Abstract
Developmental processes are remarkably well conserved among species, and among the most highly conserved developmental regulators are transcription factor families. The Onecut transcription factor family consists of three members known for their single “cut” DNA-binding domain and an aberrant homeodomain. The three members of the Onecut family are highly conserved from Drosophila to humans and have significant roles in regulating the development of diverse tissues derived from the ectoderm or endoderm, where they activate a number of gene families. Of note, the genetic interaction between Onecut family members and Neurogenin genes appears to be essential in multiple tissues for proper specification and development of unique cell types. This review highlights the importance of the Onecut factors in cell fate specification and organogenesis, highlighting their role in vertebrates, and discusses their role in the maintenance of cell fate and prevention of disease. We cover the essential spatial and temporal control of Onecut factor expression and how this tight regulation is required for proper specification and subsequent terminal differentiation of multiple tissue types including those within the retina, central nervous system, liver and pancreas. Beyond development, Onecut factors perform necessary functions in mature cell types; their misregulation can contribute to diseases such as pancreatic cancer. Given the importance of this family of transcription factors in development and disease, their consideration in essential transcription factor networks is underappreciated.
Keywords: onecut, pancreas, liver, nervous system, transcription factor
1. Introduction to Onecut factors
The history of the Onecut (Oc) family of transcription factors begins, as does the history of many transcription factors, in Drosophila. Work carried out by Jan and colleagues discovered that mutations in the cut locus in Drosophila resulted in the transformation of external sensory organs into chordotonal organs during embryonic development [1, 2]. They subsequently showed that the protein product of the cut locus was a nuclear homeodomain-containing protein that was necessary for the development of sensory precursor cells within the extrasensory organ [1, 3]. The Cut protein also contained a DNA-binding domain distinct from, and unrelated to, the homeodomain, thereafter called a “cut” domain. Since its initial discovery, multiple transcription factors containing cut domains have been identified, but many of those factors contain multiple cut repeats. This review will focus on the Onecut family of transcription factors, all of which contain a single cut domain. While Onecut proteins have been identified and studied in many model systems since their discovery, this review will focus on their role in mammalian systems.
The first identified mammalian paralogs of the Drosophila cut domain were the murine Clox (Cut like homeobox) factors, which contain three cut domains in addition to a homeodomain and as such are not Onecut factors [4]. However, much of the earliest work on Onecut factors in mammals focused on their role in the liver. During studies of liver-enriched transcription factors, a protein was identified that could bind to the 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase promoter with high affinity. It was named Hepatic nuclear factor 6 (Hnf6) based on its expression pattern and unique DNA-binding characteristics, which separated it from previously identified hepatic nuclear factors such as Hnf1α and β, Hnf3α and β (FoxA1 and 2, respectively), and Hnf4α. Characterization of the Hnf6 protein revealed that it contained a single domain homologous to the Drosophila cutdomain at the N-terminus and a novel, divergent homeodomain at the C-terminus [5, 6]. Based on homology to Hnf6, a second and third Onecut factor were identified in the liver: Onecut 2 (Oc2) and Onecut 3 (Oc3), respectively [7, 8]. Hnf6 has since been renamed Onecut 1 (Oc1). The expression patterns of Oc2 and Oc3 frequently overlap with Oc1 and they have some of the same transcriptional targets, but the relationship between these factors is context-dependent and will be covered in greater detail in sections below.
Two Hnf6/Oc1 variants were identified in the rat, namely Hnf6α and Hnf6β. Hnf6β contains an additional 26 amino acids in the linker region between the cut domain and the homeodomain that are not present in Hnf6α. The resulting structural difference does confer a slight variation in DNA-binding affinity in vitro, but the presence of more than one Oc1 isoform has not been identified in other organisms. Therefore the significance of the Hnf6β isoform in the rat is unclear [9]. Further investigation into the function of the Oc factors revealed that the homeodomain was dispensable for binding to the DNA of some, but not all, transcriptional targets. Conversely, binding to some targets of Oc1 does not require the cut domain and instead relies upon the homeodomain. In many circumstances, the non-DNA bound DNA-binding domain participates in the recruitment of transcriptional co-factors such as the CREB-binding protein (CBP) or CCAAT/enhancer-binding protein α (C/EBPα) for transcriptional activation [10, 11]. Interestingly, acetylation of the Oc1 protein itself by CBP is necessary for increased Oc1 protein stability and transcriptional activity and hence this recruitment of CBP by Oc1 is necessary for its function [12]. Together, these data indicate that the function of the Oc factors is complex and their role at a given target gene is promoter context-specific [13].
As will become evident, this unique family of transcription factors has an important role in the development of several different organs. The endodermally-derived hepatobiliary tract as well as the pancreas both rely on the Oc factors for proper differentiation of many mature cell types (Figure 1). Likewise, the ectodermally-derived retina and motor neurons require Oc factors for development of full function. This review will discuss the importance of these factors in each context as well as the similarities and differences between each system.
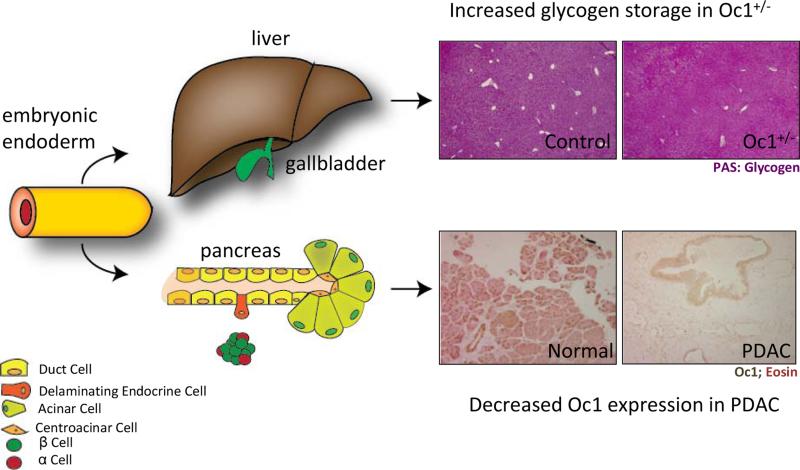
Figure 1. Implications of Oc1 loss in disease.
The pancreas and liver, both endodermally-derived organs, are impacted by loss of Oc1 during development and disease. Top: Oc1 heterozygosity causes defects in glycogen breakdown, resulting in increased glycogen stores in the liver, as shown by Periodic Acid Schiff staining in 3-week old mouse livers (pinkish purple). Bottom: Oc1 is expressed in the nuclei of normal, healthy ducts and acini of human pancreas. Its expression is lost entirely from lesions of pancreatic ductal adenocarcinoma (PDAC).
2. Oc factors control development of the hepatobiliary tract
The hepatobiliary tract is composed of the liver, gall bladder and associated duct network. The liver performs vital functions in fetal hematopoesis, xenobiotic detoxification, metabolism, glycogen storage and glucose mobilization. The gallbladder stores the bile produced by the liver prior to its use in lipid digestion. The primary cell type of the liver, the hepatocyte, performs many of the functions essential to the liver, but the other cell types also play vital roles including regeneration and bile transport. The Oc factors are expressed in hepatocytes as well as in the other primary cell type in the liver, cholangiocytes, which make up the hepatic bile duct [14]. A substantial body of work in the liver has contributed to our knowledge of the expression and function of Oc1, including identification of both direct and indirect transcriptional targets. The consensus DNA binding sequence for Oc1 was identified through its activity in binding to the FoxA2 (formerly Hnf3β) promoter, which in turn regulates other liver-enriched transcription factors [15]. Subsequently, Oc1 has been shown to be a regulatory factor for many genes regulating hepatic development and function, thus implicating it as a critical factor regulating hepatocyte and cholangiocyte identity (Figure 2) [16, 17]. This section will focus on the important role of the Oc factors in development and disease of the hepatobiliary system.
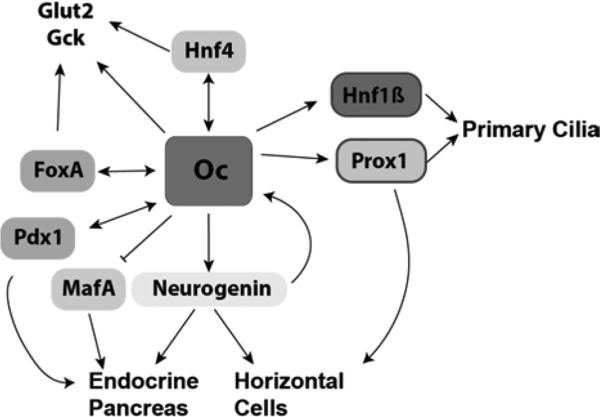
Figure 2. Network of Oc factor targets and associated processes.
Oc factors regulate a common network of transcription factors in different tissues during development to promote differentiation of multiple different mature cell types. This regulation carries over to function of mature cells in the liver through regulation of glucose-processing enzymes.
2.1. Liver development
The mouse liver is specified from the definitive foregut endoderm via signals the derived from cardiac mesoderm and septum transversum mesenchyme (STM) at approximately e8.5. At that time, the endodermal cells initiate a program of hepatic gene expression that includes Oc1/2, FoxA2 and Hnf4α amongst others. Early hepatoblasts in the primordial liver bud proliferate and expand into the surrounding mesenchyme. Oc1/2 perform partially redundant roles in this process as inactivation of both factors resulted in a hypoplastic liver by e9.5 in spite of normal hepatoblast numbers between e8.5 and e9.5. Rather, it appears that Oc1/2-deficient livers fail to expand due to impaired hepatoblast delamination and invasion of STM. Indeed, delayed degradation of the basal lamina surrounding the hepatic bud was evident at e9.5 and possibly explained by increased expression of Thrombospondin-4 (a pro-adhesion glycoprotein) and reduced expression of osteopontin (a pro-migration glycoprotein) [18]. By e11.5, hepatoblasts do begin to invade the STM, but degradation of the basal lamina never reaches the same extent as controls.
In addition to regulating genes associated with hepatoblast migration, Oc1/2 regulate many of the genes necessary for differentiation of hepatoblasts into hepatocytes and further regulate their mature function (Figure 2). Indeed, Oc1/2 activate expression of other hepatic nuclear factor (Hnf) transcription factors essential for liver development (although these are not, in fact, structurally related proteins). These include the winged helix transcription factors FoxA1 and 2 (Hnf3α and β, respectively) and the fatty acid-binding nuclear receptor Hnf4α [15, 19]. Oc1 directly binds to and activates the promoters of FoxA2 and Hnf4α while Oc2 binds to and activates the promoter of Hnf4α [16, 20]. Together, these direct Oc transcriptional targets regulate the transcription factor network necessary for hepatocyte differentiation. Oc1 can also physically interact with both FoxA2 and Hnf4α, but its activity is not dependent upon those interactions [21].
Importantly, Oc1 can also act as a transcriptional repressor in the liver. Work in cell lines derived from Oc1-null embryonic mouse livers revealed increased expression of FoxA1 and some TGF-β response genes as well as increased expression of TGF-β receptor II [22, 23]. These data are particularly interesting since they implicate Oc1 not only as a direct transcriptional regulator, but also an indirect modulator of intracellular signaling. Further, Oc1 also impacts gene expression through regulation of microRNAs. Indeed, the liver-specific microRNA miR-122 is substantially reduced in e15.5 Oc1-null livers. While little is known about the targets of miR-122 during liver development, miR-122-null mice have severe impairments in the process of hepatoblast differentiation to hepatocytes, thus indicating the importance of the Oc1-miR-122 axis [24]. Thus, the Onecut factors, and Oc1 in particular, are vital to proper hepatocyte differentiation and development.
An interesting role for Oc1 is as a key regulator of the response to growth hormone (GH) in the differentiation and proliferation of developing hepatocytes. GH signaling in vivo or treatment of rat liver nuclear extracts with GH stimulation in vitro increases Oc1 transcript levels. GH reduces expression of the liver-enriched transcription factor C/EBPα, alleviating repression of Oc1 and allowing for a rapid increase in Oc1 expression [25]. GH-mediated Oc1 activation in hepatocytes increases expression of some cytochrome P450 genes in a female-selective manner [26, 27]. The importance of Oc1 in female-specific liver function remains unclear.
2.2. Liver function
In addition to other transcription factors, Oc factors regulate expression of many genes that are essential for liver function (Figure 2). Among the most important of these are glucokinase (Gck) and the glucose transporter Glut2 [28, 29]. Oc1 binds to and activates the glucokinase promoter in hepatocytes; loss of Oc1 results in a 50% reduction in glucokinase levels [28]. Likewise, over-expression of Oc1 in hepatocytes increases expression of Glut2, thereby increasing the ability of those cells to take up glucose [29]. The role of Oc1 in regulating hepatocyte function is thus most important for the regulation of glucose homeostasis and hepatic glucose output. In brief, hepatocytes store excess glucose in the form of glycogen. Glycogen can be broken down to free glucose in times of need (eg. fasting, exercise, etc.) for elevation of systemic glucose levels. Both glucokinase and Glut2 have essential roles in this glycogen/glucose flux. In the absence of Oc1, glycogen is not properly metabolized to glucose and remains stored at relatively elevated levels in hepatocytes, leading to hypoglycemia (Figure 1). This role for Oc1 in regulation of genes associated with glucose homeostasis has larger implications for systemic diseases such as diabetes.
2.3. Biliary tract development and function
The gallbladder, the intrahepatic and extrahepatic bile ducts (IHBD and EHBD, respectively), and the primary bile duct constitute the biliary tract whose function is storing and transporting bile produced by hepatocytes to the duodenum. The entire biliary tract develops from the same early progenitors as the liver; thus the transcription factors regulating differentiation and development of the biliary tract largely overlap with those of the liver. Differentiated cells of the bile ducts are known as cholangiocytes. The Oc factors are also necessary for proper differentiation of cholangiocytes. Indeed, in the absence of Oc1, development of the biliary tract fails, there is no gall bladder, and both the IHBD and EHBD are malformed. This duct malformation may be due to decreased expression of the Oc1 target, Hnf1β, which is essential for proper bile duct development and formation of primary cilia (Figure 2) [30, 31]. Additionally, Oc1-null mice lack primary cilia in the biliary epithelial cells, which could further explain the duct defects, since primary cilia serve as extracellular sensors and are an integral component of cellular signaling.
An interesting interaction has been observed between Oc1 and Notch signaling in promotion of IHBD development. Notch signaling has an integral role in IHBD development and in part regulates expression of Oc1 [32]. Loss of Oc1 and Notch signaling within the bipotential hepatoblast progenitor cells (which give rise to both hepatocytes and biliary epithelial cells) resulted in substantial IHBD malformation including a decreased epithelial cell pool and reduced ductal branching that was more severe than inactivation of either component alone [33]. Interestingly, C/EPBα represses Oc1 in biliary progenitors just as it does in developing hepatocytes. Inactivation of C/EBPα in embryonic liver is sufficient to up-regulate Oc1 and convert early hepatoblasts to a biliary fate [34]. Oc1 functions through activation of a transcription factor network that includes Hnf1β to promote bile duct development. However, the Oc factors appear to be necessary exclusively during development of the biliary tract since neither Oc1 nor Notch signaling are necessary for regeneration of adult cholangiocytes following injury [35].
Although no studies to date have examined the role of Oc3 in the development of the biliary tract in mammals, there is evidence for a role for Oc3 in zebrafish. Onecut3 has been determined to be the functional ortholog of mammalian Oc1 in zebrafish since it serves a nearly identical function [36]. Complicating the matter, there is also a zebrafish hnf6 gene expressed in the developing biliary tract, which operates in a regulatory loop with onecut3. The exact roles of these factors are not yet fully elucidated, but loss of either factor (hnf6 or onecut3) does result in malformation of the zebrafish bile ducts [36]. Clearly the role for the Onecut factors in the development of the biliary tract is an essential and conserved process.
2.4. Hepatobiliary disease
Given the importance of the Onecut factors for development of the entire hepatobiliary system, it is not surprising that developmental defects could result in liver disease. As noted above, one of the most important roles of Oc1 is to direct development of the IHBD and EHBD. The phenomenon of ductal plate malformations (the ductal plate is composed of biliary epithelial cell progenitors and the associated portal vein mesenchyme), or persistence of fetal biliary structures postnatally, is attributed to improper development of the biliary tract and contributes to both Jeune Syndrome and Meckel Syndrome in humans [37]. Oc1 and its downstream target Hnf1β are necessary for biliary tract development and in the absence of either factor, ductal plate malformations including hepatic artery malformations occur [30, 37, 38]. This malformation may be in part due to the failure of ductal plates to contribute to vasculogenesis in the portal mesenchyme [39].
Oc1 also plays a role in cancer of the liver in humans. The direct Oc1 target miR-122 has antitumor effects and prevents hepatocellular carcinoma through repression of tumorigenic genes such as cyclin G1, A disintegrin and metalloprotease 10, and insulin-like growth factor-1 receptor [40]. Oc1 is also capable of preventing replication of the hepatitis B virus in hepatoma cells, which is significant since chronic hepatitis B infection is a leading risk factor for liver cancer [41]. Oc1 may also contribute to hepatocyte recovery following liver diseases such as hepatitis C infection or hepatic necrosis. Indeed, human biliary epithelial cells re-express OC1 following liver injury in a process that is thought to contribute to hepatocyte regeneration [42]. This data is further supported by the fact that Oc1 over-expression in hepatocytes stimulates expression of cyclins and tumor growth factor (TGF) α to promote entry into S phase of the cell cycle and thereby promote hepatocyte regeneration following injury [43]. In contrast, expression of OC1 in the HepG2 human hepatoma cell line results in cell cycle arrest [44]. These data suggest that Oc1 promotes hepatocyte terminal differentiation and may act as a tumor suppressor, but is also important for tissue regeneration.
3. Oc1 regulation of pancreas development and disease
The pancreas is both an endocrine and exocrine organ with dual roles in regulation of blood glucose homeostasis and production of digestive enzymes (Figure 1). The endocrine compartment, composed of the islets of Langerhans, makes up 2% of the adult pancreas by mass and is responsible for sensing blood glucose levels and secreting endocrine hormones to maintain glucose homeostasis. The exocrine compartment constitutes the remaining 98% of pancreatic mass and is predominantly composed of the digestive enzyme-secreting acinar cells as well as the pancreatic ducts, which transport those enzymes to the rostral duodenum. All pancreatic cell types are specified from endodermally-derived multipotent pancreatic progenitor cells (MPCs) during development. Several excellent detailed reviews describe pancreas specification and development [45-47]. Here we focus on the role of the Oc factors in different stages of pancreas development and the implications for adult pancreas function.
3.1. Pancreas specification
The pancreas is specified from the definitive posterior foregut endoderm at approximately e8.5 in the mouse, with the dorsal pancreatic bud emerging first. Cells within the dorsal bud are marked by the joint expression of the transcription factors Pancreatic and duodenal homeobox 1 (Pdx1), Pancreas-specific transcription factor 1a (Ptf1a), SRY (sex-determining region-Y)-box 9 (Sox9), and Oc1 amongst others [45]. These factors operate within a co-regulatory network to promote pancreas specification, but are also dependent on each other for activation. Oc1 has a critical role in this capacity, especially with respect to activation of Pdx1. Pdx1 is absolutely critical for pancreas development and in its absence pancreatic agenesis occurs [48-51]. In addition to Pdx1, Oc1 positively regulates several transcription factors involved in pancreas development including Hnf1β, Hnf4α and FoxA2 (Figure 2) [20, 52, 53]. Oc2 expression largely overlaps with Oc1 during pancreas specification, but its expression within the whole pancreas progressively decreases following e12.5 [54]. Oc3 expression completely overlaps with Oc1 in the developing pancreas and liver, but its expression appears to be entirely dependent on activation by Oc1 as Oc1-null animals do not express Oc3 at any stage. The reliance of Oc3 on Oc1 for expression is in contrast to Oc2 expression, which is independent of Oc1 [55]. Importantly, Oc2 and Oc3 are not fully redundant with Oc1 as these two factors cannot fully compensate for the loss of Oc1 during development, and combined inactivation of Oc2/Oc3 does not impair pancreas development [56]. Additionally, the pancreatic phenotype in Oc1 null mutants is not exacerbated by the additional inactivation of either Oc2 or Oc3, indicating that those factors play a less significant role in pancreas specification [7, 9, 56, 57].
3.2. Endocrine differentiation
The islets of Langerhans contain multiple different hormone-secreting cells that regulate glucose homeostasis. These are predominantly the insulin-secreting β cells and glucagon-secreting α cells, which function to lower or raise blood glucose levels respectively. All pancreatic endocrine cells arise from a common pool of endocrine progenitor cells that express the transcription factor Neurogenin3 (Neurog3) [58]. There is evidence that Neurog3-expressing cells are unipotent and predominantly give rise to only one of the five endocrine cell types, but it is unclear whether Oc factors have a role in directing endocrine progenitors toward a particular endocrine fate [59]. Oc1 is necessary for proper induction of Neurog3, thus initiating endocrine specification (Figure 2); Oc1 inactivation results in a near complete loss of Neurog3-positive cells [57, 60]. Although Oc1 alone is capable of activating Neurog3 transcription, it acts cooperatively with Pdx1 to increase Neurog3 transcript levels in vitro, indicating the importance of these two factors working together to specify the endocrine lineage [61]. A few hormone-positive cells persist in the absence of Oc1; however, these cells do not express markers of mature endocrine cells suggesting that Oc1 is required for endocrine maturation [57, 60]. Although conservation is high between the Oc factors, their lack of functional redundancy in the endocrine lineage is highlighted by the inability of Oc2/3 to promote Neurog3 expression and endocrine specification in the absence of Oc1. Of note, Oc2 is capable of binding and activating a Neurog3 promoter element in vitro, yet there is no rescue of Neurog3 expression in the absence of Oc1 [56]. In addition, pancreata from Oc2/3-double null mutants have normal Neurog3 protein expression. Oc2 and Oc3 are expressed in the developing enteroendocrine cells of the stomach and intestine where Oc1 is never expressed. Oc2 and Oc3 are co-expressed with Neurog3 during enteroendocrine differentiation. However, results of Oc2/3 dual gene inactivation studies reveal that they are also dispensable for enteroendocrine differentiation [56].
Following endocrine specification, the role of Oc factors becomes more nuanced. In addition to Oc1 activating the endocrine lineage program through regulation of Neurog3, continued Oc1 activity is required to ensure endocrine differentiation. Deletion of Oc1 from committed endocrine cells using a Neurog3-Cre driver results in some endocrine progenitor cells being diverted to the exocrine lineage [57]. Yet, Oc1 expression is silenced later in the endocrine lineage and is not detected in hormone-positive cells at any time [52, 57]. Indeed, our group has shown that this down-regulation of Oc1 is necessary for proper differentiation and maturation of β cells. Maintenance of Oc1 expression in the endocrine lineage results in increased expression of Neurog3 and increased numbers of endocrine cells, but defects in β-cell maturation. Sustained Oc1 expression in the β-cell lineage represses the expression of the β-cell maturity markers MafA and Glut2, leading to impaired β-cell function as indicated by impaired glucose-stimulated insulin secretion and insulin granule biosynthesis [62-64]. Activation of Oc1 in differentiated β cells using the insulin promoter also results in decreased insulin production and development of diabetes. However, in this model there was increased β-cell apoptosis and decreased β-cell mass that was not observed when Oc1 was expressed earlier in the endocrine lineage [65]. These data indicate that Oc1 is critical for endocrine specification, but that it acts only in the initial stages of specification and commitment and in fact becomes detrimental to endocrine cells at later stages of differentiation. Temporal regulation and function of Oc2 and Oc3 has not yet been analyzed.
3.3. Exocrine differentiation
Oc1 expression is maintained at a high level in ducts and a low level in acinar cells throughout development and adulthood [57, 66]. Although the role of Oc1 in differentiation of the acinar cells is not fully elucidated, it clearly plays a role in proper differentiation of ducts. Oc1 promotes the duct cell fate by acting upstream of the definitive duct marker Hnf1β (Figure 2). Indeed, loss of Oc1 results in a greater than 2-fold reduction in Hnf1β transcript levels during early duct differentiation; a partial recovery of Hnf1β occurs later in gestation. The increase in Hnf1β later in development in the absence of Oc1 is possibly due to up-regulation of Oc2 in an attempt to compensate for the loss of Oc1 [52].
Although Oc1 is important for duct development, it does not affect differentiation of all types of pancreatic ducts equally. Loss of Oc1 does not affect intercalated ducts (the smallest ducts within the pancreas), but impairs interlobular and intralobular ducts. As early as e12.5 ductal branching is impaired in Oc1 mutants and dilated ductal lumens as well as ductal cysts are apparent by e15.5 [66]. Proliferation is increased in the ductal epithelium in Oc1-null mutant mice and the normal cuboidal squamous architecture is lost, resulting in a multilayered epithelium that has lost its polarity [57, 66]. The exact mechanism of the ductal dysmorphogenesis is not yet fully elucidated, but it is likely due in part to the loss of primary cilia in duct cells that have lost Oc1 expression. Just as in the hepatobiliary system, Oc1 is part of a transcriptional regulatory pathway that includes Hnf1β and Prox1, and regulates the transcription of genes involved in the formation of primary cilia, such as Pkhd1 and Cys1 (Figure 2). Expression of both Hnf1β and Prox1 is reduced in the Oc1-null pancreatic ductal epithelium [57, 66]. Oc2 and Oc3 cannot compensate for Oc1 with respect to primary cilia formation, as at no point during development do those structures develop in the ductal epithelial cells. Additionally, Oc2-null animals have normal duct and cilia formation indicating that Oc1 is the primary Oc factor regulating exocrine development. Interestingly, these results and regulatory networks are very similar to those of the developing IHBD, suggesting commonalities in function.
3.4. Pancreatic disease
Given the importance of Oc1 for the development of β cells and pancreatic ducts, it is not surprising that loss or mis-expression of Oc1 could predispose one to disease. Oc1 dysfunction could contribute to defects in human pancreas development through its regulation of Pdx1 expression [48, 67]. Loss of Pdx1 expression results in pancreas agenesis in humans and mice, but some instances of human pancreatic hypoplasia or agenesis linked to impaired Pdx1 expression show no alterations in the Pdx1 coding region. In these cases, decreased Pdx1 expression could result from changes in the binding sites for, or the activity of, upstream regulatory factors such as Oc1, although this has not yet been confirmed.
Oc1 also regulates transcription factors and functional genes associated with diabetes, including transcription factor genes associated with monogenic forms of diabetes known as maturity onset diabetes of the young (MODY). Oc1 directly regulates Pdx1 (MODY 4), Hnf4α (MODY 1), and in the liver, glucokinase (MODY 2), and participates in a network regulating Hnf1β (MODY5) [20, 28, 50, 52]. In addition, decreased or prematurely silenced Oc1 expression in the endocrine lineage would be predicted to result in fewer differentiated endocrine cells, potentially predisposing one to diabetes later in life.
A stronger connection has been drawn between Oc1 and exocrine pancreas disease. Inactivation of Oc1 in the developing pancreatic epithelium results in ductal hyperplasia, ductal cysts and periductal hemorrhaging. Further, acinar-to-ductal metaplasia (ADM), an injury response by acinar cells, was prominent and was similar in many respects to human pancreatitis [68-70]. Histological analysis revealed that OC1 is up-regulated in human pancreatic acinar cells undergoing ADM, but OC1 expression is reduced in pre-cancerous pancreatic intraepithelial neoplasia (PanIN) lesions. Likewise, mouse models of ADM show a transient up-regulation of Oc1, but expression becomes reduced when the lesions progress to PanINs (Figure 1). These results suggest a threshold level of Oc1 between normal acini and ducts with higher levels of Oc1 being required for the duct phenotype [71, 72]. Unexpectedly, the transient up-regulation of Oc1 in ADM occurs independently of the pro-duct transcription factor Sox9. Rather, Oc1 up-regulation in ADM seems to be due in part to loss of micro-RNA-mediated Oc1 repression. Loss of micro-RNAs (through Dicer inactivation) in acini results in development of ADM, and this is dependent on Oc1 activity [73]. The Jacquemin group has also shown that over-expression of Oc1 in acinar cells is sufficient to drive ADM onset [72]. These results indicate that Oc1 (or its downstream effectors) is necessary for development of a ductal phenotype, and that different threshold levels of Oc1 regulate an acinar rather than duct phenotype [72]. ADM is considered by many to be a precursor lesion for PanINs, which are very commonly precursors to pancreatic ductal adenocarcinoma (PDAC). As mentioned above, decreasing Oc1 expression correlates with increasing severity of PanINs in mice and humans. Indeed, OC1 is nearly undetectable in samples of human PDAC (Figure 1) [71]. These results are particularly interesting given that Oc1 has been shown to act through p53 to prevent epithelial-to-mesenchymal transition in lung cancer cells, setting a precedent for its role as a tumor-suppressor [74]. Together, these results demonstrate that Oc factors, especially Oc1, may have a role in maintaining the differentiated state of the exocrine pancreas, and that loss of Oc1 leads to diseases of the exocrine pancreas.
3.5. Directed differentiation
Of particular interest to the pancreas field is the directed differentiation of either embryonic or induced pluripotent stem cells to a β-cell fate. These protocols attempt to mimic the signaling that normally occurs during in vivo differentiation. With respect to directed differentiation of β cells, embryonic or induced pluripotent stem cells are manipulated in a step-wise fashion using activators and inhibitors of different growth factor signaling pathways through the following stages: from definitive endoderm, through posterior foregut, pancreatic progenitor, endocrine progenitor and finally, β cell [75]. Given that Oc1 regulation plays critical roles throughout this progression, it is surprising that it has not been utilized in protocols for in vitro differentiation of β cells. However, it has been used as a marker of effective differentiation down the posterior foregut pathway. Indeed, effective induction of Pdx1 and thus differentiation to definitive endoderm is often measured by expression of Oc1 [76]. Signaling molecules including retinoic acid, activin A, FGF and BMP are all capable of inducing an Oc1-expressing definitive endoderm, and in many cases even more highly differentiated cell types [77-79].
4. Role of Oc factors in neural development and function
A role for the Oc factors in neuronal development has been identified in many model systems indicating an important conserved function. While the discovery of the cut locus in Drosophila indicated its function in differentiation of the external sensory organs, the protein produced from that locus in fact contained three cut repeats. A paralog of mammalian Oc1 was identified in Drosophila named D-Onecut, role which has a unique in regulation of photoreceptor cell differentiation [80]. Indeed, Oc orthologs regulate neural cell specification and differentiation in ascidians, zebrafish, Xenopus and C. elegans [6, 81-83]. Thus, the various cell types of the nervous system may represent the broadest and most diverse population where the Oc factors regulate cell lineage specification and differentiation.
4.1. Retina
The retina serves as the light-sensing part of the eye and is a direct extension of the central nervous system. It is a multilayered network of neurons that ultimately feeds sensory information to the optic nerve, which in turn relays signals directly to the brain. There are seven mature cell types within the neural retina, all of which differentiate from retinal progenitor cells (RPCs) in a sequential manner as directed by specific transcription factor cues [84]. In the mouse, retinal differentiation takes place between e11.5 and P8. A microarray performed on e14.5 retinas identified Oc1 amongst the transcription factors expressed during retinal differentiation. Interestingly, many of the other transcription factors identified in retinal development are also part of the Oc1 regulatory network in pancreas development (Figure 3). These include Neurogenin-2, Pax6, NeuroD, Isl2 and Sox9 [85]. Of particular note, a homolog of Neurogenin-2, Neurogenin-3, is a direct target of Oc1 in the developing pancreas. This connection brings attention to how transcription factor families and gene regulatory networks can be connected and co-opted during differentiation of otherwise unrelated cell types during development.
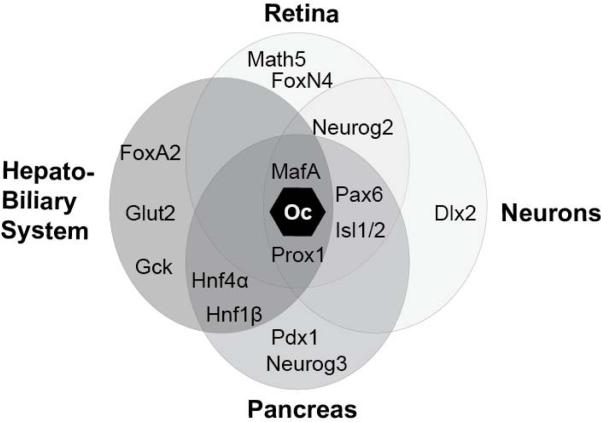
Figure 3. Common targets and co-factors of Oc factors.
The Oc factors operate within common gene expression networks in multiple different tissue types. Shown here, the ectodermally-derived tissues (retina and neurons; light gray) and endodermally-derived tissues (hepatobiliary and pancreas; darker gray) share many of the same downstream targets that promote development of their respective cell types.
The role of Oc factors in promoting specific cell fates during retinal development is a rather recent focus in the field. Oc1 and Oc2 have overlapping expression patterns early in development, which, for the most part, persist into the perinatal period. Oc1 and Oc2 appear to promote retinal ganglion cell development from RPCs through enhancing expression of Math5, Isl1 and Pou4f2 [86]. The other major retinal cell type promoted by Oc1 and Oc2 is the horizontal cell. These cells serve to connect the photoreceptors of the retina and propagate signals laterally within the inner nuclear layer. Interestingly, retinal-specific Oc1 gene inactivation results in an 80% reduction in the horizontal cell population, but no other cell types are substantially affected [87]. Inactivation of both Oc1 and Oc2 results in a complete absence of horizontal cells and more modest reductions in cones, retinal ganglion cells and starburst amacrine cells [87, 88]. The network of transcription factors implicated in horizontal cell differentiation from RPCs has striking similarities to specification of pancreas from the endoderm. Indeed, during the differentiation of horizontal cells from RPCs, Oc1 acts in parallel with Ptf1a and in conjunction with Otx2 to promote expression of Prox1 and Lim1, thereby driving a horizontal cell fate [87, 89]. In the pancreas, Oc1 also acts in parallel with Ptf1a to activate gene expression programs for the development of the exocrine cell types. These include Prox1 in duct development as well as many others. However, Oc1 and Oc2 are downstream effectors of Pax6 in horizontal cells whereas Oc1 acts upstream of Pax6 in the pancreatic endocrine lineage [90]. The parallels between the retinal transcription factor network and that of the developing pancreas should not be too surprising given the similarities in function between neurons that package and secrete neurotransmitters in response to cell depolarization, and endocrine cells that package and secrete hormones in response to cell depolarization. Only 20 years ago, it was thought the pancreatic endocrine cells an originated from ectodermally- or neuronally-derived lineage (such as the neural crest) that migrated into the pancreas. Lineage tracing studies revealed that pancreatic endocrine cells are derived from the endoderm, just like the exocrine cells [91].
4.2. Motor neurons
Oc factors show high conservation of function to specify neuronal cell types in multiple model organisms [81-83, 92]. As previously noted, the Oc factors tend to operate within regulatory pathways containing similar families of transcription factors, regardless of cell type or germ layer (Figures 2 and 3). Indeed, in ascidians Neurogenin activates Oc, which in turn acts in an autoregulatory loop to promote expression of both factors, indicating that these parallels in developmental transcription factor networks are not limited to mammalian or even vertebrate systems [93].
Much of the initial in-depth work investigating the neuronal function of Oc factors was in the setting of motor neuron development. Motor neurons differentiate from a region within the spinal cord called the progenitor motor neuron (pMN) domain. In the pMN domain, Oc factors are expressed early and participate in a network with other transcription factors such as Neurogenin-2, Pax6, Nkx6.1 and Isl1/2 [94, 95]. Of note, while all three Oc factors are expressed in the developing pMN domain, they follow the same temporal expression pattern observed in the endoderm, with Oc1 expression activated first and most highly expressed followed by Oc2 and Oc3 at progressively lower levels. As development proceeds, and motor neurons born from the pMN domain mature, the Oc expression pattern changes with Oc1 becoming reduced, Oc3 becoming undetectable and Oc2 having a modest increase in expression [95]. The decrease in Oc1 expression can in part be explained by an increase in expression of miR-9, which is capable of repressing Oc1 expression both in vitro and in vivo [96]. Oc1 also regulates the formation of neuromuscular junctions formed by motor neurons. In the absence of Oc1, motor neuron atrophy occurs and neuromuscular junctions fail to form properly [97]. However, the Oc factors are not limited to regulating the development of motor neurons in mice; many other types of neurons rely upon this family of factors. For example, Oc1 is also necessary for proper organization of cerebellar Purkinje cells as well as differentiation of Renshaw cell interneurons, both of which are essential for proper locomotion [98, 99].
4.3. Dopaminergic neurons
The Oc factors function in the development of a diverse set of neurons within both the central and peripheral nervous systems. The mesodiencephalon is a nucleus of dopaminergic neurons controlling motor function and cognitive ability. All Oc family members are expressed in the mesodiencephalon early in development, but Oc1 expression is lost by e12.5 whereas Oc2/3 expression is maintained. Loss of Oc1 results in a reduction in the number of Th (tyrosine hydroxylase)-positive neurons (which convert L-tyrosine to the dopamine precursor L-DOPA) in the mesodiencephalon. Loss of all three Oc factors further reduced the Th-positive neuron population indicating a partially redundant function in development of those cells [100]. Oc1 appears to affect the differentiation of Th+ cells through direct regulation of the transcription factor Lmx1a, which in turn promotes expression of Neurog2 and Nkx6.1 [101]. Oc factors regulate the development of many other dopaminergic cell types. Oc2 is expressed in developing trigeminal neurons, which innervate the face, and in its absence there is loss of projections from those neurons [102]. Further, there is complete loss of neurons in the rhombencephalic mesencephalic trigeminal nucleus in the absence of any Oc factors indicating that they are indispensible for differentiation of those cells [103]. In another dopaminergic nucleus, the A13 dopaminergic nucleus, all three Oc factors are expressed during development with Oc1 having the highest and most prolonged expression. A13 dopaminergic neurons still differentiated in Oc1/2 compound mutants, but they were not maintained properly and they aberrantly spread into other regions. Interestingly, Oc1/2 again operate within a network including the transcription factors Pax6 and Isl1, further indicating the importance of these shared developmental networks among vastly different organ systems [104].
While no direct connections have been made between Oc factors and neuronal disease, it is quite possible that differentiation defects in any one of a number of different neuronal cell types could predispose an individual to disease development. Indeed, the mesodiencephalic dopaminergic neurons, whose differentiation is regulated by Oc1, are associated with development of Parkinson's disease [100, 105]. Additionally, the multifaceted regulation of neuromuscular development by Oc factors could contribute to impairments in locomotion and muscle function if a loss of the Oc factors were to occur. Taken together, it is clear that the Oc family is vital for establishing and maintaining many different neuron populations, and that this regulation largely is within the same network of transcription factor families also important to development of other organ systems.
5. Conclusion
Although the Oc factors are expressed in a broad array of tissues, they serve similar functions in each of them (Figure 3). They are capable of promoting differentiation and maturation of a multitude of different cell types derived from both the endoderm and ectoderm. The Oc factors, especially Oc1, operate within very similar gene networks to perform this function with common cofactors and effectors such as Neurogenin2/3, Pax6, Prox1, Hnf1β and others. The unique environments of the progenitor cells in which these networks are active likely confer the specificity leading to the vastly different mature cell types. While Oc factors are predominantly expressed during development, they also clearly have a function in maintaining the mature differentiated state of multiple cell types, thereby conferring protection from disease. This unique family of transcription factors thus provides a perfect example of how regulation of developmental processes can have longstanding effects on adult disease.
ACKNOWLEDGEMENTS
We would like to thank the Gannon lab for helpful discussions. This work was supported by NIH/NIDDK grant R01DK105689 to M. G. P. A. K. was supported in part by the Vanderbilt University Training Program in Stem Cell and Regenerative Developmental Biology (T32 HD05702).
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors have no conflicts to declare.
REFERENCES
- 1.Blochlinger K, Bodmer R, Jan LY, Jan YN. Genes Dev. 1990;4(8):1322–31. doi: 10.1101/gad.4.8.1322. [DOI] [PubMed] [Google Scholar]
- 2.Bodmer R, Barbel S, Sheperd S, Jack JW, Jan LY, Jan YN. Cell. 1987;51(2):293–307. doi: 10.1016/0092-8674(87)90156-5. [DOI] [PubMed] [Google Scholar]
- 3.Blochlinger K, Bodmer R, Jack J, Jan LY, Jan YN. Nature. 1988;333(6174):629–35. doi: 10.1038/333629a0. [DOI] [PubMed] [Google Scholar]
- 4.Andres V, Nadal-Ginard B, Mahdavi V. Development. 1992;116(2):321–34. doi: 10.1242/dev.116.2.321. [DOI] [PubMed] [Google Scholar]
- 5.Lemaigre FP, Durviaux SM, Rousseau GG. J. Biol. Chem. 1993;268(26):19896–905. [PubMed] [Google Scholar]
- 6.Lemaigre FP, Durviaux SM, Truong O, Lannoy VJ, Hsuan JJ, Rousseau GG. Proc. Natl. Acad. Sci. USA. 1996;93(18):9460–4. doi: 10.1073/pnas.93.18.9460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacquemin P, Lannoy VJ, Rousseau GG, Lemaigre FP. J. Biol. Chem. 1999;274(5):2665–71. doi: 10.1074/jbc.274.5.2665. [DOI] [PubMed] [Google Scholar]
- 8.Vanhorenbeeck V, Jacquemin P, Lemaigre FP, Rousseau GG. Biochem. Biophys. Res. Commun. 2002;292(4):848–54. doi: 10.1006/bbrc.2002.6760. [DOI] [PubMed] [Google Scholar]
- 9.Lannoy VJ, Burglin TR, Rousseau GG, Lemaigre FP. J. Biol. Chem. 1998;273(22):13552–62. doi: 10.1074/jbc.273.22.13552. [DOI] [PubMed] [Google Scholar]
- 10.Lannoy VJ, Rodolosse A, Pierreux CE, Rousseau GG, Lemaigre FP. J. Biol. Chem. 2000;275(29):22098–103. doi: 10.1074/jbc.M000855200. [DOI] [PubMed] [Google Scholar]
- 11.Yoshida Y, Hughes DE, Rausa FM, 3rd, Kim IM, Tan Y, Darlington GJ, Costa RH. Hepatology. 2006;43(2):276–86. doi: 10.1002/hep.21044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rausa FM, 3rd, Hughes DE, Costa RH. J. Biol. Chem. 2004;279(41):43070–6. doi: 10.1074/jbc.M407472200. [DOI] [PubMed] [Google Scholar]
- 13.Landry C, Clotman F, Hioki T, Oda H, Picard JJ, Lemaigre FP, Rousseau GG. Dev. Biol. 1997;192(2):247–57. doi: 10.1006/dbio.1997.8757. [DOI] [PubMed] [Google Scholar]
- 14.Lemaigre F, Zaret KS. Curr. Opin. Genet. Dev. 2004;14(5):582–90. doi: 10.1016/j.gde.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Samadani U, Costa RH. Mol. Cell Biol. 1996;16(11):6273–84. doi: 10.1128/mcb.16.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, Fraenkel E, Bell GI, Young RA. Science. 2004;303(5662):1378–81. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng W, Guo L, Zhang Z, Soo HM, Wen C, Wu W, Peng J. Dev. Biol. 2006;294(2):482–96. doi: 10.1016/j.ydbio.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 18.Margagliotti S, Clotman F, Pierreux CE, Beaudry JB, Jacquemin P, Rousseau GG, Lemaigre FP. Dev. Biol. 2007;311(2):579–89. doi: 10.1016/j.ydbio.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 19.Rausa F, Samadani U, Ye H, Lim L, Fletcher CF, Jenkins NA, Copeland NG, Costa RH. Dev. Biol. 1997;192(2):228–46. doi: 10.1006/dbio.1997.8744. [DOI] [PubMed] [Google Scholar]
- 20.Briancon N, Bailly A, Clotman F, Jacquemin P, Lemaigre FP, Weiss MC. J. Biol. Chem. 2004;279(32):33398–408. doi: 10.1074/jbc.M405312200. [DOI] [PubMed] [Google Scholar]
- 21.Rubins NE, Friedman JR, Le PP, Zhang L, Brestelli J, Kaestner KH. Mol. Cell Biol. 2005;25(16):7069–77. doi: 10.1128/MCB.25.16.7069-7077.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clotman F, Jacquemin P, Plumb-Rudewiez N, Pierreux CE, Van der Smissen P, Dietz HC, Courtoy PJ, Rousseau GG, Lemaigre FP. Genes Dev. 2005;19(16):1849–54. doi: 10.1101/gad.340305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plumb-Rudewiez N, Clotman F, Strick-Marchand H, Pierreux CE, Weiss MC, Rousseau GG, Lemaigre FP. Hepatology. 2004;40(6):1266–74. doi: 10.1002/hep.20459. [DOI] [PubMed] [Google Scholar]
- 24.Laudadio I, Manfroid I, Achouri Y, Schmidt D, Wilson MD, Cordi S, Thorrez L, Knoops L, Jacquemin P, Schuit F, Pierreux CE, Odom DT, Peers B, Lemaigre FP. Gastroenterology. 2012;142(1):119–29. doi: 10.1053/j.gastro.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 25.Rastegar M, Rousseau GG, Lemaigre FP. Endocrinology. 2000;141(5):1686–92. doi: 10.1210/endo.141.5.7478. [DOI] [PubMed] [Google Scholar]
- 26.Delesque-Touchard N, Park SH, Waxman DJ. J. Biol. Chem. 2000;275(44):34173–82. doi: 10.1074/jbc.M004027200. [DOI] [PubMed] [Google Scholar]
- 27.Gardmo C, Mode A. J. Mol. Endocrinol. 2006;37(3):433–41. doi: 10.1677/jme.1.02116. [DOI] [PubMed] [Google Scholar]
- 28.Lannoy VJ, Decaux JF, Pierreux CE, Lemaigre FP, Rousseau GG. Diabetologia. 2002;45(8):1136–41. doi: 10.1007/s00125-002-0856-z. [DOI] [PubMed] [Google Scholar]
- 29.Tan Y, Hughes D, Wang X, Costa RH. Hepatology. 2002;35(1):30–9. doi: 10.1053/jhep.2002.30317. [DOI] [PubMed] [Google Scholar]
- 30.Clotman F, Lannoy VJ, Reber M, Cereghini S, Cassiman D, Jacquemin P, Roskams T, Rousseau GG, Lemaigre FP. Development. 2002;129(8):1819–28. doi: 10.1242/dev.129.8.1819. [DOI] [PubMed] [Google Scholar]
- 31.Coffinier C, Gresh L, Fiette L, Tronche F, Schutz G, Babinet C, Pontoglio M, Yaniv M, Barra J. Development. 2002;129(8):1829–38. doi: 10.1242/dev.129.8.1829. [DOI] [PubMed] [Google Scholar]
- 32.Falix FA, Weeda VB, Labruyere WT, Poncy A, de Waart DR, Hakvoort TB, Lemaigre F, Gaemers IC, Aronson DD, Lamers WH. Dev. Biol. 2014;396(2):201–13. doi: 10.1016/j.ydbio.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 33.Vanderpool C, Sparks EE, Huppert KA, Gannon M, Means AL, Huppert SS. Hepatology. 2012;55(1):233–43. doi: 10.1002/hep.24631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamasaki H, Sada A, Iwata T, Niwa T, Tomizawa M, Xanthopoulos KG, Koike T, Shiojiri N. Development. 2006;133(21):4233–43. doi: 10.1242/dev.02591. [DOI] [PubMed] [Google Scholar]
- 35.Walter TJ, Vanderpool C, Cast AE, Huppert SS. Am. J. Pathol. 2014;184(5):1479–88. doi: 10.1016/j.ajpath.2014.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matthews RP, Lorent K, Pack M. Dev. Dyn. 2008;237(1):124–31. doi: 10.1002/dvdy.21407. [DOI] [PubMed] [Google Scholar]
- 37.Clotman F, Libbrecht L, Gresh L, Yaniv M, Roskams T, Rousseau GG, Lemaigre FP. J. Hepatol. 2003;39(5):686–92. doi: 10.1016/s0168-8278(03)00409-4. [DOI] [PubMed] [Google Scholar]
- 38.Raynaud P, Tate J, Callens C, Cordi S, Vandersmissen P, Carpentier R, Sempoux C, Devuyst O, Pierreux CE, Courtoy P, Dahan K, Delbecque K, Lepreux S, Pontoglio M, Guay-Woodford LM, Lemaigre FP. Hepatology. 2011;53(6):1959–66. doi: 10.1002/hep.24292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fabris L, Cadamuro M, Libbrecht L, Raynaud P, Spirli C, Fiorotto R, Okolicsanyi L, Lemaigre F, Strazzabosco M, Roskams T. Hepatology. 2008;47(2):719–28. doi: 10.1002/hep.22015. [DOI] [PubMed] [Google Scholar]
- 40.Nakao K, Miyaaki H, Ichikawa T. J. Gastroenterol. 2014;49(4):589–93. doi: 10.1007/s00535-014-0932-4. [DOI] [PubMed] [Google Scholar]
- 41.Hao R, He J, Liu X, Gao G, Liu D, Cui L, Yu G, Yu W, Chen Y, Guo D. J. Virol. 2015;89(8):4345–55. doi: 10.1128/JVI.03094-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Limaye PB, Alarcon G, Walls AL, Nalesnik MA, Michalopoulos GK, Demetris AJ, Ochoa ER. Lab. Invest. 2008;88(8):865–72. doi: 10.1038/labinvest.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan Y, Yoshida Y, Hughes DE, Costa RH. Gastroenterology. 2006;130(4):1283–300. doi: 10.1053/j.gastro.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lehner F, Kulik U, Klempnauer J, Borlak J. PLoS One. 2010;5(10):e13344. doi: 10.1371/journal.pone.0013344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pan FC, Wright C. Dev. Dyn. 2011;240(3):530–65. doi: 10.1002/dvdy.22584. [DOI] [PubMed] [Google Scholar]
- 46.Gittes GK. Dev. Biol. 2009;326(1):4–35. doi: 10.1016/j.ydbio.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 47.Wilding L, Gannon M. Diabetes Metab. Res. Rev. 2004;20(2):114–23. doi: 10.1002/dmrr.429. [DOI] [PubMed] [Google Scholar]
- 48.Stoffers DA, Zinkin NT, Stanojevic V, Clarke WL, Habener JF. Nat. Genet. 1997;15(1):106–10. doi: 10.1038/ng0197-106. [DOI] [PubMed] [Google Scholar]
- 49.Fujitani Y, Fujitani S, Boyer DF, Gannon M, Kawaguchi Y, Ray M, Shiota M, Stein RW, Magnuson MA, Wright CV. Genes Dev. 2006;20(2):253–66. doi: 10.1101/gad.1360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jacquemin P, Lemaigre FP, Rousseau GG. Dev. Biol. 2003;258(1):105–16. doi: 10.1016/s0012-1606(03)00115-5. [DOI] [PubMed] [Google Scholar]
- 51.Jonsson J, Carlsson L, Edlund T, Edlund H. Nature. 1994;371(6498):606–9. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 52.Maestro MA, Boj SF, Luco RF, Pierreux CE, Cabedo J, Servitja JM, German MS, Rousseau GG, Lemaigre FP, Ferrer J. J. Hum. Mol. Genet. 2003;12(24):3307–14. doi: 10.1093/hmg/ddg355. [DOI] [PubMed] [Google Scholar]
- 53.Poll AV, Pierreux CE, Lokmane L, Haumaitre C, Achouri Y, Jacquemin P, Rousseau GG, Cereghini S, Lemaigre FP. Diabetes. 2006;55(1):61–9. [PubMed] [Google Scholar]
- 54.Jacquemin P, Pierreux CE, Fierens S, van Eyll JM, Lemaigre FP, Rousseau GG. Gene Expr. Patterns. 2003;3(5):639–44. doi: 10.1016/s1567-133x(03)00110-8. [DOI] [PubMed] [Google Scholar]
- 55.Pierreux CE, Vanhorenbeeck V, Jacquemin P, Lemaigre FP, Rousseau GG. J. Biol. Chem. 2004;279(49):51298–304. doi: 10.1074/jbc.M409038200. [DOI] [PubMed] [Google Scholar]
- 56.Vanhorenbeeck V, Jenny M, Cornut JF, Gradwohl G, Lemaigre FP, Rousseau GG, Jacquemin P. Dev. Biol. 2007;305(2):685–94. doi: 10.1016/j.ydbio.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 57.Zhang H, Ables ET, Pope CF, Washington MK, Hipkens S, Means AL, Path G, Seufert J, Costa RH, Leiter AB, Magnuson MA, Gannon M. Mech. Dev. 2009;126(11-12):958–73. doi: 10.1016/j.mod.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gu G, Dubauskaite J, Melton DA. Development. 2002;129(10):2447–57. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- 59.Johansson KA, Dursun U, Jordan N, Gu G, Beermann F, Gradwohl G, Grapin-Botton A. Dev. Cell. 2007;12(3):457–65. doi: 10.1016/j.devcel.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 60.Jacquemin P, Durviaux SM, Jensen J, Godfraind C, Gradwohl G, Guillemot F, Madsen OD, Carmeliet P, Dewerchin M, Collen D, Rousseau GG, Lemaigre FP. Mol. Cell Biol. 2000;20(12):4445–54. doi: 10.1128/mcb.20.12.4445-4454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oliver-Krasinski JM, Kasner MT, Yang J, Crutchlow MF, Rustgi AK, Kaestner KH, Stoffers DA. J. Clin. Invest. 2009;119(7):1888–98. doi: 10.1172/JCI37028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gannon M, Ray MK, Van Zee K, Rausa F, Costa RH, Wright CV. Development. 2000;127(13):2883–95. doi: 10.1242/dev.127.13.2883. [DOI] [PubMed] [Google Scholar]
- 63.Tweedie E, Artner I, Crawford L, Poffenberger G, Thorens B, Stein R, Powers AC, Gannon M. Diabetes. 2006;55(12):3264–70. doi: 10.2337/db06-0090. [DOI] [PubMed] [Google Scholar]
- 64.Wilding Crawford L, Tweedie Ables E, Oh YA, Boone B, Levy S, Gannon M. PLoS One. 2008;3(2):e1611. doi: 10.1371/journal.pone.0001611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hara M, Shen J, Pugh W, Polonsky KS, Le Beau MM, Bell GI. Exp. Clin. Endocrinol. Diabetes. 2007;115(10):654–61. doi: 10.1055/s-2007-982514. [DOI] [PubMed] [Google Scholar]
- 66.Pierreux CE, Poll AV, Kemp CR, Clotman F, Maestro MA, Cordi S, Ferrer J, Leyns L, Rousseau GG, Lemaigre FP. Gastroenterology. 2006;130(2):532–41. doi: 10.1053/j.gastro.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 67.Chen R, Hussain K, Al-Ali M, Dattani MT, Hindmarsh P, Jones PM, Marsh P. Pediatrics. 2008;121(6):e1541–7. doi: 10.1542/peds.2007-3543. [DOI] [PubMed] [Google Scholar]
- 68.Zhu L, Shi G, Schmidt CM, Hruban RH, Konieczny SF. Am. J. Pathol. 2007;171(1):263–73. doi: 10.2353/ajpath.2007.061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Strobel O, Dor Y, Alsina J, Stirman A, Lauwers G, Trainor A, Castillo CF, Warshaw AL, Thayer SP. Gastroenterology. 2007;133(6):1999–2009. doi: 10.1053/j.gastro.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schmid RM. J. Clin. Invest. 2002;109(11):1403–4. doi: 10.1172/JCI15889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pekala KR, Ma X, Kropp PA, Petersen CP, Hudgens CW, Chung CH, Shi C, Merchant NB, Maitra A, Means AL, Gannon MA. Lab. Invest. 2014;94(5):517–27. doi: 10.1038/labinvest.2014.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Prevot PP, Simion A, Grimont A, Colletti M, Khalaileh A, Van den Steen G, Sempoux C, Xu X, Roelants V, Hald J, Bertrand L, Heimberg H, Konieczny SF, Dor Y, Lemaigre FP, Jacquemin P. Gut. 2012;61(12):1723–32. doi: 10.1136/gutjnl-2011-300266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Prevot PP, Augereau C, Simion A, van den Steen G, Dauguet N, Lemaigre FP, Jacquemin P. Gastroenterology. 2013;145(3):668–78. e3. doi: 10.1053/j.gastro.2013.05.016. [DOI] [PubMed] [Google Scholar]
- 74.Yuan XW, Wang DM, Hu Y, Tang YN, Shi WW, Guo XJ, Song JG. J. Biol. Chem. 2013;288(43):31206–16. doi: 10.1074/jbc.M113.480285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pagliuca FW, Millman JR, Gurtler M, Segel M, Van Dervort A, Ryu JH, Peterson QP, Greiner D, Melton DA. Cell. 2014;159(2):428–39. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miyazaki S, Yamato E, Miyazaki J. Diabetes. 2004;53(4):1030–7. doi: 10.2337/diabetes.53.4.1030. [DOI] [PubMed] [Google Scholar]
- 77.Xu X, Browning VL, Odorico JS. Mech. Dev. 2011;128(7-10):412–27. doi: 10.1016/j.mod.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mfopou JK, Chen B, Mateizel I, Sermon K, Bouwens L. Gastroenterology. 2010;138(7):2233–45. 2245, e1–14. doi: 10.1053/j.gastro.2010.02.056. [DOI] [PubMed] [Google Scholar]
- 79.Micallef SJ, Janes ME, Knezevic K, Davis RP, Elefanty AG, Stanley EG. Diabetes. 2005;54(2):301–5. doi: 10.2337/diabetes.54.2.301. [DOI] [PubMed] [Google Scholar]
- 80.Nguyen DN, Rohrbaugh M, Lai Z. Mech. Dev. 2000;97(1-2):57–72. doi: 10.1016/s0925-4773(00)00431-7. [DOI] [PubMed] [Google Scholar]
- 81.Sasakura Y, Makabe KW. Mech. Dev. 2001;104(1-2):37–48. doi: 10.1016/s0925-4773(01)00352-5. [DOI] [PubMed] [Google Scholar]
- 82.Hong SK, Kim CH, Yoo KW, Kim HS, Kudoh T, Dawid IB, Huh TL. Mech. Dev. 2002;112(1-2):199–202. doi: 10.1016/s0925-4773(01)00647-5. [DOI] [PubMed] [Google Scholar]
- 83.Haworth KE, Latinkic B. Int. J. Dev. Biol. 2009;53(1):159–62. doi: 10.1387/ijdb.072472ke. [DOI] [PubMed] [Google Scholar]
- 84.Heavner W, Pevny L. Cold Spring Harb. Perspect. Biol. 2012;4(12):a008391. doi: 10.1101/cshperspect.a008391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mu X, Zhao S, Pershad R, Hsieh TF, Scarpa A, Wang SW, White RA, Beremand PD, Thomas TL, Gan L, Klein WH. Nucleic Acids Res. 2001;29(24):4983–93. doi: 10.1093/nar/29.24.4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu FG, Sapkota D, Li RZ, Mu XQ. Journal of Comparative Neurology. 2012;520(5):952–969. doi: 10.1002/cne.22741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu F, Li R, Umino Y, Kaczynski TJ, Sapkota D, Li S, Xiang M, Fliesler SJ, Sherry DM, Gannon M, Solessio E, Mu X. J. Neurosci. 2013;33(32):13053–65. 13065a. doi: 10.1523/JNEUROSCI.0116-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sapkota D, Chintala H, Wu F, Fliesler SJ, Hu Z, Mu X. Proc. Natl. Acad. Sci. USA. 2014;111(39):E4086–95. doi: 10.1073/pnas.1405354111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Emerson MM, Surzenko N, Goetz JJ, Trimarchi J, Cepko CL. Dev. Cell. 2013;26(1):59–72. doi: 10.1016/j.devcel.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Klimova L, Antosova B, Kuzelova A, Strnad H, Kozmik Z. Dev. Biol. 2015;402(1):48–60. doi: 10.1016/j.ydbio.2015.02.023. [DOI] [PubMed] [Google Scholar]
- 91.Slack JM. Development. 1995;121(6):1569–80. doi: 10.1242/dev.121.6.1569. [DOI] [PubMed] [Google Scholar]
- 92.Kudoh T, Tsang M, Hukriede NA, Chen X, Dedekian M, Clarke CJ, Kiang A, Schultz S, Epstein JA, Toyama R, Dawid IB. Genome Res. 2001;11(12):1979–87. doi: 10.1101/gr.209601. [DOI] [PubMed] [Google Scholar]
- 93.Pezzotti MR, Locascio A, Racioppi C, Fucci L, Branno M. Dev. Biol. 2014;390(2):273–87. doi: 10.1016/j.ydbio.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 94.Roy A, Francius C, Rousso DL, Seuntjens E, Debruyn J, Luxenhofer G, Huber AB, Huylebroeck D, Novitch BG, Clotman F. Development. 2012;139(17):3109–19. doi: 10.1242/dev.078501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Francius C, Clotman F. Neuroscience. 2010;165(1):116–29. doi: 10.1016/j.neuroscience.2009.09.076. [DOI] [PubMed] [Google Scholar]
- 96.Luxenhofer G, Helmbrecht MS, Langhoff J, Giusti SA, Refojo D, Huber AB. Dev. Biol. 2014;386(2):358–70. doi: 10.1016/j.ydbio.2013.12.023. [DOI] [PubMed] [Google Scholar]
- 97.Audouard E, Schakman O, Rene F, Huettl RE, Huber AB, Loeffler JP, Gailly P, Clotman F. PLoS One. 2012;7(12):e50509. doi: 10.1371/journal.pone.0050509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Audouard E, Schakman O, Ginion A, Bertrand L, Gailly P, Clotman F. Mol. Cell Neurosci. 2013;56:159–68. doi: 10.1016/j.mcn.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 99.Stam FJ, Hendricks TJ, Zhang J, Geiman EJ, Francius C, Labosky PA, Clotman F, Goulding M. Development. 2012;139(1):179–90. doi: 10.1242/dev.071134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chakrabarty K, Von Oerthel L, Hellemons A, Clotman F, Espana A, Groot Koerkamp M, Holstege FC, Pasterkamp RJ, Smidt MP. Biol. Open. 2012;1(8):693–704. doi: 10.1242/bio.20121230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yuan J, Lei ZN, Wang X, Deng YJ, Chen DB. Brain Res. 2015;1608:40–50. doi: 10.1016/j.brainres.2015.02.046. [DOI] [PubMed] [Google Scholar]
- 102.Hodge LK, Klassen MP, Han BX, Yiu G, Hurrell J, Howell A, Rousseau A, Lemaigre F, Tessier-Lavigne M, Wang F. Neuron. 2007;55(4):572–86. doi: 10.1016/j.neuron.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 103.Espana A, Clotman F. Mol. Cell Neurosci. 2012;50(1):93–102. doi: 10.1016/j.mcn.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 104.Espana A, Clotman F. Journal of Comparative Neurology. 2012;520(7):1424–41. doi: 10.1002/cne.22803. [DOI] [PubMed] [Google Scholar]
- 105.Smidt MP, Burbach JP. Nat. Rev. Neurosci. 2007;8(1):21–32. doi: 10.1038/nrn2039. [DOI] [PubMed] [Google Scholar]