Abstract
Rhodopsin is the prototypical G protein-coupled receptor, responsible for detection of dim light in vision. Upon absorption of a photon, rhodopsin undergoes structural changes, characterised by distinct photointermediates. Currently, only the ground-state structure has been described. We have determined a density map of a photostationary state highly enriched in metarhodopsin I, to a resolution of 5.5 Å in the membrane plane, by electron crystallography. The map shows density for helix 8, the cytoplasmic loops, the extracellular plug, all tryptophan residues, an ordered cholesterol molecule and the β-ionone ring. Comparison of this map with X-ray structures of the ground state reveals that metarhodopsin I formation does not involve large rigid-body movements of helices, but there is a rearrangement close to the bend of helix 6, at the level of the retinal chromophore. There is no gradual build-up of the large conformational change known to accompany metarhodopsin II formation. The protein remains in a conformation similar to that of the ground state until late in the photobleaching process.
Keywords: electron crystallography, G protein-coupled receptor, membrane protein, metarhodopsin I, visual pigment
Introduction
Rhodopsin is the dim-light photoreceptor, and is a prototypical member of the G protein-coupled receptor (GPCR) family (Bockaert and Pin, 1999). It consists of a 348-amino-acid protein, opsin, which binds the chromophore 11-cis-retinal via a protonated Schiff base linkage to Lys296, giving the ground state of the protein an absorption maximum at 498 nm (Menon et al, 2001). Absorption of a photon by 11-cis-retinal triggers its isomerisation to the all-trans form, converting light energy into atomic motion (Wald, 1968). Rhodopsin then thermally relaxes through a series of distinct photointermediates, each with characteristic UV/visible absorption maxima (λmax). The early photointermediates include bathorhodopsin (batho, λmax=543 nm), a blue-shifted intermediate (BSI) and lumirhodopsin (lumi, λmax=497 nm) (Lewis and Kliger, 1992). An equilibrium is formed between the later photointermediates, metarhodopsin I (meta I, λmax=480 nm) and metarhodopsin II (meta II, λmax=380 nm). Meta II corresponds to the fully activated receptor, which binds to and activates the heterotrimeric G protein transducin (Menon et al, 2001). Several isochromic metarhodopsin intermediates have been proposed, including meta I380, an early intermediate with a deprotonated Schiff base (Thorgeirsson et al, 1992), and sequential meta II species separated by proton uptake, called meta IIa and meta IIb (Arnis and Hofmann, 1993).
To date, rhodopsin is the only GPCR amenable to direct structural analysis. The first 3D density maps of rhodopsin were obtained by electron crystallography of two-dimensional (2D) crystals of bovine (Unger and Schertler, 1995) and frog rhodopsin (Unger et al, 1997), revealing the arrangement of the transmembrane helices. This information was combined with structural constraints obtained by amino-acid sequence analysis of several hundred GPCRs to build a model for the Cα backbone of the rhodopsin family of GPCRs (Baldwin et al, 1997). The ground state of rhodopsin has been solved at high resolution by X-ray crystallography from 3D crystals with a P41 space group (Palczewski et al, 2000; Teller et al, 2001; Okada et al, 2002), and a P31 space group (Li et al, in press). Overall, the structures agree well. There are, however, differences at the cytoplasmic surface, where G protein binding and activation occur: in the P31 crystal form, helix 5 is longer, and the cytoplasmic loops between helices 5 and 6 and helices 3 and 4 adopt a different position.
To understand the molecular details of visual transduction and GPCR activation, we need to study the structure of photoactivated states of rhodopsin. Our attempts to look at photoactivated states in 3D crystals have so far proved unsuccessful, with diffraction markedly decreasing following exposure to light, which suggests a large conformational change. However, a 3D structure of the ground state of bovine rhodopsin has been obtained by electron crystallography of 2D crystals with a p22121 plane-group symmetry (Krebs et al, 2003). We have therefore used UV/visible and Fourier transform infrared (FTIR) difference spectroscopy (Vogel et al, 2004) to characterise the photointermediates formed in similar 2D crystals. Rhodopsin forms photointermediates up to meta I within 2D crystals. We have focused on studying the later photointermediate, meta I, by freeze-trapping and electron cryomicroscopy. Meta I is difficult to trap in the presence of most detergents, and FTIR difference spectroscopy has shown that meta I conformations in detergent solution differ from those in a more native, lipid environment (Beck et al, 1998). 2D crystals, in which rhodopsin molecules are arranged in a monolayer and retain some lipid molecules, therefore offer an attractive opportunity for structural studies of meta I in a native environment.
There is currently some controversy about the nature of the conformational change accompanying formation of meta I. Meta I can form at −40°C (Yoshizawa and Wald, 1963), and at temperatures below the phase transition temperature of the surrounding lipids (Baldwin and Hubbell, 1985). Limited proteolysis of rhodopsin indicated little change to the cytoplasmic surface at meta I, but a measurable change at meta II (Kühn et al, 1982). High pressure drives the meta I–meta II equilibrium towards meta I, suggesting that meta I is more compact than meta II (Lamola et al, 1974). Electron paramagnetic resonance spectroscopy also indicated little change to the cytoplasmic region of rhodopsin following meta I formation (Resek et al, 1993). The conformational changes were therefore thought to be small, principally confined to the chromophore and its binding pocket. However, photoaffinity labelling studies have suggested that photoisomerisation of rhodopsin involves significant movement of helices 3 and 4, coupled with movement of the chromophore out of the binding pocket and conformational changes to cytoplasmic loops 2 and 3, by the stages of lumi and meta I (Borhan et al, 2000). In addition, preresonance Raman vibrational studies of the Glu181Gln mutant of rhodopsin have indicated that there is a switch in the counterion of the Schiff base from Glu113 in the ground state to Glu181 in meta I (Yan et al, 2003). The switch is proposed to occur via transfer of a proton from Glu181 to Glu113 via a hydrogen-bonding network, accompanied by modest conformational changes involving extracellular loop 2 and helix 3 (Yan et al, 2003).
The density map from this study reveals that meta I formation does not involve large rigid-body movements or rotations of rhodopsin's helices. Instead, conformational changes are localised around the chromophore binding pocket, close to kinks in the helices observed in the X-ray structures. The structure of the photostationary state has been calculated from new amplitudes and phases that are independent of any previous structural study of rhodopsin. Since meta I formation does not involve large conformational changes, the new map allows more detailed interpretation of differences in the X-ray structures of the ground state. Significantly, the map shows that there is no gradual build-up of the large conformational change known to accompany meta II formation. The protein adopts a conformation similar to the ground state until late in the photobleaching process.
Results and discussion
Characterisation of rhodopsin photointermediates in 2D crystals
2D crystals of bovine rhodopsin with p22121 plane-group symmetry were obtained by dialysis of detergent-solubilised rhodopsin isolated from bovine rod outer segment membranes (Krebs et al, 1998, 2003). Addition of cholesterol prior to dialysis increases the yield of rhodopsin crystals, allowing the preparation of 5–10 mg amounts of 2D crystals with minimal protein aggregates for spectroscopic characterisation (Mielke et al, 2002b).
We used UV/visible spectroscopy to follow the formation of rhodopsin photointermediates. Illuminated 2D crystals contained a mixture of meta I (absorbing at 480 nm) and a species absorbing at 380 nm, the proportion of which could be varied by adjusting the temperature and pH (Figure 1A and B). Lower temperatures favoured formation of meta I, as observed for rhodopsin in native membranes (Matthews et al, 1963). The 2D crystals showed an inverted pH dependency compared to native membranes (Matthews et al, 1963), with higher pH favouring formation of a species absorbing at 380 nm. At pH 7, we could use temperature alone to accumulate either of these species.
Figure 1.
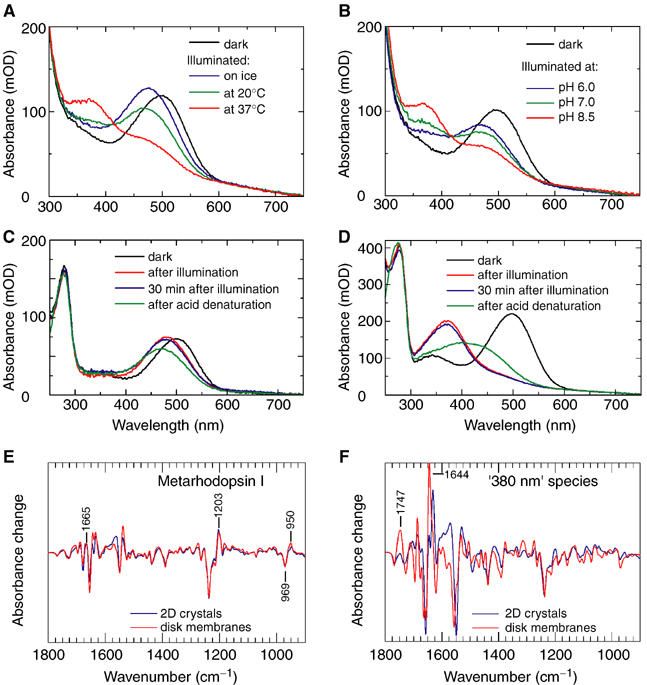
UV/visible spectroscopic analysis of photointermediate formation in 2D crystals. (A) Temperature dependency. The dark spectrum of rhodopsin 2D crystals was recorded in dialysis buffer at 20°C, pH 7.0. Spectra were then recorded after illuminating 2D crystals in the same buffer at the given temperature. (B) pH dependency. The dark spectrum was recorded in dialysis buffer at 30°C, pH 7.0. Spectra were then recorded after illumination of the sample in dialysis buffer at 30°C and a given pH. (C) Protocol for trapping meta I in 2D crystals. A suspension of 2D crystals was incubated on ice, and its spectrum was recorded at 1–2°C (black line). The crystals were illuminated on ice for 1 min with light filtered to contain only wavelengths greater than 515 nm. The absorption maximum shifts to 480 nm following illumination, corresponding to the formation of meta I (red line). Four cryoelectron microscopy grids were then prepared. A further spectrum was recorded (blue line). Finally, sulphuric acid was added to the photometer cell until the pH was between 1.0 and 3.0 (green line). (D) Formation of a species absorbing at 380 nm in 2D crystals. A suspension of 2D crystals was incubated at 45°C, and its spectrum was recorded at 45°C. The crystals were illuminated for 1 min with light filtered to contain only wavelengths greater than 515 nm. Spectroscopy reveals a shift in the absorption maximum to 380 nm following illumination. Several characteristics of this photointermediate led us to conclude that it does not correspond to meta II. (E, F) FTIR difference spectroscopy of rhodopsin in 2D crystals compared to rhodopsin in its native (disk) membrane. The absorbance change is shown at the same scale in both spectra. (E) FTIR difference spectra of meta I. The spectrum for the 2D crystals was obtained in 200 mM MES and 200 mM NaCl at 3°C, pH 7, from the same batch of crystals used in the structure determination. The spectrum for rhodopsin in disk membranes was obtained at 10°C, pH 8.5 (see Vogel et al, 2004 for a corresponding UV/visible spectrum). (F) FTIR difference spectra of species absorbing at 380 nm. The spectrum for the 2D crystals was obtained at 45°C, pH 7. The spectrum for rhodopsin in disk membranes was obtained at 20°C, pH 5, conditions that produce meta II in the membrane (see Vogel et al, 2004 for a corresponding UV/visible spectrum).
The trapping of meta I is described in Materials and methods and Figure 1C. A spectrum of the crystals recorded before illumination shows an absorption peak at 498 nm, characteristic of the ground state (Figure 1C, black line). Spectra recorded immediately after illumination show a shift in the absorption maximum to 480 nm, characteristic of meta I formation (Figure 1C, red line). Four cryoelectron microscopy grids were prepared from aliquots taken from the photometer cell, thus freeze-trapping meta I for subsequent structural studies. Preparation and manipulation of four grids typically took 20–30 min, after which a spectrum was recorded, to check that the sample had not changed significantly (Figure 1C, blue line). The sample was then acid denatured, and a further spectrum was recorded (Figure 1C, green line). The shift in absorption maximum to 470 nm, and absence of a peak at 380 nm, indicates that the retinal chromophore is still attached to the protein by a protonated Schiff base linkage (Kito et al, 1968). Control experiments showed that the red-shift in absorption maximum of the acid-denatured rhodopsin from 440 nm is due to the low temperatures used for spectroscopy (data not shown).
We adopted a similar protocol for trapping the species absorbing at 380 nm, working at 45°C (Figure 1D). This temperature ensured that no residual 480 nm species was present after illumination. The broader absorption maximum after acid denaturation (Figure 1D, green line), indicating free retinal (which absorbs at 380 nm) plus residual protonated Schiff base, and the inverted pH dependency made us doubt that this species corresponded to meta II.
We further characterised rhodopsin photoproducts formed in 2D crystals by FTIR difference spectroscopy (Vogel et al, 2004). This showed that batho, lumi and meta I photointermediates formed in 2D crystals are very similar to those obtained from rhodopsin in native disk membranes, despite the transition from lumi to meta I being shifted to a higher temperature in the 2D crystal (Vogel et al, 2004). Meta I formed in 2D crystals, under conditions used in the freeze-trapping protocol, shows very similar FTIR difference spectra to that formed in native membranes (Figure 1E). Characteristic meta I marker bands include the positive amide I band at 1665 cm−1 and the hydrogen-out-of-plane (HOOP) difference band at −969/+950 cm−1. The positive band at 1203 cm−1 is characteristic of a protonated Schiff base. At room temperature, formation of meta II is blocked in 2D crystals, and an intermediate state is formed that has a protonated Schiff base and lacks an ability to activate transducin (Vogel et al, 2004). The 380 nm species formed at 45°C and observed by UV/visible spectroscopy lacks characteristic meta II marker bands, as shown by reduced intensities of the positive band at 1747 cm−1 and of the meta II amide band at 1644 cm−1 (Vogel et al, 2004). Instead, this species represents a non-native state, showing some spectral similarity to an alkaline denatured state obtained in disk membranes at 30°C, pH 9 (Vogel and Siebert, 2002) (Figure 1F).
Further UV/visible spectroscopy experiments showed that the photostationary state consisted of 60–70% meta I and 30–40% of cis photoproducts (see Materials and methods). The high meta I content in the 2D crystals, and its similarity to native meta I, made this an ideal photointermediate to characterise structurally.
Electron microscopy and image processing
Images of 2D crystals of the photostationary state were recorded at liquid nitrogen temperatures using a field emission gun electron microscope operating at 300 kV. We were especially interested in collecting good data from tilted specimens, to give good resolution perpendicular to the membrane plane. Poor vertical resolution can be due to lack of specimen flatness (Butt et al, 1991). For this reason, 2D crystals were prepared on molybdenum grids (Vonck, 2000), the carbon support film was prepared by multiple steps of carbon evaporation (Butt et al, 1991; Fujiyoshi, 1998), and images were recorded using the spot-scan technique (Bullough and Henderson, 1987). This allowed us to collect data at 45 and 60° specimen tilts. Furthermore, the program TTBOXK was used to correct for the gradient of defocus across tilted images and read-out amplitudes and phases (Henderson et al, 1990). For many images, this led to an increase in the number of spots and spot quality compared to processing with MMBOXA (Henderson et al, 1986).
Figure 2 shows representative lattice lines fitted to the experimentally determined amplitude and phase data. The density map was calculated to 5.5 Å resolution using a 0.005 Å−1 sampling of the lattice lines. Table I summarises electron crystallographic statistics from the structure determination.
Figure 2.
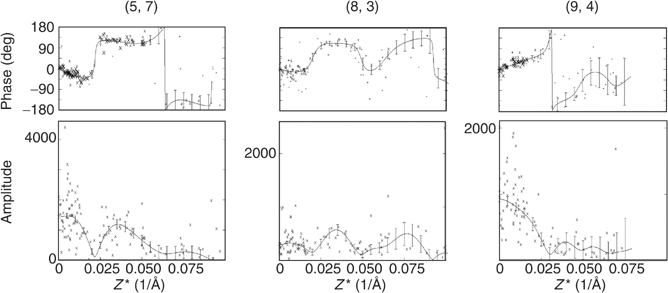
Lattice line data. Plots of experimental amplitudes (lower panel) and phases (upper panel) together with the curves produced by weighted least-squares fitting, and the associated errors (generated by the program LATLINEK).
Table 1.
Electron crystallographic data
| Plane group | p22121 |
| Cell dimensions | |
| a (Å) | 58.8 |
| b (Å) | 83.7 |
| c (Å) | 200.0 (arbitrary) |
| α=β=γ (deg) | 90.0 |
| Number of imagesa | 87 |
| Range of defocus (Å) | 2700–14 100 |
| Effective resolution of 3D data setb (Å) | |
| In-plane | 5.5 |
| Perpendicular to the membrane | 11.7 |
| Average temperature factor (Bxy)c | 200±95 |
| Total number of observed amplitudes and phases | 22 099 |
| Number of unique structure factors | 2073 |
| Completeness (%) | |
| 0–45° | 83.2 |
| 0–60° | 68.2 |
| 0–90° | 59.0 |
| Overall weighted R-factord (%) | 34.9 |
| Overall weighted phase residuald (deg) |
24.0 |
| aThe number of images are 22 at 0°, 16 at 20°, 12 at 35°, 26 at 45° and 11 at 60°. | |
| bAs calculated from the point-spread function. | |
| cAverage temperature factor to scale image amplitudes against bacteriorhodopsin electron diffraction data, correcting for high-resolution image amplitude fall-off. | |
| dFrom the program LATLINEK. | |
Three-dimensional density map of the photostationary state
Figure 3A and B shows two rhodopsin molecules within the electron density map of the photostationary state. This map is a weighted average of the structures of meta I and some residual isorhodopsin and ground state, but meta I predominates (60–70%). The density map reveals all seven transmembrane helices in the meta I state. For the first time in a density map of rhodopsin determined by electron cryomicroscopy, there is clear continuous density for helix 8, which lies parallel to the membrane plane. There is also density for the extracellular plug and for the cytoplasmic loops, including continuous density for cytoplasmic loop 2, which connects helices 3 and 4. These features are the most difficult to resolve by electron crystallography, and indicate the quality of data used to determine the map. There is also density for at least the first N-acetyl glucosamine group in the oligosaccharides attached to Asn2 and Asn15. Figure 5A–Cshows slices through the electron density map of a single molecule.
Figure 3.
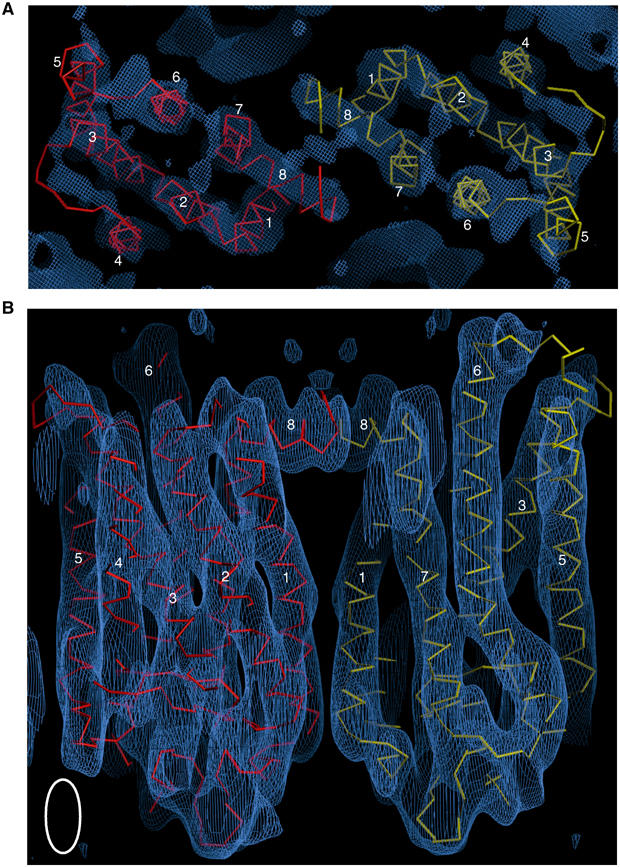
Views of the density map, showing two rhodopsin molecules. Helices are numbered. (A) Section through the cytoplasmic side, at the level of helix 8, showing clear continuous density for cytoplasmic loop 2, which connects helices 3 and 4. Contoured at the 0.6σ level. (B) Side view along the membrane plane of the density contoured at 1σ, showing two rhodopsin molecules. The maps were analysed, and the coordinates of the ground state of rhodopsin determined by X-ray crystallography (Li et al, in press) were modelled into the density by manual fitting followed by real-space rigid-body refinement implemented in the program O (Jones et al, 1991). The white ellipse shows the point-spread function of the data set, which is used to determine the vertical resolution of the density map (Henderson et al, 1990; Unger and Schertler, 1995).
Figure 5.
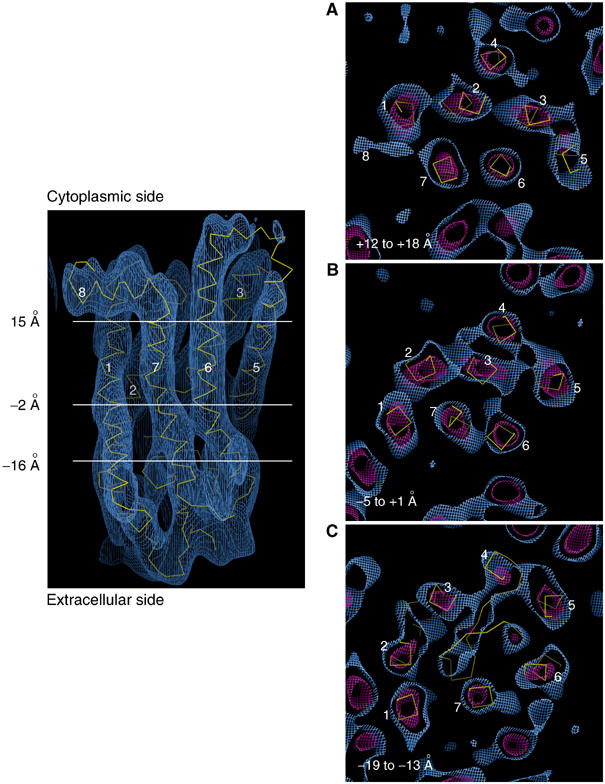
Sections (6 Å) through the density map perpendicular to the membrane plane, viewed from the cytoplasmic side. Helices are numbered. (A) Section at +12 to +18 Å, towards the cytoplasmic side. (B) Section at the position of the chromophore (−5 to +1 Å). (C) Section at the level of the extracellular plug (−19 to −13 Å), showing density associated with several extracellular loops. The side view is shown at the 1σ contour level. For (A–C), the blue density is at the 0.6σ level, and the red density is at the 3.6σ level.
The density map is a significant improvement on the density map of the ground state of rhodopsin determined by electron cryomicroscopy (Krebs et al, 2003), which lacks strong density for helix 5 and also density for helix 8, cytoplasmic loops and the extracellular plug. While it is possible that these differences reflect conformational changes accompanying meta I formation, it is more likely that they are due to better quality image data being used in the calculation of the meta I map. The higher quality of data is attributable to improvements in the 2D crystallisation procedure (Mielke et al, 2002b) and in image processing, and data collection on a later generation electron microscope operating at a higher accelerating voltage. Since so many features are well resolved in the density map, we have fitted the coordinates of the X-ray structure of rhodopsin in its ground state (Li et al, in press; PDB code 1GZM) to the map by hand, followed by real-space refinement in the program O (Jones et al, 1991), treating the entire ground-state structure as a rigid body. The Cα trace of the ground-state structure fitted to the density map is shown in Figures 3, 4, 5 , 6 and 7.
Figure 4.
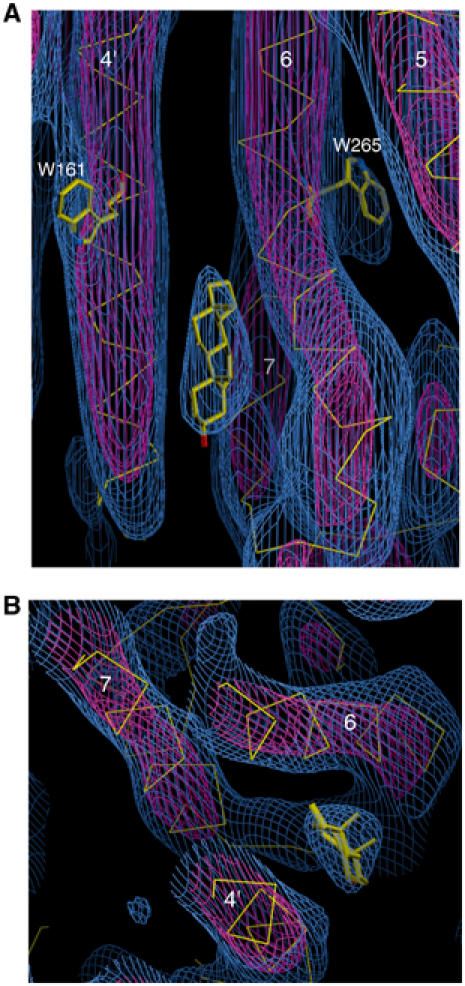
Two views of a density feature assigned to cholesterol. The red density is contoured at 3.2σ, and the blue density at 1σ. (A) Side view, showing cholesterol, helix 4 of one rhodopsin molecule (with Trp161) and helices 5, 6 (with Trp265) and 7 of the adjacent rhodopsin molecule. These two molecules face in opposite directions: the extracellular side of the molecule on the left and the cytoplasmic side of the molecule on the right are towards the bottom of the page. (B) Top view, showing cholesterol sitting in a pocket between the two rhodopsin molecules.
Figure 6.
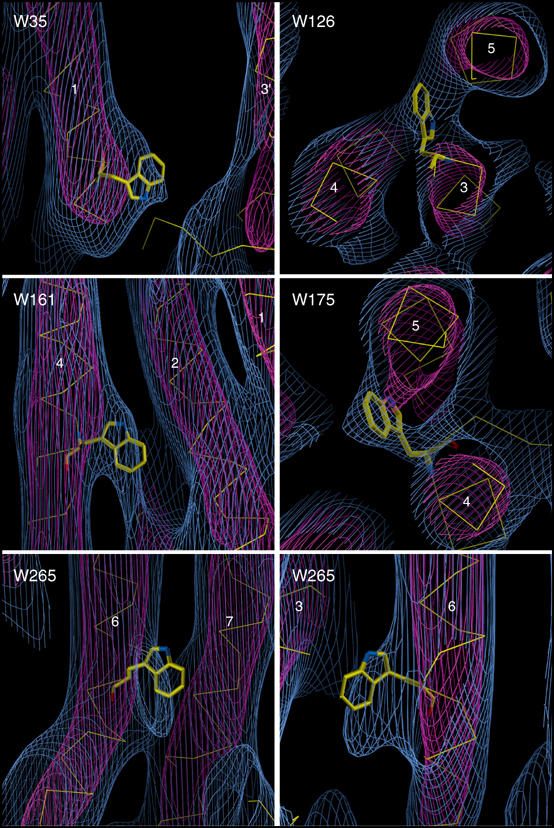
Density features in the density map around the positions of tryptophan residues known from the ground-state rhodopsin structure. Helices are numbered, the red density is contoured at 3.2σ and the blue density is contoured at the 0.6σ level.
Figure 7.
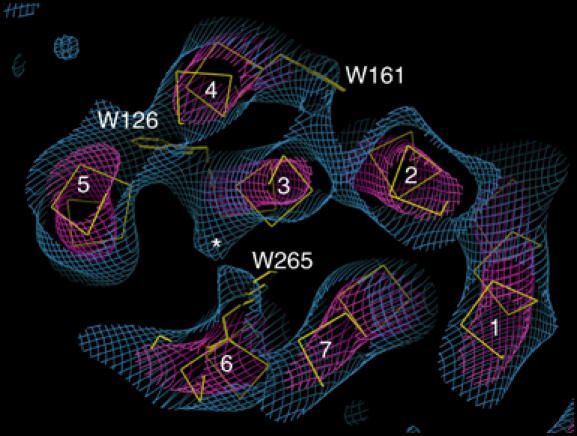
8 Å section through the density map, tilted slightly to view from the extracellular side along helix 3, showing the transmembrane helices (numbered), density features close to the position of Trp126, Trp161 and Trp265, and density that we attribute to the β-ionone ring (marked with an asterisk).
Arrangement of molecules within the crystal
Two of the four rhodopsin molecules within the unit cell show the same orientation in the membrane, and are involved in close crystal contacts, forming a dimer (Figure 3). Dimer formation is mediated partly by interactions between helix 1 in the two molecules (Figure 3B), although the high crossing angle of these helices would make this interaction relatively weak. There is also a close interaction between helix 8 in both molecules, which lie parallel to each other (Figure 3A). This dimeric arrangement of rhodopsin monomers has been seen in several other 2D crystals of rhodopsin—p2221 crystals of bovine rhodopsin (Schertler et al, 1993), and p2 and p22121 crystals of frog rhodopsin (Schertler and Hargrave, 1995)—indicating that interactions between helices 1 and 8 on two rhodopsin molecules are sufficiently strong to promote formation of this dimer under a range of different crystallisation conditions.
We detect a strong density feature in a pocket between those molecules that lie along the in-plane two-fold screw axes. This lies near the extracellular side of helices 6 and 7 of one molecule and the intracellular side of helix 4 of the adjacent molecule. A possible candidate for this density is cholesterol. Cholesterol helps to improve the reliability and yield of crystallisation. It is therefore reasonable to expect a cholesterol molecule to bind tightly to a specific molecular interface, so that it would be relatively well ordered and present in high occupancy. In the density map, we see features for tryptophans (see below and Figure 6), which have bicyclic fused ring systems, and would expect to be able to detect the tetracyclic fused ring system of cholesterol. The density feature is too bulky to be an ordered phospholipid molecule. A cholesterol molecule (minus its hydrocarbon side chain) can be fitted into the density feature (Figure 4A and B). It is oriented with its tetracyclic fused ring system oriented normal to the plane of the crystal, which would be equivalent to an orientation normal to a membrane bilayer. This orientation, and the distance of the cholesterol hydroxyl group from the bilayer centre, are very similar to those determined by X-ray and neutron scattering from artificial lipid bilayers and native membranes (Caspar and Kirschner, 1971; Franks and Lieb, 1979). The hydroxyl group of the cholesterol is modelled pointing towards the extracellular side, and is located near polar protein residues and where the phospholipid headgroups would be. One face of the cholesterol ring system is exposed to interact with lipids present in the 2D crystal, which are less ordered and no density is visible for them in the map at this resolution.
Why is meta II formation inhibited in 2D crystals?
There are two possible reasons for inhibition of meta II formation: crystallographic packing or the lipid environment. Meta II formation is thought to involve a rigid-body movement of helix 6 (Hubbell et al, 2003). In the p22121 2D crystals, helix 6 of one molecule interacts with helices 4 and 5 of a neighbouring molecule. These close crystal contacts may prevent meta II formation. In addition, meta II formation in membranes is dependent on the lipid environment, with the meta I–meta II equilibrium shifting towards meta I when rhodopsin is reconstituted into liposomes containing increasing amounts of lipids with saturated fatty acyl chains or of cholesterol (Mitchell et al, 1990, 1992). We have checked that crystals grown without addition of cholesterol do not allow meta II formation, both by UV/visible spectroscopy (data not shown) and FTIR difference spectroscopy (Vogel et al, 2004). However, it is possible that the delipidating chromatography steps used to purify rhodopsin before dialysis strip away some of the highly unsaturated lipids that are essential for meta II formation.
Comparison between the density map and the ground-state X-ray structure
The cytoplasmic surface of molecules in the density map, at the level of helix 8, is very similar to ground-state rhodopsin (Figure 3A). Meta I formation therefore does not involve large conformational changes at the cytoplasmic surface. Cytoplasmic loop 1, connecting helices 1 and 2, is in the same position as in the crystal structures of the ground state.
Cytoplasmic loop 2, connecting helices 3 and 4, has changed orientation compared to that in the P31 crystal structure and lies closer to the position in molecule A of the P41 structures (Figure 3A). This is most likely due to the different packing in 2D and 3D crystals: the orientation of the loop in the P31 structure leads to a clash with residues at the amino-terminal end of helix 1 of the next molecule in the p22121 2D crystal. Cytoplasmic loop 2 may have some intrinsic mobility, which may be relevant to its interaction with transducin following meta II formation (Franke et al, 1990, 1992). Indeed, altered mobility of this loop following formation of meta II has been detected in time-resolved fluorescence depolarisation experiments (Mielke et al, 2002a). The L-shaped appearance of cytoplasmic loop 2 is conserved between the X-ray structures and the 2D crystal, suggesting that mobility is confined to the ends of helices 3 and 4, which may act like hinges (Li et al, in press).
Helix 6 in the density map extends from the membrane surface at the cytoplasmic side (Figures 3B and 5), as seen in the P31 and P41 crystal structures. Experimental evidence suggests that meta II formation is accompanied by movement of helix 6 (Hubbell et al, 2003), and it is likely that movement of the cytoplasmic extension of helix 6 is involved in transducin binding and activation. The P31 crystal structure shows that helical periodicity continues into cytoplasmic loop 3, which links helices 5 and 6, in agreement with spin-labelling studies (Altenbach et al, 1996). The end of helix 5 and the position of cytoplasmic loop 3 are considerably different in the P31 and P41 crystal structures. We do not see continuous density for cytoplasmic loop 3 (Figure 3A); however, the density is fully consistent with the position and temperature factors of helices 5 and 6, and cytoplasmic loop 3 in the P31 crystal structure. Indeed, the position of cytoplasmic loop 3 in the P41 structures would lead to a steric clash with residues at the extracellular side of helix 4 of the neighbouring molecule in the 2D crystal.
The density map shows that meta I formation is not accompanied by large movement of helix 8 (Figure 3), which also interacts with transducin (Ernst et al, 2000; Marin et al, 2000) and is thought to move during meta II formation (Fritze et al, 2003). A density feature close to the position of Thr242 is attributed to part of the C-terminus, in agreement with a cysteine mutagenesis study (Cai et al, 1997), but in contrast to the position of the C-terminus in the P41 crystal structures.
The fit of the ground-state structure to the density map reveals that there are no rigid-body motions of entire helices of greater than 2 Å following meta I formation (Figure 5). The density map shows that kinks in helices 2, 6 and 7, and the link between helices 7 and 8, follow the ground-state structure very well. The conformational change therefore does not involve large rotations of these helices, or a movement perpendicular to the membrane plane. There are density bulges close to the positions of all five of the bulky tryptophan residues in the ground-state structure (Figure 6). Of these, Trp35 is located at the extracellular end of helix 1, Trp126 lies near the middle of helix 3, Trp161 lies near the middle of helix 4 and Trp175 lies in extracellular loop 2, close to the extracellular end of helix 4. Since these residues have associated density features in the map, it is possible to rule out large rotations of helices 1, 3 and 4.
Meta I formation therefore involves localised changes in specific regions of rhodopsin. Since the map is of a photostationary state (60–70% meta I), the magnitude of these localised changes is slightly reduced. Sections through the density map, parallel to the membrane plane, reveal further information about the nature of these localised changes (Figure 5A–C). Looking along the helix bundle, just below the position of helix 8, the density fits the Cα trace of the ground-state structure closely (Figure 5A). Just above the retinal chromophore, we detect a small misfit between the density and the ground-state structure: close to the kink in helix 6 (Figure 5B). There is a density bulge in the middle of helix 6, on the side facing the β-ionone ring, which lies close to the ground-state position of Trp265. This density feature significantly deviates from the position of Trp265 in the ground-state structures, suggesting the possibility of movement of Trp265 (Figure 6). The kink in the helix in this region, and hydrogen bonds from Cys264, Tyr268 and Pro291 to a water molecule (Okada et al, 2002; Li et al, in press), might allow a localised rearrangement of this part of the helix, without moving the distal ends. We attribute this movement to meta I formation, although there is some contribution from cis photoproducts to the photostationary state. For other transmembrane helices, the density fits the ground-state structure very closely, looking along the helix axes.
Density for transmembrane helices close to the extracellular plug region fits the ground-state structure well (Figure 5C). There is density associated with some parts of the extracellular plug, although the β sheets are not resolved, which indicates that the plug has not changed position significantly.
Correlation of the present structural analysis of meta I with previous studies
Electron paramagnetic resonance spectroscopy of site-directed rhodopsin mutants has shown that no light-induced changes are observed following meta I formation when spin labels are introduced at Cys140 (cytoplasmic end of helix 3) and Cys316 (helix 8) (Resek et al, 1993). This agrees with the present study (Figure 3A).
Photoaffinity labelling studies have suggested that photoisomerisation of rhodopsin involves significant movement of helices 3 and 4, coupled with movement of the chromophore out of the binding pocket and conformational changes to cytoplasmic loops 2 and 3, by the lumi stage (Borhan et al, 2000). In contrast, we see no gross changes in the structure of rhodopsin at the meta I state. Therefore, it is unlikely that there were any major movements of helices in the previously formed lumi state. This is consistent with small volume changes upon lumi formation detected by photoacoustic calorimetry (Marr and Peters, 1991). It has been suggested that steric hindrance displaces the ring of the photoaffinity-labelled retinal used by Borhan and colleagues in the ground state, and may force different conformational changes during photoactivation (Spooner et al, 2003). Furthermore, the small conformational changes revealed here agree with the conclusions of a study of the thermodynamic properties of lumi, meta I and meta II (Imai et al, 1994).
A density bulge hanging off helix 3, close to the density for Trp265, is likely to be due to the β-ionone ring, since there are no bulky aromatic residues close to this feature (Figure 7). Density in a similar region was found in the ground-state structure determined by electron cryomicroscopy (Krebs et al, 2003). This suggests that in meta I the β-ionone ring lies close to its position in the ground state. Our result is in agreement with solid-state NMR analysis of the conformation of the β-ionone ring of the chromophore in the ground and meta I states (Spooner et al, 2003). This work has shown that the chromophore is retained within its binding pocket at the meta I stage, and that the β-ionone ring on average remains in its ground-state position.
Time-resolved UV resonance Raman studies of early rhodopsin photointermediates have detected spectroscopic changes due to tryptophan residue(s) in photorhodopsin, followed by changes to tryptophan and tyrosine residues in batho (Kim et al, 2003). From analysis of X-ray structures of the ground state, the authors suggest that Trp265 and Tyr268 are involved in these changes. Movement of Trp265 is consistent with UV/visible spectoscopic analysis of site-directed rhodopsin mutants, showing that Trp126 and Trp265 undergo conformational changes following meta II formation (Lin and Sakmar, 1996). Earlier UV resonance Raman spectroscopy studies detected absorption shifts of tyrosine residue(s), and weaker changes possibly attributable to tryptophan(s) (Kochendoerfer et al, 1997). Spectral changes to tryptophan residues would be in agreement with the small movement of Trp265 revealed by the density map.
FTIR difference spectroscopy of site-directed mutants of rhodopsin have shown that Glu122, on helix 3, changes its environment, becoming more strongly hydrogen-bonded in meta I (Beck et al, 1998). Our studies suggest that this could be due to a localised change to this part of helix 3, or to a small movement of the β-ionone ring.
A counterion switch is proposed to occur during the transition to meta I by transfer of a proton from Glu181 to Glu113 via a hydrogen-bonding network, accompanied by modest conformational changes involving extracellular loop 2 and helix 3 (Yan et al, 2003). The density map shows that changes to helix 3 involve only amino-acid side-chain rearrangements. Our data do not support the magnitude of conformational changes proposed by Yan and colleagues. However, the small conformational change that we detect in helix 6 may be linked to the counterion switch. Although the resolution of the current map is not sufficient to see density for the side chain of Tyr268, it lies close to the region of misfit between the density map and ground-state structure. X-ray structures of ground-state rhodopsin reveal that Tyr268 forms hydrogen bonds to Glu181 (Li et al, in press). A localised movement around Trp265 could free Tyr268, disrupting this hydrogen bonding, allowing the side chain of Glu181 to move closer to the Schiff base and adopt its proposed role as counterion in meta I.
Conclusions
The present work shows that meta I formation involves no large (rigid-body) movements or rotations of helices from their position in the ground state. Instead, changes seem to be localised, probably involving movement of side chains in kinked regions of helices close to the retinal-binding pocket, such as Trp265. The observed small changes could account for spectroscopic changes known to accompany meta I formation. The larger movements of helices that characterise meta II formation (Hubbell et al, 2003) are not initiated at the early photointermediate stage, but occur during the transition from meta I to meta II.
Materials and methods
2D crystallisation, spectroscopy and trapping of meta I
2D crystallisation was performed as described previously (Krebs et al, 1998; Mielke et al, 2002b), with detergent-solubilised rhodopsin being incubated with a 10:1 (mol/mol) ratio of cholesterol to rhodopsin prior to dialysis. UV/visible spectra of 2D crystal suspensions were recorded on a Shimadzu UV-2401PC spectrophotometer equipped with an integrating sphere (to reduce the effect of light scattering by the turbid samples) and temperature-controlled cell holders. The following procedure describes the trapping protocol for meta I. A suspension of rhodopsin 2D crystals in dialysis buffer (crhodopsin=0.5 mg/ml) was equilibrated in a photometer cell (optical pathlength 0.1 cm) on ice in the dark (temperature 0.1–3°C). All spectra were recorded at 2°C. After recording a spectrum in the dark, the sample was illuminated on ice for 1 min using a 150 W halogen lamp (Stocker & Yale Inc., Model 20) attached to a glass fibre optic. The excitation wavelength was selected by a cutoff filter (OG515, Schott) placed between the lamp and fibre optic. Immediately after illumination, another spectrum was recorded. The sample was then kept on ice. Grids were prepared using a Gatan Cryoplunge device, specially modified to have a temperature- and humidity-controlled chamber for holding the grids. In the dark, a small aliquot (3 μl) of sample was put on a carbon-coated and freshly glow-discharged molybdenum electron microscopy grid (Pacific Grid-Tech, San Diego, CA) held at 1–2°C and greater than 70% relative humidity. After 2 min, excess liquid was blotted for 20 s within the humidity chamber and the grid was rapidly frozen in liquid nitrogen-cooled liquid ethane. After the last grid was frozen, a control spectrum was taken. A further spectrum was recorded after acid denaturation with H2SO4 until a pH between 1.7 and 3.0 was reached. All subsequent grid manipulations were performed at liquid nitrogen temperatures. FTIR difference spectra were recorded as described by Vogel et al (2004).
Analysis of the photostationary state
Contributions of ground-state 11-cis rhodopsin and 9-cis isorhodopsin to the photostationary state were determined following a method similar to that of Lewis et al (1997) and Vogel et al (2003). Samples of 2D crystals were illuminated under the conditions used to trap meta I. Hydroxylamine and lauryl dimethylamineoxide (LDAO) were then added to the samples (to final concentrations of 20 mM and 0.1%, respectively), allowing hydroxylamine-sensitive photoproducts to bleach chemically. Spectra showed that the reaction was complete after 2 min. The amount of residual hydroxylamine-stable pigment (a mixture of 9-cis isorhodopsin and 11-cis rhodopsin) was determined by illuminating the sample for 3 min (OG515 cutoff filter), now in the presence of hydroxylamine, which completely bleaches rhodopsin molecules left with cis chromophores after the first illumination. These experiments show that the photostationary state is composed of 60–70% meta I and 30–40% of a mixture of 9-cis isorhodopsin and some 11-cis (ground state).
Cryoelectron microscopy and image processing
Images were taken at liquid nitrogen temperature using an FEI Tecnai F30 300 kV field emission gun microscope and a GATAN cold stage (Gatan Ltd, Corby, UK). Electron micrographs were recorded under low-dose conditions (10–15 electrons/Å2) at × 58 000 magnification using spot-scan illumination and a 20 μm C2 condenser aperture (Bullough and Henderson, 1987). Micrographs were recorded on Kodak SO-163 film and developed in full-strength D19 developer for 12 min at 20°C. Optical diffraction was used to select negatives showing good crystalline areas, and these were scanned using a Zeiss SCAI densitometer with a step size of 7 μm. The MRC image processing software was used to extract amplitude and phase information from each image (Henderson et al, 1986; Crowther et al, 1996). Lattice distortions were removed using an iterative three-step unbending procedure (Havelka et al, 1995). Phases were corrected for effects of the contrast transfer function against the Thon-rings of the micrographs (program CTFFIND2). For images taken at low tilt angles (0 and 20°), tilt angle and tilt axis parameters were determined from the defocus values at the corners and centre of the image, using a program (LOWTILT) written by one of us (JJR). For images taken at higher tilt angles, tilt axis and tilt angle parameters could be reliably determined from the lattice parameters (program EMTILT) (Shaw and Hills, 1981). At this stage, images taken at tilt angles of 20° and higher were reprocessed to correct for the gradient of defocus across the image (program TTBOXK) (Henderson et al, 1990). The defocus parameters for TTBOXK were refined by altering the defocus in step sizes of 100 Å, assuming no astigmatism, and comparing the number of spots extracted from the image and the spot quality. For most images, running TTBOXK led to an improvement in the number of spots extracted from the image, and an improved spot quality, compared to using MMBOXA.
To combine data from each image, cycles of merging and origin refinement were combined with cycles of tilt axis and tilt angle refinement (program ORIGTILTK). Amplitudes were scaled (program SCALIMAMP3D) using bacteriorhodopsin as a reference to restore resolution-dependent fall-off of image-derived amplitudes (Havelka et al, 1995; Unger and Schertler, 1995). Lattice lines were fitted to the continuous amplitude and phase data using LATLINEK. The lattice lines were sampled at 0.005 Å−1 intervals, and the resulting structure factors were used to calculate density maps using the CCP4 crystallographic program FFTBIG (Collaborative Computational Project, Number 4, 1994).
Fitting of the ground-state X-ray structure into the meta I density map
The 3D density map was displayed and analysed using the program O (Jones et al, 1991). The X-ray structure of bovine rhodopsin in the ground state (PDB accession code 1GZM) (Li et al, in press) was fitted into the meta I map by hand followed by rigid-body refinement using the RSR_rigid option in O. We fitted molecule B from the X-ray structure, including the one N-acetyl glucosamine group on Asn2 and Asn15, but removing all of the following groups: palmitoyl, β-D-mannose, LDAO, C8E4 and water molecules. The rigid-body refinement clearly converged to a single solution, even from slightly different starting positions. A PDB file for cholesterol was obtained from the HIC-Up database (originally from pdb1lri.ent), and edited to remove the hydrocarbon side chain. The cholesterol molecule was fitted into a strong density feature in the map by rigid-body refinement in O.
The density map has been deposited in the EMD database under accession code EMD-1079.
Acknowledgments
We thank John Berriman for help with electron microscopy, Brad Amos for constructing the optical diffractometer used in this study, and Friedrich Siebert, Richard Henderson and Jade Li for many helpful discussions. Part of the work was supported by an HFSP grant (RGP 0054/2002). JJR was supported by an MRC Predoctoral Fellowship. TM was supported by a European Commission Marie-Curie fellowship (HPMF-CT-1999-00093).
References
- Altenbach C, Yang K, Farrens DL, Farahbakhsh ZT, Khorana HG, Hubbell WL (1996) Structural features and light-dependent changes in the cytoplasmic interhelical E–F loop region of rhodopsin: a site-directed spin-labeling study. Biochemistry 35: 12470–12478 [DOI] [PubMed] [Google Scholar]
- Arnis S, Hofmann KP (1993) Two different forms of metarhodopsin II: Schiff base deprotonation precedes proton uptake and signaling state. Proc Natl Acad Sci USA 90: 7849–7853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin JM, Schertler GF, Unger VM (1997) An alpha-carbon template for the transmembrane helices in the rhodopsin family of G-protein-coupled receptors. J Mol Biol 272: 144–164 [DOI] [PubMed] [Google Scholar]
- Baldwin PA, Hubbell WL (1985) Effects of lipid environment on the light-induced conformational changes of rhodopsin. 1. Absence of metarhodopsin II production in dimyristoylphosphatidylcholine recombinant membranes. Biochemistry 24: 2624–2632 [DOI] [PubMed] [Google Scholar]
- Beck M, Sakmar TP, Siebert F (1998) Spectroscopic evidence for interaction between transmembrane helices 3 and 5 in rhodopsin. Biochemistry 37: 7630–7639 [DOI] [PubMed] [Google Scholar]
- Bockaert J, Pin JP (1999) Molecular tinkering of G protein-coupled receptors: an evolutionary success. EMBO J 18: 1723–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borhan B, Souto ML, Imai H, Shichida Y, Nakanishi K (2000) Movement of retinal along the visual transduction path. Science 288: 2209–2212 [DOI] [PubMed] [Google Scholar]
- Bullough PA, Henderson R (1987) Use of spot scan procedure for recording low-dose micrographs of beam sensitive specimens. Ultramicroscopy 21: 223–230 [Google Scholar]
- Butt HJ, Wang DN, Hansma PK, Kühlbrandt W (1991) Effect of surface roughness of carbon support films on high-resolution electron diffraction of two-dimensional protein crystals. Ultramicroscopy 36: 307–318 [Google Scholar]
- Cai K, Langen R, Hubbell WL, Khorana HG (1997) Structure and function in rhodopsin: topology of the C-terminal polypeptide chain in relation to the cytoplasmic loops. Proc Natl Acad Sci USA 94: 14267–14272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar DLD, Kirschner DA (1971) Myelin membrane structure at 10 Å resolution. Nat New Biol 231: 46–52 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Crowther RA, Henderson R, Smith JM (1996) MRC image processing programs. J Struct Biol 116: 9–16 [DOI] [PubMed] [Google Scholar]
- Ernst OP, Meyer CK, Marin EP, Henklein P, Fu WY, Sakmar TP, Hofmann KP (2000) Mutation of the fourth cytoplasmic loop of rhodopsin affects binding of transducin and peptides derived from the carboxyl-terminal sequences of transducin alpha and gamma subunits. J Biol Chem 275: 1937–1943 [DOI] [PubMed] [Google Scholar]
- Franke RR, Konig B, Sakmar TP, Khorana HG, Hofmann KP (1990) Rhodopsin mutants that bind but fail to activate transducin. Science 250: 123–125 [DOI] [PubMed] [Google Scholar]
- Franke RR, Sakmar TP, Graham RM, Khorana HG (1992) Structure and function in rhodopsin. Studies of the interaction between the rhodopsin cytoplasmic domain and transducin. J Biol Chem 267: 14767–14774 [PubMed] [Google Scholar]
- Franks NP, Lieb WR (1979) The structure of lipid bilayers and the effects of general anaesthetics. An X-ray and neutron diffraction study. J Mol Biol 133: 469–500 [DOI] [PubMed] [Google Scholar]
- Fritze O, Filipek S, Kuksa V, Palczewski K, Hofmann KP, Ernst OP (2003) Role of the conserved NPxxY(x)5,6F motif in the rhodopsin ground state and during activation. Proc Natl Acad Sci USA 100: 2290–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyoshi Y (1998) The structural study of membrane proteins by electron crystallography. Adv Biophys 35: 25–80 [DOI] [PubMed] [Google Scholar]
- Havelka WA, Henderson R, Oesterhelt D (1995) Three-dimensional structure of halorhodopsin at 7 Å resolution. J Mol Biol 247: 726–738 [DOI] [PubMed] [Google Scholar]
- Henderson R, Baldwin JM, Ceska TA, Zemlin F, Beckmann E, Downing KH (1990) Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol 213: 899–929 [DOI] [PubMed] [Google Scholar]
- Henderson R, Baldwin JM, Downing KH, Lepault J, Zemlin F (1986) Structure of purple membrane from Halobacterium halobium: recording, measurement and evaluation of electron micrographs at 3.5 Å resolution. Ultramicroscopy 19: 147–178 [Google Scholar]
- Hubbell WL, Altenbach C, Hubbell CM, Khorana HG (2003) Rhodopsin structure, dynamics, and activation: a perspective from crystallography, site-directed spin labeling, sulfhydryl reactivity, and disulfide cross-linking. Adv Protein Chem 63: 243–290 [DOI] [PubMed] [Google Scholar]
- Imai H, Mizukami T, Imamoto Y, Shichida Y (1994) Direct observation of the thermal equilibria among lumirhodopsin, metarhodopsin I, and metarhodopsin II in chicken rhodopsin. Biochemistry 33: 14351–14358 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47 (Part 2): 110–119 [DOI] [PubMed] [Google Scholar]
- Kim JE, Pan D, Mathies RA (2003) Picosecond dynamics of G-protein coupled receptor activation in rhodopsin from time-resolved UV resonance Raman spectroscopy. Biochemistry 42: 5169–5175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kito Y, Suzuki T, Azuma M, Sekoguti Y (1968) Absorption spectrum of rhodopsin denatured with acid. Nature 218: 955–957 [DOI] [PubMed] [Google Scholar]
- Kochendoerfer GG, Kaminaka S, Mathies RA (1997) Ultraviolet resonance Raman examination of the light-induced protein structural changes in rhodopsin activation. Biochemistry 36: 13153–13159 [DOI] [PubMed] [Google Scholar]
- Krebs A, Edwards PC, Villa C, Li J, Schertler GF (2003) The three-dimensional structure of bovine rhodopsin determined by electron cryomicroscopy. J Biol Chem 278: 50217–50225 [DOI] [PubMed] [Google Scholar]
- Krebs A, Villa C, Edwards PC, Schertler GF (1998) Characterisation of an improved two-dimensional p22121 crystal from bovine rhodopsin. J Mol Biol 282: 991–1003 [DOI] [PubMed] [Google Scholar]
- Kühn H, Mommertz O, Hargrave PA (1982) Light-dependent conformational change at rhodopsin's cytoplasmic surface detected by increased susceptibility to proteolysis. Biochim Biophys Acta 679: 95–100 [Google Scholar]
- Lamola AA, Yamane T, Zipp A (1974) Effects of detergents and high pressures upon the metarhodopsin I–metarhodopsin II equilibrium. Biochemistry 13: 738–745 [DOI] [PubMed] [Google Scholar]
- Lewis JW, Kliger DS (1992) Photointermediates of visual pigments. J Bioenerg Biomembr 24: 201–210 [DOI] [PubMed] [Google Scholar]
- Lewis JW, van Kuijk FJ, Carruthers JA, Kliger DS (1997) Metarhodopsin III formation and decay kinetics: comparison of bovine and human rhodopsin. Vis Res 37: 1–8 [DOI] [PubMed] [Google Scholar]
- Lin SW, Sakmar TP (1996) Specific tryptophan UV-absorbance changes are probes of the transition of rhodopsin to its active state. Biochemistry 35: 11149–11159 [DOI] [PubMed] [Google Scholar]
- Marin EP, Krishna AG, Zvyaga TA, Isele J, Siebert F, Sakmar TP (2000) The amino terminus of the fourth cytoplasmic loop of rhodopsin modulates rhodopsin–transducin interaction. J Biol Chem 275: 1930–1936 [DOI] [PubMed] [Google Scholar]
- Marr K, Peters KS (1991) Photoacoustic calorimetric study of the conversion of rhodopsin and isorhodopsin to lumirhodopsin. Biochemistry 30: 1254–1258 [DOI] [PubMed] [Google Scholar]
- Matthews RG, Hubbard R, Brown PK, Wald G (1963) Tautomeric forms of metarhodopsin. J Gen Physiol 47: 215–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon ST, Han M, Sakmar TP (2001) Rhodopsin: structural basis of molecular physiology. Physiol Rev 81: 1659–1688 [DOI] [PubMed] [Google Scholar]
- Mielke T, Alexiev U, Glasel M, Otto H, Heyn MP (2002a) Light-induced changes in the structure and accessibility of the cytoplasmic loops of rhodopsin in the activated MII state. Biochemistry 41: 7875–7884 [DOI] [PubMed] [Google Scholar]
- Mielke T, Villa C, Edwards PC, Schertler GF, Heyn MP (2002b) X-ray diffraction of heavy-atom labelled two-dimensional crystals of rhodopsin identifies the position of cysteine 140 in helix 3 and cysteine 316 in helix 8. J Mol Biol 316: 693–709 [DOI] [PubMed] [Google Scholar]
- Mitchell DC, Straume M, Litman BJ (1992) Role of sn-1-saturated,sn-2-polyunsaturated phospholipids in control of membrane receptor conformational equilibrium: effects of cholesterol and acyl chain unsaturation on the metarhodopsin I metarhodopsin II equilibrium. Biochemistry 31: 662–670 [DOI] [PubMed] [Google Scholar]
- Mitchell DC, Straume M, Miller JL, Litman BJ (1990) Modulation of metarhodopsin formation by cholesterol-induced ordering of bilayer lipids. Biochemistry 29: 9143–9149 [DOI] [PubMed] [Google Scholar]
- Okada T, Fujiyoshi Y, Silow M, Navarro J, Landau EM, Shichida Y (2002) Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography. Proc Natl Acad Sci USA 99: 5982–5987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M (2000) Crystal structure of rhodopsin: a G protein-coupled receptor. Science 289: 739–745 [DOI] [PubMed] [Google Scholar]
- Resek JF, Farahbakhsh ZT, Hubbell WL, Khorana HG (1993) Formation of the meta II photointermediate is accompanied by conformational changes in the cytoplasmic surface of rhodopsin. Biochemistry 32: 12025–12032 [DOI] [PubMed] [Google Scholar]
- Schertler GF, Hargrave PA (1995) Projection structure of frog rhodopsin in two crystal forms. Proc Natl Acad Sci USA 92: 11578–11582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertler GF, Villa C, Henderson R (1993) Projection structure of rhodopsin. Nature 362: 770–772 [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Hills GJ (1981) Tilted specimen in the electron microscope—a simple specimen holder and the calculation of tilt angles for crystalline specimens. Micron 12: 279–282 [Google Scholar]
- Spooner PJ, Sharples JM, Goodall SC, Seedorf H, Verhoeven MA, Lugtenburg J, Bovee-Geurts PH, DeGrip WJ, Watts A (2003) Conformational similarities in the beta-ionone ring region of the rhodopsin chromophore in its ground state and after photoactivation to the metarhodopsin-I intermediate. Biochemistry 42: 13371–13378 [DOI] [PubMed] [Google Scholar]
- Teller DC, Okada T, Behnke CA, Palczewski K, Stenkamp RE (2001) Advances in determination of a high-resolution three-dimensional structure of rhodopsin, a model of G-protein-coupled receptors (GPCRs). Biochemistry 40: 7761–7772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson TE, Lewis JW, Wallace-Williams SE, Kliger DS (1992) Photolysis of rhodopsin results in deprotonation of its retinal Schiff's base prior to formation of metarhodopsin II. Photochem Photobiol 56: 1135–1144 [DOI] [PubMed] [Google Scholar]
- Unger VM, Hargrave PA, Baldwin JM, Schertler GF (1997) Arrangement of rhodopsin transmembrane alpha-helices. Nature 389: 203–206 [DOI] [PubMed] [Google Scholar]
- Unger VM, Schertler GF (1995) Low resolution structure of bovine rhodopsin determined by electron cryo-microscopy. Biophys J 68: 1776–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel R, Ruprecht J, Villa C, Mielke T, Schertler GF, Siebert F (2004) Rhodopsin photoproducts in 2D crystals. J Mol Biol 338: 597–609 [DOI] [PubMed] [Google Scholar]
- Vogel R, Siebert F (2002) Conformation and stability of alpha-helical membrane proteins. 2. Influence of pH and salts on stability and unfolding of rhodopsin. Biochemistry 41: 3536–3545 [DOI] [PubMed] [Google Scholar]
- Vogel R, Siebert F, Mathias G, Tavan P, Fan G, Sheves M (2003) Deactivation of rhodopsin in the transition from the signaling state meta II to meta III involves a thermal isomerization of the retinal chromophore C=N double bond. Biochemistry 42: 9863–9874 [DOI] [PubMed] [Google Scholar]
- Vonck J (2000) Parameters affecting specimen flatness of two-dimensional crystals for electron crystallography. Ultramicroscopy 85: 123–129 [DOI] [PubMed] [Google Scholar]
- Wald G (1968) Molecular basis of visual excitation. Science 162: 230–239 [DOI] [PubMed] [Google Scholar]
- Yan EC, Kazmi MA, Ganim Z, Hou JM, Pan D, Chang BS, Sakmar TP, Mathies RA (2003) Retinal counterion switch in the photoactivation of the G protein-coupled receptor rhodopsin. Proc Natl Acad Sci USA 100: 9262–9267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa T, Wald G (1963) Pre-lumirhodopsin and the bleaching of visual pigments. Nature 197: 1279–1286 [DOI] [PubMed] [Google Scholar]


