected mice on days 1, 3, 7, 14, 28, and 56 p.i. with T. spiralis (n = 4 per time point) (Fig. 3).
TABLE 4.
Relative abundance and probable cellular location of selected transcripts
| Transcript group and name | Affymetrix probe identification no. | GenBank no. | Mean relative abundancea (n = 4) on:
|
Fold change | Pb | |
|---|---|---|---|---|---|---|
| Day 0 | Day 14 | |||||
| Goblet cell/Paneth cell transcripts | ||||||
| Siat4c | 95599_at | D28941 | 13 | 1,068 | 75.9 | 2.58 × 10−7 |
| Clca3 (Gob-5) | 162287_r_at | AV373378 | 3,728 | 7,497 | 2.0 | 0.0006 |
| Glandular kallikrein K9 | 94716_f_at | M17962 | 279 | 972 | 2.8 | 0.001 |
| Gob-4 | 101075_f_at | AB016592 | 1,284 | 1,869 | 1.5 | 0.0028 |
| Muc3 | 101390_at | AF027131 | 1,735 | 3,327 | 1.9 | 0.0003 |
| Phosphomannomutase 1 | 93360_at | AF007267 | 13 | 834 | 64.1 | 4.14 × 10−6 |
| Relmβ/FIZZ2 | 93755_at | AA611740 | 5 | 1,103 | 206.2 | 3.55 × 10−6 |
| Cryptdin-related protein 4C | 94738_s_at | M33227 | 5,137 | 791 | −6.5 | 0.001 |
| DNase 1 | 92865_at | AJ000062 | 1,728 | 130 | −13.3 | 0.006 |
| Mast cell transcripts | ||||||
| Mcpt-1 | 94728_f_at | X68803 | 39 | 2,519 | 63.8 | 0.0006 |
| Mcpt-2 | 99958_at | J05177 | 23 | 1,631 | 71.0 | 0.0003 |
| Innate/immunological transcripts | ||||||
| CD24a | 100600_at | M58661 | 506 | 2,455 | 4.8 | 0.0002 |
| Daf1 | 103617_at | D63679 | 45 | 365 | 8.2 | 0.0002 |
| RP105 (Ly64) | 98000_at | J04634 | 5,187 | 2,127 | −2.4 | 0.0005 |
| Metabolic enzymes | ||||||
| Aldolase 1, A isoform | 160090_f_at | Y00516 | 1,721 | 3,824 | 2.2 | 4.42 × 10−5 |
| Enolase 1, alpha non-neuron | 160568_at | AI841389 | 2,328 | 3,820 | 1.6 | 0.0003 |
| Lactate dehydrogenase A-4 | 96072_at | M17516 | 4,442 | 7,618 | 1.7 | 0.001 |
| Malic enzyme | 101082_at | J02652 | 27 | 909 | 33.0 | 0.0004 |
| Phosphoglycerate kinase 1 | 93346_at | M15668 | 1,211 | 1,919 | 1.6 | 0.0003 |
| Phosphoglycerate mutase 1 | 160091_at | AW125401 | 711 | 1,012 | 1.4 | 8.34 × 10−5 |
| Glutathione antioxidant screen | ||||||
| Glutathione peroxidase 2 | 99810_at | X91864 | 1,446 | 6,552 | 4.5 | 5.67 × 10−5 |
| Glutathione reductase 1 | 160646_at | AI851983 | 446 | 1,729 | 3.9 | 0.0004 |
| Glutathione S-transferase, omega 1 | 97819_at | AI843119 | 1,361 | 3,382 | 2.5 | 5.69 × 10−6 |
| Enterocyte transcripts | ||||||
| Sprr2A | 101024_i_at | AJ005559 | 293.9 | 11,158 | 38 | 7.42 × 10−7 |
| Reg3α | 103954_at | D63357 | 1,125 | 59.7 | −19 | 0.00086 |
| Reg3β | 96009_s_at | D63359 | 9,079.7 | 2,102.3 | −4.4 | 0.047 |
| Reg3γ | 96064_at | D63362 | 4,301 | 353.7 | −12.2 | 0.0078 |
The mean signal level at day 0 and day 14 is given for the selected transcripts. Values that were below the threshold level of detection (set at a mean of 50) are in boldface. For comparative purposes, the effective-fold change includes instances in which one of the values is below the threshold of detection.
P values are shown for signals that changed significantly on infection (t test with Welch correction; n = 4 [P ≤ 0.001]), with the exception of DNase I, Reg3β, and Reg3γ.
FIG. 3.
Temporal changes in expression of specific transcripts in jejunal epithelium after T. spiralis infection. (a) RT-PCR products for Mcpt-1 and Mcpt-2; Reg3α, -β, and -γ; Sprr2A; DNase I; Siat4c; Daf1; and Relmβ; with mitochondrial creatine kinase (Ckmt1) as a positive control for the RT reactions. Total RNA for RT was extracted from epithelium isolated from T. spiralis-infected BALB/c mice on days 0, 1, 3, 7, 14, 28, and 56 p.i. (n = 4/time point). (b) Histograms showing relative net intensities of the RT-PCR products in panel a normalized against the highest net intensity recorded with each primer set to visualize changes in transcript levels over time. ✽, Significantly different from uninfected levels (P ≤ 0.05).
Goblet cells and/or mucin synthesis.
There was increased expression of a majority of the goblet cell-associated genes confirming, for example, a previously reported increase in the intestinal mucin gene Muc3, which, concurrently with Muc2, is known to be upregulated during T. spiralis infection (54) (Muc2 was not represented on the gene chip used). There was also a marked increase in the expression of calcium-activated chloride channel A3 (Clca3) and Gob-5, along with Gob-4 (Table 4), which are goblet cell proteins involved in mucus secretion (29, 34, 61). Of interest was the new expression of Siat4c in infected epithelium (Table 4 and Fig. 2), which is involved in mucin sialylation (13). Transcripts for Siat4c were absent in RNA from uninfected epithelium as detected by RT-PCR but were significantly upregulated to various levels in all samples as early as day 3 of infection, i.e., shortly after establishment of infection in the small intestine. Transcripts were abundant at all subsequent time points until day 28, becoming undetectable again by day 56 p.i. (Fig. 3a and 3bviii), by which time pathological changes in the intestine are normally resolved (19). Phosphomannomutase 1, which is involved in the assembly of the oligosaccharide chains of glycoproteins, is also newly expressed in infected epithelium. In agreement with previous observations on intestinal trefoil factor expression during nematode infection (62), there was no change in the levels of transcription of trefoil factor 3 (supplementary Table 1 [www.gti.ed.ac.uk; experiment accession number GXE-00003]), one of the most abundant of the goblet cell-derived transcripts detected (25).
Expression of Relmβ (sometimes referred to as “found in inflammatory zone 2′” [FIZZ2]) also occurs de novo (Fig. 2 and 3). Recent data show Relmβ is localized to and secreted by intestinal goblet cells (22). Here we found that Relmβ is highly upregulated in the small intestinal epithelium (Fig. 2), with obvious transcripts on days 3, 7, and 14 p.i., i.e., during worm colonization and expulsion, but absent from day 28 p.i., by which time T. spiralis infections are normally resolved (14) (Fig. 3a and bx).
Paneth cell transcripts.
Paneth cells store and release a number of substances for which antimicrobial activity is well documented, such as cryptdins (alpha-defensins), phospholipase A2 and lysozyme P (2, 49). The transcripts for a number of Paneth cell products gave very high hybridization signals in both uninfected and infected intestinal epithelium, indicating they were highly expressed (Tables 3 and 4 and supplementary Table 1). Cryptdins and cryptdin-related sequences are strongly represented among the 50 most detectable genes in uninfected epithelium (Table 3), and, similarly, transcripts for matrix metalloprotease-7 (MMP-7), a Paneth cell product required for activation of cryptdin precursors in the intestine (2), and the Kazal-type serine proteinase inhibitor, Mpgc60, which is expressed by both Paneth and goblet cells (31) appear to be moderately abundant (supplementary Table 1). There are also high levels of transcripts for intelectin 1, which has been identified in murine intestinal Paneth cells (30) (intelectin 2 was not present on the gene chip used) and for the membrane-associated (group IIA) form of phospholipase A2, which is an important inflammatory mediator in the intestinal mucosa (44; see also supplementary Table 1). Despite the well-documented Paneth cell hyperplasia that is known to occur during nematode expulsion and was apparent histologically in the present study (Table 1), there was only a small, statistically nonsignificant increase in expression of many of these genes with an apparent decrease in the level of expression of lysozyme P (Table 3 and supplementary Table 1). Importantly, Paneth cells are apparently the major source of DNase I in the small intestine (55), which was downregulated 13.3-fold (Fig. 2 and Table 4). Transcripts for DNase I were significantly below uninfected levels on days 1 to 28 p.i., and this was most pronounced on day 14, i.e., during worm expulsion (Fig. 3a and bvii).
Mast cell-related transcripts.
Mast cells are rare in the intestinal mucosae of normal BALB/c mice, but nematode infection is associated with substantial intraepithelial mast cell recruitment, where mast cells migrate into the epithelium and express mucosa-specific proteases (40, 53). Mcpt-1 and -2 are expressed by mast cells in this location, and a massive upregulation of these transcripts is accordingly seen in infected samples (Fig. 2 and Table 4). There was a 60- to 70-fold increase in the expression of Mcpt-1 and -2 (Table 4), whereas Mcpt-5, -6, and -9 were upregulated marginally according to hybridization signal, and Mcpt-4 was absent (supplementary Table 1). Mcpt-5, -6, and -7 are reportedly expressed preferentially in submucosal locations (17, 18) and thus are unlikely to be highly represented in the epithelium. RT-PCR analysis confirmed that transcripts for Mcpt-1 and Mcpt-2 increased soon after worm colonization between days 3 and 28 p.i., becoming maximal on days 7 and 14 p.i., when mucosal mast cells are most abundant in the epithelium (Fig. 3a and bi and ii). Mast cells persist in the mucosa in reduced numbers for some weeks after the infection is resolved (18), so it is unsurprising to find Mcpt-1 and Mcpt-2 transcripts still above baseline levels on days 28 and 56 p.i. The mast cell growth factor, stem cell factor, was detectable at a low transcription level and increased, but not significantly, on infection (supplementary Table 1). The receptor for stem cell factor, c-kit, which is expressed by mast cells and lymphoid cells, was detectable at a low level only after infection (supplementary Table 1).
Enterocyte transcripts.
The bulk of intestinal epithelium is comprised of enterocytes, and the enterocyte-specific intestinal fatty acid-binding protein (FABPI), villin, and villin 2 all gave pronounced hybridization signal intensities in uninfected epithelium (Table 3). Concomitant with goblet cell and Paneth cell hyperplasia and villus atrophy, the relative proportion of enterocytes decreased by ca. 30% (Table 2). This was reflected in the 25 to 30% reduction in signal for enterocyte-specific transcripts, villin and villin-2/ezrin (supplementary Table 1). One of the most pronounced decreases in enterocyte transcripts was in the recently identified regulatory 3 proteins, Reg3α, -β, and -γ (Fig. 2 and Table 4). Reg proteins are associated with cell growth (5), and the Reg3 family is highly expressed in the murine small intestine (43). RT-PCR analyses of Reg3α transcripts show it is very rapidly downregulated after infection by T. spiralis, decreasing significantly by day 3 and increasing again around day 14, returning to preinfection levels by days 28 or 56 p.i. (Fig. 3a and biii [P < 0.05]). Reg3β showed a similar, but less marked alteration in expression to Reg3α (Fig. iv), but Reg3γ transcripts did not decrease significantly until day 7 p.i. (Fig. v). We also confirmed infection-associated downregulation of these transcripts in whole-gut RNA (data not shown).
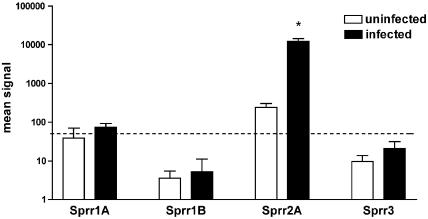
Cytoskeletal transcripts. (i) Sprr transcripts.
The change in expression of small proline-rich protein (Sprr) family members is summarized graphically in Fig. 4. Transcripts for Sprr2A (Fig. 2 and Table 4) increase massively on infection, becoming one of the most highly expressed transcripts. In contrast, transcription of Sprr1A, -1B, and -3, all of which are expressed in epidermis, is minimal (Fig. 4). RT-PCR analysis showed Sprr2A to be upregulated as early as day 1 p.i., when the infective larvae establish in the jejunum, and reaching a maximum around days 3 to 7 p.i., showing expression is triggered rapidly by worm colonization. Transcripts remained significantly above uninfected levels even on day 56 p.i. (Fig. 3a and bvi). It is clear that the Sprr response only involves the Sprr2 family since expression of Sprr1 and -3 was unchanged (Fig. 4).
FIG. 4.
Change in expression of Sprrs by jejunal epithelium in response to T. spiralis infection. The hybridization intensities for Sprrs at days 0 and 14 of infection are compared (expressed as the mean ± the standard deviation [n = 4]). Upregulation of Sprr2A was observed (P ≤ 0.001), but no change was seen for Sprr1A, -1B, or -3. The signal cutoff level of 50 is shown as a broken line.
(ii) Tight junction protein transcripts.
It has been suggested that the increase in gut epithelial permeability that occurs during parasite infection is a consequence of alterations in tight junction proteins (52). Although a decrease in transcription of the tight junction protein occludin was indicated here (supplementary Table 1), which is consistent with many similar observations for a variety of inflammatory conditions of the bowel (21, 32), there was no apparent decrease in zona occludens-1 or junctional adhesion molecule. Claudin-3 and -7 were expressed strongly by uninfected epithelium, in addition to claudin-4, whereas transcripts for claudin-1, -2, -5, and -6 were absent. A slight decrease in claudin-3 expression was seen (supplementary Table 1), but no other changes in claudin expression were observed.
Adaptive and innate immunity or inflammation.
Apart from weak transcription of interleukin-18 (IL-18), the majority of cytokines known to be active in the gut environment (tumor necrosis factor alpha [TNF-α], transforming growth factor β [TGF-β], IL-10, and IL-12) were at background levels, but IL-10Rβ, TNF-αR1α, and soluble IL-4R were all detectable and unaltered by infection (supplementary Table 1). Major histocompatibility class I and II gene transcription was generally downregulated, as was RP105, TECK, and leukotriene A4 hydrolase (Table 4 and supplementary Table 1). In contrast, Daf1 and CD24a were both substantially upregulated (Table 4). Daf1 is a membrane regulator of C3 activation, which has been identified in both airway and intestinal epithelium (35). Daf1 transcripts were found to be significantly upregulated for the duration of infection (days 1 to 28 p.i.) (Fig. 3a and bix). Daf1 is known to be expressed in dendritic cells (35), and we have detected transcripts in cultured mast cells (data not shown), which may contribute to Daf1 expression in the infected intestinal epithelium. Although the jejunal epithelium is rich in intraepithelial lymphocytes, transcript levels for granzyme A and granzyme B were apparently low and were not altered by infection (Table 4). Furthermore, the ubiquitous T-cell marker CD3 was at background levels (supplementary table I [www.gti.ed.ac.uk; experiment accession number GXE-00003]]). Previous work has shown that stripped epithelium preparations can contain low, but detectable levels of IgA transcripts, which is indicative of a low level of B-cell contamination (27). Elevated levels of IgA transcripts were noted for only one of the infected samples, with a resultant, but not statistically significant (P ≤ 0.001) increase in mean infected values (supplementary data I).
Metabolic enzymes and the glutathione antioxidant screen.
A general upregulation of transcription of enzymes of the glycolytic pathway was observed (Table 4 and supplementary data I). In addition, the upregulation of malic enzyme is particularly notable. This enzyme converts malate to pyruvate with the regeneration of NADPH, contributing to the maintenance of the reducing environment of the cell. The NADPH regenerating pentose phosphate pathway, requiring glucose-6-phosphate 1-dehydrogenase, is also apparently upregulated (supplementary Table 1). The NADPH thus provided is required for such processes as fatty acid biosynthesis and antioxidant defense, and in the latter it is utilized by glutathione reductase to convert oxidized glutathione back to glutathione. Transcripts for glutathione reductase increased 3.9-fold on infection (Table 4). The increased hybridization signals for glutathione synthetase and glutathione peroxidase 2 suggest a general increase in antioxidant activity in infected epithelium (Table 4 and supplementary Table 1).
DISCUSSION
Microarray analysis is a useful initial screening technology to monitor the pattern of gene change in a given tissue but requires large sample sizes to allow analysis of the more subtle alterations in the patterns of gene expression. Here we focused predominantly on those genes or gene families showing >8-fold variation from normal and significant at the stringent P ≤ 0.001 level for further investigation by semiquantitative RT-PCR. The transcript profile of normal jejunal epithelium recovered by the EDTA perfusion technique pioneered by Bjerknes and Cheng (6) was strongly represented by the Paneth cell-specific cryptdin family (Table 3). This is consistent with our observations (Table 2) and with the observations by Bjerknes and coworkers that EDTA exfoliates both villus and crypt epithelium. Other highly detectable transcripts included apolipoproteins, intestinal trefoil factor, polyimmunoglobulin receptor, intelectin, and galectin-6. All of these genes are gut associated (4), and many are involved in defense against pathogens. These general observations suggest that the data generated in the current experiment are consistent with the expression profile expected from the jejunal epithelial compartment and, with the exception of occasional B cells, show little contamination from the lamina propria.
Infection with T. spiralis is associated with substantial changes in the epithelium. These include infiltration with mucosal mast cells, villus atrophy, crypt hyperplasia, and goblet cell and Paneth cell hyperplasia (20, 25). The expression of mast cell granule proteases was consistent with the extensive protein and genetic analyses of mucosal mast cell gene expression (48). The assignment of gene transcripts to goblet cells and Paneth cells (Table 4) is based on literature surveys. However, these assignments must be tentative since it is not always clear whether the gene concerned is necessarily expressed in one or the other cell type, for example, the Kazal-type protease inhibitor Mpgc60 (31). Therefore, the assignments here are primarily for the genes considered to be cell specific, but it is likely that there will be overlap, especially where nematode-induced inflammation is concerned. In the case of Paneth cell transcripts, there were small (1.1- to 1.6-fold) but not statistically significant increases in transcripts for defensins, phospholipase A2, intelectin, and MMP-7, which would be consistent with Paneth cell hyperplasia, although lysozyme P expression was slightly depressed. Upregulation of goblet cell genes such as Muc-3, Gob-4, and Clca-3 is consistent with goblet cell hyperplasia, enhanced mucus secretion, and altered mucus viscosity, which has the potential to physically entrap worms and hasten their expulsion from the gut (37). Also relevant is the marked upregulation of Siat4c, since increased sialylation may be involved in the alteration of mucin solubility and increased resistance to degradation (47). Studies in rats infected with Nippostrongylus brasiliensis (26, 46) show that mucins synthesized during infection are altered by the blood group A transferase, α-N-acetylgalactosaminyltransferase, which was upregulated in the intestinal mucosa. Our earlier work showed that the quality and function of intestinal mucin changes during infection with N. brasiliensis (reviewed in reference 37). Similarly, changes in mucin function during infection with T. spiralis have been described by Appleton and coworkers (9), but the biochemical nature of these changes was not investigated. These earlier findings are strongly suggestive of biochemical changes in the mucin, even though the work was carried out in rats. The upregulation of Siat4c is thus consistent with earlier studies. Equally relevant, but a conceptually novel finding is the downregulation of DNase I, with a potential loss of capacity to break down luminal DNA, which could again increase mucus viscosity. It is notable that DNase I is used therapeutically to reduce mucus viscosity in cystic fibrosis cases by reducing the DNA content (70).
Analysis of immune and innate response genes provided rather limited information on genes that may be transcribed in low abundance, and therefore below the level of detection by this method, or are rapidly degraded, like those for the cytokine family. For example, transcripts for cytokine genes known to be expressed in jejunal epithelium (TNF-α and TGF-β) were at background levels, even though we have shown by RT-PCR that there is constitutive expression of TGF-β in this model (27, 50). The expression of IL-10Rβ is consistent with observations on the role of IL-10 in the gut (42), but we are unaware of any studies on expression of IL-10Rα and -β by jejunal epithelial cells. It was notable that RP105, a putative lipopolysaccharide coreceptor with Toll-like receptor-4 function in B cells (45), was highly expressed by normal epithelium. Unfortunately, very few of the Toll-like receptors were represented on the Affymetrix chip used, but it will be interesting to determine whether the lipopolysaccharide response mechanism in B cells also occurs in jejunal epithelium. The general depression of major histocompatibility complex class I and II gene expression, together with a downward trend in β2-microglobulin, was surprising given that enhanced expression of these proteins has been detected by immunohistochemistry in the rat, although the precise timing of the up- and downregulation of major histocompatibility complex genes varies according to the stage of infection (3) and may be different again in the mouse, although this would require extensive immunohistochemical studies for confirmation. It is also possible that posttranscriptional control elements contribute to upregulation of these genes.
Of great interest were the highly up- or downregulated genes which have not been previously associated with nematode infection but have been associated with other models of intestinal regeneration and inflammation. For example, studies on an acute colitis model in mice showed the Reg3 proteins to be upregulated in regenerated colonic epithelial cells (41). Although little is known about the biological function of Reg proteins, they are members of the C-type lectin family, which are known to participate in many cell surface recognition events associated with cell growth and differentiation (5). The pattern of expression of Reg proteins in the intestinal epithelium is therefore of interest in terms of the crypt hyperplasia and/or villus atrophy that is maximal during the expulsion phase of T. spiralis on days 13 or 14 p.i. in BALB/c mice (33). Evidence from N. brasiliensis infection in rats shows that the resultant pathology is a combination of increased apoptosis in the villi and simultaneous acceleration of cell proliferation in the crypts (24), and it may well be that alteration in expression of the different Reg proteins could reflect this balance.
In contrast, Sprr2A expression was significantly increased as early as day 3 of infection (Fig. 3b), and this is a pattern similar to that of Relmβ and Siat4c. Although the substantially increased expression of Sprrs has not previously been described for nematode infection, increased Sprr2A expression was described in regenerating intestine after bowel resection (57) and after bacterial colonization of the intestines of germfree mice (23). It was suggested in the latter study that Sprr2A participates in fortifying the intestinal epithelial barrier in response to bacterial colonization (23). Sprr gene products have a well-documented role in adaptive barrier formation in the skin (8, 64) and are upregulated in a number of skin inflammatory conditions (12). It is notable that, during nematode infection, the intestinal epithelium is flattened from its columnar shape to a cuboidal and sometimes almost squamous appearance (59). We would speculate that the Sprr2 family provide a stabilizing cornifying envelope within the altered cells.
Relmβ, which we found to be newly expressed in intestinal epithelium during T. spiralis infection, has previously been found to be restricted to goblet cells in the colon and cecum of the murine gastrointestinal tract and to be secreted into the stool (22). Preliminary experiments suggest that expression of Relmβ/FIZZ2 is indirectly regulated by STAT-6 (58) and is induced by IL-13 (22). The present observations are consistent with this, since both IL-4 and IL-13 are major regulators of mucosal responses to nematode parasites (16). Enhanced colonic expression and secretion of Relmβ is induced by colonization with normal enteric bacteria (22), and it is also rapidly induced after aerosol challenge of the airway epithelium in systemically primed mice (58). These data and our observations described here indicate Relmβ expression is induced by, and may regulate, the microenvironment of the gastrointestinal tract and is a component of the allergic inflammatory response.
The transcriptome analysis described above provides new insights into the nature of the epithelial response to nematode infection. However, when interpreting these data, it is important to understand the limitations imposed by the experimental systems used. First, the stripping of epithelium by EDTA perfusion and separation of intestinal epithelial cells from the extracellular matrix, while it provides a highly defined cell preparation, could potentially itself induce expressional changes unrelated to infection, although this was minimized by carrying out all postinfusion isolation steps at 4°C (24). Second, as mentioned above, microarray analysis, although a powerful tool for quantifying gene transcripts, is still limited with respect to the total of known murine genes and is not sensitive enough to detect low-copy-number transcripts (15), such as those for some of the low-abundance cytokines. In addition, it must be remembered that altered transcription of a gene may not alter levels of the corresponding protein in a predictable manner. Nonetheless, this analysis has highlighted the very substantial alterations in epithelial gene expression associated with the phase of immunological expulsion of T. spiralis, and we have been able to confirm temporally altered transcription profiles of a range of genes by RT-PCR analysis. The global analysis provided here clearly associates mast cell, goblet cell, and Paneth cell transcripts temporally with nematode expulsion. Importantly, the presence of newly expressed Relmβ and Sprr2A and the significantly decreased transcription of the Reg3 protein family are consistent with other reports of altered expression of these genes during intestinal regeneration and inflammation. Future work, with, for example, transgenic mice lacking cytokines or their receptors, will throw more light on the mechanisms regulating the expression of the genes described in the present study.
Acknowledgments
We thank Judith Pate, Elisabeth Thornron, Eileen Duncan, and Liz Moore for invaluable technical assistance. We also thank Thorsten Forster, Scottish Centre for Genomic Technology and Informatics, University of Edinburgh, and Gary Wu and David Arns, University of Pennsylvania, for helpful discussion.
This study was supported by The Wellcome Trust (grant 060312).
Editor: J. F. Urban, Jr.
REFERENCES
- 1.Artis, D., C. S. Potten, K. J. Else, F. D. Finkelman, and R. K. Grencis. 1999. Trichuris muris: host intestinal epithelial cell hyperproliferation during chronic infection is regulated by interferon-gamma. Exp. Parasitol. 92:144-153. [DOI] [PubMed] [Google Scholar]
- 2.Ayabe, T., D. P. Satchell, P. Pesendorfer, H. Tanabe, C. L. Wilson, S. J. Hagen, and A. J. Ouellette. 2002. Activation of Paneth cell alpha-defensins in mouse small intestine. J. Biol. Chem. 277:5219-5228. [DOI] [PubMed] [Google Scholar]
- 3.Barclay, A. N., and D. W. Mason. 1982. Induction of Ia antigen in rat epidermal cells and gut epithelium by immunological stimuli. J. Exp. Med. 156:1665-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bates, M. D., C. R. Erwin, L. P. Sanford, D. Wiginton, J. A. Bezerra, L. C. Schatzman, A. G. Jegga, C. Ley-Ebert, S. S. Williams, K. A. Steinbrecher, B. W. Warner, M. B. Cohen, and B. J. Aronow. 2002. Novel genes and functional relationships in the adult mouse gastrointestinal tract identified by microarray analysis. Gastroenterology 122:1467-1482. [DOI] [PubMed] [Google Scholar]
- 5.Bernard-Perrone, F. R., W. P. Renaud, O. M. Guy-Crotte, P. Bernard, C. G. Figarella, H. Okamoto, D. C. Balas, and F. O. Senegas-Balas. 1999. Expression of REG protein during cell growth and differentiation of two human colon carcinoma cell lines. J. Histochem. Cytochem. 47:863-870. [DOI] [PubMed] [Google Scholar]
- 6.Bjerknes, M., and H. Cheng. 1981. Methods for the isolation of intact epithelium from the mouse intestine. Anat. Rec. 199:565-574. [DOI] [PubMed] [Google Scholar]
- 7.Bozic, F., D. Forciá, R. Mazuran, A. Marinculiá, Z. Kozariá, and D. Stojceviá. 1998. γδTCR+ intestinal intraepithelial lymphocytes (i-IEL) in reaction against intestinal nematode Trichinella spiralis. Comp. Immunol. Microbiol. Infect. Dis. 21:201-214. [DOI] [PubMed] [Google Scholar]
- 8.Cabral, A., P. Voskamp, A. M. Cleton-Jansen, A. South, D. Nizetic, and C. Backendorf. 2001. Structural organization and regulation of the small proline-rich family of cornified envelope precursors suggest a role in adaptive barrier function. J. Biol. Chem. 276:19231-19237. [DOI] [PubMed] [Google Scholar]
- 9.Carlisle, M. S., D. D. McGregor, and J. A. Appleton. 1991. Intestinal mucus entrapment of Trichinella spiralis larvae induced by specific antibodies. Immunology 74:546-551. [PMC free article] [PubMed] [Google Scholar]
- 10.Coutinho, H. B., T. I. Robalinho, V. B. Coutinho, J. R. Almeida, J. T. Filho, G. King, D. Jenkins, Y. Mahida, H. F. Sewell, and D. Wakelin. 1996. Immunocytochemistry of mucosal changes in patients infected with the intestinal nematode Strongyloides stercoralis. J. Clin. Pathol. 49:717-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crowe, S. E., and M. H. Perdue. 1992. Functional abnormalities in the intestine associated with mucosal mast cell activation. Regional Immunol. 4:113-117. [PubMed] [Google Scholar]
- 12.De Heller-Milev, M., M. Huber, R. Panizzon, and D. Hohl. 2000. Expression of small proline rich proteins in neoplastic and inflammatory skin diseases. Br. J. Dermatol. 143:733-740. [DOI] [PubMed] [Google Scholar]
- 13.Delmotte, P., S. Degroote, M. D. Merten, I. Van-Seuningen, A. Bernigaud, C. Figarella, P. Roussel, and J. M. Perini. 2001. Influence of TNFα on the sialylation of mucins produced by a transformed cell line MM-39 derived from human tracheal gland cells. Glycoconjugate J. 18:487-497. [DOI] [PubMed] [Google Scholar]
- 14.deVos, T., G. Danell, and T. A. Dick. 1992. Trichinella spiralis: dose dependence and kinetics of the mucosal immune response in mice. Exp. Parasitol. 75:99-111. [DOI] [PubMed] [Google Scholar]
- 15.Evans, S. J., N. A. Datson, M. Kabbaj, R. C. Thompson, E. Vreugdenhil, E. R. De Kloet, S. J. Watson, and H. Akil. 2002. Evaluation of Affymetrix Gene Chip sensitivity in rat hippocampal tissue using SAGE analysis. Eur. J. Neurosci. 16:409-413. [DOI] [PubMed] [Google Scholar]
- 16.Finkelman, F. D., T. Shea-Donohue, J. Goldhill, C. A. Sullivan, S. C. Morris, K. B. Madden, W. C. Gause, and J. F. Urban. 1997. Cytokine regulation of host defense against parasitic gastrointestinal nematodes: lessons from studies with rodent models. Annu. Rev. Immunol. 15:505-533. [DOI] [PubMed] [Google Scholar]
- 17.Friend, D. S., N. Ghildyal, K. F. Austen, M. F. Gurish, R. Matsumoto, and R. L. Stevens. 1996. Mast cells that reside at different locations in the jejunum of mice infected with Trichinella spiralis exhibit sequential changes in their granule ultrastructure and chymase phenotype. J. Cell Biol. 135:279-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friend, D. S., N. Ghildyal, M. F. Gurish, J. Hunt, X. Hu, K. F. Austen, and R. L. Stevens. 1998. Reversible expression of tryptases and chymases in the jejunal mast cells of mice infected with Trichinella spiralis. J. Immunol. 160:5537-5545. [PubMed] [Google Scholar]
- 19.Friend, D. S., M. F. Gurish, K. F. Austen, J. Hunt, and R. L. Stevens. 2000. Senescent jejunal mast cells and eosinophils in the mouse preferentially translocate to the spleen and draining lymph node, respectively, during the recovery phase of helminth infection. J. Immunol. 165:344-352. [DOI] [PubMed] [Google Scholar]
- 20.Garside, P., M. W. Kennedy, D. Wakelin, and C. E. Lawrence. 2000. Immunopathology of intestinal helminth infection. Parasite Immunol. 22:605-612. [DOI] [PubMed] [Google Scholar]
- 21.Gassler, N., C. Rohr, A. Schneider, J. Kartenbeck, A. Bach, N. Obermuller, H. F. Otto, and F. Autschbach. 2001. Inflammatory bowel disease is associated with changes of enterocytic junctions. Am. J. Physiol. 281:G216-G228. [DOI] [PubMed] [Google Scholar]
- 22.He, W., M. L. Wang, H. Q. Jiang, C. M. Steppan, M. E. Shin, M. C. Thurnheer, J. J. Cebra, M. A. Lazar, and G. D. Wu. 2003. Bacterial colonization leads to colonic secretion of RELMβ/FIZZ2, a novel goblet cell specific protein. Gastroenterology 125:1388-1397. [DOI] [PubMed] [Google Scholar]
- 23.Hooper, L. V., M. H. Wong, A. Thelin, L. Hansson, P. G. Falk, and J. I. Gordon. 2001. Molecular analysis of commensal host-microbial relationships in the intestine. Science 291:881-884. [DOI] [PubMed] [Google Scholar]
- 24.Hyoh, Y., M. Nishida, T. Tegoshi, M. Yamada, R. Uchikawa, S. Matsuda, and N. Arizono. 1999. Enhancement of apoptosis with loss of cellular adherence in the villus epithelium of the small intestine after infection with the nematode Nippostrongylus brasiliensis in rats. Parasitology 119:199-207. [DOI] [PubMed] [Google Scholar]
- 25.Kamal, M., D. Wakelin, A. J. Ouellette, A. Smith, D. K. Podolsky, and Y. R. Mahida. 2001. Mucosal T cells regulate Paneth and intermediate cell numbers in the small intestine of T. spiralis-infected mice. Clin. Exp. Immunol. 126:117-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karlsson, N. G., F. J. Olson, P. A. Jovall, Y. Andersch, L. Enerback, and G. C. Hansson. 2000. Identification of transient glycosylation alterations of sialylated mucin oligosaccharides during infection by the rat intestinal parasite Nippostrongylus brasiliensis. Biochem. J. 350:805-814. [PMC free article] [PubMed] [Google Scholar]
- 27.Knight, P. A., S. H. Wright, J. K. Brown, X. Huang, D. Sheppard, and H. R. Miller. 2002. Enteric expression of the integrin αvβ6 is essential for nematode-induced mucosal mast cell hyperplasia and expression of the granule chymase, mouse mast cell protease-1. Am. J. Pathol. 161:771-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knight, P. A., S. H. Wright, C. E. Lawrence, Y. Y. Paterson, and H. R. Miller. 2000. Delayed expulsion of the nematode Trichinella spiralis in mice lacking the mucosal mast cell-specific granule chymase, mouse mast cell protease-1. J. Exp. Med. 192:1849-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komiya, T., Y. Tanigawa, and S. Hirohashi. 1999. Cloning of the gene gob-4, which is expressed in intestinal goblet cells in mice. Biochim. Biophys. Acta 1444:434-438. [DOI] [PubMed] [Google Scholar]
- 30.Komiya, T., Y. Tanigawa, and S. Hirohashi. 1998. Cloning of the novel gene intelectin, which is expressed in intestinal Paneth cells in mice. Biochem. Biophys. Res. Commun. 251:759-762. [DOI] [PubMed] [Google Scholar]
- 31.Krause, R., M. Hemberger, M. Messerschmid, W. Mayer, R. Kothary, C. Dixkens, and R. Fundele. 1998. Molecular cloning and characterization of murine Mpgc60, a gene predominantly expressed in the intestinal tract. Differ. Res. Biol. Diversity 63:285-294. [DOI] [PubMed] [Google Scholar]
- 32.Kucharzik, T., S. V. Walsh, J. Chen, C. A. Parkos, and A. Nusrat. 2001. Neutrophil transmigration in inflammatory bowel disease is associated with differential expression of epithelial intercellular junction proteins. Am. J. Pathol. 159:2001-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawrence, C. E., J. C. Paterson, L. M. Higgins, T. T. MacDonald, M. W. Kennedy, and P. Garside. 1998. IL-4-regulated enteropathy in an intestinal nematode infection. Eur. J. Immunol. 28:2672-2684. [DOI] [PubMed] [Google Scholar]
- 34.Leverkoehne, I., and A. D. Gruber. 2002. The murine mCLCA3 (alias gob-5) protein is located in the mucin granule membranes of intestinal, respiratory, and uterine goblet cells. J. Histochem. Cytochem. 50:829-838. [DOI] [PubMed] [Google Scholar]
- 35.Lin, F., Y. Fukuoka, A. Spicer, R. Ohta, N. Okada, C. L. Harris, S. N. Emancipator, and M. E. Medof. 2001. Tissue distribution of products of the mouse decay-accelerating factor (DAF) genes: exploitation of a Daf1 knockout mouse and site-specific monoclonal antibodies. Immunology 104:215-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.ManWarren, T., L. Gagliardo, J. Geyer, C. McVay, S. Pearce Kelling, and J. Appleton. 1997. Invasion of intestinal epithelia in vitro by the parasitic nematode Trichinella spiralis. Infect. Immun. 65:4806-4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, H. R. 1987. Gastrointestinal mucus, a medium for survival and for elimination of parasitic nematodes and protozoa. Parasitology 94:S77-S100. [DOI] [PubMed] [Google Scholar]
- 38.Miller, H. R., J. F. Huntley, G. F. Newlands, A. Mackellar, D. A. Lammas, and D. Wakelin. 1988. Granule proteinases define mast cell heterogeneity in the serosa and the gastrointestinal mucosa of the mouse. Immunology 65:559-566. [PMC free article] [PubMed] [Google Scholar]
- 39.Miller, H. R., and W. F. Jarrett. 1971. Immune reactions in mucous membranes. I. Intestinal mast cell response during helminth expulsion in the rat. Immunology 20:277-288. [PMC free article] [PubMed] [Google Scholar]
- 40.Miller, H. R., and A. D. Pemberton. 2002. Tissue-specific expression of mast cell granule serine proteinases and their role in inflammation in the lung and gut. Immunology 105:375-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mizoguchi, E., R. J. Xavier, H. C. Reinecker, H. Uchino, A. K. Bhan, D. K. Podolsky, and A. Mizoguchi. 2003. Colonic epithelial functional phenotype varies with type and phase of experimental colitis. Gastroenterology 125:148-161. [DOI] [PubMed] [Google Scholar]
- 42.Moore, K. W., R. de Waal-Malefyt, R. L. Coffman, and A. O'Garra. 2001. Interleukin-10 and the interleukin-10 receptor. Annu. Rev. Immunol. 19:683-765. [DOI] [PubMed] [Google Scholar]
- 43.Narushima, Y., M. Unno, K. Nakagawara, M. Mori, H. Miyashita, Y. Suzuki, N. Noguchi, S. Takasawa, T. Kumagai, H. Yonekura, and H. Okamoto. 1997. Structure, chromosomal localization, and expression of mouse genes encoding type III Reg, RegIIIα, RegIIIβ, and RegIIIγ. Gene 185:159-168. [DOI] [PubMed] [Google Scholar]
- 44.Nevalainen, T. J., M. M. Haapamaki, and J. M. Gronroos. 2000. Roles of secretory phospholipases A2 in inflammatory diseases and trauma. Biochim. Biophys. Acta 1488:83-90. [DOI] [PubMed] [Google Scholar]
- 45.Ogata, H., I. Su, K. Miyake, Y. Nagai, S. Akashi, I. Mecklenbrauker, K. Rajewsky, M. Kimoto, and A. Tarakhovsky. 2000. The toll-like receptor protein RP105 regulates lipopolysaccharide signaling in B cells. J. Exp. Med. 192:23-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olson, F. J., M. E. Johansson, K. Klinga-Levan, D. Bouhours, L. Enerback, G. C. Hansson, and N. G. Karlsson. 2002. Blood group A glycosyltransferase occurring as alleles with high sequence difference is transiently induced during a Nippostrongylus brasiliensis parasite infection. J. Biol. Chem. 277:15044-15052. [DOI] [PubMed] [Google Scholar]
- 47.Parker, N., H. H. Tsai, S. D. Ryder, A. H. Raouf, and J. M. Rhodes. 1995. Increased rate of sialylation of colonic mucin by cultured ulcerative colitis mucosal explants. Digestion 56:52-56. [DOI] [PubMed] [Google Scholar]
- 48.Pemberton, A. D., J. K. Brown, S. H. Wright, P. A. Knight, M. L. McPhee, A. R. McEuen, P. A. Forse, and H. R. Miller. 2003. Purification and characterization of mouse mast cell proteinase-2 and the differential expression and release of mouse mast cell proteinase-1 and -2 in vivo. Clin. Exp. Allerg. 33:1005-1012. [DOI] [PubMed] [Google Scholar]
- 49.Porter, E. M., C. L. Bevins, D. Ghosh, and T. Ganz. 2002. The multifaceted Paneth cell. Cell. Mol. Life Sci. 59:156-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosbottom, A., P. A. Knight, G. McLachlan, E. M. Thornton, S. W. Wright, H. R. Miller, and C. L. Scudamore. 2002. Chemokine and cytokine expression in murine intestinal epithelium following Nippostrongylus brasiliensis infection. Parasite Immunol. 24:67-75. [DOI] [PubMed] [Google Scholar]
- 51.Russell, D. A., and G. A. Castro. 1989. Immunological regulation of colonic ion transport. Am. J. Physiol. 256:G396-G403. [DOI] [PubMed] [Google Scholar]
- 52.Scudamore, C. L., M. A. Jepson, B. H. Hirst, and H. R. Miller. 1998. The rat mucosal mast cell chymase, RMCP-II, alters epithelial cell monolayer permeability in association with altered distribution of the tight junction proteins ZO-1 and occludin. Eur. J. Cell Biol. 75:321-330. [DOI] [PubMed] [Google Scholar]
- 53.Scudamore, C. L., L. McMillan, E. M. Thornton, S. H. Wright, G. F. Newlands, and H. R. Miller. 1997. Mast cell heterogeneity in the gastrointestinal tract: variable expression of mouse mast cell protease-1 (mMCP-1) in intraepithelial mucosal mast cells in nematode-infected and normal BALB/c mice. Am. J. Pathol. 150:1661-1672. [PMC free article] [PubMed] [Google Scholar]
- 54.Shekels, L. L., R. E. Anway, J. Lin, M. W. Kennedy, P. Garside, C. E. Lawrence, and S. B. Ho. 2001. Coordinated Muc2 and Muc3 mucin gene expression in Trichinella spiralis infection in wild-type and cytokine-deficient mice. Dig. Dis. Sci. 46:1757-1764. [DOI] [PubMed] [Google Scholar]
- 55.Shimada, O., H. Ishikawa, H. Tosaka-Shimada, T. Yasuda, K. Kishi, and S. Suzuki. 1998. Detection of deoxyribonuclease I along the secretory pathway in Paneth cells of human small intestine. J. Histochem. Cytochem. 46:833-840. [DOI] [PubMed] [Google Scholar]
- 56.Song, H. J., G. Poy, N. Darwiche, U. Lichti, T. Kuroki, P. M. Steinert, and T. Kartasova. 1999. Mouse Sprr2 genes: a clustered family of genes showing differential expression in epithelial tissues. Genomics 55:28-42. [DOI] [PubMed] [Google Scholar]
- 57.Stern, L. E., C. R. Erwin, R. A. Falcone, F. S. Huang, C. J. Kemp, J. L. Williams, and B. W. Warner. 2001. cDNA microarray analysis of adapting bowel after intestinal resection. J. Pediatr. Surg. 36:190-195. [DOI] [PubMed] [Google Scholar]
- 58.Stutz, A. M., L. A. Pickart, A. Trifilieff, T. Baumruker, E. Prieschl-Strassmayr, and M. Woisetschlager. 2003. The Th2 cell cytokines IL-4 and IL-13 regulate found in inflammatory zone 1/resistin-like molecule alpha gene expression by a STAT6 and CCAAT/enhancer-binding protein-dependent mechanism. J. Immunol. 170:1789-1796. [DOI] [PubMed] [Google Scholar]
- 59.Symons, L. E. 1976. Scanning electron microscopy of the jejunum of the rat infected by the nematode Nippostrongylus brasiliensis. Int. J. Parasitol. 6:107-111. [DOI] [PubMed] [Google Scholar]
- 60.Tavazoie, S., J. D. Hughes, M. J. Campbell, R. J. Cho, and G. M. Church. 1999. Systematic determination of genetic network architecture. Nat. Genet. 22:281-285. [DOI] [PubMed] [Google Scholar]
- 61.Timmons, P. M., C. T. Chan, P. W. Rigby, and F. Poirier. 1993. The gene encoding the calcium-binding protein calcyclin is expressed at sites of exocytosis in the mouse. J. Cell Sci. 104:187-196. [DOI] [PubMed] [Google Scholar]
- 62.Tomita, M., H. Itoh, N. Ishikawa, A. Higa, H. Ide, Y. Murakumo, H. Maruyama, Y. Koga, and Y. Nawa. 1995. Molecular cloning of mouse intestinal trefoil factor and its expression during goblet cell changes. Biochem. J. 311:293-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tomita, M., T. Kobayashi, H. Itoh, T. Onitsuka, and Y. Nawa. 2000. Goblet cell hyperplasia in the airway of Nippostrongylus brasiliensis-infected rats. Respiration 67:565-569. [DOI] [PubMed] [Google Scholar]
- 64.Turksen, K., and T. C. Troy. 2002. Permeability barrier dysfunction in transgenic mice overexpressing claudin 6. Development 129:1775-1784. [DOI] [PubMed] [Google Scholar]
- 65.Urban, J. F., L. Schopf, S. C. Morris, T. Orekhova, K. B. Madden, C. J. Betts, H. R. Gamble, C. Byrd, D. Donaldson, K. Else, and F. D. Finkelman. 2000. Stat6 signaling promotes protective immunity against Trichinella spiralis through a mast cell- and T cell-dependent mechanism. J. Immunol. 164:2046-2052. [DOI] [PubMed] [Google Scholar]
- 66.Wakelin, D. 1978. Immunity to intestinal parasites. Nature 273:617-620. [DOI] [PubMed] [Google Scholar]
- 67.Wakelin, D., and M. M. Wilson. 1977. Transfer of immunity to Trichinella spiralis in the mouse with mesenteric lymph node cells: time of appearance of effective cells in donors and expression of immunity in recipients. Parasitology 74:215-224. [DOI] [PubMed] [Google Scholar]
- 68.Wastling, J. M., P. Knight, J. Ure, S. Wright, E. M. Thornton, C. L. Scudamore, J. Mason, A. Smith, and H. R. Miller. 1998. Histochemical and ultrastructural modification of mucosal mast cell granules in parasitized mice lacking the beta-chymase, mouse mast cell protease-1. Am. J. Pathol. 153:491-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wright, K. A. 1979. Trichinella spiralis: an intracellular parasite in the intestinal phase. J. Parasitol. 65:441-445. [PubMed] [Google Scholar]
- 70.Zahm, J. M., C. Galabert, A. Chaffin, J. P. Chazalette, C. Grosskopf, and E. Puchelle. 1998. Improvement of cystic fibrosis airway mucus transportability by recombinant human DNase is related to changes in phospholipid profile. Am. J. Respir. Crit. Care Med. 157:1779-1784. [DOI] [PubMed] [Google Scholar]