Abstract
We have developed an optofluidic analysis system that processes biomolecular samples starting from whole blood and then analyzes and identifies multiple targets on a silicon-based molecular detection platform. We demonstrate blood filtration, sample extraction, target enrichment, and fluorescent labeling using programmable microfluidic circuits. We detect and identify multiple targets using a spectral multiplexing technique based on wavelength-dependent multi-spot excitation on an antiresonant reflecting optical waveguide chip. Specifically, we extract two types of melanoma biomarkers, mutated cell-free nucleic acids —BRAFV600E and NRAS, from whole blood. We detect and identify these two targets simultaneously using the spectral multiplexing approach with up to a 96% success rate. These results point the way toward a full front-to-back chip-based optofluidic compact system for high-performance analysis of complex biological samples.
I. INTRODUCTION
Cancers rank second among the leading causes of death in the United States, causing tremendous suffering and creating ever-increasing costs for diagnosis, disease monitoring, and treatment. Recently, molecular biomarkers have gained increased attention as part of the growing field of personalized and precision medicine which relies on genomic analysis for cancer diagnosis and treatment.1–3 Among these genomic biomarkers, cell-free nucleic acids (CNA) that are circulating in the blood stream are particularly promising as they occur at substantially elevated concentrations in blood (plasma and serum) and other bodily fluids.4,5 Moreover, higher concentrations of methylated CNA presented in the blood of cancer patients compared to healthy individuals make CNA detection in blood (“liquid biopsy”) a reliable diagnostic approach.6–8 For example, elevated concentrations of both total CNA and BRAFV600E mutation CNA were verified as a biomarker for patients diagnosed with melanoma comparing to controls.7 Mutated CNA has been identified in advanced lung cancer patients through plasma droplet digital PCR and compared with biopsy for tissue genotyping.8 This blood-based technique can also alleviate patient stress and pain associated with (repeat) biopsies. However, the current liquid biopsy approaches usually include state-of-the-art bench-top equipment, such as complex PCR machines, to provide sufficient sensitivity, specificity, and accuracy for the low concentration of CNA on the order of few ng/ml.7
Lab-on-chip platforms are ideal candidates for CNA testing as they require only very small sample volumes, thus minimizing testing costs and complexity, which is important for extending this technique to less developed regions. Several promising chip-based approaches to CNA detection have already been reported. A microfluidic single molecule spectroscopy system has been demonstrated by combining microfluidic chips with confocal spectroscopy, in order to detect and size CNA from blood serum.9 Another method relies on surface-enhanced Raman scattering, where silver nanoscale hexagonal columns are generated on a surface of a phosphor to detect CNAs from serum.10 Magnetoresistive sensors for detecting CNA on a biochip with immobilized DNA probes were also demonstrated.11 However, standard blood serum extraction must be performed before the detection which includes more bench-top equipment. In order to isolate CNA from blood or plasma on-chip, a dielectrophoretic isolation method to extract CNA followed by imaging readout from a fluorescence microscope was developed.12 However, PCR analysis is necessary in the identification of the CNA and the detection microscope is not portable.
In order to provide sufficient sensitivity, specificity, and accuracy, while minimizing testing costs and complexity, optofluidic devices that combine both microfluidic handling and optical analysis of a biological sample in a single system have been developed recently.13,14 Specifically, hybrid integration approaches in which sample preparation and sensing tasks are carried out in separately designed and optimized chips with direct, amplification-free detection of the target nucleic acids have emerged.13,15 Therefore, these approaches have the potential to eliminate the complexities of benchtop sample preparation steps and the amplification-based PCR process. Here, we report the use of such a hybrid optofluidic system for chip-based detection of multiple CNA cancer biomarkers starting from whole blood. Specifically, two melanoma biomarkers based on single-point DNA mutations, BRAFV600E and NRAS codon 61,7,16 were detected and discriminated. Sample preparation steps were carried out on polydimethylsiloxane (PDMS)-based microvalve-actuated chips and include blood filtration, sample extraction, target enrichment, and fluorescent labeling. During the sample extraction process, a novel two-step solid phase extraction was applied for isolation and purification of DNA from the protein-rich environment. Optical analysis of the biomarkers was implemented in a silicon-based liquid-core waveguide chip.17 A multi-mode interference (MMI) waveguide was used to create multi-spot excitation patterns in a fluidic channel where the number of spots depends on the color of the excitation light. This enables spectrally multiplexed discrimination of molecular-beacon labeled BRAF and NRAS biomarkers with excellent fidelity. This detection scheme requires only a single photodetector for multiple fluorescent objects, which minimizes the assay complexity and further reduces the footprint of the future portable chip-based analysis system. This work represents the first time that DNA biomarkers are analyzed entirely with optofluidic components starting with whole blood and constitutes a major step to complete and miniaturized front-to-back chip-based biomarker analysis.
II. MATERIALS AND METHODS
A. Principle
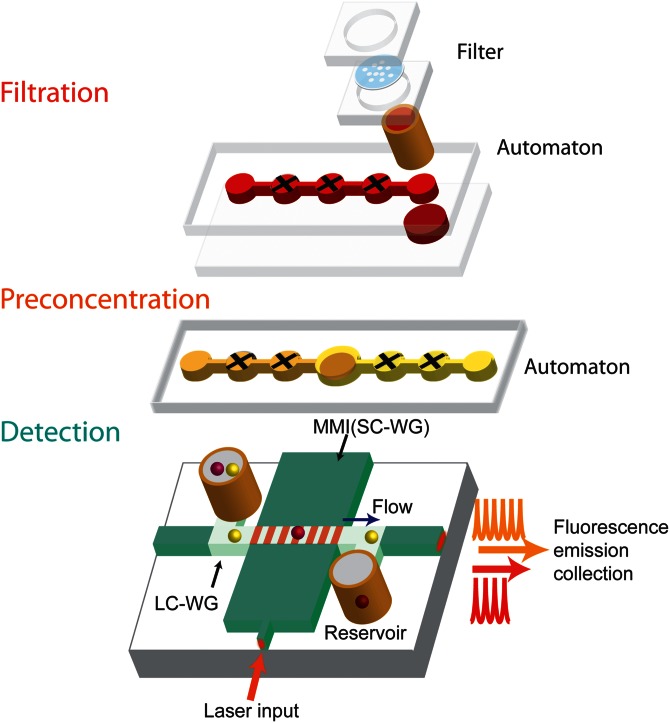
Figure 1 shows the schematic of the elements of our hybrid optofluidic system. The purpose of the microfluidic layers (upper part) is to prepare small volumes of raw sample material (here blood) for optically multiplexed detection of biomarkers. Here, sample preparation includes filtration, solid-phase extraction, and fluorescent labeling of nucleic acid targets. To implement these functions, we used an interconnected array of programmable, lifting-gate micro-valves (Automaton) as an easy and precise method to control small volumes of liquid on a chip.18,19 By implementing a sized-based particle filter on an automaton platform (filtration Automaton), the microfluidic devices are able to process whole blood and extract DNA from it. A second layer (preconcentration automaton) enables detection of low concentrations of DNA by preconcentrating and labeling the nucleic acids.
FIG. 1.
Schematic of hybrid optofluidic system. Filtration and preconcentration of PDMS chips utilize interconnected, cross-shaped lifting-gate microvalves (Automaton). The silicon-based ARROW chip for multiplex detection features interconnected solid-core (dark green) and liquid-core (light green) optical waveguides for in-flow detection of particles flowing through the liquid-core channel.
For the optofluidic layer used for multiplexed optical detection (lower part in Fig. 1), we adopted an antiresonant reflecting optical waveguide (ARROW) platform using orthogonal excitation and collection in the chip plane.20 Fiber coupled laser light was butt coupled to an ARROW solid-core excitation waveguide (SC-WG, dark green) using single mode fiber. Fluorescent targets are guided by pressure-based flow in the liquid-core waveguides (LC-WG, light green) connected with two reservoirs. When fluorescent targets pass through the excitation volume defined by the optical modes at the intersection between SC-WG and LC-WG, the generated fluorescence signal is guided inside the LC-WG and propagates orthogonally to the collection SC-WG.22 The light is collected from the SC-WG and analysed by an avalanche photodiode (APD).
Spectrally multiplexed target detection is implemented with a multi-spot excitation technique on the ARROW chip. Multiple excitation spots are generated using a multi-mode interference waveguide (MMI) section that supports multiple lateral modes.21 When an MMI is excited by a single optical mode from a narrow input waveguide (see Fig. 1), all of the MMI modes inside propagate at different phase velocities. At certain lengths L, well defined patterns composed of N distinct spots are formed along the MMI. This multi-spot pattern is illustrated by the seven red lines across the liquid channel in Fig. 1. The product of the spot number N and the wavelength λ is a constant determined by the MMI parameters length (L), effective width (w), and core refractive index (nc) and given by
| (1) |
This allows us to design an MMI section that intersects the fluidic channel, where multiple excitation spots are generated. As a fluorescent target flows past this excitation region, multi-peak signals in the time domain are produced and recorded. Since Nλ is a constant, we can generate different spot numbers at various laser wavelengths.21 As a result, we can identify different dye-labeled targets by their fluorescence pattern when they pass though the “patterned” excitation volume. This approach has recently been used to detect and identify whole influenza virus particles in buffer solution.17 Here, we apply it for the first time to the detection of nucleic acid biomarkers from whole blood.
B. Device design and parameters
Details of the fabrication procedures for each of the three chips are presented in the supplementary material. Here, we provide a brief overview of each.
1. Filtration automaton
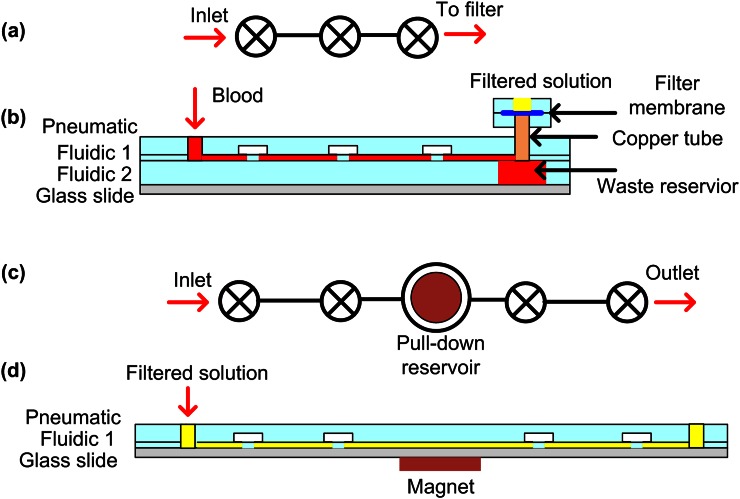
Figures 2(a) and 2(b) show the schematic of the filtration chip. It is composed of two components: a blood filter and the Automaton pumping layer. The blood filter layer consists of a pair of identical circular chambers 8 mm in diameter and 160 μm in height. The filtering membrane in between the chambers is a Whatman nuclepore polycarbonate track-etched membrane (PTEM) with pore sizes of 0.8 μm in diameter (Fisher Scientific).
FIG. 2.
Schematic of (a) top view of the aligned pneumatic layer and fluidic 1 of the filtration Automaton; (b) cross-sectional view of the filtration Automaton; (c) top view of the aligned pneumatic layer and fluidic 1 of the preconcentration Automaton; and (d) cross-sectional view of the preconcentration Automaton. For fabrication details see supplemental material.
The Automaton pumping layer consists of an array of individually addressable lifting-gate microvalves and pumps, as shown in Fig. 2(a).14,18,19 It is composed of four layers: a pneumatic layer, fluidic layer 1, fluidic layer 2, and a glass substrate, as shown in Fig. 2(b). The pneumatic layer has three individual circular chambers with a diameter of ∼3.7 mm and a height of ∼160μm. Fluidic layer 1 is a deformable membrane of 300 μm thick, containing fluidic channel patterns with a channel depth of ∼160 μm. Figure 2(a) shows the top-view of the aligned fluidic layer 1 and pneumatic layer. Fluidic layer 2 has a 6 mm × 4 mm reservoir aligned with the outlet of fluidic layer 1. The glass substrate is used to seal the Automaton. In order to have the flexibility of exchanging the blood filters when needed, a 3 mm-diameter copper tube was used to connect the Automaton and the blood filters instead of permanent bonding.
We designed a linear array of three microvalves serving as a directional pump to pass whole blood to the outlet without backflow.14 Once the whole blood reaches the outlet, it settles at the bottom of the waste reservoir at fluidic layer 2 due to gravity and slowly fills until it reaches the filtration chip. During the process, heavier cells and platelets sediment out at the bottom (fluidic layer 2), which minimizes filter clogging.
2. Preconcentration automaton
Figures 2(c) and 2(d) show the schematic of the second microfluidic sample processing chip - a preconcentration Automaton device. It has three layers: a pneumatic layer, fluidic layer 1, and a glass substrate. The device dimension is analogous to the Automaton pumping layer (filtration automaton, see supplementary material).
We designed the preconcentration Automaton to have four microvalves and one pull-down reservoir in the middle, as shown in Fig. 2(c). During the preconcentration process, a mixed solution (filtered blood and magnetic beads) was pumped from the inlet to the pull-down reservoir and held for a certain period before being pumped to the outlet. A magnet was attached below the pull-down reservoir in order to accumulate the magnetic beads from the solution to the bottom of the reservoir. The pull-down reservoir is designed to have a larger diameter (∼5 mm) than a single microvalve to accommodate a large number of accumulated beads. The magnet diameter is chosen to be slightly smaller (∼3 mm) than the pull-down reservoir to avoid magnetic beads attachment to the rough side-wall.
3. ARROW chip
The ARROW chip is defined on silicon and has an over-all dimension of ∼10 mm × 12 mm. Details for definition of the waveguiding layers and creation of the hollow channel are given in the supplementary material. The MMI SC-WG was 75 μm wide and 1.9 mm long and was interfaced to a 4 μm wide single mode input waveguide. The LC-WG cross-section was 12 μm × 5 μm. The collection SC-WG was 12 μm wide aligned with LC-WG. Two copper reservoirs were attached to the LC-WG inlets.
C. DNA and magnetic beads parameters
1. Target DNA and molecular beacons
100-bp synthetic BRAF and NRAS single-stranded DNA targets were designed according to the BRAFV600E mutation and Codon 61 mutation reported in melanoma studies.7,16 Molecular beacons fluorescing under excitation with 633 nm and 750 nm light were designed to be reverse-complimentary to the BRAF and NRAS mutation regions, respectively.
2. Pull-down DNA
We designed biotinylated pull-down oligos I and II for both BRAF and NRAS DNA extraction. Shorter pull-down oligos I were designed for a relatively low melting temperature of ∼50 to 55 °C during the release step of the two-step solid phase extraction. This reduced the probability of pull-down oligos falling off from the magnetic beads during the heating period due to unstable bonds. Longer pull-down oligos II with higher melting temperatures were designed to provide stronger binding to the target DNA. Table I summarizes the DNA and molecular beacon sequences for the solid-phase extraction and labelling.
TABLE I.
Summary of the DNA and molecular beacons sequences. Mutated DNA: underlined; Pull-down I region: bold; Pull-down II region: italic; and Molecular beacon region: gray shading.
| Name | Sequence (5′-3′) | Length |
|---|---|---|
| BRAF DNA | CTACACCTCAGATATATTTCTTCATGAAGACCTCACAGTAAAAATAGGTGATTTTGGTCTAGCTACAGAGAAATCTCGATGGAGTGGGTCCCATCAGTTT | 100 |
| BRAF pull-down I | AGATTTCTCTGTAG | 14 |
| BRAF pull-down II | CACCTATTTTTACTGTGAGGTCTTCATGAAGAAATATATCTGAGG | 45 |
| BRAF beacon | TYE665dye-cgcgcaACCCACTCCATCGAGATTTCTCTGTAGCTAGACCAtgcgcg-Quencher | 35/6stem |
| NRAS DNA | CTTACAGAAAACAAGTGGTTATAGATGGTGAAACCTGTTTGTTGGACATACTGGATACAGCTGGACGAGAAGAGTACAGTGCCATGAGAGACCAATACAT | 100 |
| NRAS pull-down I | TCTTCTCGTCCA | 12 |
| NRAS pull-down II | TATGTCCAACAAACAGGTTTCACCATCTATAACCACTTGTTTTCT | 45 |
| NRAS beacon | IRdye750-ggcggTCTTCTCGTCCAGccgcc-IRQuencher | 13/5stem |
3. Magnetic beads
We chose 1μm-sized streptavidin coated magnetic beads (Invitrogen) to immobilize the various biotinylated pull-down oligos. We prepared four sets of magnetic beads with various pull-down oligos at a stock concentration of ∼1 × 1010/ml following the suggested protocol.23
III. EXPERIMENT
A. Sample preparation
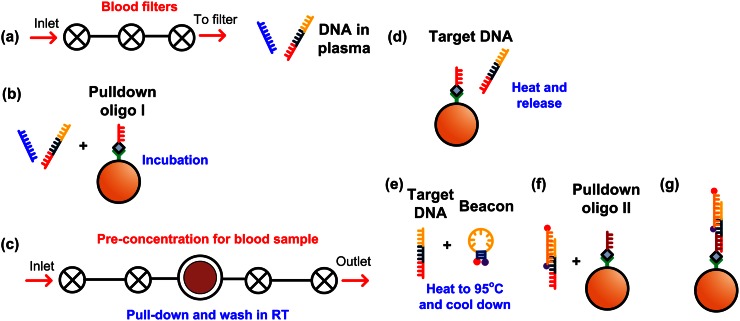
Samples for multiplex detection of cancer biomarkers were prepared by spiking 300 nM mutated BRAF and NRAS synthetic DNA into ∼200 μl of mouse whole blood (Biochemed service). The whole blood was first diluted 1:1 with phosphate-buffered saline (PBS) before processing. Figure 3 schematically illustrates the sample preparation process. The Automaton chips were sequentially used for whole blood filtering and preconcentrating magnetic beads. Prior to use, the Automaton chips were filled with 10 mg/ml bovine serum albumin (BSA) and incubated for 15 min in order to minimize nonspecific adsorption to the channel walls. Approximately 200μl of filtered solution was collected from the top of the filtration Automaton, as shown in Fig. 3(a). The first-step solid phase extraction (Fig. 3(b)) was performed before the preconcentration step (Fig. 3(c)). Due to the aggregation of the magnetic beads in protein-rich solution (plasma), a second (off-chip) solid phase extraction step was necessary (Figs. 3(d)–3(g)) to prevent the LC-WG of ARROW from clogging during detection. In the final step, the magnetic beads with cancer DNA biomarkers and fluorescent beacons attached (Fig. 3(g)) were resuspended in buffer solution (1× T50) at a final concentration of ∼5 × 106/ml for detection. Details of the preparation procedures are presented in the supplementary material.
FIG. 3.
Sample preparation process: (a) Whole blood was passed through the filtration automaton to obtain the plasma solution; (b) Incubating plasma solution and magnetic beads with pull-down oligo I; (c) The plasma-pull-down I beads solution was pre-concentrated and washed at room temperature (RT); (d) Target DNA was released through heating up to melting temperature; (e) Released target DNA was mixed with molecular beacon and heated up to 95 °C for 5 min and cooled down slowly; (f) Incubating the mixture with magnetic beads with pull-down oligo II; (g) Magnetic beads with target DNA labeled with molecular beacon.
B. Multiplex cancer biomarker optical detection
The magnetic beads labeled with different molecular beacons were put in a liquid-core ARROW platform for detection, as shown in Fig. 1. Here, 633 nm and 745 nm lasers were coupled simultaneously into the MMI single mode input waveguide. The beads holding attached target/beacon complexes were pipetted into the liquid-core ARROW channel through one of the reservoirs connecting the inlet of the channel. Negative pressure was applied to the other reservoir to enable constant flow inside the ARROW channel. As the beads passed the excitation region, the molecular beacons attached to the targets on the beads were excited; thus, fluorescence signal was generated. The fluorescent light is collected from the waveguide by a microscope objective, sent through a multiband-pass filter to remove scattered excitation light, and analyzed by an APD.
IV. RESULTS AND DISCUSSION
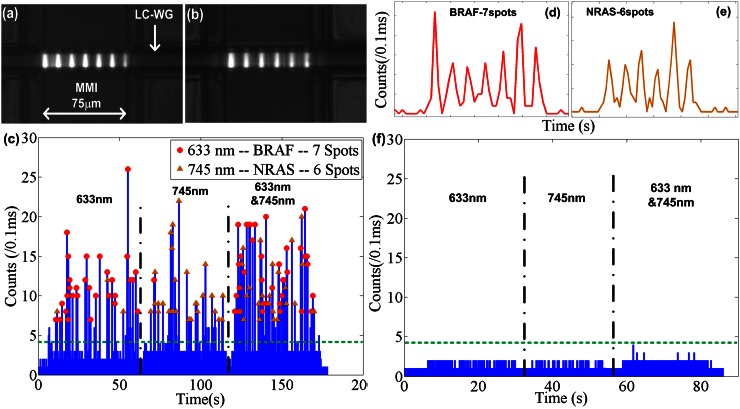
Figures 4(a) and 4(b) show microscopic images of the intersection region between MMI and the LC-WG filled with Alexa 633 and 755 dye. Upon excitation, 7 (Fig. 4(a)) and 6 (Fig. 4(b)) excitation spots are generated by the MMI in the intersecting fluidic channel by 633 nm and 745 nm laser, respectively. Figure 4(c) shows the recorded fluorescence signal from the magnetic beads. In order to ascertain proper detection at each wavelength, we first excited the MMI with only the 633 nm laser for ∼50s, and then switched to only the 745 nm laser for ∼50s. During each period, we detect a sequence of signals that have the correct number of peaks as indicated by the symbols on top of each signal. Then, we excited the MMI with both wavelengths simultaneously. The signal now consists of a mixture of multi-peak signals which were analysed as described below.
FIG. 4.
(a) and (b) Microscopy images of the MMI generated excitation pattern inside LC-WG with (a) 633 nm laser and (b) 745 nm laser. (c) Measured optical fluorescence signal at various excitation wavelengths: 633 nm (10–60s), 745 nm (65–115s), and 633 nm and 745 nm (120–180s). (d) Zoom-in view of beads with BRAF target producing 7 peaks. (e) Zoom-in view of beads with NRAS target producing 6 peaks. (f) Measured optical fluorescence signal for negative control at various excitation wavelengths: 633 nm (5–30s), 745 nm (32–55s), and 633 nm and 745 nm (60–85s).
First, we carried out a negative control experiment to demonstrate the specificity of the assay. We spiked ∼1μM mutated NRAS DNA into blood serum and mixed the solution with magnetic beads functionalized with BRAF pull-down oligos I, II and BRAF beacon. The same sample preparation process as for the positive sample was followed, and the final solution was pipetted into the same ARROW chip and optically analysed. As shown in Figure 4(f), we first excited the MMI with only the 633 nm laser for ∼25s, and then switched to only the 745 nm laser for ∼23s. Then, we excited the MMI with both wavelengths simultaneously for ∼30s. None of these periods resulted in any false positive signals. The result shows that the solid-phase extraction process is highly specific, and no notable non-specific binding between beacon and magnetic beads in the protein rich environment occurred.
We used the results of this negative control experiment to define the background threshold at 4 counts/0.1 ms (green dashed lines in Figs. 4(c) and 4(f)) for identification of a target bead in the positive sample. The highest signal to noise ratio is ∼6.7. From the single wavelength control experiment, we recorded ∼ 34 beads (from 10s-60s) with BRAF cancer DNA and 633 nm-excited beacon. Each bead generates 7 peaks, as shown in Fig. 4(d). We recorded ∼25 beads (from 65s to 115s) with NRAS cancer DNA and 750 nm-excited beacon. Each bead generates 6 peaks, as shown in Fig. 4(e). From the recorded peak spacing in the time domain, we can estimate that the flow velocity inside LC-WG is ∼1.5 cm/s.
For target identification for the positive sample, we counted the peak number for each bead and labeled the ones with 7 peaks as BRAF (red dots in Fig. 4(c)) and the ones with 6 peaks as NRAS (orange triangles in Fig. 4(c)). Note that no spectral separation of the fluorescence signal at the detector is necessary, as the spectral information is encoded in the time domain (spot number in the multi-peak pattern). According to the single wavelength controls, we successfully identified 31 out of 34 beads as BRAF with an accuracy of ∼91%, and 24 out of 25 as NRAS with an accuracy of ∼96%. Considering there is no overlap in the excitation profile between the 633 nm- and 750 nm-excited dyes, we believe that the incorrect identifications come from the non-uniform excitation profile inside the LC-WG and the positions of the beads such that one or more peaks are too small to be correctly detected. In the future, this accuracy can be further improved by better control of flow speed and particle position inside the liquid-core ARROW channel to create more uniform optical signals or by applying additional analysis algorithms that rely on the time spacing rather than the number of peaks.17
V. CONCLUSIONS
We have developed a hybrid optofluidic analysis system that processes and analyzes biomolecular samples starting from whole blood. Relevant to future blood-based diagnosis, we demonstrated detection and identification of the skin cancer melanoma DNA biomarkers (BRAF and NRAS) in a whole blood sample. Specifically, we extracted 300 nM CNA from whole blood using programmable microvalves integrated with filters, concentrated the CNAs using a microfluidic circuit, and detected the two biomarkers using a multi-spot excitation technique on an ARROW optofluidic platform with up to 96% success rate. Our optofluidic analysis system can be easily expanded into multiple types of DNA detection from whole blood through the nucleic acid-based, two-step solid phase extraction and utilizing more excitation colors or combinations thereof. In the future, we can further improve the sensitivity to a lower CNA concentration by optimizing the blood cell filters, reducing the CNA loss in the Automaton and improving the ARROW chip performance, to reach the state-of-the-art liquid biopsy standard of few ng/ml concentration range.
VI. SUPPLEMENTARY MATERIAL
The supplementary material includes a detailed description of the fabrication processes for all chips and the procedure for biological sample preparation.
ACKNOWLEDGMENTS
This work was supported by the NIH/NIAID and NSF under Grant Nos. 4R33AI100229, 1R01AI116989-01, CBET-1159453, and CBET-1159423 as well as the W. M. Keck Center for Nanoscale Optofluidics at the University of California at Santa Cruz. J. W. Parks was supported by the Eugene Cota–Robles Fellowship and an RMI Graduate Research Fellowship. We also thank Dr. T. Yuzvinsky for fruitful discussions.
References
- 1. Ludwig J. A. and Weinstein J. N., Nat. Rev. , 845 (2005). 10.1038/nrc1739 [DOI] [PubMed] [Google Scholar]
- 2. Ransohoff D. F., Nat. Rev. , 309 (2004). 10.1038/nrc1322 [DOI] [PubMed] [Google Scholar]
- 3. Jimeno A. and Hidalgo M., Mol. Cancer Ther. , 787 (2006). 10.1158/1535-7163.MCT-06-0005 [DOI] [PubMed] [Google Scholar]
- 4. Schwarzenbach H., Hoon D. S. B., and Pantel K., Nature Reviews Cancer , 426 (2011). 10.1038/nrc3066 [DOI] [PubMed] [Google Scholar]
- 5. Anker P., Mulcahy H., and Stroun M., Int. J. Cancer , 149 (2003). 10.1002/ijc.10791 [DOI] [PubMed] [Google Scholar]
- 6. Koyanagi K., Mori T., O'Day S. J., Martinez S. R., Wang H., and Hoon D. S. B., Cancer Res. , 6111 (2006). 10.1158/0008-5472.CAN-05-4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Salvianti F., Pinzani P., Verderio P., Ciniselli C. M., Massi D., Giorgi V. D., Grazzini M., Pazzagli M., and Orlando C., PLoS One , e49843 (2012). 10.1371/journal.pone.0049843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Williams P. M. and Conley B. A., JAMA Oncol. , 1003 (2016). 10.1001/jamaoncol.2016.0240 [DOI] [PubMed] [Google Scholar]
- 9. Liu K., Brock M. V., Shih I., and Wang T., J. Am. Chem. Soc. , 5793 (2010). 10.1021/ja100342q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ito H., Hasegawa K., Hasegawa Y., Nishimaki T., Hosomichi K., Kimura S., Ohba M., Yao H., Onimaru M., Inoue I., and Inoue H., Sci. Rep. , 10455 (2015). 10.1038/srep10455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dias T. M., Cardoso F. A., Martins S. A. M., Martins V. C., Cardoso S., Gaspar J. F., Monteiro G., and Freitas P. P., Anal. Methods , 119 (2016). 10.1039/C5AY01587A [DOI] [Google Scholar]
- 12. Sonnenberg A., Marciniak J. Y., Skowronski E. A., Manouchehri S., Rassenti L., Ghia E. M., Widhopf G. F. II, Kipps T. J., and Heller M. J., Electrophoresis , 1828 (2014). 10.1002/elps.201400016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cai H., Parks J. W., Wall T. A., Stott M. A., Stambaugh A., Alfson K., Griffiths A., Mathies R. A., Carrion R., Patterson J. L., Hawkins A. R., and Schmidt H., Sci. Rep. , 14494 (2015). 10.1038/srep14494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Parks J. W., Olson M. A., Kim J., Ozcelik D., H. Cai, Jr. , Carrion R., Patterson J. L., Mathies R. A., Hawkins A. R., and Schmidt H., Biomicrofluidics , 054111 (2014). 10.1063/1.4897226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Parks J. W., Cai H., Zempoaltecatl L., Yuzvinsky T. D., Leake K., Hawkins A. R., and Schmidt H., Lab Chip , 4118 (2013). 10.1039/c3lc50818h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Omholt K., Platz A., Kanter L., Ringborg U., and Hansson J., Clin. Cancer Res. , 6483 (2003), available at http://clincancerres.aacrjournals.org/content/9/17/6483.long. [PubMed] [Google Scholar]
- 17. Ozcelik D., Parks J. W., Wall T. A., Stott M. A., Cai H., Parks J. W., Hawkins A. R., and Schmidt H., Proc. Natl. Acad. Sci. , 12933 (2015). 10.1073/pnas.1511921112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jensen E. C., Bhat B. P., and Mathies R. A., Lab Chip , 685 (2010). 10.1039/B920124F [DOI] [PubMed] [Google Scholar]
- 19. Kim J., Kang M., Jensen E. C., and Mathies R. A., Anal. Chem. , 2067 (2012). 10.1021/ac202934x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yin D., Deamer D. W., Schmidt H., Barber J. P., and Hawkins A. R., Appl. Phys. Lett. , 3477 (2004). 10.1063/1.1807966 [DOI] [Google Scholar]
- 21. Soldano L. B. and Pennings E. C. M., IEEE J Lightwave Technol. , 615 (1995). 10.1109/50.372474 [DOI] [Google Scholar]
- 22. Zhao Y., Leake K., Measor P., Jenkins M., Keeley J., Schmidt H., and Hawkins A. R., IEEE Photonics Technol. Lett. , 46 (2012). 10.1109/LPT.2011.2172407 [DOI] [Google Scholar]
- 23.See https://www.thermofisher.com/order/catalog/product/65601 for information about Dynabeads MyOne Streptavidin T1 manual and protocol.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The supplementary material includes a detailed description of the fabrication processes for all chips and the procedure for biological sample preparation.