Abstract
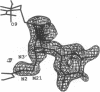
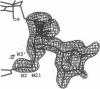
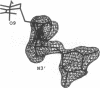
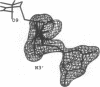
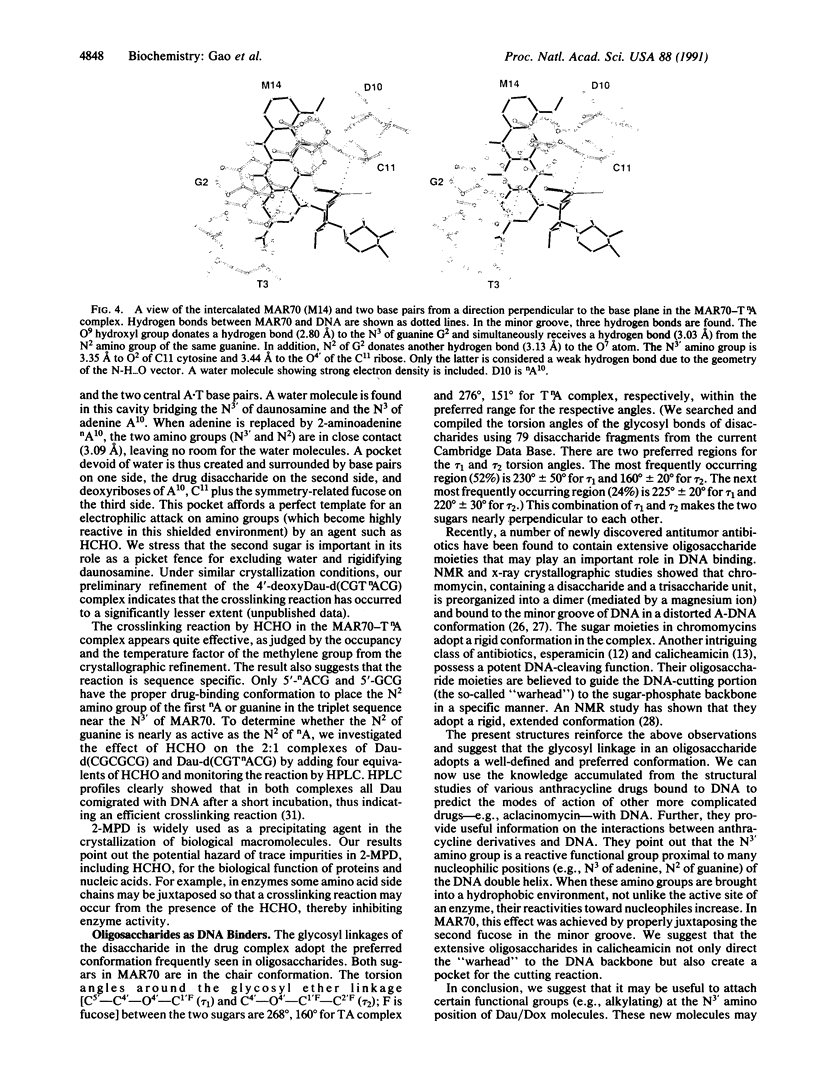
MAR70 is a synthetic derivative of the anticancer drug daunorubicin that contains an additional sugar, attached to the O4' of daunosamine. When MAR70 was crystallized with the DNA hexamer d(CGTnACG), where nA is 2-aminoadenine, a covalent methylene bridge was formed between the N3' of daunosamine and the N2 of 2-aminoadenine. This spontaneous reaction occurred through the crosslinking action of formaldehyde. The crosslink was demonstrated by the three-dimensional structure of the 2:1 adduct between MAR70 and d(CGTnACG) solved at 1.3-A resolution by x-ray diffraction analysis. The perfect juxtaposition of the two amino groups in the complex provides a template for efficient addition of formaldehyde. This adduct structure is compared with the analogous structure at 1.5-A resolution of the complex of MAR70-d(CGTACG), in which no formaldehyde addition was observed. In both complexes, two MAR70 molecules bind to the DNA hexamer double helix; the elongated aglycon chromophore is intercalated between the CpG steps and spans the G.C Watson-Crick base pairs. The disaccharides occupy nearly the entire minor groove of the distorted B-DNA hexamer double helix. The second sugar is in contact with the sugar-phosphate backbone and does not affect the binding interactions of the daunorubicin portion to DNA. The structure allows us to model the binding to DNA of drugs having more extensive oligosaccharides. In addition, it suggests that placing a reactive (e.g., alkylating) functional group at the N3' amino position of daunorubicin might be a fruitful route for designing anticancer drugs.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Denny W. A. DNA-intercalating ligands as anti-cancer drugs: prospects for future design. Anticancer Drug Des. 1989 Dec;4(4):241–263. [PubMed] [Google Scholar]
- Frederick C. A., Williams L. D., Ughetto G., van der Marel G. A., van Boom J. H., Rich A., Wang A. H. Structural comparison of anticancer drug-DNA complexes: adriamycin and daunomycin. Biochemistry. 1990 Mar 13;29(10):2538–2549. [PubMed] [Google Scholar]
- Gao X. L., Patel D. J. Solution structure of the chromomycin-DNA complex. Biochemistry. 1989 Jan 24;28(2):751–762. doi: 10.1021/bi00428a051. [DOI] [PubMed] [Google Scholar]
- Hanka L. J., Dietz A., Gerpheide S. A., Kuentzel S. L., Martin D. G. CC-1065 (NSC-298223), a new antitumor antibiotic. Production, in vitro biological activity, microbiological assays and taxonomy of the producing microorganism. J Antibiot (Tokyo) 1978 Dec;31(12):1211–1217. doi: 10.7164/antibiotics.31.1211. [DOI] [PubMed] [Google Scholar]
- Kind R., Hütter K., Zeeck A., Schmidt-Bäse K., Egert E. Viriplanin A, a new anthracycline antibiotic of the nogalamycin group. II. The structure of a novel hydroxyamino sugar from reduced viriplanin A. J Antibiot (Tokyo) 1989 Jan;42(1):7–13. doi: 10.7164/antibiotics.42.7. [DOI] [PubMed] [Google Scholar]
- Liaw Y. C., Gao Y. G., Robinson H., van der Marel G. A., van Boom J. H., Wang A. H. Antitumor drug nogalamycin binds DNA in both grooves simultaneously: molecular structure of nogalamycin-DNA complex. Biochemistry. 1989 Dec 26;28(26):9913–9918. doi: 10.1021/bi00452a006. [DOI] [PubMed] [Google Scholar]
- Robinson H., Liaw Y. C., van der Marel G. A., van Boom J. H., Wang A. H. NMR studies on the binding of antitumor drug nogalamycin to DNA hexamer d(CGTACG). Nucleic Acids Res. 1990 Aug 25;18(16):4851–4858. doi: 10.1093/nar/18.16.4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle M. S., Hall J. G., Denny W. A., Wakelin L. P. NMR studies of the interaction of the antibiotic nogalamycin with the hexadeoxyribonucleotide duplex d(5'-GCATGC)2. Biochemistry. 1988 Jun 14;27(12):4340–4349. doi: 10.1021/bi00412a022. [DOI] [PubMed] [Google Scholar]
- Sugiura Y., Uesawa Y., Takahashi Y., Kuwahara J., Golik J., Doyle T. W. Nucleotide-specific cleavage and minor-groove interaction of DNA with esperamicin antitumor antibiotics. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7672–7676. doi: 10.1073/pnas.86.20.7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasz M., Chawla A. K., Lipman R. Mechanism of monofunctional and bifunctional alkylation of DNA by mitomycin C. Biochemistry. 1988 May 3;27(9):3182–3187. doi: 10.1021/bi00409a009. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Ughetto G., Quigley G. J., Rich A. Interactions between an anthracycline antibiotic and DNA: molecular structure of daunomycin complexed to d(CpGpTpApCpG) at 1.2-A resolution. Biochemistry. 1987 Feb 24;26(4):1152–1163. doi: 10.1021/bi00378a025. [DOI] [PubMed] [Google Scholar]
- Wang J. C. The degree of unwinding of the DNA helix by ethidium. I. Titration of twisted PM2 DNA molecules in alkaline cesium chloride density gradients. J Mol Biol. 1974 Nov 15;89(4):783–801. doi: 10.1016/0022-2836(74)90053-9. [DOI] [PubMed] [Google Scholar]
- Warpehoski M. A., Hurley L. H. Sequence selectivity of DNA covalent modification. Chem Res Toxicol. 1988 Nov-Dec;1(6):315–333. doi: 10.1021/tx00006a001. [DOI] [PubMed] [Google Scholar]
- Westendorf J., Aydin M., Groth G., Weller O., Marquardt H. Mechanistic aspects of DNA damage by morpholinyl and cyanomorpholinyl anthracyclines. Cancer Res. 1989 Oct 1;49(19):5262–5266. [PubMed] [Google Scholar]
- Williams L. D., Egli M., Qi G., Bash P., van der Marel G. A., van Boom J. H., Rich A., Frederick C. A. Structure of nogalamycin bound to a DNA hexamer. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2225–2229. doi: 10.1073/pnas.87.6.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zein N., Poncin M., Nilakantan R., Ellestad G. A. Calicheamicin gamma 1I and DNA: molecular recognition process responsible for site-specificity. Science. 1989 May 12;244(4905):697–699. doi: 10.1126/science.2717946. [DOI] [PubMed] [Google Scholar]
- Zhang X. L., Patel D. J. Solution structure of the nogalamycin-DNA complex. Biochemistry. 1990 Oct 9;29(40):9451–9466. doi: 10.1021/bi00492a020. [DOI] [PubMed] [Google Scholar]