Abstract
Urological malignancies (cancers of the prostate, bladder, kidney and testes) account for 15% of all human cancers and more than 500,000 deaths worldwide each year. This group of malignancies is spread across multiple generations, affecting the young (testicular) through middle and old-age (kidney, prostate and bladder). Like most human cancers, urological cancers are characterized by widespread epigenetic insult, causing changes in DNA hypermethylation and histone modifications leading to silencing of tumor suppressor genes and genomic instability. The inherent stability yet dynamic plasticity of the epigenome lends itself well to therapeutic manipulation. Epigenetic changes are amongst the earliest lesions to occur during carcinogenesis and are essentially reversible (unlike mutations). For this reason, much attention has been placed over the past two decades on deriving pharmacological compounds that can specifically target and reverse such epi-mutations, either halting cancer on its developmental trajectory or reverting fully formed cancers to a more clinically manageable state. This review discusses DNA methyltransferase and histone deacetylase inhibitors that have been extensively studied in preclinical models and clinical trials for advanced and metastatic urological cancers.
Keywords: Epigenome, Prostate cancer, Bladder cancer, Kidney cancer, Testicular cancer, Therapeutics
1. Introduction
Worldwide, urological malignancies account for ~15% of all non-cutaneous cancers and 12% of cancer related deaths. Urological malignancies include cancers of the prostate, bladder kidney and testes. Of all urological malignancies, prostate cancer is by far the most common with nearly one million men diagnosed annually and almost 300,000 deaths from this disease (Ferlay et al., 2010; Center et al., 2012) (Table 1, Fig. 2). Wide global variation exists for prostate cancer; incidence rates vary >25-fold worldwide, with ~75% of cases occurring in developed countries (Draisma et al., 2003). Epidemiological studies support a strong role for dietary constituents in modifying prostate cancer risk, with diets rich in lycopenes and cruciferous vegetables afforded a protective effect, while large intake of dairy, saturated fat and burnt meat is associated with increased risk. Age is the strongest risk factor for prostate cancer, with the majority of tumors occurring in men over the age of 60.
Table 1.
Summary of facts and figures for urological malignancies.
| Prostate | Bladder | Kidney | Testicular | |
|---|---|---|---|---|
| Number of new cases diagnosed per yeara | 899,102 | 382,660 | 273,518 | 52,322 |
| Number of deaths per yeara | 258,133 | 150,282 | 116,368 | 9,874 |
| Median age at diagnosis (years)b | 72 | 73 | 64 | 33 |
| Median age at metastasis (years)b | 82 | 75 | 69 | 33 |
| Major treatments for localized disease | Prostatectomy Radiotherapy | Cystectomy | Nephrectomy | Orchidectomy |
| Major side effects of treatments | Impotence, incontinence | Impotence, urinary diversion | Minimal | Minimal |
| Cure-rate for treated localized disease | 80% | Superficial=85% Muscle-invasive=50% |
70% | >99% |
| Major treatments for metastatic disease | Hormonal therapy, chemotherapy | Chemotherapy | Targeted meds | Chemotherapy |
| Mean survival for metastatic disease | 22 months | 14 months | 24 months | 23 years |
Obtained from GLOBOCAN 2008.
Median age according to the US Surveillance, Epidemiology and End Results (SEER) database.
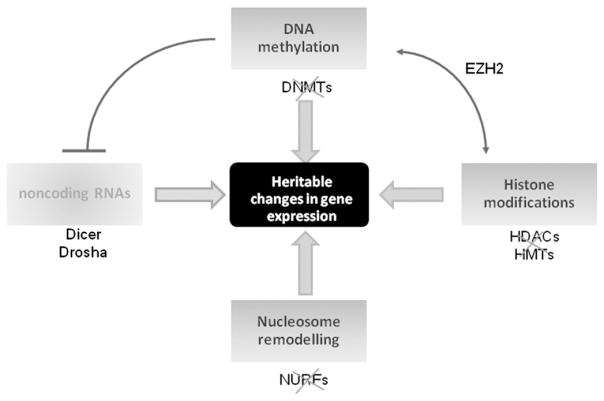
Fig. 2.
The co-operative network of epigenetic modifications. Crosstalk between DNA methylation, histone modifications, nucleosome remodeling and noncoding RNAs is the cornerstone for epigenetic gene regulation. Evidence now supports a role for small noncoding RNAs in epigenetic gene silencing; believed to be involved indirectly though post-transcriptionally inactivating key epigenetic enzymes. The microRNA class of noncoding RNAs have recently emerged as a target of DNA methylation in cancer cells.
Cancers of the kidney and bladder may develop in either sex but are significantly more common in men. Bladder cancer is the 2nd most common urological malignancy and the 9th most common malignancy worldwide, with ~330,000 new cases diagnosed and ~130,000 deaths per annum (Ferlay et al., 2010). Risk factors for the development of bladder cancer include age (50–70 years), molecular abnormalities (most notably TP63, EGFR, TP53 and RB1 genes and Ras and p21 proteins), chemical and environmental exposures (most notably cigarette smoking) and chronic irritation (Kaufman et al., 2009).
Tumors of the kidney are considerably less common than malignancies of the bladder and prostate, although the incidence has been slowly rising over the past 3 decades (Ferlay et al., 2010). Renal cell carcinoma (RCC) has the highest mortality rate among all the genitourinary tract tumors, with a third of patients dying from their disease. Risk factors for RCC include age (50–70 years), cigarette smoking, obesity, hypertension and silencing of the VHL tumor suppressor gene, which drives tumor angiogenesis through VEGF over-expression (Cohen & McGovern, 2005).
Testicular cancer is the least common of all the urological cancers, accounting for <2% of male cancers and with an estimated life-time risk of ~1 in 250. It is however, the most common solid cancer in Caucasian males aged between 15 and 35 years (Houldsworth et al., 2006; Rosen et al., 2011). Various risk factors have been identified, including a range of testicular abnormalities, however in the majority of cases there is no identifiable etiology. Genetic abnormalities including Klinefelter’s syndrome (47XXY), XY dysgenesis and Down’s syndrome (trisomy of chromosome 21) are all associated with increased risk (Horwich et al., 2006).
The pattern of disease presentation for each of these malignancies varies widely as does their natural history, clinical characteristics and prognosis. This review discusses the evidence to date for the potential role of epigenetic modalities in novel therapeutic strategies for urological cancers.
1.1. Prostate cancer therapies
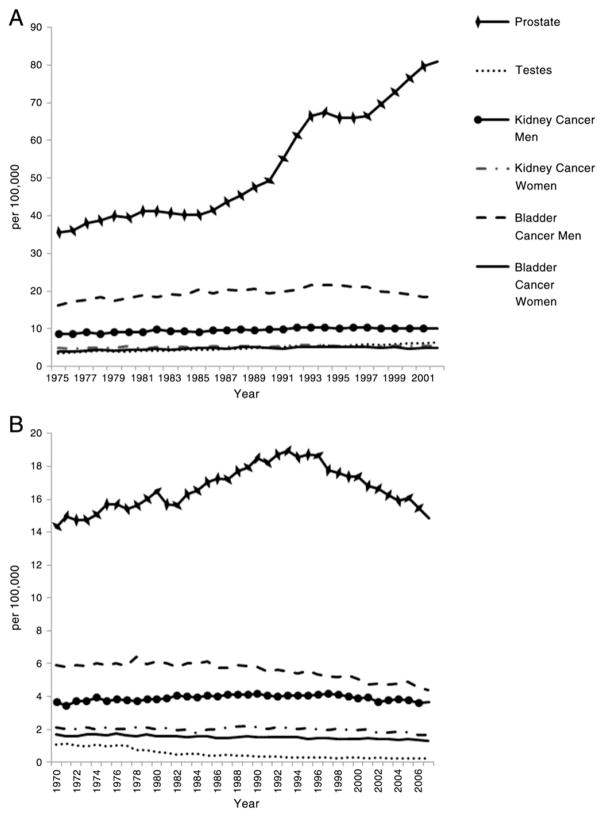
The last two decades have seen a dramatic rise in the incidence of prostate cancer, largely due to widespread serum prostate specific antigen (PSA) testing in developed countries (Fig. 1A). Prostate cancer is an extremely heterogeneous condition ranging from microscopic, well-differentiated tumors that may be clinically indolent to aggressive cancers that have a high likelihood of invasion and metastasis. The greatest challenge arising from PSA-detected prostate cancers is the substantial over-detection and hence over-treatment of clinically indolent disease. Nevertheless, the European Randomized Study of Screening for Prostate Cancer has shown a reduction in prostate cancer specific mortality due to PSA screening after 11 years of follow-up (Schroder et al., 2012).
Fig. 1.
Global facts and figures for urological cancers. A) World age-standardized incidence and B) world age-standardized mortality. Data are shown for USA, Canada, Denmark, Finland, France, Norway, Sweden, Netherlands, UK and Australia and were sourced from the WHO International Agency for Research on cancer CI5plus Cancer Incidence in Five Continents Annual Dataset (http://ci5.iarc.fr/CI5plus/ci5plus.htm) and Cancer Mortality database (http://www-dep.iarc.fr/WHOdb/WHOdb.htm).
Treatment options for localized prostate cancer are potentially curative by surgery (open, laparoscopic or robotic assisted radical prostatectomy) or radiation therapy (external-beam or brachytherapy) and have comparable long term survival rates. Conservative management through active surveillance or watchful waiting is now offered to patients who meet certain criteria for low-risk disease (Heidenreich et al., 2011; Tosoian et al., 2011). In recent years, advances have been made in minimally-invasive therapies, offering similar curative treatment rates but without the side-effects of conventional invasive procedures. Imaging techniques can provide precise tumor localization, increasing interest in “focal” therapies selectively targeting cancer foci, such as high-intensity focused ultrasound (HIFU), cryotherapy and photodynamic therapy (Eggener et al., 2010; Cordeiro et al., 2012).
Advanced prostate cancer is managed by androgen-deprivation therapy. However the therapeutic response is short-lived and the disease recurs typically within 18–24 months in a castration-resistant form. Docetaxel-based chemotherapy plus prednisone is the standard of care for metastatic castration-resistant prostate cancer (CRPC), giving a median survival benefit of ~3 months (Petrylak et al., 2004; Tannock et al., 2004). In recent years, several new agents have been developed targeting different mechanistic disease pathways fundamentally altering the treatment landscape in CRPC. These treatment options include a new cytotoxic agent (Cabazitaxel), immunotherapy (sipuleucel-T) and androgen receptor-signaling inhibitors (abiraterone acetate and MDV3100) (Sartor & Fitzpatrick, 2012). These advances highlight the significant role that targeted molecular based therapeutics can play in alleviating disease burden.
1.2. Bladder cancer therapies
More than 90% of bladder cancers are transitional cell carcinomas (TCC), 5% are squamous cell carcinomas and are largely associated with chronic urinary infection with Schistosoma haematobium, and less than 2% are adenocarcinomas (Table 2) (Kaufman et al., 2009). Of newly diagnosed TCC, ~70% are non-invasive superficial tumors that are potentially curative by local endoscopic resection and adjuvant chemotherapy. However, up to 70% of noninvasive TCC recur and between 10 and 20% of these become muscle-invasive. Muscle-invasive bladder cancer is treated by radical cystectomy to remove the entire bladder and its associated draining lymph nodes; a procedure associated with significant morbidity and a 5-year survival rate of ~50% (Stein & Skinner, 2006). The use of neo-adjuvant cisplatin-containing combination chemotherapy has been shown to modestly improve survival to 56% (Winquist et al., 2004). Alternatively, radical radiotherapy can be administered to surgically unfit candidates however outcomes are less favorable than with surgery.
Table 2.
Classification of urological malignancies at diagnosis.
| Tumor type | 5-year survival according to pathological stage at diagnosis | |
|---|---|---|
| Prostate | Epithelial (>95%) | Organ confined prostate cancer (T1–T2): 100% |
| Adenocarcinoma (95%) Urothelial Squamous Basal cell Other (<5%) |
Locally advanced prostate cancer (T3–T4): >95% Metastatic prostate cancer (N+/M+): 29% |
|
| Bladder | Epithelial (>95%) | Superficial bladder cancer & carcinoma in situ (Ta/Tis/T1): >98% |
| Transitional cell carcinoma (>90%) Squamous cell carcinoma (1–7%) Adenocarcinoma (2–3%) Other (<5%) |
Muscle-invasive bladder cancer (T2–T4): 63%–88% Metastatic bladder cancer (N+/M+): 15%–46% |
|
| Kidney | Renal cell carcinoma (>95%) | Organ confined renal cell carcinoma (T1–T2): 81% |
| Clear cell renal carcinoma (75–80%) Papillary renal cell carcinoma (10–15%) Chromophobe renal cell carcinoma (5%) Other (<5%) |
Locally advanced renal cell carcinoma (T3–T4): 53%–74% Metastatic renal cell carcinoma (N+/M+): 8% |
|
| Testicular | Germ cell tumors (90%) | Testicular cancer confined to testis and epididymis (T1–T2): 99% |
| Seminoma (48%) Non-seminomatous (42%) Mixed NSGCT (10%) Sex cord stromal tumors (3%) Other (7%) |
Locally advanced testicular cancer (T3–T4): 96% Metastatic testicular cancer (N+/M+): 72% |
Abbreviations: is=in situ and NSGCT=non seminomatous germ cell tumor.
Survival data obtained from American and European Associations of Urology guidelines
It is clear that given the limitations of current clinical treatments both in preventing recurrence of superficial bladder cancer and ensuring survival from muscle-invasive disease, novel therapeutic targets and agents are urgently required. Targeted molecular therapies for localized bladder cancer have been relatively slow in forthcoming. Overexpression of ERBB2 has been found to be an independent predictor of survival in a proportion of muscle-invasive TCC (Kruger et al., 2002). This resulted in phase I and II clinical trials investigating paclitaxel and daily radiation therapy with trastuzumab (anti-ERBB2 directed therapy) for patients with muscle-invasive TCC that over-expressed ERBB2, and who are not fit for cystectomy (Hussain et al., 2007).
1.3. Kidney cancer therapies
RCC is the most common form of kidney cancer and represents a spectrum of histological subtypes, signifying varied biology, genetics and behavior (Table 1) (Cohen & McGovern, 2005). Due to the paucity of early clinical manifestations, detection of RCC is often delayed, with one-third of patients diagnosed with advanced or metastatic disease (Castellano et al., 2012). However, earlier and incidental detection due to abdominal imaging for other medical indications has facilitated identification of tumors at more treatable and less advanced stages.
Historically, treatment has been predominately surgical, regardless of stage; RCC are inherently resistant to radiotherapy and chemotherapy. Increased understanding of the pathobiology of RCC has led to the development of targeted immunotherapies, which have revolutionized the treatment of metastatic RCC. Targeted agents, such as the oral, multi-targeted receptor tyrosine kinase inhibitors sunitinib and sorafenib, the mammalian target of rapamycin (mTOR) inhibitors temsirolimus and everolimus (Motzer et al., 2008) and the VEGF-targeted monoclonal antibody bevacizumab (given in combination with interferon-α) and pazopanib (Sternberg et al., 2010) are all well established treatments (Escudier et al., 2007a, 2007b; Rini et al., 2008; Motzer & Molina, 2009; Castellano et al., 2012).
1.4. Testicular cancer therapies
Testicular neoplasms denote an incongruent collection of tumors, which are classified according to their histological patterns (Table 2). The vast majority are classified as germ-cell tumors, derived from transformation of primordial germ cells (Houldsworth et al., 2006). Advances in the treatment of testicular cancer over the past number of decades, particularly in chemotherapeutic agents, have ensured that this disease has one of the highest cure rates of all cancers. The estimated 5 year mortality rate in the United States is 4%, ranking the multimodal management of testicular cancer the gold-standard model for the treatment of all solid tumors (Horwich et al., 2006).
Testicular tumors often progress silently until presentation with a painless or slightly aching scrotal lump. Serum tumor markers including alpha-fetoprotein (AFP), human chorionic gonadotrophin (hCG), and lactate dehydrogenase (LDH) are used for prognostication and treatment monitoring (Albers et al., 2011). The mainstay of treatment for testicular cancer is radical inguinal orchidectomy followed by adjuvant surveillance, adjuvant chemotherapy and adjuvant radiotherapy, depending on tumor histology and stage (Krege et al., 2008; Tandstad et al., 2011).
2. Epigenetic modalities
Following the conclusive unveiling of the human genome in 2001, revealing a surprisingly low constitution of ~25,000 genes, it became clear that genetics alone was not capable of explaining the vast complexity displayed by eukaryotes. Epigenetics, defined as heritable alterations in gene expression that are not accompanied by changes in our genetic code, is recognized as a key regulator of our DNA — controlling its packaging and transcription to RNA, and as a surrogate mechanism to mutation in yielding phenotypic diversity (Fig. 2). The crux upon which this epigenetic regulatory system is based lies with the manner in which DNA is compacted within the nucleus. Epigenetic “readers, writers and erasers” catalyze a variety of biochemical modifications to either the DNA (methylation) or the histone proteins (acetylation, methylation, ubiquitylation, sumoylation, phosphorylation, citrullination, ADP-ribosylation), to establish co-operative regulation of chromatin conformation (Dawson et al., 2012). The dynamic plasticity that epigenetic modifications display is paramount to enabling the complex temporal and spatial alterations in gene expression that are required for normal eukaryotic development and adult homeostasis (Gu et al., 2010).
2.1. DNA methylation and the methyltransferase family
The best characterized epigenetic mark — DNA methylation involves addition of a methyl group from the universal methyl donor S-adenosylmethionine to a nucleotide residue, which in mammals is primarily the fifth carbon position of cytosine in cytosine–phosphate–guanine (CpG) dinucleotides, converting cytosine to 5-methylcytosine or the ‘fifth base of the genome’ (Bird, 1986). However, recent studies of human embryonic stem cells have shown methylation to exist in certain restricted non-CpG contexts (Lister et al., 2009).
DNA methylation is typically associated with transcriptional silencing. This is achieved directly (by prohibiting transcription factor binding) (Mancini et al., 1999) and indirectly (by attracting methyl-binding domain proteins that interact with histone deacetylases and nucleosome remodeling factors to compact the chromatin, thus rendering the promoter inaccessible for transcription) (Jones et al., 1998; Nan et al., 1998). As such, intergenic regions and repeat elements are heavily methylated (Eden et al., 2003; Roman-Gomez et al., 2005), whereas actively transcribed genes are characterized by unmethylated promoters. De novo hypermethylation of promoter CpG islands (short 200–2000 bp stretches of DNA, heavily enriched in CpG sites) is a hallmark of most human cancers, acting as a transcriptional gene silencing mechanism (Baylin, 2005).
Humans contain three catalytically-active DNA methyltransferase enzymes (DNMT1, DNMT3a, DNMT3b) which regulate DNA methylation, in addition to an associated regulatory protein (DNMT3L) (Denis et al., 2011). DNMT1 is essential for maintaining patterns of methylation over replicative time by copying the pattern of the parent strand onto the new strand during DNA replication (Song et al., 2012). DNMT3a and DNMT3b possess the capacity to target unmethylated CpG sites and therefore function in establishing de novo methylation patterns (Rhee et al., 2002; Chedin, 2011). DNMT3L does not possess innate catalytic capabilities but does play an important role in orchestrating DNA methylation by regulating activity of the other DNMT3 members (Jurkowska et al., 2011; Van Emburgh & Robertson, 2011). Related family member DNMT2 shows poor in vitro DNA methyltransferase activity acting instead on RNA methylation (Schaefer et al., 2010; Thiagarajan et al., 2011).
DNA methylation patterns become altered during the very early transition from a normal cell phenotype to the pre-neoplastic state, with promoter hypermethylation of tumor suppressor genes (Brooks et al., 1998; Kang et al., 2004; Christoph et al., 2006b, 2007). This is followed by hypomethylation of intergenic sequences (Aporntewan et al., 2011), retroviral oncogenes (Goering et al., 2011), proto-oncogene promoters (Wang et al., 2007) and pericentromeric regions (Narayan et al., 1998), all facilitating epigenetic instability. Overall, there is significant reduction in the 5-methylcytosine content of the genome.
There is strong evidence to show that methylation patterns are clonally inherited during prostate tumor progression (Yegnasubramanian et al., 2004). However, recent advances in whole-genome methylation analysis have identified distinct subsets of genes that become hypermethylated in late stage prostate cancer and during metastatic progression, during which epigenetic changes become markedly more heterogeneous (Kim et al., 2011; Mahapatra et al., 2012). LINE1 hypomethylation is significantly enhanced in metastatic prostate cancer (Yegnasubramanian et al., 2008) and it has recently been shown that ETS-negative prostate cancers show reduced LINE1 methylation compared to ETS-positive tumors (Kim et al., 2011). It also appears that non-invasive and invasive bladder cancers may arise via distinct epigenetic pathways, in which non-invasive tumors display a distinct hypomethylated phenotype whereas invasive tumors exhibit widespread promoter hypermethylation (Wolff et al., 2010).
2.2. Histone modifications
Histone acetylation is the best studied post-translational modification of the histone octamer. Histone acetyltransferases catalyze the addition of an acetyl group from acetyl coenzyme A to a lysine typically at the histone N-terminus, which negates the positive histone charge, resulting in chromatin expansion and facilitating transcription (Marmorstein, 2001). This process is counter-balanced by the action of the histone deacetylase (HDAC) family. In removing the acetyl group, the HDACs restore the positive charge of the histone proteins, promoting chromatin compaction and transcriptional repression (Ng & Bird, 2000). Hence, acetylation is a core modulator of regulating the transition between eu- and heterochromatin.
Histone methylation is less well characterized and can have both activating (e.g. H3K4me2, H3k36me2) and repressing (e.g. H3k27me2/3 and H3K9me2/3) effects on gene expression, depending on the amino acid position and the number of methylated residues (Klose & Zhang, 2007; Crea et al., 2012b). Histone methylation is catalyzed by the histone methyltransferase (HMT) family of enzymes, with lysine undergoing mono-, di- or tri-methylation and arginine becoming di-methylated either symmetrically or non-symmetrically (Bedford & Richard, 2005; Martin & Zhang, 2005). While acetylation negates the positive histone charge, methylation alters the basicity and hydrophobicity of histones, changing the nucleosome’s affinity for a multitude of transcription factors.
The histone lysine demethylase (KDM) class of epigenetic enzymes removes both activating and repressive methyl groups from specific histone lysine residues causing chromatin reconfiguration. Of particular interest in this review are lysine-specific demethylase 1 (LSD1 or KDM1A) and members of the Jumonji domain containing family (JMJD2A, JMJD2C/GASC1, JMJD2D) that also function as p53 and nuclear receptor co-regulators to cooperatively mediate sex steroid hormone-induced transcription through their KDM activity (Metzger et al., 2005; Huang et al., 2007; Perillo et al., 2008).
During tumorigenesis, the histone methylome has been reported to undergo insult. Aberrant HMT activity has been implicated in survival and proliferation and epithelial–mesenchymal transition in prostate cancer (Ezponda et al., 2012), in bladder cancer (Kottakis et al., 2011) and in RCC (Duns et al., 2010). KDM expression is also dysregulated in human cancers, whereby KDMs may act as either tumor suppressors or oncogenes, depending on their gene regulatory function. For example, aberrant expression of LSD1 is reported throughout different stages of prostate and bladder cancer (Kauffman et al., 2011; Crea et al., 2012b). As a component of co-repressor complexes, LSD1 demethylates activating methyl marks at H3K4. In the presence of hormone-activated AR however, the LSD1–JMJD2 complex cooperate to remove the repressive H3K9 tri-methylation mark, thus facilitating AR-dependent gene expression and prostate tumor cell proliferation (Metzger et al., 2005; Wissmann et al., 2007).
2.3. On the road — epimutations as drivers of cancer
In spite of widespread heterogeneity between tumor types, it is believed that all cancers strive towards the acquisition of eight key traits: self-sufficiency in growth signals, resistance to anti-proliferative signals, apoptotic evasion, limitless replicative potential, angiogenesis, metastasis and, most recently, bioenergetic reprogramming and immune evasion (Hanahan & Weinberg, 2011). Add to this the ‘epigenetic progenitor model of cancer’(Feinberg et al., 2006), and we can begin to understand how epigenetic alterations in precursor cells can create genomic instability, which fosters additional genetic ‘hits’, such as point mutations and deletions, unwanted transcription of proviral and retrotransposon repeats, leading to disruption of their surrounding genes, further epigenetic catastrophe and rapid acquisition of the cancerous phenotype (Eden et al., 2003; Chalitchagorn et al., 2004). Virtually all tiers of the epigenetic hierarchy undergo subversion during tumor evolution.
3. DNA methylation inhibitors (DNMTi)
Endogenously, DNA methylation provides a means to rapidly and stably alter eukaryotic gene expression in response to stimuli. DNA methylation can be considered as an attractive therapeutic target because it is reversible and relatively simple to manipulate biochemically. By globally inducing hypomethylation, genes which have become aberrantly silenced, in particular tumor suppressors, may be reactivated. To date, numerous DNA methylation inhibitors (DNMTi) have been developed with varying pre-clinical results and the most promising clinical translatability for hematological malignancies with FDA approval granted for patients with myelodisplatic syndrome (Tsai et al., 2012). Notably, the efficacy of hypomethylating agents is dependent on the tumor genotype (Pulukuri & Rao, 2005; Richter et al., 2007).
3.1. Re-achieving the healthy genome with nucleoside analogs
The first class of DNMTi comprises simple nucleoside analogs: azacytidine (5-Aza-CR; trade name Vidaza), a simple cytidine derivative and 5-aza-2′-deoxycytidine (5-Aza-CdR; decitabine; trade name Dacogen), a deoxycytidine derivative. Via active demethylation of tumor suppressor genes and cytotoxicity arising from incorporation into nucleic acids, these DNMTi exert potent anti-tumorigenic activity (Jones & Liang, 2009). In vitro evidence suggests that DNMTi may also passively induce gradual demethylation by competitively binding DNMTs over replicative time (Kuo et al., 2007). A significant drawback of these agents, however, is their relative toxicity and limited potency at tolerable levels. Therefore, efforts have been made to develop novel azacytidine derivatives (Matousova et al., 2011). Zebularine, although initially developed in 1961, has recently attracted attention as a ribonucleoside DNMTi (Champion et al., 2010). It exhibits greater intrinsic stability and lower toxicity than its counterparts, thus enabling longer oral administration, facilitating maintenance of a demethylated state (Billam et al., 2010).
Multiple preclinical studies of nucleoside analogs have validated their potential use in restoring expression of a myriad of genes in in vitro models of prostate, bladder, renal and testicular cancer at a consistent rate over time (Christoph et al., 2006a). Over a decade ago, treatment of the androgen-insensitive DU145 cell line with 5,6-dihydro-5-azacytidine was shown to restore androgen receptor (AR) reactivation (Nakayama et al., 2000) as well as responsiveness to dihydrotestosterone (DTT) (Izbicka et al., 1999). Treatment with histone deacetylase inhibitor (HDACi) trichostatin A (TSA) invariably shows substantial synergistic gene reactivation in vitro when used in combination with a DNMTi (Cameron et al., 1999). In renal cancer, TGFβ receptor type II expression, in addition to TGFβ responsiveness, was restored by 5-Aza-CdR (Zhang et al., 2005). Furthermore, combined treatment with 5-Aza-CdR and genistein, a non-toxic isoflavone, significantly induced cell cycle arrest and halted proliferation across three renal carcinoma cell lines, over genistein alone (Majid et al., 2009). More recently, it was shown in T24 bladder cancer cell line that 5-Aza-CdR is capable of concomitant reactivation of host mRNA and intragenic intronic microRNA expression (Saito et al., 2009). Nucleoside analogs have also been revealed as capable of potentially targeting epithelial-to-mesenchymal transition and meta-static spread in bladder cancer with 5-Aza-CdR restoring prostasin expression in cell lines, with concomitant reactivation of E-cadherin and recovery of epithelial morphology (Chen et al., 2009).
Bladder cancer was the first malignancy to show epigenetic reactivation of a tumor suppressor gene after oral delivery of a hypomethylating agent. Following promising results by demethylating the p16 gene promoter and globally decreasing hypermethylated regions in T24 human bladder carcinoma cells, studies on zebularine were escalated to in vivo models. Using the xenograft murine model of EJ6 human bladder carcinoma culture in nude mice, zebularine was shown clinically efficient when delivered either intravenously or orally, producing an 11% average maximum weight change between test cases and controls at 1000 mg/kg dosing (Cheng et al., 2003, 2004).
3.2. Restoration of radio- and chemosensitivity
A promising avenue for the clinical utility of hypomethylating agents is their potential to promote radio- and/or chemosensitivity through reactivation of specific enzymes involved in drug metabolism (Shen et al., 2007; Zhang et al., 2010). Combined treatment of prostate cancer cell line DU145 with docetaxel (TaxotereR) and either 5-Aza-CdR or 5-Aza-CR enhanced docetaxel sensitivity through demethylation and reactivation of DNA damage response gene GADD45A (Ramachandran et al., 2009) and tumor suppressors E-cadherin and maspin and significantly reduced proliferation over either agent alone (Hurtubise & Momparler, 2004). Similarly, 5-Aza-CdR was found to enhance the apoptotic effect of cisplatinum (trade name: cisplatin) in DU145 cells (Fang et al., 2004). In RCC, cytotoxicity of vinblastine was increased both in vitro and in a Caki-1 xenograft model by co-treatment with 5-Aza-CdR (Iwata et al., 2011). Hypermethylation of the pro-apoptotic XAF1 gene has been associated with the apoptotic resistant phenotype of RCC that emerges post-immunotherapy. Rescuing XAF1 expression by either 5-Aza-CdR or a DNMT antisense oligonucleotide has been shown to restore susceptibility to interferon-induced apoptosis (Reu et al., 2006).
Cisplatinum resistance poses a key problem in the treatment of bladder cancer. 5-Aza-CdR has been shown to sensitize TCC to the effects of cisplatin (Shang et al., 2008). Hypermethylation of homeobox gene TLX3 has been associated with chemoresistance in 21% of patients and overexpression of TLX3 restored chemosensitivity (Tada et al., 2011). Similarly, it has been shown that 5-Aza-C reactivation of proapoptotic gene TMS1 restores cisplatin sensitivity in the UMUC3 bladder cancer cell line (Ramachandran et al., 2011).
Testicular gem cell tumors are characterized by hypersensitivity to cisplatin and DNMTi, which probably mechanistically relates to their high endogenous levels of DNMT3b (Beyrouthy et al., 2009). In comparison to other solid tumors, very low nanomolar concentrations of 5-Aza-CdR were capable of recovering expression of 3 hypermethylated genes (RASSF1A, MGMT, HOXA9), and activating the p21 and ATM pathways, resulting in decreased proliferation and cell survival. This study went on to demonstrate that in testicular germ cell tumors that develop cisplatin resistance, sensitivity can be restored by 5-Aza-CdR (Beyrouthy et al., 2009).
3.3. Beyond the nucleoside: alternative DNMTi
Due to the inherent toxicity and adverse effects of nucleoside analogs, efforts are ongoing to establish synthetic or dietary non-nucleoside compounds that do not become integrated into the DNA yet can still effectively exert hypomethylating activity. Procainamide, an antiarrhythmic drug with observed in vitro demethylating capacity, was shown to be capable of hypomethylating the prostate cancer stalwart, GSTP1, in the LNCaP cell line as well as in xenograft athymic nude mouse models (Lin et al., 2001). Disulfiram, a thiol-reactive compound used in the treatment of chronic alcoholism, was proven to possess intrinsic DNMTi properties, globally decreasing the quantity of 5-methylcytosine and capable of reactivating APC and RARβ through inhibition of DNMT1 activity. Treatment also resulted in decreased proliferation and increased apoptosis in cell line and xenograft prostate cancer models (Lin et al., 2011). Curcumin (diferuloylmethane), a phytochemical compound found endogenously in turmeric curry, also appears to possess demethylating properties, restoring expression of transcription factor NEUROG1 in prostate cancer LNCaP cells through preventing MeCP2 binding to the NEUROG1 promoter (Shu et al., 2011). It was recently shown that curcumin is also capable of demethylating and reactivating noncoding microRNA genes, namely tumor suppressor microRNA miR-203 in bladder cancer cell lines. This resulted in down-regulation of miR-203 target genes Akt2 and Src and culminated in decreased proliferation and increased apoptosis (Saini et al., 2011).
3.4. DNMTi in clinical trials for urological cancers
While the full mechanistic action of many potential hypomethylating agents is still being actively investigated at the preclinical stage, several notable clinical trials have been completed for urological malignancies (Table 3). Overall the outcomes have not been particularly hopeful, with perhaps the most promising results seen in metastatic CRPC patients. As early as 1998, a clinical trial took place for 5-aza-2′-deoxycytidine on men who developed metastatic prostate cancer following complete androgen blockade and flutamide withdrawal. Of the 14 men recruited, 12 were evaluable with 2 cases displaying a time to progression greater than 10 weeks. Interestingly, both men were African-American, suggesting ethnicity may be an important variable for consideration (Thibault et al., 1998). More recently, a phase II clinical trial of Vidaza in CRPC patients (n=36) revealed a significant impact upon PSA doubling time (PSA-DT), the median PSA-DT of the treated group (2.8 months) far exceeded that of the control group (1.5 months) (Sonpavde et al., 2011).
Table 3.
Clinical trials of DNMTi for urological cancers.
| Drug | Clinical trial identifier | Phase | Status | Protocol | Outcome |
|---|---|---|---|---|---|
| Prostate | |||||
| Vidaza | NCT00384839 | Phase II | Completed | CRPC cases on combined androgen blockade with PSA-DT<3 months treated with 75 mg/m2 s.c. on days 1–5 of each 28 day cycle for up to 12 cycles where tolerable. n=36 | PSA-DT>3 months achieved in 55.8% patients; median progression free survival=12.4 weeks |
| Decitabine | – | Phase II | Completed | Metastatic cases post-total blockade and flutamide withdrawal were treated with 57 mg/m2/dose i.v. as 1 h infusion every 8 h for 3 doses; cycles repeated every 5–8 weeks. n=14 | 17% had stable disease with progression time>10 weeks |
| Vidaza, docetaxel and prednisone | NCT00503984 | Phase I/II | Recruiting | CRPC patients previously treated with docetaxel. Azacitidine: i.v., days 1–5 of each 3 weekly cycle; docetaxel: i.v. on day 6 of each 3 weekly cycle; prednisone: 5 mg twice daily from day 1 to 21 of each cycle. Predicted enrolment: n=42 | NYA |
| Bladder & testes | |||||
| Decitabine | NCT00030615 | Phase I | Completed | Advanced bladder and testicular cancer patients received daily escalating doses of decitabine for 4 weeks until MTT was determined. | NYA |
| Kidney | |||||
| Vidaza, bevacizumab | NCT00934440 | Phase I/II | Recruiting | Advanced RCC patients receiving: bevacizumab at standard dose of 10 mg/kg IV every two weeks; vidaza: several different doses s.c. dependent on phase I/II. Predicted enrolment: n=23 | NYA |
| Decitabine, IFNα-2B | NCT00561912 | Phase II | Terminated | Advanced RCC patients received decitabine: 15 mg/m2 i.v. daily for 5 days; IFNα-2B s.c. twice daily continuously as of day 1 cycle 3. | Terminated due to poor accrual (n=2) |
| MG98 | NCT00003890 | Phase I/II | Terminated | Untreated metastatic RCC patients received 360 mg/m2/dose i.v. twice weekly for 3 out of 4 weeks. n=17 | No conclusive pattern of decreased DNMT1 activity in PBMCs was detected post MG98 treatment. Terminated due to toxicity. |
Abbreviations: MTT: maximum tolerated dose, PBMCs: peripheral blood mononuclear cells, and NYA: not yet available.
In bladder, renal and testicular cancers, few clinical trials have been carried out on these agents. Several have been terminated due to toxicity or poor uptake (Winquist et al., 2006). More recent trials are studying the synergistic benefit of combined epigenetic and immunotherapies (Table 3).
4. Histone deacetylase inhibitors (HDACi)
Beyond the DNA level, post-translational modifications of histone proteins also function in modulating eukaryotic gene expression and therefore may be pharmacologically targeted. Among these, histone acetylation has received most attention for its capacity to neutralize the positive charge of histones, thereby facilitating euchromatin expansion and gene transcription. Hence, a multitude of HDAC inhibitors (HDACi) have been developed, targeting various members of the HDAC family of enzymes (Khan & La Thangue, 2012).
Due to their diversity, HDACi are broadly categorized into 5 groups based on chemical composition and structure: short-chain fatty acids, hydroxamic acids, cyclic tertrapeptides, benzamides and aliphatic acids. Alternatively, HDACi may be classified based on the member(s) of the HDAC enzyme family they target, with further disparity both between and within the five structural groups (Khan & La Thangue, 2012). Some act specifically on certain HDACs, for example, entinostat (MS-275) inhibits class I HDACs, whereas others such as trichostatin A, vorinostat and panobinostat can inhibit multiple classes of HDAC. Perhaps surprisingly, it has been shown that <10% of genes are influenced by HDACi directly (Peart et al., 2005). Therefore, it appears that HDAC inhibition serves as an initial instigator in restoring normal gene expression but that phenotypic effects are perpetuated by multiple downstream mechanisms. This notion is supported by the observation that HDACs interact with numerous non-histone targets, many of which are important for driving tumorigenesis (Johnstone & Licht, 2003), such as α-tubulin (Marcus et al., 2005), important for cell motility and metastasis, bcl-6 (Dhordain et al., 1998), an oncogene involved in apoptotic regulation, and p53 (Murphy et al., 1999). Although the exact mechanisms and pathways involved in HDACi remain unclear, several recurrent anti-tumorigenic phenotypic outcomes have been observed. HDACi mediate selective tumor-cell death through several pathways, including induction of apoptosis, growth arrest and autophagy, and effects on DNA repair, mitosis, and angiogenesis (Long et al., 2009; Khan & La Thangue, 2012). Pan-HDACi belinostat potently inhibited the growth of prostate cancer cell lines by increasing expression of p21 and inducing G2/M arrest and reduced tumor growth and metastatic spread in orthotopic prostate cancer tumor models (Qian et al., 2008). Similarly it was shown in RCC cell lines, that panobinostat (LBH589) induced degradation of both Aurora A and B kinases through a proteasome-mediated pathway by targeting HDAC3 and HDAC6, resulting in G2/M arrest and apoptosis (Cha et al., 2009).
Multiple lines of evidence support a role for combining pharmacological compounds to sensitize tumors to the effects of HDACi. Interferon alpha enhances the anti-tumor effects of HDACi valproic acid (VPA) on prostate cancer growth (Hudak et al., 2012). In bladder cancer, both cisplatin and gemcitabine have been demonstrated to potentiate the antitumor effects of trichostatin A (Jeon et al., 2011). Vorinostat was found to synergistically improve the anti-cancer activity of mTOR inhibitor temsirolimus in a panel of RCC cell lines in vitro and in two xenograft models in vivo. Combined treatment significantly reduced survivin expression leading to apoptosis and a reduction in angiogenesis (Mahalingam et al., 2010). Similarly in mouse models of prostate cancer, the combination of panobinostat with the mTOR inhibitor everolimus resulted in enhanced anti-tumor activity (over either agent alone) mediated by increased p21 and p27 expression and reduced angiogenesis and tumor proliferation via androgen receptor, c-Myc and HIF-1α signaling (Ellis et al., 2011).
4.1. HDACI in clinical trials for urological cancers
The main challenge in translating HDACi into cancer therapeutics is trying to establish which molecules will yield maximum efficacy while minimizing off-target toxicity; difficult due to the pleiotropic nature of the enzymes involved. Several HDACi have entered clinical trials for the treatment of prostate and renal cancer, with mixed success (Table 4).
Table 4.
Clinical trials of HDACi for urological cancers.
| Drug | Clinical trial identifier | Phase | Status | Protocol | Outcome |
|---|---|---|---|---|---|
| Prostate | |||||
| Panobinostat, docetaxel and prednisone | NCT00663832 | Phase I | Completed | CRPC received panobinostat 10, 15 or 20 mg i.v. on days 1 and 8 in combination with docetaxel i.v. on day 1 and prednisone p.o. 5 mg bid every day of a 21-day cycle. n=44 | Determined the MTD of i.v. Panobinostat is 15 mg in combination with docetaxel and prednisone in patients with CRPC. 63% of patients showed >50% decline in PSA levels |
| Panobinostat, bicalutamide | NCT00878436 | Phase I/II | Recruiting | CRPC patients receiving panobinostat at either 60 or 120 mg per week for 2 consecutive weeks, with one week rest and bicalutamide 50 mg p.o. daily, continuously. Predicted enrolment: n=78 | Investigating safety and efficacy of combined treatment of panobinostat at 2 dose levels combined with bicalutamide for CRPC as measured by time to PSA progression and proportion of patients that achieve a ≥50% PSA decline by 9 months of therapy |
| Vorinostat | NCT00330161 | Phase II | Active, not recruiting | Metastatic CRPC patients (PSA>5 ng/ml, disease progression, adequate organ function) received 400 mg vorinostat orally each day. n=27 | 7% patients achieved an objective response rate; no PSA decline>50% observed; significant toxicities reported |
| Romidepsin | NCT00106418 | Phase II | Completed | Metastatic CRPC patients received romidepsin 13 mg/m2 i.v. over 4 h on days 1, 8 and 15 every 4 weeks. n=35 | 2 patients reached a confirmed radiological partial response of >6 months, in addition to >50% PSA decline |
| Kidney | |||||
| Panobinostat | NCT00550277 | Phase II | Completed | Metastatic RCC patients received 45 mg panobinostat twice per week for 8 week cycles with re-evaluation at the end of each cycle; n=20 | No objective response observed; generally well-tolerated with minimal side effects |
| Romidepsin | NCT00106613 | Phase II | Completed | Metastatic RCC patients received 13 mg/m2 romidepsin i.v. on days 1, 8 and 15 over 4 week cycles with disease progression analyzed every 8 weeks. n=29 | 7% experienced an objective response. Terminated due to toxicity. |
In 2009, Bradley and colleagues released the results of their phase II clinical trial of vorinostat (Suberoylanilide Hydroxamic Acid) for metastatic CRPC (Bradley et al., 2009). In this study, 27 patients with disease progression following chemotherapy were recruited. Overall survival, PSA decline rate and safety were amongst those listed as secondary endpoints. Unfortunately, only 7% patients displayed stable disease and no PSA decline of >=50% was noted. Furthermore, all participants ceased treatment before the 6 month interval due to associated toxicities. Hence, the toxicity of vorinostat at such doses is clearly not translatable to the clinic.
Molife et al. completed a phase II clinical trial in 2010, examining the potential therapeutic benefit of romidepsin (depsipeptide) in metastatic CRPC (Molife et al., 2010). Via treatment with this HDACi, it was hoped that AR signaling could be abrogated by inducing acetylation of heat shock protein 90. The primary endpoint was the absence of radiological progression at 6 months. Only two patients were reported as displaying radiological partial response (RECIST) lasting≥6 months in conjunction with PSA decline≥50%. Additionally, 11 patients had to be discontinued from trial due to adverse events. Hence, romidepsin does not appear of clinical use at such concentrations and toxicities. Results of a phase I combination trial of oral panobinostat alone in conjunction with docetaxel in 16 CRPC patients showed that docetaxel did not augment panobinostat efficacy at high dose (20 mg) but in the second arm of the study in patients receiving a lower dose of 15 mg panobinostat, 63% patients showed at least a 50% decline in PSA levels (Rathkopf et al., 2010).
Similarly, the potential to treat metastatic RCC with HDACi romidepsin and panobinostat has been disappointing showing either no response or very high levels of toxicity, respectively (Stadler et al., 2006; Hainsworth et al., 2011).
4.2. Inhibitors targeting the histone methylome
EZH2 belongs to the polycomb group of transcriptional repressors, and forms the catalytic subunit of the Polycomb repressive complex 2 (PRC2), which catalyzes trimethylation on H3K27, triggering gene silencing. It also acts as a molecular scaffold, attracting DNMTs thus bridging two distinct epigenetic modes of gene regulation: chromatin conformation and DNA methylation (Vire et al., 2006). EZH2 over-expression is associated with metastatic progression and poor prognosis in urological malignancies (Varambally et al., 2002; Raman et al., 2005; Wagener et al., 2008), although it does not appear to exert its characteristic oncogenic properties in testicular cancer (Hinz et al., 2010).
EZH2 inhibition may be employed as an anti-angiogenic and anti-metastatic strategy. One potential compound is 3-dezaneplanocin-A (DZNeP), a HMT inhibitor, which inhibits S-adenosyl-homocysteine, required for EZH2-dependent lysine methylation. DZNeP exerts anti-tumor activity in many cancer cell lines including prostate, where it also blocks cancer cell migration and invasion (Crea et al., 2012a). Several selective small molecule inhibitors of EZH2 have recently been described that decrease global H3K27me3 levels and reactivate silenced PRC2 target genes, such as GSK126 (McCabe et al., 2012) and EPZ005687 (Knutson et al., 2012).
The possibility of pharmacologically targeting KDMs has been demonstrated (Kruidenier et al., 2012). A range of KDM inhibitors (KDMi) have been identified (Rotili & Mai, 2011; Crea et al., 2012b). In prostate cancer, LSD1 remains the most promising target for KDMi, and several substrate analogs, polyamine analogs and inhibitors of the mono-amine-oxidase domain have been developed such as tranylcypromine and clorgyline. Tranylcypromine and its analogs effectively inhibit H3K4 demethylation and may inhibit progression to castration resistance (Crea et al., 2012b). Clorgyline exerts anti-proliferative and pro-differentiation activity on high-grade prostate cancer cells in part mediated by inhibiting growth factor signaling and down-regulating HMT EZH2 (Zhao et al., 2009). Namoline is a more specific LSD1 inhibitor which has been shown to reduce androgen-dependent cell proliferation and xenograft tumor growth (Willmann et al., 2012).
5. Future directions: remaining challenges
As we continue to advance our understanding of the molecular pathobiology of urological malignancies, we move towards an improved ability to predict which patients will progress to lethal disease. This in turn will enable patient-orientated individualized treatment and specific targeted therapies for patients with advanced disease.
Without doubt the greatest obstacle in translating epigenetic drugs to clinical use for solid tumors, not just urological malignancies, remains their genome-wide mode of action and thus lack of specificity and hence severe toxicity. Unless hypermethylation of individual tumor suppressor genes can be restored without the side-effects of whole genome hypomethylation, it is difficult to see how DNMTi and HDACi will significantly advance the treatment of solid tumors in their current form. Although HDACs exhibit selective toxicity against tumor cells at nanomolar concentration, their prolonged use is associated with significant toxicity (Prince et al., 2009). A number of plant-derived polyphenols have cancer preventative properties, display structural features compatible with HDAC inhibition and may be more tolerable. For example, epigallocatechin-3-gallate (the polyphenol from green tea) has been shown to cause cell cycle arrest and induce apoptosis in prostate cancer cells by upregulating p21 and Bax through H3 acetylation (increasing promoter accessibility), causing epigenetic modifications and down-regulating HDACs through increasing their proteasomal degradation (Thakur et al., 2012).
While acetylation remains the best studied histone modification to date, it in itself is only one component of the elusive histone code. Multiple other post-translational modifications have been described and are implicated in various cancers and essentially remain largely understudied. Unlike DNMTs and HDACs, for which inhibitors have been developed and discovered, the quest to establish histone methyltransferase inhibitors lies ahead. Whether these agents can be engineered with increased specificity and hence lower toxicity, remains to be seen.
Abbreviations
- CRPC
castration-resistant prostate cancer
- DNMT
DNA methyltransferase
- DNMTi
DNA methyltransferase inhibitors
- HDAC
histone deacetylase
- HDACi
histone deacetylation inhibitors
- PSA
prostate specific antigen
- RCC
renal cell carcinoma
- TCC
transitional cell carcinoma
Footnotes
Funding source: We gratefully acknowledge funding from The Prostate Cancer Foundation, The Irish Cancer Society, The Health Research Board and the National Institute on Minority Health And Health Disparities of the National Institutes of Health under award number T37MD001452. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors confirm that this manuscript has not been published and is not under consideration for publication elsewhere.
Conflict of interest statement
The authors declare no conflict of interest.
References
- Albers P, Albrecht W, Algaba F, Bokemeyer C, Cohn-Cedermark G, Fizazi K, et al. EAU guidelines on testicular cancer: 2011 update. Eur Urol. 2011;60:304–319. doi: 10.1016/j.eururo.2011.05.038. [DOI] [PubMed] [Google Scholar]
- Aporntewan C, Phokaew C, Piriyapongsa J, Ngamphiw C, Ittiwut C, Tongsima S, et al. Hypomethylation of intragenic LINE-1 represses transcription in cancer cells through AGO2. PLoS One. 2011;6:e17934. doi: 10.1371/journal.pone.0017934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2(Suppl 1):S4–S11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Beyrouthy MJ, Garner KM, Hever MP, Freemantle SJ, Eastman A, Dmitrovsky E, et al. High DNA methyltransferase 3B expression mediates 5-aza-deoxycytidine hypersensitivity in testicular germ cell tumors. Cancer Res. 2009;69:9360–9366. doi: 10.1158/0008-5472.CAN-09-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billam M, Sobolewski MD, Davidson NE. Effects of a novel DNA methyltransferase inhibitor zebularine on human breast cancer cells. Breast Cancer Res Treat. 2010;120:581–592. doi: 10.1007/s10549-009-0420-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Bradley D, Rathkopf D, Dunn R, Stadler WM, Liu G, Smith DC, et al. Vorinostat in advanced prostate cancer patients progressing on prior chemotherapy (National Cancer Institute Trial 6862): trial results and interleukin-6 analysis: a study by the Department of Defense Prostate Cancer Clinical Trial Consortium and University of Chicago Phase 2 Consortium. Cancer. 2009;115:5541–5549. doi: 10.1002/cncr.24597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks JD, Weinstein M, Lin X, Sun Y, Pin SS, Bova GS, et al. CG island methylation changes near the GSTP1 gene in prostatic intraepithelial neoplasia. Cancer Epidemiol Biomarkers Prev. 1998;7:531–536. [PubMed] [Google Scholar]
- Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- Castellano D, Ravaud A, Schmidinger M, De Velasco G, Vazquez F. Therapy management with sunitinib in patients with metastatic renal cell carcinoma: key concepts and the impact of clinical biomarkers. Cancer Treat Rev. 2012 doi: 10.1016/j.ctrv.2012.04.009. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Center MM, Jemal A, Lortet-Tieulent J, Ward E, Ferlay J, Brawley O, et al. International variation in prostate cancer incidence and mortality rates. Eur Urol. 2012;61:1079–1092. doi: 10.1016/j.eururo.2012.02.054. [DOI] [PubMed] [Google Scholar]
- Cha TL, Chuang MJ, Wu ST, Sun GH, Chang SY, Yu DS, et al. Dual degradation of aurora A and B kinases by the histone deacetylase inhibitor LBH589 induces G2-M arrest and apoptosis of renal cancer cells. Clin Cancer Res. 2009;15:840–850. doi: 10.1158/1078-0432.CCR-08-1918. [DOI] [PubMed] [Google Scholar]
- Chalitchagorn K, Shuangshoti S, Hourpai N, Kongruttanachok N, Tangkijvanich P, Thong-ngam D, et al. Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene. 2004;23:8841–8846. doi: 10.1038/sj.onc.1208137. [DOI] [PubMed] [Google Scholar]
- Champion C, Guianvarc’h D, Senamaud-Beaufort C, Jurkowska RZ, Jeltsch A, Ponger L, et al. Mechanistic insights on the inhibition of c5 DNA methyltransferases by zebularine. PLoS One. 2010;5:e12388. doi: 10.1371/journal.pone.0012388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chedin F. The DNMT3 family of mammalian de novo DNA methyltransferases. Prog Mol Biol Transl Sci. 2011;101:255–285. doi: 10.1016/B978-0-12-387685-0.00007-X. [DOI] [PubMed] [Google Scholar]
- Chen LM, Verity NJ, Chai KX. Loss of prostasin (PRSS8) in human bladder transitional cell carcinoma cell lines is associated with epithelial–mesenchymal transition (EMT) BMC Cancer. 2009;9:377. doi: 10.1186/1471-2407-9-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JC, Matsen CB, Gonzales FA, Ye W, Greer S, Marquez VE, et al. Inhibition of DNA methylation and reactivation of silenced genes by zebularine. J Natl Cancer Inst. 2003;95:399–409. doi: 10.1093/jnci/95.5.399. [DOI] [PubMed] [Google Scholar]
- Cheng JC, Weisenberger DJ, Gonzales FA, Liang G, Xu GL, Hu YG, et al. Continuous zebularine treatment effectively sustains demethylation in human bladder cancer cells. Mol Cell Biol. 2004;24:1270–1278. doi: 10.1128/MCB.24.3.1270-1278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoph F, Kempkensteffen C, Weikert S, Kollermann J, Krause H, Miller K, et al. Methylation of tumour suppressor genes APAF-1 and DAPK-1 and in vitro effects of demethylating agents in bladder and kidney cancer. Br J Cancer. 2006a;95:1701–1707. doi: 10.1038/sj.bjc.6603482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoph F, Kempkensteffen C, Weikert S, Krause H, Schostak M, Miller K, et al. Frequent epigenetic inactivation of p53 target genes in seminomatous and nonseminomatous germ cell tumors. Cancer Lett. 2007;247:137–142. doi: 10.1016/j.canlet.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Christoph F, Weikert S, Kempkensteffen C, Krause H, Schostak M, Kollermann J, et al. Promoter hypermethylation profile of kidney cancer with new proapoptotic p53 target genes and clinical implications. Clin Cancer Res. 2006b;12:5040–5046. doi: 10.1158/1078-0432.CCR-06-0144. [DOI] [PubMed] [Google Scholar]
- Cohen HT, McGovern FJ. Renal-cell carcinoma. N Engl J Med. 2005;353:2477–2490. doi: 10.1056/NEJMra043172. [DOI] [PubMed] [Google Scholar]
- Cordeiro ER, Cathelineau X, Thuroff S, Marberger M, Crouzet S, de la Rosette JJ. High-intensity focused ultrasound (HIFU) for definitive treatment of prostate cancer. BJU Int. 2012;110(9):1228–1242. doi: 10.1111/j.1464-410X.2012.11262.x. [DOI] [PubMed] [Google Scholar]
- Crea F, Fornaro L, Bocci G, Sun L, Farrar WL, Falcone A, et al. EZH2 inhibition: targeting the crossroad of tumor invasion and angiogenesis. Cancer Metastasis Rev. 2012a;31:753–761. doi: 10.1007/s10555-012-9387-3. [DOI] [PubMed] [Google Scholar]
- Crea F, Sun L, Mai A, Chiang YT, Farrar WL, Danesi R, et al. The emerging role of histone lysine demethylases in prostate cancer. Mol Cancer. 2012b;11:52. doi: 10.1186/1476-4598-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T, Huntly BJ. Targeting epigenetic readers in cancer. N Engl J Med. 2012;367:647–657. doi: 10.1056/NEJMra1112635. [DOI] [PubMed] [Google Scholar]
- Denis H, Ndlovu MN, Fuks F. Regulation of mammalian DNA methyltransferases: a route to new mechanisms. EMBO Rep. 2011;12:647–656. doi: 10.1038/embor.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhordain P, Lin RJ, Quief S, Lantoine D, Kerckaert JP, Evans RM, et al. The LAZ3(BCL-6) oncoprotein recruits a SMRT/mSIN3A/histone deacetylase containing complex to mediate transcriptional repression. Nucleic Acids Res. 1998;26:4645–4651. doi: 10.1093/nar/26.20.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draisma G, Boer R, Otto SJ, van der Cruijsen IW, Damhuis RA, Schroder FH, et al. Lead times and overdetection due to prostate-specific antigen screening: estimates from the European Randomized Study of Screening for Prostate Cancer. J Natl Cancer Inst. 2003;95:868–878. doi: 10.1093/jnci/95.12.868. [DOI] [PubMed] [Google Scholar]
- Duns G, van den Berg E, van Duivenbode I, Osinga J, Hollema H, Hofstra RM, et al. Histone methyltransferase gene SETD2 is a novel tumor suppressor gene in clear cell renal cell carcinoma. Cancer Res. 2010;70:4287–4291. doi: 10.1158/0008-5472.CAN-10-0120. [DOI] [PubMed] [Google Scholar]
- Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- Eggener S, Salomon G, Scardino PT, De la Rosette J, Polascik TJ, Brewster S. Focal therapy for prostate cancer: possibilities and limitations. Eur Urol. 2010;58:57–64. doi: 10.1016/j.eururo.2010.03.034. [DOI] [PubMed] [Google Scholar]
- Ellis L, Lehet K, Ramakrishnan S, Adelaiye R, Miles KM, Wang D, et al. Concurrent HDAC and mTORC1 inhibition attenuate androgen receptor and hypoxia signaling associated with alterations in microRNA expression. PLoS One. 2011;6:e27178. doi: 10.1371/journal.pone.0027178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudier B, Eisen T, Stadler WM, Szczylik C, Oudard S, Siebels M, et al. Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J Med. 2007a;356:125–134. doi: 10.1056/NEJMoa060655. [DOI] [PubMed] [Google Scholar]
- Escudier B, Pluzanska A, Koralewski P, Ravaud A, Bracarda S, Szczylik C, et al. Bevacizumab plus interferon alfa-2a for treatment of metastatic renal cell carcinoma: a randomised, double-blind phase III trial. Lancet. 2007b;370:2103–2111. doi: 10.1016/S0140-6736(07)61904-7. [DOI] [PubMed] [Google Scholar]
- Ezponda T, Popovic R, Shah MY, Martinez-Garcia E, Zheng Y, Min DJ, et al. The histone methyltransferase MMSET/WHSC1 activates TWIST1 to promote an epithelial-mesenchymal transition and invasive properties of prostate cancer. Oncogene. 2012 doi: 10.1038/onc.2012.297. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Zheng C, Liu Z, Ekman P, Xu D. Enhanced sensitivity of prostate cancer DU145 cells to cisplatinum by 5-aza-2′-deoxycytidine. Oncol Rep. 2004;12:523–526. [PubMed] [Google Scholar]
- Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- Goering W, Ribarska T, Schulz WA. Selective changes of retroelement expression in human prostate cancer. Carcinogenesis. 2011;32:1484–1492. doi: 10.1093/carcin/bgr181. [DOI] [PubMed] [Google Scholar]
- Gu L, Wang Q, Sun QY. Histone modifications during mammalian oocyte maturation: dynamics, regulation and functions. Cell Cycle. 2010;9:1942–1950. doi: 10.4161/cc.9.10.11599. [DOI] [PubMed] [Google Scholar]
- Hainsworth JD, Infante JR, Spigel DR, Arrowsmith ER, Boccia RV, Burris HA. A phase II trial of panobinostat, a histone deacetylase inhibitor, in the treatment of patients with refractory metastatic renal cell carcinoma. Cancer Invest. 2011;29:451–455. doi: 10.3109/07357907.2011.590568. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Heidenreich A, Bellmunt J, Bolla M, Joniau S, Mason M, Matveev V, et al. EAU guidelines on prostate cancer. Part 1: screening, diagnosis, and treatment of clinically localised disease. Eur Urol. 2011;59:61–71. doi: 10.1016/j.eururo.2010.10.039. [DOI] [PubMed] [Google Scholar]
- Hinz S, Magheli A, Weikert S, Schulze W, Krause H, Schrader M, et al. Deregulation of EZH2 expression in human spermatogenic disorders and testicular germ cell tumors. World J Urol. 2010;28:631–635. doi: 10.1007/s00345-009-0498-6. [DOI] [PubMed] [Google Scholar]
- Horwich A, Shipley J, Huddart R. Testicular germ-cell cancer. Lancet. 2006;367:754–765. doi: 10.1016/S0140-6736(06)68305-0. [DOI] [PubMed] [Google Scholar]
- Houldsworth J, Korkola JE, Bosl GJ, Chaganti RS. Biology and genetics of adult male germ cell tumors. J Clin Oncol. 2006;24:5512–5518. doi: 10.1200/JCO.2006.08.4285. [DOI] [PubMed] [Google Scholar]
- Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, et al. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- Hudak L, Tezeeh P, Wedel S, Makarevic J, Juengel E, Tsaur I, et al. Low dosed interferon alpha augments the anti-tumor potential of histone deacetylase inhibition on prostate cancer cell growth and invasion. Prostate. 2012;72:1719–1735. doi: 10.1002/pros.22525. [DOI] [PubMed] [Google Scholar]
- Hurtubise A, Momparler RL. Evaluation of antineoplastic action of 5-aza-2′-deoxycytidine (Dacogen) and docetaxel (Taxotere) on human breast, lung and prostate carcinoma cell lines. Anticancer Drugs. 2004;15:161–167. doi: 10.1097/00001813-200402000-00010. [DOI] [PubMed] [Google Scholar]
- Hussain MH, MacVicar GR, Petrylak DP, Dunn RL, Vaishampayan U, Lara PN, Jr, et al. Trastuzumab, paclitaxel, carboplatin, and gemcitabine in advanced human epidermal growth factor receptor-2/neu-positive urothelial carcinoma: results of a multicenter phase II National Cancer Institute trial. J Clin Oncol. 2007;25:2218–2224. doi: 10.1200/JCO.2006.08.0994. [DOI] [PubMed] [Google Scholar]
- Iwata H, Sato H, Suzuki R, Yamada R, Ichinomiya S, Yanagihara M, et al. A demethylating agent enhances chemosensitivity to vinblastine in a xenograft model of renal cell carcinoma. Int J Oncol. 2011;38:1653–1661. doi: 10.3892/ijo.2011.999. [DOI] [PubMed] [Google Scholar]
- Izbicka E, MacDonald JR, Davidson K, Lawrence RA, Gomez L, Von Hoff DD. 5,6 Dihydro-5′-azacytidine (DHAC) restores androgen responsiveness in androgen-insensitive prostate cancer cells. Anticancer Res. 1999;19:1285–1291. [PubMed] [Google Scholar]
- Jeon HG, Yoon CY, Yu JH, Park MJ, Lee JE, Jeong SJ, et al. Induction of caspase mediated apoptosis and down-regulation of nuclear factor-kappaB and Akt signaling are involved in the synergistic antitumor effect of gemcitabine and the histone deacetylase inhibitor trichostatin A in human bladder cancer cells. J Urol. 2011;186:2084–2093. doi: 10.1016/j.juro.2011.06.053. [DOI] [PubMed] [Google Scholar]
- Johnstone RW, Licht JD. Histone deacetylase inhibitors in cancer therapy: is transcription the primary target? Cancer Cell. 2003;4:13–18. doi: 10.1016/s1535-6108(03)00165-x. [DOI] [PubMed] [Google Scholar]
- Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10:805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Jurkowska RZ, Rajavelu A, Anspach N, Urbanke C, Jankevicius G, Ragozin S, et al. Oligomerization and binding of the Dnmt3a DNA methyltransferase to parallel DNA molecules: heterochromatic localization and role of Dnmt3L. J Biol Chem. 2011;286:24200–24207. doi: 10.1074/jbc.M111.254987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang GH, Lee S, Lee HJ, Hwang KS. Aberrant CpG island hypermethylation of multiple genes in prostate cancer and prostatic intraepithelial neoplasia. J Pathol. 2004;202:233–240. doi: 10.1002/path.1503. [DOI] [PubMed] [Google Scholar]
- Kauffman EC, Robinson BD, Downes MJ, Powell LG, Lee MM, Scherr DS, et al. Role of androgen receptor and associated lysine-demethylase coregulators, LSD1 and JMJD2A, in localized and advanced human bladder cancer. Mol Carcinog. 2011;50:931–944. doi: 10.1002/mc.20758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman DS, Shipley WU, Feldman AS. Bladder cancer. Lancet. 2009;374:239–249. doi: 10.1016/S0140-6736(09)60491-8. [DOI] [PubMed] [Google Scholar]
- Khan O, La Thangue NB. HDAC inhibitors in cancer biology: emerging mechanisms and clinical applications. Immunol Cell Biol. 2012;90:85–94. doi: 10.1038/icb.2011.100. [DOI] [PubMed] [Google Scholar]
- Kim JH, Dhanasekaran SM, Prensner JR, Cao X, Robinson D, Kalyana-Sundaram S, et al. Deep sequencing reveals distinct patterns of DNA methylation in prostate cancer. Genome Res. 2011;21:1028–1041. doi: 10.1101/gr.119347.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8:890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- Kottakis F, Polytarchou C, Foltopoulou P, Sanidas I, Kampranis SC, Tsichlis PN. FGF-2 regulates cell proliferation, migration, and angiogenesis through an NDY1/KDM2B-miR-101-EZH2 pathway. Mol Cell. 2011;43:285–298. doi: 10.1016/j.molcel.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krege S, Beyer J, Souchon R, Albers P, Albrecht W, Algaba F, et al. European consensus conference on diagnosis and treatment of germ cell cancer: a report of the second meeting of the European Germ Cell Cancer Consensus Group (EGCCCG): part I. Eur Urol. 2008;53:478–496. doi: 10.1016/j.eururo.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Kruger S, Weitsch G, Buttner H, Matthiensen A, Bohmer T, Marquardt T, et al. HER2 overexpression in muscle-invasive urothelial carcinoma of the bladder: prognostic implications. Int J Cancer. 2002;102:514–518. doi: 10.1002/ijc.10731. [DOI] [PubMed] [Google Scholar]
- Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 2012;488:404–408. doi: 10.1038/nature11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo HK, Griffith JD, Kreuzer KN. 5-Azacytidine induced methyltransferase-DNA adducts block DNA replication in vivo. Cancer Res. 2007;67:8248–8254. doi: 10.1158/0008-5472.CAN-07-1038. [DOI] [PubMed] [Google Scholar]
- Lin J, Haffner MC, Zhang Y, Lee BH, Brennen WN, Britton J, et al. Disulfiram is a DNA demethylating agent and inhibits prostate cancer cell growth. Prostate. 2011;71:333–343. doi: 10.1002/pros.21247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Asgari K, Putzi MJ, Gage WR, Yu X, Cornblatt BS, et al. Reversal of GSTP1 CpG island hypermethylation and reactivation of pi-class glutathione S-transferase (GSTP1) expression in human prostate cancer cells by treatment with procainamide. Cancer Res. 2001;61:8611–8616. [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J, Zhao J, Yan Z, Liu Z, Wang N. Antitumor effects of a novel sulfur-containing hydroxamate histone deacetylase inhibitor H40. Int J Cancer. 2009;124:1235–1244. doi: 10.1002/ijc.24074. [DOI] [PubMed] [Google Scholar]
- Mahalingam D, Medina EC, Esquivel JA, II, Espitia CM, Smith S, Oberheu K, et al. Vorinostat enhances the activity of temsirolimus in renal cell carcinoma through suppression of survivin levels. Clin Cancer Res. 2010;16:141–153. doi: 10.1158/1078-0432.CCR-09-1385. [DOI] [PubMed] [Google Scholar]
- Mahapatra S, Klee EW, Young CY, Sun Z, Jimenez RE, Klee GG, et al. Global methylation profiling for risk prediction of prostate cancer. Clin Cancer Res. 2012;18:2882–2895. doi: 10.1158/1078-0432.CCR-11-2090. [DOI] [PubMed] [Google Scholar]
- Majid S, Dar AA, Ahmad AE, Hirata H, Kawakami K, Shahryari V, et al. BTG3 tumor suppressor gene promoter demethylation, histone modification and cell cycle arrest by genistein in renal cancer. Carcinogenesis. 2009;30:662–670. doi: 10.1093/carcin/bgp042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini DN, Singh SM, Archer TK, Rodenhiser DI. Site-specific DNA methylation in the neurofibromatosis (NF1) promoter interferes with binding of CREB and SP1 transcription factors. Oncogene. 1999;18:4108–4119. doi: 10.1038/sj.onc.1202764. [DOI] [PubMed] [Google Scholar]
- Marcus AI, Zhou J, O’Brate A, Hamel E, Wong J, Nivens M, et al. The synergistic combination of the farnesyl transferase inhibitor lonafarnib and paclitaxel enhances tubulin acetylation and requires a functional tubulin deacetylase. Cancer Res. 2005;65:3883–3893. doi: 10.1158/0008-5472.CAN-04-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein R. Structure and function of histone acetyltransferases. Cell Mol Life Sci. 2001;58:693–703. doi: 10.1007/PL00000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Matousova M, Votruba I, Otmar M, Tloustova E, Gunterova J, Mertlikova-Kaiserova H. 2-Deoxy-5,6-dihydro-5-azacytidine — a less toxic alternative of 2-deoxy-5-azacytidine: a comparative study of hypomethylating potential. Epigenetics. 2011;6:769–776. doi: 10.4161/epi.6.6.16215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492(7427):108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- Molife LR, Attard G, Fong PC, Karavasilis V, Reid AH, Patterson S, et al. Phase II, two-stage, single-arm trial of the histone deacetylase inhibitor (HDACi) romidepsin in metastatic castration-resistant prostate cancer (CRPC) Ann Oncol. 2010;21:109–113. doi: 10.1093/annonc/mdp270. [DOI] [PubMed] [Google Scholar]
- Motzer RJ, Escudier B, Oudard S, Hutson TE, Porta C, Bracarda S, et al. Efficacy of everolimus in advanced renal cell carcinoma: a double-blind, randomised, placebo-controlled phase III trial. Lancet. 2008;372:449–456. doi: 10.1016/S0140-6736(08)61039-9. [DOI] [PubMed] [Google Scholar]
- Motzer RJ, Molina AM. Targeting renal cell carcinoma. J Clin Oncol. 2009;27:3274–3276. doi: 10.1200/JCO.2009.21.8461. [DOI] [PubMed] [Google Scholar]
- Murphy M, Ahn J, Walker KK, Hoffman WH, Evans RM, Levine AJ, et al. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 1999;13:2490–2501. doi: 10.1101/gad.13.19.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T, Watanabe M, Suzuki H, Toyota M, Sekita N, Hirokawa Y, et al. Epigenetic regulation of androgen receptor gene expression in human prostate cancers. Lab Invest. 2000;80:1789–1796. doi: 10.1038/labinvest.3780190. [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Narayan A, Ji W, Zhang XY, Marrogi A, Graff JR, Baylin SB, et al. Hypomethylation of pericentromeric DNA in breast adenocarcinomas. Int J Cancer. 1998;77:833–838. doi: 10.1002/(sici)1097-0215(19980911)77:6<833::aid-ijc6>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Ng HH, Bird A. Histone deacetylases: silencers for hire. Trends Biochem Sci. 2000;25:121–126. doi: 10.1016/s0968-0004(00)01551-6. [DOI] [PubMed] [Google Scholar]
- Peart MJ, Smyth GK, van Laar RK, Bowtell DD, Richon VM, Marks PA, et al. Identification and functional significance of genes regulated by structurally different histone deacetylase inhibitors. Proc Natl Acad Sci USA. 2005;102:3697–3702. doi: 10.1073/pnas.0500369102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, et al. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- Petrylak DP, Tangen CM, Hussain MH, Lara PN, Jr, Jones JA, Taplin ME, et al. Docetaxel and estramustine compared with mitoxantrone and prednisone for advanced refractory prostate cancer. N Engl J Med. 2004;351:1513–1520. doi: 10.1056/NEJMoa041318. [DOI] [PubMed] [Google Scholar]
- Prince HM, Bishton MJ, Harrison SJ. Clinical studies of histone deacetylase inhibitors. Clin Cancer Res. 2009;15:3958–3969. doi: 10.1158/1078-0432.CCR-08-2785. [DOI] [PubMed] [Google Scholar]
- Pulukuri SM, Rao JS. Activation of p53/p21Waf1/Cip1 pathway by 5-aza-2′-deoxycytidine inhibits cell proliferation, induces pro-apoptotic genes and mitogen-activated protein kinases in human prostate cancer cells. Int J Oncol. 2005;26:863–871. [PubMed] [Google Scholar]
- Qian X, Ara G, Mills E, LaRochelle WJ, Lichenstein HS, Jeffers M. Activity of the histone deacetylase inhibitor belinostat (PXD101) in preclinical models of prostate cancer. Int J Cancer. 2008;122:1400–1410. doi: 10.1002/ijc.23243. [DOI] [PubMed] [Google Scholar]
- Ramachandran K, Gopisetty G, Gordian E, Navarro L, Hader C, Reis IM, et al. Methylation-mediated repression of GADD45alpha in prostate cancer and its role as a potential therapeutic target. Cancer Res. 2009;69:1527–1535. doi: 10.1158/0008-5472.CAN-08-3609. [DOI] [PubMed] [Google Scholar]
- Ramachandran K, Gordian E, Singal R. 5-Azacytidine reverses drug resistance in bladder cancer cells. Anticancer Res. 2011;31:3757–3766. [PubMed] [Google Scholar]
- Raman JD, Mongan NP, Tickoo SK, Boorjian SA, Scherr DS, Gudas LJ. Increased expression of the polycomb group gene, EZH2, in transitional cell carcinoma of the bladder. Clin Cancer Res. 2005;11:8570–8576. doi: 10.1158/1078-0432.CCR-05-1047. [DOI] [PubMed] [Google Scholar]
- Rathkopf D, Wong BY, Ross RW, Anand A, Tanaka E, Woo MM, et al. A phase I study of oral panobinostat alone and in combination with docetaxel in patients with castration-resistant prostate cancer. Cancer Chemother Pharmacol. 2010;66:181–189. doi: 10.1007/s00280-010-1289-x. [DOI] [PubMed] [Google Scholar]
- Reu FJ, Bae SI, Cherkassky L, Leaman DW, Lindner D, Beaulieu N, et al. Overcoming resistance to interferon-induced apoptosis of renal carcinoma and melanoma cells by DNA demethylation. J Clin Oncol. 2006;24:3771–3779. doi: 10.1200/JCO.2005.03.4074. [DOI] [PubMed] [Google Scholar]
- Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- Richter E, Masuda K, Cook C, Ehrich M, Tadese AY, Li H, et al. A role for DNA methylation in regulating the growth suppressor PMEPA1 gene in prostate cancer. Epigenetics. 2007;2:100–109. doi: 10.4161/epi.2.2.4611. [DOI] [PubMed] [Google Scholar]
- Rini BI, Halabi S, Rosenberg JE, Stadler WM, Vaena DA, Ou SS, et al. Bevacizumab plus interferon alfa compared with interferon alfa monotherapy in patients with metastatic renal cell carcinoma: CALGB 90206. J Clin Oncol. 2008;26:5422–5428. doi: 10.1200/JCO.2008.16.9847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman-Gomez J, Jimenez-Velasco A, Agirre X, Cervantes F, Sanchez J, Garate L, et al. Promoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia. Oncogene. 2005;24:7213–7223. doi: 10.1038/sj.onc.1208866. [DOI] [PubMed] [Google Scholar]
- Rosen A, Jayram G, Drazer M, Eggener SE. Global trends in testicular cancer incidence and mortality. Eur Urol. 2011;60:374–379. doi: 10.1016/j.eururo.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Rotili D, Mai A. Targeting histone demethylases: a new avenue for the fight against cancer. Genes Cancer. 2011;2:663–679. doi: 10.1177/1947601911417976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini S, Arora S, Majid S, Shahryari V, Chen Y, Deng G, et al. Curcumin modulates microRNA-203-mediated regulation of the Src-Akt axis in bladder cancer. Cancer Prev Res (Phila) 2011;4:1698–1709. doi: 10.1158/1940-6207.CAPR-11-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Friedman JM, Chihara Y, Egger G, Chuang JC, Liang G. Epigenetic therapy upregulates the tumor suppressor microRNA-126 and its host gene EGFL7 in human cancer cells. Biochem Biophys Res Commun. 2009;379:726–731. doi: 10.1016/j.bbrc.2008.12.098. [DOI] [PubMed] [Google Scholar]
- Sartor AO, Fitzpatrick JM. Urologists and oncologists: adapting to a new treatment paradigm in castration-resistant prostate cancer (CRPC) BJU Int. 2012;110:328–335. doi: 10.1111/j.1464-410X.2011.10818.x. [DOI] [PubMed] [Google Scholar]
- Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24:1590–1595. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder FH, Hugosson J, Roobol MJ, Tammela TL, Ciatto S, Nelen V, et al. Prostate-cancer mortality at 11 years of follow-up. N Engl J Med. 2012;366:981–990. doi: 10.1056/NEJMoa1113135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang D, Liu Y, Matsui Y, Ito N, Nishiyama H, Kamoto T, et al. Demethylating agent 5-aza-2′-deoxycytidine enhances susceptibility of bladder transitional cell carcinoma to Cisplatin. Urology. 2008;71:1220–1225. doi: 10.1016/j.urology.2007.11.029. [DOI] [PubMed] [Google Scholar]
- Shen L, Kondo Y, Ahmed S, Boumber Y, Konishi K, Guo Y, et al. Drug sensitivity prediction by CpG island methylation profile in the NCI-60 cancer cell line panel. Cancer Res. 2007;67:11335–11343. doi: 10.1158/0008-5472.CAN-07-1502. [DOI] [PubMed] [Google Scholar]
- Shu L, Khor TO, Lee JH, Boyanapalli SS, Huang Y, Wu TY, et al. Epigenetic CpG demethylation of the promoter and reactivation of the expression of Neurog1 by curcumin in prostate LNCaP cells. AAPS J. 2011;13:606–614. doi: 10.1208/s12248-011-9300-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Teplova M, Ishibe-Murakami S, Patel DJ. Structure-based mechanistic insights into DNMT1-mediated maintenance DNA methylation. Science. 2012;335:709–712. doi: 10.1126/science.1214453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonpavde G, Aparicio AM, Zhan F, North B, Delaune R, Garbo LE, et al. Azacitidine favorably modulates PSA kinetics correlating with plasma DNA LINE-1 hypomethylation in men with chemonaive castration-resistant prostate cancer. Urol Oncol. 2011;29:682–689. doi: 10.1016/j.urolonc.2009.09.015. [DOI] [PubMed] [Google Scholar]
- Stadler WM, Margolin K, Ferber S, McCulloch W, Thompson JA. A phase II study of depsipeptide in refractory metastatic renal cell cancer. Clin Genitourin Cancer. 2006;5:57–60. doi: 10.3816/CGC.2006.n.018. [DOI] [PubMed] [Google Scholar]
- Stein JP, Skinner DG. Radical cystectomy for invasive bladder cancer: long-term results of a standard procedure. World J Urol. 2006;24:296–304. doi: 10.1007/s00345-006-0061-7. [DOI] [PubMed] [Google Scholar]
- Sternberg CN, Davis ID, Mardiak J, Szczylik C, Lee E, Wagstaff J, et al. Pazopanib in locally advanced or metastatic renal cell carcinoma: results of a randomized phase III trial. J Clin Oncol. 2010;28:1061–1068. doi: 10.1200/JCO.2009.23.9764. [DOI] [PubMed] [Google Scholar]
- Tada Y, Yokomizo A, Shiota M, Tsunoda T, Plass C, Naito S. Aberrant DNA methylation of T-cell leukemia, homeobox 3 modulates cisplatin sensitivity in bladder cancer. Int J Oncol. 2011;39:727–733. doi: 10.3892/ijo.2011.1049. [DOI] [PubMed] [Google Scholar]
- Tandstad T, Smaaland R, Solberg A, Bremnes RM, Langberg CW, Laurell A, et al. Management of seminomatous testicular cancer: a binational prospective population-based study from the Swedish norwegian testicular cancer study group. J Clin Oncol. 2011;29:719–725. doi: 10.1200/JCO.2010.30.1044. [DOI] [PubMed] [Google Scholar]
- Tannock IF, de Wit R, Berry WR, Horti J, Pluzanska A, Chi KN, et al. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med. 2004;351:1502–1512. doi: 10.1056/NEJMoa040720. [DOI] [PubMed] [Google Scholar]
- Thakur VS, Gupta K, Gupta S. Green tea polyphenols causes cell cycle arrest and apoptosis in prostate cancer cells by suppressing class I histone deacetylases. Carcinogenesis. 2012;33:377–384. doi: 10.1093/carcin/bgr277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiagarajan D, Dev RR, Khosla S. The DNA methyltranferase Dnmt2 participates in RNA processing during cellular stress. Epigenetics. 2011;6:103–113. doi: 10.4161/epi.6.1.13418. [DOI] [PubMed] [Google Scholar]
- Thibault A, Figg WD, Bergan RC, Lush RM, Myers CE, Tompkins A, et al. A phase II study of 5-aza-2′deoxycytidine (decitabine) in hormone independent metastatic (D2) prostate cancer. Tumori. 1998;84:87–89. doi: 10.1177/030089169808400120. [DOI] [PubMed] [Google Scholar]
- Tosoian JJ, Trock BJ, Landis P, Feng Z, Epstein JI, Partin AW, et al. Active surveillance program for prostate cancer: an update of the Johns Hopkins experience. J Clin Oncol. 2011;29:2185–2190. doi: 10.1200/JCO.2010.32.8112. [DOI] [PubMed] [Google Scholar]
- Tsai HC, Li H, Van Neste L, Cai Y, Robert C, Rassool FV, et al. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell. 2012;21:430–446. doi: 10.1016/j.ccr.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Emburgh BO, Robertson KD. Modulation of Dnmt3b function in vitro by interactions with Dnmt3L, Dnmt3a and Dnmt3b splice variants. Nucleic Acids Res. 2011;39:4984–5002. doi: 10.1093/nar/gkr116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Wagener N, Holland D, Bulkescher J, Crnkovic-Mertens I, Hoppe-Seyler K, Zentgraf H, et al. The enhancer of zeste homolog 2 gene contributes to cell proliferation and apoptosis resistance in renal cell carcinoma cells. Int J Cancer. 2008;123:1545–1550. doi: 10.1002/ijc.23683. [DOI] [PubMed] [Google Scholar]
- Wang Q, Williamson M, Bott S, Brookman-Amissah N, Freeman A, Nariculam J, et al. Hypomethylation of WNT5A, CRIP1 and S100P in prostate cancer. Oncogene. 2007;26:6560–6565. doi: 10.1038/sj.onc.1210472. [DOI] [PubMed] [Google Scholar]
- Willmann D, Lim S, Wetzel S, Metzger E, Jandausch A, Wilk W, et al. Impairment of prostate cancer cell growth by a selective and reversible lysine-specific demethylase 1 inhibitor. Int J Cancer. 2012;131:2704–2709. doi: 10.1002/ijc.27555. [DOI] [PubMed] [Google Scholar]
- Winquist E, Kirchner TS, Segal R, Chin J, Lukka H. Neoadjuvant chemotherapy for transitional cell carcinoma of the bladder: a systematic review and meta-analysis. J Urol. 2004;171:561–569. doi: 10.1097/01.ju.0000090967.08622.33. [DOI] [PubMed] [Google Scholar]
- Winquist E, Knox J, Ayoub JP, Wood L, Wainman N, Reid GK, et al. Phase II trial of DNA methyltransferase 1 inhibition with the antisense oligonucleotide MG98 in patients with metastatic renal carcinoma: a National Cancer Institute of Canada Clinical Trials Group investigational new drug study. Invest New Drugs. 2006;24:159–167. doi: 10.1007/s10637-006-5938-1. [DOI] [PubMed] [Google Scholar]
- Wissmann M, Yin N, Muller JM, Greschik H, Fodor BD, Jenuwein T, et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol. 2007;9:347–353. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
- Wolff EM, Chihara Y, Pan F, Weisenberger DJ, Siegmund KD, Sugano K, et al. Unique DNA methylation patterns distinguish noninvasive and invasive urothelial cancers and establish an epigenetic field defect in premalignant tissue. Cancer Res. 2010;70:8169–8178. doi: 10.1158/0008-5472.CAN-10-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yegnasubramanian S, Haffner MC, Zhang Y, Gurel B, Cornish TC, Wu Z, et al. DNA hypomethylation arises later in prostate cancer progression than CpG island hypermethylation and contributes to metastatic tumor heterogeneity. Cancer Res. 2008;68:8954–8967. doi: 10.1158/0008-5472.CAN-07-6088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yegnasubramanian S, Kowalski J, Gonzalgo ML, Zahurak M, Piantadosi S, Walsh PC, et al. Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res. 2004;64:1975–1986. doi: 10.1158/0008-5472.can-03-3972. [DOI] [PubMed] [Google Scholar]
- Zhang P, Singh A, Yegnasubramanian S, Esopi D, Kombairaju P, Bodas M, et al. Loss of Kelch-like ECH-associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Mol Cancer Ther. 2010;9:336–346. doi: 10.1158/1535-7163.MCT-09-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Rubenstein JN, Jang TL, Pins M, Javonovic B, Yang X, et al. Insensitivity to transforming growth factor-beta results from promoter methylation of cognate receptors in human prostate cancer cells (LNCaP) Mol Endocrinol. 2005;19:2390–2399. doi: 10.1210/me.2005-0096. [DOI] [PubMed] [Google Scholar]
- Zhao H, Flamand V, Peehl DM. Anti-oncogenic and pro-differentiation effects of clorgyline, a monoamine oxidase A inhibitor, on high grade prostate cancer cells. BMC Med Genomics. 2009;2:55. doi: 10.1186/1755-8794-2-55. [DOI] [PMC free article] [PubMed] [Google Scholar]