Abstract
IL37 is a member of IL-1 cytokine family but conveys anti-inflammatory functions. The biological characteristic and genetic heterogeneity of IL37 are not fully understood yet. Here using the whole-genome sequencing data from 1000 Genomes Project, we performed population and evolutionary genetic analysis of human IL37 gene. First, 2184 IL37 gene sequences from different human populations were retrieved. The IL37 protein sequences were inferred from the coding DNA sequences and multiple species alignment was made. Then, the phylogenetic tree of IL37 was built and dN/dS ratios were calculated for each evolutionary branch, the classic McDonald and Kreitman test was also performed. Next, we conducted intraspecific evolutionary genetic analysis and built the genealogy network of 116 unique IL37 haplotypes through median-joining network analysis. Finally, we compared IL37 sequences between the modern and archaic humans. Our results for the first time provide solid evidence that common IL37 variants other than NCBI reference sequence are present worldwide. Our data also supports that IL37 variants are shaped and maintained by selection instead of neutral evolution. We further identified that human IL37 variants consist of two major haplogroups and their presence in archaic humans corroborates its ancient origin in hominid evolution. In conclusion, these data indicate that common IL37 variants are maintained among human populations by selective force, suggesting their potential involvements in immune regulation and human diseases. In addition, the ancient history of IL37 variants reveals interesting insight into the complicated human evolutionary history.
Introduction
IL37, a novel member of the IL-1 family also known as IL-1F7, was discovered by several independent groups in 2000.1 Its function is not completely understood yet. Interestingly, distinct from most IL-1 family members, which have been characterized as proinflammatory, IL37 has emerged as a fundamental immune inhibitor.2 A total of five spliced variants (IL37a–e) exist as IL37 transcripts. Among them, IL37b is the largest member with 218 amino acids and encoded by five exons spanning the IL37 gene, of which exons 1 and 2 encode the prodomain and exons 3–5 encode the 12 putative β-strands forming β-trefoil structure, which is characteristic of the IL-1 family molecules.1, 3 The IL37b is a precursor, which is thought to be processed by caspase-1 into the mature form and translocates actively into the cell nucleus.4 IL37b mRNA has been found in multiple human tissues, including the lymph node, thymus, bone marrow, placenta, lung and testis.5 IL37 protein level in PBMCs and dendritic cells is upregulated when stimulated by TLR ligands or proinflammatory cytokines.1 In vitro overexpression of IL37 in macrophages or epithelial cells could greatly dampen production of major proinflammatory cytokines, including IL-1α, IL-1β, TNF-α, IL-6 and MIP-2.1, 2, 6 In vivo, IL37 transgene protects mice from LPS-induced shock, chemical-induced colitis.7, 8 However, the role of IL37 in physiological conditions and its involvement in human diseases remained largely unknown.
With the development of DNA sequencing and genotyping technology, genetic methods become more powerful at unveiling the function of human genes. Among these methods, evolutionary genetics deals with variations within human populations. The basis for this approach is that evolution will clear the deleterious mutations and select the beneficial ones among human populations, so it leaves behind signal that could be detected using population genetic variation data and statistic tools.9 As a complement to clinical and epidemiological genetic approaches, the evolutionary genetic approach has increased our understanding of the evolutionary forces that shape the human genome and provided important insights into the function of selected genes.10 Human immune system as an interface between the body and external world is especially prone to be affected by selective pressure, for example, various pathogens and other immune disorders. Indeed, genome-wide scanning of selection signals has often found immune genes as preferred candidates.11 Identifying the extents and types of natural selection acting upon genes involved in immunity-related processes has already revealed insights of host defense mediated by them, as well as delineated those genes being essential in host defenses. So far, human pattern recognition receptors and interferons have been analyzed using this approach, attesting its effectiveness in revealing functional aspects of immune genes.12, 13, 14 However, the extent to which human IL37 have been subject to natural selection remains largely unknown. In addition, we are interested by the fact that IL37 gene has been lost in human's closest relative chimpanzee during the evolutionary process.15 Here utilizing data from 1000 Genomes Project,16 we report that IL37 variant proteins other than reference sequence are common in human populations. Evolutionary genetic investigation of IL37 variations reveals its significant deviation from neutrality. We present that modern human IL37 variants consist of two major haplogroups and have an early origin during hominid evolution.
Materials and methods
Variant data set and sequences
The 1000 Genomes project phase1 data were downloaded from the data repository (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase1/analysis_results/ integrated_call_sets/). This data set contains the phased high-quality variant calls of both single-nucleotide polymorphisms (SNPs) and short insertions and deletions for 1092 individuals from four major populations, including 379 Europeans (EUR), 286 East Asians (ASN), 246 Africans (AFR) and 181 admixed Americans (AMR). The high-quality whole-genome sequence data of archaic human (including Neanderthal and Denisova) were downloaded from http://cdna.eva.mpg.de/. Ancestral sequences of human and gorilla were downloaded from Ensembl (http://www.ensembl.org). In addition, the low-coverage Neanderthal sequences representing 72% of IL37 gene locus were retrieved from EBI ftp site (ftp://ftp.ebi.ac.uk/pub/databases/ensembl/neandertal).
Among the five IL37 transcripts, transcript isoform 1 (IL37b) is the largest and best characterized one, which was chosen for the current study with no further indication. IL37 NCBI RefSeq protein (NP_055254.2) and cDNA (NM_014439.3) orthologous sequences for 10 non-human primates and Canis were downloaded from Ensembl website.
Human IL37 gene sequence reconstruction and linkage disequilibrium block inference
The variant information in human genome region chr2.hg19:g.113670547_113676458 corresponding to transcribed region of IL37b were extracted from the downloaded data. The exons were numbered like in NG_029219.1. IL37 genomic sequences for 1092 individuals were inferred by replacing bases of reference sequence with corresponding variant bases borrowing the same logic as FastaAlternateReferenceMaker tool of GATK software package (https://www.broadinstitute.org/gatk/gatkdocs/org_broadinstitute_gatk_tools_walkers_fasta_FastaAlternateReferenceMaker.php). Haploview 4.2 program was used to infer linkage disequilibrium (LD) patterns and values of the target region as indicated in the figure using polymorphisms with minor allele frequencies >1%.
DNA polymorphism analysis and variant annotation
The DNA polymorphic parameters including the number of segregation site, number of haplotypes, haplotype diversity and nucleotide diversity for human IL37 gene were analyzed by DnaSP5.10.01 (Barcelona, Spain). The variants in target region were annotated by ANNOVAR.17 For variant effect prediction, the SIFT (Genomic Institute of Singapore) and Polyphen (Boston, MA, USA) scores were acquired from Ensembl website.
Sequence alignment and phylogenetic analysis
Multiple species alignments of IL37 protein sequences were performed using Clustal-omega-1.2.0 (Dublin, Ireland) followed by manual curation. The curated protein alignments were then conversed to corresponding codon alignments by PAL2NAL online web server.18 Phylogenetic tree was constructed by SeaView 4.5.0 (Villeurbanne, France) using the method of neighbor joining based on IL37 cDNA sequence alignments. dN, dS calculation and phylogenetic tree manipulations were performed using python package ETE2.2,19 which calls codeml of PAML4 package20 with branch model for dN and dS calculation at the backend.
Haplotype network analysis
Haplotype network was constructed using NETWORK 4.6.1.2 (Fluxus)21 with method of median joining (MJ). To root the haplotype network, gorilla IL37 sequence and human–gorilla ancestral sequence were added to 1000 Genomes data set. As the large divergence between Gorilla and human sequences poses challenge for inferring the correct genealogy network, the most recent common ancestor (MRCA) of all human haplotypes were added to facilitate MJ network construction. This MRCA sequence was deduced by a parsimonious way in that all shared derived alleles in 1000 Genomes samples were assigned derived alleles, whereas all other sites should be ancestral alleles. The segregation time of two major human IL37 haplogroups from MRCA were estimated based on the lineage specific molecular clock calibrated by the divergence time 10 million years ago (Mya) and 110 divergent sites between gorilla and human in 5.9-kb genomic sequence of IL37 genes, which is exactly located in the LD block.
McDonald and Kreitman test
McDonald and Kreitman (MK) test was performed by online server tool MKT at http://mkt.uab.es with non-synonymous and synonymous changes selected as the types of analysis.
Intraspecific evolutionary genetic analysis
The statistics of Tajima's D, Fu's Fs, Fu and Li's F* and D* for the studied genes were calculated using population variation data by DnaSP5.10.01. Fst values among different populations were calculated using VCFtools (http://vcftools.sourceforge.net/).
The statistical significance of each test score was evaluated among an empirical distribution of the corresponding test scores of 1114 known RefSeq immune genes, which are listed in Supplementary Table S5. More detailed information about the methods can be found on the Supplementary Materials.
Results
General population genetic statistics
The IL37 gene and its 15 kb flanking sequences were retrieved from 1000 Genomes Project data as described in the methods. As many analyses can be performed only effectively on regions that have experienced little or no recombination, we therefore investigated the LD structure of the region and identified a 17 kb LD block that contained the entire transcribed region of IL37 gene (Supplementary Figure S1). All subsequent genetic analyses were then confined to the transcribed region located in this LD block. In our samples of 2184 chromosomes, we detected in total 114 SNPs and analyzed the summary population statistics (Table 1). We found that both nucleotide and haplotype diversity were much higher in AFR compared with EUR and ASN, consistent with the recent African origin hypothesis of modern human. Our data also showed 14 non-synonymous and 1 nonsense substitutions in IL37 genes (Supplementary Table S1). Supplementary Table S2 shows the numbers and distributions of missense and nonsense substitutions in IL37 among different populations.
Table 1. Population genetic parameters of human IL37 gene.
| Population | n | S | Hap | Hd | π |
|---|---|---|---|---|---|
| AFR | 492 | 84 | 65 | 0.904 | 0.00249 |
| AMR | 362 | 65 | 29 | 0.748 | 0.001385 |
| ASN | 572 | 61 | 24 | 0.773 | 0.000775 |
| EUR | 758 | 64 | 37 | 0.737 | 0.001545 |
| Global | 2184 | 115 | 115 | 0.826 | 0.001729 |
Abbreviations: AFR, Africans; AMR, Admixed Americans; ASN, Asians; EUR, Europeans; Hap, number of haplotypes; Hd, haplotype diversity; n, number of chromosomes; π, nucleotide diversity; S, number of segregation sites.
5.9-kb-transcribed region of human IL37 genes were analyzed.
IL37 variant proteins and their distributions among human populations
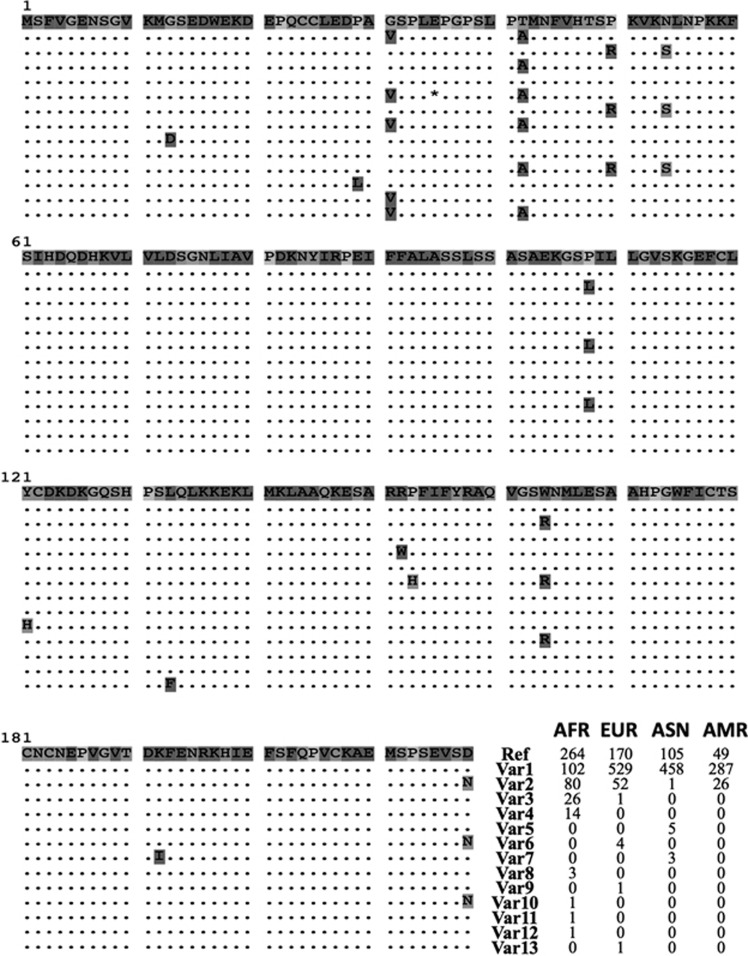
The fact that many non-synonymous mutations are common polymorphisms (>5%) indicates that IL37 variant proteins are common among different human populations. Therefore, we decided to find how many variant proteins there are and how they distributed geographically. Through deducing the protein sequences from the coding DNA sequences, we found in total 14 protein variants among human populations including one truncated protein caused by an early stop codon (Figure 1). Three major variants together account for >97% of all the sequences. Interestingly, the NCBI reference (NP_055254.2) is only dominant in AFR and a variant (IL37-Var1) with c.92G>T (p.(Gly31Val)) and c.124A>G (p.(Thr42Ala)) substitutions dominates in non-AFR. The third major variant (IL37-Var2), which is roughly 16% in AFR and 7% globally, differs at five sites with other major variants, including c.149C>G (p.(Pro50Arg)), c.161A>G (p.(Asn54Ser)), c.324G>A (p.(Pro108Leu)), c.490T>C (p.(Trp164Arg)) and c.652G>A (p.(Asp218Asn)). Notably, all these five non-synonymous SNPs are in a perfect LD with r2=1 in all of our samples (Supplementary Figure S1E). To further reveal their functional consequences, we used scores from both Polyphen and SIFT programs to predict the effects of non-synonymous changes on IL37 protein (Supplementary Table S1). According to Polyphen scores, five non-synonymous sites were predicted to be possibly damaging (0.453–0.956) and two sites were probably damaging (≥0.957). According to SIFT prediction, two and three sites predicted by Polyphen to be probably and possibly damaging respectively were also predicted to be deleterious (≤0.05).
Figure 1.
IL37 variant proteins and their distributions among human populations. Fourteen unique protein sequences including the reference IL37 protein sequence (NP_055254.2) from NCBI were identified in 1000 Genomes samples. These protein sequences were aligned by Clustal-omega followed by manual curation. Frequencies of protein variants among different populations are shown to the right of the sequences.
Multiple species alignment of IL37 proteins
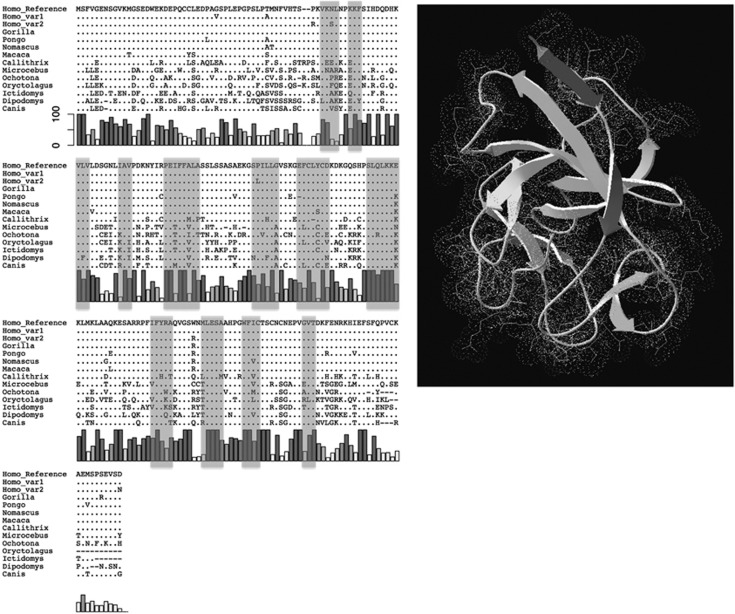
To assess the evolutionary processes, we downloaded 11 mammalian IL37 protein orthologues from Ensemble database and aligned them with three major human variants (Figure 2). We found that IL37 is conserved from macaque, apes to humans. For example, human reference sequence only differs at two and nine sites with gorilla and macaque sequences, respectively. However, human Var1 and Var2, the two major human variants, differ at seven positions with each other. Clearly, this is a much large divergence than expected.
Figure 2.
Sequence alignment of IL37 proteins from multiple species. Three major IL37 variant sequences identified in 1000 Genomes samples and 11 non-human mammalian orthologous sequences (10 primates and Canis) were aligned by Clustal-omega. Nucleotide positions identical to human reference IL37 sequence are indicated as dots. Grey bars under the sequence alignment depict the conservation profile of the corresponding amino-acid residues, with 100 indicates complete conservation. Shaded regions correspond to predicted secondary structure of β-sheet in 3D protein structure (right panel), which constitute the basic structural framework of IL-1 family members. The cartoon view in the right panel with predicted 3D protein structure of IL37 was created by SWISS-MODEL (http://swissmodel.expasy.org).
Evolutionary genetics analysis of human IL37 genes
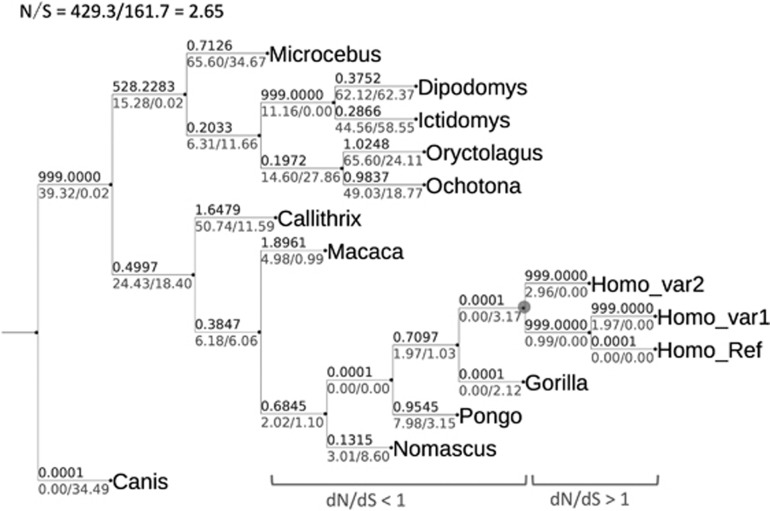
One of the key aims of evolutionary genetic study is to find whether the observed genetic variations are shaped by neutrality or selection. For this purpose, we first constructed the phylogenetic tree based on IL37 cDNA sequences using neighbor-joining method. Next, we computed the synonymous (dS) and non-synonymous (dN) substitution rate along each branch of the tree (Figure 3). Our data showed that most dN/dS ratios of the primate lineages, except macaque are less than one indicating a prevalent purifying selection. Interestingly, this purifying selection mode is, however, reversed (indicated by dN/dS>1 in Figure 3) in most recent lineages leading to modern humans with accumulation of many non-synonymous substitutions and few synonymous ones.
Figure 3.
The dN/dS ratios of different evolutionary lineages in primates. Phylogenetic tree was constructed using neighbor-joining method using IL37 orthologous sequences from multiple species. dN and dS for each evolutionary branch was calculated by codeml of PAML4 with branch model. The values above represent dN/dS ratios and the values below correspond to dN and dS, respectively. The enlarged internal node indicates the MRCA of modern humans. Distinct primate evolutionary phases, labeled as dN/dS <1 and dN/dS >1, are indicated before and after this node.
We then used another classical method, the MK test, to confirm the evolutionary forces shaping IL37 diversity after the divergence of the human and gorilla. Under the MK test, the ratio of non-synonymous:synonymous polymorphisms within humans was compared with ratio of fixed non-synonymous:synonymous divergences between human and gorilla, using Fisher's exact test. The ratios of polymorphism and divergence are expected to be equal under neutrality. Our analysis of interspecific divergence between gorilla and humans found 5.12 and 1.00 (Jukes and Cantor corrected) fixed differences at synonymous and non-synonymous sites, respectively. Table 2 contrasts the interspecific divergence with the degree of intraspecific polymorphism found in our samples using the MK test. The significant result (P=0.01467) confirmed that human IL37 genetic varieties deviated from neutrality.
Table 2. Contingency table of MK test for IL37 gene.
| Polymorphism | Divergencea | Total | |
|---|---|---|---|
| Synonymous | 4 | 5.12 | 9.12 |
| Non-synonymous | 14b | 1.00 | 15 |
| Total | 18 | 6.12 | 24.12 |
Abbreviation: MK, McDonald and Kreitman.
Corrected by Jukes and Cantor method.
Exclude the nonsense mutation. P-value=0.01467 (Fisher's exact test).
To further reveal the signals of recent selection and local adaptation of IL37 gene, we used classic tests such as Tajima's D, Fu's Fs, Fu and Li's F* and D* and Fst based on analyzing intraspecific polymorphisms among different human populations. We calculated the statistic values of 1114 RefSeq immune genes and used the empirical approach to evaluate the significance of corresponding statistics for IL37 gene (Supplementary Table S3 and Supplementary Figure S2). Our data showed that Tajima's D for AFR were within the 4% of the positive side of the empirical distribution, whereas Fu and Li's D* and F* for ASN were significantly negative (P=0.01) according to both coalescent simulation (data not shown) and empirical distribution (Supplementary Table S3), indicating excess of rare alleles in ASN. Pairwise population genetic differentiation (measured by Fst tests) showed that many variant sites of IL37 gene displayed large genetic differences in AFR vs ASN comparison, including the five non-synonymous sites defining the two major haplogroups of human IL37 gene (described in the following part of this study), which showed large Fst values (Supplementary Figure S2).
MJ network analysis of human IL37 haplotypes
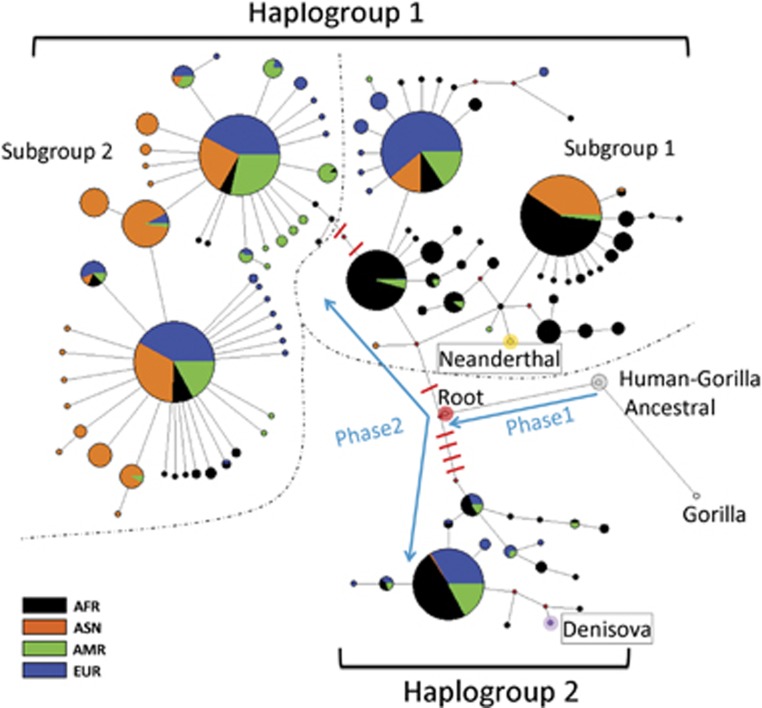
In our data, 114 haplotypes of human IL37 were identified (Supplementary Figure S3 and Supplementary Table S4). A MJ network was then constructed to show the genealogical relationships between the inferred haplotypes of the LD block (Figure 4). The topology of this network undoubtedly shows that two major clades (haplogroups 1 and 2) are separated by long branch length, each containing one haplogroup. Within haplogroup 1, there are two subgroups. Subgroup 1 is substantially of AFR origin, whereas subgroup 2 is dominant in Eurasians. Interestingly, we also noticed two distinct evolutionary phases after the split between human and gorilla (Figure 4). The first phase includes just synonymous mutations and the second phase includes many non-synonymous mutations, suggesting that the human IL37 variants were shaped by selection during the second phase of human evolution. We next estimated the time to MRCA of the haplogroups using phylogeny-based methods measuring ρ, the average distance of descendant haplotypes from a specified root. On the basis of 110 divergences between human and Gorilla IL37 sequences where the split time is assumed to be 10 Mya, we estimated the haplogroups 1 and 2 began to diverge around 3.6 Mya.
Figure 4.
Haplotype network of archaic and modern human IL37 haplotypes. A total of 119 haplotypes, including 114 modern human, 1 common ancestor of modern human, 2 archaic human, 1 gorilla and 1 human–gorilla ancestor haplotypes, were included for network construction. The sequence of common ancestor of modern human was deduced as described in method section and included to facilitate median-joining network construction. Gorilla and human–gorilla ancestral haplotypes were used to calibrate the molecular clock for evolution time estimation. Node labeled as ROOT (expanded red colored) corresponds to the reconstructed human common ancestor. The human haplotypes are divided into haplogroups 1 and 2 by a long branch with five non-synonymous substitutions. Dashed lines delineate two subgroups of haplogroup 1, which is separated by two non-synonymous substitutions. Phase 1 and phase 2 indicate two phases of hominid evolution after human–gorilla split. Non-synonymous substitutions only occurred at phase 2. Other nodes are labeled as indicated.
Two major IL37 haplogroups in modern humans segregated earlier during hominid evolution
To further confirm the early divergent time between human IL37 variants, we took advantage of the newly published archaic human sequences, both the Neanderthals and Denisova. In agreement with the early origin of two IL37 haplogroups, we found that the two archaic human IL37 sequences exactly belong to haplogroup 1 and 2, respectively (Supplementary Figure S3 and Figure 4). These data strongly suggest that the human IL37 variants originated from a common ancestor very earlier during hominid evolution and passed down to all the different human lineages ever since.
Discussion
Human IL37, a unique member of IL-1 family cytokines, exerts anti-inflammatory rather than proinflammatory functions. This characteristic endows IL37 as one of the few known inhibitory cytokines that balance immune reactions.2 Although its biological function is not elucidated completely, many studies indicate a protective role of IL37 on immunopathogenesis through reducing anti-inflammatory cytokines from innate immune cells.22 Here to better understand this novel cytokine, we investigated its variation patterns among different populations using evolutionary approaches and made a few novel findings: (1) IL37 variant proteins different from reference sequence are common among various human populations; (2) evolutionary genetic analysis suggests that human IL37 variants have evolved by selection and deviate from neutrality and (3) human IL37 variants consist of two major haplogroups which segregated anciently and were maintained in ancient hominid lineages leading to Homo sapiens. The identification of common IL37 variant proteins has two important implications for future research. First, a major IL37 variant (Var2) differs from reference sequence at five non-synonymous sites all located in the core region forming the characteristic IL-1 β-Trefoil structure, likely leading to a different function. Therefore, we propose that the biological functions of IL37 variants need to be verified and compared with earlier experimental results using NCBI reference protein.4, 8, 22 Second, our data provides valuable guidance on picking SNPs for disease association studies. Previous reports suggested that IL37 was involved in the development of various inflammatory conditions, including ankylosing spondylitis and rheumatoid arthritis.23, 24, 25 However, all these studies were based on genotyping rs3811047:A>G, which result in c.124A>G (p.(Thr42Ala)) substitution and does not differentiate between major IL37 variants identified in this study. On the basis of our data, we recommend genotyping any one of the five non-synonymous SNPs, rs2708943:C>G, rs2723183:A>G, rs2723187:C>T, rs2708947:C>T or rs2723192:A>G, to differentiate between IL37 haplogroups 1 and 2, which differ at least at five non-synonymous sites and probably result in different susceptibility to human diseases.
Interestingly, IL37 gene was lost owing to deletion in Chimpanzee during evolution while conserved in other great apes and modern humans.15 The complete loss of IL37 in Chimps without pathological consequences indicates a nonessential role of IL37 in overall fitness of Chimpanzee. However, its function in other species or diseased settings should not be excluded concomitantly. As recent data suggested, there are human disease causing loci but rather fixed in gorillas with normal phenotype.26 Indeed, our data supports that IL37 has an important role in immune regulation in humans and other primates. First, the dN/dS ratios in gibbon, orangutan and gorilla all indicate a purifying selection suggesting a longstanding critical role of IL37 along with primate evolution. Second, the dN/dS ratios of human IL37 variants and MK test all show that human IL37 variations deviate from neutrality. Third, we noticed that the five non-synonymous sites defining the IL37-Var2 all congregate in the predicted functional region of the protein, consistent with that human IL37 variation is shaped by selection and not neutral changes. During modern human evolution, pathogens constantly represent as an important selective force and therefore the adaptation of the immune system has been influenced by these pressures. Many studies have revealed that immune genes were indeed enriched for adaptive selection.9, 10 On the basis of the fact that human IL37 genes consist of two clearly separated haplogroups, we postulated that this could be formed by balancing selection when earlier hominid faced new challenges, such as novel infectious agents or inflammatory diseases. Although it is still hard to predict the true selective forces and the relation between genotype and phenotype, we are attempted to think a counterbalance between the beneficial and deleterious effects of different variants leading to IL37 selection, as this mechanism has been recently suggested to be involved in shaping the diversity of many other innate immune genes.27, 28, 29
We believed that our genealogical analysis of IL37 variants revealed interesting fact about the genome and human evolution. We calculated the time to MRCA (tMRCA) of human IL37 and certainly it is an outlier in the human genome with 3.6 million years compared with the reported average 800 000 years.30 This unusually long tMRCA supports the proposed balancing selection of human IL37 gene. However, this could also be regarded as a relic of an ancient admixture event in Africa, which resulted in a deep divergence of the two IL37 haplogroups, in agreement with the genetic communications among African archaic humans that gains more support recently.31, 32 From the MJ network analysis, we immediately noticed the two phases of IL37 evolution divided by a branching point. The first phase is characterized with no amino-acid changing substitutions after the split of gorilla and human ancestor, indicating a purifying selection phase and probably reflecting a stable environment during this stage of hominid evolution. The second phase is characterized by the accelerated non-synonymous changes with two haplogroups segregated eventually. The two-phase scenario is similar to the evolutionary selection patterns revealed by dN/dS ratios that support accelerated accumulation of non-synonymous substitutions during late phase of human evolution. We affirmed that these two haplogroups were maintained in human ancestors over million years as extinct Neanderthals and Denisova also harbored the corresponding IL37 variants inherited from ancient ancestors. We speculate that coexistence of both the IL37 haplogroups could benefit the survival of our African ancestors and it will be interesting to test whether there is a heterozygous advantage when our ancestors facing new environment challenges.
Finally, we found that IL37 haplogroup 2 had almost been completely lost in East ASN and the subgroup 2 in haplogroup 1 was now the dominant version in Eurasians. Recent positive selection and local adaptation to specific environment can result in such changes. In agreement with this speculation, we have detected signals of decreased genetic diversity in ASN and relatively high Fst scores of many IL37 SNPs between ASN and AFR, including the five non-synonymous sites defining the two haplogroups, supporting the potential local adaptation in ASN. However, the complex demographic history in ASN, such as numerous migrations, bottleneck events and subsequent enhanced genetic drifts can easily result in loss of the minor haplogroup. This complexity prevented us to make a definite conclusion about the recent selection and local adaptation of IL37 at this point.
Acknowledgments
This study was supported by NSFC grant 81371823 (SZ). We thank Dr Wen Zhang for discussion and suggestions, and thank Dr Ruoyu Luo for the technique support.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Boraschi D, Lucchesi D, Hainzl S et al: IL-37: a new anti-inflammatory cytokine of the IL-1 family. Eur Cytokine Netw 2011; 22: 127–147. [DOI] [PubMed] [Google Scholar]
- Banchereau J, Pascual V, O'Garra A: From IL-2 to IL-37: the expanding spectrum of anti-inflammatory cytokines. Nat Immunol 2012; 13: 925–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garlanda C, Dinarello CA, Mantovani A: The interleukin-1 family: back to the future. Immunity 2013; 39: 1003–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulau AM, Nold MF, Li S et al: Role of caspase-1 in nuclear translocation of IL-37, release of the cytokine, and IL-37 inhibition of innate immune responses. Proc Natl Acad Sci USA 2014; 111: 2650–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Risser P, Mao W et al: IL-1H, an interleukin 1-related protein that binds IL-18 receptor/IL-1Rrp. Cytokine 2001; 13: 1–7. [DOI] [PubMed] [Google Scholar]
- Dinarello CA: Overview of the interleukin-1 family of ligands and receptors. Semin Immunol 2013; 25: 389–393. [DOI] [PubMed] [Google Scholar]
- McNamee EN, Masterson JC, Jedlicka P et al: Interleukin 37 expression protects mice from colitis. Proc Natl Acad Sci USA 2011; 108: 16711–16716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nold MF, Nold-Petry CA, Zepp JA, Palmer BE, Bufler P, Dinarello CA: IL-37 is a fundamental inhibitor of innate immunity. Nat Immunol 2010; 11: 1014–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manry J, Quintana-Murci L: A genome-wide perspective of human diversity and its implications in infectious disease. Cold Spring Harb Perspect Med 2013; 3: a12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L, Clark AG: Population genetic tools for dissecting innate immunity in humans. Nat Rev Immunol 2013; 13: 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova JL, Abel L, Quintana-Murci L: Immunology taught by human genetics. Cold Spring Harb Symp Quant Biol 2013;78:157–172. [DOI] [PubMed] [Google Scholar]
- Quach H, Wilson D, Laval G et al: Different selective pressures shape the evolution of Toll-like receptors in human and African great ape populations. Hum Mol Genet 2013; 22: 4829–4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasseur E, Boniotto M, Patin E et al: The evolutionary landscape of cytosolic microbial sensors in humans. Am J Hum Genet 2012; 91: 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manry J, Laval G, Patin E et al: Evolutionary genetic dissection of human interferons. J Exp Med 2011; 208: 2747–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman TL, Tuzun E, Morrison VA et al: A genome-wide survey of structural variation between human and chimpanzee. Genome Res 2005; 15: 1344–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Auton A, Brooks LD et al: An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H: ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010; 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P: PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 2006; 34: W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Dopazo J, Gabaldon T: ETE: a python Environment for Tree Exploration. BMC Bioinformatics 2010; 11: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z: PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 2007; 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Rohl A: Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 1999; 16: 37–48. [DOI] [PubMed] [Google Scholar]
- Teng X, Hu Z, Wei X et al: IL-37 ameliorates the inflammatory process in psoriasis by suppressing proinflammatory cytokine production. J Immunol 2014; 192: 1815–1823. [DOI] [PubMed] [Google Scholar]
- Pei B, Xu S, Liu T, Pan F, Xu J, Ding C: Associations of the IL-1F7 gene polymorphisms with rheumatoid arthritis in Chinese Han population. Int J Immunogenet 2013; 40: 199–203. [DOI] [PubMed] [Google Scholar]
- Ge R, Pan F, Liao F et al: Analysis on the interaction between IL-1F7 gene and environmental factors on patients with ankylosing spondylitis: a case-only study. Mol Biol Rep 2011; 38: 2281–2284. [DOI] [PubMed] [Google Scholar]
- Pan F, Liao F, Xia G et al: Association of IL-1F7 gene with susceptibility to human leukocyte antigen-B27 positive ankylosing spondylitis in Han Chinese population. Clin Chim Acta 2010; 411: 124–126. [DOI] [PubMed] [Google Scholar]
- Scally A, Dutheil JY, Hillier LW et al: Insights into hominid evolution from the gorilla genome sequence. Nature 2012; 483: 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casals F, Sikora M, Laayouni H et al: Genetic adaptation of the antibacterial human innate immunity network. BMC Evol Biol 2011; 11: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M, Pozzoli U, Cagliani R et al: Parasites represent a major selective force for interleukin genes and shape the genetic predisposition to autoimmune conditions. J Exp Med 2009; 206: 1395–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Admetlla A, Bosch E, Sikora M et al: Balancing selection is the main force shaping the evolution of innate immunity genes. J Immunol 2008; 181: 1315–1322. [DOI] [PubMed] [Google Scholar]
- Garrigan D, Hammer MF: Reconstructing human origins in the genomic era. Nat Rev Genet 2006; 7: 669–680. [DOI] [PubMed] [Google Scholar]
- Lachance J, Vernot B, Elbers CC et al: Evolutionary history and adaptation from high-coverage whole-genome sequences of diverse African hunter-gatherers. Cell 2012; 150: 457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer MF, Woerner AE, Mendez FL, Watkins JC, Wall JD: Genetic evidence for archaic admixture in Africa. Proc Natl Acad Sci USA 2011; 108: 15123–15128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.