Abstract
Leucine-rich nuclear export signals (NESs) mediate rapid nuclear export of proteins via interaction with CRM1. This interaction is stimulated by RanGTP but remains of a relatively low affinity. In order to identify strong signals, we screened a 15-mer random peptide library for CRM1 binding, both in the presence and absence of RanGTP. Under each condition, strikingly similar signals were enriched, conforming to the NES consensus sequence. A derivative of an NES selected in the absence of RanGTP exhibits very high affinity for CRM1 in vitro and stably binds without the requirement of RanGTP. Localisation studies and RNA interference demonstrate inefficient CRM1-mediated export and accumulation of CRM1 complexed with the high-affinity NES at nucleoporin Nup358. These results provide in vivo evidence for a nuclear export reaction intermediate. They suggest that NESs have evolved to maintain low affinity for CRM1 to allow efficient export complex disassembly and release from Nup358.
Keywords: CRM1/exportin 1, leptomycin B, nuclear pore complex, nuclear transport, Nup358/RanBP2
Introduction
Nucleocytoplasmic transport occurs through large protein complexes that fenestrate the nuclear envelope (NE), termed nuclear pore complexes (NPCs) (reviewed in Suntharalingam and Wente, 2003). Nucleocytoplasmic transport is accomplished by soluble transport receptors that interact with both cargo and NPC. Importins mediate import of several different classes of proteins, while exportins mediate nuclear exit of proteins, tRNAs, U snRNAs and other RNAs, with the exception of mRNAs (reviewed in Görlich and Kutay, 1999).
The small GTPase Ran functions as a switch that governs directionality of importin- and exportin-mediated transport (reviewed in Görlich and Kutay, 1999). Nuclear Ran is predominantly bound to GTP, whereas cytoplasmic Ran is loaded with GDP. Nuclear RanGTP dissociates importin/cargo complexes providing direction to nuclear import (Görlich et al, 1996). Exportin/cargo heterodimers require the cooperative binding of RanGTP as RanGTP/exportin/cargo heterotrimeric complexes are several orders of magnitude more stable (Fornerod et al, 1997a; Kutay et al, 1997; 1998). After translocation through the nuclear pore complex, export complexes are destabilised by RanBP1 or RanBP1-like domains in RanBP2/Nup358. The export reaction is completed by hydrolysis of Ran-bound GTP, stimulated by RanGAP1 (reviewed in Görlich and Kutay, 1999).
One well-characterised example of the exportin class is CRM1/exportin 1, which exports proteins exposing a leucine-rich nuclear export signal (NES) (Fornerod et al, 1997a; Fukuda et al, 1997; Stade et al, 1997). Similar to other importin β-like receptors, CRM1 has been suggested to translocate through the NPC by multiple low-affinity hydrophobic interactions with FG repeat-containing nucleoporins (Ribbeck and Görlich, 2001; Rout and Aitchison, 2001). In addition to these weak interactions, the FG repeat region of Nup214/CAN, a nucleoporin located at the cytoplasmic side of the NPC, forms a particularly strong interaction with CRM1, which is further stimulated by RanGTP and NES cargo (Fornerod et al, 1997b; Askjaer et al, 1999; Kehlenbach et al, 1999). Another nucleoporin that associates with CRM1 is Nup358/RanBP2. This protein forms large fibrillar structures that emanate from the NPC into the cytoplasm (Wu et al, 1995; Yokoyama et al, 1995; Delphin et al, 1997; Walther et al, 2002). CRM1 binds Nup358 in an empty state, which suggested that Nup358 serves as a docking site for recycling CRM1 (Singh et al, 1999; Bernad et al, 2004). The majority of importins and exportins mediate nuclear transport of one or a few structurally related substrates (for examples, see Görlich and Kutay, 1999). Notable exceptions are the importin α/β heterodimeric import receptor and CRM1, which transport a great variety of proteins and ribonucleoprotein particles across the NPC. This promiscuity in transport substrates likely evolved because these receptors recognise short ubiquitous peptide sequences. The leucine-rich NES recognised by CRM1 was first identified in the viral HIV-1 Rev protein (Fischer et al, 1995) and in the cellular protein A phosphorylation inhibitor (PKI) (Wen et al, 1995). Both sequences contain a stretch of four regularly spaced leucines. Numerous studies have contributed to the definition of a leucine-rich NES consensus sequence as Φ-X2–3-Φ-X2–3-Φ-X-Φ (Φ: L, I, F, V, M; X: any amino acid) (Bogerd et al, 1996; Zhang and Dayton, 1998; Henderson and Eleftheriou, 2000; la Cour et al, 2003). The presence of leucine residues is not a prerequisite for NESs and several NESs have been identified that diverge from this postulated consensus sequence (see Fornerod and Ohno, 2002 for review). Following the currently ill-defined NES consensus sequence, most proteins are predicted to harbour NES consensus sequences. This hampers the annotation of valid export signals and their characterisation in vivo.
Although currently characterised NESs differ to some extent in their capacity to bind CRM1, each possesses a rather low affinity for CRM1 (Askjaer et al, 1999; Paraskeva et al, 1999). This is not characteristic of all exportins, as exportin-t and CAS/exportin 2 bind their cargo in the low nM range (Kutay et al, 1997; 1998). Two strong CRM1 interactors have been reported: NMD3 and snurportin 1, which bind CRM1 with 100-fold higher affinity as compared to the well-studied Rev protein (Paraskeva et al, 1999; Thomas and Kutay, 2003). The interaction with snurportin 1 is mediated through a large domain of at least 159 amino acids, while the domain for strong interaction of Nmd3 is unknown.
In order to select for strong NESs and to chart NES diversity, we have screened a 15-mer random peptide library for CRM1-binding peptides. Surprisingly, both in the presence and absence of RanGTP, highly similar sequences were selected. In vitro, a derivative of one of these NESs bound CRM1 with high affinity, bypassing the requirement for RanGTP. In vivo, nuclear export of this signal was ineffective, as it accumulated at Nup358. We suggest that physiological NESs must maintain a low affinity for CRM1 to allow efficient disassembly from CRM1 and release from Nup358.
Results
In vitro selection of synthetic NESs
To identify high-affinity peptide interactors of CRM1, we screened a fUSE5 15-mer random peptide library (Nishi et al, 1993) with a complexity of 2 × 108. Z-tagged CRM1 was immobilised on IgG-sepharose columns and affinity selections of 1 × 109 infectious phage were performed in the presence or absence of RanGTP. To increase selectivity, phages selected in the presence of RanGTP were eluted through the combined action of RanGAP and RanBP1. Three to four selection rounds were performed through recursive cycles of phage amplification and affinity selection. As shown in Figure 1A, clear increases in the number of affinity-selected phages were apparent under both selection conditions. Sequence analysis revealed strong enrichment of a unique signal under each selection condition. A single phage bearing the peptide sequence VDLLSSLFSGFSVGG was enriched to near-homogeneity after three selection rounds in the presence of RanGTP and was termed Powerphage or P phage. This phage contained the NES consensus sequence LSSLFSGFSV, hereafter to be referred to as P0. After four selection rounds in the absence of RanGTP, a single phage was highly enriched, termed Starphage or S phage, bearing the sequence DVSDLARLFSALGVS. Surprisingly, as NES peptides were not expected to bind in the absence of RanGTP, this peptide contains an NES consensus sequence LARLFSALGV very similar to P0 (Figure 2A). This signal will hereafter be referred to as S0. Next, purified P and S phages were tested individually for their ability to specifically bind CRM1 in the absence or presence of RanGTP. No binding of either P or S phages was observed to z-tagged transportin 1 columns (Figure 1B), confirming the specificity of the phage display selection. As expected, P phages bound CRM1 in the presence of RanGTP, and S phages in the absence of RanGTP (Figure 1B). Binding of S phages was enhanced six-fold by RanGTP, compared to a stimulation of approximately 250-fold for P phages (Figure 1C). These data suggest that both in the presence and absence of RanGTP, leucine-rich-type NESs were affinity-selected on CRM1 from a random peptide pool.
Figure 1.
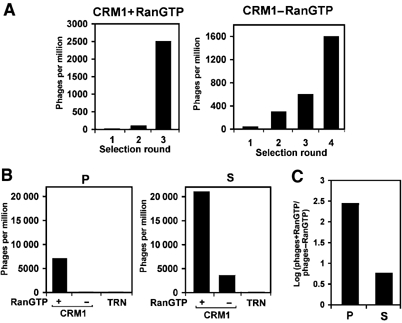
Selection of CRM1-binding peptides from a random peptide library. (A) A phage library displaying 15-mer random peptides was affinity selected on CRM1 columns in the presence or absence of RanGTP. The number of selected phages in each recursive selection round is expressed as colony forming phages selected per million of input. (B) Purified P (P0) and S (S0) phages were affinity selected on CRM1 and transportin 1 (TRN) columns in the presence or absence of RanGTP as indicated. Selected phages were compared as above. (C) S phages are less responsive to RanGTP than P phages. Log ratios of phages selected on CRM1 columns in the presence (phages+RanGTP) and absence (phages−RanGTP) of RanGTP are calculated for P (P0) or S (S0) phages.
Figure 2.
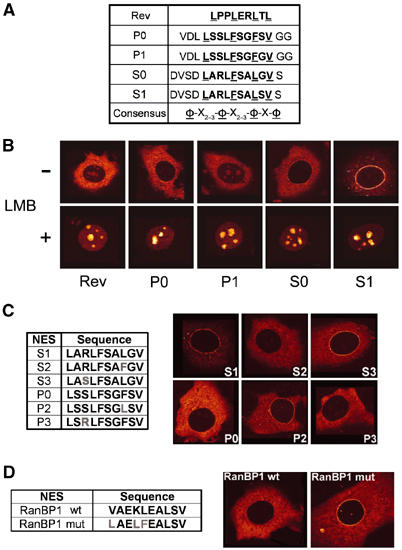
Permutations of selected peptides mediate distinct CRM1-mediated subcellular localisation. (A) Amino-acid sequences of P0, P1, S0 and S1 15-mer peptides compared to the HIV-1 Rev NES and the NES consensus sequence. NES sequences are in bold and consensus hydrophobic amino acids (φ) are underlined. One-letter amino-acid abbreviations are used. (B) Shuttling reporter proteins containing GFP, the export-incompetent Rev(1.4) variant and peptide sequences from (A) were transiently expressed in MCF7 cells and subcellular localisation was detected by green fluorescence 24 h post-transfection. The effect of 50 nM LMB was monitored 3 h after addition. (C) Identification of the critical amino-acid residues in S1 NES. Positions 3 and 8 in P0 and S1 NESs were interchanged (left), and subcellular localisation of the GFP reporter plasmids was analysed as in (B) (right). (D) Mutagenesis of the natural RanBP1 NES. A peptide corresponding to the natural RanBP1 NES was mutated to conform the high-affinity NES consensus (left). Subcellular localisation was analysed as above (right).
In vivo activities of synthetic NESs
Even though the potential NESs P0 and S0 conform to the ‘3-2-1' spacing of hydrophobic amino acids, we noted an unusual glycine residue between the third and the fourth hydrophobic amino acid of the S0 sequence (Figure 2A). A glycine at this position is known to abolish the activity of the Rev NES (Zhang and Dayton, 1998). We therefore mutated this glycine into a serine in S0 and replaced in P0 the serine at this position for a glycine (Figure 2A). We named these second-generation NES sequences S1 and P1, respectively. To test the export activities of the peptides in vivo, we inserted these into a reporter construct that has previously been used to compare NES activity (Henderson and Eleftheriou, 2000). This reporter consists of green fluorescent protein (GFP) fused to a mutant form of the HIV-1 Rev protein, Rev(1.4), that provides importin β-mediated import and nucleolar retention but lacks export activity. When fused to the strongest NESs, this reporter localises completely to the cytoplasm (Henderson and Eleftheriou, 2000). The HIV-1 Rev NES sequence was used as a positive control. As shown in Figure 2B, both P0 and S0 localise the reporter protein only to the cytoplasm, indicating that they confer strong export capacities, as the export signals completely overcome import activity as well as nucleolar retention. P1-GFP exhibits faint nucleolar staining similar to the Rev control NES. Interestingly, S1-GFP exhibited a prominent staining at the NE. This localisation is also appreciable for S0-GFP, although to a lesser extent. All NES fusion proteins accumulate in the nucleus upon treatment with the CRM1 inhibitor leptomycin B (LMB) (Figure 2B). This demonstrates that the cytoplasmic localisation mediated by the P0, P1, S0 and S1 sequences, and the nuclear rim staining of S0-GFP and S1-GFP are CRM1-dependent.
The difference in localisation of the S1 and P0 NESs could be explained by their penultimate hydrophobic position, a critical position in Rev-type NESs. To determine if this is the case, we mutated this position into leucine in P0, creating P2, and into phenylalanine in S1, creating S2 (Figure 2C). When introduced in the GFP reporter plasmid and expressed in cells as above, P2 mediated a clear nuclear rim staining, whereas S2 did not (Figure 2C). No effect was observed when the arginine of S1 was changed to serine (S3), or the corresponding serine in P0 was changed to arginine (P3) (Figure 2C). These data indicate that the amino-acid sequence LXXLFXXLSL can mediate CRM1-dependent NE localisation. When the hydrophobic residues of a naturally occurring NES present in RanBP1 (Zolotukhin and Felber, 1997) were mutated conforming this consensus, this NES promoted clear nuclear rim localisation (Figure 2D).
S1 NES binds to CRM1 with high affinity and stably binds without the requirement of RanGTP
In order to understand the striking localisation of the S1-like NESs, we analysed the CRM1-binding characteristics of this NES in RanGAP and pull-down assays. We expressed NES peptides as GFP fusion proteins in bacteria and purified the proteins using affinity chromatography. The CRM1 RanGAP assay is based on the fact that RanGTP is protected from RanGAP-mediated hydrolysis when present in export complexes, and can therefore be used to compare NES affinities (Kutay et al, 1997; Askjaer et al, 1999; Paraskeva et al, 1999). As references for standard NES strength, we used GFP fused to the PKI or Rev NES (Fischer et al, 1995; Wen et al, 1995), whereas the z-tagged form of Nmd3 was used as a reference for a high-affinity CRM1 interaction in this assay (Thomas and Kutay, 2003). As shown in Figure 3A, the protein containing the S1 NES showed an approximately 100-fold higher affinity for CRM1 than the standard NESs. In fact, the affinity of S1 for CRM1 was comparable to the 2z-Nmd3 protein. The S0 NES showed an affinity in between S1 and the standard NESs. To further evaluate the affinity of S1 for CRM1, we tested the LMB sensitivity of the S1/CRM1/RanGTP complex. Increasing concentrations of LMB were added to preformed NES/CRM1/RanGTP complexes and subjected to RanGAP-stimulated RanGTP hydrolysis. Under these conditions, CRM1 complexes containing the S1 NES were resistant to LMB in contrast to standard NESs from PKI or MVM NS2 (Figure 3B). The sensitivity of 2z-Nmd3-containing complexes was intermediate. When CRM1 was preincubated with LMB before the addition of the S1 NES, the protective effect was lost (Figure 3B). In this assay, the high concentration of RanGAP (100 nM) ensures that once RanGTP is released from CRM1, RanGTPase is immediately activated before RanGTP can rebind to CRM1 (Bischoff and Görlich, 1997). Therefore, the assay mainly measures off-rates, indicating that high-affinity binding of S1 to CRM1 is accomplished by a slow off-rate.
Figure 3.
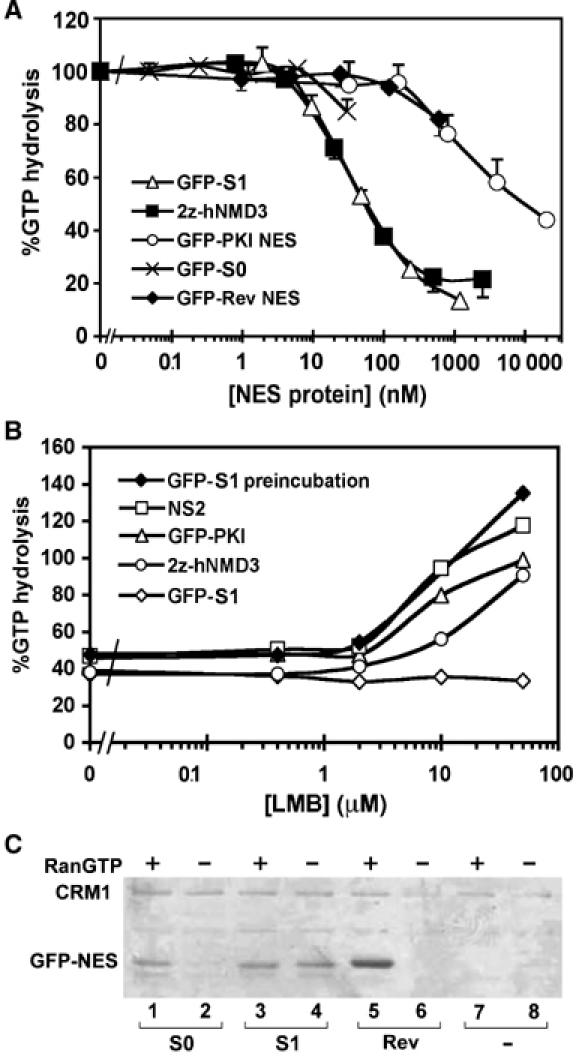
S1 NES binds to CRM1 with high affinity and independently of RanGTP. (A) CRM1 affinities of S0 and S1 peptides in recombinant GFP3 fusion proteins were measured using the CRM1 RanGAP assay, which measures their ability in the presence of CRM1 to protect RanGTP from RanGAP-stimulated hydrolysis as a function of NES concentration. Regular strength NESs of PKI and HIV-1 Rev, and the high-affinity interaction of 2z-Nmd3 served as references. Error bars denote standard errors of three independent experiments. (B) Differences in LMB sensitivity of export complexes. Different concentrations of LMB were added to RanGTP/CRM1 complexes containing GFP3-S1, GFP-PKI, the MVM NS2 peptide or 2z-Nmd3, and stability was measured using the CRM1 RanGAP assay as above. (C) RanGTP-independent binding to CRM1 of S1 NES. IgG-sepharose columns containing 1.5 μM z-tagged CRM1 were incubated in the absence (−) or presence (+) of 4.5 μM RanGTP as indicated, and in the absence (−) or presence of 1 μM GFP3-S1 (S1), GFP3-S0 (S0) or GFP3-Rev (Rev). Bound fractions are visualised using SDS–PAGE and Coomassie staining. A fraction of z-tagged CRM1 is coeluted and indicated on the left.
To assess the capability of S1 to bind CRM1 in the absence of RanGTP, a CRM1 pull-down assay was performed. CRM1 columns were incubated with various GFP3-tagged NESs in the presence or absence of RanGTP, after which eluted fractions were analysed by Coomassie staining (Figure 3C). While binding of S0 NES- and Rev NES-containing proteins to CRM1 was greatly stimulated by RanGTP, GFP3-S1 bound both in the presence and absence of RanGTP (Figure 3C). From these data, we conclude that the S1 NES exhibits a ‘supraphysiological' affinity (i.e. greater or stronger than normally present in the cell) for CRM1 such that stable binding to this export receptor takes place in the absence of RanGTP.
S1 NES localises at Nup358
To further investigate the prominent NE signal of the S1 NES reporter protein, we determined the localisation of S1-GFP. Fixed cells were permeabilised with digitonin, which permeabilises the cell membrane but leaves the NE intact. Anti-GFP antibodies continued to stain the NE, albeit weaker than direct GFP fluorescence (Figure 4B). In cells permeabilised with Triton X-100, allowing antibody access to the inside of the NE, the nuclear rim staining was the same (data not shown), suggesting that antigen accessibility explains the difference with direct GFP fluorescence. To study S1-GFP localisation at higher resolution, S1-GFP and RevNES-GFP were localised by immunogold staining on ultrathin cryosections using anti-GFP antibodies and 10 nm protein A-conjugated gold. As shown in Figure 4A, S1-GFP predominantly localises to the cytoplasmic side of the NPC, at the position of the cytoplasmic filaments of the NPC. RevNES-GFP did not show significant NPC localisation. Considering the EM localisation of S1-GFP, we selected Nup358 as a candidate for mediating S1-GFP accumulation. Short hairpin interfering RNA to Nup358 were expressed in HeLa cells together with S1-GFP. Cells were analysed 72 h after transfection when Nup358 protein levels are reduced by up to 90% (Bernad et al, 2004). As illustrated in Figure 4C, upon knockdown of Nup358, S1-GFP disappeared almost completely from the NE. Control transfections showed no reduction of NPC-associated S1-GFP. Another nucleoporin that could mediate S1/CRM1 interaction at the cytoplasmic face of the NPC is Nup214 (Askjaer et al, 1999; Kehlenbach et al, 1999). However, removal of Nup214 from the NPC by RNAi did not effect S1 localisation (Figure 4C). To confirm that the S1/CRM1 complex physically interacts with Nup358, we affinity-selected proteins from Xenopus interphase egg extract on immobilised S1 or Rev NES peptides. In these extracts, Ran is almost exclusively in the GDP-bound form. Under these conditions, a significant fraction of Nup358 stably associates with CRM1 to the S1 NES affinity column, but not to the Rev NES column (Figure 4D). We conclude that S1 NES accumulation at the NPC is directly mediated by Nup358.
Figure 4.
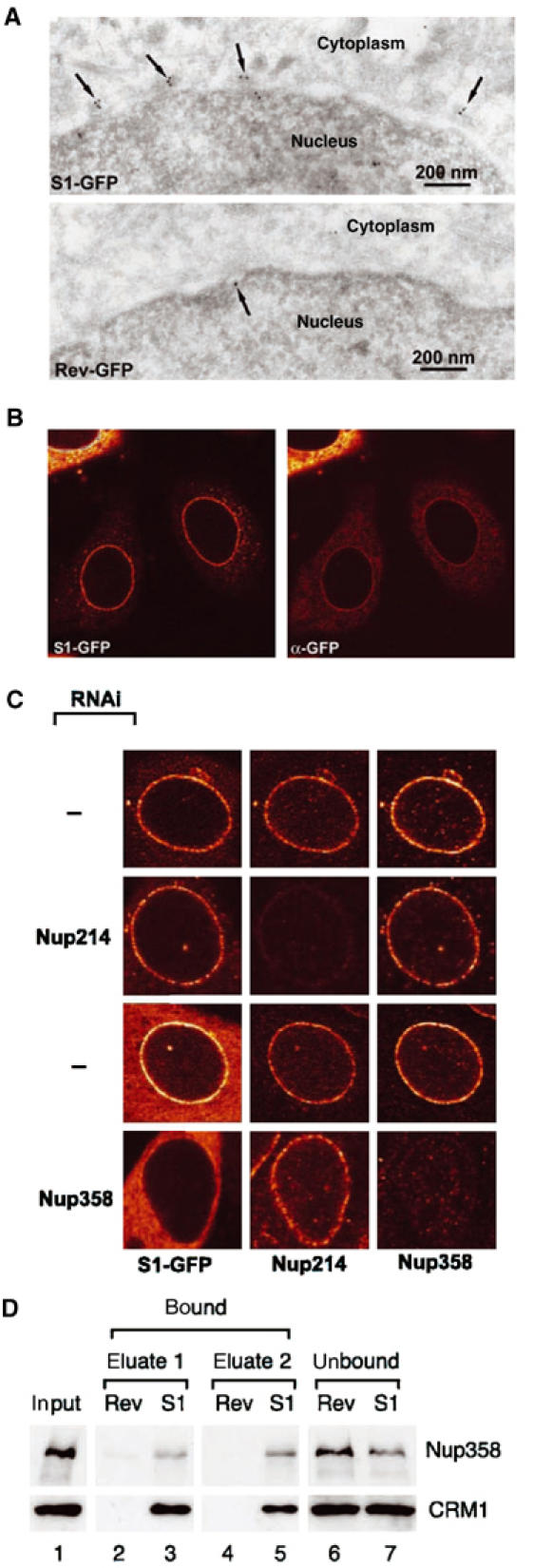
S1 NES localises at Nup358. (A) Immunoelectron microscopy. Cryosections of MCF-7 cells transfected with S1-GFP- or RevNES-GFP-containing reporter constructs (see Figure 2) were labelled with anti-GFP antibodies followed by protein A gold. In cells expressing S1-GFP, protein gold (arrows) decorates the outer aspect of the nuclear envelope at NPCs. (B) Immunofluorescence. Cells as in (A) were permeabilised with low concentrations of digitonin such that the nuclear membrane remained intact and labelled with anti-GFP antibodies. Anti-GFP antibodies stain the NE and colocalise largely with the signal from GFP. (C) Knockdown of Nup358 by RNAi removes S1-GFP from the NE. HeLa cells were cotransfected with a plasmid expressing shRNAs targeting Nup358 or Nup214 and an S1-GFP-containing reporter plasmid. Cells were analysed 72 h post-transfection for Nup358 and Nup214 levels and S1-GFP by indirect immunofluorescence and direct GFP fluorescence, respectively. A strong knockdown of Nup358, but not of Nup214, reduces S1-GFP from the NPC. (D) S1 NES physically interacts with Nup358. Proteins from Xenopus interphase egg extracts were affinity selected on immobilised biotinylated Rev or S1 NES peptides. Starting material (lane 1) and bound (lanes 2–5) and unbound (lanes 6 and 7) fractions were analysed for the presence of Nup358 and CRM1 by Western blotting.
The S1/CRM1 complex arrests at Nup358 and S1 is an inhibitor of CRM1
We have recently shown that CRM1 localises to Nup358 in vivo in an LMB-insensitive way (Bernad et al, 2004) and proposed this could represent empty CRM1 before recycling into the nucleus. Conceivably, S1 NES attaches to this population of Nup358-bound CRM1. Alternatively, the S1/CRM1 complex could attach de novo at Nup358. In this case, additional CRM1, stoichiometric to the S1 cargo, would be expected to localise at the NE. Therefore, we investigated whether CRM1 would accumulate with GFP in S1-GFP-transfected cells. Untransfected cells show a predominantly nuclear and NE CRM1 staining (Figure 5A). Expression of S1-GFP at the NE in transfected cells causes a clear NE accumulation of CRM1 (Figure 5A). To assess changes in nucleocytoplasmic distribution of CRM1, staining intensities in nuclear and cytoplasmic compartments were determined in S1-GFP- or Rev-GFP-expressing cells. Transfection of S1-GFP induced a 35% increase of cytoplasmic CRM1 (Figure 5B). CRM1 localisation was not influenced by expression of RevNES-GFP. These data indicate that the S1/CRM1 complex arrests at Nup358 upon NPC translocation and that S1 remains bound to CRM1 in the cytoplasm.
Figure 5.
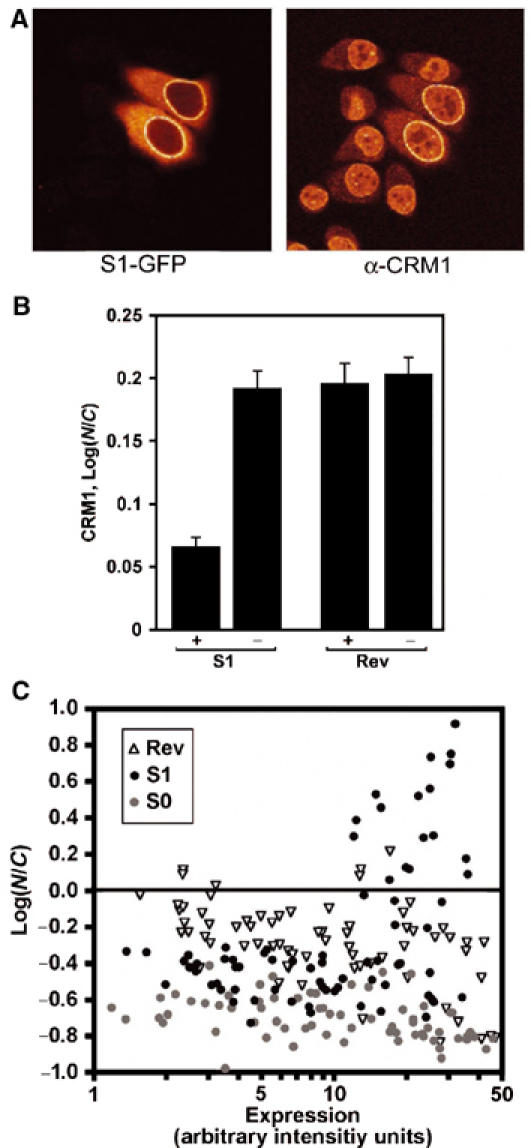
S1 NES sequesters CRM1 at the NE (A) and in the cytoplasm (B) and can inhibit its own export (C). (A) HeLa cells were transfected with the S1-GFP-containing reporter construct and GFP was detected together with CRM1 with direct GFP fluorescence and indirect immunofluorescence, respectively. (B) Cells were transfected with the S1 NES (S1) or Rev NES (Rev)-containing GFP reporter plasmids as in (A) and nuclear and cytoplasmic CRM1 immunofluorescence signals were measured in confocal sections of 15 transfected (+) or untransfected (−) cells. Log ratios of the means are significantly reduced in S1-GFP-expressing cells, but not in RevNES-GFP-expressing cells. Error bars denote standard errors. (C) Single cells expressing different levels of S0 (gray circles), S1 (black circles) and Rev NES (white triangles) GFP reporter proteins were analysed for nuclear and cytoplasmic GFP levels. S0 NES and Rev NES mediate cytoplasmic localisation irrespective of expression level. In contrast, the S1 NES promotes nuclear export at low expression levels but not at high expression levels.
The sequestering of CRM1 by the S1 NES suggests that expression of the S1 could lead to an inhibition of CRM1 function. To test this, we expressed S0, S1 and Rev-GFP proteins transiently for 24 h in MCF-7 cells and measured their subcellular localisation as a function of cellular protein expression level. As shown in Figure 5C, S0 and Rev NESs can promote nuclear export of the shuttling GFP reporter, irrespective of the expression level. In contrast, S1-GFP only promotes cytoplasmic accumulation when expressed at low to moderate levels, whereas at high expression S1-GFP accumulates in the nucleoplasm (Figure 5C). This indicates that by sequestering CRM1, the S1 NES acts as an inhibitor of CRM1 function.
S1/CRM1/RanGTP complexes display normal sensitivity to RanBP1
Our data suggest that the S1 NES remains bound to Nup358 as a consequence of its ability to bind CRM1 without RanGTP. Alternatively, S1/CRM1/RanGTP complexes could fail to dissociate at Nup358 because they are insensitive to RanBP1-like activity. In a CRM1 RanGAP assay, low concentrations of RanBP1 strongly promote RanGTP hydrolysis (Askjaer et al, 1999), presumably by loosening the RanGTP/CRM1 interaction (Bischoff and Görlich, 1997). As shown in Figure 6A, all NESs tested in this assay responded similarly to RanBP1 addition after export complex formation. The RanBP1 concentration at which RanGTP hydrolysis has recovered by 50% diverged no more than three-fold from S1 NES to the standard NESs. These data predict that, unlike CRM1 (Figure 5A), Ran does not accumulate at the cytoplasmic face of the NPC upon S1-GFP expression. To allow visualisation of a potential Ran enrichment at the cytoplasmic side of the nuclear pore, S1-GFP-transfected cells were permeabilised with digitonin and stained with an anti-Ran antibody. As shown in Figure 6B, Ran was not enriched at the NE upon expression of S1-GFP, nor was it increased in the cytoplasm.
Figure 6.
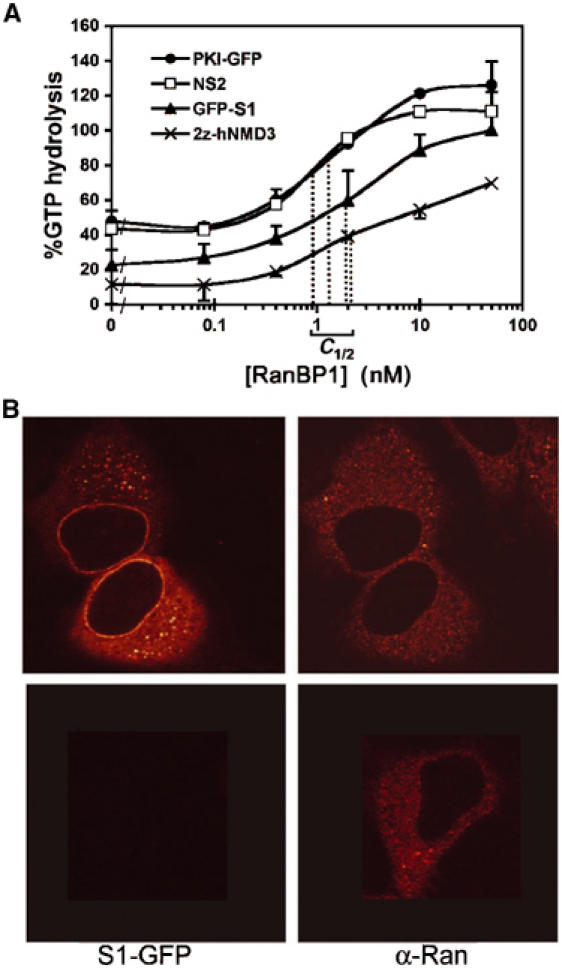
RanGTP can leave the S1/CRM1/RanGTP complex at Nup358. (A) GFP3-S1/CRM1/RanGTP complexes display normal sensitivity to RanBP1. Trimeric NES/CRM1/RanGTP complexes were assembled and incubated with increasing concentrations of recombinant RanBP1, and CRM1 RanGAP assays were performed as in Figure 3. Regular strength NESs of PKI and Rev and the high-affinity interaction with 2z-Nmd3 served as references. RanBP1 concentrations of half-maximum release of protection of GTP hydrolysis (C1/2) are indicated. (B) Ran does not accumulate at the NE in S1-GFP-expressing cells. HeLa cells were transiently transfected with the S1-GFP-containing reporter construct and permeabilised with low concentrations of digitonin to ensure intactness of the nuclear membrane. S1-GFP and Ran were detected by direct GFP fluorescence (left) and indirect immunofluorescence (right), respectively.
Discussion
In this study, we identify signals exhibiting high-affinity interactions with the widely studied export receptor CRM1. The results presented here bear relevance to the understanding of the mechanism of CRM1-mediated export and the evolution of leucine-rich NES motifs.
Supraphysiological NESs provide in vivo evidence for a novel nuclear export intermediate
An unexpected outcome of our peptide selection was that a shuttling substrate containing the highest affinity S1 NES accumulated at the NE. This accumulation represents CRM1/NES export complexes because the localisation is LMB sensitive and CRM1 accumulates with the S1 NES. Immunoelectron microscopy showed S1 NES accumulation at the cytoplasmic face of the NPC. Nup214 binds strongly to CRM1 in vitro in a RanGTP- and NES-stimulated way, therefore this nucleoporin represented a likely candidate to mediate S1/CRM1 NPC localisation (Fornerod et al, 1997b; Askjaer et al, 1999; Kehlenbach et al, 1999). However, RNAi experiments showed that the NE localisation was dependent on Nup358. This effectively rules out a potential role of Nup214 in the NE accumulation of S1/CRM1 as Nup214 is not affected by the removal of Nup358 (Bernad et al, 2004).
An LMB-insensitive interaction between CRM1 and Nup358 has been reported in vitro (Singh et al, 1999) and in vivo (Bernad et al, 2004) and most likely represents the empty state of CRM1. The S1 NES-dependent CRM1/Nup358 interaction is cargo dependent and LMB sensitive, and must therefore represent a different binding site. Co-immunoprecipitation of CRM1 with Nup358 from Xenopus egg extracts is greatly stimulated by RanQ69LGTP, a nonhydrolysable form of RanGTP, in contrast to importin β, importin 5 or importin 7 (Walther et al, 2003). This observation supports the idea of a cargo-dependent CRM1 interaction site on Nup358.
Why does the high-affinity NES accumulate at Nup358, while standard NESs do not show this behaviour? Biochemical analyses revealed that S1 possesses an affinity for CRM1 two orders of a magnitude higher than standard NESs. Our in vitro data further show that S1, unlike standard NESs, is able to interact stably with CRM1 in the absence of RanGTP. Thus, a likely explanation for the accumulation of S1 at Nup358 is that this reflects a failure of the S1/CRM1/RanGTP complex to dissociate, thereby keeping CRM1 in the export complex conformation. Because Nup358 contains four RanBP1-like RanGTP-binding domains (RBDs), the S1/CRM1 accumulation might be bridged by RanGTP. However, our biochemical data indicate that S1/CRM1/RanGTP complexes are fully sensitive to destabilisation by RanBP1. In addition, no accumulation of Ran was observed at the cytoplasmic face of the NE, indicating that Ran is able to leave the S1/CRM1/RanGTP complex. We conclude that the S1/CRM1 complex remains bound to Nup358 via CRM1, mimicking an export complex just prior to RanBP1-like RBD and RanGAP assisted complex disassembly. As illustrated in Figure 7A, we propose that Nup358 functions as the CRM1 export complex disassembly site at the NPC. To facilitate this process, Nup358 contains binding sites for export complexes, RanGTP hydrolysing cofactors as well as binding sites for RanGDP and empty CRM1. An additional effect of the export complex binding site of Nup358 would be to decrease reverse export of CRM1 export complexes that would form in the cytoplasm. Under normal conditions, these complexes are unlikely to form because of the cytoplasmic activity of RanGAP and RanBP1 (Becskei and Mattaj, 2003; Görlich et al, 2003). However, under conditions where cytoplasmic RanGTP is relatively high, for example by a decrease of RanGAP activity at lower temperatures (Görlich et al, 2003), capture of cytoplasmic export complexes may contribute to nuclear exclusion of NES proteins. Consistent with this idea, NES cargoes and CRM1 accumulate prominently at the nuclear rim when added in vitro to permeabilised cells in combination with RanQ69L (Kehlenbach et al, 1999; Nachury and Weis, 1999). Proteins containing NESs such as S1 that bind to CRM1 without RanGTP are predicted to be more susceptible to reverse export. However, S1 is clearly absent from the nuclear compartment, indicating that export is more efficient than reverse export. This most likely reflects the higher affinity of S1 NES for CRM1 in the nucleus through the cooperative binding of RanGTP. We have previously shown that reduction of Nup358 leads to a moderate reduction of Rev-NES-mediated nuclear export (Bernad et al, 2004), which we have proposed is due to a decrease in CRM1 recycling to the nucleus. We have not measured the effect of Nup358 depletion on cytoplasmic accumulation of S1 NES cargoes, but it is conceivable that at low expression levels the S1 NES is able to efficiently compete for limiting amounts of CRM1, making its nuclear export relatively unaffected by Nup358 depletion.
Figure 7.
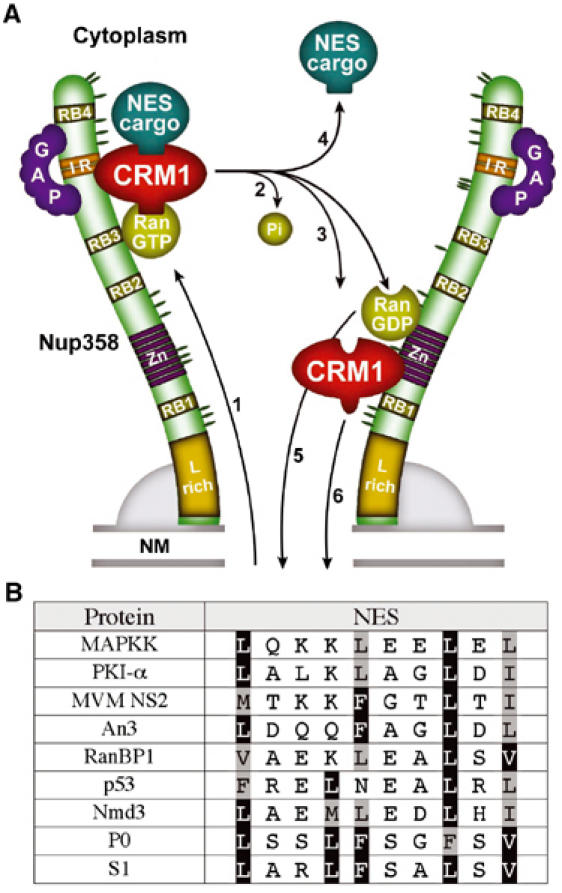
(A) Model of CRM1 export complex disassembly at the cytoplasmic face of the NPC. Nup358 is depicted as a filamentous protein (Delphin et al, 1997) with the different domains indicated. The orientation is suggested by immunoelectron microscopy studies (Walther et al, 2002) and the localisation of the ALK-Nup358 oncoprotein (Ma et al, 2003). (1) NES/CRM1/RanGTP complexes are translocated through the core NPC and bind to a cargo-dependent CRM1-binding site on Nup358; this binding may be cooperative with RanGTP/Ran-binding domain (RBD, RB1–4) interaction; (2) RanGTP hydrolysis stimulated by Nup358-bound RanGAP (Mahajan et al, 1997; Matunis et al, 1998) and the RBDs of Nup358 (Wu et al, 1995; Yokoyama et al, 1995; Vetter et al, 1999); (3) CRM1 can be released into the cytoplasm, as is the NES cargo protein (4) or bind to the LMB-insensitive CRM1-binding site on Nup358 that is located in the zinc-finger region (Singh et al, 1999; Bernad et al, 2004); likewise, RanGDP may bind to the zinc-finger RBD (Yaseen and Blobel, 1999); (5) RanGDP and (6) CRM1 recycle to the nucleus. NM, nuclear membranes; Zn, zinc-finger domains; L-rich, leucine-rich domain. (B) Natural NESs deviate at hydrophobic residues from highest affinity NES sequence. Alignment of the sequence of previously identified natural NESs from MAPKK (Fukuda et al, 1996), PKI (Wen et al, 1995), MVM NS2, Xenopus An3 (Askjaer et al, 1999), RanBP1 (Plafker and Macara, 2000), p53 (Stommel et al, 1999) and Nmd3 (Thomas and Kutay, 2003) and the artificial P0 and S1 NESs. Consensus hydrophobic residues are shaded, and hydrophobic residues identical to the S1 NES are boxed.
High-affinity NESs are ‘too good to be optimal'
We set out to identify signals exhibiting high-affinity interactions with CRM1 by screening a library of 15-mer peptide motifs in a phage display set-up. The number of representations for a random 15-mer peptide encompasses 3 × 1019 unique sequences. The complexity of the 15-mer peptide library employed was many orders of magnitude smaller at 2 × 108 unique sequences. The probability of retrieving a consensus NES, defined by four hydrophobic amino acids spaced in 3-2-1, 2-3-1 or 2-2-1 (without intervening hydrophobic amino acids), in our library is approximately 0.02. Therefore, roughly 4 × 106 different consensus NES sequences are expected in the library. Remarkably, under RanGTP selection conditions, which favoured export complex formation, a unique signal was enriched after just three rounds. This phage contained the highly active P0 NES that conforms to the consensus NES sequence. This indicates that the phage display selection conditions allowed us to enrich high-affinity NESs and that these are rarely encountered in the library. In the absence of RanGTP, a unique signal was selected, which displays a robust NES sequence of a striking similarity to the P0 signal. This experimental outcome suggests that CRM1 contains one major peptide-binding site, which corresponds to the NES-binding site.
We obtained a quantitative measure of CRM1 interaction by comparing the S1 NES to a z-tagged version of Nmd3 (Thomas and Kutay, 2003). This protein displays a high affinity for CRM1 comparable to that of snurportin 1 (see below). S1-GFP and 2z-Nmd3 possessed a similar affinity for CRM1 that was approximately 100-fold higher than standard NESs. Even though a short Rev-type NES has been proposed in human Nmd3 that is required for CRM1 interaction, this is unlikely to be sufficient for the high-affinity binding, as the untagged version of Nmd3 has a much lower affinity for CRM1 (see Thomas and Kutay, 2003 for discussion). Interestingly, CRM1 is less sensitive to LMB when bound to S1 as compared to standard NESs or 2z-Nmd3. LMB covalently binds to Cys528 of hCRM1 (Kudo et al, 1998; Neville and Rosbash, 1999), suggesting that access to Cys528 is masked by a tight NES interaction.
In vitro-measured affinities between CRM1 and NES cargoes are low in comparison to interactions of other exportins with their cargo (Kutay et al, 1997; 1998; Askjaer et al, 1999). Snurportin 1, a natural high-affinity cargo for CRM1, does not contain a short Rev-type NES but requires a large domain for CRM1 interaction (Paraskeva et al, 1999). This was taken to suggest that high-affinity CRM1 interaction could not be accomplished by small leucine-rich type NESs, and that CRM1 required a cofactor RanBP3 to boost NES–CRM1 affinity (Englmeier et al, 2001; Lindsay et al, 2001). In contrast, our data now demonstrate that high-affinity CRM1 binding can be accomplished by leucine-rich NESs, but is ineffective in vivo, because high-affinity NESs interact with CRM1 without RanGTP. As a consequence, illustrated by the S1 NES, export complexes accumulate at Nup358 and in the cytoplasm. This suggests that the large CRM1 interaction domain of snurportin 1 and perhaps Nmd3 are required for efficient release from CRM1. Sequence alignment of 58 published high-confidence NESs displays a high level of variation, even within the consensus hydrophobic amino acids (la Cour et al, 2003). As illustrated in Figure 7B, natural NESs only show a subset of the hydrophobic residues of the high-affinity NESs. When we replaced the consensus hydrophobic residues of a natural NES, derived from RanBP1 protein, into the high-affinity NES hydrophobic residues, the mutated version showed high-affinity behaviour, as it was targeted to the NPC. These data advocate that natural Rev-type NESs are selected to bind CRM1 but counterselected to bind with high affinity.
In conclusion, selection and analysis of high-affinity NES sequences provide clues to understanding the low-affinity nature and complexity of natural NESs. The highest affinity NESs are novel inhibitors of the CRM1 pathway and may be useful for structural characterisation of the CRM1/NES complex. The approach to select supraphysiological cargoes for nuclear transport receptors could be more widely applicable to study discrete steps of nucleocytoplasmic communication in vivo.
Materials and methods
Antibodies
Anti-GFP antibodies for immunofluorescence were from Abcam, and that for immunoelectron microscopy were from Roche. Antibodies to Nup358 (Walther et al, 2003), CRM1 (Fornerod et al, 1997b), Nup214 (Bernad et al, 2004) and Ran (Hetzer et al, 2000) were described previously.
Plasmid construction
For in vivo transport assays, phage inserts were provided with BglII and AgeI sites and inserted into the AgeI and BamHI sites of Rev(1.4)-GFP (Henderson and Eleftheriou, 2000). Second-generation mutations were introduced by PCR. For bacterial expression, three copies of GFP were placed in front of the NES, and introduced into the XmaI/PstI sites of pQE30 (Qiagen). pSuper-214, the shRNA expression plasmid to Nup214, targets nt 3828–3850 of the Nup214 ORF.
Recombinant protein expression and purification
Z-tagged CRM1 and transportin 1 were expressed as previously described (Görlich et al, 1997; Askjaer et al, 1999). Ran, RanBP1 and Rna1p were expressed and purified according to Izaurralde et al (1997). Ran was loaded with GTP according to the method described previously (Bischoff et al, 1994), and 2z-hNMD3 was a gift from U Kutay (Thomas and Kutay, 2003). NS2 peptide was described previously (Askjaer et al, 1999). GFP3-NESs, GFP-PKI (Siebrasse et al, 2002) and CRM1 (Englmeier et al, 1999) were purified on Ni-NTA agarose (Qiagen). CRM1 was further purified on a Resource Q column. zz-CRM1 and zz-transportin columns were prepared as published (Askjaer et al, 1999).
Phage display
The 15-mer phage library (kind gift from T Schumacher) represents a secondary amplification of a library initially created by Nishi et al (1993) using the filamentous phage vector fUSE5 (Scott and Smith, 1990). Zz-CRM1 columns were blocked for 2 h at 4°C in phage binding buffer (PBB; TBS, 0.01% Tween 20, 1 mM MgCl2) containing 1% BSA. After two washes with PBB, 5 μl of phage library containing 1 × 109 infectious phages was added to CRM1 columns, either in the presence or absence of 4.5 μM RanGTP in 50 μl PBB plus 0.1% BSA. Phages were bound for 2 h at 4°C. Columns were washed three times with PBB and eluted for 5 min at RT in 50 μl PBB containing 180 nM RanBP1 and 430 nM Rna1p or PBB alone. Selected phage pools were amplified by using Escherichia coli K91-Kan as previously described (Smith and Scott, 1993). After each selection round, 0.5–1 μl eluted phages and 1 μl of input phages were used for titration as described (Smith and Scott, 1993). After each selection round starting from the second round, phages were isolated and amplified as described. A 0.75 μl volume of phage suspension was directly used for sequencing, using primer 5′-TGAATTTTCTGTATGAGG.
CRM1 RanGAP assays
CRM1 RanGAP assays were performed as described (Askjaer et al, 1999). For LMB assays, increasing concentrations of LMB in 5 μl Ran buffer were mixed with 10 μl of 100 nM Rna1 and added to assembled complexes containing 1 μM CRM1, 200 pM Ran[γ-32P]GTP and either 480 nM GFP-S1, 380 nM 2z-hNMD3, 20 μM PKI-GFP or 5 μM NS2 present in 35 μl Ran buffer. Preincubation of LMB was performed by addition of LMB to 1 μM CRM1 and 200 pM Ran[γ-32P]GTP, 5 min prior to addition of 480 nM GFP3-S1. Assays testing RanBP1 sensitivity were performed by incubation of export complexes containing 360 nM CRM1 and concentrations as described for other GAP assays. Increasing concentrations of RanBP1 in 5 μl PBS/8.7% glycerol together with 10 μl 100 pM Rna1 in Ran buffer were used for Ran[γ-32P]GTP hydrolysis.
Pull-down assays
To detect recombinant CRM1/NES interaction, z-tagged CRM1 columns were incubated with 1 μM GFP-NES protein and 2.5 μM RanGTP when indicated. Binding reactions were performed in 50 mM HEPES–KOH (pH 7.9), 200 mM NaCl and 8.7% glycerol (buffer B) containing 0.1 mM DTT. After slowly shaking for 2 h at 4°C, beads were washed three times with buffer B and eluted for 10 min at RT with 50 μl buffer B. Samples were fractionated on SDS–polyacrylamide gels and visualised by Coomassie staining. To detect interaction with Nup358, 0.5 μmol of biotinylated S1 or Rev (GVPLQLPPLERLTLDC) NES peptide was immobilised on 5 μl streptavidin agarose beads (Sigma). NES beads were blocked for 1 h in 1% BSA and incubated for 3 h with 100 μl of Xenopus interphase egg extract diluted 1:1 with 10 mM HEPES (pH 7.4), 100 mM KOAc, 3 mM MgOAc, 5 mM EGTA, 150 mM sucrose and 1 mM DTT (acetate buffer). Beads were washed three times with acetate buffer and bound proteins were eluted in 0.2 and 2% SDS. Bound and unbound fractions were separated on 6% SDS–PAGE and blotted.
Cell culture and transfections
MCF-7 cells were transfected using electroporation as previously described (Agami and Bernards, 2000). HeLa cells were transfected by using Fugene-6 (Roche) according to the manufacturer's instructions. Both cell lines were transfected with 1 μg of pRev(1.4)-GFP plasmids on glass coverslips in 35 mm diameter dishes. Cells were fixed 24 h post-transfection. When required, 50 nM LMB was added 3 h prior to fixation. For RNAi assays, 1 μg of pSuper-358 (Bernad et al, 2004) or pSuper-214 was cotransfected and cells were cultured for 72 h before analysis.
Immunofluorescence stainings and image analysis
Indirect immunofluorescence was performed as previously described (Bernad et al, 2004). Images were recorded with a Leica TCS SP2 confocal microscope. For CRM1 localisation analysis, Image J software was used to measure the nuclear and cytoplasmic intensities of 15 cells. For measuring nuclear export as a function of GFP-NES expression level, total cellular and nuclear GFP signals were recorded using large pinhole confocal microscopy. To cover the complete range of expression, fields of cells were recorded with different PMT settings (250–550 V), and pixel values were combined using PMT to pixel value calibration curves.
Cryoimmunogold electron microscopy
Transfected MCF-7 cells were fixed, sectioned, immunolabelled and imaged as described (Calafat et al, 1997).
Acknowledgments
We thank Hans Janssen and Nico Ong for their expert technical assistance with electron microscopy, Ton Schumacher for the phage library and advice on phage display technology, Tassos Perrakis for assistance with protein purification, Ulrike Kutay for the generous gift of recombinant 2z-Nmd3, Josean Rodriguez and Beric Henderson for Rev(1.4)-GFP plasmid, Daniel Bilbao-Cortes and Iain Mattaj for anti-Ran antiserum, Reuven Agami for pSuper plasmids and advice, Lauran Oomen and Lenny Brocks for assistance with confocal microscopy, Tobias Walther, Marnix Jansen, Judith Boer and Helen Pickersgill for discussions and critically reading the manuscript and Marnix Jansen for suggesting the word supraphysiological. RB was supported by a grant from the Dutch Science foundation NWO-ALW.
References
- Agami R, Bernards R (2000) Distinct initiation and maintenance mechanisms cooperate to induce G1 cell cycle arrest in response to DNA damage. Cell 102: 55–66 [DOI] [PubMed] [Google Scholar]
- Askjaer P, Bachi A, Wilm M, Bischoff FR, Weeks DL, Ogniewski V, Ohno M, Niehrs C, Kjems J, Mattaj IW, Fornerod M (1999) RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol Cell Biol 19: 6276–6285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becskei A, Mattaj IW (2003) The strategy for coupling the RanGTP gradient to nuclear protein export. Proc Natl Acad Sci USA 100: 1717–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad R, Van Der Velde H, Fornerod M, Pickersgill H (2004) Nup358/RanBP2 attaches to the nuclear pore complex via association with Nup88 and Nup214/CAN and plays a supporting role in CRM1-mediated nuclear protein export. Mol Cell Biol 24: 2373–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff FR, Görlich D (1997) RanBP1 is crucial for the release of RanGTP from importin beta-related nuclear transport factors. FEBS Lett 419: 249–254 [DOI] [PubMed] [Google Scholar]
- Bischoff FR, Klebe C, Kretschmer J, Wittinghofer A, Ponstingl H (1994) RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc Natl Acad Sci USA 91: 2587–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Fridell RA, Benson RE, Hua J, Cullen BR (1996) Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol Cell Biol 16: 4207–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calafat J, Janssen H, Stahle-Backdahl M, Zuurbier AE, Knol EF, Egesten A (1997) Human monocytes and neutrophils store transforming growth factor-alpha in a subpopulation of cytoplasmic granules. Blood 90: 1255–1266 [PubMed] [Google Scholar]
- Delphin C, Guan T, Melchior F, Gerace L (1997) RanGTP targets p97 to RanBP2, a filamentous protein localized at the cytoplasmic periphery of the nuclear pore complex. Mol Biol Cell 8: 2379–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englmeier L, Fornerod M, Bischoff FR, Petosa C, Mattaj IW, Kutay U (2001) RanBP3 influences interactions between CRM1 and its nuclear protein export substrates. EMBO Rep 2: 926–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englmeier L, Olivo JC, Mattaj IW (1999) Receptor-mediated substrate translocation through the nuclear pore complex without nucleotide triphosphate hydrolysis. Curr Biol 9: 30–41 [DOI] [PubMed] [Google Scholar]
- Fischer U, Huber J, Boelens WC, Mattaj IW, Lührmann R (1995) The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82: 475–483 [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M (2002) Exportin-mediated nuclear export of proteins and ribonucleoproteins. Results Probl Cell Differ 35: 67–91 [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW (1997a) CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90: 1051–1060 [DOI] [PubMed] [Google Scholar]
- Fornerod M, van-Deursen J, van-Baal S, Reynolds A, Davis D, Murti KG, Fransen J, Grosveld G (1997b) The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J 16: 807–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E (1997) CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390: 308–311 [DOI] [PubMed] [Google Scholar]
- Fukuda M, Gotoh I, Gotoh Y, Nishida E (1996) Cytoplasmic localization of mitogen-activated protein kinase kinase directed by its NH2-terminal, leucine-rich short amino acid sequence, which acts as a nuclear export signal. J Biol Chem 271: 20024–20028 [DOI] [PubMed] [Google Scholar]
- Görlich D, Dabrowski M, Bischoff FR, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E (1997) A novel class of RanGTP binding proteins. J Cell Biol 138: 65–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Kutay U (1999) Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol 15: 607–660 [DOI] [PubMed] [Google Scholar]
- Görlich D, Pante N, Kutay U, Aebi U, Bischoff FR (1996) Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J 15: 5584–5594 [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Seewald MJ, Ribbeck K (2003) Characterization of Ran-driven cargo transport and the RanGTPase system by kinetic measurements and computer simulation. EMBO J 22: 1088–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson BR, Eleftheriou A (2000) A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp Cell Res 256: 213–224 [DOI] [PubMed] [Google Scholar]
- Hetzer M, Bilbao-Cortes D, Walther TC, Gruss OJ, Mattaj IW (2000) GTP hydrolysis by Ran is required for nuclear envelope assembly. Mol Cell 5: 1013–1024 [DOI] [PubMed] [Google Scholar]
- Izaurralde E, Kutay U, von Kobbe C, Mattaj IW, Görlich D (1997) The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J 16: 6535–6547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehlenbach RH, Dickmanns A, Kehlenbach A, Guan T, Gerace L (1999) A role for RanBP1 in the release of CRM1 from the nuclear pore complex in a terminal step of nuclear export. J Cell Biol 145: 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo N, Wolff B, Sekimoto T, Schreiner EP, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M (1998) Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res 242: 540–547 [DOI] [PubMed] [Google Scholar]
- Kutay U, Bischoff FR, Kostka S, Kraft R, Görlich D (1997) Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell 90: 1061–1071 [DOI] [PubMed] [Google Scholar]
- Kutay U, Lipowsky G, Izaurralde E, Bischoff FR, Schwarzmaier P, Hartmann E, Görlich D (1998) Identification of a tRNA-specific nuclear export receptor. Mol Cell 1: 359–369 [DOI] [PubMed] [Google Scholar]
- la Cour T, Gupta R, Rapacki K, Skriver K, Poulsen FM, Brunak S (2003) NESbase version 1.0: a database of nuclear export signals. Nucleic Acids Res 31: 393–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay ME, Holaska JM, Welch K, Paschal BM, Macara IG (2001) Ran-binding protein 3 is a cofactor for Crm1-mediated nuclear protein export. J Cell Biol 153: 1391–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Hill DA, Collins MH, Morris SW, Sumegi J, Zhou M, Zuppan C, Bridge JA (2003) Fusion of ALK to the Ran-binding protein 2 (RANBP2) gene in inflammatory myofibroblastic tumor. Genes Chromosomes Cancer 37: 98–105 [DOI] [PubMed] [Google Scholar]
- Mahajan R, Delphin C, Guan T, Gerace L, Melchior F (1997) A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88: 97–107 [DOI] [PubMed] [Google Scholar]
- Matunis MJ, Wu J, Blobel G (1998) SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J Cell Biol 140: 499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV, Weis K (1999) The direction of transport through the nuclear pore can be inverted. Proc Natl Acad Sci USA 96: 9622–9627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville M, Rosbash M (1999) The NES–Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J 18: 3746–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi T, Tsurui H, Saya H (1993) Construction and application of a phage random peptide library. Exp Med 11: 1759–1764, (in Japanese) [Google Scholar]
- Paraskeva E, Izaurralde E, Bischoff FR, Huber J, Kutay U, Hartmann E, Lührmann R, Görlich D (1999) CRM1-mediated recycling of snurportin 1 to the cytoplasm. J Cell Biol 145: 255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plafker K, Macara IG (2000) Facilitated nucleocytoplasmic shuttling of the ran binding protein RanBP1 [In Process Citation]. Mol Cell Biol 20: 3510–3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K, Görlich D (2001) Kinetic analysis of translocation through nuclear pore complexes. EMBO J 20: 1320–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD (2001) The nuclear pore complex as a transport machine. J Biol Chem 276: 16593–16596 [DOI] [PubMed] [Google Scholar]
- Scott JK, Smith GP (1990) Searching for peptide ligands with an epitope library. Science 249: 386–390 [DOI] [PubMed] [Google Scholar]
- Siebrasse JP, Coutavas E, Peters R (2002) Reconstitution of nuclear protein export in isolated nuclear envelopes. J Cell Biol 158: 849–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh BB, Patel HH, Roepman R, Schick D, Ferreira PA (1999) The zinc finger cluster domain of RanBP2 is a specific docking site for the nuclear export factor, exportin-1. J Biol Chem 274: 37370–37378 [DOI] [PubMed] [Google Scholar]
- Smith GP, Scott JK (1993) Libraries of peptides and proteins displayed on filamentous phage. Methods Enzymol 217: 228–257 [DOI] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K (1997) Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90: 1041–1050 [DOI] [PubMed] [Google Scholar]
- Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM (1999) A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J 18: 1660–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntharalingam M, Wente SR (2003) Peering through the pore: nuclear pore complex structure, assembly, and function. Dev Cell 4: 775–789 [DOI] [PubMed] [Google Scholar]
- Thomas F, Kutay U (2003) Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J Cell Sci 116: 2409–2419 [DOI] [PubMed] [Google Scholar]
- Vetter IR, Nowak C, Nishimoto T, Kuhlmann J, Wittinghofer A (1999) Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature 398: 39–46 [DOI] [PubMed] [Google Scholar]
- Walther TC, Askjaer P, Gentzel M, Habermann A, Griffiths G, Wilm M, Mattaj IW, Hetzer M (2003) RanGTP mediates nuclear pore complex assembly. Nature 424: 689–694 [DOI] [PubMed] [Google Scholar]
- Walther TC, Pickersgill HS, Cordes VC, Goldberg MW, Allen TD, Mattaj IW, Fornerod M (2002) The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J Cell Biol 158: 63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Meinkoth JL, Tsien RY, Taylor SS (1995) Identification of a signal for rapid export of proteins from the nucleus. Cell 82: 463–473 [DOI] [PubMed] [Google Scholar]
- Wu J, Matunis MJ, Kraemer D, Blobel G, Coutavas E (1995) Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J Biol Chem 270: 14209–14213 [DOI] [PubMed] [Google Scholar]
- Yaseen NR, Blobel G (1999) Two distinct classes of Ran-binding sites on the nucleoporin Nup-358. Proc Natl Acad Sci USA 96: 5516–5521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama N, Hayashi N, Seki T, Pante N, Ohba T, Nishii K, Kuma K, Hayashida T, Miyata T, Aebi U (1995) A giant nucleopore protein that binds Ran/TC4. Nature 376: 184–188 [DOI] [PubMed] [Google Scholar]
- Zhang MJ, Dayton AI (1998) Tolerance of diverse amino acid substitutions at conserved positions in the nuclear export signal (NES) of HIV-1 Rev. Biochem Biophys Res Commun 243: 113–116 [DOI] [PubMed] [Google Scholar]
- Zolotukhin AS, Felber BK (1997) Mutations in the nuclear export signal of human ran-binding protein RanBP1 block the Rev-mediated posttranscriptional regulation of human immunodeficiency virus type 1. J Biol Chem 272: 11356–11360 [DOI] [PubMed] [Google Scholar]


