Abstract
Brucella spp. are gram-negative intracellular facultative pathogens that are known to produce 2,3-dihydroxybenzoic acid (DHBA), a catechol siderophore that is essential for full virulence in the natural host. The mechanism of DHBA entry into Brucella and other gram-negative bacteria is poorly understood. Using mini-Tn5Kmcat mutagenesis, we created a transposon library of Brucella melitensis 16M and isolated 32 mutants with a defect in iron acquisition or assimilation. Three of these transposon mutants are deficient in utilization of DHBA. Analysis of these three mutants indicated that the ExbB, DstC, and DugA proteins are required for optimal assimilation of DHBA and/or citrate. ExbB is part of the Ton complex, and DstC is a permease homologue of an iron(III) ABC transporter; in gram-negative bacteria these two complexes are involved in the uptake of iron through the outer and inner membranes, respectively. DugA is a new partner in iron utilization that exhibits homology with the bacterial conserved GTPase YchF. Based on this homology, DugA could have a putative regulatory function in iron assimilation in Brucella. None of the three mutants was attenuated in cellular models or in the mouse model of infection, which is consistent with the previous suggestion that DHBA utilization is not required in these models.
In various environments, iron is present in two states, and its availability is affected by pH and aeration. Fe(III) oxides present in aerobic conditions are very insoluble at neutral pH (recently reviewed in references 5 and 29). In contrast, Fe(II) is relatively soluble, and obtaining iron is a much easier task for bacteria growing anaerobically. The problems that bacteria face in acquiring sufficient iron from their surroundings are particularly acute for pathogens. Bacteria are able to deal with this iron restriction imposed by their environment, e.g., through the mechanism of ferric iron capture. They may produce iron(III)-scavenging siderophores or directly bind heme- and iron-containing compounds from the host.
In an aerobic environment, the concentration of available iron(III) is so low that energy is required for transport of the iron. An energized high-affinity iron(III) (as well as heme and vitamin B12) transporter is conserved in many gram-negative bacteria. This system is composed of different outer membrane-specific receptors, all of which are linked to an inner membrane complex that contains at least the TonB, ExbB, and ExbD proteins (35), and of a specific periplasmic binding protein associated with an ABC transporter. It has been proposed that the binding of a siderophore to the gated outer membrane receptor enhances the interaction of this receptor with TonB (11, 48). The energy required for TonB's gatekeeper activity is provided by the proton motive force, and ExbB and ExbD proteins are essential for this function (1, 13, 39). After this, the ferrisiderophore is transported into the cytoplasm by a specific periplasmic binding protein associated with an ABC transporter. In the cytoplasm, iron is finally dissociated from the siderophore (e.g., following iron reduction). Little is known about the other steps of the pathway, and nothing is known about the recycling, storage, and modification of the siderophore; however, one pathway for secretion of the enterobactin of Escherichia coli has recently been shown to be mediated by the membrane exporter EntS protein (28).
Brucella spp. are facultative intracellular gram-negative bacteria that are responsible for worldwide zoonotic diseases. These pathogens induce abortion and sterility in domestic mammals, and some of the species, including Brucella melitensis, are responsible for a chronic fever in humans (25). Brucella can replicate in macrophages and in nonprofessional phagocytes. During its life cycle, Brucella probably uses different siderophores and host iron-containing compounds in the various environments that it encounters. The catecholic siderophores 2,3-dihydroxybenzoic acid (DHBA) and brucebactin, a complex siderophore structurally based on DHBA, have been reported to be produced and utilized as iron sources by Brucella (7, 31, 45). In addition, Brucella abortus also has the ability to acquire the most abundant source of iron in mammals, heme (2). The mechanism of entry of ferric DHBA through the Brucella envelope has not been studied, and compared to the more complex catechol-based siderophore uptake, the capture of ferric DHBA has only been marginally studied in other bacteria (32, 61). Moreover, B. abortus does not use enterochelin, deferoxamine (DFA), ferrichrome, or rhizobactin as an exogenous siderophore, and except for heme, nothing is known about utilization of host iron-containing compounds by Brucella spp. (2).
The role of iron metabolism in Brucella virulence is not well understood. Brucella can grow within macrophages in which iron has been depleted if gamma interferon or DFA is added (37, 38). The ent operon encoding DHBA and brucebactin synthesis does not appear to play an essential role in intracellular survival and in virulence in the mouse model (7, 31). However, in pregnant cattle, DHBA production is essential for complete maternal and fetal colonization (8). The fbpA gene, a homologue of a gene encoding an iron binding protein, may be involved in iron assimilation in the host, as it was shown to be expressed in macrophages (26).
In this report, we describe isolation and characterization of B. melitensis DHBA uptake transposon mutants. Our results indicate that the exbB, dst, and dugA genes are implicated in the use of iron sources. This report provides the first description of an iron utilization complex in Brucella; we describe the Ton system, an iron(III) ABC transporter, and a new partner, DugA, a probable GTPase. Since mutants with mutations in iron acquisition pathways are often described as attenuated (3, 22, 36, 57), we evaluated the survival of these mutants in bovine macrophages and in HeLa cells. The three mutants used were also tested for virulence in BALB/c mice. In these ex vivo and in vivo assays, the mutants were not attenuated compared to the wild-type strain.
MATERIALS AND METHODS
Bacterial strains and plasmids.
B. melitensis 16M Nalr, a spontaneous nalidixic acid-resistant mutant, is the wild-type strain that was used throughout this study; this strain was received from J.-M. Verger, Laboratoire de Pathologie Infectieuse et d'Immunologie, Institut National de Recherche Agronomique, Nouzilly, France (71). E. coli XL1-Blue (Stratagene) and strain S17-1 were also used in this study (63). The vectors used in this work were pUTmini-Tn5Kmcat, a plasmid that contains the transposase gene separated from the transposon and is unable to replicate in Brucella spp. (18); pBBR1MCS4, a replicative vector in Brucella (41); pCAT19, a pUC19 derivative containing a chloramphenicol acetyltransferase-encoding cassette (27); and plasmids pARBV, pBBRdugA1, and pBBRdugA2, which are described below.
Media and antibiotics.
Brucella strains were grown on tryptic soy agar containing 0.1% yeast extract, in 2YT medium (10% yeast extract, 10 g of tryptone per liter, 5 g of NaCl per liter), and in Luria-Bertani (LB) medium (liquid or solid [13 g of agar per liter]). E. coli strains were manipulated as described by Sambrook et al. (25). To make LB agar iron limiting, we added the iron chelator 2,2′-dipyridyl (DIP) (0.2 μM; Sigma) or the ferric iron chelator ethylenediamine di(o-hydroxyphenylacetic acid) (EDDHA) (0.65 mM; Sigma). At these concentrations wild-type strain B. melitensis 16M was not inhibited. DIP and EDDHA are routinely used to identify and characterize iron acquisition mutants of a variety of bacteria, particularly brucellae (31, 55, 62).
The antibiotic concentrations used were as follows: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; nalidixic acid, 25 μg/ml; gentamicin, 50 μg/ml, and tetracycline, 12.5 μg/ml.
Iron utilization assays.
Liquid growth assays to test the ability of B. melitensis mutants to utilize DHBA as a sole source of iron were performed as follows. Cells were grown in liquid LB medium containing the iron chelator DFA (500 μM; Sigma) alone or with DHBA (500 μM) (44). At this concentration of DFA, the growth of the wild-type strain was greatly reduced. All bacterial cultures were incubated at least in triplicate at 37°C; samples were taken at different times, and growth was determined by measuring the optical density at 590 nm.
Plate bioassay experiments were also performed with Brucella strains. Brucella cultures grown to the mid-log phase were plated on LB agar containing 1 mM EDDHA (at this concentration of EDDHA, parental strain B. melitensis 16M did not grow). The siderophores to be tested [10 μl of DHBA (without iron; 0.4 M) or 10 μl of iron(III) citrate (100 mM; Sigma)] were added to sterile paper disks and placed onto the solid medium. The plates were checked for the presence of a halo of growth after 4 days of incubation at 37°C.
Transposon mutagenesis.
The transposon mini-Tn5Kmcat, a derivative of transposon mini-Tn5Km1 (19) bearing the kanamycin resistance gene and the chloramphenicol acetyltransferase reporter gene, was constructed (the resulting transposon was 2,409 bp long) and used to mutagenize B. melitensis 16M (18, 20, 30). Briefly, pUTmini-Tn5Kmcat, an ampicillin-resistant plasmid containing the transposon (without the transposase gene) was introduced into B. melitensis 16M Nalr by conjugation and incubated for 1 h without selection. Transconjugants resistant to kanamycin and sensitive to ampicillin were selected (18). A total of 3,040 clones were individually stored in 2YT medium containing 30% glycerol at −80°C in microtiter plates. Chromosomal DNA from 30 different mutants (including F13, F15, and F32) was prepared, digested with EcoRV or HindIII, and analyzed by Southern blot hybridization with the kanamycin or chloramphenicol resistance genes as a probe. The results demonstrated that each mutant contained a single transposon insertion. In this work, we screened 800 mutants belonging to this library of stable and independent mini-Tn5 mutants of B. melitensis 16M for an iron assimilation defect.
Molecular techniques and sequence analysis.
DNA manipulation was performed by using standard techniques (5a). Restriction enzymes were purchased from Roche and were used as described by the manufacturer.
Inverse PCRs (iPCR) were performed as described previously by Delrue et al. (20) with the transposon mutants after restriction of genomic DNA with TaqI. The oligonucleotides used were based on the cat gene sequence and on the km gene. The sequences of the oligonucleotides were 5′-GCATTCATCAGGCGGGCAAGAATGTGAAT-3′, 5′-GCCATCACGACTGTGCTGGTCAT-3′, 5′-GCCATCACGACTGTGCTGGTCAT-3′, and 5′-GATTCCGACTCGTCCAACATCA-3′. The PCR products were separated on agarose gels and were purified by using a High Pure PCR product kit as described by the manufacturer (Boehringer Mannheim, Roche). The sequencing reactions were performed by using an ABI DNA sequencer with a Big Dye terminator kit (Perkin-Elmer Cetus) and an ABI 377A sequencer. The oligonucleotides used for sequencing were the previously described primers used for iPCR amplification.
To obtain more genomic sequence near the site of transposition, chromosomal DNA from the F32 mutant was prepared, digested with SalI and with EcoRV (there were no SalI or EcoRV sites in the Tn5 derivative), and ligated with pBBRMCS4 previously digested with SalI and EcoRV, respectively. Each ligation mixture was transformed into E. coli XL1-Blue and plated on LB medium supplemented with kanamycin. A plasmid from kanamycin-resistant colonies was prepared, and the insert was sequenced.
The iPCR with the F15 mutant was used to screen a B. abortus genomic library (9). One positive clone was isolated and characterized.
All the sequences obtained were analyzed by using the National Center for Biotechnology Information BLAST server, the Conserved Domain database, and GenBank (4, 46). Multiple alignment was performed by using CLUSTAL W (1.74) (66). Secondary structures were predicted by using PHD (58, 59), and PSORT (49, 50) was used to examine the signal sequence. The B. melitensis 16M genome was analyzed by using the server at http://serine.urbm.fundp.ac.be/∼seqbruce/GENOMES/Brucella_melitensis/ (C. Lambert, E. Depiereux, J.-J. Letesson, and X. De Bolle, unpublished data).
To obtain a wild-type copy of dugA in the F15 mutant, the intact predicted coding sequence (CDS) of dugA was amplified from B. melitensis DNA by using the oligonucleotides 5′-CTATGACGCATAGCCACAA-3′ and 5′-TCAGGTATTGAAGCGGAACA-3′ (sequences corresponding to start and stop codons are underlined) and cloned at the EcoRV site of pBBR1MCS4 (41). This resulted in constructs pBBRdugA1 and pBBRdugA2, in which the dugA CDS was cloned under control of the E. coli lac promoter, which allowed constitutive expression of this CDS in Brucella (24, 41), and in the opposite direction (no expression of dugA), respectively.
For Southern hybridization, the DNA probe (819 bp) specific to the chloramphenicol acetyltransferase (cat) gene was obtained after TaqI restriction of the pCAT19 plasmid and purification from an agarose gel with a JET sorb kit (Genomed). The probe was fluorescein labeled with a Tropix kit. Southern blotting on a positively charged nylon membrane (Tropix) was performed with a Hybaid vacuum blotter (Biozym). Membrane prehybridization, hybridization, and washing were performed under highly stringent conditions by using the Southern-Star protocol (Tropix). Hybridization was detected by autoradiography.
Intracellular survival.
Survival of mutants was evaluated in an immortalized cell line of bovine peritoneal macrophages (65) and in HeLa cells by using previously described procedures (20). Briefly, brucellae grown in 2YT medium for 24 h were resuspended in a complete cell culture medium for inoculation. For the F32 mutant, inocula were also prepared from bacteria grown to the mid-log phase in LB broth containing 5 to 30 μg of EDDHA per ml, which induced sufficient iron starvation to reduce, but not stop, the growth of Brucella. Brucellae were added to cells at a multiplicity of infection of 300:1, and culture plates were centrifuged for 10 min at 1,100 rpm at room temperature in a Jouan BR4i centrifuge. Gentamicin (50 μg/ml) was added 1 h after infection. The number of viable brucellae was estimated 3, 24, and/or 48 h postinfection (54) after the monolayers were washed with complete culture medium and lysed with 0.1% Triton X-100 in deionized water. Serial dilutions were plated on 2YT agar to determine the average number of CFU per well. The data presented below are means for at least three culture wells and are representative of two experiments.
In vivo survival.
Groups of 7-week-old BALB/c female mice (n = 4) were inoculated intraperitoneally (i.p.) with 104 CFU of either B. melitensis 16M or a mutant or intravenously (i.v.) with the F32 mutant or the wild-type strain (53). At 1 and 4 weeks postinoculation, four mice from each group were sacrificed, and spleens were collected. The spleens were homogenized in 2 ml of phosphate-buffered saline with a Stomacher 80 homogenizer. Serial dilutions of the homogenized spleens were plated on tryptic soy agar containing 0.1% yeast extract to determine bacterial counts.
Nucleotide sequence accession number.
The B. melitensis sequence data reported in this paper have been deposited in the GenBank nucleotide sequence database under accession numbers AF325201 (for the exbB, exbD, and tonB genes), AY028973 (for the pth and dugA genes), and AF358662 (for the tol genes).
RESULTS
Isolation of B. melitensis 16M mutants sensitive to an iron chelator.
We constructed a library of B. melitensis 16M mini-Tn5Kmcat transposon mutants. More than 800 of these mutants were individually screened for deficient growth on iron-poor medium. Thirty-two B. melitensis 16M mutants appeared to be deficient for growth on iron-poor DIP-containing medium. In standard media, the mutants showed no obvious growth defect. The defect was confirmed for all 32 mutants (designated F1 to F32) on medium supplemented with another iron chelator, EDDHA. These 32 mutants were screened for DHBA siderophore uptake.
Iron source utilization deficiency of the F15, F13, and F32 mutants.
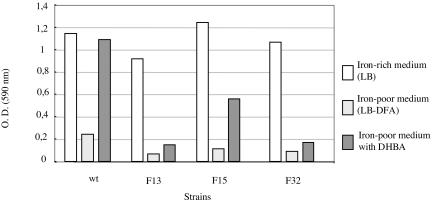
By utilizing an iron-deficient growth medium, we tested the ability of the 32 transposon mutants to use DHBA as the sole iron source. A marked increase in the growth of the wild-type strain was observed when DHBA was added to the iron-deficient medium. The growth of 29 mutants was also completely restored. In contrast, addition of DHBA did not fully restore growth of the F13 and F32 mutant strains (Fig. 1). For the F15 strain, the ability of the organism to use DHBA was also reduced, but it was reduced less than it was for the F13 and F32 mutants. In addition, the wild-type strain and the F15 mutant could grow on an iron-deficient medium when iron(III) citrate was provided in a disk assay. In contrast, growth of the F32 and F13 mutants in a plate bioassay was significantly reduced when ferric citrate or DHBA was used as the iron source compared to growth of the wild type. The diameters of growth observed in this assay were as follows: wild type, 53 to 54 mm; F15, 52 to 54 mm; F32, 21 to 22 mm; and F13, 24 to 26 mm. These results showed that B. melitensis 16M is able to use ferric citrate directly or indirectly as a sole iron source.
FIG. 1.
Typical cell density 48 h postinoculation of B. melitensis wild-type and mutant strains in the absence of DFA (LB) or in the presence of DFA (500 μM) (LB-DFA) with or without DHBA (500 μM). O.D. (590 nm), optical density at 590 nm; wt, wild type.
Identification of the mutated genes in the F13, F15, and F32 mutants.
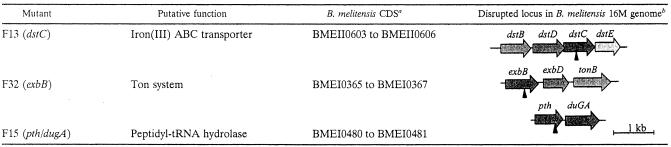
Southern blot hybridization demonstrated that each mutant contained a single transposon insertion (data not shown). iPCRs were used to clone genomic DNA flanking the transposon, and the PCR products were then sequenced. DNA and protein database searches were performed by using the flanking sequences, and the results are shown in Table 1.
TABLE 1.
B. melitensis genes essential for ferric DHBA utilization
CDS of B. melitensis genome on the National Center for Biotechnology Information Entrez Genome website (http://www.ncbi.nlm.nih.gov/PMGifs/Genomes/micr.html).
An arrowhead indicates the transposon localization.
A genomic DNA fragment containing the transposon from the F32 mutant (renamed the exbB mutant) was subcloned into pBBR1MCS4, and the DNA sequence of the regions flanking the transposon was determined by using PCR products. The sequence was determined for both strands by using two independent PCR products. Examination of this nucleotide sequence revealed three CDSs homologous to exbB, exbD, and tonB separated by 5 and 2 bp. Such a tight organization suggests that these three CDSs are part of an operon (60), and the transposon is inserted in the first CDS, exbB (Table 1). The three encoded proteins constitute the Ton system. Like its two homologues in Pseudomonas putida and Bordetella pertussis (56), Brucella ExbB (320 amino acids) has an N-terminal extension compared to E. coli ExbB. This extension contains an additional predicted transmembrane helix and an Ala-Pro-rich region with low complexity. A multiple alignment of four ExbB proteins showed that the sequence conservation occurs mainly in the three transmembrane segments common to all ExbB proteins. The 175-amino-acid B. melitensis ExbD protein has a hydropathy profile similar to that of E. coli. Like their E. coli homologues, the B. melitensis 16M ExbB and ExbD proteins also showed homology to the Tol proteins (12, 14). The TolQ and TolR sequences of B. melitensis 16M were obtained previously (67), and their amino acids were 44 and 41% identical to the amino acids of B. melitensis 16M ExbB and ExbD, respectively. As described previously for E. coli (10), the E/P-rich region and a K/P-rich region (at residues 82 and 119, respectively) of TonB (272 amino acids), encoded by the third CDS, are predicted to span the periplasm, and the protein is predicted to be anchored in the inner membrane via an N-terminal transmembrane segment (68). The predicted transmembrane sequence contains a conserved His residue involved in TonB activity in E. coli. The conserved predicted topology and sequence suggest that the Ton system of Brucella has a membrane arrangement similar to that of E. coli.
The deduced polypeptide sequence of the disrupted B. melitensis gene in the F13 mutant (designated the dstC mutant) is homologous to fatC (ferric anguibactin iron transport system), which encodes a permease of the anguibactin siderophore ABC transporter of Vibrio anguillarum (Table 1). The entire sequence of the locus designated dstBDCE (DHBA siderophore transport system) was obtained from the recently sequenced B. melitensis 16M genome (21). The polypeptide sequences deduced from the dstBDCE CDSs exhibit similarity with a periplasmic binding protein, with two permeases, and with the ATPase protein of an iron(III) ABC transporter. In the B. melitensis dstBDCE locus, all four genes are oriented in the same direction and overlap or have very short intergenic sequences (up to 59 bp long), suggesting that they are part of a single transcriptional unit encoding the proteins of a binding protein-dependent ABC transporter.
Interestingly, in the F15 mutant (designated the pth/dugA mutant), the homology of the disrupted sequence and pth does not suggest a direct link with iron assimilation in bacteria. The transposon insertion is located 190 bp upstream of the stop codon of the homologue of the pth coding sequence (753 bp), which encodes peptidyl-tRNA hydrolase, an enzyme that cleaves peptidyl-tRNA that has been abortively released from the ribosome during protein synthesis. We cloned the B. abortus pth homologue, as well as the gene located 41 bp downstream that we designated dugA and encodes a probable YchF homologue (a GTP binding protein), as described in Materials and Methods (Table 1). This genomic context is maintained in about 50% of diverse bacterial genomes (15), such as the E. coli (17), Mesorhizobium loti, and Bradyrhizobium japonicum genomes. DugA (DHBA utilization GTPase homologue A) possesses a domain homologue with the GTP1/OBG domain and contains the highly conserved E. coli YchF GTPase signature motifs G1 to G4 (Fig. 2) (16). Analysis of the B. melitensis 16M genome (data not shown) demonstrated that as in other bacteria, a core of 11 GTPases is universally conserved. These 11 GTPases are proposed to regulate special aspects of ribosome function and are probably RNA binding proteins (15, 16). DugA of Brucella belongs to the 11th conserved GTPase group, designated YchF, which has no predicted function.
FIG. 2.
CLUSTAL W (1.74) multiple-sequence alignment of the amino acid sequences of the DugA protein of B. melitensis (Bru) and the YchF proteins of E. coli (E.coli) and Haemophilus influenzae (Hinfl). Identical amino acids are indicated by asterisks. The GTPase signature motifs G1 to G4 conserved in YchF of E. coli are enclosed in boxes (16).
Infectivity of B. melitensis mutants in bovine macrophages and in HeLa cells.
We studied the ability of the exbB, dstC, and pth/dugA mutants to infect professional and nonprofessional phagocytes in order to focus attention on the importance of the iron utilization mechanism for survival in these cells. Only minor differences in intracellular growth in bovine macrophages and HeLa cells between the mutant and wild-type strains were observed (Table 2). Similar results were obtained after infection of cells with an exbB bacterial inoculum that had been grown in iron-poor medium (Table 2).
TABLE 2.
Differences in log CFU between the wild-type strain and B. melitensis 16M mutants in cellular infections
| Conditions | Time postinfection (h) | Differences in bacterial countsa
|
|||||
|---|---|---|---|---|---|---|---|
| Bovine macrophages
|
HeLa cells
|
||||||
| exbB mutant | dstC mutant | pth/dugA mutant | exbB mutant | dstC mutant | pth/dugA mutant | ||
| Iron rich | 48 | 0.18 ± 0.16 | 0.31 ± 0.34 | −0.18 ± 0.50 | −0.32 ± 0.10 | 0.01 ± 0.13 | −0.15 ± 0.01 |
| Iron restricted | 48 | 0.17 ± 0.03 | NDb | ND | 0.33 ± 0.12 | ND | ND |
Differences were calculated as follows: (total mean log CFU of wild type) − (total mean log CFU of mutant). The experiments were performed with bovine macrophages and HeLa cells at 48 h postinfection, and bacterial inocula were grown in iron-rich or iron-restricted conditions. The data are means ± standard deviations; the standard deviations for the wild-type strain were ±0.18 and ±0.20 for the two cellular models tested. Cellular infection experiments were performed twice in triplicate.
ND, not determined.
Survival of the mutants in BALB/c mice.
We compared the numbers of bacteria in the spleens of mice infected i.p. with the wild-type strain and with the dstC, pth/dugA, and exbB mutants, as well as the numbers of bacteria in the spleens of mice infected i.v. with the wild-type strain and with the exbB mutant (Table 3). No significant difference in spleen colonization between the mutants and the wild-type strain was observed at 1 and 4 weeks after infection, whether the i.p. or i.v. route was used for the exbB mutant. This indicates that the exbB, dstC, and pth/dugA mutants were not attenuated in the BALB/c mice after i.p. infection.
TABLE 3.
Differences in bacterial counts between the wild-type and mutants in BALB/c mouse spleens
| Time postinfection (wk) | Differences in bacterial countsa
|
|||
|---|---|---|---|---|
| i.p. infection
|
i.v. infection with exbB mutant | |||
| exbB mutant | dstC mutant | pth/dugA mutant | ||
| 1 | 0.06 ± 0.07 | 0.16 ± 0.22 | −0.11 ± 0.13 | −0.55 ± 0.54 |
| 4 | 0.10 ± 0.11 | 0.69 ± 0.17 | 0.13 ± 0.14 | 0.10 ± 0.03 |
Differences were calculated as follows: (total mean log CFU of wild type) − (total mean log CFU of mutant). Samples (n = 4) were examined 1 and 4 weeks after i.p. or i.v. infection.
Implication of the dugA gene in DHBA assimilation by B. melitensis 16M.
In contrast to the dstC and exbB mutants, we were unable to explain the iron assimilation defect of the pth/dugA mutant on the basis of the pth mutation. Moreover, this gene is essential in E. coli, and recently, it was shown that ychF is cotranscribed with pth (17, 40). We hypothesized that transposon insertion in the 3′ end of pth affects expression of dugA. We tested this hypothesis by providing a wild-type copy of the dugA gene in trans in the pth/dugA mutant.
The pth/dugA mutant containing plasmid pBBRdugA1 with dugA under control of the lac promoter, which allowed expression of dugA, was able to grow as well as the wild-type strain in the presence of DHBA (Table 4). In contrast, the pth/dugA mutant transformed with plasmid pBBRdugA2, which did not allow expression of DugA (because the dugA CDS and the lac promoter are in opposite orientations), manifested no complementation of the phenotype. This result demonstrated that expression of dugA led to complementation of the pth/dugA mutant. We propose that transposon insertion in pth has a polar effect on dugA expression and that the pth/dugA mutant is deficient in iron assimilation because of altered expression of the dugA gene.
TABLE 4.
Liquid growth assay in iron-poor medium (LB medium with 500 μM DFA) for utilization of the DHBA siderophore (500 μM) by B. melitensis wild-type strain 16M and the exbB mutant as controls and the pth/dugA mutant transformed with pBBRdugA1 and pBBRdugA2
| Medium | Growth of strainsa
|
|||
|---|---|---|---|---|
| exbB | Wild type | pth/dugA with pBBRdugA1 | pth/dugA with pBBRdugA2 | |
| Iron-rich medium (LB medium) | + | + | + | + |
| Iron-poor (LB medium with DFA) | − | + | + | − |
| Iron-poor medium with DHBA | − | + | + | − |
+, growth equivalent to wild-type growth; −, growth much less than wild-type growth. Growth was examined 48 h after inoculation.
DISCUSSION
We identified a set of 32 B. melitensis transposon mutants that appear to be defective in iron acquisition or assimilation. When three of these mutants (the exbB, dstC, and pth/dugA mutants) were screened for DHBA uptake, the results allowed identification of components of a DHBA utilization pathway or a DHBA-ferric citrate utilization pathway, the Ton system and an iron(III) binding protein-dependent ABC transporter described for other bacteria and also a homologue of YchF GTPase.
Three B. melitensis mutants identified in this work, the dstC, pth/dugA, and exbB mutants, exhibited altered growth under in vitro iron-limiting conditions. In addition, growth of these three mutants in iron-depleted medium was not rescued by addition of DHBA (or by addition of ferric citrate for the exbB and dstC mutants). These results indicated that the gene disrupted in the dstC, pth/dugA, and exbB mutants is involved in DHBA and/or ferric citrate utilization. Iron(III) citrate has not been described previously as an iron source for Brucella, and our results show that utilization of this compound by Brucella is ExbBD-TonB complex and Dst ABC transporter dependent.
Brucella iron capture via DHBA is still not clearly understood. In E. coli, the Ton system and the ferrous uptake system seem to be partially implicated in ferric DHBA capture, and in Pseudomonas aeruginosa, DHBA transport is neither iron repressible nor strongly energy dependent, unlike other siderophore uptake mechanisms (32, 33, 61). In B. abortus, Lopez-Goni et al. (45) did not observe any induction of outer membrane protein expression under low-iron conditions and proposed that DHBA uses a receptor-independent pathway or an unusual non-iron-repressible receptor. Our results demonstrate that the Ton protein complex and an ABC transporter (Dst) are implicated in utilization of Fe(III)-DHBA by Brucella. In other bacteria, this iron uptake pathway is also energy and receptor dependent. We propose that the ExbBD-TonB complex and the Dst binding protein-dependent ABC transporter are implicated in DHBA (and ferric citrate) assimilation by B. melitensis 16M in low-iron conditions, probably at the stage of outer membrane and inner membrane crossing, respectively. The Dst transporter could probably also be utilized by brucebactin, a more complex siderophore for which DHBA probably serves as a precursor. In E. coli, one iron(III) ABC transporter is also used by more than one siderophore belonging to the same structure family (hydroxamates, catecholates, or ferricitrate), and many siderophores have a specific receptor (except, for example, for DHBA and dihydroxybenzoylserine). In the B. melitensis genome, no sequence encoding a potential TonB-dependent receptor was found near the dst genes (BMEII0604 to BMEII0607). However, three CDSs encoding homologues of iron receptors were detected: BMEII0105/II0297 and BMEI0657. One part of this sequence is adjacent to CDSs encoding an iron(III) ABC transporter (BMEI0657 to BMEI0660). In addition to this ABC transporter (BMEI0657 to BMEI0660) and to the Dst transporter (BMEII0604 to BMEII0607), four other putative iron(III) ABC transporters determined by homology were detected in the B. melitensis genome: BMEII0535 to BMEII0537/BMEII0583 to BMEII0585/BMEII1120 to BMEII1123 and BMEII0565 to BMEII0567.
The exbB, dstC, and pth/dugA mutants of B. melitensis exhibited no defect in intracellular infection. Bellaire et al. (7) and González Carreró et al. (31) obtained similar results with B. abortus entC and entE mutants unable to synthesize DHBA and brucebactin, respectively. DHBA is dispensable for intracellular survival of Brucella. In an intracellular environment, the source of iron is not clearly identified, but a siderophore is not essential for survival, as demonstrated for Shigella flexneri (43, 51) and Mycobacterium sp. (42). However, the Ton complex, which is absent in the exbB mutant, is probably implicated in the acquisition of iron sources other than DHBA, like all putative endogenous-exogenous siderophores and some host iron-containing compounds. Despite this, the Ton system appears to be dispensable for intracellular growth of Salmonella enterica (69), as it is for Brucella. The strategy used by intracellular pathogens to obtain iron could be to multiply within intracellular sites where available iron is stored, and Brucella seems to behave in this way (43, 69). In addition, it seems that Brucella does not possess a second Ton complex (or protein), as described for Vibrio cholerae and P. aeruginosa (52, 73). In fact, only one copy of the exbBD-tonB genes was detected in the genome of B. melitensis 16M. However, tol genes are present, and in E. coli, TolQ/R proteins partially complement the absence of ExbB/D proteins (12, 64). Finally, brucellae could use Ton-independent iron uptake mechanisms, as suggested for heme capture by Neisseria gonorrhoeae and Haemophilus ducreyi (23, 70, 72).
Iron availability varies in the different environments encountered by a pathogen in the host, implying that the mechanisms of iron acquisition differ according to the infection route. An S. enterica tonB mutant is attenuated in an intragastric infection but not in an i.p. infection (69). No significant difference in spleen colonization between the Brucella exbB, dstC, and pth/dugA mutants and the wild-type strain was observed after i.p. infection. These results are also consistent with the findings of Bellaire et al. (7), who showed that biosynthesis of DHBA is not required for chronic infection in the mouse model. After i.p. inoculation, Brucella is probably internalized by peritoneal macrophages and rapidly transported to the spleen, where it multiplies. Our observations in cell line infection experiments suggest that the Ton complex, the Dst transporter, and the DugA protein are not required for intracellular Brucella replication. Following i.v. inoculation, bacteria are for a while located in the blood, in which iron is withheld by iron(III) binding proteins, and after this bacteria are also targeted to the spleen. However, with regard to the model of Brucella pathogenesis, blood is not a critical replication site. We concluded that the Ton complex, Dst transporter, and DugA GTPase are not required for survival or growth of B. melitensis in the compartments targeted in our animal models of infection. Our routes of infection bypass mucosal surfaces where the initial phase of infection takes place in a natural host. Therefore, we cannot exclude the possibility that the system altered in the mutants contributes to growth at sites, such as the mucosal epithelium, the submucosal environment, or the peripheral lymph nodes, that are not encountered by Brucella when the organism is administered i.p. or i.v.
Despite these results, DHBA plays a role in virulence, as demonstrated by significant attenuation of the entC mutant in pregnant cattle (8). A possible explanation for the requirement for DHBA in the host was the state of gestation. Indeed, erythritol, a preferred carbon source for Brucella produced by placental trophoblasts (but not produced in mice), stimulates the production of DHBA (6). Perhaps brucellae use sources of iron other than DHBA during infection but also need the DHBA (and brucebactin) siderophore after a change in the iron status in a different gestational stage. Consequently, exbB, dstC, and pth/dugA mutant virulence should be further evaluated in the natural host, and the effects of infection with the mutants on gestation should also be investigated.
Our data also demonstrate that a GTPase homologue is implicated in DHBA utilization. FeoB is the only G protein that has been described as a protein that is involved in iron assimilation (34, 47). This GTPase is an essential membrane protein for iron(II) uptake in E. coli; however, there is no similarity between DugA and FeoB. Moreover, no FeoB homologue was detected in the B. melitensis 16M genome sequence. DugA is the homologue of YchF, the 11th universally conserved GTPase in bacteria (16), and thus may be a regulator. Indeed, Caldon et al. (16) hypothesized that the core of 11 GTPases is necessary to regulate ribosome function or transmit signals from the ribosome to downstream effector pathways. This regulating function through interaction with RNA and/or ribosomes could explain the genomic position of dugA next to pth (encoding peptidyl-tRNA hydrolase). Finally, the proposed function of the DugA protein is a starting point for studies of the role of its homologues.
In addition to characterization of the Ton system, Dst transporter, and DugA GTPase in Brucella, further genetic and phenotypic analyses of other selected mutants should provide valuable insights into the mechanisms of Brucella iron assimilation and into their involvement in virulence.
Acknowledgments
We gratefully acknowledge P. Michel, Centre d'Etude et de Recherche Vétérinaire et Agrochimique, Brussels, Belgium, for his help with the mouse model. We thank the laboratory of L. G. Adams, Department of Veterinary Pathobiology, Texas A&M University, College Station, for providing the bovine peritoneal macrophage line.
Isabelle Danese, Rose-May Delrue, and Valérie Haine received a specialization grant from the Fonds pour la Formation à la Recherche dans l'Industrie et dans l'Agriculture. This work was supported by the Commission of the European Communities (contract QLK2-CT-1999-00014).
Editor: V. J. DiRita
REFERENCES
- 1.Ahmer, B. M., M. G. Thomas, R. A. Larsen, and K. Postle. 1995. Characterization of the exbBD operon of Escherichia coli and the role of ExbB and ExbD in TonB function and stability. J. Bacteriol. 177:4742-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almiron, M., M. Martinez, N. Sanjuan, and R. A. Ugalde. 2001. Ferrochelatase is present in Brucella abortus and is critical for its intracellular survival and virulence. Infect. Immun. 69:6225-6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Tawfiq, J. A., K. R. Fortney, B. P. Katz, A. F. Hood, C. Elkins, and S. M. Spinola. 2000. An isogenic hemoglobin receptor-deficient mutant of Haemophilus ducreyi is attenuated in the human model of experimental infection. J. Infect. Dis. 181:1049-1054. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 5a.Ausubel, F. M., R. Brent, R. E. Kingston, D. E. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1991. Current protocols in molecular biology. Green Publishing Associates, New York, N.Y.
- 6.Bellaire, B. H., P. H. Elzer, C. L. Baldwin, and R. M. Roop II. 2003. Production of the siderophore 2,3-dihydroxybenzoic acid is required for wild-type growth of Brucella abortus in the presence of erythritol under low-iron conditions in vitro. Infect. Immun. 71:2927-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bellaire, B. H., P. H. Elzer, C. L. Baldwin, and R. M. Roop II. 1999. The siderophore 2,3-dihydroxybenzoic acid is not required for virulence of Brucella abortus in BALB/c mice. Infect. Immun. 67:2615-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellaire, B. H., P. H. Elzer, S. Hagius, J. Walker, C. L. Baldwin, and R. M. Roop II. 2003. Genetic organization and iron-responsive regulation of the Brucella abortus 2,3-dihydroxybenzoic acid biosynthesis operon, a cluster of genes required for wild-type virulence in pregnant cattle. Infect. Immun. 71:4-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bellefontaine, A. F., C. E. Pierreux, P. Mertens, J. Vandenhaute, J. J. Letesson, and X. De Bolle. 2002. Plasticity of a transcriptional regulation network among alpha-proteobacteria is supported by the identification of CtrA targets in Brucella abortus. Mol. Microbiol. 43:945-960. [DOI] [PubMed] [Google Scholar]
- 10.Braun, V. 1995. Energy-coupled transport and signal transduction through the gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol. Rev. 16:295-307. [DOI] [PubMed] [Google Scholar]
- 11.Braun, V. 1998. Pumping iron through cell membranes. Science 282:2202-2203. [DOI] [PubMed] [Google Scholar]
- 12.Braun, V. 1989. The structurally related exbB and tolQ genes are interchangeable in conferring tonB-dependent colicin, bacteriophage, and albomycin sensitivity. J. Bacteriol. 171:6387-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braun, V., S. Gaisser, C. Herrmann, K. Kampfenkel, H. Killmann, and I. Traub. 1996. Energy-coupled transport across the outer membrane of Escherichia coli: ExbB binds ExbD and TonB in vitro, and leucine 132 in the periplasmic region and aspartate 25 in the transmembrane region are important for ExbD activity. J. Bacteriol. 178:2836-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braun, V., and C. Herrmann. 1993. Evolutionary relationship of uptake systems for biopolymers in Escherichia coli: cross-complementation between the TonB-ExbB-ExbD and the TolA-TolQ-TolR proteins. Mol. Microbiol. 8:261-268. [DOI] [PubMed] [Google Scholar]
- 15.Caldon, C. E., and P. E. March. 2003. Function of the universally conserved bacterial GTPases. Curr. Opin. Microbiol. 6:135-139. [DOI] [PubMed] [Google Scholar]
- 16.Caldon, C. E., P. Yoong, and P. E. March. 2001. Evolution of a molecular switch: universal bacterial GTPases regulate ribosome function. Mol. Microbiol. 41:289-297. [DOI] [PubMed] [Google Scholar]
- 17.Cruz-Vera, L. R., J. M. Galindo, and G. Guarneros. 2002. Transcriptional analysis of the gene encoding peptidyl-tRNA hydrolase in Escherichia coli. Microbiology 148:3457-3466. [DOI] [PubMed] [Google Scholar]
- 18.Danese, I., A. Tibor, P. A. Denoel, V. Weynants, F. Godfroid, and J.-J. Letesson. 1996. Transposition mutagenesis of Brucella melitensis 16M with a mini-Tn5Kmcat and evaluation of the reporter gene expression. Arch. Physiol. Biochem. 104:45. [Google Scholar]
- 19.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delrue, R. M., M. Martinez-Lorenzo, P. Lestrate, I. Danese, V. Bielarz, P. Mertens, X. De Bolle, A. Tibor, J. P. Gorvel, and J. J. Letesson. 2001. Identification of Brucella spp. genes involved in intracellular trafficking. Cell Microbiol. 3:487-497. [DOI] [PubMed] [Google Scholar]
- 21.DelVecchio, V. G., V. Kapatral, P. Elzer, G. Patra, and C. V. Mujer. 2002. The genome of Brucella melitensis. Vet. Microbiol. 90:587-592. [DOI] [PubMed] [Google Scholar]
- 22.De Voss, J. J., K. Rutter, B. G. Schroeder, H. Su, Y. Zhu, and C. E. Barry III. 2000. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc. Natl. Acad. Sci. USA 97:1252-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elkins, C., P. A. Totten, B. Olsen, and C. E. Thomas. 1998. Role of the Haemophilus ducreyi Ton system in internalization of heme from hemoglobin. Infect. Immun. 66:151-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elzer, P. H., R. W. Phillips, M. E. Kovach, K. M. Peterson, and R. M. Roop II. 1994. Characterization and genetic complementation of a Brucella abortus high-temperature-requirement A (htrA) deletion mutant. Infect. Immun. 62:4135-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Enright, F. M., L. N. Araya, P. H. Elzer, G. E. Rowe, and A. J. Winter. 1990. Comparative histopathology in BALB/c mice infected with virulent and attenuated strains of Brucella abortus. Vet. Immunol. Immunopathol. 26:171-182. [DOI] [PubMed] [Google Scholar]
- 26.Eskra, L., A. Canavessi, M. Carey, and G. Splitter. 2001. Brucella abortus genes identified following constitutive growth and macrophage infection. Infect. Immun. 69:7736-7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuqua, W. C. 1992. An improved chloramphenicol resistance gene cassette for site-directed marker replacement mutagenesis. BioTechniques 12:223-225. [PubMed] [Google Scholar]
- 28.Furrer, J. L., D. N. Sanders, I. G. Hook-Barnard, and M. A. McIntosh. 2002. Export of the siderophore enterobactin in Escherichia coli: involvement of a 43 kDa membrane exporter. Mol. Microbiol. 44:1225-1234. [DOI] [PubMed] [Google Scholar]
- 29.Genco, C. A., and D. W. Dixon. 2001. Emerging strategies in microbial haem capture. Mol. Microbiol. 39:1-11. [DOI] [PubMed] [Google Scholar]
- 30.Godfroid, F., B. Taminiau, I. Danese, P. Denoel, A. Tibor, V. Weynants, A. Cloeckaert, J. Godfroid, and J. J. Letesson. 1998. Identification of the perosamine synthetase gene of Brucella melitensis 16M and involvement of lipopolysaccharide O side chain in Brucella survival in mice and in macrophages. Infect. Immun. 66:5485-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.González Carreró, M. I., F. J. Sangari, J. Aguero, and J. M. Garcia Lobo. 2002. Brucella abortus strain 2308 produces brucebactin, a highly efficient catecholic siderophore. Microbiology 148:353-360. [DOI] [PubMed] [Google Scholar]
- 32.Hancock, R. E., K. Hantke, and V. Braun. 1977. Iron transport in Escherichia coli K-12. 2,3-Dihydroxybenzoate-promoted iron uptake. Arch. Microbiol. 114:231-239. [DOI] [PubMed] [Google Scholar]
- 33.Hantke, K. 1987. Ferrous iron transport mutants in Escherichia coli K12. FEM Microbiol. Lett. 44:53-57. [Google Scholar]
- 34.Hantke, K. 2003. Is the bacterial ferrous iron transporter FeoB a living fossil? Trends Microbiol. 11:192-195. [DOI] [PubMed] [Google Scholar]
- 35.Higgs, P. I., P. S. Myers, and K. Postle. 1998. Interactions in the TonB-dependent energy transduction complex: ExbB and ExbD form homomultimers. J. Bacteriol. 180:6031-6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janakiraman, A., and J. M. Slauch. 2000. The putative iron transport system SitABCD encoded on SPI1 is required for full virulence of Salmonella typhimurium. Mol. Microbiol. 35:1146-1155. [DOI] [PubMed] [Google Scholar]
- 37.Jiang, X., and C. L. Baldwin. 1993. Effects of cytokines on intracellular growth of Brucella abortus. Infect. Immun. 61:124-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang, X., and C. L. Baldwin. 1993. Iron augments macrophage-mediated killing of Brucella abortus alone and in conjunction with interferon-gamma. Cell. Immunol. 148:397-407. [DOI] [PubMed] [Google Scholar]
- 39.Karlsson, M., K. Hannavy, and C. F. Higgins. 1993. ExbB acts as a chaperone-like protein to stabilize TonB in the cytoplasm. Mol. Microbiol. 8:389-396. [DOI] [PubMed] [Google Scholar]
- 40.Kim, S., M. Watarai, Y. Kondo, J. Erdenebaatar, S. Makino, and T. Shirahata. 2003. Isolation and characterization of mini-Tn5Km2 insertion mutants of Brucella abortus deficient in internalization and intracellular growth in HeLa cells. Infect. Immun. 71:3020-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop I I, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 42.Lambrecht, R. S., and M. T. Collins. 1993. Inability to detect mycobactin in mycobacteria-infected tissues suggests an alternative iron acquisition mechanism by mycobacteria in vivo. Microb. Pathog. 14:229-238. [DOI] [PubMed] [Google Scholar]
- 43.Lawlor, K. M., P. A. Daskaleros, R. E. Robinson, and S. M. Payne. 1987. Virulence of iron transport mutants of Shigella flexneri and utilization of host iron compounds. Infect. Immun. 55:594-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leonard, B. A., I. Lopez-Goni, and C. L. Baldwin. 1997. Brucella abortus siderophore 2,3-dihydroxybenzoic acid protects brucellae from killing by macrophages. Vet. Res. 28:87-92. [PubMed] [Google Scholar]
- 45.Lopez-Goni, I., I. Moriyon, and J. B. Neilands. 1992. Identification of 2,3-dihydroxybenzoic acid as a Brucella abortus siderophore. Infect. Immun. 60:4496-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marchler-Bauer, A., A. R. Panchenko, B. A. Shoemaker, P. A. Thiessen, L. Y. Geer, and S. H. Bryant. 2002. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 30:281-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marlovits, T. C., W. Haase, C. Herrmann, S. G. Aller, and V. M. Unger. 2002. The membrane protein FeoB contains an intramolecular G protein essential for Fe(II) uptake in bacteria. Proc. Natl. Acad. Sci. USA 99:16243-16248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moeck, G. S., and J. W. Coulton. 1998. TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport. Mol. Microbiol. 28:675-681. [DOI] [PubMed] [Google Scholar]
- 49.Nakai, K., and P. Horton. 1999. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24:34-35. [DOI] [PubMed] [Google Scholar]
- 50.Nakai, K., and M. Kanehisa. 1991. Expert system for predicting protein localization sites in Gram-negative bacteria. Proteins 11:95-110. [DOI] [PubMed] [Google Scholar]
- 51.Nassif, X., M. C. Mazert, J. Mounier, and P. J. Sansonetti. 1987. Evaluation with an iuc::Tn10 mutant of the role of aerobactin production in the virulence of Shigella flexneri. Infect. Immun. 55:1963-1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Occhino, D. A., E. E. Wyckoff, D. P. Henderson, T. J. Wrona, and S. M. Payne. 1998. Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbD genes. Mol. Microbiol. 29:1493-1507. [DOI] [PubMed] [Google Scholar]
- 53.Pardon, P., and J. Marly. 1976. Resistance of Brucella abortus infected mice to intravenous or intraperitoneal Brucella reinfection. Ann. Immunol. 127C:57-70. [PubMed] [Google Scholar]
- 54.Pizarro-Cerda, J., S. Meresse, R. G. Parton, G. van der Goot, A. Sola-Landa, I. Lopez-Goni, E. Moreno, and J. P. Gorvel. 1998. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect. Immun. 66:5711-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pope, C. D., W. O'Connell, and N. P. Cianciotto. 1996. Legionella pneumophila mutants that are defective for iron acquisition and assimilation and intracellular infection. Infect. Immun. 64:629-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pradel, E., N. Guiso, F. D. Menozzi, and C. Locht. 2000. Bordetella pertussis TonB, a Bvg-independent virulence determinant. Infect. Immun. 68:1919-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reeves, S. A., A. G. Torres, and S. M. Payne. 2000. TonB is required for intracellular growth and virulence of Shigella dysenteriae. Infect. Immun. 68:6329-6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rost, B., and C. Sander. 1993. Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol. 232:584-599. [DOI] [PubMed] [Google Scholar]
- 59.Rost, B., C. Sander, and R. Schneider. 1994. PHD—an automatic mail server for protein secondary structure prediction. Comput. Appl. Biosci. 10:53-60. [DOI] [PubMed] [Google Scholar]
- 60.Salgado, H., G. Moreno-Hagelsieb, T. F. Smith, and J. Collado-Vides. 2000. Operons in Escherichia coli: genomic analyses and predictions. Proc. Natl. Acad. Sci. USA 97:6652-6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Screen, J., E. Moya, I. S. Blagbrough, and A. W. Smith. 1995. Iron uptake in Pseudomonas aeruginosa mediated by N-(2,3-dihydroxybenzoyl)-l-serine and 2,3-dihydroxybenzoic acid. FEMS Microbiol. Lett. 127:145-149. [DOI] [PubMed] [Google Scholar]
- 62.Sigel, S. P., J. A. Stoebner, and S. M. Payne. 1985. Iron-vibriobactin transport system is not required for virulence of Vibrio cholerae. Infect. Immun. 47:360-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 10:783-791. [Google Scholar]
- 64.Skare, J. T., and K. Postle. 1991. Evidence for a TonB-dependent energy transduction complex in Escherichia coli. Mol. Microbiol. 5:2883-2890. [DOI] [PubMed] [Google Scholar]
- 65.Stabel, J. R., and T. J. Stabel. 1995. Immortalization and characterization of bovine peritoneal macrophages transfected with SV40 plasmid DNA. Vet. Immunol. Immunopathol. 45:211-220. [DOI] [PubMed] [Google Scholar]
- 66.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tibor-de Walque, A. 1999. Caractérisation de trois protéines de la membrane externe de Brucella abortus. Ph.D. thesis. University of Namur, Namur, Belgium.
- 68.Traub, I., S. Gaisser, and V. Braun. 1993. Activity domains of the TonB protein. Mol. Microbiol. 8:409-423. [DOI] [PubMed] [Google Scholar]
- 69.Tsolis, R. M., A. J. Baumler, F. Heffron, and I. Stojiljkovic. 1996. Contribution of TonB- and Feo-mediated iron uptake to growth of Salmonella typhimurium in the mouse. Infect. Immun. 64:4549-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Turner, P. C., C. E. Thomas, C. Elkins, S. Clary, and P. F. Sparling. 1998. Neisseria gonorrhoeae heme biosynthetic mutants utilize heme and hemoglobin as a heme source but fail to grow within epithelial cells. Infect. Immun. 66:5215-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Verger, J. M., M. Grayon, E. Chaslus-Dancla, M. Meurisse, and J. P. Lafont. 1993. Conjugative transfer and in vitro/in vivo stability of the broad-host-range IncP R751 plasmid in Brucella spp. Plasmid 29:142-146. [DOI] [PubMed] [Google Scholar]
- 72.Wandersman, C., and I. Stojiljkovic. 2000. Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores. Curr. Opin. Microbiol. 3:215-220. [DOI] [PubMed] [Google Scholar]
- 73.Zhao, Q., and K. Poole. 2000. A second tonB gene in Pseudomonas aeruginosa is linked to the exbB and exbD genes. FEMS Microbiol. Lett. 184:127-132. [DOI] [PubMed] [Google Scholar]