Abstract
Glioblastoma (GBM) is an invariably fatal brain tumor in which a small subpopulation of self-renewable glioma stem cells (GSCs) contributes to tumor propagation and relapse. Targeting GSCs could therefore have a significant clinical impact for GBM. Telomestatin is a naturally-occurring compound that preferentially impairs GSC growth by perturbing transcription and inducing a DNA damage response. Telomestatin stabilizes G-quadruplexes (G4s), which are guanine-rich four-strand nucleic acid structures observed in vitro and in vivo. However, the mechanism underlying the GSC-selective nature of the DNA damage response remains unknown. Here we demonstrate that GSCs are more susceptible to telomestatin-induced telomere dysfunction and replication stress when compared with GSC-derived non-stem glioma cells (NSGCs). Telomestatin induced dissociation of the telomere-capping protein TRF2 from telomeres, leading to telomeric DNA damage in GSCs–but not in NSGCs. BIBR1532, a telomerase catalytic inhibitor, did not preferentially inhibit GSC growth, suggesting that telomestatin promotes telomere dysfunction in a telomerase-independent manner. GSCs and NSGCs had comparable levels of G4s in their nuclei, and both responded to telomestatin with phosphorylation of RPA2 at Ser33–a hallmark of replication stress. However, activation of the checkpoint kinase Chk1, induction of a DNA damage response, and subsequent growth inhibition occurred only in telomestatin-treated GSCs. These observations suggest that telomestatin impairs GSC growth through removal of TRF2 from telomeres and potent activation of the replication stress response pathway. Therefore, a novel G4-directed therapeutic strategy could specifically target cancer stem cells in GBM.
Keywords: DNA damage response, Glioma stem cells, G-quadruplex, Replication stress, Telomere
1. Introduction
Glioblastoma (GBM) is the most common type of primary brain tumor in adults. Despite standard therapies comprising surgical resection followed by radiotherapy and temozolomide-based chemotherapy, GBM patients survive for only 12–15 months on average [1–3]. Recent studies have revealed small subpopulations of glioma stem cells (GSCs) in GBM lesions, and these cells contribute to tumor propagation and relapse [4,5]. GSCs express stem cell markers—such as CD133, SOX2, and nestin–and can self-renew and establish tumors in vivo [6,7]. Moreover, GSCs contribute to resistance to radiotherapy and chemotherapy [4,5]. Therefore, GSCs have been suggested as a novel therapeutic target for GBM.
Some guanine-rich nucleic acid sequences can form four-stranded DNA structures called G-quadruplexes (G4s) [8,9]. G4 sequence motifs are present in telomeric DNA [10,11] and in gene regulatory regions, such those of proto-oncogenes [12–15]. Telomestatin is a naturally-occurring compound isolated from Streptomyces anulatus 3533-SV4 [16] and is referred to as G4 ligand because it potently stabilizes the G4 structures [17,18]. We have previously reported that telomestatin downregulates the proto-oncogene c-Myb, which is expressed strongly in GSCs but is absent from CD133-negative non-stem glioma cells (NSGCs). Therefore, telomestatin preferentially impairs the growth of GSCs compared with NSGCs [19]. Meanwhile, the telomestatin-induced DNA damage response occurs in GSCs but not in NSGCs [19]. While this DNA damage response would explain, at least in part, the preferential deleterious effect of telomestatin on GSCs, why telomestatin induces a DNA damage response only in GSCs remains unknown.
Here we investigated the differential responses of GSCs and NSGCs to telomestatin. GSCs were more sensitive than NSGCs to drug-induced telomere dysfunction and replication stress, which induced a DNA damage response.
2. Materials and methods
2.1. Cell culture, growth assay, and flow cytometry
GBM146 and 157 GSCs were maintained in serum-free sphere medium, and NSGCs were obtained from these GSCs as previously described [19,20]. Cell proliferation was evaluated using a CellTiter-Glo Luminescent Cell Viability Assay (Promega, Fitchburg, WI, USA). Flow cytometry was performed as described [20].
2.2. Western blot analysis
Cell lysates were prepared and western blot analysis was performed as previously described [21] with the following primary antibodies: rabbit anti-SOX2 (3579S, 0.02 μg/mL; Cell Signaling Technology, Danvers, MA, USA), rabbit anti-nestin (MAB5326, 2 μg/mL; Millipore, Darmstadt, Germany), mouse anti-Chk1 (2360S, 0.1 μg/mL; Cell Signaling Technology), rabbit anti-Ser317-phospho-Chk1 (2344S, 0.2 μg/mL; Cell Signaling Technology), or mouse anti-β-actin (A5441, 1:5000; Sigma–Aldrich, St. Louis, MO, USA).
2.3. Immunofluorescence staining
Cells were grown on poly-l-lysine–treated coverslips (Matsunami Glass, Osaka, Japan) and fixed with 2% paraformaldehyde in phosphate-buffered saline (PBS), and then permeabilized with 0.5% Nonidet P-40/PBS. The fixed cells were blocked in PBS containing 1% bovine serum albumin and incubated with rabbit anti-53BP1 (4937S, 0.67 μg/mL; Cell Signaling Technology), mouse anti-TRF2 (NB100-56506, 5 μg/mL; Novus, Littleton, CO, USA), or rabbit anti-Ser33-phospho-RPA2 (NB100-544, 0.5 μg/mL; Novus). These antibodies were detected using the Alexa 488-conjugated anti-rabbit immunoglobulin (IgG) or Alexa 488-conjugated anti-mouse IgG (Thermo Fisher Scientific, Waltham, MA USA). DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI). To detect G4s, cells were fixed with 100% methanol for 10 min at −20 °C and dried. Fixed cells were blocked in PBS containing 1% BSA and incubated with FLAG-tagged anti-G4 (BG4, 0.1 μg/mL, kindly provided by Dr. Shankar Balasubramanian, University of Cambridge) [22], rabbit anti-FLAG (2368S, 0.13 μg/mL; Cell Signaling Technology), and Alexa 488-conjugated anti-rabbit IgG. For enzyme treatment, the fixed cells were incubated with 120 U of Turbo DNase (Thermo Fisher Scientific) for 60 min at 37 °C. The iFISH assay was performed as described [19]. To classify the cell cycle, cells were labeled with EdU, which was detected by immunofluorescence staining with the anti-EdU antibody (Thermo Fisher Scientific). The EdU-positive cells were classified as in S phase of the cell cycle. Cells were also counterstained with DAPI, and the EdU-negative cells were classified into G1 and G2/M, depending on the intensity and the area of DAPI signal. These analyses were performed with the IN Cell Analyzer 6000 (GE Healthcare, Little Chalfont, UK), which allowed us to evaluate the cell cycle-specific incidence of DNA damage foci in quantitative and high-throughput manners.
2.4. Enzyme-linked immunosorbent assay (ELISA)
Biotinylated oligonucleotides were annealed in 100 mM KCl or 100 mM NaCl by heating to 95 °C for 10 min, then cooled slowly to room temperature and stored at 4 °C before use. Avidin-coated plates (Sumitomo Bakelite, Tokyo, Japan) were then exposed to biotinylated oligonucleotides, followed by incubation with BG4. Samples were then incubated with a horseradish peroxidase (HRP)-conjugated anti-FLAG antibody (Abcam, Cambridge, UK) and the HRP substrate TMB (Roche, Mannheim, Germany), followed by measurement of absorbance at 450 nm. Circular dichroism spectra for confirmation of G-quadruplex formation were produced as described [23]. Sequences of oligonucleotides (oligos) used are as follows: KIT2 (G4 motif) 5′-CGGGCGGGCGCGAGGGAGGGG-3′; hTELO (G4 motif) 5′-GG(TTAGGG)x4TTAG-3′; and control DNA (non-G4) 5′-GGCATAGTGCGTGGGCG-3′.
3. Results
3.1. Telomestatin induces telomere dysfunction and inhibits GSC growth
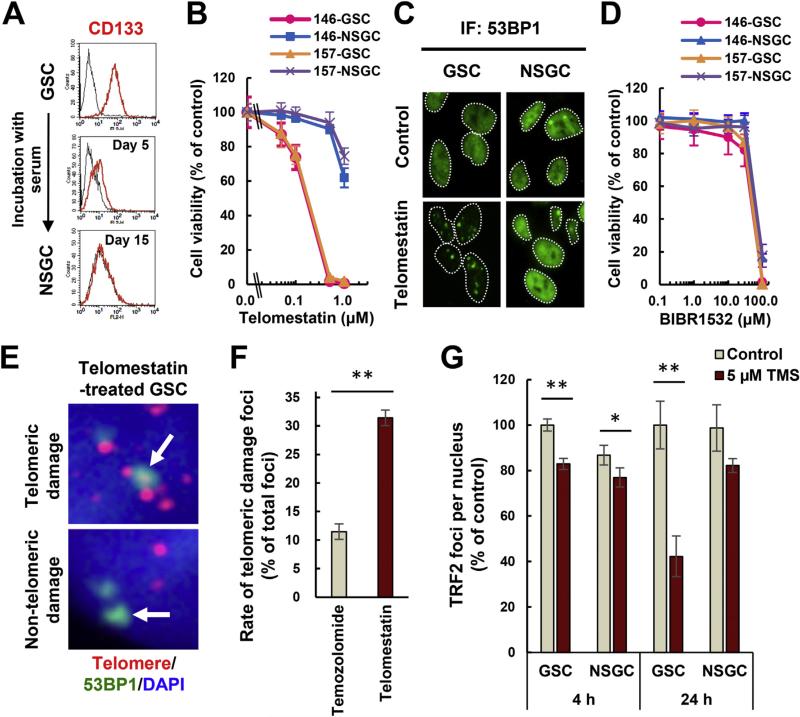
GBM146 and GBM157 are established GSC models which express the GSC markers CD133, SOX2, and Nestin under sphere culture conditions. Upon serum exposure, these cells underwent differentiation to NSGSs with reduced or absent GSC marker expression (Fig. 1A and Supplemental Fig. 1A). Consistent with our previous report [19], GSCs were susceptible to telomestatin-induced DNA damage and growth inhibition (IC50 values of approximately 0.2 μM) but became resistant upon differentiation to NSGCs (Fig. 1B, C and Supplemental Fig. 1B). While telomestatin inhibits telomerase activity [24], this mechanism of action does not explain the preferential effect on GSCs because BIBR1532—a telomerase inhibitor without G4-stabilizing activity [25]–had no preferential effect on GSC growth (Fig. 1D).
Fig. 1.
Telomestatin induces telomere dysfunction and inhibits GSC growth. (A) Measurement of CD133 levels in GBM146 cells by flow cytometry. Black histogram indicates normal immunoglobulin as negative control. (B) Effect of telomestatin on growth of GSCs and NSGCs. GBM146 and GBM157 cells were treated with telomestatin for 144 h. Error bar, standard deviation. (C) DNA damage foci induced by telomestatin in GSCs. GBM146 cells were treated with 5 μM telomestatin in serum-free medium for 24 h and subjected to immunofluorescence staining with anti-53BP1 antibody. (D) Effect of telomerase inhibitor BIBR1532 on growth of GSCs and NSGCs. GBM146 and GBM157 cells were treated with indicated concentrations of BIBR1532 for 144 h (E) iFISH assay. GSCs were treated with 1 μM telomestatin in serum-free medium for 96 h. Representative images of telomeric and non-telomeric DNA damage foci are shown (arrows in upper and lower panels, respectively). Red, telomere; green, 53BP1; blue, DAPI staining for nuclear DNA. (F) The rate of telomeric 53BP1 damage foci in telomestatin- or temozolomide-treated GSCs. GSCs were treated with 1 μM telomestatin or 10 μM temozolomide in serum-free medium for 96 h (these drug concentrations inhibited the cell growth to equivalent extents: data not shown). The rate of telomeric 53BP1 foci among all 53BP1 foci was calculated. (G) TRF2 immunofluorescence staining. GBM146 cells were treated with 5 μM telomestatin in serum-free medium for 4 or 24 h. The numbers of TRF2 foci per nucleus were counted and normalized against those in GSC/control cells. Statistical evaluations were performed using the Welch t-test. *, P < 0.05; **, P < 0.01. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The degree of telomeric DNA damage induced by telomestatin was higher than that induced by the alkylating agent temozolomide (Fig. 1E and F). Given that telomeric DNA can form G4, these results support that notion that telomestatin recognizes intracellular G4s and elicits DNA damage responses at those genomic loci. Telomestatin-treated GSCs had markedly reduced numbers of TRF2 foci compared with non-treated cells and NSGCs (Fig. 1G and Supplemental Fig. 1C: since TRF2 binds telomeres, it can be detected as nuclear foci). By using immunofluorescence staining and chromatin immunoprecipitation, we previously confirmed that telomestatin-induced reduction of the TRF2 immunofluorescence signal is equivalent to the drug-induced dissociation of TRF2 from telomeres [26]. Since TRF2 binds to and protects telomeres from DNA damage responses, these observations suggest that telomestatin induces telomeric DNA damage by promoting TRF2 dissociation from telomeres [26]. Additionally, the DNA-damaging agents temozolomide and camptothecin induced 53BP1 foci in NSGCs (Supplemental Fig. 1D and data not shown), excluding the possibility that resistance of NSGCs to telomestatin-induced DNA damage results from dormancy or dysfunction of the DNA damage response pathway in the cells.
3.2. Telomestatin-induced DNA damage depends on replication and transcription
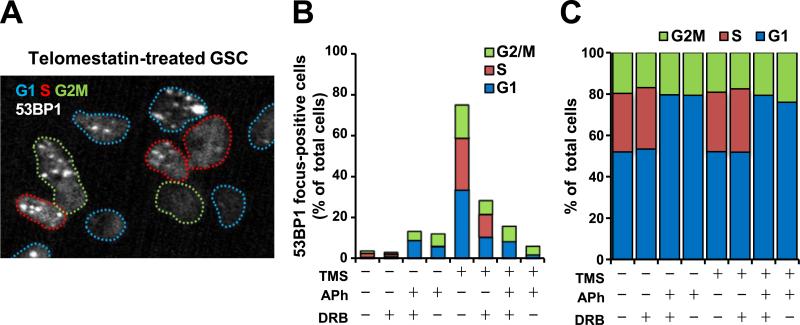
Accumulating evidence indicates that G4 structures also form at non-telomeric loci when these regions are single-stranded during DNA replication or transcription [12]. Therefore, such loci could be targets of telomestatin. We next examined whether telomestatin-induced DNA damage in GSCs occurred at particular stages of the cell cycle by monitoring 53BP1 foci in conjunction with EdU staining to detect S phase and DAPI staining to classify G1 and G2/M phases. Telomestatin induced DNA damage at each phase of the cell cycle (Fig. 2A and B). Aphidicolin and 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB), which inhibit DNA replication and transcription, respectively, prevented telomestatin-induced 53BP1 foci (Fig. 2B). Although DRB partially diminished the number of foci, aphidicolin almost completely abolished them, suggesting that aphidicolin had some leaky, dominant effect even in non-S phase cells. DRB did not affect the cell cycle distribution whereas aphidicolin completely abolished the population of EdU-positive cells (Fig. 2C). These data indicate that telomestatin induces a DNA damage response in GSCs through replication- and transcription-dependent mechanisms. The EdU-positive populations in GSCs and NSGCs were 14.0% of 6.3%, respectively (data not shown). This low abundance of EdU-positive cells in NSGCs may partially explain the decreased formation of 53BP1 foci induced by telomestatin in these cells.
Fig. 2.
Telomestatin-induced DNA damage is dependent on replication and transcription. (A) Cell cycle analysis of DNA damage foci in telomestatin-treated GSCs. Cells were treated with 5 μM telomestatin for 4 h and subjected to anti-53BP1 immunofluorescence staining. Cells in S phase were detected by EdU labeling. Colored dotted lines indicate the nuclei boundaries and the cell-cycle phase. A representative image depicting telomestatin-induced DNA damage in G1-, S-, and G2/M-phase cells is shown. Blue, G1; red, S; green, G2/M. (B) Effects of replication/transcription inhibition on telomestatin-induced DNA damage foci in GSCs. Cells were pretreated with 0.5 μM aphidicolin (APh, an inhibitor of DNA replication) for 2 h and/or 50 μM 6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB, an inhibitor of transcription) for 1 h before treatment with 5 μM telomestatin for 4 h. Cells were classified as focus-positive or -negative according to the numbers of punctate nuclear 53BP1 foci (n > 2). (C) Cell cycle distribution as in B. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. G4s exist equivalently in GSCs and NSGCs
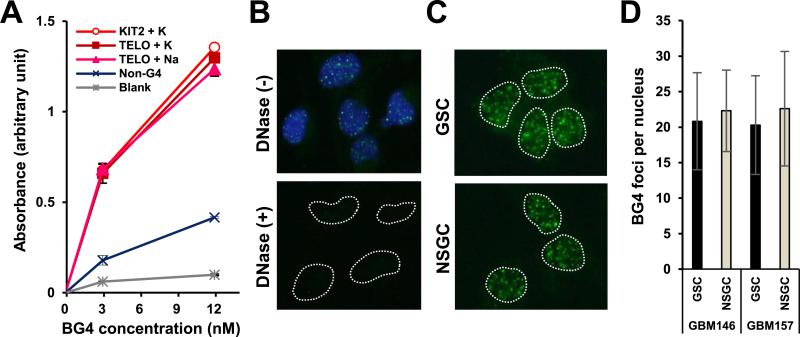
We next investigated the intracellular distribution of G4 in GSCs and NSGCs using the G4-specific antibody BG4 [22]. ELISA was used to confirm the specificity of BG4 for G4. Incubation of the KIT2 oligo in KCl, and the telomeric oligo (hTELO) in NaCl and in KCl formed G4s of parallel propeller, anti-parallel propeller, and mixed parallel–anti-parallel propeller structures, respectively (Supplemental Fig. 2). BG4 exhibited higher affinity for these G4-forming oligos compared with the non-G4-forming control oligo, and this affinity was comparable between different G4 conformations (Fig. 3A). Immunofluorescence staining using BG4 as a primary antibody visualized nuclear G4 foci, which disappeared after DNase treatment (Fig. 3B). These observations demonstrate that BG4 has a high affinity for G4s and can identify intracellular G4s [22]. Because GSCs were more sensitive to telomestatin than NSGCs, we predicted that GSCs possessed more G4s than NSGCs. However, GSCs and NSGCs contained similar numbers of G4s (Fig 3C and D).
Fig. 3.
GSCs and NSGCs possess equivalent numbers of G4s. (A) ELISA verification of the affinity of the anti-G4 antibody BG4 for G4-forming oligos (see Supplemental Fig. 2). (B) Immunofluorescence staining of GSCs with BG4. Blue, DAPI staining of nuclear DNA. Dotted lines indicate nuclear boundaries. Lower panel indicates disappearance of nuclear BG4 foci after DNase treatment. (C) BG4 foci in GSCs and NSGCs. (D) Quantification of BG4 foci numbers per nucleus in C.
3.4. GSCs are sensitive to telomestatin-induced replication stress
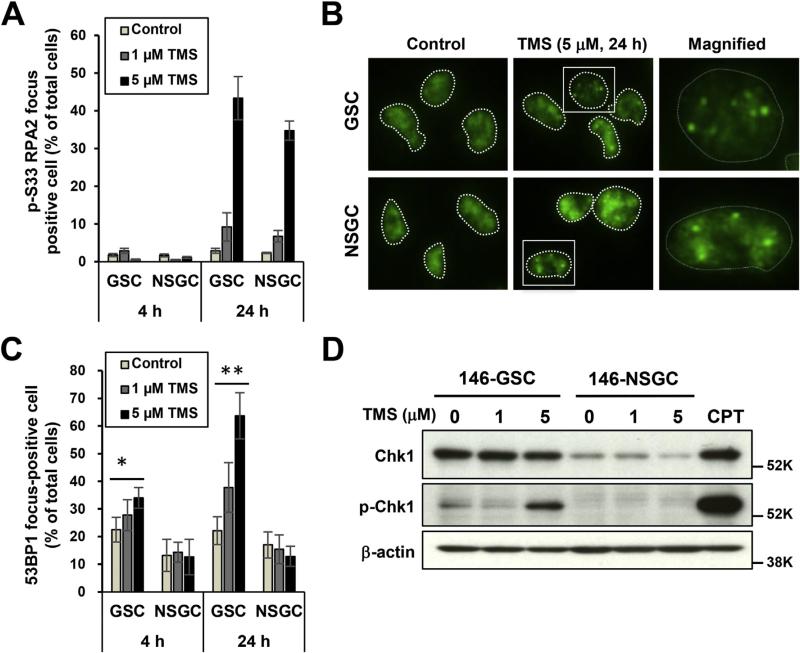
Because telomestatin induced DNA damage through a DNA replication-dependent mechanism, we hypothesized that telomestatin induces replication stress in cells. Replication protein A (RPA), a single-strand DNA-binding protein, is necessary for DNA replication and phosphorylation of RPA2 at Ser33 occurs in response to replication stress [27]. Telomestatin induced phosphorylation of RPA2-Ser33 in both GSCs and NSGCs (Fig. 4A and B). However, telomestatin-induced nuclear 53BP1 foci were observed only in GSCs (Fig. 4C). These findings indicate that while telomestatin induces DNA damage only in GSCs, it stimulates replication stress in both GSCs and NSGCs. Therefore, the mechanism determining the preferential effect of telomestatin on GSCs is likely within the replication stress-response pathway.
Fig. 4.
GSCs are sensitive to telomestatin-induced replication stress. (A) Immunofluorescence staining of telomestatin-treated GSCs and NSGCs with anti-p-RPA2-Ser33 antibody. GBM146 cells were treated with 1 or 5 μM telomestatin in serum-free medium for 4 or 24 h. Cells were classified as focus-positive or -negative according to numbers of punctate nuclear foci of p-RPA2-Ser33 staining (n > 3). (B) Representative image of A. (C) Immunofluorescence of telomestatin-induced 53BP1 foci in GSCs and NSGCs. GBM146 cells were treated as in A and classified according to the numbers of punctate nuclear 53BP1 foci (n > 4). Statistical evaluations were performed using the Welch t-test. *, P < 0.05; **, P < 0.01. (D) Western blot analysis. GBM146 cells were treated with 1 or 5 μM telomestatin for 24 h. Cell lysates were prepared and subjected to western blot analysis. CPT, GSCs were treated with 2 μM camptothecin for 1 h as positive control of p-Chk1.
Phosphorylation of RPA2-Ser33 is mediated by Ataxia telangiectasia and Rad3-related protein (ATR), an essential checkpoint protein kinase activated by accumulation of RPA-coated single-stranded DNA—a characteristic feature of replication stress. Activated ATR further phosphorylates Chk1 protein kinase, leading to blockade of cell-cycle progression [28]. The basal level of Chk1 protein in GSCs was much higher than that in NSGCs, and telomestatin induced phosphorylation of Chk1 only in GSCs (Fig. 4D). These observations indicate that Chk1 is more reactive to telomestatin in GSCs than in NSGCs, and this may explain the selective anti-proliferative effect of telomestatin on GSCs. Additionally, the ATR inhibitor VE-821 induced DNA damage only in GSCs and preferentially inhibited the growth of GSCs (Supplemental Fig. 3). These data suggest that ATR pathway plays an important role in GSCs, and perturbation of this pathway either by G4 stabilization or ATR inhibition produces a GSC-selective anti-proliferative effect.
4. Discussion
We have demonstrated that telomestatin induces replication stress in both GSCs and NSGCs, whereas telomestatin-induced telomeric and non-telomeric DNA damage responses occurred predominantly in GSCs. Although telomestatin can inhibit telomerase activity [16,24], at least two lines of evidence indicate that neither this reduced telomerase activity nor subsequent telomere shortening account for the deleterious effects of telomestatin on GSCs. First, the telomerase inhibitor BIBR1532—which has no G4-stabilizing activity—did not exhibit selectivity for GSCs. Second, there is no lag time between telomestatin treatment and induction of DNA damage. Such a delay would be present because multiple cell cycles are required for the telomerase inhibition-mediated end replication problem to induce sufficient telomere shortening and trigger a DNA damage response at chromosome ends. Instead, telomestatin appears to uncap telomeres by dissociating TRF2, eliciting a prompt DNA damage response in GSCs. Telomeric G4s can be formed at DNA replication. Because the percentage of EdU-positive populations was higher in GSCs than NSGCs, GSCs may have a larger numbers of telomeric G4s than NSGCs. If it were the case, GSCs would be more susceptible to telomestatin-induced TRF2 dissociation from telomeres. At a molecular level, TRF2 forms a homodimer and binds double-strand telomeric DNA via the two Myb domains derived from each TRF2 molecule. Therefore, G4 structures do not allow TRF2 binding via the Myb domains, and G4 stabilization by telomestatin would facilitate TRF2 dissociation from telomeres. The precise mechanism underlying the GSC-selective nature of TRF2 dissociation from telomeres remains unknown. However, GSCs—and not NSGCs—are tumorigenic in vivo. Consistently, TRF2 dissociation from telomeres has been specifically observed in cancer cells, but not normal cells [26].
Telomestatin induced telomeric DNA damage responses more readily than did temozolomide, supporting the notion that telomestatin recognizes G4s in intact cells. Additionally, we used immunofluorescence staining to reveal that numbers of G4s—i.e., telomestatin targets—were comparable in GSCs and NSGCs. This means that the abundance of targets is not a critical determinant of telomestatin sensitivity. Consistently, telomestatin induced formation of p-RPA2-Ser33 foci—a hallmark of replication stress—in GSCs and NSGCs at comparable frequencies. Given that telomestatin elicited DNA damage responses in a replication-dependent manner, it is likely that G4 structures at a replication fork are stabilized by telomestatin and the progression of replication is disturbed. This results in ATR-dependent phosphorylation of RPA2.
Despite formation of comparable foci of p-RPA2-Ser33 in GSCs and NSGCs, only GSCs underwent Chk1 phosphorylation and exhibited a DNA damage response upon telomestatin stimulation. A recent study has revealed that expression levels of DNA damage response proteins, including ATR and Chk1, are high in GSCs [29]. Consistently, we found that levels of Chk1 protein were higher in GSCs compared with NSGCs. These findings suggest that the ATR-Chk1 pathway is relatively easily activated in GSCs under conditions of telomestatin-induced replication stress, and offer an explanation as to why telomestatin preferentially elicits a DNA damage response in GSCs. Furthermore, there are a large number of 53BP1 foci after 4-h treatment with telomestatin despite that the majority of TRF2 still remains bound telomeres and p-RPA2-Ser33 foci have not yet been formed (Fig. 2B). We speculate that most of these acute 53BP1 foci may be derived from the transcription sites with G4-forming sequences. Consistent with this idea, telomestatin-induced acute 53BP1 foci were significantly reduced by DRB, an inhibitor of transcription (Fig. 2B).
Activation of the ATR-Chk1 pathway promotes the repair of DNA damage by cell-cycle arrest, which also confers resistance to radiotherapy [29]. In contrast, potent activation of the ATR-Chk1 pathway causes sustained cell-cycle arrest and eventual cell death [30]. Additionally, the G4 ligand NSC746364 induces cell-cycle arrest through activation of ATR-Chk1 pathway, and induces death of human lung cancer cells [31]. Interestingly, inhibition of ATR kinase by VE-821 causes a preferential anti-proliferative effect in GSCs. Taken together, these observations indicate that GSCs are more sensitive to imbalances in the ATR-Chk1 pathway, suggesting this pathway may be a useful therapeutic target for GBM.
We have shown that telomestatin promotes TRF2 removal from telomeres and potent activation of the replication stress pathway in GSCs. These observations may assist development of a novel therapeutic strategy for GBM.
Supplementary Material
Acknowledgments
We thank Dr. Shankar Balasubramanian for providing the BG4 antibody. This work was supported in part by KAKENHI from the Japan Society for the Promotion of Science (JSPS) to H.S. (25290060).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Transparency document
Transparency document related to this article can be found online at http://dx.doi.org/10.1016/j.bbrc.2016.01.176.
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.bbrc.2016.01.176.
References
- 1.Wen PY, Kesari S. Malignant gliomas in adults. N. Engl. J. Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, Hau P, Brandes AA, Gijtenbeek J, Marosi C, Vecht CJ, Mokhtari K, Wesseling P, Villa S, Eisenhauer E, Gorlia T, Weller M, Lacombe D, Cairncross JG, Mirimanoff RO. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet. Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 4.Cheng L, Bao S, Rich JN. Potential therapeutic implications of cancer stem cells in glioblastoma. Biochem. Pharmacol. 2010;80:654–665. doi: 10.1016/j.bcp.2010.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF. A restricted cell population propagates glioblastoma growth after chemo-therapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 7.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 8.Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. U. S. A. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 10.Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 11.Yu HQ, Miyoshi D, Sugimoto N. Characterization of structure and stability of long telomeric DNA G-quadruplexes. J. Am. Chem. Soc. 2006;128:15461–15468. doi: 10.1021/ja064536h. [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez R, Miller KM, Forment JV, Bradshaw CR, Nikan M, Britton S, Oelschlaegel T, Xhemalce B, Balasubramanian S, Jackson SP. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 2012;8:301–310. doi: 10.1038/nchembio.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lemarteleur T, Gomez D, Paterski R, Mandine E, Mailliet P, Riou JF. Stabilization of the c-myc gene promoter quadruplex by specific ligands' inhibitors of telomerase. Biochem. Biophys. Res. Commun. 2004;323:802–808. doi: 10.1016/j.bbrc.2004.08.150. [DOI] [PubMed] [Google Scholar]
- 14.Wei D, Todd AK, Zloh M, Gunaratnam M, Parkinson GN, Neidle S. Crystal structure of a promoter sequence in the B-raf gene reveals an intertwined dimer quadruplex. J. Am. Chem. Soc. 2013;135:19319–19329. doi: 10.1021/ja4101358. [DOI] [PubMed] [Google Scholar]
- 15.Dai J, Chen D, Jones RA, Hurley LH, Yang D. NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Res. 2006;34:5133–5144. doi: 10.1093/nar/gkl610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin-ya K, Wierzba K, Matsuo K, Ohtani T, Yamada Y, Furihata K, Hayakawa Y, Seto H. Telomestatin, a novel telomerase inhibitor from Streptomyces anulatus. J. Am. Chem. Soc. 2001;123:1262–1263. doi: 10.1021/ja005780q. [DOI] [PubMed] [Google Scholar]
- 17.Kim MY, Vankayalapati H, Shin-Ya K, Wierzba K, Hurley LH. Telomestatin, a potent telomerase inhibitor that interacts quite specifically with the human telomeric intramolecular g-quadruplex. J. Am. Chem. Soc. 2002;124:2098–2099. doi: 10.1021/ja017308q. [DOI] [PubMed] [Google Scholar]
- 18.Gomez D, Paterski R, Lemarteleur T, Shin-Ya K, Mergny JL, Riou JF. Interaction of telomestatin with the telomeric single-strand overhang. J. Biol. Chem. 2004;279:41487–41494. doi: 10.1074/jbc.M406123200. [DOI] [PubMed] [Google Scholar]
- 19.Miyazaki T, Pan Y, Joshi K, Purohit D, Hu B, Demir H, Mazumder S, Okabe S, Yamori T, Viapiano M, Shin-ya K, Seimiya H, Nakano I. Telomestatin impairs glioma stem cell survival and growth through the disruption of telomeric G-quadruplex and inhibition of the proto-oncogene, c-Myb. Clin. cancer Res. official J. Am. Assoc. Cancer Res. 2012;18:1268–1280. doi: 10.1158/1078-0432.CCR-11-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ouchi R, Okabe S, Migita T, Nakano I, Seimiya H. Senescence from glioma stem cell differentiation promotes tumor growth. Biochem. Biophys. Res. Commun. 2016;470:275–281. doi: 10.1016/j.bbrc.2016.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirashima K, Migita T, Sato S, Muramatsu Y, Ishikawa Y, Seimiya H. Telo-mere length influences cancer cell differentiation in vivo. Mol. Cell. Biol. 2013;33:2988–2995. doi: 10.1128/MCB.00136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirashima K, Seimiya H. Telomeric repeat-containing RNA/G-quadruplex-forming sequences cause genome-wide alteration of gene expression in human cancer cells in vivo. Nucleic Acids Res. 2015;43:2022–2032. doi: 10.1093/nar/gkv063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tauchi T, Shin-ya K, Sashida G, Sumi M, Okabe S, Ohyashiki JH, Ohyashiki K. Telomerase inhibition with a novel G-quadruplex-interactive agent, telomestatin: in vitro and in vivo studies in acute leukemia. Oncogene. 2006;25:5719–5725. doi: 10.1038/sj.onc.1209577. [DOI] [PubMed] [Google Scholar]
- 25.Damm K, Hemmann U, Garin-Chesa P, Hauel N, Kauffmann I, Priepke H, Niestroj C, Daiber C, Enenkel B, Guilliard B, Lauritsch I, Muller E, Pascolo E, Sauter G, Pantic M, Martens UM, Wenz C, Lingner J, Kraut N, Rettig WJ, Schnapp A. A highly selective telomerase inhibitor limiting human cancer cell proliferation. EMBO J. 2001;20:6958–6968. doi: 10.1093/emboj/20.24.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tahara H, Shin-Ya K, Seimiya H, Yamada H, Tsuruo T, Ide T. G-Quadruplex stabilization by telomestatin induces TRF2 protein dissociation from telomeres and anaphase bridge formation accompanied by loss of the 3' telomeric overhang in cancer cells. Oncogene. 2006;25:1955–1966. doi: 10.1038/sj.onc.1209217. [DOI] [PubMed] [Google Scholar]
- 27.Vassin VM, Anantha RW, Sokolova E, Kanner S, Borowiec JA. Human RPA phosphorylation by ATR stimulates DNA synthesis and prevents ssDNA accumulation during DNA-replication stress. J. cell Sci. 2009;122:4070–4080. doi: 10.1242/jcs.053702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shechter D, Costanzo V, Gautier J. Regulation of DNA replication by ATR: signaling in response to DNA intermediates. DNA Repair (Amst) 2004;3:901–908. doi: 10.1016/j.dnarep.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 29.Ahmed SU, Carruthers R, Gilmour L, Yildirim S, Watts C, Chalmers AJ. Selective Inhibition of Parallel DNA Damage Response Pathways Optimizes Radiosensitization of Glioblastoma Stem-like Cells. Cancer Res. 2015;75:4416–4428. doi: 10.1158/0008-5472.CAN-14-3790. [DOI] [PubMed] [Google Scholar]
- 30.Zhao Y, Wu Z, Zhang Y, Zhu L. HY-1 induces G(2)/M cell cycle arrest in human colon cancer cells through the ATR-Chk1-Cdc25C and Weel pathways. Cancer Sci. 2013;104:1062–1066. doi: 10.1111/cas.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung YL, Pan CH, Liou WH, Sheu MJ, Lin WH, Chen TC, Huang HS, Wu CH. NSC746364, a G-quadruplex-stabilizing agent, suppresses cell growth of A549 human lung cancer cells through activation of the ATR/Chk1-dependent pathway. J. Pharmacol. Sci. 2014;124:7–17. doi: 10.1254/jphs.13096fp. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.