ABSTRACT
In the present study we addressed the function of the transcriptional activator Zrf1 in the generation of the 3 germ layers during in vitro development. Currently, Zrf1 is rather regarded as a factor that drives the expression of neuronal genes. Here, we have employed mouse embryonic stem cells and P19 cells to understand the role of Zrf1 in the generation of mesoderm-derived tissues like adipocytes, cartilage and heart. Our data shows that Zrf1 is essential for the transcriptional activation of genes that give rise to mesoderm and in particular heart development. In both, the mESC and P19 systems, we provide evidence that Zrf1 contributes to the generation of functional cardiomyocytes. We further demonstrate that Zrf1 binds to the transcription start sites (TSSs) of heart tissue-specific genes from the first and second heart field where it drives their temporal expression during differentiation. Taken together, we have identified Zrf1 as a novel regulator of the mesodermal lineage that might facilitate spatiotemporal expression of genes.
KEYWORDS: Zrf1, mesoderm, cardiomyocyte, heart fields, E14 cells, P19 cells
Introduction
Mouse embryonic stem cells (mESCs) derived from embryonic blastocysts at day 3.5 (E3.5) are self-renewing, pluripotent cells capable of generating all cell types of the embryo.1 After removal of the factors that maintain pluripotency ES cells differentiate into derivatives of the 3 embryonic germ layers: ectoderm, mesoderm and endoderm.2 The mesoderm derived heart is the first organ to develop in the growing embryo. During early gastrulation, cardiac progenitor cells arising from the anterior lateral mesoderm form the cardiac crescent at the midline of the embryo. Later they migrate in anterior-posterior direction from the primitive streak to generate 2 heart fields on both sides of the midline.3-5 Whereas the first heart field is responsible for the formation of the atria, left ventricle and the nodal conduction system, the second heart field is responsible for the formation of the right ventricle, outflow tract and part of the atria.6 The proper development of heart tissue involves complex network of signaling pathways and cardiogenic transcription factors, most of which are evolutionarily conserved. Due to the close proximity to the underlying endoderm, cardiac crescent is exposed to different signaling pathways during cardiomyogenesis.5 Nodal induced mesoderm formation is maintained by BMP signaling, which triggers Wnt signaling in the proximal epiblast.5 Wnt induces the expression of mesoendodermal markers, Brachyury and Eomes, which function upstream of Mesp1.7,8 Mesp1 represents one of the earliest transcription factors for cardiac progenitor specification. It is transiently expressed in the mesoderm and is required for the anterior-posterior movement of the embryo.9,10 Downstream of Mesp1 distinct cardiogenic transcription factors operate to facilitate the development of both heart fields. Nkx2.5, the mammalian homolog of the Drosophila gene Tinman11,12 is expressed in the early heart progenitor cells.4 Although Nkx2.5 knockout mice are still able to commit to the cardiac lineage Nkx2.5 plays a dominant role for the hierarchical cascade of cardiac transcription factors.13-15 While Hand1,16,17 Tbx518,19 and Gata420,21 are related to the formation of first heart field derived structures, Isl1,22,23 Hand2,16,17,24 Mef2c,25,26 MyocD27,28 are related to the formation of second heart field derived structures. In cardiac differentiation the spatiotemporal control of both pluripotency and differentiation genes is the key for proper development. In this regard, epigenetic regulation plays a fundamental role. Polycomb group repressor complexes (PRCs) were demonstrated to be regulators of gene expression as they partake in the establishment and maintenance of embryonic stem cell fates.29-31 Polycomb repressive complexes come in at least 2 flavors.32,33 PRC2 catalyzes the tri-methylation of histone H3 at lysine 27 (H3K27me3), which constitutes a hallmark of transcriptional repression.34 PRC1 establishes a mono-ubiquitylation of histone H2A at lysine 119 (H2AK119ub) by the action of the E3 ubiquitin ligase subunit Ring1A or Ring1B, respectively.35 PRC1 is thought to repress transcription through direct interaction with the general transcription machinery36 and likely independent of the H2AK119ub mark by chromatin condensation.37-40 H2A-ubiquitylation seems to be involved in the derepression of Polycomb target genes as it provides a recruitment platform for the chromatin component Zrf1.41 During differentiation of human teratocarcinoma cells Zrf1 is recruited to promoters of Polycomb target genes where it replaces PRC1.41 Further, it was shown that Zrf1 is required to establish neural progenitor cells (NPCs) from mESCs and to maintain NPC self-renewal.42 Neuronal differentiation triggered by Zrf1 is partially regulated by Id1, which interacts with Zrf1 and blocks its recruitment to chromatin.43 Furthermore, the Zrf1 C. elegans ortholog dnj11 is involved in asymmetric cell division of neurosecretory motoneuron neuroblasts.44 Taken together, Zrf1 describes a regulator of neuronal differentiation and contributes to the generation of ectoderm-derived lineages. However, not much is known about its involvement in the regulation of the other germ layers (endoderm and mesoderm). Given the high abundance of H2A-ubiquitylation and the genome-wide distribution of PRC1 complexes at genes of all 3 germ layers,31 we reasoned that Zrf1 might control the differentiation of the other germ layers. Previous studies pointed at a potential role of Zrf1 in the mesoderm-derived haematopoietic lineage as elevated Zrf1 expression was found in leukemic blasts.45
In the present study we analyzed the function of Zrf1 during differentiation to mesoderm-derived lineages including cartilage, adipocyte and cardiac lineages. To this end we examined the expression of marker genes of all germ layers in differentiating Zrf1 mESC knockdown cells. We observed a significant impact of Zrf1 at mesodermal marker genes during embryoid body (EB) formation, which is reflected in the deformation of the adipocyte, cartilage and cardiomyocyte lineages at later stages of in vitro development. Re-establishing Zrf1 expression in Zrf1 knockdown cell lines indicates that it is directly involved in the formation of these tissues. In particular, Zrf1 is essential for proper development of cardiac tissue as experiments with mESCs and P19 cells46,47 indicate. Mechanistically, Zrf1 binds to the transcriptional start sites (TSSs) of heart tissue-specific genes where it drives their temporal expression during differentiation. Taken together, we have identified Zrf1 as a novel regulator of the mesodermal lineage.
Results
Knockdown of Zrf1 in mESC provokes deformation of the mesoderm
To assess a potential function for Zrf1 during the generation of the 3 germ layers we generated mESCs either expressing a non-specific shRNA (Control) or shRNA targeting Zrf1 (shZrf1) by viral infection (Fig. 1A). We next used both cell lines for the generation of embryoid bodies (EBs) and analyzed the mRNA levels of selected marker genes of all germ layers during the first 6 days of EB formation (Fig. S1A). We observed that depletion of Zrf1 had an impact on the expression of ectodermal and endodermal genes but a more pronounced effect on mesodermal marker genes. Remarkably, we noted that the expression of mesodermal marker genes, such as Runx1, Mixl1 and Flk1 was affected drastically by Zrf1 knockdown. These data suggest that Zrf1 might play a role in the regulation of genes from all germ layers but that early during development it is particularly important to facilitate the generation of mesoderm. We next examined EBs derived from control and Zrf1 knockdown mESCs at later stages of development (Fig. 1B). After 16 days, control cells differentiated into EBs with structures including cystic cavities similar to the yolk-sac and an outer endodermal layer analogous to primitive endoderm (white arrows). In contrast, Zrf1 depleted cells failed to develop these structures. To gain a better understanding of the structural impairments observed in shZrf1 cells, we performed hematoxylin and eosin (H&E) staining of EBs derived from control and Zrf1 knockdown cells (Fig. 1C). Notably, whereas control cells readily formed important features of the 3 germ layers (black arrows indicate neural rosette, fibrous connective tissue and gut like epithelium, respectively), Zrf1 knockdown cells failed to form these tissues. In particular, we noticed that the generation of connective tissue, which is indicative of mesoderm development, was impaired in Zrf1 depleted cells. In agreement with these findings, we observed a drastic reduction of nuclear Brachyury consistent with its diminished protein levels in Zrf1 knockdown cells (Figs. 1D and S1B). Taken together, our data point at a critical function for Zrf1 in mesoderm development during EB formation.
Figure 1.
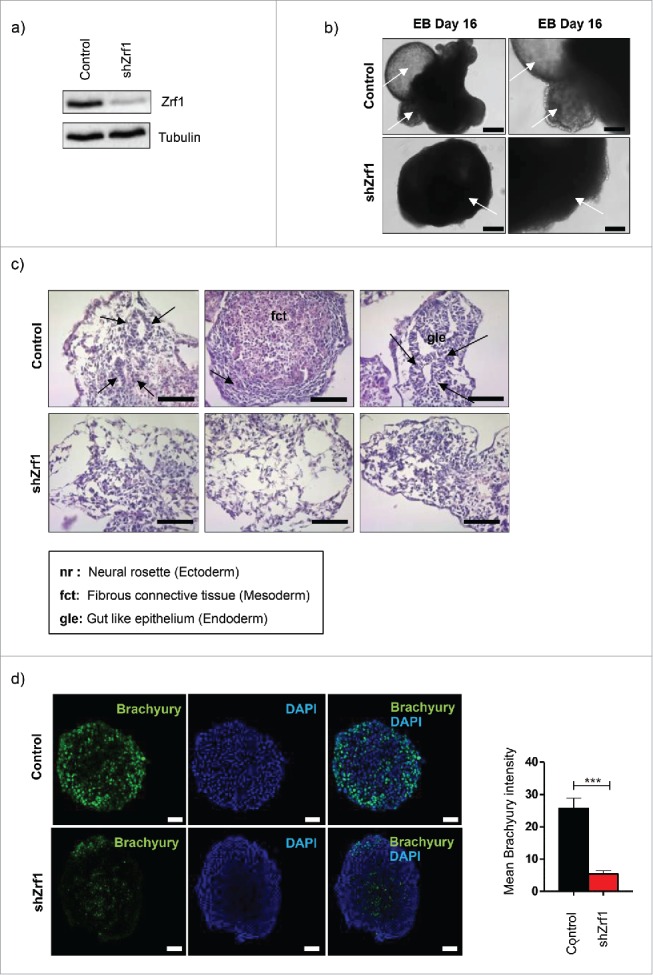
Embryoid bodies (EBs) derived from Zrf1 depleted cells show abnormal differentiation. (A) Western blot for Zrf1 after transfection of E14 cells with control and shRNA. Alpha tubulin was used as a loading control. (B) Brightfield images of control and shZrf1 derived EBs were taken at 10x (left panel) and 20x (right panel) magnification after 16 days of differentiation. White arrows indicate the presence or absence of structures similar to yolk-sac and external primitive endoderm in control EBs or Zrf1 depleted EBs, respectively. Scale bars: 200 µm (left panel) and 100 µm (right panel). (C) Representative images of hematoxylin and eosin stainings of control and Zrf1 knockdown EBs were taken at 40X magnification after 16 days of differentiation. Black arrows in the upper panel indicate the neural rosette, fibrous connective tissue and gut like epithelium, respectively. Scale bar, 50 µm. (D) Representative immunofluorescence images and analyses of control and Zrf1 knockdown EBs using Brachyury antibody (mesoderm marker) after 4 days of differentiation. Data represent the mean Brachyury (green) intensity of 3 independent experiments. ***p < 0.001 as calculated by 2-tailed unpaired t test. Scale bars, 50 µm.
Re-establishing Zrf1 expression in Zrf1 knockdown mESCs rescues the mesoderm phenotype
To ensure that the observed phenotype was due to the knockdown of Zrf1 we conducted functional rescue experiments. We restored Zrf1 expression in shZrf1 cells by transfecting plasmids encoding for human FLAG-tagged Zrf1. After antibiotic selection we established mESCs stably expressing FLAG-Zrf1 (Rescue) (Fig. 2A). We generated EBs from the 3 cell lines (control, shZrf1 and rescue) and monitored their phenotypes during 8 days (Fig. 2B). Starting from day 4, rescue cells exhibited a proper spherical structure similar to control cells and constituted a homogenous population as judged by their respective diameters throughout in vitro development (Fig. S2A). In comparison to Zrf1 knockdown cells the rescue cells continued to outgrow forming cystic cavities (Fig. 2B, white arrows). Next, we analyzed whether the previously observed mesoderm phenotypes were rescued by re-expression of Zrf1. To this end we generated histological sections of control, shZrf1 and rescue EBs and performed specific stainings for chondrogenesis and adipogenesis (Fig. 2C). To visualize mesodermal tissues such as cartilage (blue) and bone marrow (dark blue) we performed alcian blue stainings. Our data indicate that restoring Zrf1 levels caused a partial re-establishment of chondrogenic cells in agreement with our previous findings (Fig. 2C; upper panel). To further support a specific function for Zrf1 in mesoderm formation we analyzed its impact on the generation of adipocytes by performing oil-red-O stainings (Fig. 2C; lower panel). Whereas control cells have intense big red lipid droplets, as a marker of differentiated adipocytes, Zrf1 knockdown cells have significantly less lipid droplets with a comparably smaller size. Rescue cells, on the other hand, partially restore this defect exhibiting a more pronounced oil-red-O staining. These data indicate that Zrf1 is important during adipocyte differentiation and hence during mesoderm formation. Further, we assessed the generation of cardiomyocytes by their spontaneous beating activity (Fig. 2D and Supplemental Videos 1-3). In agreement with our previous findings we observed a drastic reduction of beating cell clusters upon Zrf1 knockdown, which was partially restored in rescue cell lines. Moreover FACS analysis of contraction-associated protein cTnT (cardiac troponin T) (Fig. S2B) performed with control, Zrf1 knockdown and rescue cells confirmed the beating phenotype. Taken together, our data implies that Zrf1 fulfills an essential function during mesoderm development.
Figure 2.
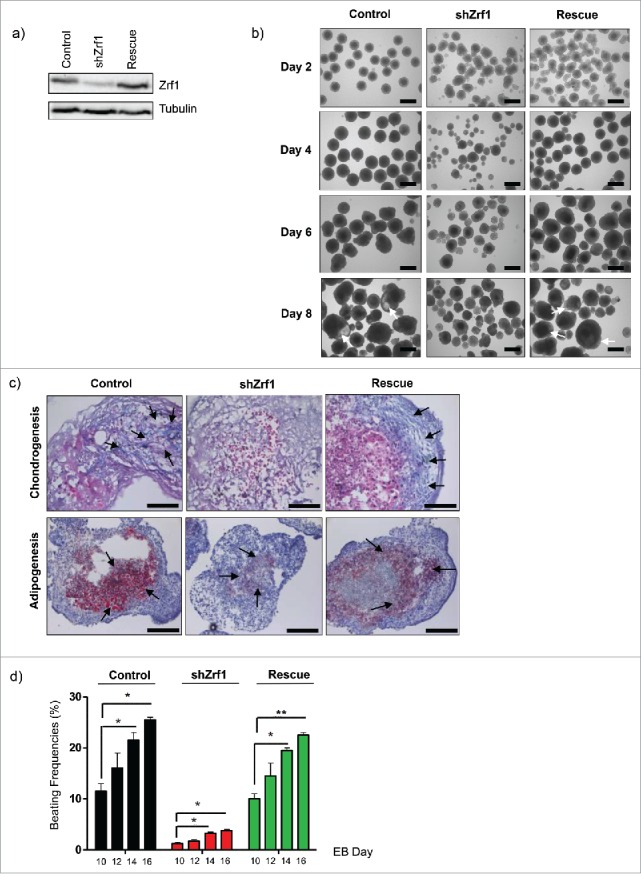
Restoration of Zrf1 expression in Zrf1 knockdown ES cells rescues the mesoderm phenotype. (A) Western blot analyzing Zrf1 levels in control, shZrf1 and rescue cells. Alpha tubulin was used as a loading control. (B) Brightfield images of control, shZrf1 and rescue cells derived EBs were taken at 4x magnification at days 0, 4, 6 and 8. White arrows indicate the cystic cavities in control and rescue cell derived EBs. Scale bars, 500 µm. (C) Representative images of alcian blue (upper panel) and oil-red-O (lower panel) stainings of control, Zrf1 knockdown and rescue EBs were taken at either 40x or 20x magnification after 16 days of differentiation. Black arrows indicate mesoderm derived tissues in alcian blue staining. Cartilage: blue, purple. Bone marrow: dark blue. Black arrows indicate mesoderm derived lipid droplets in oil-red-O staining. Scale bars, upper panel 50 µm, lower panel 100 µm. (D) The numbers of spontaneously beating EBs were counted under an inverted-light microscope on Day 10, 12, 14 and 16 of the culture. Data represent the average of 3 experiments, +/− SEM *p < 0.5, **p < 0.01, as calculated by 2-tailed unpaired t test.
Zrf1 controls the temporal expression of cardiomyogenesis specific genes
Based on our data and a previously published report showing the importance of PRC1 in cardiac mesoderm differentiation48 we decided to focus on the role of Zrf1 in cardiac differentiation. First we analyzed specific signaling genes important for cardiogenic mesoderm induction, which represents the first step of cardiomyogenesis (Fig. 3A). We observed that the expression of Nodal and BMP4 were slightly decreased in Zrf1 knockdown cells. In contrast, Wnt3a expression levels were not drastically affected. Next we analyzed 2 mesoendodermal markers, Brachyury and Eomes, together with one of the earliest cardiac transcription factors, Mesp1. We observed a dramatic and specific reduction of Brachyury and Mesp1 expression upon Zrf1 depletion, whereas Eomes levels did not significantly change. When examining the expression levels of important transcription factors of both heart fields we noticed that almost all were affected by Zrf1 depletion (Fig. 3A and B). To further explore the effect of Zrf1 depletion in adult cardiomyocytes we analyzed specific gene groups for atrium, ventricle and the nodal conduction system.49,50 We observed that the expression of nearly all the genes tested were affected in Zrf1 knockdown cells (Fig. S3A). These data suggest that Zrf1 is essential for the proper expression of genes that drive cardiomyogenesis but that it has no bias toward cardiac subtypes.
Figure 3.
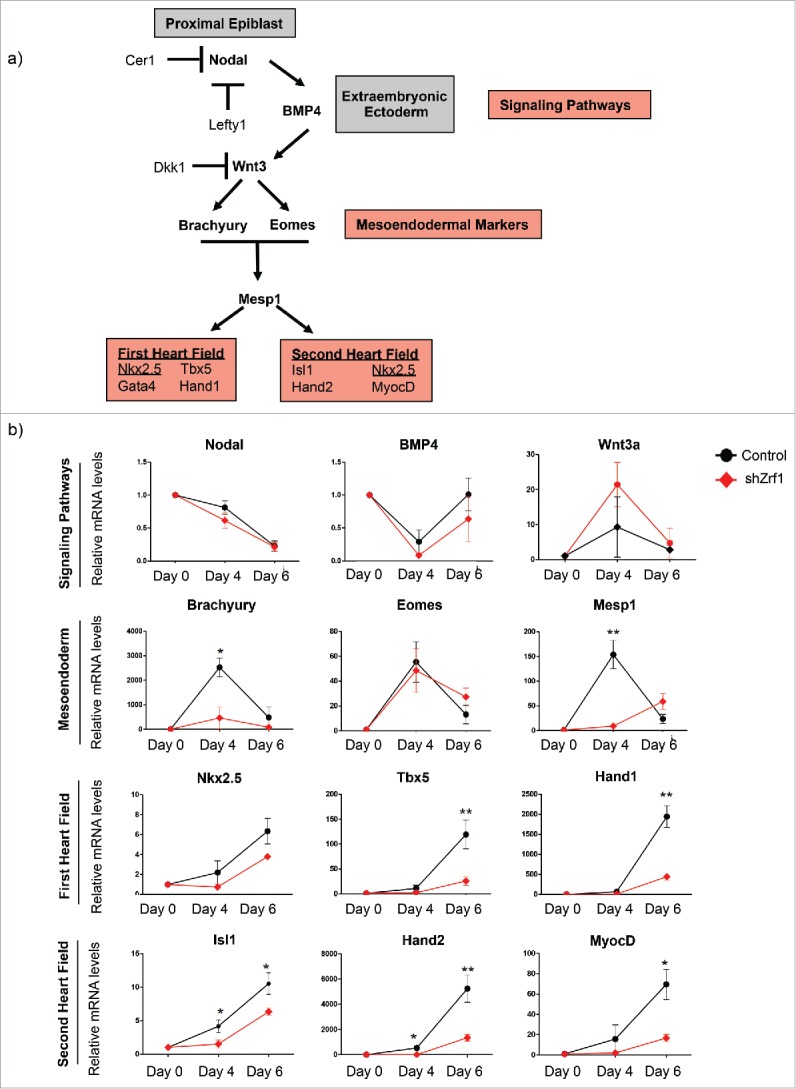
Zrf1 controls the temporal expression of the genes related to cardiomyogenesis. (A) Schematic illustration of cardiomyogenesis.6 Cardiomyogenesis begins with mesoderm induction which is triggered by Nodal signaling in the proximal epiblast. Nodal signaling maintains BMP4 expression in the extraembryonic ectoderm. BMP4 acts by inducing Wnt3 expression in the proximal epiblast. Nodal and Wnt signaling are restricted to the posterior epiblast by specific antagonists (Lefty1 and Cer1; Dkk1, respectively). Wnt induces the expression of mesoendodermal markers Brachyury and Eomes. Then Brachyury and Eomes together induce the expression of Mesp1. Downstream of Mesp1, a complex network of transcription factors, which belong to first and second heart fields, tightly control the proper development of cardiac tissues. (B) Real-time qPCR of signaling pathway, mesoendoderm, Mesp1, first and second heart field related genes at days 0, 4 and 6 of EB differentiation. Expression was normalized to the housekeeping gene S18. Data represent the average of 5 experiments, +/− SEM *p < 0.5, **p < 0.01, ***p < 0.001 as calculated by 2-tailed unpaired t test.
To further demonstrate a specific role for Zrf1 during cardiac development we assessed the expression of selected cardiomyogenesis genes in the rescue cell lines (Fig. 4A). We noticed a partial or complete restoration of the expression of first and second heart field related genes. However, genes involved in the signaling pathways, Brachyury or Mesp1 (data not shown) were either not affected by Zrf1 depletion or showed enhanced expression in rescue cells. Collectively, these data show that re-expression of Zrf1 is not sufficient to rescue the expression of signaling genes (Fig. 3A), yet it plays a direct role in the transcriptional activation of genes essential for the formation of the heart fields. To investigate the direct relationship between Zrf1 and heart field related genes, we carried out ChIP experiments at days 2, 4 and 6 after EB generation utilizing a mESC line expressing a FLAG-tagged Zrf1 fusion protein. We observed that Zrf1 is highly enriched at the TSSs of selected genes at day 4, which seems to be a critical time point in Zrf1 mediated gene expression (Fig. S4A). Likewise, in ChIP experiments employing Zrf1 antibodies we found Zrf1 specifically occupying the TSSs of Nkx2.5, Tbx5, Hand1 and Hand 2 pointing at a function in activating these genes, similar to the Zrf1 dependent activation of neuronal lineage genes as previously reported41 (Fig. 4B).
Figure 4.
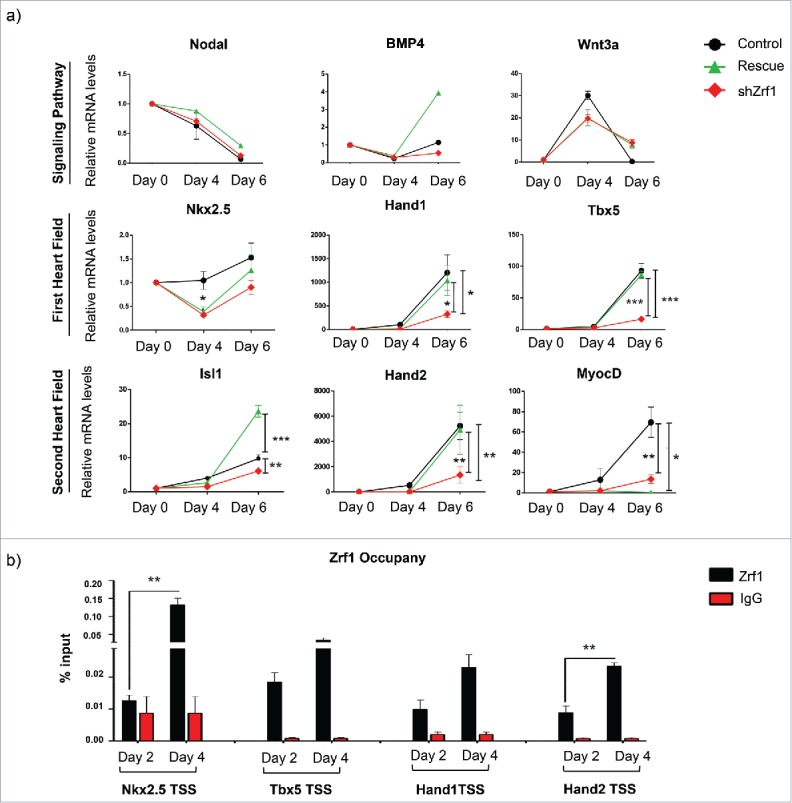
Zrf1 plays a direct role in the transcriptional activation of some cardiomyogenesis specific genes. (A) Real-time qPCR of control, shZrf1 and rescue EBs for cardiomyogenesis specific genes at days 0, 4 and 6 of EB differentiation. Expression was normalized to the housekeeping gene S18. Data represent the average of 3 experiments, +/− SEM *p < 0.5, **p < 0.01, *** p < 0.001 as calculated by 2-tailed unpaired t test. (B) Zrf1 ChIP-qPCR of selected genes (Nkx2.5, Tbx5, Hand1 and Hand2) in FLAG-tagged Zrf1 cells derived EBs after 2 and 4 days of differentiation. Values are expressed as percentage of input. Data represent the average of 3 experiments, +/− SEM **p < 0.01, as calculated by 2-tailed unpaired t test.
Collectively these data suggest that Zrf1 binds the promoters of cardiomyogenesis specific genes early during development. Zrf1 dependent activation of its target genes causes the temporal expression of genes and thereby contributes to faithful cardiomyocyte formation.
Zrf1 is essential for cardiomyocyte differentiation in P19 cells
During murine embryonic development Zrf1 plays a role in regulating genes that contribute to the generation of the first and second heart fields. To further emphasize our findings we next investigated a potential Zrf1 function in P19 cells. P19 cells are a pluripotent cell line derived from a teratocarcinoma induced in C3H/HeHa mice,46,47,51 which are able to differentiate into a variety of cell types representative of all 3 germ layers when induced by chemical agents.47 In particular, P19 cells provide an excellent cell differentiation model system that mimics the events of early cardio-embryogenesis.52 Thus, we generated P19 cell lines by stably integrating plasmids encoding short hairpin RNAs targeting Zrf1 (shZrf1) or a non-specific sequence (shControl). After verifying the knockdown levels in 2 Zrf1 knockdown clones (Fig. 5A), we next assessed the Zrf1 protein levels after supplementing the cells with 1% DMSO, which causes spontaneous differentiation.52 In line with our previous data, we observed that Zrf1 expression was increasing until day 4 after DMSO treatment (Fig. S5A). Beyond this time point Zrf1 expression was decreasing stepwise suggesting a function early during P19 differentiation comparable to the Zrf1 function during stem cell differentiation. To further investigate the effect of Zrf1 depletion in adult cardiomyocytes, we analyzed the expression of genes, which are present in functional cardiomyocytes such as cardiac troponins (cTnT and cTnI), myosin heavy chains (α-MHC and β-MHC) and myosin light chains (MLC2a and MLC2v). Moreover, we assessed the expression levels of genes encoding for gap junction proteins (Cx40 and Cx43) and genes that are responsible for the pacemaker activity of the heart (HCN4 and ANP) (Fig. 5B). We observed that Zrf1 had an impact on the expression of all of the genes tested. Particularly the expression of genes, which are essential for cardiac muscle formation (cTnT, cTnI, α-MHC, β-MHC, MLC2a), showed a strong dependence on Zrf1. Moreover, we performed cardiac troponin I staining to confirm the presence of contractile units in control cells (Fig. S5B). These data suggest that Zrf1 plays a role in the generation of the murine heart. To further support this hypothesis we assessed the beating rate of control and Zrf1 knockdown cell lines at days 12 and 15 of cardiac differentiation. When analyzing areas of both monolayer cultures we observed in control conditions about 14% (Day 12) and 26 % (Day 15) beating activity (Fig. 5C and Supplemental Videos 4 and 5). In contrast, when analyzing comparable monolayer regions of Zrf1 knockdown cells we nearly did not detect any beating activity. Collectively, these data show that Zrf1 is essential for the proper development of cardiac tissue.
Figure 5.
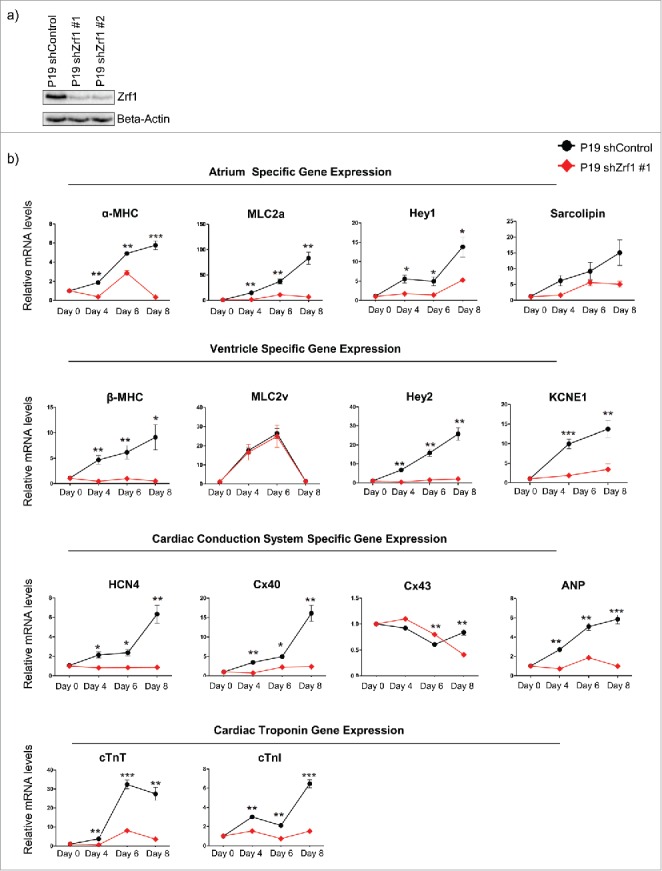
Zrf1 is essential for cardiomyocyte differentiation in P19 cells. (A) Western blot for Zrf1 after transfection of P19 cells with 2 different shRNAs. Beta actin was used as a loading control. (B) Real-time qPCR of atrium, ventricle, nodal conduction system specific genes together with cardiac troponin genes at days 0, 4, 6 and 8 days of cardiac differentiation. Expression was normalized to the housekeeping gene RPO. Data represent the average of 3 experiments, +/− SEM *p < 0.5, **p < 0.01, ***p < 0.001 as calculated by 2-tailed unpaired t test. (C) Representative images and quantifications of beating areas of control and Zrf1 knockdown P19 cells after 12 and 15 days of cardiac differentiation. The spontaneous beating areas in P19 cells were recorded with either a Leica DM-IL or Leica AF7000 microscope and quantified by using ImageJ program. Data represent the average of 3 experiments, +/− S.E.M. *p < 0.5, **p < 0.01, ***p < 0.001 as calculated by two-tailed unpaired t test. Scale bars, 100 µm.
Discussion
Cardiovascular diseases (CVDs) are the leading cause of death worldwide. Whether caused by congenital defects or acquired injuries, CVDs usually result in loss of terminally differentiated cardiomyocytes. Adult human cardiomyocytes have limited self-renewal capacity. For this reason, stem cell-based regenerative therapy emerges as one of the most promising approaches to fight CVDs. Regenerative therapy aims at the replacement of injured or defective cardiac tissues by healthy ones making use of various cell sources.53 Still, faithful cardiac regenerative therapy will rely on understanding the underlying molecular mechanisms of cardiac development.54 The transcriptional regulation of cardiac development requires precise spatiotemporal control of gene expression. Hence, epigenetic regulators play an essential role by establishing a cell-type specific chromatin pattern which is essential for cell commitment and differentiation.55 Among many epigenetic players, Trithorax and Polycomb group proteins have attracted particular attention for their ability to activate and repress transcription. Recently, Mel18, a subunit of PRC1 was shown to impact on mesoderm differentiation by directly controlling the expression of transcription factors essential for early cardiac mesoderm cell specification.48 Still, many open questions remain as to how PRCs regulate transcription during cardiomyogenesis.
In the present study, we elucidated a specific function of Zrf1 during the early stages of mesoderm development. The H2A-ubiquitin binding Zrf1 protein replaces PRC1 from promoters of its targets genes thereby contributing to transcriptional activation.41,56 So far, Zrf1 was shown to primarily operate during differentiation into the neuronal lineage.41-43 Notably, PRC1 silences a wide range of genes generating the 3 germ layers during development.31 Hence, taking into account the broad spectrum of PRC1 target genes and the high abundance of H2A-ubiquitylation, we speculated that Zrf1 played additional roles in the formation of other germ layers. The phenotype of Zrf1 depleted EBs and the deregulation of key genes from the 3 germ layers during in vitro development suggest a broader role of Zrf1 during development. We observed a particularly pronounced effect during the generation of mesoderm as the expression of one of the key factors, Brachyury, was drastically impaired in Zrf1 knockdown cells. Moreover, we noticed a significant reduction of the expression of Flk1, Runx1 and Mixl1 genes, which are important mesodermal markers for hematopoiesis. In agreement with these data it was previously shown that expression of Zrf1 is upregulated in acute and chronic myeloid leukemia.45 Analyzing Zrf1 knockdown derived EBs we observed malformation of 3 different mesoderm derived tissues (adipocytes, cartilage and cardiac tissue). Importantly, re-establishing Zrf1 expression in the knockdown cells led to a significant rescue of the phenotypes. Taken together, these results point at an essential regulatory role for Zrf1 in the generation of mesoderm derived tissues.
Focusing on differentiation of mESC and P19 cells we found that Zrf1 plays a prominent role in cardiomyocyte differentiation. Analyzing the transcription of selected cardiac-specific genes in control and Zrf1 depleted EBs, we provide evidence that Zrf1 affects the expression of almost every tested gene. However, rescue experiments indicated a specific involvement of Zrf1 only in the transcriptional activation of first and second heart field related genes (Fig. 4A). Hand1, Hand2, Tbx5 are essential transcription factors for left-right patterning of the heart. Their expression is influenced by the transcription factor Nkx2.5, which represents a critical regulator of cardiomyogenesis.16-18 Notably, Zrf1 shows the strongest enrichment at the TSS of Nkx2.5 thereby controlling the main transcriptional regulator driving the development of both heart fields (Figs. 4B and S4A). Hence, the restoration of gene expression of other target genes (Hand1, Hand2 and Tbx5) in the rescue cell line could partially be a consequence of Zrf1 dependent Nkx2.5 expression.
At the physiological level we observed a marked decrease of the beating activity in E14 derived Zrf1 knockdown cells, which was partially rescued by re-establishing Zrf1 expression. Since differentiation of E14 cells establishes a mixed population of differentiated cells we used a well-characterized model system of cardiac differentiation. Employing P19 cells we confirmed that Zrf1 is essential for the proper development of cardiomyocytes, which exhibit contractile units of reminiscent of heart muscles and rhythmic beating activities as judged by RT-PCR, microscopy and beating assay results of control and Zrf1 knockdown cells (Figs. 5B, C and S5B).
In conclusion, Zrf1 contributes to the generation of the mesoderm layer but in particular it promotes the generation of the cardiac lineage during in vitro development. Hence, the function of Zrf1 needs to be considered when addressing molecular mechanisms of cardiovascular diseases.
Materials and methods
Plasmids used in this study
The lentiviral shRNA#1/2 and pCBA-3xFlag plasmids were kind gifts from Dr. Luciano di Croce. The sequences of lentiviral shRNA #1/2 are listed in Supplemental Table S1. Non-Mammalian shControl was purchased from Sigma.
Generation of stable mESC and P19 knockdown cultures
For the production of lentiviruses containing shRNA, HEK293T cells were grown overnight in 10 cm dishes and the next day the medium was replaced with OPTIMEM low serum medium (Gibco), 2 hours prior to transfection. For the transfection, 15 μg shRNA, 5 μg pMDLg/pRRe (Addgene), 5 μg pRSV-Rev (Addgene) and 5 μg pMD2.G (Addgene) plasmids were mixed with 500 μl OPTIMEM and 43 μg Lipofectamine 2000 (Life Technologies) with OPTIMEM. 5 min later, plasmids were mixed with Lipofectamine and 10 min later the mixture was added to the HEK293T cells. 9 hours post-transfection, the OPTIMEM was replaced with 6 ml complete stem cell medium and the following day the medium containing the viral particles was collected and passed through a 0.45 μm filter (Millipore). 2 ml of filtered medium containing the viral particles was added for transduction of 2 × 105 stem cells in a 6-well plate (pre-incubated with 8 μg/ml polybrene (Sigma) for 2 hours) and the plates were centrifuged for 90 min at 1200 rpm at room temperature. 2ml fresh medium was added per well and cells were incubated overnight. The same procedure was repeated 3 more times. Subsequently the cells were left to recover for 48 hours. Knockdown cells were selected with 2 μg/ml puromycin (Sigma) for 3 days and then frozen or analyzed for RNA or protein expression. For rescue experiments, the pCBA-3xFlag vector was transfected into shZrf1 cells using Lipofectamine 2000 according to the manufacturer's protocol. shRNA #1/2 and non-mammalian control plasmids were transfected into P19 cells with Lipofectamine 2000 as well. A total of 3 × 105 cells were transfected with 5 µg plasmid. 5 hours later, cells were split in 2 10 cm plate and left to recover overnight. Next day, the medium was changed and cells were selected with 4 μg/ml puromycin for 4 days.
Cell culture and differentiation
Embryonic stem cells (E14Tg2A) were cultured in feeder-free plates, coated for at least 30 min with 0.1% Gelatin, ready to use (Millipore) or in powder form (Sigma), dissolved in MQ water after overnight stirring at 75°C. Cells were cultured in Glasgow Minimum Essential Medium (Sigma) supplemented with 15% PanSera ES Bovine Serum (PAN-Biotech), Non-Essential Amino Acids (Gibco), Sodium Pyruvate (Gibco), L-Glutamine (Gibco), Penicillin/Streptomycin (Gibco), β-mercaptoethanol (Sigma) and Leukemia Inhibitory Factor (LIF). E14Tg2A embryoid bodies (EBs) were formed with the hanging-drop method, starting with 103 mESCs / 30 μL drop, in medium without LIF. 48 hours after the drop formation, EBs were collected and cultured from that point on in petri dishes. The medium was replaced every second day. Images were acquired with a Leica DM-IL microscope. EB size distributions were quantified with ImageJ analysis by measuring the Feret's diameter of each EB. Murine P19 ECCs were kind gift from Dr. Vijay Tiwari. The P19 cells were cultured in DMEM (Gibco) supplemented with 10% fetal bovine serum (Gibco), L-Glutamine (Gibco) and Penicillin/Streptomycin (Gibco). To induce cardiac differentiation, a total of 3 × 105 P19 cells were aggregated to form embryoid bodies in the presence of 1 % DMSO (Sigma) for 4 days. The formed EBs were then transferred to gelatin coated 6-well culture plates and cultured in complete medium free of DMSO for an additional 11 days. The medium was replenished every 48 hours.
Histological stainings
EBs were collected after 16 days of differentiation and fixed with 4% PFA at room temperature for 30 min. Fixed EBs were treated with 10% and 20% sucrose for 30 min each and stored in 30% sucrose overnight at 4 °C. The next day EBs were embedded in OCT (VWR) and frozen at −80°C. Frozen blocks were sectioned at an 8 μm thickness and H&E staining was performed according to standard protocols. For alcian blue staining, sections were stained with alcian and hematoxylin dyes together for 10 minutes. Afterwards they were counterstained with eosin. For oil red O staining, sections were firstly stained with oil red O dye for 20 minutes, then counterstained with hematoxylin. Pictures were acquired with a Leica DM2500 microscope.
Western blot analysis
Cell pellets and EB samples referring to the aferomentioned time points were collected and washed with PBS. They were resuspended in 2x Laemmli buffer according to the pellet size, sonicated for 15 cycles (30 sec on/30 sec off) at high setting using a Diagenode Bioruptor plus. After boiling for 20 min at 95°C, samples were centrifuged for 15 at RT maximum speed and separated on 12% SDS-PAGE for Western Blot analysis. Zrf1 (Novus), Brachyury (Santa Cruz), Gata4 (Sigma) and H2A (Abcam) antibodies were used to detect the proteins.
Chromatin immunoprecipitations
ChIPs were carried out as previously described57 with the following modifications: A) Differentiated EBs were collected, washed with PBS and then dissociated with Accutase (Gibco) enzyme for 3 minutes. After spin down, single-cell suspension were fixed in fixation buffer (11% freshly added formaldehyde / 0.1M NaCl/1mM EDTA/50mM HEPES-KOH pH 7.6) containing 1% formaldehyde for 15 min at room temperature and quenched with 0.125 M Glycine for additional 5 min at room temperature. B) Nuclear extracts were sonicated for 30 cycles (30 sec on/30 sec off) in a Bioruptor Plus. C) Sonicated chromatin was incubated for 16 hours at 4°C with either 5 μg Zrf1 (Novus) or Flag (Sigma) antibody together with magnetic beads (Milipore). 1 μg rabbit or mouse IgG was used as negative control.
RNA extraction and cDNA synthesis
RNA used for qPCR was extracted with Trizol Reagent. 1 μg of total RNA was used for cDNA, using the First Strand cDNA Synthesis Kit (Fermentas). The primers used in the reverse transcription qPCR assays are listed in the Supplemental Table S1.
Immunofluorescence stainings and analysis
After differentiation in hanging drops, E14 cells derived EBs were fixed in 4% PFA for 30 minutes, treated with sucrose, embedded in OCT and frozen at −80°C. The next day frozen blocks were sectioned at an 8μm thickness. Differentiated P19 cells, which were grown on gelatin coated coverslips, were fixed in 4% PFA for 20 minutes. Both tissue sections and cells on coverslips were permeabilized with 0.5% Triton X-100 for 10 min, washed 3 times with 1x PBS, blocked with 5% fetal bovine serum in PBS for one hour. Sections were incubated either with Brachyury (Santa Cruz) or Cardiac Troponin I (Abcam) antibodies overnight at 4°C. The following day the sections were washed 3 times with PBS containing 0.2% Triton X-100 (wash buffer) for 3 times and incubated with secondary antibodies (Alexa Fluor 488) for 1.5 hours. Subsequently they were washed 3 times with wash buffer and mounted with Vectashield with DAPI. Fluorescence images were acquired on a Leica SP5 microscope. Quantification of Brachyury stainings in EBs is performed like this: As it was not possible to discern the individual nuclei to quantify the Brachyury staining per nucleus, the mean Brachyury staining intensity (intensity per area) in the entire EB nuclear area was measured instead. The DAPI staining image was smoothed with a Gaussian filter and the Nilblack local threshold was applied to segment the nuclei. The holes in the segmented image were filled, the outlines were smoothed with the binary open operation and objects smaller than 20 µm were excluded. The mean Brachyury staining intensity was measured inside the segmented nuclei.
Beating assays
The numbers of spontaneously beating EBs were counted under an inverted-light microscope at the respective time points. For each experiment, at least 100 EBs from each cell type were counted. The spontaneous beating areas in P19 cells were recorded with either a Leica DM-IL or Leica AF7000 microscope. To quantify the beating areas in P19 cells, differences between the consecutive frames in each movie were calculated. The differences between images were smoothed with a Gaussian filter and the triangle method threshold was applied to segment into beating and not-beating parts. A maximum projection image was calculated to account for the beating areas from the entire movie. The resulting image was smoothed with a minimum filter and beating and not-beating areas were measured. The analysis was performed with ImageJ software.
Flow cytometry
After 13 days of differentiation, EBs from control, Zrf1 knockdown and rescue mESCs were collected and washed twice with PBS. Following the washes, EBs were incubated in %0.25 Trypsin-EDTA for 5 minutes at 37°C. Enzymatic activity was stopped using 50% FBS in PBS and dissociation was completed employing mechanical force using a 20G needle. Dissociated EBs were fixed and permeabilized 20 minutes on ice using a BD Cytofix/Cytoperm kit. Prior to cTnT-PE (BD-Pharmingen) staining, cells were blocked with 1%BSA for 15 minutes. Staining was performed on ice for 2 hours in the dark. During each step, cells were washed twice with wash buffer contained in the aforementioned kit. After the last wash, cells were resuspended in staining buffer (1% FBS in PBS) and analyzed with an LSRFortessa cell analyzer (BD Biosciences). The cTnT positive cells (cTnT+) sorted from the 3 cell lines (control, Zrf1 knockdown and rescue) are represented as a relative ratio (Fig. S2b). cTnT+ cells from Zrf1 knockdown and rescue cells are normalized to cTnT+ cells in control cells and the values were calculated as fold change.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to the IMB microscopy and flow cytometry facilities for their expert help and assistance. We would like to thank Dr. Luciano Di Croce for shRNA plasmids and Dr. Vijay Tiwari for the P19 cell line used in this study. We thank the members of the Richly laboratory for comments on the manuscript.
Funding
This work was supported by grants from the Boehringer Ingelheim Foundation.
Author contributions
AK and HR conceived and designed the experiments. AK performed all experiments and analyzed the data. AK and HR wrote the paper.
ORCID
Holger Richly http://orcid.org/0000-0002-7711-0350
References
- [1].Niwa H. How is pluripotency determined and maintained? Development 2007; 134:635-46; PMID:17215298; http://dx.doi.org/ 10.1242/dev.02787 [DOI] [PubMed] [Google Scholar]
- [2].Smith AG. Embryo-derived stem cells: of mice and men. Annu Rev Cell Dev Biol 2001; 17:435-62; PMID:11687496; http://dx.doi.org/ 10.1146/annurev.cellbio.17.1.435 [DOI] [PubMed] [Google Scholar]
- [3].Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet 2005; 6:826-35; PMID:16304598; http://dx.doi.org/ 10.1038/nrg1710 [DOI] [PubMed] [Google Scholar]
- [4].Abu-Issa R, Kirby ML. Heart field: from mesoderm to heart tube. Annu Rev Cell Dev Biol 2007; 23:45-68; PMID:17456019; http://dx.doi.org/ 10.1146/annurev.cellbio.23.090506.123331 [DOI] [PubMed] [Google Scholar]
- [5].Burridge PW, Keller G, Gold JD, Wu JC. Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell stem cell 2012; 10:16-28; PMID:22226352; http://dx.doi.org/ 10.1016/j.stem.2011.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Dyer LA, Kirby ML. The role of secondary heart field in cardiac development. Dev Biol 2009; 336:137-44; PMID:19835857; http://dx.doi.org/ 10.1016/j.ydbio.2009.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Costello I, Pimeisl IM, Dräger S, Bikoff EK, Robertson EJ, Arnold SJ. The T-box transcription factor Eomesodermin acts upstream of Mesp1 to specify cardiac mesoderm during mouse gastrulation. Nat Cell Biol 2011; 13:1084-91; PMID:21822279; http://dx.doi.org/ 10.1038/ncb2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].David R, Jarsch VB, Schwarz F, Nathan P, Gegg M, Lickert H, Franz WM. Induction of MesP1 by Brachyury(T) generates the common multipotent cardiovascular stem cell. Cardiovasc Res 2011; 92:115-22; PMID:21632880; http://dx.doi.org/ 10.1093/cvr/cvr158 [DOI] [PubMed] [Google Scholar]
- [9].Bondue A, Lapouge G, Paulissen C, Semeraro C, Iacovino M, Kyba M, Blanpain C. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell 2008; 3:69-84; PMID:18593560; http://dx.doi.org/ 10.1016/j.stem.2008.06.009 [DOI] [PubMed] [Google Scholar]
- [10].Bondue A, Blanpain C. Mesp1: a key regulator of cardiovascular lineage commitment. Circ Res 2010; 107:1414-27; PMID:21148448; http://dx.doi.org/ 10.1161/CIRCRESAHA.110.227058 [DOI] [PubMed] [Google Scholar]
- [11].Azpiazu N, Frasch M. tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Gen Dev 1993; 7:1325-40. [DOI] [PubMed] [Google Scholar]
- [12].Bodmer R. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development 1993; 118:719-29; PMID:7915669 [DOI] [PubMed] [Google Scholar]
- [13].Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Gene Dev 1995; 9:1654-66; PMID:7628699 [DOI] [PubMed] [Google Scholar]
- [14].Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development 1999; 126:1269-80; PMID:10021345 [DOI] [PubMed] [Google Scholar]
- [15].Prall OW, Menon MK, Solloway MJ, Watanabe Y, Zaffran S, Bajolle F, Biben C, McBride JJ, Robertson BR, Chaulet H, et al.. An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell 2007; 128:947-59; PMID:17350578; http://dx.doi.org/ 10.1016/j.cell.2007.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].McFadden DG, Barbosa AC, Richardson JA, Schneider MD, Srivastava D, Olson EN. The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development 2005; 132:189-201; PMID:15576406; http://dx.doi.org/ 10.1242/dev.01562 [DOI] [PubMed] [Google Scholar]
- [17].Srivastava D. HAND proteins: molecular mediators of cardiac development and congenital heart disease. Trends Cardiovasc Med 1999; 9:11-18; PMID:10189962 [DOI] [PubMed] [Google Scholar]
- [18].Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner DA, Gessler M, Nemer M, Seidman CE, et al.. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 2001; 106:709-21; PMID:11572777 [DOI] [PubMed] [Google Scholar]
- [19].Takeuchi JK, Ohgi M, Koshiba-Takeuchi K, Shiratori H, Sakaki I, Ogura K, Saijoh Y, Ogura T. Tbx5 specifies the left/right ventricles and ventricular septum position during cardiogenesis. Development 2003; 130:5953-64; PMID:14573514; http://dx.doi.org/ 10.1242/dev.00797 [DOI] [PubMed] [Google Scholar]
- [20].Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Gen Dev 1997; 11:1048-60; PMID:9136932 [DOI] [PubMed] [Google Scholar]
- [21].Zhou P, He A, Pu WT. Regulation of GATA4 transcriptional activity in cardiovascular development and disease. Curr Top Dev Biol 2012; 100:143-69; PMID:22449843; http://dx.doi.org/ 10.1016/B978-0-12-387786-4.00005-1 [DOI] [PubMed] [Google Scholar]
- [22].Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell 2003; 5:877-89; PMID:14667410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Laugwitz KL, Moretti A, Lam J, Gruber P, Chen Y, Woodard S, Lin LZ, Cai CL, Lu MM, Reth M, et al.. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature 2005; 433:647-53; PMID:15703750; http://dx.doi.org/ 10.1038/nature03215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Srivastava D, Thomas T, Lin Q, Kirby ML, Brown D, Olson EN. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat Genet 1997; 16:154-60; PMID:9171826; http://dx.doi.org/ 10.1038/ng0697-154 [DOI] [PubMed] [Google Scholar]
- [25].Edmondson DG, Lyons GE, Martin JF, Olson EN. Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development 1994; 120:1251-63; PMID:8026334 [DOI] [PubMed] [Google Scholar]
- [26].Lin MH, Bour BA, Abmayr SM, Storti RV. Ectopic expression of MEF2 in the epidermis induces epidermal expression of muscle genes and abnormal muscle development in Drosophila. Dev Biol 1997; 182:240-55; PMID:9070325; http://dx.doi.org/ 10.1006/dbio.1996.8484 [DOI] [PubMed] [Google Scholar]
- [27].Small EM, Warkman AS, Wang DZ, Sutherland LB, Olson EN, Krieg PA. Myocardin is sufficient and necessary for cardiac gene expression in Xenopus. Development 2005; 132:987-97; PMID:15673566; http://dx.doi.org/ 10.1242/dev.01684 [DOI] [PubMed] [Google Scholar]
- [28].Huang J, Min Lu M, Cheng L, Yuan LJ, Zhu X, Stout AL, Chen M, Li J, Parmacek MS. Myocardin is required for cardiomyocyte survival and maintenance of heart function. Proc Natl Acad Sci U S A 2009; 106:18734-39; PMID:19850880; http://dx.doi.org/ 10.1073/pnas.0910749106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 2009; 10:697-708; PMID:19738629; http://dx.doi.org/ 10.1038/nrm2763 [DOI] [PubMed] [Google Scholar]
- [30].Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell 2010; 7:299-313; PMID:20804967; http://dx.doi.org/ 10.1016/j.stem.2010.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer 2009; 9:773-84; PMID:19851313; http://dx.doi.org/ 10.1038/nrc2736 [DOI] [PubMed] [Google Scholar]
- [32].Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell 2007; 128:735-45; PMID:17320510; http://dx.doi.org/ 10.1016/j.cell.2007.02.009 [DOI] [PubMed] [Google Scholar]
- [33].Morey L, Helin K. Polycomb group protein-mediated repression of transcription. Trend Biochem Sci 2010; 35:323-32; PMID:20346678; http://dx.doi.org/ 10.1016/j.tibs.2010.02.009 [DOI] [PubMed] [Google Scholar]
- [34].Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002; 298:1039-43; PMID:12351676; http://dx.doi.org/ 10.1126/science.1076997 [DOI] [PubMed] [Google Scholar]
- [35].Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004; 431:873-78; PMID:15386022; http://dx.doi.org/ 10.1038/nature02985 [DOI] [PubMed] [Google Scholar]
- [36].Simon JA, Kingston RE. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell 2013; 49:808-24; PMID:23473600; http://dx.doi.org/ 10.1016/j.molcel.2013.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science 2004; 306:1574-77; PMID:15567868; http://dx.doi.org/ 10.1126/science.1100576 [DOI] [PubMed] [Google Scholar]
- [38].Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi AI, Wutz A, et al.. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell 2010; 38:452-64; PMID:20471950; http://dx.doi.org/ 10.1016/j.molcel.2010.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell 2005; 20:845-54; PMID:16359901; http://dx.doi.org/ 10.1016/j.molcel.2005.12.002 [DOI] [PubMed] [Google Scholar]
- [40].Illingworth RS, Moffat M, Mann AR, Read D, Hunter CJ, Pradeepa MM, Adams IR, Bickmore WA. The E3 ubiquitin ligase activity of RING1B is not essential for early mouse development. Genes Dev 2015; 29:1897-902; PMID:26385961; http://dx.doi.org/ 10.1101/gad.268151.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Richly H, Rocha-Viegas L, Ribeiro JD, Demajo S, Gundem G, Lopez-Bigas N, Nakagawa T, Rospert S, Ito T, Di Croce L. Transcriptional activation of polycomb-repressed genes by ZRF1. Nature 2010; 468:1124-28; PMID:21179169; http://dx.doi.org/ 10.1038/nature09574 [DOI] [PubMed] [Google Scholar]
- [42].Aloia L, Di Stefano B, Sessa A, Morey L, Santanach A, Gutierrez A, Cozzuto L, Benitah SA, Graf T, Broccoli V, et al.. Zrf1 is required to establish and maintain neural progenitor identity. Gen Dev 2014; 28:182-97; PMID:24449271; http://dx.doi.org/ 10.1101/gad.228510.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Aloia L, Gutierrez A, Caballero JM, Di Croce L. Direct interaction between Id1 and Zrf1 controls neural differentiation of embryonic stem cells. EMBO Rep 2015; 16:63-70; PMID:25361733; http://dx.doi.org/ 10.15252/embr.201439560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hatzold J, Conradt B. Control of apoptosis by asymmetric cell division. PLoS biology 2008; 6:771-84; http://dx.doi.org/ 10.1371/journal.pbio.0060084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Greiner J, Ringhoffer M, Taniguchi M, Hauser T, Schmitt A, Döhner H, Schmitt M. Characterization of several leukemia-associated antigens inducing humoral immune responses in acute and chronic myeloid leukemia. Int J Cancer 2003; 106:224-31; PMID:12800198; http://dx.doi.org/ 10.1002/ijc.11200 [DOI] [PubMed] [Google Scholar]
- [46].van der Heyden MA, Defize LH. Twenty one years of P19 cells: what an embryonal carcinoma cell line taught us about cardiomyocyte differentiation. Cardiovasc Res 2003; 58:292-302; PMID:12757864 [DOI] [PubMed] [Google Scholar]
- [47].McBurney MW. P19 embryonal carcinoma cells. Int J Dev Biol 1993; 37:135-40; PMID:8507558 [PubMed] [Google Scholar]
- [48].Morey L, Santanach A, Blanco E, Aloia L, Nora EP, Bruneau BG, Di Croce L. Polycomb regulates mesoderm cell fate-specification in embryonic stem cells through activation and repression mechanisms. Cell stem cell 2015; 17:300-15; PMID:26340528; http://dx.doi.org/ 10.1016/j.stem.2015.08.009 [DOI] [PubMed] [Google Scholar]
- [49].Ng SY, Wong CK, Tsang SY. Differential gene expressions in atrial and ventricular myocytes: insights into the road of applying embryonic stem cell-derived cardiomyocytes for future therapies. Am J Physiol Cell Physiol 2010; 299, C1234-1249; PMID:20844252; http://dx.doi.org/ 10.1152/ajpcell.00402.2009 [DOI] [PubMed] [Google Scholar]
- [50].Liang X, Evans SM, Sun Y. Insights into cardiac conduction system formation provided by HCN4 expression. Trend Cardiovasc Med 2015; 25:1-9; http://dx.doi.org/ 10.1016/j.tcm.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].McBurney MW, Rogers BJ. Isolation of male embryonal carcinoma cells and their chromosome replication patterns. Dev Biol 1982; 89:503-08; PMID:7056443 [DOI] [PubMed] [Google Scholar]
- [52].Paquin J, Danalache BA, Jankowski M, McCann SM, Gutkowska J. Oxytocin induces differentiation of P19 embryonic stem cells to cardiomyocytes. Proc Natl Acad Sci U S A 2002; 99:9550-55; PMID:12093924; http://dx.doi.org/ 10.1073/pnas.152302499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Aguirre A, Sancho-Martinez I, Izpisua Belmonte JC. Reprogramming toward heart regeneration: stem cells and beyond. Cell Stem Cell 2013; 12:275-84; PMID:23472869; http://dx.doi.org/ 10.1016/j.stem.2013.02.008 [DOI] [PubMed] [Google Scholar]
- [54].Alexander JM, Bruneau BG. Lessons for cardiac regeneration and repair through development. Trends Mol Med 2010; 16:426-34; PMID:20692205; http://dx.doi.org/ 10.1016/j.molmed.2010.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Dobreva G, Braun T. When silence is broken: polycomb group proteins in heart development. Circ Res 2012; 110:372-74; PMID:22302750; http://dx.doi.org/ 10.1161/CIRCRESAHA.111.263145 [DOI] [PubMed] [Google Scholar]
- [56].Richly H, Di Croce L. The flip side of the coin: role of ZRF1 and histone H2A ubiquitination in transcriptional activation. Cell Cycle 2011; 10:745-50; PMID:21311219 [DOI] [PubMed] [Google Scholar]
- [57].Mazzoni EO, Mahony S, Closser M, Morrison CA, Nedelec S, Williams DJ, An D, Gifford DK, Wichterle H. Synergistic binding of transcription factors to cell-specific enhancers programs motor neuron identity. Nat Neurosci 2013; 16:1219-27; PMID:23872598; http://dx.doi.org/ 10.1038/nn.3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


