ABSTRACT
Guanine nucleotide binding protein-like 3-like (GNL3L) is an evolutionarily conserved putative nucleolar GTPase belonging to the HSR1-MMR1 family. In the present study, using protein-protein interaction assays, we show that Leucine Zipper Down-regulated in Cancer-1 (LDOC1) is a novel interacting partner of GNL3L. Furthermore, our results reveal that ectopic expression of LDOC1 destabilizes endogenous GNL3L levels and down modulates GNL3L-induced cell proliferation, in contrast, the knockdown of LDOC1 potentiates cell proliferation upon GNL3L expression. Interestingly, GNL3L upregulates NF-κB dependent transcriptional activity by modulating the expression of NF-κB subunit p65, which is reversed upon co-expression of LDOC1 with GNL3L. GNL3L also potentiates TNF-α mediated NF-κB activity. In addition, anti-apoptotic function of GNL3L is impaired upon p65 knockdown, suggesting its critical role in GNL3L mediated cell proliferation/survival. An inverse correlation of GNL3L and LDOC1 expression profiles in various tumor tissues from BioXpress database indicate their critical role in cancer. Collectively, our data provides evidence that GNL3L-LDOC1 interplay regulates cell proliferation through the modulation of NF-κB pathway during tumorigenesis.
KEYWORDS: Apoptosis, cell signaling, GNL3L, LDOC1, NF-κB transcription factor, proliferation, protein-protein interaction
Introduction
Human GNL3L is a 582 amino acid long putative nucleolar GTPase and is conserved among eukaryotes. GNL3L contains an N-terminal basic domain and a central GTP-binding domain in which the GTP-binding motifs G1-G5 are arranged in the order G5-G4-G1-G2-G3, a characteristic feature of circularly permutated GTPases.1 In addition to the novel lysine-rich nucleolar localization signal (NoLS) present in the N-terminus, the GTP binding motifs also play an important role in nucleolar localization of GNL3L.1,2 GNL3L also shuttles between nucleus and cytoplasm in CRM1-dependent manner and the inhibition of nuclear export process is found to significantly promote ‘S’-phase progression of the cell cycle.3 GNL3L interacts with karyopherin–α and karyopherin-β through its lysine-rich NoLS and nuclear localization signal (NLS), respectively and is transported to the nucleolar compartment.1 GNL3L interacts with MDM2 in the nucleoplasm and prevents its ubiquitination.4 GNL3L competes with steroid receptor co-activators (SRCs) for binding to estrogen-related receptor (ERR) and blocks SRC-mediated co-activation of ERR.5 GNL3L also interacts with Telomere repeat binding factor (TRF1) and modulates mitotic progression.6 More recently, GNL3L has been identified as one among the factors that are responsible for the maintenance of tumorigenic property of tumor initiating cells.7
Nuclear factor-kappa B (NF-κB) comprises of a family of transcription factors8 that have been widely implicated in various cellular processes including immunity, development, and oncogenesis. NF-κB is the downstream target for variety of oncogenes such as c-Myc and mutated Ras.9,10 Evidences also link NF-κB activation to anti-apoptosis, induction of cell proliferation,11 invasion12-14 and angiogenesis15-17 with implications in cancer progression.18,19 In mammals, the NF-κB superfamily consists of 2 subfamilies: the ‘NF-κB’ proteins and the ‘Rel’ proteins. The Rel subfamily consists of RelA/p65, RelB, c-Rel and the Drosophila proteins Dorsal and Dif. The p50/p65 heterodimer is the most abundant and ubiquitously found NF-κB complex in cells.20 In this study, we report that GNL3L promotes NF-κB dependent transcriptional activity by stabilizing p65 protein levels. In addition, we have characterized Leucine Zipper Downregulated in Cancer-1 (LDOC1) as a novel interacting partner of GNL3L that can modulate the stimulatory effect exerted on NF-κB by GNL3L. LDOC1 is a 17-kDa protein that is down-regulated in most pancreatic, gastric and cervical cancer cell lines21,22 and is a negative regulator of NF-κB.23 The present investigation provides evidence that LDOC1- mediated GNL3L destabilization leads to decreased total cellular p65 pool, which in-turn results in reduced cell proliferation and apoptosis.
Results
GNL3L physically interacts with LDOC1
Proteome-scale mapping of human binary protein-protein interactions suggested that GNL3L associated with LDOC1,24 but the functional consequences of this interaction remain unknown. In the current investigation, various in vitro analyses were used to validate and characterize the interaction between GNL3L and LDOC1. Toward this end, GST-tagged LDOC1 was purified as described in Materials and Methods and used in a pull down assay with HEK293T cell lysates expressing pcDNA3-Flag or Flag-GNL3L. Results in Fig. 1A clearly indicate that Flag-GNL3L efficiently interacted with GST-LDOC1 (lane 3). The absence of interaction between the vector and GST-LDOC1 as well as between GST and Flag-GNL3L indicated that the interaction between GNL3L and LDOC1 was specific. The integrity of purified GST and GST-LDOC1 proteins used for the pull down assay are shown in Fig. 1B. In addition, we performed co-immunoprecipitation assays to confirm this interaction within mammalian cell lines. To this end, Flag-GNL3L and LDOC1-HA were co-expressed in both HEK293T and LDOC1-negative SiHa cells. Immunoprecipitation was carried out with anti-Flag antibody followed by western blot analysis using anti-HA antibody. The efficiency of immunoprecipitation was verified by western blot analysis using anti-Flag antibody. Results in Fig. 1C indicate that Flag-GNL3L specifically interacts with LDOC1-HA in both these cell lines (lane 4). To confirm further, LDOC1-HA was transfected in a panel of mammalian cells lines and the interaction of endogenous GNL3L with LDOC1-HA was determined by co-immunoprecipitation (Fig. 1D). Our results clearly demonstrate that endogenous GNL3L specifically interacts with LDOC1-HA in all the cell lines tested (Fig. 1D; lanes 2, 4 and 6). Collectively, these data provide evidence that LDOC1 is a novel interacting partner of GNL3L.
Figure 1.
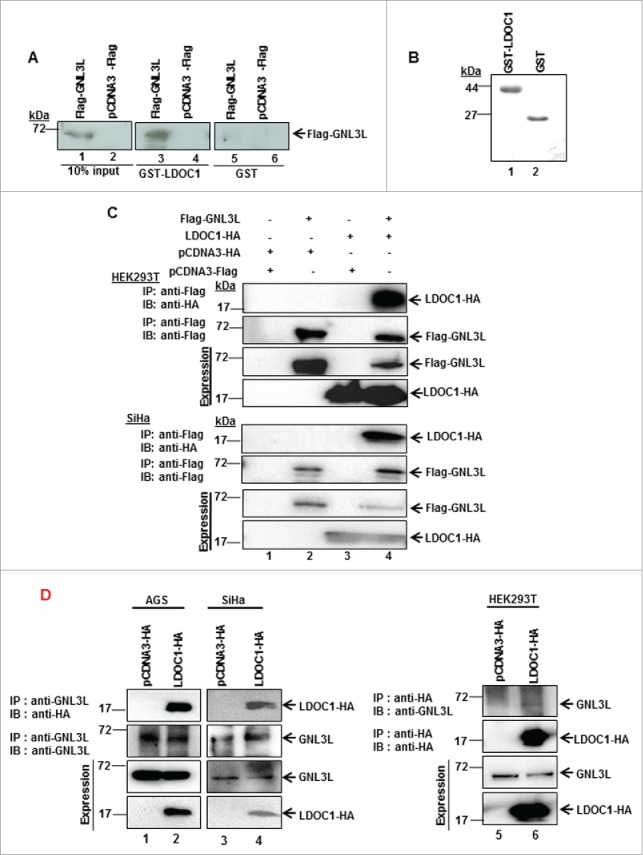
GNL3L interacts with LDOC1. HEK293T cells were transfected with Flag-GNL3L or the control vector using PEI. After 48 hrs of transfection, cell lysates were prepared and GST pull-down assay (A) was performed with GST-LDOC1 followed by western blot analysis using anti-Flag antibody. GST was used as negative control. The expression of Flag-GNL3L was confirmed by western blot analysis using anti-Flag antibody. (B) The expression of GST-LDOC1 and GST was confirmed using Coomassie blue staining. (C) HEK293T and SiHa cell lysates expressing Flag-GNL3L and LDOC1-HA were subjected to co-immunoprecipitation using anti-Flag antibody. Complexes were eluted and separated on SDS-12%PAGE followed by western blot analysis with anti-HA antibody. The efficiency of immunoprecipitation was confirmed by western blot analysis using anti-Flag antibody. (D) HEK293T cell lysates expressing LDOC1-HA were subjected to co-immunoprecipitation using anti-HA antibody whereas SiHa or AGS cells expressing LDOC1-HA were co-immunoprecipitated with anti-GNL3L antibody. Complexes were eluted and separated on SDS-12%PAGE followed by western blot analysis with anti-GNL3L or anti-HA antibodies.
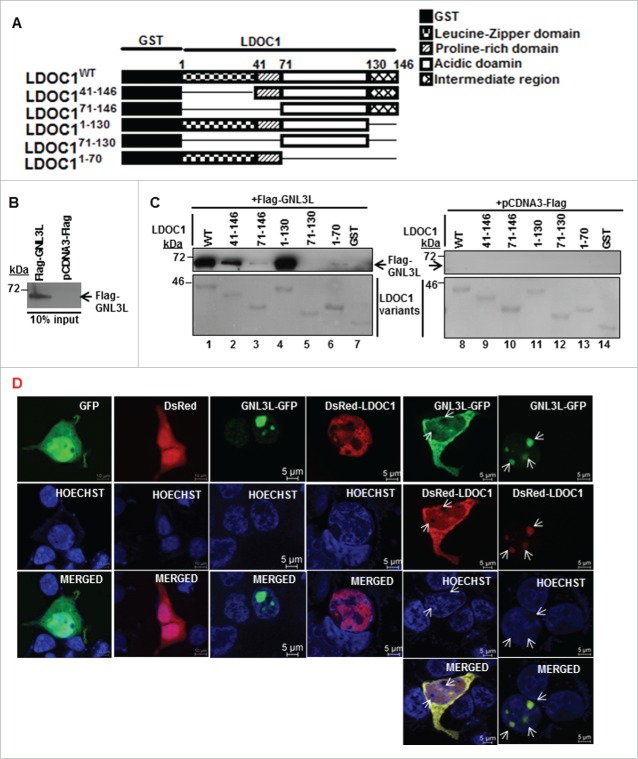
In order to identify the domain in LDOC1 required for its interaction with GNL3L, various GST-tagged deletion constructs of LDOC1 were generated (Fig. 2A) and in vitro pull-down assay was performed with HEK293T cell lysates overexpressing pcDNA3-Flag or Flag-GNL3L (Fig. 2B). The purity and integrity of various LDOC1 mutant proteins were checked using Ponceau-S stain. Western blot analysis of the pulled-down protein complexes using anti-Flag antibody revealed that GNL3L specifically interacted with LDOC1WT and LDOC141-146 as well as LDOC11-130 mutants (Fig. 2C; lane 1, 2 and 4). It is worth noting that the deletion of leucine zipper and proline-rich regions in LDOC1 resulted in reduced interaction with GNL3L (Fig. 2C; lane 3). The importance of this domain was also evident from the fact that the LDOC171-130 construct was unable to interact with GNL3L (Fig. 2C; lane 5). Yet, the fact that LDOC171-146 and LDOC11-70 are unable to interact with GNL3L indicates that the LDOC1-GNL3L interaction could also be conformation-dependent.
Figure 2.
Identification of GNL3L interaction domain in LDOC1. (A) Schematic representation of wild type and deletion mutants of LDOC1. (B) HEK293T cells were transfected with plasmid encoding Flag-GNL3L or control vector and the expression of GNL3L was confirmed by western blot analysis using anti-Flag antibody. (C) HEK293T cells lysates containing Flag-GNL3L were used in a GST pull-down assay with wild type or indicated variants of GST-LDOC1. GST served as the negative control. Specific interaction between Flag-GNL3L and LDOC1proteins was confirmed by western blot analysis using anti-Flag antibody. Ponceau-S staining indicated the integrity and purity of LDOC1 variants. (D) SiHa cells were transfected with GNL3L-GFP, DsRed-LDOC1 or indicated control vectors and their sub-cellular localization was analyzed using confocal microscopy. The arrow marks represent nucleoli.
Recent reports indicated that GFP-LDOC1 localizes to the nucleus with very weak cytoplasmic distribution25 and GNL3L is predominantly localized in nucleolus with weak nuclear staining.1 We next sought to define whether GNL3L and LDOC1 co-localize inside the cell. Toward this end, SiHa cells were co-transfected with GNL3L-GFP and DsRed-LDOC1- expression plasmids and their localization was determined as described in Materials and Methods. Surprisingly, 2 patterns of localization were observed for LDOC1-DsRed and GNL3L-GFP; first, in which both the proteins were co-localized in the nucleolus and in the second, they were co-localized in the nucleus and cytoplasm but excluded from the nucleolus (Fig. 2D; indicated by arrow). GFP and DsRed were used as controls and both proteins were found to be diffusely distributed throughout the cells (Fig. 2D). These results suggest that the interaction of LDOC1 with GNL3L specifically alters the nucleolar retention of GNL3L and could be one of the possible mechanism(s) by which LDOC1 modulates GNL3L function during cell proliferation.
LDOC1 downregulates GNL3L
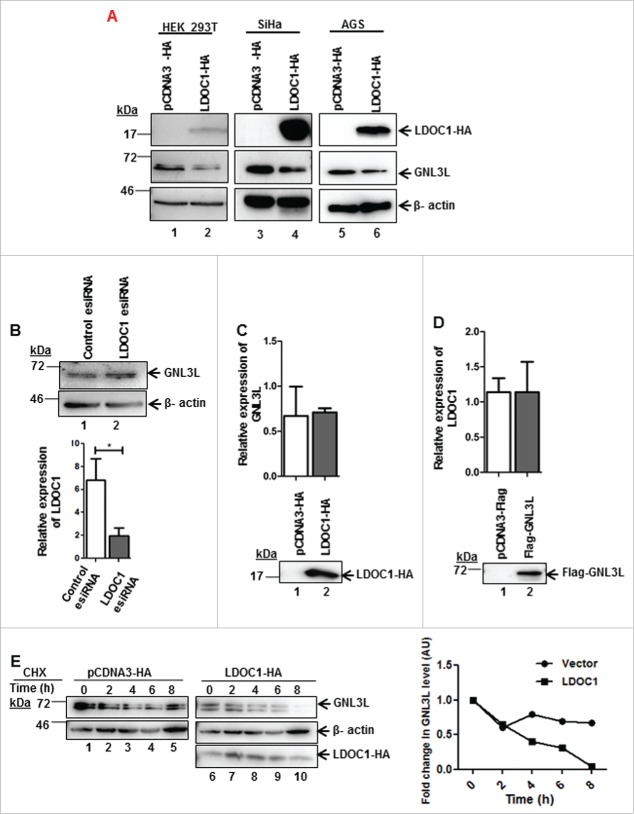
In order to understand the functional significance of LDOC1-GNL3L interaction, we first chose to analyze the endogenous GNL3L protein levels upon ectopic expression of LDOC1 in HEK293T, SiHa and AGS cell lines. Ectopic expression of LDOC1 resulted in marked reduction of endogenous GNL3L protein levels in all the cell lines tested (Fig. 3A; middle panel; lane 2, 4 and 6), in contrast, depletion of LDOC1 in HEK293T cells marginally increased endogenous GNL3L levels (Fig. 3B; upper panel, lane 2). The efficiency of LDOC1 knockdown was assessed using RT-qPCR (Fig. 3B). In order to understand the mechanism of LDOC1 mediated down-modulation of GNL3L, we first analyzed the levels of GNL3L mRNA upon LDOC1 overexpression in HEK293T cells by RT-qPCR. No significant change in the GNL3L mRNA levels was observed (Fig. 3C), suggesting that LDOC1 modulates GNL3L at translational or post-translational level. Similarly, the ectopic expression of GNL3L had no effect on LDOC1 mRNA levels (Fig. 3D). Further, to understand the mechanism of LDOC1 mediated downregulation of GNL3L, we performed a cycloheximide (CHX) chase assay. Results in Fig. 3E indicate a steady decline of endogenous GNL3L levels upon ectopic expression of LDOC1 in a time-dependent manner as compared to the vector transfected cells. Taken together, these data suggest that LDOC1 downregulates GNL3L by promoting protein destabilization.
Figure 3.
LDOC1 downregulates GNL3L. (A) HEK293T or SiHa or AGS cells were transfected with LDOC1-HA or the corresponding control vector. After 48 hrs of transfection, the expression levels of ectopically expressed LDOC1 and endogenous GNL3L were determined by western blot analysis using anti-HA and anti-GNL3L antibodies, respectively. (B) HEK293T cells were transfected with 80 pM of LDOC1 esiRNA or the universal negative control esiRNA and endogenous GNL3L expression was assessed by western blot analysis using anti-GNL3L antibody. Knockdown of LDOC1 was confirmed by RT-qPCR analysis using specific primers. Total RNA isolated from HEK293T cells transfected with LDOC1-HA (C) or Flag-GNL3L (D) was reverse-transcribed and RT-qPCR was carried out using specific primers to analyze the endogenous mRNA expression of GNL3L and LDOC1, respectively. (E) HEK293T cells transfected with LDOC1-HA or the control vector were treated with cycloheximide (CHX) for indicated time periods. The expression of endogenous GNL3L was determined by western blot using anti-GNL3L antibody. Beta-actin levels served as the internal control. Densitometry analysis of a representative western blot depicting the endogenous GNL3L levels at various time points.
LDOC1 inhibits GNL3L function
It is known that LDOC1 inhibits cell proliferation and induces apoptosis in a cell type specific manner.23,25,26 In order to define the effect of LDOC1 on GNL3L function during cell proliferation, we performed various in vitro functional assays. Flag-GNL3L and LDOC1-HA were transiently expressed in HEK293T cells and the rate of cell proliferation was analyzed using MTT assay. Results depicted in Fig. 4A clearly indicate that GNL3L overexpression led to an increase in cell proliferation; in contrast, LDOC1 expression inhibited cell growth. Interestingly, the co-expression of LDOC1-HA with Flag-GNL3L resulted in significant reduction of GNL3L-induced cell proliferation (Fig. 4A). Results from the Annexin-V staining assay indicate that ectopic expression of LDOC1 led to a significant reduction in live cell number with a corresponding increase in the apoptotic cell population (Fig. 4B). The ectopic expression of GNL3L was found to induce a modest increase in the live cell percentage while reducing the apoptotic cell population, thus confirming its growth promoting function (Fig. 4B). Importantly, co-expression of LDOC1 with GNL3L led to a decrease in the live cell percentage and increase in apoptotic cell percentage as compared to GNL3L. These data suggest that LDOC1 may negatively regulate GNL3L function. To further confirm that LDOC1 indeed modulates GNL3L function, apoptosis and cell proliferation assays were performed upon Flag-GNL3L overexpression under LDOC1 knockdown condition. Results in Fig. 4C indicate that LDOC1 depletion resulted in significant increase in the percentage of live cell population while apoptotic cell population was decreased. This was in contrast to the condition where GNL3L and LDOC1 were co-expressed (Fig. 4B). Results from the MTT assay indicated that LDOC1 knockdown led to a significant increase in cell proliferation (Fig. 4D). Expression of Flag-GNL3L was analyzed using western blot employing anti-Flag antibody (Fig. 4E). LDOC1 expression was determined by RT-qPCR (Fig. 4E). Collectively, these data suggest that LDOC1 negatively regulates GNL3L function during cell proliferation.
Figure 4.
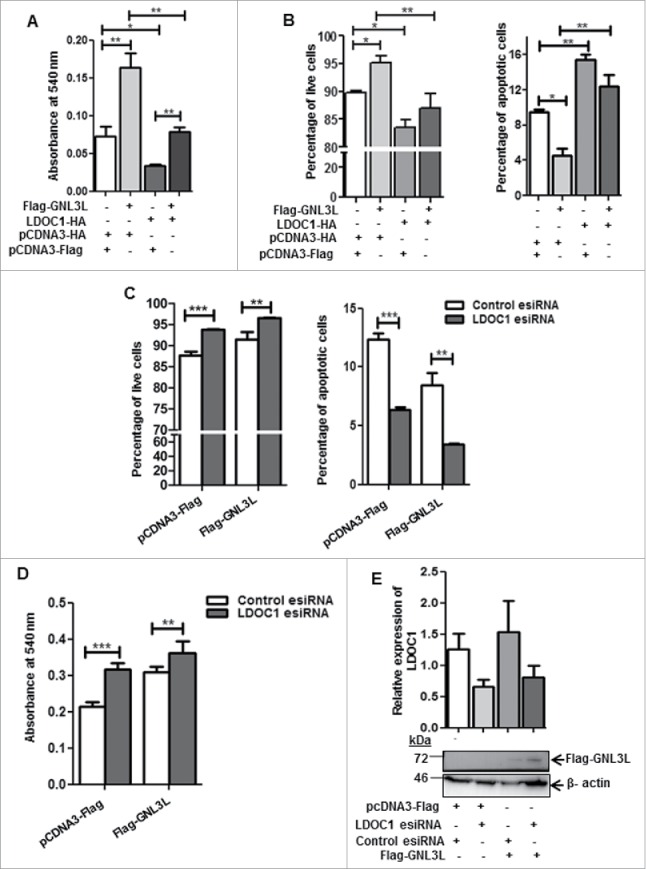
LDOC1 inhibits GNL3L function. (A) MTT and (B) Annexin-V binding assays were carried out after 48 hrs of transfection of HEK293T cells with Flag-GNL3L, LDOC1-HA or corresponding control vectors. HEK293T cells were transfected with Flag-GNL3L or control vector along with LDOC1 esiRNA or control esiRNA. After 48 hrs of transfection, (C) Annexin-V binding as well as (D) MTT assays were performed. The knockdown of LDOC1 was confirmed by RT-qPCR analysis using specific primers. (E) The expression of Flag-GNL3L was analyzed by western blot analysis using anti-Flag antibody.
LDOC1 and GNL3L exhibit opposing effects on NF-κB dependent transcriptional activity
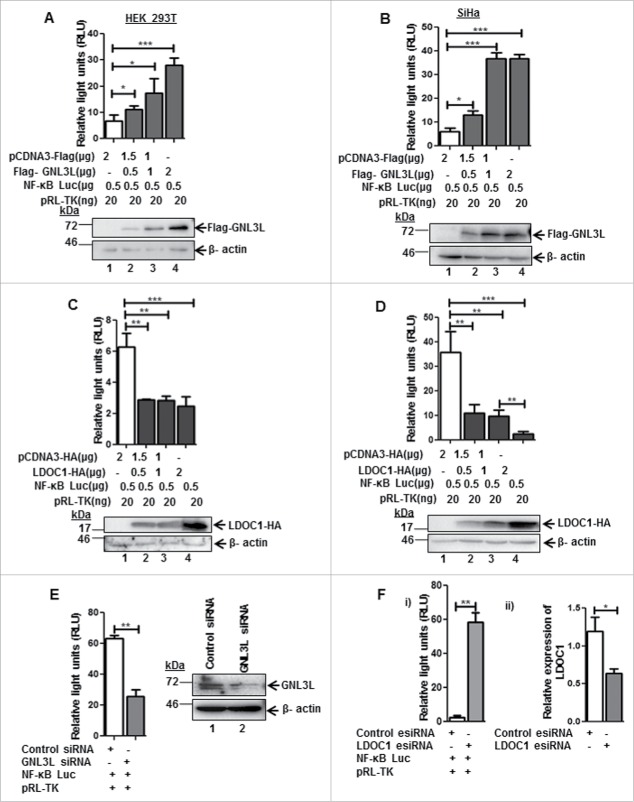
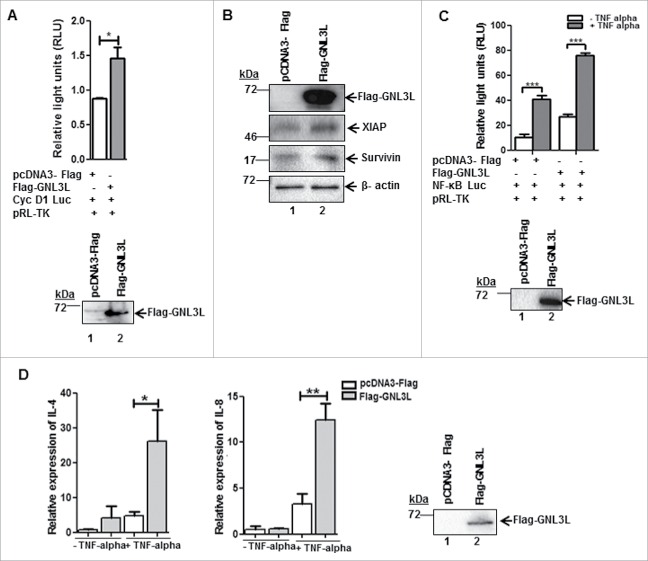
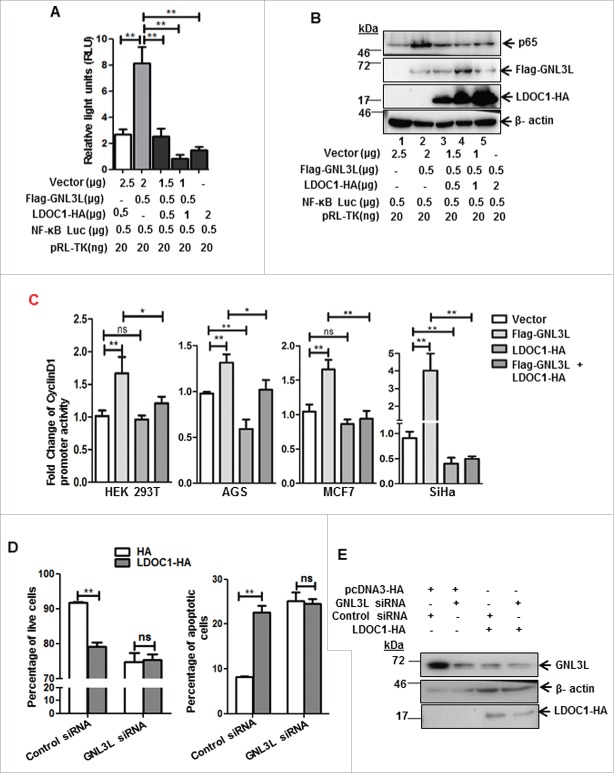
LDOC1 is known to negatively modulate NF-κB activity.23 Results from a genome wide siRNA screen indicate that GNL3L is one of the possible NF-κB mediators.27 It is well known that NF-κB plays a critical role in cell proliferation during tumorigenesis but the cross talk between NF-κB, GNL3L, and LDOC1 during cell proliferation is not known. Toward this end, we first chose to analyze the role of GNL3L and LDOC1 on NF-κB dependent transcriptional activity. NF-κB dependent promoter-luciferase plasmid was co-transfected with different amounts of Flag-GNL3L in HEK293T cells and the relative luciferase activity was measured at the end of 48 hours. Renilla luciferase activity served as the internal control. Results in Fig. 5A reveal that GNL3L promoted NF-κB dependent luciferase activity in a dose dependent manner. Western blot analysis using anti-Flag antibody confirmed the dose-dependent expression of Flag-GNL3L (Fig. 5A). Beta actin levels were used as loading control. Similar results were also observed in SiHa cells (Fig. 5B). We next tested the effect of LDOC1 on NF-κB dependent transcriptional activity. Dose dependent inhibition of NF-κB activity was observed upon LDOC1 expression both in HEK293T as well as in SiHa cells (Fig. 5C, D). In addition, the knockdown of GNL3L by siRNA resulted in a significant reduction of NF-κB dependent transcriptional activity, confirming the specificity of GNL3L mediated NF-κB activity (Fig. 5E). On the contrary, knockdown of LDOC1 resulted in a significant increase in NF-κB dependent transcriptional activity (Fig. 5F-i). The extent of LDOC1 knockdown was assessed by RT-qPCR (Fig. 5F-ii). The transcriptional activity of cyclin-D1 promoter was also analyzed upon overexpression of Flag-GNL3L, as cyclin D1 is a direct transcriptional target of NF-κB.28 The results in Fig. 6A clearly indicate that there is a significant up-regulation of cyclin-D1 transcription upon overexpression of Flag-GNL3L. Furthermore, the ectopic expression of GNL3L resulted in upregulation of anti-apoptotic NF-κB targets such as XIAP and Survivin (Fig. 6B, lane 2). In order to define the role of GNL3L on NF-κB signaling further, we analyzed NF-κB activity upon GNL3L expression in the presence of TNF-α, which promotes the classical NF-κB signaling cascade.20 Interestingly, we observed potentiation of TNF-α induced NF-κB activity upon GNL3L expression (Fig. 6C). In addition, expression levels of well-known inflammatory cytokines and the targets of NF-κB28 such as IL-4 and IL-8 were significantly high in Flag-GNL3L expressing cells treated with TNF-α as compared to the control (Fig. 6D). Together, these data provide evidence that GNL3L positively regulates NF-κB dependent transcriptional activity which in turn resulted in up-regulation of its anti-apoptotic and inflammatory targets.
Figure 5.
LDOC1 and GNL3L exhibit opposing effects on NF-κB dependent transcriptional activity. GNL3L upregulates NF-κB dependent transcriptional activity. (A) HEK293T and (B) SiHa cells were co-transfected with Flag-GNL3L or control vector along with pGL4.32[luc2P/NF-κB-RE/Hygro] and pRL-TK-Renilla luciferase (internal control) plasmids. After 48 hrs of transfection, luciferase assay was performed to check the NF-κB dependent transcriptional activity. LDOC1 downregulates NF-κB dependent transcriptional activity. (C) HEK293T and (D) SiHa cells were co-transfected with LDOC1-HA or control vector along with pGL4.32[luc2P/NF-κb-RE/Hygro] and pRL-TK-Renilla luciferase (internal control) plasmids. After 48 hrs of transfection, luciferase assay was performed to check basal level of NF-κB dependent transcriptional activity. The expression of Flag-GNL3L and LDOC1-HA was confirmed by western blot analysis using anti-Flag and anti-HA antibodies, respectively. Beta-actin was used as internal control. (E) HEK293T cells were transfected with appropriate amount of GNL3L siRNA or control siRNA along with pGL4.32[luc2P/NF-κB-RE/Hygro] and pRL-TK-Renilla luciferase plasmids. Luciferase assay was performed after 48 hrs of transfection to analyze NF-κB dependent transcriptional activity. (F-i) HEK293T cells were transfected with LDOC1 esiRNA or control esiRNA along with pGL4.32[luc2P/NF-κB-RE/Hygro] and pRL-TK-Renilla luciferase plasmids. Luciferase assay was performed after 48 hrs of transfection to analyze NF-κB dependent transcriptional activity. (F-ii) Knockdown of LDOC1 was analyzed by RT-qPCR using specific primers.
Figure 6.
GNL3L alters the expression of NF-κB target genes. (A) HEK293T cells were co-transfected with Flag-GNL3L or control vector along with cyclin D1-promoter luciferase construct and pRL-TK-Renilla luciferase plasmids. After 48 hrs of transfection, luciferase assay was performed to check the cyclin D1 promoter activity. (B) GNL3L upregulates anti-apoptotic NF-κB target proteins. SiHa cells were transfected with Flag-GNL3L or control vector and the expression levels of endogenous XIAP and Survivin were analyzed using specific antibodies. Beta-actin was used as the loading control. (C) HEK293T cells were co-transfected with Flag-GNL3L or control vector along with pGL4.32[luc2P/NF-κB-RE/Hygro] and pRL-TK-Renilla luciferase plasmids. After 42 hrs of transfection, TNF-α was added (20 ng/mL final concentration) and luciferase assay was performed at the end of 48 hrs to check the NF-κB dependent transcriptional activity. (D) The mRNA expression levels of IL-4 and IL-8 were analyzed post TNF-α treatment by RT-qPCR in HEK293T cells transfected with Flag-GNL3L or control vector.
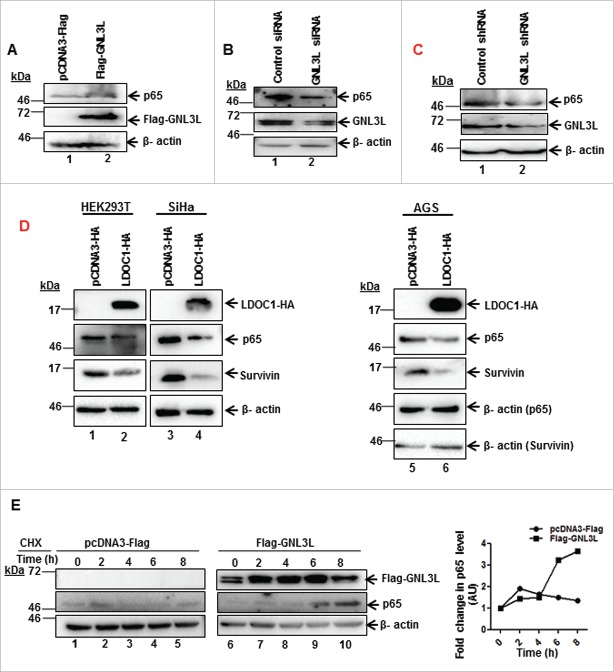
GNL3L modulates the expression and stability of NF-κB subunit p65
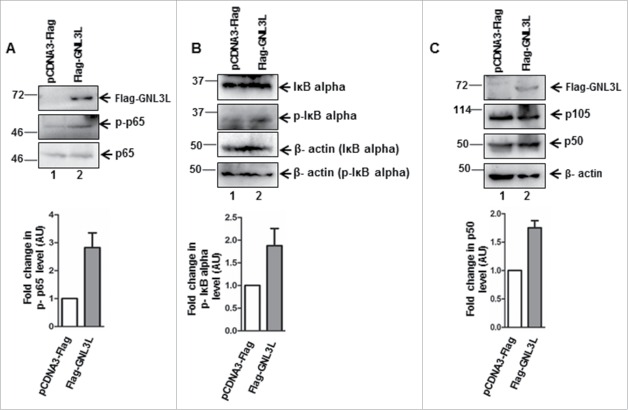
Toward understanding the mechanism by which GNL3L could modulate NF-κB dependent transcriptional activity, we analyzed the expression levels of important molecular players in the classical NF-κB pathway. Ectopic expression of GNL3L significantly up-regulates the levels of NF-κB subunit, p65 (Fig. 7A). In contrast, knockdown of GNL3L by siRNA as well as shRNA resulted in decreased p65 levels in HEK293T cells (Fig. 7B and C). Furthermore, ectopic expression of LDOC1 in HEK293T, SiHa and AGS cell lines resulted in reduction of total p65 protein levels (Fig. 7D; lane 2, 4 and 6) as well as NF-κB target gene survivin levels (Fig. 7D; lane 2, 4 and 6). Beta actin was used as an internal control. We next designed experiments to specifically understand the functional significance of GNL3L mediated p65 regulation. Toward this end, cycloheximide chase assay was performed to check whether the increase of p65 protein levels observed upon GNL3L overexpression was due to increased protein stability. Results in Fig. 7E clearly indicate that p65 protein was stabilized upon GNL3L expression as compared to the vector. We next checked whether GNL3L alter the phosphorylation status of NF- κB subunit p65. Results in Fig. 8 clearly demonstrate that ectopic expression of GNL3L up-regulates levels of phospho-p65 (Fig. 8A; lane 2), phospho-IκB α (Fig. 8B; lane 2), and p50 (Fig. 8C; lane 2). These data clearly emphasize that GNL3L modulates NF- κB pathway.
Figure 7.
GNL3L modulates the expression and stability of NF-κB subunit p65. GNL3L upregulates p65 protein levels. (A) HEK293T cells were transfected with Flag-GNL3L or control vector and the expression levels of endogenous p65 and ectopically expressed Flag-GNL3L were confirmed by western blot analysis using anti-p65 and anti-Flag antibodies, respectively. GNL3L specific siRNA (B) or shRNA (C) or control siRNA or shRNA were transfected into HEK293T cells and the expression levels of endogenous p65 and GNL3L were confirmed by western blot analysis using anti-p65 and anti-GNL3L antibodies, respectively. (D) LDOC1 downregulates p65 protein levels. HEK293T or SiHa or AGS cells were transfected with LDOC1-HA or control vector and the expression levels of endogenous p65, survivin and ectopically expressed LDOC1-HA were confirmed by western blot analysis using anti-p65, anti-survivin, and anti-HA antibodies, respectively. (E) HEK293T cells transfected with Flag-GNL3L or the control vector were treated with cycloheximide (CHX) for different time periods. The expression of Flag-GNL3L and endogenous p65 was determined by western blot using anti-Flag and anti-p65 antibodies, respectively. Densitometry analysis of a representative western blot shows the endogenous p65 levels at different time points.
Figure 8.
GNL3L modulates p65 phosphorylation levels. HEK293T cells were transfected with Flag-GNL3L or control vector and the expression levels of Flag-GNL3L, endogenous p65, and phospho-p65 (A); IκB-α (B) and phospho-IκB-α and p105 and p50 (C) were analyzed by western blot using appropriate antibodies. Quantitative measurements of respective western blots by densitometry analysis are provided below.
To further understand whether the stabilization of p65 protein level during GNL3L overexpression was responsible for the observed anti-apoptotic function of GNL3L, an apoptosis assay was performed in cells overexpressing Flag-GNL3L under p65 knockdown condition. Interestingly, we observed a significant decrease in the total apoptotic cell population with a corresponding increase in the live cell population in Flag-GNL3L transfected cells (Fig. 9A). Upon partial depletion of p65 using shRNA (Fig. 9B and C), GNL3L was not able to produce any significant change in the levels of live and apoptotic cell population (Fig. 9A). Expression of all the indicated proteins was analyzed by western blot using respective antibodies (Fig. 9B) and the extent of p65 knockdown is depicted in Fig. 9C. Collectively, these data suggest that GNL3L stabilizes p65 protein levels and p65 plays a critical role in anti-apoptotic function of GNL3L.
Figure 9.
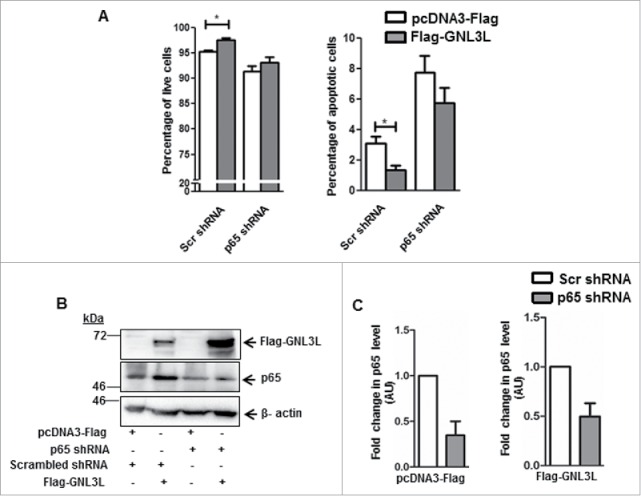
Anti-apoptotic function of GNL3L is p65-dependent. (A) Annexin-V binding assay was performed in HEK293T cells co-transfected with Flag-GNL3L or vector along with an equal concentration of p65 specific or scrambled shRNA constructs. Scr: scrambled. (B) Expression of Flag-GNL3L as well as the extent of p65 knockdown was analyzed by western blot analysis using anti-Flag and anti-p65 antibodies, respectively. Beta-actin was used as the loading control in all these experiments. (C) Densitometry analysis of the western blot was performed to quantitate the extent of p65 knockdown.
LDOC1 downregulates GNL3L-mediated NF-κB dependent transcriptional activity through modulation of p65 levels
Given the fact that GNL3L modulates p65 levels, we attempted to analyze the effect of LDOC1 on GNL3L mediated NF-κB dependent transcriptional activity and p65 protein levels. HEK293T cells were co-transfected with different concentrations of LDOC1-HA and Flag-GNL3L in combination with NF-κB dependent promoter-luciferase. Relative luciferase activity was measured 48 hours after transfection. Renilla luciferase was used as internal control. As described above, Flag-GNL3L overexpression significantly increased NF-κB dependent transcriptional activity (Fig. 10A). Interestingly, the ectopic expression of LDOC1-HA inhibited the GNL3L induced NF-κB dependent transcriptional activity in a dose dependent manner (Fig. 10A). Further, we analyzed whether the reduced luciferase activity was due to reduction in the total protein levels of p65. Results shown in Fig. 10B clearly indicate that Flag-GNL3L overexpression resulted in increase of p65 levels, which was reversed by LDOC1 in a dose dependent manner. To further understand the interplay between GNL3L and LDOC1 on NF-κB dependent transcriptional activity, cyclin D1 promoter (a natural gene target of NF-κB) activity was measured upon co-expression of GNL3L and LDOC1 in HEK293T, SiHa, AGS and MCF7 cell lines. Results in Fig. 10C reveal that GNL3L expression induces the cyclin D1 promoter activity and is reversed by LDOC1 co-expression. These data suggest that LDOC1 downmodulate GNL3L mediated NF-κB target gene expression. Finally, we tested whether LDOC1 inhibits anti-apoptotic function of GNL3L in cells overexpressing LDOC1 under GNL3L depleted condition. Ectopic expression of LDOC1 promoted apoptosis as evidenced by the increase in apoptotic cell population (Fig. 10D). Interestingly, LDOC1 overexpression was not able to induce any further significant change in apoptotic cell percentage upon GNL3L knockdown, suggesting that LDOC1 exerts its apoptotic function by blocking GNL3L activity. Western blot was performed using anti-HA and anti-GNL3L antibodies to determine LDOC1 overexpression and endogenous levels of GNL3L (Fig. 10E). Collectively, these data provide evidence that NF-κB activity is critical for GNL3L mediated cell proliferation.
Figure 10.
LDOC1 downregulates GNL3L-mediated NF-κB dependent transcriptional activity by modulating p65 expression. (A) HEK293T cells were co-transfected with Flag-GNL3L and increasing amounts of LDOC1-HA along with pGL4.32[luc2P/NF-κB-RE/Hygro] and pRL-TK-Renilla luciferase (internal control) plasmids. The NF-κB dependent transcriptional activity was analyzed by luciferase assay after 48 hrs of transfection. (B) The expression levels of Flag-GNL3L, LDOC1-HA and endogenous p65 were assessed by western blot analysis using specific antibodies. LDOC1 mediated inhibition of apoptosis is GNL3L-dependent. (C) HEK293T or AGS or MCF-7 or SiHa cells were co-transfected with Flag-GNL3L or LDOC1-HA in combination with cyclin D1-promoter luciferase and pRL-TK-Renilla luciferase plasmids. Luciferase assay was performed at the end of 48 hours of transfection to assess the cyclin D1 promoter dependent transcriptional activity. (D) Annexin-V binding assay was performed in HEK293T cells co-transfected with LDOC1-HA or vector along with GNL3L siRNA or control siRNA. (E) The expression of LDOC1-HA and the level of GNL3L knockdown was analyzed by western blot analysis using anti-HA and anti-GNL3L antibodies, respectively.
Discussion
Various studies have positioned GNL3L as a nucleolar GTP-binding protein harboring multiple signals that govern its intracellular distribution.1-3 Yet, the relatively less repertoire regarding its cellular functions and regulation prompted us to undertake this study. The present investigation provides evidence that LDOC1 is a novel functional interacting partner for GNL3L. In addition, we show that LDOC1 negatively regulates GNL3L function, possibly by protein destabilization and altering its sub-cellular distribution. Expression of LDOC1 inhibits GNL3L mediated cell proliferation. Furthermore, we show that GNL3L promotes NF-κB dependent transcriptional activity, which in-turn results in upregulation of anti-apoptotic NF-κB targets such as XIAP and Survivin as well as the inflammatory cytokines IL-4 and IL-8. Together, our data suggests that NF-κB subunit p65 plays a critical role in GNL3L mediated cell proliferation, which is reversed by LDOC1.
GNL3L, being a nucleolar protein was found to complement the rRNA processing defect in Grn1-null S. pombe cells.29 The nucleolar localization of GNL3L was also found to be critical for the progression of ‘G2/M’ phase of the cell cycle.3,4 Depletion of intracellular GTP pool and mutations in the conserved residues within G-domains has also been reported to alter the nucleolar localization of GNL3L.1,2 The observed altered localization of GNL3L upon LDOC1 expression suggests that the physical interaction of LDOC1 with GNL3L may be a pre-requisite to alter GNL3L function during cell proliferation. LDOC1 localizes to the nucleus despite not having any identifiable canonical nuclear localization signal. This leads to the speculation that GNL3L being a nucleo-cytoplasmic shuttling protein, might facilitate the localization of LDOC1 in the nucleus/nucleolus by a piggyback or novel receptor mediated mechanism. Results from the current investigation clearly indicate that ectopic expression of LDOC1 down-regulates GNL3L by protein destabilization. LDOC1 is ubiquitously expressed in normal tissues.21 Interestingly, while LDOC1 is downregulated in most pancreatic and gastric cancer cell lines,21 human colorectal and gastric cancers express high levels of GNL3L.4 These data suggest the possibility that the increased expression of GNL3L, particularly in gastric cancer, may be due to the absence of inhibitory activity of LDOC1 owing to promoter hyper- methylation.22,30 Furthermore, analysis of in vivo expression pattern of GNL3L and LDOC1 in various primary cancers from BioXpress database indicates an inverse correlation (Fig. 11), supporting the notion that the interplay between GNL3L and LDOC1 may be critical for cell growth control. Together, these results suggest that GNL3L and LDOC1 play an important role during tumorigenesis and further experiments are warranted to understand the molecular mechanism(s) regulating GNL3L and LDOC1 function to control cell proliferation.
Figure 11.
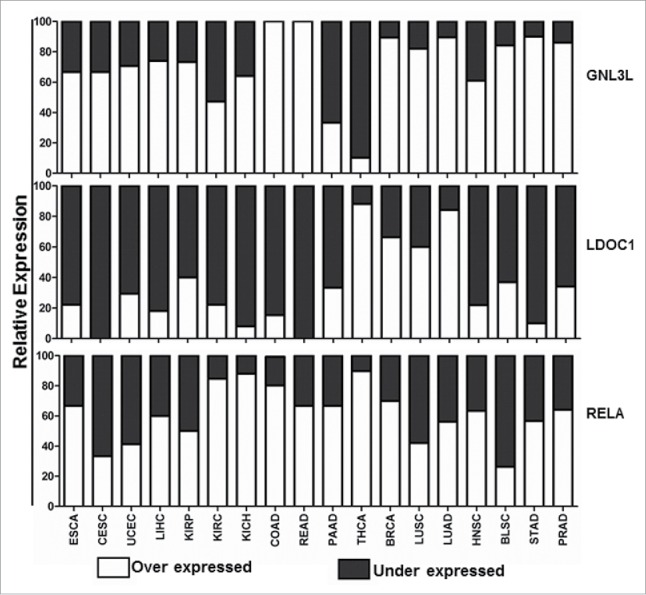
Expression profile of GNL3L, LDOC1 and RELA in different cancers. Gene expression analysis of GNL3L, LDOC1 and RELA were carried out from data set of 18 cancers and respective normal tissue expression profiles available in BioXpress database (https://hive.biochemistry.gwu.edu/tools/bioxpress/).
Previous reports from others and our group indicate that GNL3L promotes mammalian cell proliferation4,30 whereas LDOC1 induces apoptosis.23,25,26 The observed down-regulation of GNL3L by LDOC1 may significantly influence cell proliferation and apoptosis in mammalian cells. Recent reports suggest that LDOC1 is a novel regulator of NF-κB activity23 and GNL3L acts as a mediator of NF- κB pathway,27 but the mechanism remains unknown. The present study provides evidence that GNL3L promotes NF- κB dependent transcriptional activity and the knock-down of GNL3L reverses such an effect. It is well known that NF-κB regulates cell survival via up-regulation of anti-apoptotic genes such as c-IAP1, c-IAP2, XIAP and Survivin.31-37 Interestingly, GNL3L increases the expression of anti-apoptotic NF- κB targets such as XIAP and Survivin, suggesting that GNL3L exerts its anti-apoptotic function possibly via activation of NF-κB. This is further supported by the activation of NF-κB dependent transcriptional activity upon GNL3L expression.
Activation of NF-κB is associated with cell proliferation, progression, and prognosis of various cancers.38-40 Upregulation of NF-κB subunit p65 has also been reported to contribute toward neoplastic phenotype and development of immune diseases.41-43 As GNL3L is required for mammalian cell proliferation, one can speculate that the modulation of NF- κB signaling by GNL3L may play an essential role during tumorigenesis. This is supported by the positive correlation of expression patterns of GNL3L and NF- κB subunit p65 (Rel-A) in various tumor tissue samples from BioXpress database (Fig. 11).44 NF-κB has been hailed as a therapeutic target for cancer treatment owing to its ability to modulate inflammatory responses apart from the other major hallmarks of cancer.18,45,46 The modulation of IL-4 and IL-8 by GNL3L could promote NF-κB-mediated inflammation in addition to its anti-apoptotic function. LDOC1 mediated downregulation of GNL3L-induced NF-κB dependent transcriptional activity reveals a molecular event through which LDOC1-GNL3L interaction could affect cell proliferation (Fig. 12). Recent evidences suggest that Rho GTPases such as Rac1, Cdc42 and RhoA play an important role on TNF-α mediated activation of canonical NF-κB pathway.47,48,49 In addition, non-canonical NF- κB pathway is also found to be instrumental in the pathogenesis of various cancers as well as immune disorders.50 The current investigation suggests that GNL3L, putative nucleolar GTPase upregulates the expression of p50 which is a key mediator of the non-canonical NF- κB pathway. Furthermore, accumulating evidences suggest the cross-talk of Akt and ERK/MAPK signaling on NF- κB pathway51 but the role of GNL3L and LDOC1 on Akt/ERK/MAPK signaling pathway remains an open question. Collectively, the current investigation provides evidence that cross-talk between GNL3L and LDOC1 regulates NF-κB signaling during tumorigenesis and may act as a potential target for developing anti-cancer therapies.
Figure 12.
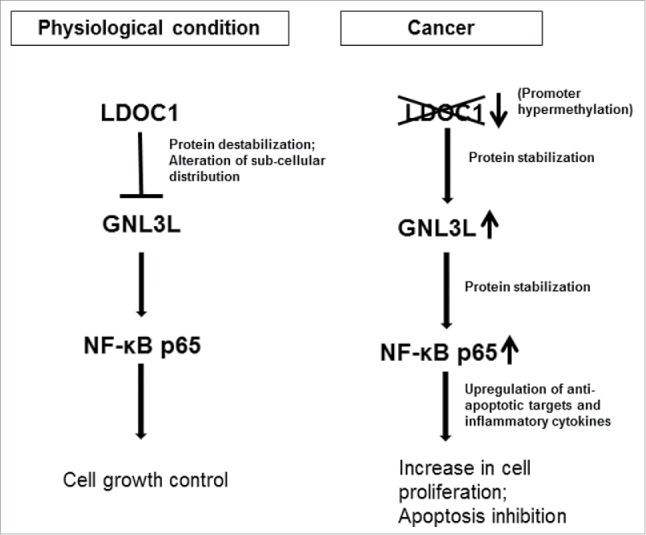
Functional significance of LDOC1-GNL3L interaction during cell proliferation. Under normal physiological conditions, LDOC1 exerts an inhibitory control over GNL3L. LDOC1 alters GNL3L protein stability and subcellular localization to modulate NF-κB signaling to balance the cell growth and apoptosis. In cancerous conditions, GNL3L is relieved from the inhibitory action of LDOC1 possibly due to its promoter hyper methylation. This results in up-regulation of NF-κB dependent transcriptional activity by GNL3L through p65 protein stabilization. The upregulation of NF-κB specific targets may resulted in increased cell proliferation and inhibition of apoptosis.
Materials and methods
Plasmid construction
Flag or GFP tagged GNL3L plasmids were generated as described previously.3 LDOC1 open reading frame was amplified from HEK293T cDNA library using specific primers (Table 1) and cloned into PGEX4T1, pCDNA3-DsRed and pCDNA3-HA. LDOC1 variants were generated in PGEX4T1 vector using appropriate primers listed in Table 1. The luciferase construct containing NF-κB response elements (pGL4.32[luc2P/NF-κB-RE/Hygro]), cyclin D1 promoter luciferase construct was a kind gift from Dr. Richard Pestell, Sydney Kimmel Cancer Center, Thomas Jefferson University, Center City, Philadelphia, Pennsylvania, USA.52 pRL-TK (Renilla luciferase-Thymidine kinase) was obtained from Promega Corporation. DNA sequencing was performed to validate the integrity of all the plasmids.
Table 1.
Primers used for cloning and RT-qPCR analysis.
| Primers used for cloning | Sequence (5′-3′) |
|---|---|
| LDOC1 Kpn1+ | TGCGACGGTACCGCCGCCACCATGGTGGATGAGTTGGT |
| LDOC1 BamHI+ | TGCGACGGATCCGCCGCCACCATGGTGGATGAGTTG GTGCTG |
| LDOC1 XhoI− | CACCATCTCGAGCTAATAATCATCCTCCTCTTCTTCG |
| LDOC1 EcoRV − | CACCATGATATCATAATCATCCTCCTCTTCTTCGTC |
| LDOC1 41BamHI+ | TGCGACGGATCCGCCGCCACCATGCTGCGCCAGGTA CGT |
| LDOC1 71BamHI+ | TGCGACGGATCCGCCGCCACCATGACGGCGTCTTAC ATGCTC |
| LDOC1 70 Xho1− | CACCATCTCGAGCTACTGCACGATAAACTCGGGGAG |
| LDOC1 130XhoI− | CACCATCTCGAGCTAGCCAAAGCACTGTTTCATCTC |
| Primers used for RT-qPCR | |
| GNL3L RT− | TTCAAGTGGGTCCGTGTCA |
| GNL3L RT+ | ACTCTGCCCACCCATCTCA |
| LDOC1 RT− | ACTGTTTCATCTCATCGAGGA |
| LDOC1 RT+ | ACGAGAACCGATTCTGCAA |
| IL-4 RT+ | CAGCCTCACAGAGCAGAAGA |
| IL-4 RT− | AGCGAGTGTCCTTCTCATGG |
| IL-8 RT+ | CAAGAGCCAGGAAGAAACCA |
| IL-8 RT− | AGCACTCCTTGGCAAAACTG |
| Beta-Actin + | ATGAAGTGTGACGTGGACAT |
| Beta-Actin − | GGCCAGGTACTGATGGTC |
Antibodies
Anti-Flag (F-3165, mouse monoclonal, 1:1500 dilution, Sigma-Aldrich, USA), anti-HA (H3663, mouse monoclonal, 1:1000 dilution, Sigma-Aldrich, USA), anti-GNL3L (ab94862, rabbit polyclonal, 1:500 dilution, Abcam), anti-p65 (F6 SC-8008, mouse monoclonal, 1:1000 dilution, Santa Cruz), anti-β actin (A1978, mouse monoclonal, 1:4000 dilution, Sigma-Aldrich), anti-XIAP (3B6 2045S, rabbit polyclonal, 1:500 dilution, Cell Signaling Technology), anti-Survivin (71G4B7 2808S, rabbit polyclonal, 1:500 dilution, Cell Signaling Technology), anti-phospho-p65 (S536 93H1, rabbit polyclonal, 1:1000 dilution, Cell Signaling Technology), anti-IκB α (L35A5 N-terminal, mouse monoclonal, 1:1000 dilution, Cell Signaling Technology), anti-phospho-IκB α (S32/36 5A5, mouse monoclonal, 1:1000 dilution, Cell Signaling Technology) and anti-NF-κB p50 (H119 SC-7178X, rabbit polyclonal, 1:1000 dilution, Santa Cruz) antibodies were used for analyzing the expression of respective proteins according to manufacturer's protocols.
Cell culture, transfection and western blot analysis
HEK293T, AGS, MCF7 and SiHa cells were maintained in Dulbecco's Modified Eagle's Medium (DMEM) (Invitrogen Life Technologies, USA) supplemented with 10% Fetal Bovine Serum (FBS) and 1% antibiotic-antimycotic (Thermo Fisher Scientific Inc., USA). Cells were grown to 60% confluence and transfection was carried out using polyethyleneimine (PEI) or lipofectamine 2000 (Thermo Fisher Scientific Inc., USA) according to manufacturer's protocol. Prior to western blotting, the transfected cells were lysed using 1X cell lysis buffer (25 mM Tris-HCl, pH7.4, 150 mM KCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton-X100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 0.4 mM PMSF, 1 mM NaF, 1 mM Na3VO4 and 1 μg/ml each of aprotinin, leupeptin and pepstatin). The proteins were separated using SDS-PAGE, transferred onto Hybond-P membrane (GE Healthcare, Sweden) and probed with appropriate antibodies. The protein-antibody complexes were incubated with horseradish peroxidase (HRP)-conjugated specific secondary antibodies (1:3000 dilution, Bio-Rad Laboratories, USA) and detected using the Enhanced Chemi-luminescence Prime detection system (GE healthcare, Sweden).
RNA interference
GNL3L siRNA (L-015743-01-0005, ON-TARGET plus SMARTpool, Human GNL3L (54552), Thermo Fisher Scientific Inc., USA) and LDOC1 esiRNA (EHU156611, Sigma-Aldrich, USA) were used to transiently knockdown GNL3L and LDOC1 expression in HEK293T cells. The universal negative control siRNA (MISSION® siRNA Universal Negative Control #1, SIC001, Sigma-Aldrich, USA) was used as a control for the knockdown studies. For shRNA knockdown studies Control shRNA (SHC016, Sigma-Aldrich, USA) and GNL3L shRNA (TRCN0000160382, Sigma-Aldrich, USA) were used. The scrambled and specific p65 shRNA constructs were a kind gift from Dr. K. Boumediene, Faculty of Medicine, University of Caen Normandy, France.53
Expression and purification of GST tagged wild type and variants of LDOC1
Plasmids encoding wild type and the indicated variants of LDOC1 were transformed into Escherichia coli BL21-DE3 strain and grown overnight at 37°C. A single colony was inoculated into ampicillin containing medium and the protein expression was induced for 12-14 hours at 18°C with 1mM IPTG. Cells were harvested and lysed in bacterial cell lysis buffer by sonication. The clarified cell lysates were incubated with glutathione-sepharose beads (GE Healthcare, Sweden) for 4 hours at 4°C. The bound proteins were eluted in elution buffer (10 mM reduced glutathione in 50 mM Tris-CL, pH 7.4) and the expression of GST tagged fusion proteins were analyzed by SDS-12%PAGE followed by staining with Coomassie blue.
GST pull-down assay
HEK293T cells were transfected with empty vector or Flag-GNL3L expression constructs as described above. Cell lysates were incubated with glutathione-sepharose beads pre-bound with wild type or variants of LDOC1 for 4 hours at 4°C. Glutathione-sepharose beads containing the bound protein complexes were washed and eluted by boiling with SDS sample loading buffer. Equal amount of elutes were resolved in SDS-12%PAGE followed by western blot analysis using anti-Flag antibody.
Protein-protein interaction assays
For co-immunoprecipitation experiments, equal amounts of cell lysates containing Flag-GNL3L and LDOC1-HA were incubated with anti-HA antibody and the bound protein complexes were eluted and resolved on SDS-12%PAGE followed by western blot analysis with anti-Flag antibody. For endogenous co-immunoprecipitation experiments, equal amounts of cell lysates containing pcDNA3-HA and LDOC1-HA were incubated with anti-GNL3L antibody and the bound protein complexes were eluted and resolved on SDS-12%PAGE followed by western blot analysis with anti-HA antibody.
Fluorescence microscopy
SiHa cells grown on chamber slides (BD Biosciences, USA) were transfected with pCDNA3-GFP, pCDNA3-DsRed, GNL3L-GFP or DsRed-LDOC1 expression plasmids using PEI. For determining the subcellular distribution of these proteins, transfected cells were fixed using 3% (w/v) paraformaldehyde at 48 hours post transfection. Hoechst 33342 was used to stain the nuclei at a final concentration of 1μg/ml. Samples were then viewed with LSM710 laser scanning confocal microscope (Carl Zeiss, Germany) and image acquisition was performed using Zen 2009 software (Carl Zeiss, Germany),
Cycloheximide chase assay
HEK293T cells were transfected with pCDNA3-HA or LDOC1-HA encoding plasmids using PEI. After 48 hours of transfection, the cells were treated with 50μg/ml of cycloheximide (CHX) and harvested at various time intervals. Equal amount of total protein was loaded on SDS-12%PAGE followed by western blot analysis using anti-GNL3L, anti-HA and anti-β actin antibodies.
Luciferase assay
HEK293T or SiHa or AGS or MCF7 cells were co-transfected with reporter plasmids (pGL4.32[luc2P/NF-κB-RE/Hygro]/ cyclin D1 promoter-luciferase) and pRL-TK-Renilla luciferase plasmids in combination with expression plasmids encoding Flag-GNL3L and LDOC1-HA. pRL-TK-Renilla was used as internal control. After 48 hours of transfection, the culture medium was removed and the cells were incubated with lysis buffer (0.1% Triton X-100, 9 mM KH2PO4 (pH 5.1), 90 mM K2HPO4 (pH 8), 1mM DTT) for 20-30 minutes with vigorous shaking. Cell lysate (50 μl) was transferred to an optiplate and the assay was performed with luciferase/Renilla assay buffer as detailed elsewhere54 using Luminometer (Perkin Elmer, USA).
MTT assay
HEK293T cells were transfected with Flag-GNL3L or LDOC1-HA expression plasmids. After 48 hours of transfection, MTT (12 mM) solution (Vybrant MTT Cell Proliferation assay kit, Thermo Fisher Scientific Inc., USA) was added, incubated for 4 hours at 37°C as per manufacturer's protocol and the absorbance was measured at 540 nm using Enspire Multimode Reader (Perkin Elmer, USA).
Annexin-V binding assay
HEK293T cells were transfected with indicated expression/shRNA plasmids or siRNA. The cells were trypsinized to obtain a single cell suspension at the end of 48 hours of transfection and incubated with Annexin-V binding buffer (0.1M Hepes/ 1.4 M NaCl, 25Mm CaCl2) for 15 minutes at room temperature. The samples were analyzed using a flow cytometer (FACSCantoII, BD Biosciences, USA) and the results were processed employing FACS Diva software.
RT-qPCR analysis
Total RNA from cells transfected with different expression plasmids was isolated using TRIzol reagent (Thermo Fisher Scientific Inc., USA). Total RNA (2 μg) was reverse transcribed using ImProm-II kit (Promega Corporation, USA) according to manufacturer's instructions. Quantitative PCR analysis was carried out using SYBR-Green mix (Roche, USA) on Realplex cycler (Eppendorf, Germany). All reactions were carried out in triplicates and the expression levels of various genes relative to β-actin were analyzed using ΔCτ values according to manufacturer's directions. The primers used for the qPCR analyzes are given in Table 1.
Gene expression profiling from dataset
RNA-Seq data of 920 pairs of tumor were compared with those of normal tissue samples, which are available in the BioXpress database44 (https://hive.biochemistry.gwu.edu/tools/bioxpress/) obtained from The Cancer Genome Atlas (TCGA: http://cancergenome.nih.gov/) for the following cancers; Esophageal carcinoma [ESCA], Cervical squamous cell carcinoma [CESC], Uterine corpus endometrial carcinoma [UCEC], Liver hepatocellular carcinoma [LIHC], Kidney renal papillary cell carcinoma [KIRP], Kidney renal clear cell carcinoma [KIRC], Kidney chromophobe [KICH], Colon adenocarcinoma [COAD], Rectum adenocarcinoma [READ], Pancreatic adenocarcinoma [PAAD], Thyroid carcinoma [THCA], Breast invasive carcinoma [BRCA], Lung squamous cell carcinoma [LUSC], Lung adenocarcinoma [LUAD], Head and neck squamous cell carcinoma [HNSC], Bladder squamous cell carcinoma [BLSC], Stomach adenocarcinoma [STAD], and Prostate adenocarcinoma [PRAD].
Statistical analysis
Statistical analysis was carried out using Graph Pad Prism 5.0 software. The experiments were repeated thrice with 3 biological replicates. Error bars representing mean ± S.D are drawn from a representative experiment with biological triplicates. The ‘p’ values indicative of statistical significance were obtained by performing student's ‘t’ test (unpaired). Image J software was used for densitometry analysis of western blots.
Abbreviations
- CHX
cycloheximide
- c-IAP1
cellular-Inhibitor of Apoptosis-1
- c-IAP2
cellular-Inhibitor of Apoptosis-2
- ERR
Estrogen Related Receptor-1
- GNL3L
Guanine Nucleotide binding protein-like 3-like
- LDOC1
Leucine Zipper Downregulated in Cancer-1
- MDM2
murine double minute-2
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NF-κB
Nuclear Factor-kappa B
- RT-qPCR
Reverse Transcription-quantitative Polymerase Chain Reaction
- TNF-α
Tumor Necrosis Factor-α
- TRF1
Telomeric Repeat binding Factor-1
- XIAP
X-linked Inhibitor of Apoptosis
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grant from Department of Science and Technology [VI-D&P/411/2012-2013/TDT (G)] Govt. of India, to SM; graduate fellowships from Council of Scientific and Industrial Research (CSIR), Govt. of India to IJT and KR; graduate fellowship from Ministry of Human Resource Development (MHRD), Govt. of India to KA.
Author contributions
IJT designed and performed experiments, analyzed the data and wrote the paper. KR performed the experiments shown in Figs. 1D, 5F, 6A, 7C, 7D, 8, 9 and 10C and analyzed the data. KA performed the experiment shown in Figs. 1D, 3A, 6D and 11 and analyzed the data. SM conceived and coordinated the study and wrote the paper.
References
- [1].Subba Rao MRK, Kumari G, Balasundaram D, Sankaranarayanan R, Mahalingam SA. Novel lysine-rich domain and gtp binding motifs regulate the nucleolar retention of human guanine nucleotide binding protein, GNL3L. J Mol Biol 2006; 364:637-54; PMID:17034816; http://dx.doi.org/ 10.1016/j.jmb.2006.09.007 [DOI] [PubMed] [Google Scholar]
- [2].Meng L, Zhu Q, Tsai R. Nucleolar trafficking of nucleostemin family proteins: common versus protein-specific mechanisms. Mol Cell Biol 2007; 27:8670-82; PMID:17923687; http://dx.doi.org/ 10.1128/MCB.00635-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Thoompumkal IJ, Subba Rao MRK, Kumaraswamy A, Krishnan R, Mahalingam S. GNL3L is a nucleo-cytoplasmic shuttling protein-Role in cell cycle regulation. PLoS One 2015; 10(8):e0135845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Meng L, Hsu JK, Tsai RYL. GNL3L depletion destabilizes MDM2 and induces p53-dependent G2/M arrest. Oncogene 2010; 30:1716-26; PMID:21132010; http://dx.doi.org/ 10.1038/onc.2010.550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Yasumoto H, Meng L, Lin T, Zhu Q, Tsai R. GNL3L inhibits activity of estrogen-related receptor γ by competing for co-activator binding. J Cell Sci 2007; 120:2532-43; PMID:17623774; http://dx.doi.org/ 10.1242/jcs.009878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhu Q, Meng L, Hsu J, Lin T, Teishima J, Tsai R. GNL3L stabilizes the TRF1 complex and promotes mitotic transition. J Cell Biol 2009; 185:827-39; PMID:19487455; http://dx.doi.org/ 10.1083/jcb.200812121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Okamoto N, Yasukawa M, Nguyen C, Kasim V, Maida Y, Possemato R, Shibata T, Ligon LK, Fukami K, Hahn CW, Masutomi K. Maintenance of tumor initiating cells of defined genetic composition by Nucleostemin. PNAS 2011; 181:20388-93; http://dx.doi.org/ 10.1073/pnas.1015171108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Singh H, Sen R, Baltimore D, Sharp PA. A nuclear factor that binds to a conserved sequence motif in transcriptional control elements of immunoglobulin genes. Nature 1986; 319:154-58; PMID:3079885; http://dx.doi.org/ 10.1038/319154a0 [DOI] [PubMed] [Google Scholar]
- [9].Kim DW, Gazourian L, Quadri SA, Romieu-Mourez R, Sherr DH, Sonenshein GE. The RelA NF- κB subunit and the aryl hydrocarbon receptor (AhR) cooperate to transactivate the c-myc promoter in mammary cells. Oncogene 2000; 19:5498-506; PMID:11114727; http://dx.doi.org/ 10.1038/sj.onc.1203945 [DOI] [PubMed] [Google Scholar]
- [10].Mayo MW, Norris JL, Baldwin AS. Ras regulation of NF-kappa B and apoptosis. Methods Enzymol 2001; 333:73-87; PMID:11400356; http://dx.doi.org/ 10.1016/S0076-6879(01)33046-X [DOI] [PubMed] [Google Scholar]
- [11].Baldwin AS. Control of oncogenesis and cancer therapy resistance by the transcription factor NF-κB. J Clin Invest 2001; 107:241-46; PMID:11160144; http://dx.doi.org/ 10.1172/JCI11991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Baldwin AS. The NF-κB and IκB proteins: new discoveries and insights. Annu Rev Immunol 1996; 14:649-81; PMID:8717528; http://dx.doi.org/ 10.1146/annurev.immunol.14.1.649 [DOI] [PubMed] [Google Scholar]
- [13].Ghosh S, May MJ, Kopp EB. NF-κB and Rel proteins: Evolutionarily conserved mediators of immune responses. Annu Rev Immunol 1998; 16:225-60; PMID:9597130; http://dx.doi.org/ 10.1146/annurev.immunol.16.1.225 [DOI] [PubMed] [Google Scholar]
- [14].Tsujii M, Kawano S, Tsuji S, Sawaoka H, Hori M, DuBois RN. Cyclooxygenase regulates angiogenesis induced by colon cancer cells. Cell 1998; 93:705-16; PMID:9630216; http://dx.doi.org/ 10.1016/S0092-8674(00)81433-6 [DOI] [PubMed] [Google Scholar]
- [15].Huang S, DeGuzman A, Bucana CD, Fidler IJ. Nuclear factor-κB activity correlates with growth, angiogenesis, metastasis of human melanoma cells in nude mice. Clin Cancer Res 2000; 6:2573-81; PMID:10873114 [PubMed] [Google Scholar]
- [16].Andela VB, Schwarz EM, Puzas JE, O'Keefe RJ, Rosier RN. Tumor metastasis and the reciprocal regulation of prometastatic and antimetastatic factors by nuclear factor κB. Cancer Res 2000; 60:6557-62; PMID:11118032 [PubMed] [Google Scholar]
- [17].Huang S, Pettaway CA, Uehara H, Bucana CD, Fidler IJ. Blockade of NF-kappaB activity in human prostate cancer cells is associated with suppression of angiogenesis, invasion, metastasis. Oncogene 2001; 20:4188-97; PMID:11464285; http://dx.doi.org/ 10.1038/sj.onc.1204535 [DOI] [PubMed] [Google Scholar]
- [18].Yamamoto Y, Gaynor R. Therapeutic potential of inhibition of the NF-κB pathway in the treatment of inflammation and cancer. J Clin Invest 2001; 107:135-42; PMID:11160126; http://dx.doi.org/ 10.1172/JCI11914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Orlowski RZ, Baldwin AS. NF-kappaB as a therapeutic target in cancer. Trends Mol Med 2002; 8:385-89; PMID:12127724; http://dx.doi.org/ 10.1016/S1471-4914(02)02375-4 [DOI] [PubMed] [Google Scholar]
- [20].Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev 2004; 18:2195-224; PMID:15371334; http://dx.doi.org/ 10.1101/gad.1228704 [DOI] [PubMed] [Google Scholar]
- [21].Nagasaki K, Manabe T, Hanzawa H, Maass N, Tsukada T, Yamaguchi K. Identification of a novel gene, LDOC1, down-regulated in cancer cell lines. Int J Cancer 1999; 140:227-34. [DOI] [PubMed] [Google Scholar]
- [22].Buchholtz ML, Jückstock J, Weber E, Mylonas I, Dian D, Brüning A. Loss of LDOC1 expression by promoter methylation in cervical cancer cells. Cancer Invest 2013; 31:571-77; PMID:24125169; http://dx.doi.org/ 10.3109/07357907.2013.845671 [DOI] [PubMed] [Google Scholar]
- [23].Nagasaki K, Schem C, Von Kaisenberg C, Biallek M, Rösel F, Jonat W, Maass N. Leucine-zipper protein, LDOC1, inhibits NF-κB activation and sensitizes pancreatic cancer cells to apoptosis. Int J Cancer 2003; 105:454-58; PMID:12712434; http://dx.doi.org/ 10.1002/ijc.11122 [DOI] [PubMed] [Google Scholar]
- [24].Rual JF, Kavitha V, Hao T, Hirozane-Kishikawa T, Dricot A. Towards a proteome-scale map of the human protein-protein interaction network. Nature 2005; 437:1173-78; PMID:16189514; http://dx.doi.org/ 10.1038/nature04209 [DOI] [PubMed] [Google Scholar]
- [25].Mizutani K, Koike D, Suetsugu S, Takenawa T. WAVE3 functions as a negative regulator of LDOC1. J Biochem 2005; 138:639-46; PMID:16272576; http://dx.doi.org/ 10.1093/jb/mvi160 [DOI] [PubMed] [Google Scholar]
- [26].Inoue M, Takahashi K, Niide O, Shibata M, Fukuzawa M, Ra C. LDOC1, a novel MZF-1 interacting protein, induces apoptosis. FEBS Letters 2005; 579:604-08; PMID:15670815; http://dx.doi.org/ 10.1016/j.febslet.2004.12.030 [DOI] [PubMed] [Google Scholar]
- [27].Gewurz EB, Towfic F, Mar JC, Shinners NP, Takasaki K, Zhao B, Cahir-McFarland DE, Quackenbush J, Xavier JR, Kieff E. Genome wide siRNA screen for mediators of NF-kappa B activation. PNAS 2011; 109:2467-72; http://dx.doi.org/ 10.1073/pnas.1120542109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Pahl HL.. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene 1999; 18:6853-66; PMID:10602461; http://dx.doi.org/ 10.1038/sj.onc.1203239 [DOI] [PubMed] [Google Scholar]
- [29].Du X, Subba Rao M, Chen X, Wu W, Mahalingam S, Balasundaram D. The homologous putative GTPases Grn1p from fission yeast and the human GNL3L are required for growth and play a role in processing of nucleolar Pre-rRNA. Mol Biol Cell 2006; 17:460-74; PMID:16251348; http://dx.doi.org/ 10.1091/mbc.E05-09-0848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lee CH, Wong TS, Chan JYW, Lu SC, Lin P, Cheng AJ, Chang JY. Epigenetic regulation of the X-linked tumour suppressors BEX1 and LDOC1 in oral squamous cell carcinoma. J Pathol 2013; 230:298-309; PMID:23362108; http://dx.doi.org/ 10.1002/path.4173 [DOI] [PubMed] [Google Scholar]
- [31].Stehlik C, de Martin R, Kumabashiri I, Schmid JA, Binder BR, Lipp J. Nuclear Factor (NF)-κB-regulated X-chromosome-linked IAP gene expression protects endothelial cells from tumor necrosis factor α-induced apoptosis. J Exp Med 1998; 188:211-16; PMID:9653098; http://dx.doi.org/ 10.1084/jem.188.1.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang CY, Mayo MW, Korneluk RG, Goeddel DV, Baldwin AS. NF-κB antiapoptosis: Induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science 1998; 281:1680-83; PMID:9733516; http://dx.doi.org/ 10.1126/science.281.5383.1680 [DOI] [PubMed] [Google Scholar]
- [33].Rothe M, Pan MG, Henzel WJ, Ayres TM, Goeddel DV. The TNFR2-TRAF signaling complex contains two novel proteins related to baculoviral inhibitor of apoptosis proteins. Cell 1995; 83:1243-52; PMID:8548810; http://dx.doi.org/ 10.1016/0092-8674(95)90149-3 [DOI] [PubMed] [Google Scholar]
- [34].Duckett CS, Nava VE, Gedrich RW, Clem RJ, Van Dongen JL, Gilfillan MC, Thompson CB. A conserved family of cellular genes related to the baculovirus iap gene and encoding apoptosis inhibitors. EMBO J 1996; 15:2685-94; PMID:8654366 [PMC free article] [PubMed] [Google Scholar]
- [35].Liston P, Roy N, Tamai K, Lefebvre C, Baird S, Cherton- Horvat G, Farahani R, McLean M, Ikeda J, MacKenzie A, Korneluk RG. Suppression of apoptosis in mammalian cells by NAIP and a related family of IAP genes. Nature 1996; 379:349-53; PMID:8552191; http://dx.doi.org/ 10.1038/379349a0 [DOI] [PubMed] [Google Scholar]
- [36].Ambrosini G, Adida C, Altieri D. A novel anti- apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med 1997; 3:917-21; PMID:9256286; http://dx.doi.org/ 10.1038/nm0897-917 [DOI] [PubMed] [Google Scholar]
- [37].Hauser HP, Bardroff M, Pyrowolakis G, Jentsch S. A giant ubiquitin-conjugating enzyme related to IAP apoptosis inhibitors. J Cell Biol 1998; 141:1415-22; PMID:9628897; http://dx.doi.org/ 10.1083/jcb.141.6.1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Viatour P, Merville MP, Bours V, Chariot A. Phosphorylation of NF-kappa B and Ikappa B proteins: implications in cancer and inflammation. Trends Biochem Sci 2015; 30:43-52; http://dx.doi.org/ 10.1016/j.tibs.2004.11.009 [DOI] [PubMed] [Google Scholar]
- [39].Shukla S, MacLennan GT, Fu P, Patel J, Marengo SR, Resnick ML, Gupta S. Nuclear factor-κB/p65 (Rel A) is constitutively activated in human prostate adenocarcinoma and correlates with disease progression. Neoplasia 2004; 6:390-400; PMID:15256061; http://dx.doi.org/ 10.1593/neo.04112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yu L, Yu H, Yu J, Luo H, Xu X, Li J. Nuclear factor- κ B p65 (RelA) transcription factor is constitutively activated in human colorectal carcinoma tissue. World J Gastroenterol 2004; 10:3255-60; PMID:15484295; http://dx.doi.org/ 10.3748/wjg.v10.i22.3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chen CD, Sawyers CL. NF-kappa B activates prostate-specific antigen expression and is up-regulated in androgen-independent prostate cancer. Mol Cell Biol 2002; 22:2862-70; PMID:11909978; http://dx.doi.org/ 10.1128/MCB.22.8.2862-2870.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Visconti R, Cerutti J, Battista S, Fedele M, Trapasso F, Zeki K, Fusco A. Expression of the neoplastic phenotype by human thyroid carcinoma cell lines requires NF-kappaB p65 protein expression. Oncogene 1997; 15:1987-94; PMID:9365245; http://dx.doi.org/ 10.1038/sj.onc.1201373 [DOI] [PubMed] [Google Scholar]
- [43].Wong HK, Kammer GM, Dennis G, Tsokos GC. Abnormal NF-κB activity in T lymphocytes from patients with systemic lupus erythematosus is associated with decreased p65-RelA protein expression. J Immunol 1999; 163:1682-89; PMID:10415075 [PubMed] [Google Scholar]
- [44].Wan Q, Dingerdissen H, Fan Y, Gulzar N, Pan Y, Wu TJ, Yan C, Zhang H, Mazumder R.. BioXpress: an integrated RNA-seq-derived gene expression database for pan-cancer analysis. Database 2015; 2015:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis 2009; 30:1073-81; PMID:19468060; http://dx.doi.org/ 10.1093/carcin/bgp127 [DOI] [PubMed] [Google Scholar]
- [46].Baud V, Karin M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat Rev Drug Discov 2009; 8:33-40; PMID:19116625; http://dx.doi.org/ 10.1038/nrd2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Tong L, Tergaonkar V. Rho protein GTPases and their interactions with NFκB: crossroads of inflammation and matrix biology. Biosci Rep 2014; 34:e00115; PMID:24877606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Montaner S, Perona R, Saniger L, Lacal JC. Multiple signalling pathways lead to the activation of the nuclear factor kappaB by the Rho family of GTPases. J Biol Chem 1998; 273:12779-85; PMID:9582304; http://dx.doi.org/ 10.1074/jbc.273.21.12779 [DOI] [PubMed] [Google Scholar]
- [49].Perona R, Montaner S, Saniger L, Sanchez-Perez I, Bravo R, Lacal JC. Activation of the nuclear factor-kappaB by Rho, CDC42, Rac-1 proteins. Genes Dev 1997; 11:463-75; PMID:9042860; http://dx.doi.org/ 10.1101/gad.11.4.463 [DOI] [PubMed] [Google Scholar]
- [50].Cildir G, Low KC, Tergaonkar V. Noncanonical NF-κB signaling in health and disease. Trends Mol Med 2016; 22:414-29; PMID:27068135; http://dx.doi.org/ 10.1016/j.molmed.2016.03.002 [DOI] [PubMed] [Google Scholar]
- [51].Dey A, Wong E, Kua N, Teo HL, Tergaonkar V, Lane D. Hexamethylene Bisacetamide (HMBA) simultaneously targets AKT and MAPK pathway and represses NF-κB activity: Implications for cancer therapy. Cell Cycle 2008; 7:3759-67; PMID:19029824; http://dx.doi.org/ 10.4161/cc.7.23.7213 [DOI] [PubMed] [Google Scholar]
- [52].Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell RG. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J Biol Chem 2008; 6:23589-97. [DOI] [PubMed] [Google Scholar]
- [53].Bauge C, Attia J, Leclercq S, Pujol JP, Gale´ra P, Boume´diene K. Interleukin-1β up-regulation of Smad7 via NF-Κb activation in human chondrocytes. Arthritis Rheum 2008; 58:221-26; PMID:18163503; http://dx.doi.org/ 10.1002/art.23154 [DOI] [PubMed] [Google Scholar]
- [54].Dyer BW, Ferrer F, Klinedinst DK, Rodriguez R. A noncommercial dual luciferase enzyme assay system for reporter gene analysis. Anal Biochem 2000; 282:158-61; PMID:10860516; http://dx.doi.org/ 10.1006/abio.2000.4605 [DOI] [PubMed] [Google Scholar]