ABSTRACT
The autoimmune regulator gene (AIRE) plays a fundamental role in tolerance by promoting the expression of tissue-specific antigens in medullary thymic epithelial cells (mTECs). Recently, AIRE expression was detected also in human keratinocytes and in tumors originating in stratified epithelia. Here, we tested whether AIRE is expressed in cancer cells. We analyzed AIRE expression in cancer cases from The Cancer Genome Atlas (TCGA) RNA-seq dataset and we found association with better outcome. AIRE protein expression was verified by immunohistochemistry in a cohort of 39 human breast cancer specimens and its prognostic relevance was confirmed in microarray-based gene expression data set NKI-295 and KM-Plotter. Both in the RNA-seq and gene expression datasets analyzed, AIRE expression was an independent strong prognostic factor for relapse-free survival (RFS), particularly in estrogen receptor-positive tumors. Enrichment of translation-related pathways was observed in AIRE-expressing tumors by Ingenuity Pathway Analysis and a significant increase of cells in G1 phase and activation of caspase cascades was induced by AIRE transfection in breast cancer luminal cell lines, suggesting that AIRE-induced over-translation of proteins lead to cycle arrest and apoptosis. These data are the first to identify AIRE expression in breast cancer and an association with prognosis.
KEYWORDS: AIRE, Apoptosis, Breast Cancer, Cell-cycle arrest, TCGA, Transcription Factor
Introduction
Central tolerance is achieved by purging self-reactive lymphocytes via the promiscuous expression of genes encoding self-antigens by medullary thymic epithelial cells (mTECs).1-5 mTECs express membrane either proteins normally expressed in the thymus and proteins expressed in peripheral organs, called tissue-specific antigens (TSAs). The transcription factor autoimmune regulator (AIRE) gene (NM_000383.3), by promoting the ectopic expression of thousands of genes encoding TSAs in mTECs, plays a fundamental role in the tolerance process.6 Accordingly, defects in the AIRE gene cause autoimmune polyendocrinopathy candidiasis ectodermal dystrophy (APECED),7 a disease characterized by a loss of self-tolerance to multiple organs. Since individual mTECs transcribe TSA genes stochastically to favor a continuous turnover of presented antigen peptides, AIRE supports mTEC apoptosis.8-10 Moreover, AIRE acts in the tolerance process by favoring the development of regulatory T cells.11,12
Not only is AIRE highly expressed in mTECs, but there is evidence of AIRE expression in murine peripheral lymphoid organs.13-15 The characteristics and functions of extra-thymic AIRE- expressing cells (eTACs) have recently been described; they are a CD45low bone marrow-derived antigen-presenting population, mediating tolerance primarily by inducing inactivation of effectors cells through a mechanism involving deficient co-stimulation.13 Very recently, AIRE expression was reported in human and mouse keratinocytes and in tumors originating in stratified and pseudostratified epithelia, as head and neck squamous carcinomas.16,17 Thus, the shared feature of promiscuous expression of some genes by mTECs and cancer cells18,19 raises the possibility that analogous mechanisms of transcriptional regulation operate in both cell types and that AIRE, as part of or a consequence of such mechanisms, is ectopically activated in some solid human tumors.
To test whether AIRE is expressed in tumors, we analyzed AIRE expression in an RNA-seq database focused on breast cancer patients, for whom the RNA-seq public data set is the largest among all tumors, and in 2 gene expression datasets. We detected AIRE expression in one-third of tumors specimens and found that AIRE-positive tumors had a better relapse-free survival (RFS) rate compared with those AIRE-negative. These data suggest an unexpected involvement of AIRE in the cancer scenario.
Results
AIRE is expressed in human breast cancer and is associated with a good prognosis
We analyzed The Cancer Genome Atlas (TCGA) database (http://cancergenome.nih.gov/), in which whole-genome transcription is analyzed by RNA-seq, for AIRE expression. In the breast cancer cohort (n = 1036) included in the TCGA data set, AIRE expression levels were divided into 2 sample groups according to AIRE counts (see Materials and Methods): AIRE-negative represents 66.6% of the cohort describing 690 cases and AIRE-positive represents 33.3% of the cohort describing 346 cases (Fig. 1A). Kaplan-Meier analysis of cases for whom survival information was available (n = 787) showed that patients with AIRE-positive tumors had a better Relapse Free Survival (RFS) than those with AIRE-negative tumors (Fig. 1B; Table 1). Association between AIRE expression and better prognosis was observed also in pancreatic and head and neck tumors (Fig. S1). Focusing on breast cancer, AIRE expression was investigated in 2 public gene expression breast cancer datasets, NKI-29520 and KM-Plotter,21 in which the presence of AIRE transcript was detected using technology based on the mRNA hybridization to specific probe sets. Multivariate analysis of AIRE expression and covariates significantly associated with prognosis in univariate analysis (p < 0.05) indicated that AIRE was an independent prognostic factor for RFS (Table 1).
Figure 1.
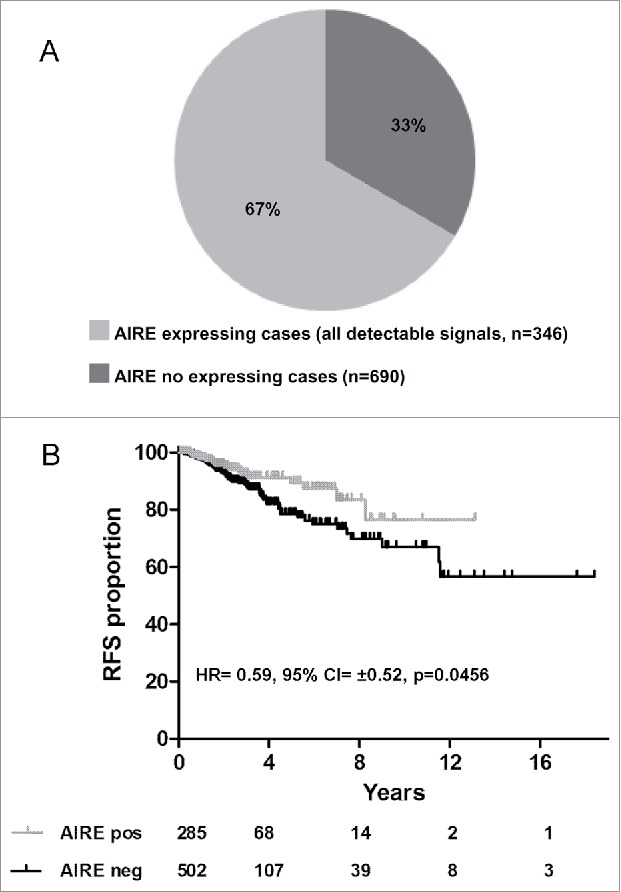
AIRE expression analysis in TCGA breast cancer dataset and clinical association of AIRE expression status with RFS. Frequency of AIRE transcript in breast cancer cases from the TCGA data set (n = 1036) (A). Kaplan-Meier relapse-free survival curves of TCGA breast cancer patients are stratified by their expression levels of AIRE: cases were classified as AIRE positive or -negative according to whether the expression of AIRE gene was revealed in the dataset (A). Log-rank test p-values were: 0.0456 considering all cases (B).
Table 1.
Univariate and multivariate analysis of risk factors associated with relapse-free survival.
| TCGA data set | ||||||
|---|---|---|---|---|---|---|
| Univariate analysis |
Multivariate analysis |
Multivariate analysis in ER-positive |
||||
| Variable | HRb (95% CIc±) | p-value | HRb (95% CIc±) | p-value | HRb (95% CIc±) | p-value |
| AIRE positivity | 0.59 (0.52) | 0.0456 | 0.46 (0.58) | 0.0092 | 0.39 (0.77) | 0.0181 |
| Tumor size >2 cm | 1.98 (0.59) | 0.0243 | 1.54 (1.03) | 0.4145 | 4.25 (1.99) | 0.1548 |
| Tumor stage II,III,IV | 3.57 (0.91) | 0.0062 | 1.45 (1.42) | 0.6119 | 0.44 (2.28) | 0.4842 |
| ER positivity | 0.61 (0.49) | 0.0457 | 0.51 (0.51) | 0.0097 | ||
| N positivity | 2.51 (0.50) | 0.0004 | 2.21 (0.57) | 0.0062 | 2.09 (0.69) | 0.034 |
| NKI-295 dataset | ||||||
| Univariate analysis |
Multivariate analysis |
Multivariate analysis in ER-positive |
||||
| Variable |
HRb (95% CIc±) |
p-value |
HRb (95% CIc±) |
p-value |
HRb (95% CIc±) |
p-value |
| AIRE positivity | 0.63 (0.41) | 0.0260 | 0.54 (0.42) | 0.0039 | 0.52 (0.54) | 0.0168 |
| Tumor size >2 cm | 1.71 (0.37) | 0.0041 | 1.41 (0.38) | 0.0772 | 1.43 (0.45) | 0.1221 |
| ER positivity | 0.51 (0.39) | 0.0006 | 0.59 (0.43) | 0.0165 | ||
| Grade III | 2.02 (0.36) | 0.0002 | 1.65 (0.41) | 0.0178 | 1.92 (0.46) | 0.0054 |
| KM-Plotter | ||||||
| Univariate analysis |
Multivariate analysisd |
Multivariate analysis in ER-positived |
||||
| Variable |
HRb (95% CIc±) |
p-value |
HRb (95% CIc±) |
p-value |
HRb (95% CIc±) |
p-value |
| AIRE positivity | 0.83 (0.13) | 0.0033 | 0.84 (0.74–0.96) | 0.0081 | 0.79 (0.65–0.97) | 0.0221 |
| ESR1 positivitye | 0.65 (0.12) | <10−6 | 0.64 (0.56–0.74) | <10−16 | ||
| MKI67 positivitye | 1.43 (0.11) | <10−6 | 1.25 (1.1–1.42) | 0.0033 | 1.4 (1.16–1.69) | 0.0005 |
Multivariate Cox proportional hazards regression analysis was performed using WinSTAT version 2009.1 for Microsoft Excel with direct elimination, and all predictors significantly associated with prognosis in univariate analysis (p < 0.05) were considered.
Hazard ratio (HR) estimated from Cox proportional hazard regression model.
Confidence interval of the estimated HR.
Multivariate analysis of this data set was performed online on KM-Potter website considering AIRE expression and that of the only available covariates: ESR1 and MKI67 expression.
median expression cut-off.
AIRE expression was analyzed in primary breast cancer specimens from the NKI-295 data set (n = 295) profiled on Agilent Hu25k microarray, and from the KM-Plotter breast cancer database (n = 3554) profiled on Affymetrix HG-U133A microarrays. To divide positive and negative AIRE tumor groups we took advantage of the AIRE data from TCGA, which defined 33% of the breast cancer cases as AIRE-positive; cases were then divided into tertiles and the third-quantile containing cases with higher AIRE expression levels was defined as AIRE-positive and compared with the remaining AIRE-negative cases. Kaplan-Meier analysis again indicated a better RFS in patients with AIRE-positive tumors compared with AIRE-negative patients in both database (Fig. 2A, B) and a significant association with a low risk of relapse in multivariate analysis (Table 1).
Figure 2.
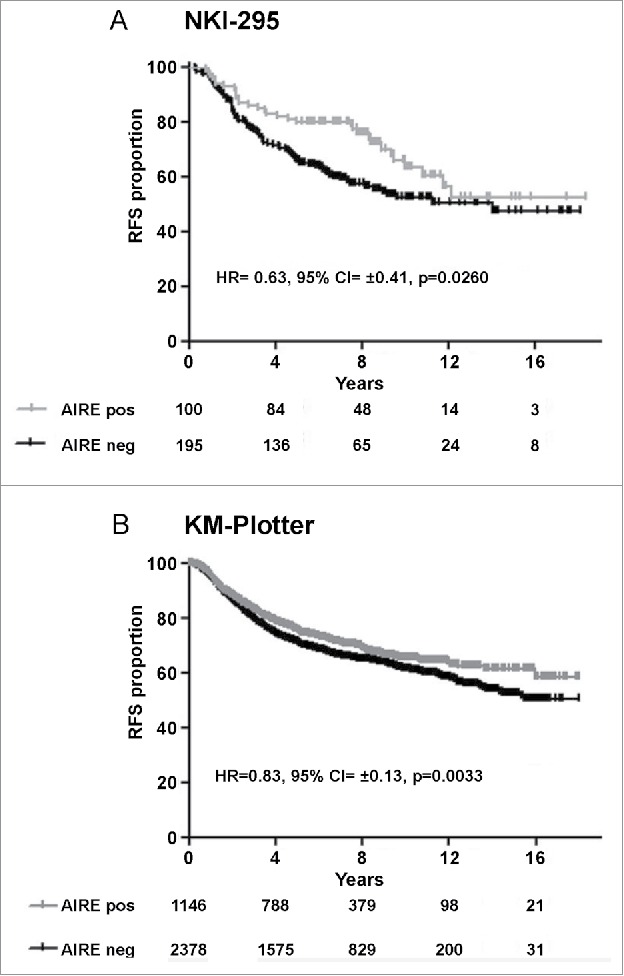
Clinical association of AIRE expression status with prognosis in gene expression breast cancer data set. Kaplan-Meier plots of relapse-free survival (RFS) of breast cancer patients in NKI-295 dataset (A) and KM-plotter data set (B).
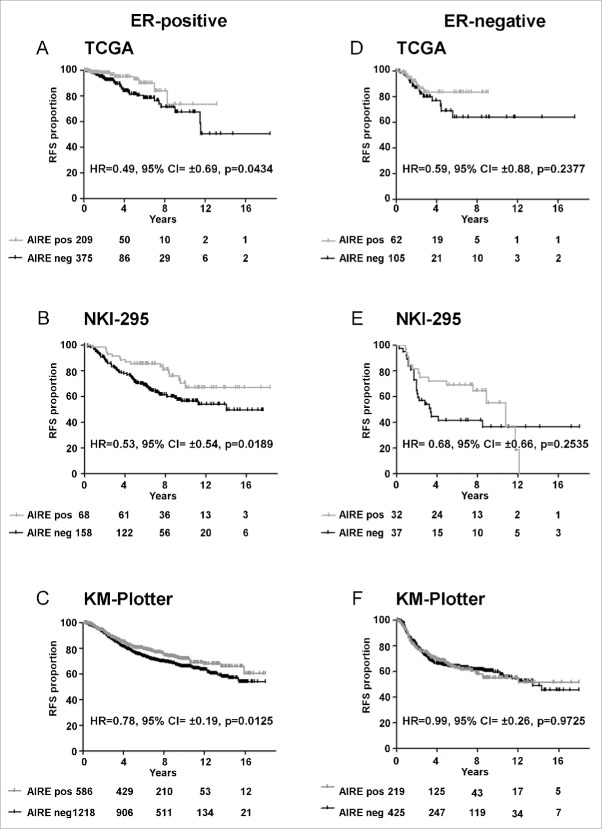
The prognostic significance of AIRE expression was evaluated in the 2 main common types of breast cancer disease, estrogen receptor (ER)-positive and ER-negative, representing very distinct diseases with different prognosis; patients of all the 3 datasets were stratified for ER immunohistochemical expression and a statistically significant association were revealed between AIRE expression and good prognosis only in ER-positive breast cancer patients (Fig. 3A, B, C). However, ER-negative samples size was too small to test the relation between AIRE expression with outcome from these patients (Fig. 3D, E, F).
Figure 3.
Prognostic significance of AIRE expression in ER-positive and –negative breast tumor. Kaplan-Meier plots of relapse-free survival (RFS) of breast cancer patients in TCGA, NKI-295 and KM-plotter dataset according with ER IHC expression.
We analyzed the expression of AIRE protein in a cohort of 39 human ER-positive primary breast cancer specimens. Formalin-fixed, paraffin-embedded (FFPE) sections were analyzed by immunohistochemistry (IHC) with an antibody recognizing AIRE protein. 28% (11/39) of cases were AIRE-positive (Fig. 4A, B), AIRE expression in tumors resulted not diffuse but focal (tumors were considered positive when ≥5 % of tumor cells showed nuclear reactivity; mean AIRE IHC-positive tumor cells/section = 27%, range 5–50%). Specificity of the staining was assessed on normal thymus tissue sections, where it was observed the ability of the antibody to detect AIRE protein expressed in mTECs (Fig. 4C).
Figure 4.
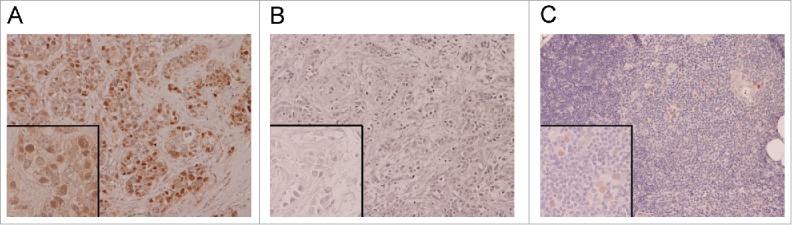
AIRE protein expression in human breast cancer. FFPE ER-positive breast cancer sections were immunostained with anti-AIRE monoclonal antibody and nuclear staining intensity was scored as positive (A) or negative (B). Normal human thymus section was used as positive control for MECs staining (C). Magnification 20X (insets 40X).
Application of Ingenuity Pathways Analysis (IPA) to explore functional networks and pathways in AIRE-positive breast cancers of the NKI-295 data set indicated that the top canonical pathways differentially modulated in AIRE-positive versus -negative patients were related to ribosome-mediated translation-related processes (Fig. S2) and composed mainly of eukaryotic translation initiation factors, ribosomal and ribonucleoproteins, poly(A) binding proteins, cytoplasmic RNA binding motif and zinc-finger proteins (Table S1 for a full list of significant canonical pathways).
The upregulation of oxidative phosphorylation and mitochondrial dysfunction pathways observed in AIRE-positive patients is consistent with reported AIRE involvement in generation of apoptotic signals from genotoxic or oxidative stress in mTECs. These data are consistent with AIRE activity as a transcription inducer and suggest that AIRE is functionally active in breast cancer.
Of note, ER-positive patients showed the same enriched canonical pathways emerged by the analysis of the entire dataset (Fig. S2), whereas in ER-negative patients, the low number of differentially expressed genes between AIRE-positive and -negative patients precluded IPA analysis.
AIRE expression reduces aggressiveness features in luminal breast cancer cell lines
To investigate the biological impact of AIRE expression on ER-positive breast cancers, MCF-7 and MDA-MB-361 cells, prototypes of the breast cancer molecular subtypes luminal A and luminal B, respectively, were transiently transfected with AIRE or empty vector for 72 hr and tested by cell cycle analysis and apoptosis assay (Fig. 5A).
Figure 5.
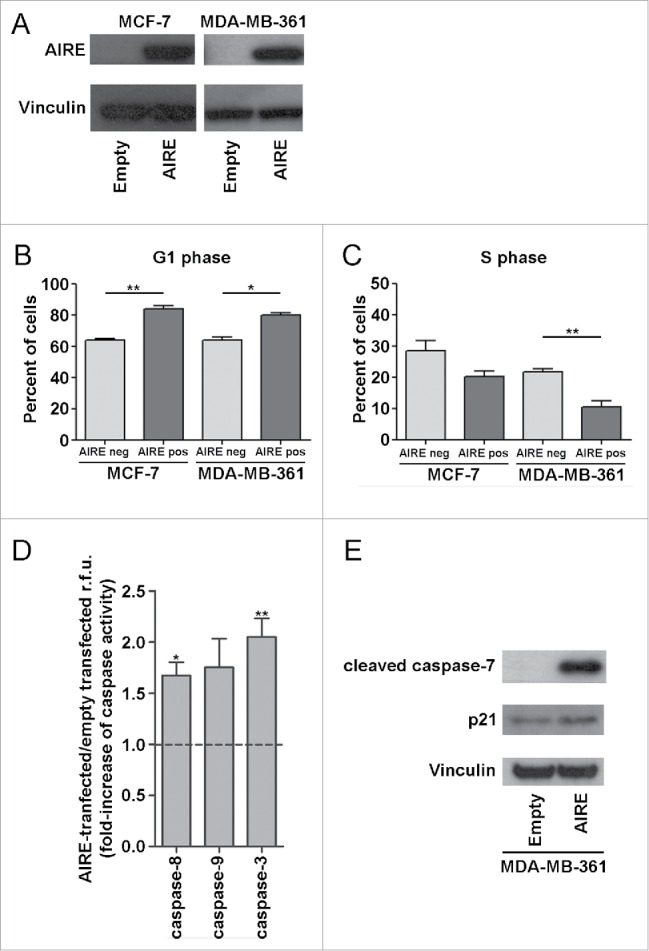
Effects of AIRE expression on cell cycle and apoptosis of luminal breast cancer cell lines. (A) AIRE protein expression was evaluated in MCF-7 and MDA-MB-361 luminal breast cancer cells transiently transfected with AIRE-expressing or empty vector after 72 h by Western blot with anti-AIRE mouse monoclonal antibody. Vinculin was used to normalize total protein loading using anti-vinculin. The distribution of AIRE-transfected cells in phases of the cell cycle was evaluated by staining with anti-AIRE antibody and propidium iodide. In MCF-7 and MDA-MB-361 cells 72 h after transfection with AIRE, the percentage of AIRE-positive cells in G1 (B) and S phases (C), as assessed by propidium iodide assay, was determined as compared to AIRE-negative cells. Data represent the mean ± s.e.m. of at least 3 independent experiments. *p < 0.05, **p < 0.01. D) Adherent cells were trypsinized and the catalytic activity of caspase-9, −3, and −8 was measured as described in Methods. Data are expressed as fold-increase with respect to cells transfected with empty vector. E) MDA-MB-361 cells transiently transfected with AIRE-expressing or empty vector were analyzed for cleaved caspase-7 and p21 protein expression after 72 h by Western blot by using anti-cleaved caspase-7 rabbit polyclonal antibody and anti-p21 polyclonal antibody. Vinculin was used to normalize total protein loading.
Both in MCF-7 and MDA-MB-361 cells expressing AIRE, flow cytometry analysis revealed a significant increase in the percentage of G1-phase cells and a corresponding reduction in the percentage of S-phase cells (Fig. 5B, C). Analysis of the effect of AIRE expression on apoptosis showed that caspase-3, −8 and −9 catalytic activity was significantly increased in MDA-MB-361 cells transiently transfected with AIRE as compared with empty vector-transfected cells (Fig. 5D). Activation of the caspase pathways in MDA-MB-361 AIRE-expressing cells was confirmed by the increased amount of cleaved caspase-7 and p21 proteins level (Fig. 5E).
Discussion
Our analyses revealed that AIRE is ectopically activated in a percentage of breast cancer specimens and that its expression correlates with good prognosis. Results of the TCGA RNA-seq data analysis of 1036 breast cancer cases, the largest RNA-seq breast cancer data set currently available, indicated that 33% of breast tumor specimens display AIRE mRNA, consistent with results from analysis of the 2 public microarray datasets NKI-29520 and KM-Plotter breast cancer database22 comprising 295 and ≥3000 patients, respectively. Moreover, we show for the first time AIRE protein expression in breast cancer specimens, and frequency of AIRE protein expression resemble mRNA detection by in silico analysis.
Analysis in all 3 data sets stratified for ER immunohistochemical expression revealed a statistically significant association between AIRE expression and good prognosis in ER-positive breast cancer patients but not in ER-negative tumors. However, the number of ER-negative tumors in the analyzed databases is too small to completely exclude any relationship between AIRE expression and outcome in these patients. Neither in the TCGA nor in the NKI dataset AIRE expression level was different between ER-positive and –negative patience.
In our 39-breast cancer cohort we analyzed for AIRE protein expression, there were only 2 relapsing patients, as expected since they were all ER-positive, and both were negative for AIRE expression.
Considering that ER and AIRE are both transcription factors, the association of AIRE expression prognostic significance with ER expression might rest in a role for ER protein in regulation of transcriptional activity exerted by AIRE. Consistent with this hypothesis, in ER-negative patients were found very few differentially expressed genes between AIRE-positive and –negative tumors. Whereas all of the in silico data sets analyzed indicated AIRE expression in breast cancer cells, we cannot exclude the possibility that the AIRE mRNA observed in tumor specimens derive from immune cells infiltrating the tumor. Indeed, analysis of the NKI-295 dataset indicated an association between MHC II and AIRE expression (data not shown), which might reflect infiltration of tumors by dendritic cells, consistent with the reported expression of human leukocyte antigen-DR in extrathymic AIRE-positive immune cells.15 However, the putative presence of these tolerogenic AIRE-expressing immune cells within the tumor contrasts with good prognosis of breast cancer patients expressing AIRE. In any case, the association of AIRE expression with good prognosis suggests that AIRE expression in breast tumors does not promote self-tolerance as it does in mTECs and eTACs. Most important, AIRE protein nuclear expression was confirmed on FFPE breast cancer specimens with a frequency of AIRE protein expression resemble mRNA detection by in silico analysis.
Our in vitro analysis of MCF-7 and MDA-MB-361, prototypes of ER-positive breast cancer subtypes luminal A and B, respectively, after transfection with an AIRE-expressing vector showed that AIRE expression in these cells affects cell cycle progression and apoptosis. The involvement of AIRE in proliferation and apoptosis has been demonstrated in thymic medullary epithelial cells (MECs), which reportedly show less proliferation but rapid turnover due to apoptosis compared to AIRE non-expressing MECs.10 Accordingly, loss of AIRE was observed in both human and murine thymomas.23,24 Other studies indicated a role of AIRE in increasing apoptosis when expressed in thymic and thyroid epithelial models8–9, suggesting that overproduction of TSAs rather than induction of an apoptotic program might indirectly induce cellular stress leading to death. Such a role does not conflict with the nature of AIRE as a transcription factor, since it favors a continuous turnover of antigen-presenting mTECs and promote cross-presentation of tissue-specific antigens to developing thymocytes.25 Our analysis of the NKI-295 data set strongly confirmed the association between AIRE expression and activation of translation-related processes and upregulation of pathways involved in the oxidative cell stress response.
Our findings of a positive role for AIRE in breast cancer contrast with its pro-inflammatory and pro-tumorigenic role reported in keratinocytes.16 AIRE-mediated expression of pro-inflammatory transcripts in skin tumors strictly depends on KRT17 and hnRNP K expression. KRT17 and hnRNP K expression did not differ significantly between AIRE-positive and –negative breast tumors neither in the TCGA nor the NKI-295 dataset (data not shown). Evidences demonstrated that genes expression, modulated by AIRE transcriptional activity, is influenced by a vast repertoire of partners and that the cellular environment has been shown to govern which genes are susceptible to AIRE regulation, mainly through epigenetic processes. Indeed, when AIRE is introduced into different cell lines, such as U937 monocytic human leukemic lymphoma cells, HEK293 human embryonic kidney epithelial cells, pancreatic β cells or the 1C6 TEC line, the large repertoire of AIRE-induced genes varies by cell type.26-29 Moreover, AIRE transcriptional activity can be also affected by genetic polymorphisms as recently reported by Conteduca et al.30
In conclusion, our results reveal for the first time AIRE expression in breast cancers and its association with better RFS, providing compelling evidence for the relevance of this unusual transcription factor as a new marker for breast carcinoma. Analysis of pathways modulated by AIRE expression and investigation in luminal cell line models of AIRE expression effects highlight AIRE involvement in cell-cycle arrest and apoptosis, raising the possibility that AIRE-induced over-translation of proteins lead to cycle arrest and apoptosis, thus contributing to a better prognosis in AIRE-expressing patients.
Materials and methods
AIRE expression in human breast cancer samples evaluated with RNA-seq
To define the distribution of AIRE expression within breast tumors, we examined the RNA-seq data from the TCGA data set, analyzing 1036 tumors. RSEM normalized gene expression data (level 3) were downloaded from the Cancer Genome Browser (https://genome-cancer.ucsc.edu/; January 11 2015), together with the clinical information for the cohort. The expression level of AIRE in the cohort was assessed using the following strategies: samples were filtered based on their expression values, considering samples with negative or 0 values as non-expressing, while samples with expression values greater than 0 were considered as expressing. The results shown here are based on data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
AIRE expression in human breast cancer samples evaluated with gene expression microarray
Public gene expression data from the NKI breast cancer dataset containing 295 cases20 were used to investigate AIRE expression based on hybridization of cDNA and oligonucleotide DNA chips. Microarray and clinical data were retrieved from the Computational Cancer Biology website present at The Netherlands Cancer Institute (http://ccb.nki.nl/data/). Gene annotation was performed using the SOURCE Database (http://source-search.princeton.edu/).
AIRE expression was assessed in a large meta-analysis of breast cancer data sets performed on Affymetrix platform.21 The online KM-Plotter database (http://www.kmplot.com), which currently includes information on 22,277 genes and their influence on survival in 4,142 breast cancer patients, was used for survival analysis.
Statistical analyses
The association between AIRE expression and categorical patho-biologic variables was assessed using the Fisher's exact test. Clinical data were accessed when available. Two-sided p < 0.05 was considered statistically significant. Relapse-free survival (RFS) was defined as the time elapsed from the date of surgery to the date of the first event. Univariate analysis was carried out based on the Kaplan-Meier curves for all categorical predictors using the log-rank test; multivariate Cox proportional hazards regression analysis was performed using WinSTAT version 2009.1 for Microsoft Excel with direct elimination, and all predictors with p ≤ 0.05 were considered. To define possible associations with RFS, tumors were stratified according to the ER immunohistochemical status listed in the clinical information database.
Patients
Samples from 39 breast cancer patients diagnosed during 2007 in our Institute (Fondazione IRCCS Istituto Nazionale dei Tumori) were selected based on IHC criteria as ER-positive (>1 % cell positivity for estrogen receptor, progesterone receptor) and availability of follow-up. All patients gave written consent to use their biological materials for future investigations and research purposes, and the study did not require further Institutional approval from the Ethics Committee. All data were analyzed anonymously and all experiments were in compliance with the Helsinki Declaration of 1975. The median follow-up of the cohort of 39 patients was 5.3 y.
Immunohistochemistry
Expression of AIRE was analyzed by IHC on 3-µm formalin-fixed, paraffin-embedded (FFPE) tumor sections, using rat monoclonal anti-AIRE antibody (Ebioscience, 14-9534-80, http://www.ebioscience.com/human-aire-antibody-purified-tm-724.htm) after antigen retrieval carried out by heating slides for 5 min at 96°C in 10 mM citrate buffer, pH 6.0. Immunoreactions were visualized using streptavidin-biotin-peroxidase (Dako-Agilent, P0397, http://www.dako.com/it/ar38/p235649/prod_products.htm), 3,31-diaminobenzidine (Dako-Agilent, K3468, http://www.dako.com/it/ar38/p107380/prod_products.htm) used as a chromogenic substrate, and sections were counterstained with hematoxylin. Images were acquired on an ECLIPSE TE2000-S inverted microscope (Nikon, Eclipse-TE2000, https://www.nikoninstruments.com/en_EU/Products/Inverted-Microscopes/Eclipse-TE2000) at 20X and 40X magnifications. Reactivity of anti-AIRE antibody was scored as positive when ≥5 % of tumor cells showed nuclear staining.
Cell lines, plasmids and transfection
Human breast cancer cell lines MCF-7 (ATCC, ATCC HTB-22, https://www.lgcstandards-atcc.org/Products/All/HTB-22.aspx) and MDA-MB-361 (ATCC, ATCC HTB-27, https://www.lgcstandards-atcc.org/Products/All/HTB-27.aspx), prototypes of molecular subtypes luminal A and luminal B, respectively, were purchased from ATCC (Rockville, MD). The cell lines were authenticated every years by the Fragment Analysis Facility at INT, using the GenePrint 10 System (Promega, B9510, https://ita.promega.com/products/cell-authentication-sample-identification/cell-line-authentication/geneprint-10-system/). Profiles were matched to their original profiles in the cell line database at ATCC. Cells were maintained in RPMI 1640 (Thermo Fisher Scientific Inc., 31870082, https://www.thermofisher.com/order/catalog/product/31870082) or Dulbecco's modified Eagle's medium (Thermo Fisher Scientific Inc., 41965062, https://www.thermofisher.com/order/catalog/product/41965062) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific Inc., 16000036, https://www.thermofisher.com/order/catalog/product/16000036) and 2 mM glutamine (Sigma-Aldrich, G7513, http://www.sigmaaldrich.com/catalog/product/sigma/g7513?lang=itandregion=IT) at 37°C in a 5% CO2 atmosphere.
The plasmid pcDNA3 encoding human AIRE and used to stably or transiently transfect parental cells was produced by amplifying full-length human AIRE cDNA from human thymus total RNA (Clontech,636549,http://www.clontech.com/IT/Products/cDNA_Synthesis_and_Library_Construction/RNA/Total_RNA_Human/Internal_Organ?sitex=10023:22372:US) and subcloning the fragment in the HindIII (New England Biolabs, R0104S, https://www.neb.com/products/r0104-hindiii) and ApaI (New England Biolabs, R0114S, https://www.neb.com/products/r0114-apai) sites of pcDNA3. Cells plated in 6-well plates were transfected with expression plasmids using Lipofectamine® 2000 Transfection Reagent (Thermo Fisher Scientific Inc., 11668019, https://www.thermofisher.com/order/catalog/product/11668019). To obtain stably transfected cells for gene expression analyses, bulk cultures of pcDNA3/AIRE- and pcDNA3 empty vector-transfected MCF-7 and MDA-MB-361 cells were selected and maintained in culture medium supplemented with 500 and 800 mg/ml G418 (Sigma-Aldrich, A1720, http://www.sigmaaldrich.com/catalog/product/sigma/a1720?lang=itandregion=IT), respectively. Transiently transfected cells were analyzed 72 hr after transfection.
Ingenuity pathway analysis (IPA)
Differentially expressed genes in AIRE-positive vs. AIRE-negative patients from the NKI-295 dataset were identified by Partek's Two-Way ANOVA, incorporating AIRE expression (positive vs. negative) as a factor in the analysis. Differentially expressed genes were filtered by the Benjamini-Hochberg false-discovery rate procedure (P < 0.10). Functional gene network analysis was conducted using QIAGEN's IPA (QIAGEN, Redwood City, CA; www.qiagen.com/ingenuity), which transforms large data sets into a group of relevant networks with direct and indirect relationships between genes based on interactions known in the literature. Pathways significantly regulated in gene expression between AIRE-positive vs. AIRE-negative patients were examined using the Core Analysis function included in IPA.
Antibodies
Biochemical analysis used anti-vinculin (Sigma-Aldrich, V9131, http://www.sigmaaldrich.com/catalog/product/sigma/v9131?lang=itandregion=IT), anti-AIRE mouse monoclonal antibody (Santa cruz Biotechnology, sc-373703, http://www.scbt.com/datasheet-373703-aire-1-c-2-antibody.html), anti-cleaved caspase-7 rabbit polyclonal antibody (Cell Signaling Technology, 9491, https://www.cellsignal.com/products/primary-antibodies/cleaved-caspase-7-asp198-antibody/9491) and anti-p21 polyclonal antibody (Cell Signaling Technology, 2947, https://www.cellsignal.com/products/primary-antibodies/p21-waf1-cip1-12d1-rabbit-mab/2947). Cytofluorimetric analysis used anti-AIRE mouse monoclonal antibody (Santa Cruz) and Alexa Fluor 488-conjugated goat anti-mouse IgG (Thermo Fisher Scientific Inc., A-11001, https://www.thermofisher.com/antibody/product/Goat-anti-Mouse-IgG-H-L-Secondary-Antibody-Polyclonal/A-11001).
Western blotting
Crude cell lysates were obtained by lysis in TNTG buffer containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100 (Sigma-Aldrich, T8787, http://www.sigmaaldrich.com/catalog/product/sigma/t8787?lang=itandregion=IT), 10% (v/v) glycerol, 2 mM Na-orthovanadate, and protease inhibitor cocktail (Sigma-Aldrich, 000000011836153001,http://www.sigmaaldrich.com/catalog/product/roche/11836153001?lang=itandregion=IT) for 20 min at 4°C. Insoluble material was removed by 10-min centrifugation at 13000 rpm at 4°C. Protein concentrations were determined using Coomassie protein assay (Thermo Fisher Scientific Inc., 23236, https://www.thermofisher.com/order/catalog/product/23236). Samples were separated on NuPage SDS-Bis-Tris gels (Thermo Fisher Scientific Inc., NP0315BOX, https://www.thermofisher.com/order/catalog/product/NP0315BOX), transferred to PVDF membranes (Millipore, IPVH00010, https://www.merckmillipore.com/IT/it/product/Membrana-Immobilon-P%2C-PVDF%2C-0%2C45%C2%A0%C2%B5m%2C-rotolo-da26%2C5%C2%A0cmx%C2%A03%2C75%C2%A0m,MM_NF-IPVH00010?bd=1#anchor_DS) and examined with ECL and ECL Plus detection reagents (Genespin, STS-E500, http://www.genespin.com/www.genespin.com/PDS.html).
Flow cytometry and cell cycle analysis
The distribution of AIRE-transfected cells in phases of the cell cycle was evaluated by staining with anti-AIRE antibody and propidium iodide. Adherent cells were trypsinized, fixed and permeabilized with Foxp3/Transcription Factor Staining Buffer Set (Ebioscience, 00-5523-00, http://www.ebioscience.com/foxp3-staining-buffer-set.htm) adapted for AIRE staining, and analyzed on a FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ). The percentage of cells in S and G phases was determined using ModFit LT software (Verity Software House, Topsham, ME).
Apoptosis analysis
Adherent cells were trypsinized and the catalytic activity of caspase-9, -3, and -8 was measured as the ability to cleave the specific substrates N-acetyl-Leu-Glu-His-Asp-AMC (LEHD-AMC, MBL, 4810, https://www.mblintl.com/products/4810), N-acetyl-Asp-Glu-Val-Asp-AMC (DEVD-AMC, MBL, 4800, https://www.mblintl.com/products/4800) and N-acetyl-Ile-Glu-Thr-Asp-AMC (IETD-AMC, MBL, 4805, https://www.mblintl.com/products/4805) using the respective APCYTO/caspase assay kits (MBL International). Hydrolysis of the specific substrates for the different caspases was monitored by spectrofluorometry with 380-nm excitation and 460-nm emission filters.
Supplementary Material
Abbreviations
- AIRE
autoimmune regulator gene
- mTECs
medullary thymic epithelial cells
- TCGA
The Cancer Genome Atlas
- RFS
relapse-free survival
- OS
overall survival
- IPA
Ingenuity Pathway Analysis
- TSAs
tissue-specific antigens
- eTACs
extra-thymic AIRE- expressing cells
- ER
estrogen receptor
- FFPE
Formalin-fixed, paraffin-embedded
- IHC
immunohistochemistry
- hr
hours
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank: Mrs. G. Abolafio and Dr. I. Muradore, staff members of the Flow Cytometry Core Facilities; Dr. M. Pennati for helpful data discussion; Mrs. C. Ghirelli and Mrs. P. Aiello for technical assistance and Dr. C. Vinci and Dr. C. Storti for technical contribution; Mrs. L. Mameli for manuscript preparation (Fondazione IRCCS Istituto Nazionale Tumori).
Funding
This work was supported by AIRC (Associazione Italiana per la Ricerca sul Cancro) under Grant 15190. Michele Sommariva is supported by a fellowship from the “Fondazione Umberto Veronesi”
ORCID
Francesca Bianchi http://orcid.org/0000-0001-5197-5279
Michele Sommariva http://orcid.org/0000-0002-7622-0996
Lucia Sfondrini http://orcid.org/0000-0003-0350-5402
References
- [1].Aschenbrenner K, D'Cruz LM, Vollmann EH, Hinterberger M, Emmerich J, Swee LK, Rolink A, Klein L. Selection of Foxp3+ regulatory T cells specific for self antigen expressed and presented by Aire+ medullary thymic epithelial cells. Nat Immunol 2007; 8:351–8; PMID:17322887; http://dx.doi.org/ 10.1038/ni1444 [DOI] [PubMed] [Google Scholar]
- [2].Metzger TC, Anderson MS. Control of central and peripheral tolerance by Aire. Immunol Rev 2011; 241:89–103; PMID:21488892; http://dx.doi.org/ 10.1111/j.1600-065X.2011.01008.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cowan JE, McCarthy NI, Parnell SM, White AJ, Bacon A, Serge A, Irla M, Lane PJ, Jenkinson EJ, Jenkinson WE, et al.. Differential requirement for CCR4 and CCR7 during the development of innate and adaptive alphabetaT cells in the adult thymus. J Immunol 2014; 193:1204–12; PMID:24990081; http://dx.doi.org/ 10.4049/jimmunol.1400993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zhu ML, Nagavalli A, Su MA. Aire deficiency promotes TRP-1-specific immune rejection of melanoma. Cancer Res 2013; 73:2104–16; PMID:23370329; http://dx.doi.org/ 10.1158/0008-5472.CAN-12-3781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Yang S, Fujikado N, Kolodin D, Benoist C, Mathis D. Immune tolerance. Regulatory T cells generated early in life play a distinct role in maintaining self-tolerance. Science 2015; 348:589–94; PMID:25791085; http://dx.doi.org/ 10.1126/science.aaa7017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von BH, Bronson R, Dierich A, Benoist C, et al.. Projection of an immunological self shadow within the thymus by the aire protein. Science 2002; 298:1395–401; PMID:12376594; http://dx.doi.org/ 10.1126/science.1075958 [DOI] [PubMed] [Google Scholar]
- [7].Nagamine K, Peterson P, Scott HS, Kudoh J, Minoshima S, Heino M, Krohn KJ, Lalioti MD, Mullis PE, Antonarakis SE, et al.. Positional cloning of the APECED gene. Nat Genet 1997; 17:393–8; PMID:9398839; http://dx.doi.org/ 10.1038/ng1297-393 [DOI] [PubMed] [Google Scholar]
- [8].Liiv I, Haljasorg U, Kisand K, Maslovskaja J, Laan M, Peterson P. AIRE-induced apoptosis is associated with nuclear translocation of stress sensor protein GAPDH. Biochem Biophys Res Commun 2012; 423:32–7; PMID:22613203; http://dx.doi.org/ 10.1016/j.bbrc.2012.05.057 [DOI] [PubMed] [Google Scholar]
- [9].Colome N, Collado J, Bech-Serra JJ, Liiv I, Anton LC, Peterson P, Canals F, Jaraquemada D, Alvarez I. Increased apoptosis after autoimmune regulator expression in epithelial cells revealed by a combined quantitative proteomics approach. J Proteome Res 2010; 9:2600–9; PMID:20218732; http://dx.doi.org/ 10.1021/pr100044d [DOI] [PubMed] [Google Scholar]
- [10].Gray D, Abramson J, Benoist C, Mathis D. Proliferative arrest and rapid turnover of thymic epithelial cells expressing Aire. J Exp Med 2007; 204:2521–8; PMID:17908938; http://dx.doi.org/ 10.1084/jem.20070795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Malchow S, Leventhal DS, Nishi S, Fischer BI, Shen L, Paner GP, Amit AS, Kang C, Geddes JE, Allison JP, et al.. Aire-dependent thymic development of tumor-associated regulatory T cells. Science 2013; 339:1219–24; PMID:23471412; http://dx.doi.org/ 10.1126/science.1233913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lei Y, Ripen AM, Ishimaru N, Ohigashi I, Nagasawa T, Jeker LT, Bosl MR, Hollander GA, Hayashi Y, Malefyt RW, et al.. Aire-dependent production of XCL1 mediates medullary accumulation of thymic dendritic cells and contributes to regulatory T cell development. J Exp Med 2011; 208:383–94; PMID:21300913; http://dx.doi.org/ 10.1084/jem.20102327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gardner JM, Metzger TC, McMahon EJ, Au-Yeung BB, Krawisz AK, Lu W, Price JD, Johannes KP, Satpathy AT, Murphy KM, et al.. Extrathymic Aire-expressing cells are a distinct bone marrow-derived population that induce functional inactivation of CD4(+) T cells. Immunity 2013; %19;39:560–72; PMID:23993652; http://dx.doi.org/ 10.1016/j.immuni.2013.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gardner JM, Devoss JJ, Friedman RS, Wong DJ, Tan YX, Zhou X, Johannes KP, Su MA, Chang HY, Krummel MF, et al.. Deletional tolerance mediated by extrathymic Aire-expressing cells. Science 2008; 321:843–7; PMID:18687966; http://dx.doi.org/ 10.1126/science.1159407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Poliani PL, Kisand K, Marrella V, Ravanini M, Notarangelo LD, Villa A, Peterson P, Facchetti F. Human peripheral lymphoid tissues contain autoimmune regulator-expressing dendritic cells. Am J Pathol 2010; 176:1104–12; PMID:20093495; http://dx.doi.org/ 10.2353/ajpath.2010.090956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hobbs RP, DePianto DJ, Jacob JT, Han MC, Chung BM, Batazzi AS, Poll BG, Guo Y, Han J, Ong S, et al.. Keratin-dependent regulation of Aire and gene expression in skin tumor keratinocytes. Nat Genet 2015; 47:933–8; PMID:26168014; http://dx.doi.org/ 10.1038/ng.3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Khammanivong A, Anandharaj A, Qian X, Song JM, Upadhyaya P, Balbo S, Bandyopadhyay D, Dickerson EB, Hecht SS, Kassie F. Transcriptome profiling in oral cavity and esophagus tissues from (S)-N'-nitrosonornicotine-treated rats reveals candidate genes involved in human oral cavity and esophageal carcinogenesis. Mol Carcinog 2016. [epub ahead of print]; PMID:26785143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gotter J, Brors B, Hergenhahn M, Kyewski B. Medullary epithelial cells of the human thymus express a highly diverse selection of tissue-specific genes colocalized in chromosomal clusters. J Exp Med 2004; %19;199:155–66; PMID:14734521; http://dx.doi.org/ 10.1084/jem.20031677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer 2005; 5:615–25; PMID:16034368; http://dx.doi.org/ 10.1038/nrc1669 [DOI] [PubMed] [Google Scholar]
- [20].Van de Vijver MJ, He YD, Van'T Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al.. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 2002; 347:1999–2009; PMID:12490681; http://dx.doi.org/ 10.1056/NEJMoa021967 [DOI] [PubMed] [Google Scholar]
- [21].Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010; 123:725–31; PMID:20020197; http://dx.doi.org/ 10.1007/s10549-009-0674-9 [DOI] [PubMed] [Google Scholar]
- [22].Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE 2013; 8: e82241; PMID:24367507; http://dx.doi.org/ 10.1371/journal.pone.0082241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Marx A, Hohenberger P, Hoffmann H, Pfannschmidt J, Schnabel P, Hofmann HS, Wiebe K, Schalke B, Nix W, Gold R, et al.. The autoimmune regulator AIRE in thymoma biology: autoimmunity and beyond. J Thorac Oncol 2010; 5: S266–S272; PMID:20859117; http://dx.doi.org/ 10.1097/JTO.0b013e3181f1f63f [DOI] [PubMed] [Google Scholar]
- [24].Liang CC, Lu TL, Yu YR, You LR, Chen CM. beta-catenin activation drives thymoma initiation and progression in mice. Oncotarget 2015; 6:13978–93; PMID:26101855; http://dx.doi.org/ 10.18632/oncotarget.4368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Klamp T, Sahin U, Kyewski B, Schwendemann J, Dhaene K, Tureci O. Expression profiling of autoimmune regulator AIRE mRNA in a comprehensive set of human normal and neoplastic tissues. Immunol Lett 2006; 106:172–9; PMID:16876259; http://dx.doi.org/ 10.1016/j.imlet.2006.06.006 [DOI] [PubMed] [Google Scholar]
- [26].Abramson J, Giraud M, Benoist C, Mathis D. Aire's partners in the molecular control of immunological tolerance. Cell 2010; 140:123–35; PMID:20085707; http://dx.doi.org/ 10.1016/j.cell.2009.12.030 [DOI] [PubMed] [Google Scholar]
- [27].Abramson J, Goldfarb Y. AIRE: From promiscuous molecular partnerships to promiscuous gene expression. Eur J Immunol 2015; 46:22–33; PMID:26450177; http://dx.doi.org/ 10.1002/eji.201545792 [DOI] [PubMed] [Google Scholar]
- [28].Guerau-de-Arellano M, Mathis D, Benoist C. Transcriptional impact of Aire varies with cell type. Proc Natl Acad Sci U S A 2008; 105:14011–6; PMID:18780794; http://dx.doi.org/ 10.1073/pnas.0806616105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Andriani F, Bertolini G, Facchinetti F, Baldoli E, Moro M, Casalini P, Caserini R, Milione M, Leone G, Pelosi G, et al.. Conversion to stem-cell state in response to microenvironmental cues is regulated by balance between epithelial and mesenchymal features in lung cancer cells. Mol Oncol 2016; 10:253–71; PMID:26514616; http://dx.doi.org/ 10.1016/j.molonc.2015.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Conteduca G, Fenoglio D, Parodi A, Battaglia F, Kalli F, Negrini S, Tardito S, Ferrera F, Salis A, Millo E, et al.. AIRE polymorphism, melanoma antigen-specific T cell immunity, and susceptibility to melanoma. Oncotarget 2016. [epub ahead of print]; 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.