Abstract
The yes-associated protein (YAP) is a key effector of the mammalian Hippo signaling pathway. YAP has eight known alternately spliced isoforms and these are widely expressed across multiple tissues. Variable effects have been ascribed to different YAP isoforms by inducing their expression in cells, but whether these differences are due to variability in the transcriptional potency of individual YAP isoforms has not been addressed. Indeed a systematic comparison of the transcriptional potencies of YAP isoforms has not been done. To address this, using overexpression and transcriptional reporter analyses we investigated the transcriptional activities of several human YAP isoforms and determined the effects of the splice variant insertions within the transactivation domain on its transcriptional potency. Utilising full-length coding sequence constructs we determined that the number of WW domains and disruption of the leucine zipper motif within YAP’s transactivation domain both contribute to transcriptional activity. Notably, disruption of YAP’s leucine zipper had a greater effect on transcriptional activity than the absence of the second WW domain. Using GAL4-YAP transcriptional activation domain fusion proteins we found that disruption of the leucine zipper significantly decreased YAP’s transcriptional activity in several cell lines. Our data indicates that expression of different YAP isoforms with varying transcriptional potencies may enable fine control of Hippo pathway signaling. Furthermore the specific isoform being utilised should be taken into consideration when interpreting published data or when designing experiments to ascribe YAP’s function.
Abbreviations: YAP, yes-associated protein; TAD, transcriptional activation domain; hYAP:, human YAP; aa:, Amino acid; mYAP:, mouse YAP; WT, wild-type; Con, control; EV, empty vector
Keywords: Yes-associated protein (YAP), Alternate splicing, Transcriptional co-activator, Luciferase assay
Highlights
-
•
Transcriptional activities of yes-associated protein (YAP) isoforms were compared.
-
•
YAP’s WW domains and leucine zipper motif both contribute to transcriptional activity.
-
•
Absence of YAP’s second WW domain weakens transcriptional potency.
-
•
Disruption of YAP’s leucine zipper weakens the transactivation domain (TAD).
-
•
Potency of the TAD from YAP α, β, γ, δ isoforms is cell-context dependent.
1. Introduction
Yes-associated protein (YAP) is a transcriptional co-activator [1] that functions as an effector for the mammalian Hippo signaling pathway [2], [3], [4], [5]. YAP promotes growth and cell survival by regulating genes involved in proliferation including cyclin D1 [6], survivin [2], connective tissue growth factor [7], and amphiregulin [8]. YAP interacts with many proteins via several protein-interaction domains to facilitate nuclear localisation, DNA-binding, and recruitment of transcription factors (reviewed in [9]).
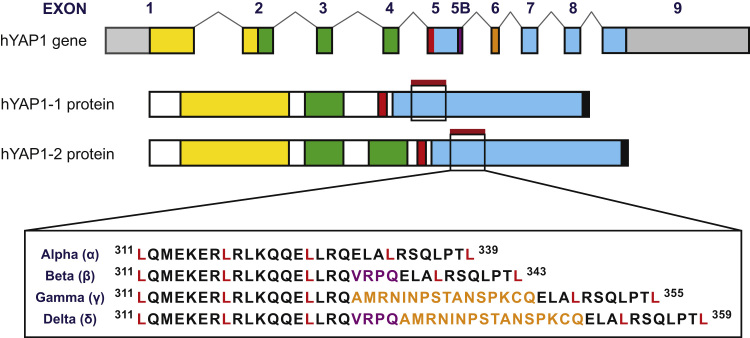
The human YAP1 gene comprises nine exons, generating at least eight alternatively spliced isoforms, all of which are detectable as mRNA in several human tissues [10]. The YAP protein comprises multiple domains that enable binding to a variety of proteins. Exons 1–3 encode the N-terminal region including the TEAD-binding and first WW domains, whereas YAP’s second WW domain, which is only present in hYAP1-2 isoforms (Fig. 1), is encoded by exon 4. Since YAP does not harbour an intrinsic DNA-binding domain it relies on association with DNA-binding transcription factors to co-activate target genes. TEAD proteins (1–4) are the major DNA-binding factors for YAP, associating with YAP via its TEAD-binding domain [11], [12]. However several other factors have been identified that utilise YAP’s WW domains for binding e.g., p73 [13] and ErbB-4 [14].
Fig. 1.
Gene structure and transcriptional activation domain variations of human YAP isoforms. The eight human YAP1 isoforms are encoded by nine exons. Exons 1–3 encode the N-terminal region including the TEAD-binding (yellow) and first WW domain (green), whereas the second WW domain, present only in hYAP1-2 isoforms, is encoded by exon 4. Exons 5–9 encode the SH3-binding region (red) and the C-terminus of YAP, with exons 5B (extended transcript of exon 5, purple) and 6 (orange) encoding an additional 4 and 16 amino acids, respectively. The presence or absence of these additional amino acids within the leucine zipper motif (crimson bar) of the transcriptional activation domain (blue), give rise to the α, β, γ, and δ isoforms as indicated. Inset: the position of the five leucine residues that form the leucine zipper are shown in red.
As a co-activator of transcription, YAP must localize to the nucleus and recruit general transcription factors to activate gene expression. The C-terminal region of YAP, rich in serine, threonine and acidic amino acids, acts as a strong transcription activation domain (TAD) reminiscent of herpesvirus VP16 [1]. YAP does not possess a traditional nuclear localisation signal, thus relies on association with other proteins via its PDZ-binding motif to mediate nuclear localisation [15], [16]. YAP’s C-terminal TAD and PDZ-binding motifs are encoded by exons 5–9.
Exon 5 has an alternate splice donor site, generating an extended transcript (exon 5B) encoding an additional four amino acids (VRPQ), whereas exon 6 specifies an additional 16 amino acids (AMRNINPSTANSPKCQ). Exons 5B and 6, which either alone or in combination are present in six out of eight human YAP (hYAP) isoforms, insert within the leucine zipper motif (also known as the coiled-coil motif) in YAP’s TAD to generate the β, γ, and δ isoforms [10]. In contrast, the α isoform does not contain any insertions in the TAD region. The leucine zipper motif, comprising five highly-conserved leucine residues at every seventh position, mediates protein-protein interactions [17].
Standardised nomenclature for hYAP isoforms was proposed by Gaffney et al. [10], and further simplified [18], as it was acknowledged that publications reporting use of “hYAP cDNA” for functional studies actually used one of several isoforms, making it difficult to compare across studies. For example, numerous publications reported using hYAP1-2γ (also referred to as YAP1-504 aa) [2], [3], [4], [5], [19], [20] and one study used hYAP1-2α (YAP1-488 aa) [21]. This is significant since overexpression of hYAP1-2γ promoted cellular proliferation, EMT, colony formation, protection from apoptosis in MCF10A cells in vitro [3], [5], [20], and liver overgrowth in vivo [2], whereas overexpression of hYAP1-2α in the UMSCC-11A squamous cell carcinoma line increased cell death [21]. Whilst the differences in YAP function may be due to cellular context, it cannot be discounted that the specific YAP isoform utilised, or the combination, may contribute to this result.
Other studies directly compared hYAP isoforms to draw conclusions about the functional importance of different YAP domains. For example, comparison of their transcriptional activities revealed that hYAP1-2α is a stronger co-activator than hYAP1-1β [14]. This was attributed to a higher affinity for ErbB-4 mediated by hYAP1-2α’s second WW domain. Similarly, significant differences were identified between hYAP1-1 and -2 with regards to promotion of apoptosis measured by PARP cleavage and p73 stabilisation [22]. The contribution of YAP’s TAD sequence to transcriptional activity was not evaluated in either study.
There is sufficient evidence indicating that YAP’s protein-protein interaction domains contribute significantly to its transcriptional activity. Multiple studies have shown that critical mutations within one or both WW domains affect YAP’s transcriptional activity [14], [23], [24], which may prevent YAP association with DNA-binding transcription factors such as ErbB-4 and Runx2 [24] or its interaction with other transcriptional modulators [25]. Subsequent studies utilising Yorkie (YAP’s Drosophila orthologue) revealed that the WW domains can recruit transcription enhancing factors including Wbp2 [23]. It is generally accepted that YAP co-activates transcription by recruiting members of the basal transcription machinery, potentially via leucine zipper mediated interactions. Notably, there are conflicting data on the functional requirement of Yorkie and YAP C-terminus. In particular, Yorkie TAD was shown to be superfluous to drive tissue growth in Drosophila, and YAP TAD is not required to promote anchorage-independent growth or resistance to contact inhibition in vitro [26]. Others have shown that deletion of YAPs C-terminus decreased oncogenic functions e.g., reduced xenograft expansion of ovarian cancer cells [27], abolished EMT-like morphological changes induced by active YAP in MCF10A cells [24], and failure to induce cellular proliferation in the mouse retina in vivo [28].
Whilst previous studies have postulated that the insertion of additional amino acids within the C-terminal TAD of YAP may impair its transcriptional activity [10], [14] a comprehensive analysis of the relative transcriptional potency of C-terminal TAD variants has not yet been done. To address this we analysed the transcriptional activities of alternately spliced hYAP isoforms and determined the effects of the C-terminal insertions in the TAD on its transcriptional potency. This allowed us to define the contribution of both the number of WW domains and an intact leucine zipper to YAP’s transcriptional activity. Our data revealed that disruption to the leucine zipper and the WW domains play key roles in determining YAP transcriptional activity. Our results clearly demonstrate that care must be taken when interpreting data generated using different hYAP isoforms in multiple cell lines.
2. Materials and methods
2.1. Plasmids and cDNAs
cDNAs for hYAP1-2α (YAP1-488 aa) and YAP1-1β (YAP1-454 aa) [10] and mouse YAP (mYAP, NCBI NM_009534.3) were obtained from Marius Sudol (National University of Singapore). hYAP1-2γ (YAP1-504 aa) was provided by Kieran Harvey (Peter MacCallum Cancer Centre, VIC). pGT4Tluc Firefly luciferase and pCI-HA-TEAD-2 plasmids [29] were provided by Melvin DePamphilis (NICHHD/NIH). GFP/TetR/VP16 [30] was used as template to generate GAL4–VP16.
mYAP constructs were generated by PCR using primers: 5′GTAGGATCCATGGAGCCCGCGCAACA and 5′GTGTCTAGACTATAACCACGTGAGAAATGGGCTCTGGGGAGCCAAGGGT for ΔTAD, 5′GTGTCTAGACTAGCTTTCTTTATCTAGCTTGGTG for ΔPDZ, and 5′GTAGGATCCATGCCTGCAGCT CAGCATCTC and 5′GTGTCTAGACTATAACCACGTGAGAAAGCTTTC for ΔTEAD.
YAP C-termini comprising the TAD and PDZ-binding motif of mYAP, and hYAP isoforms α, β and γ were amplified by PCR using primers: 5′GTAGGATCCCAGGGAGGCGTCCTGGGTGGA and 5′GTGTCTAGACTATAACCACGTGAGAAAGCTTTC for mYAP or 5′GTAGGATCCCAGGGAGGCGTCATGGGTGGCA and 5′GTGTCTAGACTATAACCATGTAAGAAAGCT for hYAP. hYAPδ C-terminus was generated by overlapping PCR of hYAP1-2γ using the same hYAP primers in combination with δ-specific primers: 5′GTGAGGCCACAGGCAATGCGGAATATCAATCCCAGCACAGC and 5′GATATTCCGCATTGCCTGTGGCCTCACCTGCCGAAGCAGTTCTTGCTG.
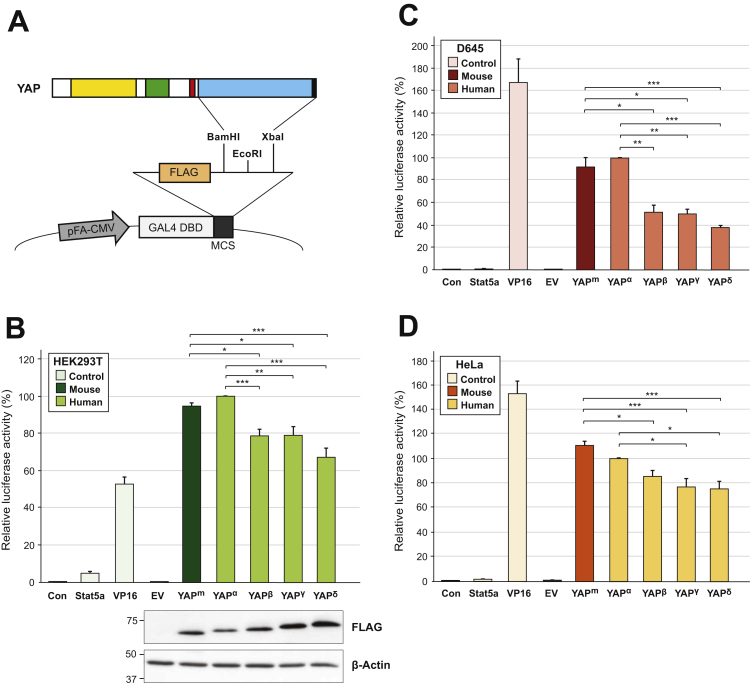
The pFA-CMV-FLAG vector was generated by sub-cloning annealed oligos encoding the FLAG sequence (DYKDDDDK) and an additional BamHI recognition sequence 5′ to the EcoRI site, in-frame into the pFA-CMV [31] vector using BglII and EcoRI. This generated a new unique BamHI site 3′ of the FLAG sequence (Fig. 4A). The TAD’s for all hYAP isoforms, mYAP and VP16 (amino acids 413–490) were cloned in-frame into the pFA-CMV-FLAG vector using unique BamHI/XbaI sites (Fig. 4A). GAL4–Stat5 was described previously [31].
Fig. 4.
Decreased transcriptional activity via disruption of YAP’s leucine zipper motif is not dependent on insert length. (A) Schematic illustration of the FLAG-tagged GAL4-YAP TAD fusion constructs. The transactivation domain (blue) and PDZ-binding motif (black) for hYAP isoforms (α, β, γ and δ) and mYAP, were cloned 3′ to the FLAG sequence using unique BamHI/XbaI sites. HEK293T (B), D645 (C) and HeLa (D) cells were transfected with pFR-Luc (Con), together with FLAG-tagged empty vector (EV), or the C-terminal region of Stat5a, VP16, mYAP (YAPm), or the four hYAP isoforms (YAPα, YAPβ, YAPγ or YAPδ) fused to the GAL4 DNA-binding domain. After 24 h cells were harvested and luciferase activity was determined. Firefly luciferase activity was normalised to Renilla luciferase activity. The average luciferase activities were then normalised to YAPα, which was set to 100%. Data is presented as mean+SEM from three (B) or four (C and D) independent experiments. *p<0.05, **p<0.01, ***p<0.001. Lower panel (B): Cell lysates were separated by SDS-PAGE, transferred to membrane and immunoblotted for FLAG and β-Actin as indicated. Size markers are shown in kilodaltons.
2.2. Cell culture
HEK293T cells were provided by David Vaux (Walter and Eliza Hall Institute (WEHI), VIC). HeLa and D645 cells were obtained from David Huang (WEHI) and John Silke (WEHI), respectively. Cells were maintained in DMEM (Life Technologies, #11885) supplemented with 10% (v/v) FBS, 50 U/mL penicillin G/50 µg/mL streptomycin, and 2 mM l-glutamine in a humidified atmosphere of 10% CO2 at 37 °C.
2.3. Luciferase assay
TEAD-dependent luciferase assays were performed by transfecting sub-confluent 12-well plates of HEK293T cells with 0.1 µg pGT4Tluc, which harbours the firefly luciferase gene driven by four tandem copies of the GTIIC site 30-mer containing the TEF-1 DNA binding sites found in the polyomavirus F101 enhancer [29] (Fig. 2A), 0.3 µg pF-5xUAS-MCS-W–SV40puro [32], [33] expressing full-length (entire coding sequence) wild-type (WT) or mutant YAP constructs, 0.05 µg pRL-TK Renilla luciferase (internal control), with or without 0.05 µg pCI-HA-TEAD-2, which can associate with full-length mYAP and hYAP isoforms via their TEAD-binding domains, and pUC13 to a total of 0.5 µg DNA per well using Effectene (Qiagen #301425).
Fig. 2.
YAP transactivation, PDZ-binding, and TEAD-binding domains are required for TEAD-mediated transcriptional activity. (A) Schematic illustration of the pGT4Tluc luciferase TEAD reporter. HEK293T cells were transfected with pGT4Tluc (Control), FLAG-tagged wild-type (WT), ΔTAD, ΔPDZ or ΔTEAD mYAP constructs (B), and pCI-HA-TEAD-2 (HA-TEAD2) as indicated. (C) After 24 h cells were harvested and luciferase activity was determined. Firefly luciferase activity was normalised to Renilla luciferase activity. The average luciferase activities were normalised to WT mYAP without HA-TEAD2, which was set to 100%. Data is presented as mean+SEM from three independent experiments. *p<0.05, **p<0.01, ***p<0.001. (D) Cell lysates from (C) were separated by SDS-PAGE, transferred to membrane and immunoblotted for FLAG and β-Actin as indicated. Size markers are shown in kilodaltons. (E) HEK293T cells were transfected with pGT4Tluc (Control), mYAP or hYAP1-2α constructs, and pCI-HA-TEAD-2 (HA-TEAD2) as indicated. After 24 h cells were harvested and processed as in (B). Cell lysates were immunoblotted for YAP and β-Actin as indicated. Size markers are shown in kilodaltons. Vertical line indicates the samples were blotted on separate gels.
For GAL4 fusion experiments, sub-confluent 12-well plates of HEK293T cells were transfected with 0.1 µg pFR-Luc (Stratagene) [31], 0.05 µg pFA-CMV-GAL4-FLAG YAP-TAD fusion constructs (Fig. 4A), 0.1 µg pRL-TK, and pUC13 to a total of 0.5 µg DNA per well using Effectene. For HeLa and D645 cell lines, cells were transfected with 0.4 µg pFR-Luc, 0.2 µg pFA-CMV-FLAG YAP-TAD fusion constructs, and 0.4 µg pRL-TK per well using ViaFect (Promega).
Luciferase activity was measured after 24 h using the Dual Luciferase Reporter Assay (Promega). Statistical significance was calculated by performing a one-way ANOVA with Tukey’s multiple comparison test from at least three independent experiments.
2.4. Antibodies and immunoblotting
Anti-FLAG (#F1804) and Anti-β-actin (#A1978) were purchased from Sigma-Aldrich (Castle Hill, NSW). Anti-HA (#2999) was purchased from CST (Genesearch, Arundel, QLD). Lysates (20 µL) were separated by SDS-PAGE and transferred to Hybond C membrane (GE, Castle Hill, NSW). Membranes were immunoblotted with antibodies and detected with chemiluminescence as described [34].
3. Results
3.1. YAP mutants lacking TAD and PDZ-binding domain have reduced transcriptional activity
To confirm that YAP’s TAD, PDZ-binding domain and TEAD-binding domain are necessary for TEAD-mediated transcriptional activity, mYAP deletion constructs (Fig. 2B) were utilised. Since TEAD is the best-characterized oncogenic partner of YAP, the activity of the mutants was assessed using a TEAD-dependent luciferase assay [29] (Fig. 2A). Transfection of WT mYAP significantly increased luciferase activity 16-fold (p<0.001) compared to the luciferase construct alone (Control) (Fig. 2C). Despite comparable expression by Western blot (Fig. 2D) the ΔTAD and ΔPDZ mutants did not increase luciferase activity above basal levels (Fig. 2C). As expected, ΔTEAD failed to increase luciferase activity above control levels and importantly this was not increased by co-transfection of TEAD2 indicating that luciferase expression with this reporter is specific for YAP-TEAD association. In contrast co-transfecting cells with TEAD2 significantly increased luciferase activity for the WT (3.6-fold, p<0.001) and ΔPDZ (5.9-fold, p<0.05) mYAP constructs (Fig. 2C). Notably, although luciferase activity for ΔPDZ increased in the presence of TEAD2 it remained significantly lower (p<0.001) than WT.
To confirm that mYAP mutants may be used to infer information about hYAP transcriptional activity, we compared the transcriptional potency of mYAP to that of hYAP1-2α since they share significant amino acid sequence homology; TEAD-binding (96%), PDZ-binding (100%) domains and TAD region (92%). No significant difference was observed in the relative luciferase activity between mYAP and hYAP1-2α either in the presence of absence of co-transfected TEAD2 (Fig. 2E). Together this data indicates that YAP’s TAD, PDZ-binding and TEAD-binding domains are required for TEAD-mediated transcriptional activity, and that exogenously expressed TEAD2 can functionally associate with both mYAP and hYAP to activate transcription.
3.2. YAP’s tandem WW domains and leucine zipper motif contribute to transcriptional activity
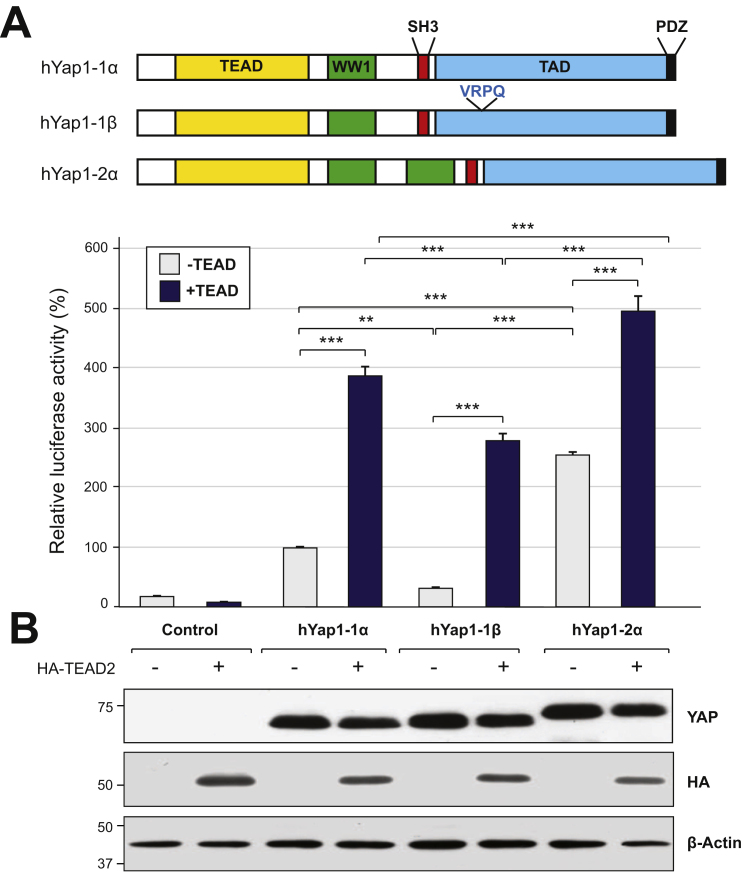
To determine whether the number of WW domain/s (single versus tandem) and interruption of the leucine zipper motif impacts YAP’s transcriptional activity, three hYAP isoforms with combinations of these features were compared using the TEAD reporter assay (Fig. 3). Transfection of hYAP1-1α and hYAP1-2α significantly increased luciferase activity 5.6-fold (p<0.001) and 14.3-fold (p<0.001) respectively compared to luciferase alone (Control). In contrast, hYAP1-1β alone failed to increase luciferase activity above control levels. Despite comparable expression levels (Fig. 3B) the relative activity of hYAP1-1β was significantly lower than hYAP1-1α (3.2-fold lower p<0.01) and hYAP1-2α (8.2-fold p<0.001) (Fig. 3A). Notably endogenous YAP was undetectable in the HEK293T cells, thus our data is specific to the exogenous YAP constructs.
Fig. 3.
YAP’s tandem WW domains and leucine zipper motif contribute to TEAD-mediated transcriptional activity. (A) HEK293T cells were transfected with pGT4Tluc (Control), hYAP1-1α, hYAP1-1β or hYAP1-2α constructs, and pCI-HA-TEAD-2 (HA-TEAD2) as indicated. After 24 h cells were harvested and luciferase activity was determined. Firefly luciferase activity was normalised to Renilla luciferase activity. The average luciferase activities were normalised to hYAP1-1α without HA-TEAD2, which was set to 100%. Data is presented as mean+SEM from three independent experiments. **p<0.01, ***p<0.001. (B) Cell lysates from (A) were separated by SDS-PAGE, transferred to membrane and immunoblotted for YAP, HA and β-Actin as indicated. Size markers are shown in kilodaltons.
As shown above, the TEAD reporter is specific for YAP-TEAD association (Fig. 2). As expected, transfection of the luciferase reporter with TEAD2 resulted in a negligible increase in luciferase activity, whereas co-transfection of TEAD2 significantly increased activity for hYAP1-2α (1.9-fold), hYAP1-1α (3.8-fold) and hYAP1-1β (8.9-fold) (Fig. 3A). Despite the relatively large increase in luciferase activity for hYAP1-1β with TEAD2, this was still significantly lower than that measured for both hYAP1-1α (1.4-fold) and hYAP1-2α (1.8-fold).
Notably, hYAP1-2α luciferase activity was consistently higher than that of hYAP1-1α with activity 2.5-fold higher without TEAD2 and 1.3-fold with TEAD2. Interestingly, this difference was not as pronounced as that between hYAP1-1α and hYAP1-1β which was 3.2-fold higher in the absence of TEAD2 but only 1.4-fold in its presence (Fig. 3A). These data indicate that both a single WW domain and disruption of the leucine zipper reduce YAP’s transcriptional activity. Furthermore, that disruption of the leucine zipper within YAP’s TAD has a possibly greater effect than the absence of tandem WW domains especially when TEAD is limiting.
3.3. Disruption of YAP’s leucine zipper motif reduces its transcriptional co-activator function
To directly examine the effect of a disrupted leucine zipper motif on YAP’s transcriptional potency, the C-terminal region for each hYAP isoform (α, β, γ and δ) and mYAP comprising the TAD and PDZ-binding motif were fused to a FLAG-tagged GAL4-DNA binding domain construct (Fig. 4A) and GAL4-dependent luciferase reporter assays were performed in several cell lines (Fig. 4). Transfection of the GAL4-YAP TAD fusion constructs significantly increased luciferase activity above the luciferase alone (Con) and empty-vector (EV) negative controls and the Stat5a positive control [31]. Luciferase activity stimulated by the VP16 TAD [1] was comparable with that of the YAP-fusions but more variable, being relatively lower than the YAP-fusions in HEK293T cells whilst being higher in D645 (Fig. 4C) and HeLa cells (Fig. 4D). Transfection of YAPα resulted in significantly higher luciferase activity compared to YAPβ (1.3-fold p<0.001), YAPγ (1.3-fold p<0.01) and YAPδ (1.5-fold p<0.001) in HEK293T cells (Fig. 4B). Interestingly, YAPm, the mouse equivalent of YAPα, increased luciferase activity to a similar extent. Western blot analyses indicated that YAPα and to some extent YAPm and YAPβ were expressed at lower levels than YAPγ and YAPδ (Fig. 4B), which may be due to differences in the stability of the various isoforms, suggesting that differences in transcriptional activity between YAPα and other isoforms with disrupted leucine zipper motifs are underestimated here. It is also worth noting that none of the GAL4-fusion constructs were detected by Western blot in either D645 or HeLa cells.
Consistent with HEK293T data, YAPα and YAPm induced luciferase activity to a similar extent and both were significantly higher than YAPβ, YAPγ and YAPδ in D645 (Fig. 4C) and HeLa (Fig. 4D) cells, with the exception that YAPβ was not significantly different to YAPα in HeLa cells. In general the differences in activity for YAPβ, YAPγ and YAPδ compared to YAPα were more pronounced in the D645 cells (2.0- to 2.6-fold decrease) compared to the HEK293Ts (1.3- to 1.5-fold decrease), and less so in HeLa cells (1.3-fold decrease). Notably, no significant difference was observed between YAPβ, YAPγ and YAPδ in all cell lines tested (Fig. 4B and C). Together, these data support our findings obtained using the entire coding sequence of YAP whereby disruption of YAP’s leucine zipper motif decreases its transcriptional co-activator activity (Fig. 3A).
Intriguingly, the difference in luciferase activity between full-length proteins (hYAP1-1α versus hYAP1-1β) (Fig. 3A) was more pronounced than for the GAL4-YAP TAD fusions (YAPα versus YAPβ) (Fig. 4B) (3.2-fold compared to 1.3-fold decrease in activity in HEK293T cells), despite both varying only by insertion of four amino acids within the leucine zipper motif. This may reflect differences in the comparative expression of the different constructs or the luciferase system used.
4. Discussion
Using overexpression and transcriptional reporter analyses, this study represents the first detailed analysis of the transcriptional potency of the TAD from alternately spliced C-terminal YAP isoforms. The existence of eight mammalian YAP isoforms suggests that they have non-redundant roles within the cell. Indeed, structural differences between YAP isoforms are predicted to affect transcriptional activity and function [1], [10], [35], and this is supported by numerous publications that have identified differences between hYAP isoforms, for example in the promotion of p73 association and PARP cleavage [22], Erb-B4 binding [14], transcriptional activity [14], [22], [24], angiomotin association [36], and SHP2 binding [37].
Here we definitively demonstrate that the number of WW domains and disruption of the leucine zipper both contribute to YAP’s transcriptional activity. Interestingly, disruption of the leucine zipper in the full-length proteins caused a relatively greater loss in activity than the absence of a second WW domain. Notably, the combination of these two factors had an additive effect on YAP’s activity. This permits a range of YAP isoforms with varying transcriptional potencies and abilities to associate with protein partners via WW and leucine zipper domain-mediated interactions.
YAP’s WW domains have two defined functions: to bind non-TEAD DNA-binding transcription factors e.g., p73 and ErbB-4 [13], [14], and to recruit enhancers e.g., Wbp2 [23] or repressors [25] of transcription. In this study a TEAD-dependent reporter was utilised to assess transcriptional activity of full-length YAP. This system is unable to assess non-TEAD mediated transcription, in which the presence of single or tandem WW domains could have a more pronounced effect on YAP’s transcriptional activity due to altered efficiency of association with DNA-binding proteins.
Since a functional leucine zipper structure is dependent on precise spacing of leucine residues [17], it is expected that any interruption of the leucine zipper should effectively disrupt the motif. This is supported by the observation that the transcriptional activities of YAPβ, YAPγ and YAPδ, with 4, 16 and 20 amino acid insertions respectively, were similarly decreased in the cell lines tested. It is important to note that a leucine zipper motif is not essential for transactivation since neither TAZ nor Yorkie possess one in their TADs. Interestingly, C-terminal homology between Yorkie and YAP is low, although Yorkie and TAZ do harbour a coiled-coil motif in their putative TAD (predicted using COILS: http://embnet.vital-it.ch/software/COILS_form.html) via other amino acids e.g., I-I-L-L-E in TAZ. Furthermore, as mentioned above, YAP’s TAD is not required for anchorage-independent growth of MCF10A and NIH3T3 cells in vitro [26]. However, YAP’s TAD is necessary for particular oncogenic functions including cell migration and invasion in vitro [26] and in vivo [27], [28]. It is therefore conceivable that YAP’s leucine zipper functions, together with other domains, to enhance transcriptional activity, enabling fine-tuning of YAP-mediated gene transcription by recruiting, or stabilising the association with one or more co-factors, which is dispensable for Yorkie in Drosophila [26].
Data obtained here using full-length YAP constructs suggests the leucine zipper might stabilise the interaction with transcriptional co-factors, possibly recruited via other domains e.g., WW domains. Thus when using GAL4-TAD fusions these regulatory factors would not be recruited into the transcriptional complex and the effects of a disrupted leucine zipper are less obvious. It is also worth noting that differences between the full-length YAP isoforms examined were less apparent when TEAD2 was co-expressed. Here it is likely that exogenous YAP and TEAD2 are the major contributors to transactivation of the luciferase reporter. However, when TEAD proteins are limiting e.g., at endogenous level, other regulatory co-factors may contribute significantly to transactivation. Under these conditions the effect of leucine zipper disruption in addition to the presence of single or tandem WW domains may be more pronounced. It is possible that the observed differences in the absence of exogenous TEAD2 may also be contributed to by subtle differences in binding affinities with endogenous TEAD proteins, or their relative abundance. However, this is unlikely to account for the observed differences since all YAP constructs were transfected at the same time and therefore the abundance of all four endogenous TEAD proteins will be equivalent across all samples.
The biological significance of this data assumes that all YAP isoforms are expressed as functional protein in vivo. Whilst mRNAs of all hYAP isoforms are detectable across a range of tissues and organs except leukocytes [10], it remains to be determined whether all isoforms are functionally expressed and in relative abundance. The differences in transcriptional activity revealed in this study are notable, and suggests the expression of different YAP isoforms may permit fine-tuning of complex transcriptional programmes that may result in different biological outcomes. Future studies could profile the specific expression of hYAP isoforms, for example during development, to determine whether this correlates with biologically relevant outputs e.g., target gene expression.
In conclusion, using overexpression and transcriptional reporter analyses this study demonstrates that hYAP isoforms have significantly different transcriptional activities. The presence of a single or tandem WW domain combined with an intact or disrupted leucine zipper domain contributes to YAP transcriptional activity. Combined with the likelihood that these structural variations will affect participation in a range of protein interactions, this data confirms that one cannot assume YAP’s eight known isoforms function equivalently. Careful selection and comparison of YAP isoforms and cellular context should be undertaken when characterising or ascribing YAP’s function.
Acknowledgements
We thank Marius Sudol, Kieran Harvey and Melvin DePamphilis for providing cDNAs, and David Vaux, David Huang and John Silke for providing cell lines. This work was supported by Grant 09-0084 from Worldwide Cancer Research (formerly the Association of International Cancer Research) and a Research Fellowship and Project grants (APP1010749 and APP1067515) from the Cancer Council Western Australia, Australia awarded to BAC. MLF received an Australian Postgraduate Award and top-up scholarships from the University of Western Australia (UWA) and the National Stem Cell Foundation of Australia. RPS is a recipient of a UWA Prescott Postgraduate Scholarship. JSC is a recipient of an Australian Postgraduate Award and top-up scholarship from UWA.
Footnotes
Transparency Document associated with this article can be found in the online version at doi:10.1016/j.bbrep.2016.02.015.
Contributor Information
Megan L. Finch-Edmondson, Email: phsmlf@nus.edu.sg.
Robyn P. Strauss, Email: Robyn.Strauss@research.uwa.edu.au.
Joshua S. Clayton, Email: Joshua.Clayton@research.uwa.edu.au.
George C. Yeoh, Email: George.Yeoh@uwa.edu.au.
Bernard A. Callus, Email: Bernard.Callus@uwa.edu.au.
Appendix A. Supplementary material
Supplementary material
References
- 1.Yagi R., Chen L.F., Shigesada K. A WW domain-containing yes-associated protein (YAP) is a novel Transcriptional co-activator. EMBO J. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong J., Feldmann G., Huang J. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hao Y.W., Chun A., Cheung K. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J. Biol. Chem. 2008;283:5496–5509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- 4.Huang J.B., Wu S., Barrera J. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Zhang J., Smolen G.A., Haber D.A. Negative regulation of YAP by LATS1 underscores evolutionary conservation of the Drosophila Hippo pathway. Cancer Res. 2008;68:2789–2794. doi: 10.1158/0008-5472.CAN-07-6205. [DOI] [PubMed] [Google Scholar]
- 6.Cao X., Pfaff S.L., Gage F.H. YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes Dev. 2008;22:3320–3334. doi: 10.1101/gad.1726608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao B., Ye X., Yu J. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1–10. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J., Ji J.Y., Yu M. YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. Nat. Cell Biol. 2009;11:1444–1450. doi: 10.1038/ncb1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sudol M., Shields D.C., Farooq A. Structures of YAP protein domains reveal promising targets for development of new cancer drugs. Semin. Cell Dev. Biol. 2012;23:827–833. doi: 10.1016/j.semcdb.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaffney C.J., Oka T., Mazack V. Identification, basic characterization and evolutionary analysis of differentially spliced mRNA isoforms of human YAP1 gene. Gene. 2012;509:215–222. doi: 10.1016/j.gene.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L., Chan S.W., Zhang X. Structural basis of YAP recognition by TEAD4 in the Hippo pathway. Genes Dev. 2010;24:290–300. doi: 10.1101/gad.1865310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z., Zhao B., Wang P. Structural insights into the YAP and TEAD complex. Genes Dev. 2010;24:235–240. doi: 10.1101/gad.1865810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strano S., Munarriz E., Rossi M. Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J. Biol. Chem. 2001;276:15164–15173. doi: 10.1074/jbc.M010484200. [DOI] [PubMed] [Google Scholar]
- 14.Komuro A., Nagai M., Navin N.E. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J. Biol. Chem. 2003;278:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- 15.Oka T., Remue E., Meerschaert K. Functional complexes between YAP2 and ZO-2 are PDZ domain-dependent, and regulate YAP2 nuclear localization and signalling. Biochem. J. 2010;432:461–472. doi: 10.1042/BJ20100870. [DOI] [PubMed] [Google Scholar]
- 16.Oka T., Sudol M. Nuclear localization and pro-apoptotic signaling of YAP2 require intact PDZ-binding motif. Genes Cells. 2009;14:607–615. doi: 10.1111/j.1365-2443.2009.01292.x. [DOI] [PubMed] [Google Scholar]
- 17.Landschulz W.H., Johnson P.F., McKnight S.L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 18.Sudol M. YAP1 oncogene and its eight isoforms. Oncogene. 2013;32 doi: 10.1038/onc.2012.520. 3922-3922. [DOI] [PubMed] [Google Scholar]
- 19.Liu X., Yang N., Figel S.A. PTPN14 interacts with and negatively regulates the oncogenic function of YAP. Oncogene. 2013;32:1266–1273. doi: 10.1038/onc.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Overholtzer M., Zhang J., Smolen G.A. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc. Natl. Acad. Sci. USA. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ehsanian R., Brown M., Lu H. YAP dysregulation by phosphorylation or ΔNp63-mediated gene repression promotes proliferation, survival and migration in head and neck cancer subsets. Oncogene. 2010;29:6160–6171. doi: 10.1038/onc.2010.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oka T., Mazack V., Sudol M. Mst2 and Lats kinases regulate apoptotic function of yes kinase-associated protein (YAP) J. Biol. Chem. 2008;283:27534–27546. doi: 10.1074/jbc.M804380200. [DOI] [PubMed] [Google Scholar]
- 23.Zhang X., Milton C.C., Poon C.L.C. Wbp2 cooperates with Yorkie to drive tissue growth downstream of the Salvador–Warts–Hippo pathway. Cell Death Differ. 2011;18:1346–1355. doi: 10.1038/cdd.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao B., Kim J., Ye X. Both TEAD-binding and WW domains are required for the growth stimulation and oncogenic transformation activity of Yes-associated protein. Cancer Res. 2009;69:1089–1098. doi: 10.1158/0008-5472.CAN-08-2997. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X., Milton C.C., Humbert P.O. Transcriptional output of the Salvador/Warts/Hippo pathway is controlled in distinct fashions in Drosophila melanogaster and mammalian cell lines. Cancer Res. 2009;69:6033–6041. doi: 10.1158/0008-5472.CAN-08-4592. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X., Grusche F.A., Harvey K.F. Control of tissue growth and cell transformation by the Salvador/Warts/Hippo pathway. PLoS One. 2012;7:e31994. doi: 10.1371/journal.pone.0031994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia Y., Chang T., Wang Y. Yap promotes ovarian cancer cell tumorigenesis and is indicative of a poor prognosis for ovarian cancer patients. PLoS One. 2014;9:e91770. doi: 10.1371/journal.pone.0091770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H., Deo M., Thompson R.C. Negative regulation of Yap during neuronal differentiation. Dev. Biol. 2012;361:103–115. doi: 10.1016/j.ydbio.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vassilev A., Kaneko K.J., Shu H.J. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001;15:1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Callus B.A., Mathey-Prevot B. Rapid selection of tetracycline-controlled inducible cell Lines using a green fluorescent-transactivator fusion protein. Biochem. Biophys. Res. Commun. 1999;257:874–878. doi: 10.1006/bbrc.1999.0558. [DOI] [PubMed] [Google Scholar]
- 31.Callus B.A., Mathey-Prevot B. Hydrophobic residues Phe751 and Leu753 are essential for Stat5 transcriptional activity. J. Biol. Chem. 2000;275:16954–16962. doi: 10.1074/jbc.M909976199. [DOI] [PubMed] [Google Scholar]
- 32.Callus B.A., Ekert P.G., Heraud J.E. Cytoplasmic p53 is not required for PUMA-induced apoptosis. Cell Death Differ. 2008;15:213–215. doi: 10.1038/sj.cdd.4402245. [DOI] [PubMed] [Google Scholar]
- 33.Dunning C.J.R., McKenzie M., Sugiana C. Human CIA30 is involved in the early assembly of mitochondrial complex I and mutations in its gene cause disease. EMBO J. 2007;26:3227–3237. doi: 10.1038/sj.emboj.7601748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Finch M.L., Passman A.M., Strauss R.P. Sub-cellular localisation studies may spuriously detect the Yes-associated protein, YAP, in nucleoli leading to potentially invalid conclusions of its function. PLoS One. 2015;10:e0114813. doi: 10.1371/journal.pone.0114813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDonald C.B., McIntosh S.K.N., Mikles D.C. Biophysical analysis of binding of WW domains of the YAP2 transcriptional regulator to PPXY motifs within WBP1 and WBP2 adaptors. Biochemistry. 2011;50:9616–9627. doi: 10.1021/bi201286p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oka T., Schmitt A.P., Sudol M. Opposing roles of angiomotin-like-1 and Zona Occludens-2 on pro-apoptotic function of YAP. Oncogene. 2012;31:128–134. doi: 10.1038/onc.2011.216. [DOI] [PubMed] [Google Scholar]
- 37.Tsutsumi R., Masoudi M., Takahashi A. YAP and TAZ, Hippo signaling targets, act as a rheostat for nuclear SHP2 function. Dev. Cell. 2013;26:658–665. doi: 10.1016/j.devcel.2013.08.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material