Abstract
Entamoeba histolytica, the highly phagocytic protozoan causative of human amoebiasis lacks the machinery to synthesize cholesterol. Here, we investigated the presence of NPC1 and NPC2 proteins in this parasite, which are involved in cholesterol trafficking in mammals. Bioinformatics analysis revealed one Ehnpc1 and two Ehnpc2 genes. EhNPC1 appeared as a transmembrane protein and both EhNPC2 as peripheral membrane proteins. Molecular docking predicted that EhNPC1 and EhNPC2 bind cholesterol and interact with each other. Genes and proteins were identified in trophozoites. Serum pulse-chase and confocal microscopy assays unveiled that after trophozoites sensed the cholesterol source, EhNPC1 and EhNPC2 were organized around the plasma membrane in a punctuated pattern. Vesicles emerged and increased in number and size and some appeared full of cholesterol with EhNPC1 or EhNPC2 facing the extracellular space. Both proteins, but mostly EhNPC2, were found out of the cell associated with cholesterol. EhNPC1 and cholesterol formed networks from the plasma membrane to the nucleus. EhNPC2 appeared in erythrocytes that were being ingested by trophozoites, co-localizing with cholesterol of erythrocytes, whereas EhNPC1 surrounded the phagocytic cup. EhNPC1 and EhNPC2 co-localized with EhSERCA in the endoplasmic reticulum and with lysobisphosphatidic acid and EhADH (an Alix protein) in phagolysosomes. Immunoprecipitation assays confirmed the EhNPC1 and EhNPC2 association with cholesterol, EhRab7A and EhADH. Serum starved and blockage of cholesterol trafficking caused a low rate of phagocytosis and incapability of trophozoites to produce damage in the mouse colon. Ehnpc1 and Ehnpc2 knockdown provoked in trophozoites a lower intracellular cholesterol concentration and a diminished rate of phagocytosis; and Ehnpc1 silencing also produced a decrease of trophozoites movement. Trafficking of EhNPC1 and EhNPC2 during cholesterol uptake and phagocytosis as well as their association with molecules involved in endocytosis strongly suggest that these proteins play a key role in cholesterol uptake.
Author Summary
NPC1 and NPC2 proteins are involved in cholesterol trafficking in mammals. Using different approaches, we have detected the orthologues EhNPC1 and EhNPC2 proteins in Entamoeba histolytica. Trophozoites are particularly rich in membranes and vacuoles, but they do not possess the machinery to synthetize cholesterol. Thus, they are completely dependent on molecules able to “fish” cholesterol from the medium. The relevance of our findings lies in the fact that cholesterol is fundamental for endocytosis and motility; and, phagocytosis is an important nutritional and virulence factor for E. histolytica. In silico and experimental strategies, using U18666A to arrest cholesterol trafficking, as well as, knockdown mutants, showed that EhNPC1 and EhNPC2 participate in cholesterol uptake and trafficking in this parasite. They are secreted by trophozoites and directly involved in erythrophagocytosis and motility. Our findings revealed E. histolytica as one of the first protozoa in which these proteins are being characterized. Moreover, E. histolytica provides an excellent and less complicated model to elucidate the intricate event of cholesterol trafficking in eukaryotic cells. The relevance of cholesterol transport for the parasite virulence and the involvement of EhNPC1 and EhNPC2 in this process, make these proteins promising targets for therapy strategies development against the parasite.
Introduction
Entamoeba histolytica is the protozoan responsible for intestinal and hepatic amoebiasis, considered the third leading cause of death worldwide due to parasites [1]. E. histolytica trophozoites are highly dynamic cells with active movement and voracious phagocytosis. High cholesterol concentration in the medium enhances their virulence [2–4] and trophozoites loaded with cholesterol showed an enrichment of the Gal/GalNAc lectin in rafts and an increase of amoeba adherence to target cells [5]. Cholesterol is fundamental for vesicle formation and lipid rafts arrangements, and both are crucial events for movement and endocytosis [4]. However, E. histolytica lacks the machinery to synthesize cholesterol [6]. Cells ingested by the parasite, including erythrocytes, are a natural cholesterol source, but, trophozoites can also uptake it from the serum-supplemented culture medium [7, 8].
Mammalian cells synthesize cholesterol through a complex pathway, in which at least 30 enzymes participate [9]. Cholesterol homeostasis is controlled by feedback regulation of its biosynthesis and uptake through receptor-mediated endocytosis by low density lipoproteins (LDL) [10]. Failures in cholesterol storage in humans cause the Niemann-Pick type C (NPC) disease, which is linked to mutations in NPC1 or NPC2 proteins that are directly involved in cholesterol trafficking [11, 12]. NPC1 (1278 amino acids) is a polytopic endosomal membrane glycoprotein required for efflux of cholesterol from endosomes [13]. It has 13 transmembrane domains, four luminal and six cytoplasmic loops, a C-terminal cytoplasmic tail and a sterol sensing domain (SSD) [14]. NPC2 (151 amino acids) is a soluble lysosomal protein with a MLD domain (ML [MD-2 (myeloid differentiation factor-2)]-related lipid-recognition) [15, 16] that regulates cholesterol trafficking from lysosomes to the endoplasmic reticulum (ER) [17, 18]. NPC2 possesses positively charged regions that facilitate its interaction with negatively charged membranes [17]. NPC2 binds to NPC1; and NPC1 binds to cholesterol by the SSD domain in an acidic milieu [18]. Moreover, cholesterol is transferred from the N terminus domain (NTD) of NPC1 to NPC2 in a bidirectional manner [19]. Based on this, Infante et al. [19] proposed the “hand-off” working model, which assumes that NPC2 takes cholesterol in the lysosomal lumen and transports it to membrane-bound NPC1 for exportation to the ER [19–21]. In late endosomes, lysobisphosphatidic acid (LBPA) regulates cholesterol, under the control of Alix protein [22, 23]. The molecular mechanisms of this regulation are not completely understood.
In E. histolytica, neither LDL receptors, nor NPC1 and NPC2 have been identified yet. The TMK39 protein participates in cholesterol uptake, however, it does not have cholesterol binding domains, suggesting that it could associate to cholesterol through other trophozoite molecules [24]. As in mammals, E. histolytica LBPA binds to EhADH (an E. histolytica ALIX family protein) [25] inside phagolysosomes and multivesicular bodies (MVB) [26]. Though, we do not discard the participation of other Alix proteins involved in endomembrane trafficking [27, 28]. Thus, it is plausible to assume that the “hand-off” model, “NPC2-NPC1-cholesterol-NPC2-NPC1” proposed for mammalian cells, could also be functioning in trophozoites to first carry exogenous cholesterol inside the cell and then, to transport it to distinct organelles. Here, we searched for NPC1 and NPC2 orthologues in E. histolytica and studied their participation in cholesterol uptake and trafficking, as well as their association with molecules involved in phagocytosis. We also provide evidence that cholesterol depletion and trafficking arrest as well as Ehnpc1 and Ehnpc2 genes knockdown, affect virulence, particularly erythrophagocytosis and cell motility.
Results
E. histolytica possesses one npc1 (Ehnpc1) and two npc2 (Ehnpc2a, Ehnpc2b) genes and their respective proteins bind cholesterol
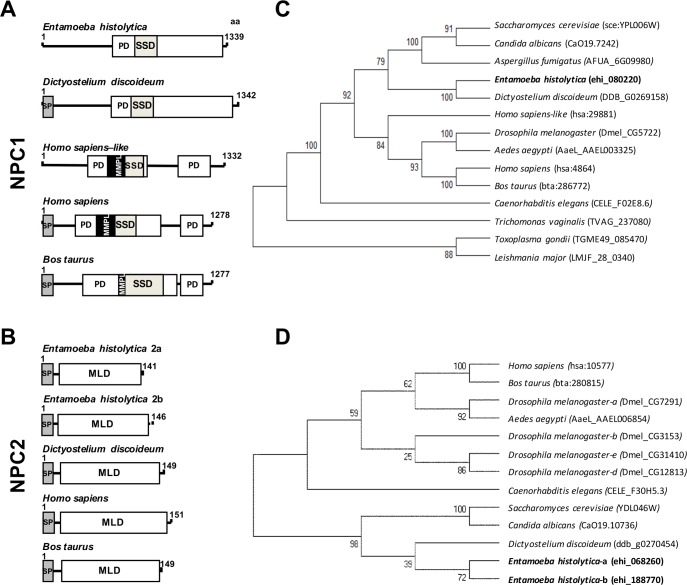
Our exploration in the AmoebaDB (http://amoebadb.org/amoeba/) revealed the presence of a 1339 amino acid sequence (EHI_080220) with 19.7 to 35% identity to NPC1 proteins of different species (Fig 1A, S1 Table). The putative EhNPC1 protein exhibited the patched (PD) and the SSD domains present in all reported NPC1 proteins [29]. However, it did not show the MMPL domain, found in bacteria as a putative integral membrane protein domain [30]. This domain is also absent in Dictyostelium discoideum NPC1 [31] (Fig 1A). We also found two sequences of 141 (EHI_068260) (EhNPC2a) and 146 (EHI_188770) (EhNPC2b) amino acids containing the MLD domain, which is a NPC2 signature [15] (Fig 1B). These sequences showed 29.4% identity between them and 11 to 26% with other NPC2 proteins (S2 Table).
Fig 1. Structural domains and phylogenetic trees of EhNPC1, EhNPC2a and EhNPC2b.
(A, B) Schemes show the main structural characteristics of NPC1 (A) and NPC2 (B) from distinct organisms. PD: patched domain, SSD: sterol sensing domain, MMPL: putative integral membrane domain, SP: signal peptide, MLD: MD-2 related lipid recognition domain. Numbers at the right correspond to the amino acids forming the proteins. (C, D) Phylogenetic tree indicating the position of E. histolytica NPC1 (C), NPC2a and NPC2b (D) proteins among different species. Numbers on horizontal lines in the trees indicate the confidence percentages of the tree topology from bootstrap analysis of 1000 replicates.
The full length amino acid sequences of EhNPC1, EhNPC2a and EhNPC2b were compared with NPC1 and NPC2 protein sequences from other organisms to construct phylogenetic trees using the MEGA 5.05 software. EhNPC1 was grouped in a single clade with D. discoideum NPC1 in the branch of slime molds, yeasts and filamentous fungus and far from hypothetical NPC1s from protozoa such as Trichomonas vaginalis, Toxoplasma gondii and Leishmania major (Fig 1C). Interestingly, EhNPC2a and EhNPC2b also displayed a close relationship with D. discoideum NPC2 in the branch of slime molds and yeasts (Fig 1D).
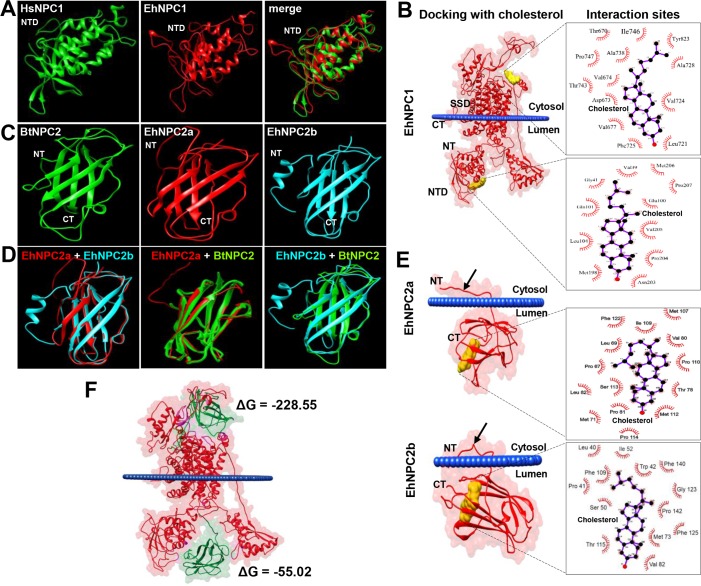
To obtain further evidence on the structural relationship of EhNCP1, EhNPC2a and EhNPC2b with their respective orthologues, we constructed their 3D models using RaptorX server (http://raptorx.uchicago.edu). The predicted 3D structure of NTD EhNPC1 (1–248 amino acids) presented 92% structural identity to the Homo sapiens NTD NPC1 crystal (23 to 252 amino acids) [20]; and their merged images extensively overlapped (Fig 2A). EhNPC1 appeared as a membrane protein with 16 transmembrane domains, two structural regions located in the endosomal lumen, where the NTD is found, as described for other organisms [20], and a single larger domain in the cytosolic side (Fig 2B).
Fig 2. 3D structures of NTD-EhNPC1, EhNPC2a and EhNPC2b and molecular dockings between them and with cholesterol.
(A) NTD-EhNPC1 model (1 to 250 amino acids) predicted by RaptorX server was compared with the crystal of human NTD-NPC1 protein (HsNPC1) (23 to 254 amino acids). (B) Docking simulation of EhNPC1 with cholesterol performed using the AutoDock Tools V1.5.6 program. SSD: sterol sensing domain. NTD: amino terminal domain. (C, D) The EhNPC2a and EhNPC2b 3D structures (full-length amino acid sequences) were compared between them and with the NPC2 B. taurus crystal (BtNPC2). (E) Docking simulation of EhNPC2a and EhNPC2b with cholesterol. Arrows: amino acids tail at the amino terminus that it is not present in the crystal of Btaurus NPC2. Squares at the right in B and E show the amino acids involved in the protein-cholesterol interaction. (F) Docking simulation of EhNPC1 and EhNPC2a binding. ΔG: binding energy. Blue lines in dockings: plasmatic membrane. NT: amino terminus. CT: carboxy terminus.
Docking analysis revealed two main interaction sites of EhNPC1 with cholesterol. According to in silico predictions, one site was located in the cytosol and the other at the NTD, in the endosomal lumen (Fig 2B). Results evidenced hydrophobic interactions of cholesterol with 13 amino acids of EhNPC1 at cytoplasmic domain (Thr670, Asp673, Val671, Val677, Leu721, Val724, Phe725, Ala728, Ala738, Thr743, Ile746, Pro747 and Tyr823); and with eleven residues of the NTD (Val39, Gly41, Glu100, Gln101, Leu104, Met198, Asn203, Pro204, Val205, Met206 and Pro207) (Fig 2B). The binding energies for these sites were ΔG ═ -8.6 and ΔG ═ -7.3 Kcal/mol, respectively, indicating a weak binding.
The 3D structures of EhNPC2a and EhNPC2b proteins presented 83.2% structural identity between themselves, and 88.6 and 86% with the Bos taurus crystal [32], respectively. EhNPC2 proteins exhibited seven beta strands (Fig 2C and 2D), as described for other NPC2 [32]. Interestingly, EhNPC2a and EhNPC2b displayed an extra amino acid tail at the N-terminus formed by 14 and 18 residues, respectively (Fig 2C–2E) that have not been reported in other NPC2 orthologues. The Orientation of Proteins in Membranes (OPM) database (http://opm.phar.umich.edu/) predicted that in EhNPC2a, Leu11, Phe12, Ala13 and Ala14 residues were attached to the membranes (Fig 2E); whereas in EhNPC2b the residues in contact with membranes were Thr19, Ala18, Leu17, Met16, Leu12 and Phe6 (Fig 2E), these results suggest that both EhNPC2 could be peripheral membrane proteins, which allow them to cross the membranes during cholesterol transport. The EhNPC2a interaction with cholesterol was predicted to be carried out by hydrophobic bonds corresponding to Pro67, Leu69, Met71, Thr78, Val80, Pro81, Leu82, Met107, Ile109, Pro110, Met112, Ser113 and Phe122 residues (Fig 2E). By contrast, EhNPC2b binds to cholesterol through Leu40, Pro41, Trp42, Ser50, Ile52, Met73, Val82, Thr115, Gly123, Phe125, Phe140 and Pro142 residues (Fig 2E). Amino acids in bold are in comparable positions and separated by the same number of residues in both proteins. The cholesterol binding energies to EhNPC1 and EhNPC2 were also low: ΔG ═ -9 and -10.15 Kcal/mol, respectively.
We also performed docking analysis using EhNPC1 and EhNPC2. Interestingly, results showed that EhNPC1 presents two putative interaction sites with EhNPC2, in cytosol and endosomal lumen (Fig 2F), with ΔG ═ -228.5 and -55.02 Kcal/mol, respectively. ΔG values reflect a stronger binding between the proteins compared to those with cholesterol. However, these values are still comparably low, corresponding to rather short-lived interactions. In summary, our bioinformatics analysis revealed that EhNPC1, EhNPC2a and EhNPC2b are NPC1 and NPC2 orthologues [21] that interact between them and with cholesterol, suggesting that they could play an important role in cholesterol transport.
Ehnpc1, Ehnpc2a and Ehnpc2b genes are expressed in trophozoites
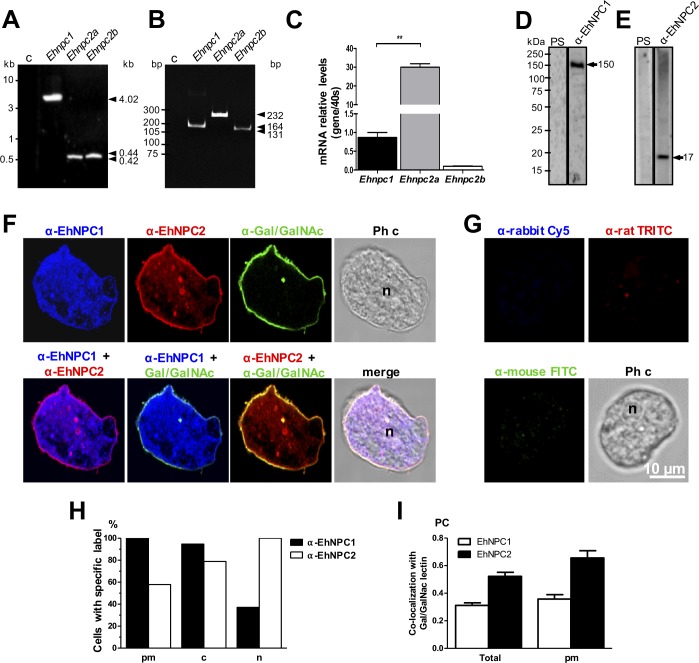
To confirm that the genes found in silico were bona fide genes in the parasite, we designed specific primers (S3 Table) using sequences obtained from the AmoebaDB. PCR amplification from genomic DNA gave 4.0, 0.42 and 0.44 kbp fragments, corresponding to the expected size of Ehnpc1, Ehnpc2a and Ehnpc2b full length genes, respectively (Fig 3A). Sequencing of the three genes revealed full open reading frames, no introns and 100% identity to the genes annotated in the AmoebaDB. RT-PCR assays evidenced bands at the expected size, indicating that genes are transcribed (Fig 3B). Intriguingly, RT-qPCR assays showed that Ehnpc2a was expressed 30 folds more than Ehnpc1 gene, whereas, Ehnpc2b was poorly expressed (Fig 3C).
Fig 3. Expression and localization of EhNPC1 and EhNPC2 in trophozoites.
(A) Ehnpc1, Ehnpc2a and Ehnpc2b full-length genes were PCR amplified using specific primers and genomic DNA. (B) RT-PCR amplification of transcript fragments using specific primers and cDNA. c: Controls without gDNA or with mRNA as template. (C) The relative expression of the three genes was measured by RT-qPCR in trophozoites, using as a control the 40s ribosomal S2 protein gene. ** p<0.01. (D, E) Total extracts of E. histolytica were separated by 10% SDS-PAGE and analyzed by western blot assays using pre-immune serum (PS) or rabbit α-EhNPC1 (D) or rat α-EhNPC2 (E) antibodies. (F) Representative images of laser confocal microscopy of PFA-fixed trophozoites using rabbit α-EhNPC1 or rat α-EhNPC2 or mouse α-Gal/GalNAc lectin antibodies. (G) Controls using only secondary antibodies. Ph c: phase contrast images. (H) Protein localization of EhNPC1 and EhNPC2 in plasma membrane (pm), cytoplasm (c) or nucleus (n). Counts were performed in 50 cells. (I) Pearson coefficient (PC) correlation measured in at least 15 confocal images, indicating co-localization of EhNPC1 or EhNPC2 with Gal/GalNAc lectin in the entire cell and in the plasma membrane. Laser sections = 0.5 μm.
We generated an antibody against an EhNPC1 specific polypeptide (766-DEQPMYDKDGQYVPVEKRLE-785) that detected in western blot assays the expected single 150 kDa band in trophozoites samples (Fig 3D). Antibodies against EhNPC2a were produced in rats using recombinant proteins. They revealed the expected 17 kDa band (Fig 3E) and it was used as a pan antibody because they recognized both EhNPC2 recombinant proteins (S1 Fig), furthermore, the Ehnpc2a gene is transcribed more efficiently than Ehnpc2b. Pre-immune sera did not recognize any band (Fig 3D and 3E).
Confocal images showed that in basal conditions (trophozoites cultured in TYI-S medium, without stimulus), 100 and 60% of trophozoites were labeled in plasma membrane by α-EhNPC1 and α-EhNPC2 antibodies, respectively (Fig 3F and 3H), co-localizing with the α-Gal/GalNAc lectin antibody, used as a plasma membrane marker (Fig 3F). However, Pearson coefficient (PC) showed a higher co-localization of Gal/GalNAc lectin with EhNPC2 (Fig 3I). 100 and 80% of trophozoites showed α-EhNPC1 and α-EhNPC2 antibodies label in the cytoplasm, and only 40 and 100% of trophozoites, respectively, presented fluorescence in the nuclei (Fig 3F and 3H). We do not know yet the function, if any, of EhNPC1 and EhNPC2 in the nucleus. In all experiments reported here, we used as controls only the secondary antibodies, which always gave negative results (Fig 3G).
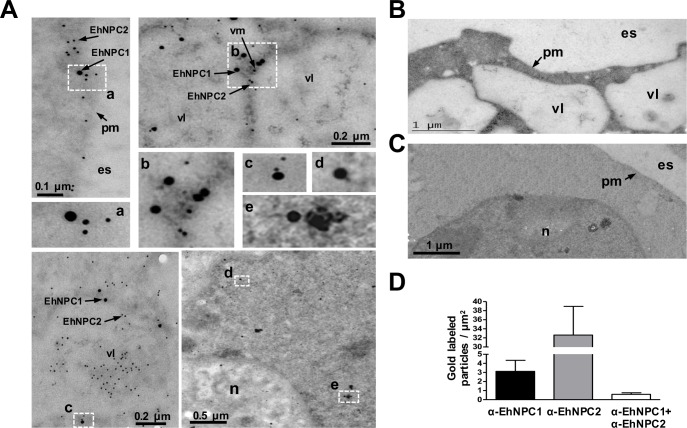
TEM gold immunolabeling assays confirmed the polytopic cellular location of these proteins (Fig 4A). Images evidenced that in basal conditions, the majority of EhNPC1 and EhNPC2 proteins appeared separated, some of them localized close each other and only few appeared together (Fig 4A–4D). EhNPC1 and EhNPC2 were localized at plasma membrane, in the vesicle lumen and membranes. The labeled vesicles could correspond to endosomes; in there EhNPC1 was frequently surrounding them and EhNPC2 appeared inside (Fig 4A). Controls using only the secondary antibodies gave none or scarce signals (Fig 4B and 4C). These results together evidenced that Ehnpc1 and Ehnpc2 genes are transcribed and translated and their proteins are localized in several cellular regions, indicating that they are highly mobile proteins, which is in agreement with their hypothetical function as cholesterol transporters.
Fig 4. Localization of EhNPC1 and EhNPC2 in trophozoites analyzed by TEM.
(A) Thin sections of trophozoites were incubated with rabbit α-EhNPC1 and rat α-EhNPC2 antibodies, followed by incubation with gold labeled α-rabbit and α-rat secondary antibodies (20 and 10 nm gold particles, respectively). Squares indicate the magnified areas marked with the corresponding lower case letters. pm: plasma membrane, vl: vesicle lumen, vm: vesicle membrane, es: extracellular space, n: nucleus. (B, C) Controls using only secondary antibodies. (D) Graph showing number of EhNPC1 and EhNPC2 molecules recognized by the respective gold-labeled antibodies and their co-localization.
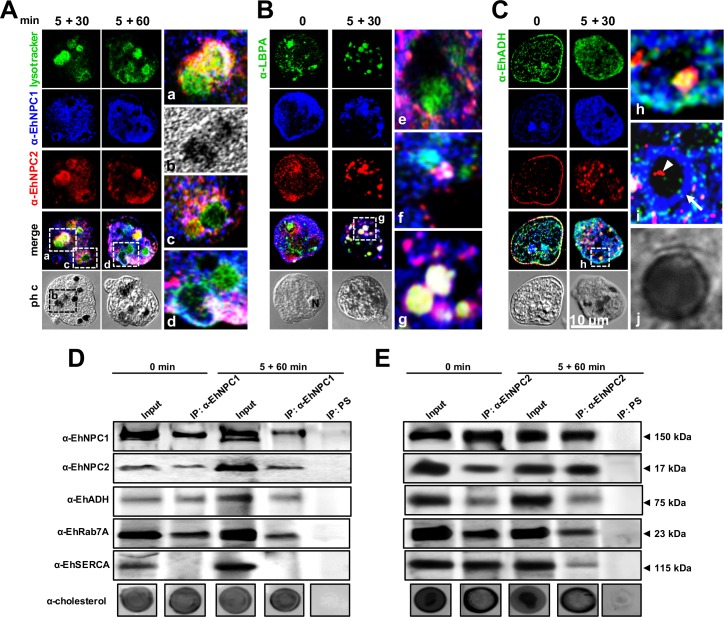
After serum stimulus, EhNPC1 and EhNPC2 co-localize with cholesterol
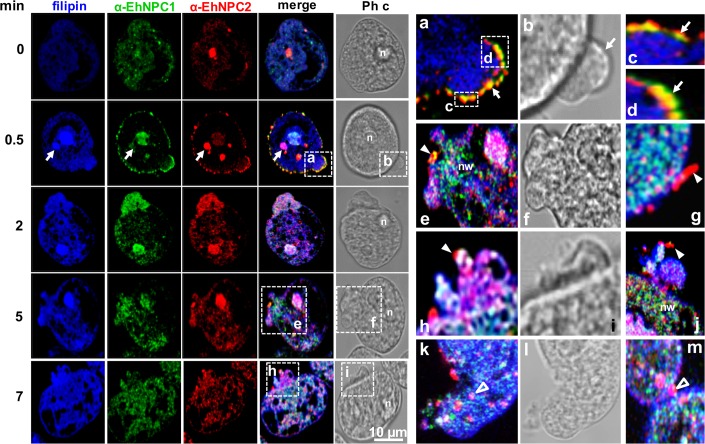
The main source of cholesterol in trophozoites in culture comes from adult bovine serum (ABS). We explored the participation of EhNPC1 and EhNPC2 in cholesterol trafficking after a serum stimulus. After serum starving of trophozoites, confocal images revealed that both proteins were barely present in the plasma membrane and cholesterol was faintly stained by filipin, a highly specific compound for cholesterol detection (Fig 5). Immediately after ABS addition (0.5 min), filipin detected cholesterol in the cytoplasm and nucleus, together with EhNPC1 and EhNPC2 (Fig 5). At this time, EhNPC1 and EhNPC2 co-localized in vesicles and in a uniform punctuate pattern at the plasma membrane (Fig 5A–5D). In most cases a yellow area was evident in membrane protrusions, indicating co-localization of both proteins (Fig 5A–5D). We also observed both proteins out of the cell (Fig 5G and 5J). After 2 to 5 min, membrane protrusions or putative vesicles appeared carrying cholesterol, and, in most of them, EhNPC2 was outwardly (Fig 5E and 5K), strengthening the hypothesis on the mobilization of both proteins during cholesterol capture. Number and size of membrane protrusions or putative vesicles increased through incubation time, and both proteins were close to cholesterol-containing vesicles outside and inside of the cell (Fig 5A, 5B, 5E, 5F, 5H and 5I). In some trophozoites, membrane protrusions or putative vesicles appeared at a cellular pole, in a “cap” model (Fig 5, 5 min), but they were also seen in other parts of the plasma membrane (Fig 5, 7 min). Outside of the cell, joined or not to the plasma membrane, EhNPC2 was revealed in vacuoles with rod and sphere shaped forms (Fig 5G–5M). Network-like structures mainly formed by EhNPC1 and cholesterol were observed inside the cells, adjacent to the networks formed mainly by EhNPC1 and cholesterol, and inside EhNPC2-containing spheres (1.5 to 2 μm) also appeared there (Fig 5E–5J). The spherical structures may correspond to the vesicles detected by TEM (Fig 4). These findings strongly suggest that cholesterol promotes the mobilization and secretion of EhNPC1 and EhNPC2.
Fig 5. Localization of EhNPC1 and EhNPC2 in trophozoites after an ABS pulse.
Trophozoites were serum starved for 12 h by culturing in TYI medium on coverslips. Then, ABS was added as a cholesterol source at 37°C for 0.5 to 7 min. Cells were washed, fixed and incubated with filipin, rabbit α-EhNPC1 and rat α-EhNPC2 antibodies, secondary antibodies, and examined by laser confocal microscopy. Arrows: cytoplasmic dots. Squares marked with lower case letters are magnified at the right. (a-d) Membrane vacuoles containing EhNPC1 and EhNPC2. Arrows: show the plasma membrane with EhNPC2 or EhNPC1 facing the extracellular space. (e) Networks stained by α-EhNPC1 antibody. (e-j) rod and spherical structures (arrowheads) facing the extracellular space, stained mainly by α-EhNPC2 antibody. (k-m) Spherical structures of 1 to 2 μm (empty arrowheads) inside the cell. n: nucleus, Ph c: phase contrast images, nw: networks like structures.
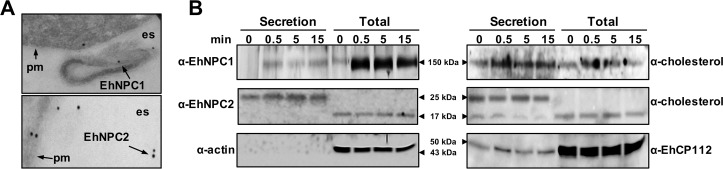
In TEM images, we detected EhNPC1 and EhNPC2 in the extracellular space of trophozoites in basal conditions (Fig 6A). To explore whether the proteins were secreted, we performed secretion assays, giving an ABS pulse for different times to serum-starved trophozoites. In agreement with the confocal microscopy images (Fig 5, 0 min), EhNPC1 and EhNPC2 were poorly detected in secreted products of starved trophozoites. However, they were found in the supernatants of trophozoites challenged with ABS for 0.5 min (Fig 6B), indicating that they were secreted in response to the ABS pulse. Surprisingly, the α-EhNPC2 antibody did not reveal the expected 17 kDa band, but only a 25 kDa band was detected in the secreted products. This aberrant migration could be due to the binding of EhNPC2 to cholesterol present in the serum. To get further evidence on this, the nitrocellulose membranes were re-blotted using an α-cholesterol antibody. This antibody detected a band with similar migration than EhNPC1 and two bands of 17 and 25 kDa, corresponding to those detected by the α-EhNPC2 antibody. EhCP112, used as a positive secretion control, was found in both, supernatant and trophozoites extracts at all times, whereas actin, used as a negative control for secretion, was only detected in trophozoites samples. These results strongly suggest that these proteins are associated with cholesterol. A hypothesis could be that vesicles containing EhNPC1 and EhNPC2 come from and back to the cell, possibly carrying cholesterol from outside.
Fig 6. Secretion of EhNPC1 and EhNPC2 after ABS stimulus.
(A) TEM of trophozoites under basal condition showing EhNPC1 and EhNPC2 in the extracellular space. pm: plasma membrane, es: extracellular space. (B) After the ABS stimulus, supernatants were collected and trophozoites were lysed at the indicated times. Secreted products and total extracts were analyzed by SDS-PAGE and western blot assays using rabbit α-EhNPC1, rat α-EhNPC2, rabbit α-cholesterol, rabbit α-EhCP112 and mouse α-actin antibodies and corresponding secondary antibodies. Actin was used as a control of cell integrity and EhCP112 as a secretion control.
EhNPC1 and EhNPC2 co-localize with cholesterol during phagocytosis
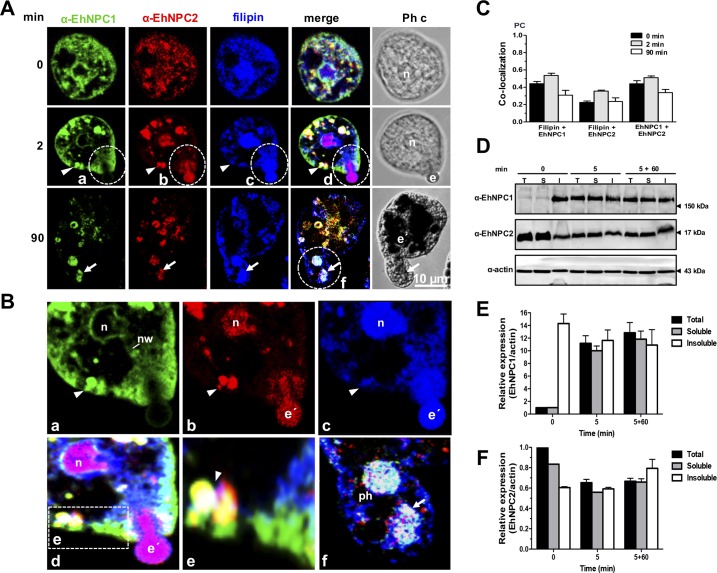
As phagocytosis is one of the main mechanisms for nutrients uptake and virulence expression in the parasite and trophozoites have an intense membrane synthesis during the event, we analyzed the EhNPC1 and EhNPC2 behavior when cells ingest erythrocytes. Fig 7 shows images of the most relevant facts observed in different experiments. Under basal conditions, both proteins and cholesterol were found at the plasma membrane, cytoplasm, and nuclei (Fig 7A, 0 min). Immediately after sensing the presence of erythrocytes, EhNPC1 and EhNPC2 proteins moved to the contact sites at the plasma membrane (Fig 7A and 7B); and they were present in cytoplasmic vesicles with cholesterol inside (Fig 7, 2 min). Interestingly, EhNPC2 co-localized with cholesterol of phagocytosed erythrocytes (Fig 7A and 7B). Both proteins were observed in trophozoites as a continuous dot pattern from the partially ingested erythrocytes to the cytoplasm (Fig 7A, 2 min; 7Ba-d). EhNPC1 was seen in the external part of the phagocytic cup (Fig 7A and 7B). Between the nucleus and erythrocytes, networks-like structures were observed decorated mainly by the α-EhNPC1 antibody and filipin, and spots recognized by α-EhNPC2 antibodies (Fig 7A, 2 min; 7B). At 90 min, cholesterol appeared distributed at the plasma membrane, and in vacuoles inside other huge vacuoles that may correspond to phagolysosomes and MVBs containing digested erythrocytes, and co-localizing with EhNPC2 and EhNPC1 (Fig 7A, 90 min; 7Bf). Co-localization of both proteins was not higher than 0.59 (Fig 7C), suggesting that they do not interact all the time. Recently, we have identified LBPA and EhADH inside phagolysosomes and MVBs after 60 and 90 min of phagocytosis [26]. By their size and appearance at late times of phagocytosis, we presume that they correspond to the ones detected here containing cholesterol, EhNPC1 and EhNPC2.
Fig 7. Localization of EhNPC1, EhNPC2 and cholesterol during phagocytosis.
(A) Trophozoites were incubated with erythrocytes (1:25) at 37°C, for the indicated times. Then, samples were prepared for laser confocal microscopy and stained with filipin, rabbit α-EhNPC1 and rat α-EhNPC2 antibodies and secondary antibodies. Ph c: phase contrast images, e’: erythrocytes, n: nucleus. Arrow head: cytoplasmic vesicles. (B) (a-d) Dotted circles: Magnification of an erythrocyte that is being internalized. (e) Magnification of vesicles close to the plasma membrane. (f) Magnification of a huge phagosome (ph) containing digested erythrocytes (arrows). nw: networks. (C) PC of the co-localization between filipin and EhNPC1 or EhNPC2 or EhNPC1 with EhNPC2 at distinct phagocytosis time. (D) Trophozoites were incubated with erythrocytes at 37°C for 5 min, then adherent and non-ingested erythrocytes were removed by washing with a mixture of water-TYI medium. Later, trophozoites were incubated at 37°C again for 60 min with TYI to continue the process. Trophozoites were lysed and also cellular fractionation was carried out. Samples were processed for 12% SDS-PAGE and western blot assays. Representative blots of three experiments are shown. T: total extracts, S: soluble fraction, I: insoluble fraction. (E,F) Densitometry of the bands in (D) corresponding to EhNPC1 (E) and EhNPC2 (F) using actin as loading control.
Then, we performed erythrophagocytosis assays in a pulse-chase model to detect protein mobilization by western blot assays. In these experiments, trophozoites were incubated with erythrocytes for 5 min, then, free and adhered erythrocytes were removed by washing samples; and the phagocytic process of the already ingested erythrocytes continued at 37°C for different times. Total proteins and the soluble and insoluble fractions of lysed trophozoites taken at 0, 5 and 5+60 min of phagocytosis were identified by western blot assays. In basal conditions (0 min), EhNPC1 was detected enriched in the insoluble fraction. However, after 5 and 5+60 min of phagocytosis the protein was similarly distributed in the three samples (Fig 7D). By contrast, under basal conditions, EhNPC2 was more abundant in the soluble fraction (Fig 7D). Densitometry analysis using α-actin antibodies as loading control, confirmed this (Fig 7E and 7F). These findings suggest that in basal conditions EhNPC1 is more abundant in membranes, whereas EhNPC2 is concentrated in the cytoplasm; while during phagocytosis they are more homogenously distributed in the entire cells, evidencing the dynamic cholesterol trafficking.
EhNPC1 and EhNPC2 are located in the ER
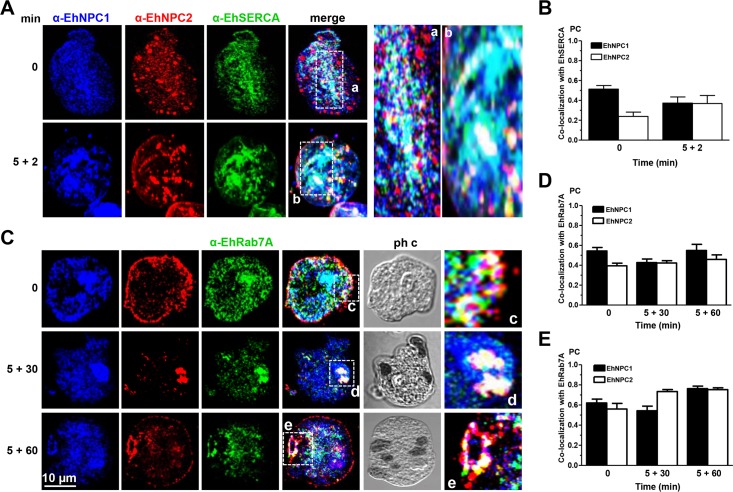
In mammalian cells, NPC1 and NPC2 are located in the ER from where they distribute cholesterol to different organelles. Here we searched for EhNPC1 and EhNPC2 in the ER. Confocal images of trophozoites in basal conditions showed that both proteins co-localized in networks and vacuoles with EhSERCA, an ER marker [33] (Fig 8Aa,b). EhNPC2 also appeared as 1.5 to 2 μm spheres in the cytoplasm (Fig 8A). After 5+2 min phagocytosis, co-localization did not vary significantly (Fig 8A), suggesting that EhNPC1 and EhNPC2 proteins maintain a constant presence in ER, as it has been described for other eukaryotes [34]. PC values of co-localization of EhNPC1 and EhNPC2 with EhSERCA were between 0.25 and 0.51 (Fig 8B). These findings corroborate that the proteins detected around the nuclei are indeed in the ER.
Fig 8. Co-localization of EhNPC1 and EhNPC2 with EhSERCA and EhRab7A.
Trophozoites were incubated with erythrocytes at 37°C and treated as Fig 7. Samples were processed for confocal microscopy using α-EhNPC1, α-EhNPC2 and (A) α-EhSERCA or (C) α-EhRab7A antibodies. (a-e) Magnification of the white squares. (B, D, E) PC of the co-localization between EhNPC1 or EhNPC2 with EhSERCA (B), or with EhRab7A in the whole cell (D) or in cellular structures stained by the three antibodies (E).
EhNPC1 and EhNPC2 co-localize with EhRab7A in endosomes.
The Nozaki group has found that EhRab7A is involved in the retrograde transport from phagosomes to the Golgi apparatus and it is located mainly in late endosomes [35, 36]. Based on their elegant work, we explored whether spherical structures containing EhNPC2 detected in trophozoites, corresponded to endosomes. By laser confocal microscopy, the α-EhRab7A antibody recognized these cytoplasmic structures, which increased in size throughout the chase-time phagocytosis assays (Fig 8C), evidencing that they are endosomes. In basal conditions and throughout phagocytosis, EhRab7A was associated with EhNPC1 and EhNPC2; frequently forming donut-like structures that may be phagolysosomes and MVBs (Fig 8Cc-e). PC values for EhNPC1 and EhNPC2 with EhRab7A were 0.39 to 0.55 in the entire cell (Fig 8D) and increased to 0.47 to 0.78 in the donut-like structures (Fig 8E). These results strengthen the assumption that EhNPC1 and EhNPC2 are also in endosomes.
EhNPC1 and EhNPC2 associate with endosomal molecules in acidic vesicles
In mammals, NPC1 binds to cholesterol in late endosomes in an acidic milieu where LBPA and Alix protein participate in cholesterol homeostasis [22]. We hypothesized that cholesterol containing vesicles inside phagolysosomes, together with EhNPC1 and EhNPC2, also contain LBPA and EhADH [25, 26]. Thus, we investigated the nature of these vesicles during phagocytosis using Lysotracker, a marker of acidic vesicles. After 5+30 and 5+60 min of erythrophagocytosis, more than 90% of the phagosomes were positive for α-EhNPC1, α-EhNPC2 antibodies and Lysotracker (Fig 9A). Lysotracker appeared in phagolysosomes and MVBs, containing partially digested erythrocytes (Fig 9Aa-d). These results show that EhNPC1 and EhNPC2 are in an acidic milieu characteristic of phagolysosomes.
Fig 9. Co-localization and association of EhNPC1 and EhNPC2 with cholesterol and endosomal molecules.
Trophozoites were incubated with erythrocytes as in Fig 7 and processed for confocal microscopy after incubation with α-EhNPC1 and α-EhNPC2 antibodies and (A) Lysotracker or (B) α-LBPA or (C) α-EhADH antibody and corresponding secondary antibodies. (a-d,g,h) Magnification of white squares. (e,f,i,j) Magnifications from other images. Ph c: phase contrast images, arrow in I: EhNPC1 outside endosomes, arrowhead: EhNPC2 inside endosomes. D, E) Trophozoites in basal condition or after 5+60 min of erythrophagocytosis were lysed and immunoprecipitation assays (IP) were performed using α-EhNPC1 (D) or α-EhNPC2 (E) antibodies or preimmune serum (PS). Immunoprecipitated proteins of trophozoites were analyzed by western blot and dot blot experiments, using α-EhNPC1, α-EhNPC2, α-EhADH, α-EhRab7A, α-EhSERCA and α-cholesterol antibodies.
Besides, at 5+30 min, all phagolysosomes showed EhNPC1, EhNPC2 and LBPA (Fig 9Be-g). Moreover, EhADH co-localized with EhNPC1 or EhNPC2 under basal conditions, mainly in the plasma membrane, and, during phagocytosis, in phagolysosomes and MVBs (Fig 9Ch-j). Immunoprecipitation assays using α-EhNPC1 antibody confirmed that this protein is associated under basal conditions and during phagocytosis with EhNPC2, EhADH and EhRab7A, but not with EhSERCA (Fig 9D), However, α-EhNPC2 antibody precipitated all of these proteins including EhSERCA, suggesting that at some point these proteins are interacting (Fig 9E). Cholesterol was also detected in dot blots in both immunoprecipitates (Fig 9D and 9E). These results evidenced that cholesterol, LBPA, EhADH, EhRab7A, EhNPC1 and EhNPC2 co-localize in phagolysosomes and MVBs, probably cooperating to facilitate the phagocytosis-digestion processes; and our data also reinforce the association of EhNPC1 and EhNPC2 with cholesterol.
Trophozoites with stocked cholesterol diminish their rate of phagocytosis and do not disturb the intestinal barrier
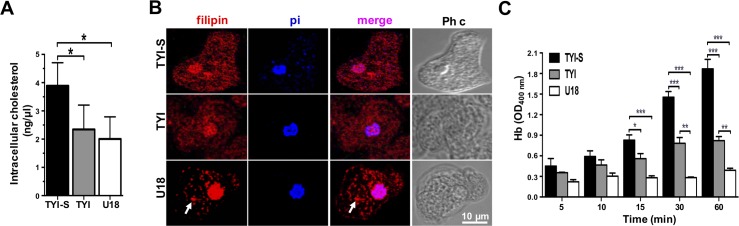
To evaluate the effect of cholesterol trafficking on phagocytosis, we used the U18666A (3-β-[(2-diethyl-amino)ethoxy]androst-5-en-17-one) (U18) drug that binds to NPC1 in the SSD, blocking the cholesterol traffic [37]. For these experiments, trophozoites cultured without serum (TYI), to eliminated part of the cholesterol source, showed less than 50% of the cholesterol concentration that the cells cultured in basal conditions (with serum). Trophozoites cultured without serum and in the presence of U18 presented slight differences in cholesterol concentration (Fig 10A). By confocal microscopy, U18 treated trophozoites, showed filipin-positive abundant clumps in the cytoplasm suggesting cholesterol accumulation (Fig 10B). These results suggest that in E. histolytica, U18 also interferes with the cholesterol trafficking, but not with the cholesterol amount inside the trophozoites (Fig 10A and 10B). However, U18 treated cells ingested at 60 min, 2 erythrocytes/trophozoite, whereas trophozoites cultured in TYI medium engulfed about 6 erythrocytes, and trophozoites cultured in TYI-S medium ingested a mean of 18 erythrocytes (Fig 10C). These findings suggested that the cholesterol arresting by U18 interferes with the rate of phagocytosis.
Fig 10. Erythrophagocytosis of trophozoites cultured in TYI-S, TYI and TYI plus U18.
Trophozoites were incubated ON at 37°C in TYI-S, TYI and TYI plus U18. (A) Cholesterol concentration in the respective trophozoites was measured as described in material and methods. (B) Laser confocal microscopy showing the morphology and the cholesterol localization by filipin staining. Nuclei were counterstained with propidium iodide (pi). Images were false colored to obtain a better contrast. Ph c: phase contrast images, arrows: cytoplasmic dots. (C) Rate of erythrophagocytosis spectrophotometrically measured by hemoglobin (Hb) concentration inside trophozoites.
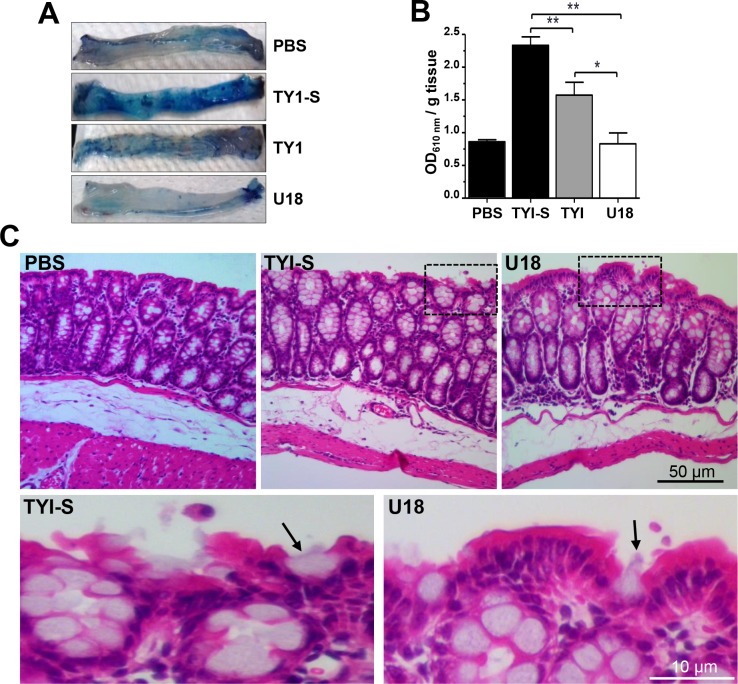
We also analyzed the effect of the cholesterol trafficking arrest by using U18 in the amoebiasis intestinal mice model (strain C57BL/6) [38]. In this model, virulent trophozoites compromise the intestinal epithelial barrier, which could be monitored by Evans blue dye permeability [39]. The intestinal epithelium of mice inoculated with U18-treated trophozoites did not suffer damage, giving similar results to the negative controls (mice inoculated only with PBS), and showing that the epithelial barrier was not disrupted (Fig 11A and 11B). By contrast, trophozoites cultured in TYI induced 62% permeability in colonic epithelium compared to trophozoites cultured in TYI-S medium, that was taken as 100% (Fig 11A and 11B). Thus, while cholesterol depletion only ameliorated the capacity of trophozoites to induce intestinal epithelial permeability, U18 treatment completely inhibited it. Tissue sections of colon epithelium stained with hematoxylin-eosin showed that trophozoites grown in TYI-S medium provoked discontinuity in epithelial layer, cells rounding, and a higher number of cells layers, cellular infiltration and swelling, whereas animals inoculated with U18 treated trophozoites presented normal epithelium, similar to the negative controls (Fig 11C). Our results showed that cholesterol is necessary for trophozoites to ingest red blood cells, and to impair the intestinal epithelial barrier.
Fig 11. In vivo virulence of trophozoites cultured in TYI-S, TYI and TYI plus U18.
Capability of trophozoites to impair the mouse intestinal barrier. (A) Distal parts of the mouse colons after treatment with trophozoites or with PBS. Intestinal barrier impairment was measured as the ability of the Evans blue dye to permeate the mouse intestinal epithelium after contact with trophozoites. n = 5. (B) Data represent the mean ± standard error. PBS: mice undergoing surgery, but not inoculated with trophozoites. * p<0.05, ** p<0.01. (C) Hematoxylin-eosin staining of tissues. Squares were magnified in the corresponding lower panels. Arrows: trophozoites.
Knockdown of Ehnpc1 and Ehnpc2 genes diminishes rate of phagocytosis and cell motility
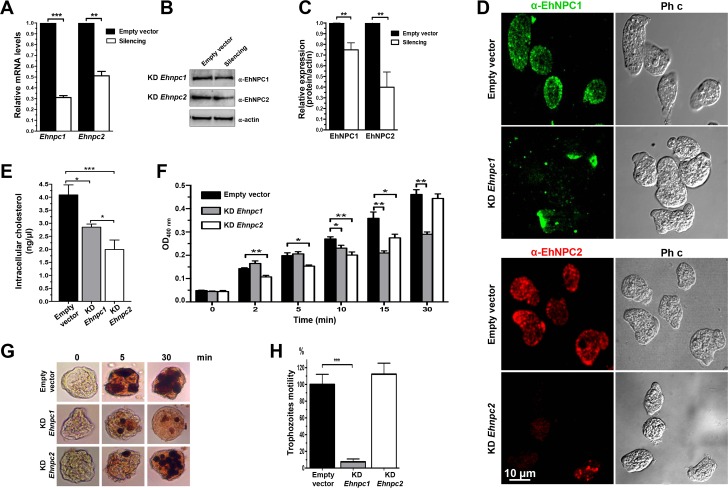
Phagocytosis and cell motility require cholesterol for membrane synthesis and fluidity. To get further evidence that EhNPC1 and EhNPC2 proteins are responsible for transporting the cholesterol for these events, we knocked down trophozoites in the Ehnpc1 and Ehnpc2 genes, using trophozoites of clone G3 and the psAP-2 vector to transfect them [40]. Both types of silenced trophozoites (KD Ehnpc1 and KD Ehnpc2) expressed 30 and 50% of Ehnpc1 and Ehnpc2 mRNA, respectively, compared with the G3 trophozoites transfected only with the empty vector (Fig 12A). Protein expression was also diminished in silenced trophozoites in basal conditions; although, surprisingly, protein level decreased more in EhNPC2 than in EhNPC1 (Fig 12B and 12C), even when E. histolytica has two Ehnpc2 genes that are transcribed (Fig 3), and only Ehnpc2a was silenced. These suggest mechanisms for a fine regulation in the level or in the half-life of the proteins that might be elucidated in the future. Confocal microscopy images confirmed the decrease of both proteins in mutant trophozoites in basal conditions; and showed that proteins were re-localized (Fig 12D). In KD Ehnpc1 trophozoites, EhNPC1 protein was polarized in the plasma membrane, whereas, in KD Ehnpc2 cells, EhNPC2 was scarcely found in plasma membrane and it was detected as few cytoplasmic dots (Fig 12D). KD Ehnpc2 trophozoites presented a lesser amount of intracellular cholesterol (1.9 ng/μl) than KD Ehnpc1 (2.8 ng/μl) whereas, the trophozoites transfected with the empty vector presented 4.0 ng/μl (Fig 12E), strongly suggesting the participation of these proteins in cellular cholesterol uptake.
Fig 12. Silencing of Ehnpc1 and Ehnpc2 genes in E. histolytica.
(A) Trophozoites clone G3 were transfected with psAP-2, psAP-2Ehnpc1 or psAP-2Ehnpc2a plasmids and stable populations were selected with 4 μg/ml G-418. RT-qPCR assays were performed using mRNA from transfected trophozoites (empty vector, KD Ehnpc1 and KD Ehnpc2), using specific primers for Ehnpc1 and Ehnpc2a genes and as a housekeeping the 40s ribosomal S2 protein gene. (B) Western blot assays of transfected trophozoites extracts, using α-EhNPC1 and α-EhNPC2 antibodies and respectively secondary antibody. As a loading control, the same membrane was reblotted with α-actin antibodies. (C) Densitometry analysis of bands showed in (B) normalized again actin protein. (D) Confocal microscopy of transfected trophozoites in basal conditions using α-EhNPC1 or α-EhNPC2 antibodies. Ph c: phase contrast. (E) Cholesterol concentration in the transfected trophozoites was measured as described in material and methods. (F) Rate of erythrophagocytosis of transfected trophozoites. * p<0.05, ** p<0.01. (G) Diaminobenzidine-stained trophozoites that ingested erythrocytes for different times. (H) Motility assays of transfected trophozoites cultured in transwell inserts. *** p<0.001.
The rate of erythrophagocytosis was also modified in silenced trophozoites. In early times of phagocytosis (2 to 5 min), KD Ehnpc1 and control trophozoites ingested the same amount of erythrocytes (6 erythrocytes/trophozoite). However, after 5 min, KD Ehnpc1 trophozoites arrived to a plateau, ingesting a mean of 10 erythrocytes/trophozoite, in comparison with control trophozoites that continued erythrocytes ingestion, reaching 18 erythrocytes/trophozoite at 30 min (Fig 12F and 12G). On the other hand, from 2 to 15 min of phagocytosis, the KD Ehnpc2 trophozoites ingested a mean of 4 to 10 erythrocytes/trophozoite in comparison with control trophozoites, which ingested a mean of 5.6 to 14.4 erythrocytes/trophozoite. However, at 30 min of phagocytosis, KD Ehnpc2 trophozoites recuperated the rate of phagocytosis and ingested a mean of 18 erythrocytes/trophozoite, similar to control trophozoites (Fig 12F and 12G). These results indicate that in the KD Ehnpc2 trophozoites, the rate of erythrophagocytosis was affected only in early times, whereas, in KD Ehnpc1 trophozoites, the erythrophagocytosis was affected after 10 min of contact with erythrocytes.
To evaluate the impact of EhNPC1 and EhNPC2 proteins on motility, we placed the transfected trophozoites in the upper chamber of transwell inserts and counted the number of trophozoites that were able to move toward the lower chamber, following a serum stimulus. Interestingly, after 3 h incubation, KD Ehnpc1 trophozoites remained in the upper chamber, showing migration incapacity, whereas, KD Ehnpc2 trophozoites showed similar motility than the control. (Fig 12H). These findings altogether revealed that EhNPC1 and EhNPC2 proteins have no redundant functions but they cooperate during cholesterol trafficking.
Discussion
Using in silico analysis and distinct experimental approaches, we have shown here that the early emerging protozoan E. histolytica has one Ehnpc1 and two Ehnpc2 genes. In contrast, most eukaryotes have two npc1 (npc1 and npc1l1) and one npc2 genes [14, 29, 32]. Molecular docking, co-localization and immunoprecipitation assays revealed that EhNPC1 and EhNPC2 proteins can interact between them and with cholesterol. Moreover, they associate with EhRab7A and EhADH proteins, whereas only EhNPC2 interacts with EhSERCA, although both proteins co-localized, immunoprecipitation assays showed no interaction between them at the times explored. Experiments with silenced trophozoites in Ehnpc1 and Ehnpc2 genes gave evidence that their corresponding proteins do not carry out redundant functions, but they cooperate in the cholesterol trafficking. The relevance of our findings lies in three main facts: firstly, cholesterol is fundamental for endocytosis and motility; both important for nutrition and virulence of E. histolytica [2–4, 8]. Secondly, cholesterol is not synthesized by trophozoites and very little is known on cholesterol trafficking in protozoa. In particular, the mechanism of cholesterol uptake and trafficking in E. histolytica trophozoites was almost unknown, except for the role of TMK39 protein in LDL proteins uptake [24], which could function in association with other proteins, such as EhNPC1 and EhNPC2, because it has not the canonical cholesterol binding sites [24]. In protozoa, except for a NPC1 like protein identified in T. gondii [41], no other references were found by us about the presence and function of NPC1 and NPC2; their genes only appear in the databases, but no experimental characterization has been done. This makes E. histolytica the first protozoa in which both genes and proteins has been identified and characterized. Thirdly, E. histolytica has a simple cholesterol trafficking, compared to other eukaryotes, because it depends on exogenous cholesterol, providing an excellent and less complicated model to elucidate this intricate event in eukaryotic cells.
NPC1 and NPC2 are ancient proteins, highly conserved throughout eukaryotic evolution [42]; to the extent that human and yeast npc1 genes are genetically and functionally interchangeable [42]. Our results fortify this assumption by the high structural homology presented by human NTD-NPC1 and the B. taurus NPC2 crystals with EhNPC1 and EhNPC2 3D structures, respectively. The recent publication elucidating the 3D structure of the complete NPC1 protein in human [37] gives support to our model. In contrast to other eukaryotes, EhNPC2a and EhNPC2b present an amino acids tail with several residues that could be in contact with membranes. This, together with the positively charged protein regions [17], gives to EhNPC2 proteins the possibility of translocation across the plasma membrane and other organelle membranes, as our experimental approaches have suggested. These characteristics give further support to the hypothesis of the "hand-off" model, which can be working also in E. histolytica for cholesterol uptake and trafficking [19]. Immunoprecipitation assays using α-EhNPC1 or α-EhNPC2 antibodies confirmed that both proteins are associated, among them and with cholesterol, in trophozoites in basal conditions and during phagocytosis although sometimes they also appeared separated, supporting the "hand-off" model.
In this study we also asked: How do E. histolytica trophozoites bring exogenous cholesterol to their distinct organelles without a canonical LDL receptor?
Now, have evidences that EhNPC1 and EhNPC2 proteins carry cholesterol and participate in all processes from the sensing of external cholesterol sources to the digestion of the endocytosed products in phagolysosomes. Proximity and co-localization of EhNPC1 and EhNPC2 in the trophozoites surface immediately after ABS pulse and at the beginning of erythrophagocytosis, as well as their detection in secreted products, suggest that they are in charge of cholesterol capturing in E. histolytica. Our results do not discard the participation of other proteins, such as TMK39 protein.
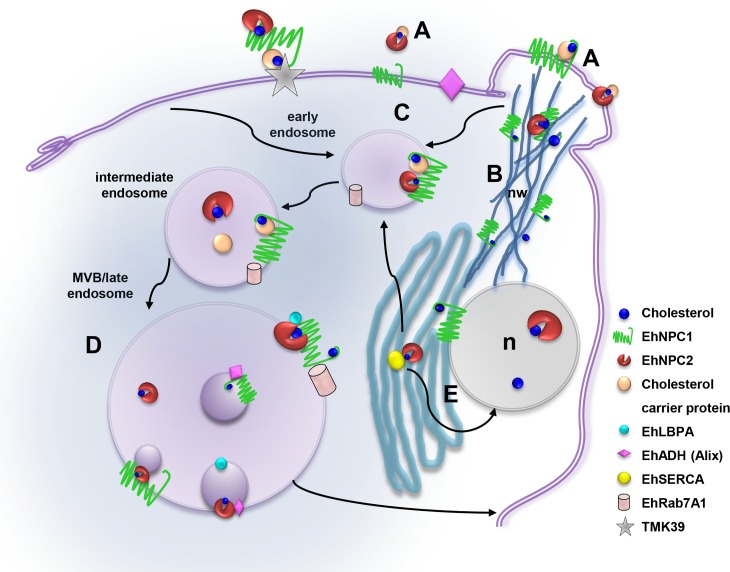
Experiments using serum-starved trophozoites suggest that the external cholesterol is taken up into the cell by EhNPC1 and EhNPC2 in several steps: i) Immediately after sensing the cholesterol, EhNPC1 and EhNPC2 are organized in the plasma membrane in a punctuated pattern, co-localizing in some regions, but staying close and uncomplexed in some areas. It is plausible to speculate that EhNPC1 facilitates the transport of EhNPC2 to the plasma membrane to uptake exogenous cholesterol, but more experiments are necessary to probe this. ii) Next, small membrane projections or putative vacuoles that grow in size and amount throughout the endocytosis process appear at early times of endocytosis that are full of EhNPC1 and EhNPC2. iii) Then, EhNPC2-containing vesicles appear outside the cell, assuming rod- and sphere-like shapes that are in contact with vesicles full of cholesterol. It was also possible to distinguish in them small spots recognized by α-EhNPC1. Secretion assays confirmed that EhNPC1 and EhNPC2 are indeed released into the medium, probably to capture cholesterol. iv) EhNPC2 appears in vesicles co-localizing with cholesterol inside trophozoites, frequently making contact with smaller EhNPC1 containing vesicles. v) EhNPC1 and cholesterol formed networks from the plasma membrane to the nucleus and the ER; and also EhNPC2 appears there. vi) EhNPC1 and EhNPC2 accumulate in the ER and the nucleus. We do not know yet the significance of these proteins in the nucleus. vii) Cholesterol was driven to the plasma membrane and to vacuoles that are surrounded by EhNPC1 with EhNPC2 inside. All these steps are summarized in the cartoon of Fig 13.
Fig 13. Working model of the EhNPC1 and EhNPC2 participation in cholesterol trafficking in E. histolytica.
(A) Cholesterol uptake: EhNPC1 and EhNPC2 capture the cholesterol in the extracellular space and in the plasma membrane probably following the “hand-off” model (EhNPC1-cholesterol-EhNPC2-cholesterol-EhNPC1) and with the TMK39 participation. (B) Network-like structures formation: EhNPC1 and cholesterol form networks from cholesterol-containing membrane protrusions to ER, endosomes and nucleus that possibly facilitate cholesterol trafficking through EhNPC2 in a “hand-off” model. (C) Cholesterol influx: EhNPC1 and EhNPC2 associate with cholesterol and are internalized in EhRab-7A containing endosomes under basal conditions and during erythrophagocytosis. (D) EhNPC1 and EhNPC2 associate with phagolysosomal and MBVs molecules, particularly with LBPA and EhADH, which probably regulate cholesterol trafficking. (E) EhNPC1 and EhNPC2 accumulate in nucleus (n) and ER and may be distributed from there to other organelles.
Through erythrophagocytosis, most of these steps are also evident with some particularities. i) Co-localization of EhNPC1 and EhNPC2 is starting after two minutes contact or before. ii) Adhered, but not yet ingested, erythrocytes appear covered by EhNPC2 protein before target cell ingestion is finished. Trophozoites presented a channel, with partially ingested erythrocytes strongly stained by the α-EhNPC2 antibody and filipin, whereas the external part of the phagocytic cup was detected by α-EhNPC1 antibody. iii) At 30 and 90 min after phagocytosis, cholesterol appeared together with EhNPC1 and EhNPC2 in vacuoles, some with a relative uniform size, inside of phagolysosomes or MVBs. Interestingly, EhRab7A, LBPA, EhADH and cholesterol co-localized in late endosomes with EhNPC1 and EhNPC2, suggesting that they may participate in the cholesterol trafficking and homeostasis, as in mammalian cells [22]. vi) Cholesterol returned to plasma membrane following a similar movement than EhNPC1 and EhNPC2. It is possible that EhNPC1 and EhNPC2 tasks do not end until the digestion process is finished and no more cholesterol is available in the phagocytosed erythrocytes (Fig 13).
The phagocytosis process relies on cholesterol trafficking through the distinct type of endosomes formation. Here we used two strategies to evaluate this: i) we arrested the cholesterol trafficking by U18 drug which binds to the SSD sites in NPC1 [37, 43]. In our experiments we confirmed that the intracellular cholesterol concentration did not vary in comparison with TYI cultures trophozoites, but it accumulates in clumps and severely affected phagocytosis and in vivo virulence. Mice inoculated with U18 treated trophozoites did not present damage in the colon epithelium. ii) Experiments with trophozoites knocked down in Ehnpc1 and Ehnpc2a genes gave strong support to the fact that both genes are important in cholesterol uptake. The diminish expression of proteins, provoked a reduction in the rate of phagocytosis. Interestingly, Ehnpc1 knocked down trophozoites presented a dramatic decrease in motility, strongly suggesting a role of EhNPC1 protein in this function. We do not speculate on the role of EhNPC2 protein, because the presence of two genes and a possible compensatory mechanism in the cell could promote the transcription of the Ehnpc2b gene after the stimulus given by serum or erythrocytes. Another possibility is a half-life increase of the EhNPC2 protein. It is currently under study in our laboratory.
In conclusion, we provide the first evidence that E. histolytica presented two cholesterol transporting proteins, EhNPC1 and EhNPC2, that participate in exogenous cholesterol uptake and in its cellular trafficking, impacting in motility and phagocytosis. Future studies need to be addressed to fully elucidate remaining questions: How do EhNPC1 and EhNPC2 arrive to the plasma membrane and leave the cell? Combining our findings with results in other eukaryotes [44] we propose that EhNPC2 is transported by EhNPC1 to these places. How does EhNPC2 enter to the phagocytosed erythrocytes? We hypothesized that the “hand-off” model proposed for mammalian cells could be also applicable to E. histolytica. This implies that EhNPC2 moves to the plasma membrane, together with EhNPC1 and from there, cholesterol is “handed-off” from one protein to another, avoiding cholesterol trafficking through the hydrophobic medium, efficiently reaching its target organelles and facilitating the expression of virulence properties that require membrane synthesis and vacuoles fusion. The importance of cholesterol transport for the virulence and the involvement of EhNPC1 and EhNPC2 in this process, make these proteins promising targets for developing better strategies to defeat the amoebiasis.
Materials and Methods
E. histolytica cultures
Trophozoites of E. histolytica, clone A, strain HM1:IMSS [45] (already-existing collection) and clone G3 [40] were axenically grown at 37°C in TYI-S-33 medium (TYI-S) and harvested at logarithmic growth phase by chilling the culture flasks at 4°C [46]. For some experiments, trophozoites were cultured in TYI medium without serum during 12 h (TYI) or in TYI supplemented with 3 μg/ml of U18 during 12 h (Sigma-Aldrich). Then, fresh medium with or without the drug was added to proceed with the experiments. All experiments presented here were performed at least three times in duplicate.
In silico analysis and phylogenetic trees construction
Human NPC1 (access number: hsa 4864) and bovine NPC2 (access number: bta 280815) protein sequences retrieved from the KEGG database (http://www.genome.jp/kegg/) were used as query to search putative E. histolytica EhNPC1 and EhNPC2 proteins and their orthologues. Structural domains were identified using the SMART genomics server (http://smart.embl-heidelberg.de/). Identity and e-value between EhNPC1 and EhNPC2 hypothetical proteins, compared with their orthologues, were determined using the Expert Protein Analysis System (ExPASy) of the Proteomic Analysis Server from the NCBI Blast service program (http://www.expasy.org/). The predicted amino acid sequences of EhNPC1, EhNPC2a and EhNPC2b (ehi_080220, ehi_068260 and ehi_188770, respectively) were aligned with orthologues sequences by ClustalW and data were submitted to phylogenetic analysis by UPGMA using MEGA 5.05 software [47]. Bootstrapping was performed for 1000 replicates.
3D structure modeling and molecular docking
The amino acid sequences of EhNPC1, EhNPC2a and EhNPC2b were analyzed using the RaptorX server (http://raptorx.uchicago.edu/) and predicted structures and orientation were obtained via the OPM database (http://opm.phar.umich.edu/). The 3D NTD EhNPC1 model (1 to 250 amino acids) was compared with the human NTD-NPC1 (3GKI) (23 to 254 amino acids) crystal, whereas the EhNPC2 3D structure (full amino acid sequence) was compared with the B. taurus NPC2 crystal (2HKA). Both crystals were retrieved from the Protein Data Bank (PDB). Templates of other NPC1 and NPC2 proteins, located at PDB, were used to obtain the EhNPC1, EhNPC2a and EhNPC2b 3D structures, which were visualized using the UCSF Chimera software. Molecular docking with cholesterol was performed for the three proteins using the AutoDockTools V1.5.6 program, and the interaction sites were analyzed by Ligplotv.4.5.3 program (http://www.ebi.ac.uk/thorntonsrv/software/LIGPLOT/). The grid size was 126x126x126 points with a 0.8 Å spacing. Dockings were performed with the empiric free energy function and the Lamarckian genetic algorithm. Number of GA runs was 200, using a maximum number for evaluation of 2x106. Cholesterol structure was obtained from the ZINC12 database (zinc.docking.org) in PDB format and energy minimization was obtained through PRODGR (davapc1.boich.dundee.ac.us/prodrg/) and the CHIMERAV1.10.1 software.
PCR and RT-PCR assays
Genomic DNA and total RNA were isolated from trophozoites using the Wizard Genomic DNA Purification kit (Promega) and Trizol reagent (Invitrogen), respectively, according to the manufacturer’s recommendations. cDNA was synthesized using oligo dT primers and the Superscript II reverse transcriptase (Invitrogen). PCR amplifications were carried out using 200 ng of DNA or cDNA as template and specific primers for Ehnpc1 or Ehnpc2a or Ehnpc2b genes (S3 Table). We used 20 μl reaction volume containing 0.5 μM each primer, 2 mM MgCl2, 200 μM dNTPs, 1X Taq buffer and 1 U Taq DNA polymerase (Invitrogen). Cycling conditions included an initial denaturing step at 94°C for 1 min, followed by 30 cycles of 94°C for 1 min, 50 or 55°C (according to respective Tm) for 1 min, and 72°C for 3 min, with a final extension step at 72°C for 7 min. Products were separated by electrophoresis in 1% agarose gels and then, cloned and sequenced. As controls, for PCR amplification of DNA we omitted the DNA in the reaction mixture, and for RT-PCR we used DNAse-treated RNA as template. For RT-PCR assays, 100 ng of cDNA, 0.15 μM of each primer (S3 Table) and the KAPA SYBR FAST PCR Master Mix (Kapa Biosystems) were used in a StepOneTM Real-Time PCR System (Applied Biosystem). Data from three independent cDNA preparations were analyzed using the 2−ΔΔCt method with ribosomal 40s S2 protein as house-keeping gene.
Production of α-EhNPC1 and α-EhNPC2 antibodies
The DEQPMYDKDGQYVPVEKRLE polypeptide from EhNPC1 (776 to 785 amino acids) was synthesized together with the KLH (Keyhole Limpet Hemocyanin) tag to increase its immunogenicity (GenScript). New Zeland rabbits (already-existing collection) were immunized first with100 μg of this polypeptide resuspended in Titermax Gold adjuvant (1:1) (Sigma) and then, with two more weekly doses of 50 μg each, to generate the α-EhNPC1 antibody.
The Ehnpc2a and Ehnpc2b full-length genes were PCR-amplified using cDNA as template and specific primers, which introduced unique BamHI and SalI restriction enzyme sites, in the sense and antisense primers, respectively (underlined in S3 Table). Genes were cloned into the pGEX6P-1 plasmid to generate pGEX6P-Ehnpc2a and pGEX6P-Ehnpc2b constructs. E. coli pLys-S bacteria were transformed with the plasmids, then, recombinant proteins were obtained by 0.1 mM IPTG induction. Recombinant proteins were electro-eluted and purified by size exclusion chromatography using a PD-10 column (GE Healthcare). Then, 60 μg of each purified recombinant protein (rEhNCP2a and rEhNPC2b) were emulsified in Titer-Max Gold adjuvant (Sigma) and subcutaneously and intramuscularly inoculated in Wistar rats (already-existing collection) and BALB/C mice (already-existing collection), respectively. Two more doses (30 μg) of each protein were injected at 20 day intervals followed by bleeding to obtain antibodies. In all experiments, we used the rat α-EhNPC2a antibody (α-EhNPC2) as pan antibody, because it detected both EhNPC2a and EhNPC2b recombinant proteins. Pre-immune serum was obtained before immunizations.
Cell fractionation and western and dot blot assays
Trophozoite extracts (30 μg) were centrifuged 10 min at 13,000xg, and, soluble (supernatant) and insoluble (pellet) fractions were obtained. Samples were separated by 10 or 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membranes and probed with rabbit α-EhNPC1 (1:3000) or rat α-EhNPC2 (1:3000) or rabbit α-EhADH (1:500) [24] or rabbit α-EhRab7A (1:200) (kindly provided by Dr. Tomoyoshi Nozaki, National Institute of Infectious Diseases, Tokyo, Japan) [36] or rabbit α-EhCP112 (1:3000) [48] or rabbit α-SERCA (1:200) [32] or mouse α-actin (1:200) [49] antibodies. Membranes were washed and incubated with the species-specific HRP-labeled secondary antibodies (Zymed; 1:10000), and developed with ECL Prime detection reagent (GE-Healthcare). Pre-immune sera were used as controls. For dot blots analysis, samples were prepared as described [26], dripped on nitrocellulose membranes and treated with rabbit α-cholesterol antibodies (1:100). (Cloud Clone Co) followed by HRP-labeled secondary antibodies.
Laser confocal microscopy experiments
Trophozoites (grown in coverslips) in basal conditions or after phagocytosis were fixed with 4% paraformaldehyde (PFA) at 37°C for 1 h, permeabilized with 0.2% Triton X-100 and blocked with 10% fetal bovine serum (FBS) in PBS. Then, preparations were incubated at 37°C for 1 h with α-EhNPC1 (1:100) or α-EhNPC2 (1:100) or α-EhADH (1:500) or α-EhRab7A (1:500) or α-Gal/GalNAc lectin (1:50) (kindly provided by Dr. W. Petri, University of Virginia, Charlottesville, USA) [50] or α-LBPA (1:30) [23, 26] or α-EhSERCA (1:1000) antibodies; followed by extensive washing and incubation for 1 h with species-specific FITC-, TRITC- or Cy5- labeled secondary antibodies (Zymed; 1:100) as appropriate. In some cases, nuclei were counterstained with propidium iodide (0.1 μg/ml) (Sigma) for 5 min. For cholesterol detection, cells were stained with 250 μg/ml filipin (Sigma). For acidic vacuoles detection, fixed trophozoites were incubated with 2 μg/ml of Lysotracker (Molecular Probes) for 2 h at 37°C. We also performed experiments using 12 h ABS-starved trophozoites (incubated in TYI medium), cultured on coverslips, and challenged with 100 μl of ABS for 0.5 to 7 min at 37°C, then, samples were processed as above. All preparations were preserved using Vecta Shield antifade reagent (Vector), examined using a Carl Zeiss LMS 700 confocal microscope and processed with ZEN 2009 Light Edition Software (Zeiss). To evaluate the co-localization between molecules, Pearson coefficients (PC) were obtained from at least 15 confocal independent images (laser sections: 0.5 μm) using the ImageJ 1.45v software and the JACoP plugin [51].
Transmission electron microscopy (TEM)
For immunogold-labeling experiments, trophozoites in basal conditions were fixed with 4% PFA and 0.5% glutaraldehyde in PBS for 1 h at room temperature (RT). Samples were embedded in LR White resin (London Resin Co) and polymerized under UV at 4°C for 48 h. Thin sections (60 nm) were mounted on formvar-covered nickel grids followed by overnight (ON) incubation with α-EhNPC1 or α-EhNPC2 antibodies (1:20) and, then, incubated ON, with the respective secondary antibodies (1:60) conjugated to 20 and 10 nm gold particles, respectively (Ted Pella Inc.). Thin sections were contrasted with uranyl acetate and lead citrate and observed with a Jeol JEM-1011 transmission electron microscope. Number of gold particles was counted in 12 independent images of 10 μm2 each.
Cholesterol quantification
Trophozoites (5x105) were re-suspended in a mixture of chloroform: isopropanol:Nonidet P40 (7:11:1) and centrifuged at 13,000xg for 10 min. The organic phase was collected and dried at 50°C. Lipid samples were processed according to the Cholesterol Quantitation kit (Sigma)
Secretion assay
Trophozoites were grown in TYI medium for 12 h and incubated with 100 μl of ABS for 0.5, 5 and 15 min at 37°C. Then, medium was collected in the presence of 1 mg/ml of E64 (Sigma) and centrifuged at 13,000xg for 10 min to obtain the secreted molecules in the supernatant fraction. To obtain the trophozoite extracts, cells were washed and lysed with proteases inhibitors as reported [52]. Samples were submitted to 12% SDS-PAGE and western blot assays using α-EhNPC1, or α-EhNPC2, or α-cholesterol, or α-EhCP112 or α-actin antibodies as described above.
Immunoprecipitation assays
Trophozoites were lysed in the presence of 10 mM Tris-HCl, 50 mM NaCl and protease inhibitors by freeze-thawing cycles and vortexing. Immunoprecipitation assays were performed using 200 μl of protein G-agarose (Invitrogen) and α-EhNPC1 or α-EhNPC2 antibodies as described [53]. Immunoprecipitates were analyzed by SDS-PAGE and western blot assays using α-EhNPC1 or α-EhNPC2 or α-EhADH or α-EhRab7A or α-EhSERCA or α-cholesterol antibodies, as described [24].
Phagocytosis assays
Trophozoites were incubated with human erythrocytes (already-existing collection) (1:25) at 37°C (for phagocytosis assays) for 5 to 60 min. For pulse-chase experiments, incubation was carried out for 5 min at 37°C. Then, preparations were quickly washed three times with TYI-water (1:1) to remove the adhered and non-ingested erythrocytes. Adhered and non-ingested erythrocytes were lysed by incubation in distilled water for 10 min at RT. Then, trophozoites and ingested erythrocytes were lysed using absolute formic acid and the hemoglobin was quantified by spectrophotometry at 400 nm [54]. In some experiments ingested erythrocytes were stained by Novikoff solution [55] and samples were observed through the light microscope (Axiolab, Zeiss).
In vivo virulence assays
Mice C57BL/6 strain (already-existing collection, 8–12 week old in a weight range of 22–30 g each), were anesthetized by an intraperitoneal injection of 25 mg/ml of ketamine and 2.5 mg/ml of xylazine in PBS. Then, 106 trophozoites were anally inoculated (with previously glycerol lubrication) in five mice per condition. As a control, mice were inoculated with PBS. After 30 min, laparotomy was performed, a cannula was inserted through a small incision in the proximal colon and the colon was washed extensively with PBS. After this, using the same cannula, Evans blue dye was administrated into the colon and mice were left at RT for 15 min. Then, anesthetized mice were euthanized by cervical dislocation. Subsequently, the colon was flushed with abundant PBS to remove the dye, followed by washing with 1 mM N-acetylcysteine to remove dye from mucus. Colon was dissected, weighted and the leaked dye was extracted with gentle shaking in 2 ml dimethyl formamide and spectrophotometrically measured at 620 nm. Permeability was calculated as OD620 per g tissue.
Hematoxylin and eosin staining
Colon samples were fixed in 10% phosphate-buffered formalin and processed for conventional embedding paraffin, then, 4–6 μm sections were stained with hematoxylin and eosin [56]. Samples were analyzed using a light microscope (Axiolab, Zeiss).
Plasmid construction for silencing experiments
The first 400 or 420 bp from the 5′-end of Ehnpc1 and Ehnpc2a genes, respectively, were PCR amplified using specific primers (S3 Table) and cloned into the vector psAP-2 downstream of the 5′ upstream segment (473 bp) of the ap-a gene [40].
Transfection assays
Trophozoites of clone G3 were transfected as described [57]. Briefly, G3 trophozoites were cultured in 35-mm Petri dishes and transfected with 20 μg of the corresponding plasmid: psAP-2-Ehnpc1 (1–400 bp) or psAP-2-Ehnpc2a (1–420 bp) containing gene fragments, using SuperFect (Qiagen) reagent. The transfected parasites were incubated for 48 h at 37°C, selected by 4 μg/ml of G-418 (Sigma-Aldrich) and maintained as a stable cell line. Then, Ehnpc1 and Ehnpc2a silencing was confirmed by RT-qPCR, western blot analysis and immunofluorescence.
Migration assays
Serum-starved (3 h) trophozoites (7.5x104) were placed in the upper chamber of transwell inserts (5 μm pore size, 24 well, Costar) and 500 μl of ABS were added to the lower chamber. Trophozoites were then incubated for 3 h at 37°C. At the end of the incubation, the inserts and media were removed, and trophozoite migration was determined by counting the number of trophozoites that were attached to the lower chamber of the well.
Statistical analysis
Values of all assays were expressed as mean ± standard error of three independent experiments, each in duplicate. Statistical analyzes were carried out using the GraphPad Prism V 5.01 software by Anova or Studentˈs test.
Ethic Statements
The Institutional Animal Care and Use Committee (IACUC) ethics committee reviewed and approved the animal care and use of mice, rabbits and rats to produce antibodies and mice used in virulence in vivo experiments (Protocol Number 0313–06) by the document CICUAL 001, in which is specified that our institute fulfils the NOM-062-ZOO-1999 that deals with the Technical Specifications for Production, Care and Use of Laboratory Animals given by the General Direction of Animal Health of the Minister of Agriculture (SAGARPA), that verify the fulfil of the international regulations/guidelines for the use and care of animals used in laboratory and has verified and approved the animal care at CINVESTAV (Verification Approval Number: BOO.02.03.02.01.908).
Supporting Information
Western blot assays of rEhNPC2a and rEhNPC2b recombinant proteins immunodetected with rat α-EhNPC2a antibodies. Trophozoites lysates (ET) were used as a positive control.
(TIF)
(DOCX)
(DOCX)
BamHI and SalI restriction enzyme sites are underlined in forward and reverser primers, respectively.
(DOCX)
Acknowledgments
Authors are deeply grateful to Dr. Mineko Shibayama (Department of Infectomics and Molecular Pathogenesis, CINVESTAV-IPN) and María Eugenia Mendoza (Department of Physiology, Biophysics and Neurosciences, CINVESTAV-IPN) for their valuable advises and help for the animal experiments. The authors thank Tomás Sánchez and Alejandrina Reyes for technical support.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the National Council of Science and Technology of Mexico (CONACYT; Grant number 220049). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Mortimer L, Chadee K. The immunopathogenesis of Entamoeba histolytica. Exp Parasitol. 2010;126(3):366–80. Epub 2010/03/23. S0014-4894(10)00090-1 [pii]. 10.1016/j.exppara.2010.03.005 [DOI] [PubMed] [Google Scholar]
- 2.Serrano-Luna J, Gutierrez-Meza M, Mejia-Zepeda R, Galindo-Gomez S, Tsutsumi V, Shibayama M. Effect of phosphatidylcholine-cholesterol liposomes on Entamoeba histolytica virulence. Can J Microbiol. 2010;56(12):987–95. Epub 2010/12/18. w10-088 [pii]. 10.1139/W10-088 [DOI] [PubMed] [Google Scholar]
- 3.Das SR, Singh BN. Virulence of Strains of Entamoeba Histolytica to Rats & Guinea-Pigs, & Effect of Cholesterol on Virulence. Indian J Exp Biol. 1965;3:106–9. Epub 1965/04/01. [PubMed] [Google Scholar]
- 4.Mittal K, Welter BH, Temesvari LA. Entamoeba histolytica: lipid rafts are involved in adhesion of trophozoites to host extracellular matrix components. Exp Parasitol. 2008;120(2):127–34. Epub 2008/07/01. S0014-4894(08)00146-X [pii]. PubMed Central PMCID: PMC2615684. 10.1016/j.exppara.2008.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Welter BH, Goldston AM, Temesvari LA. Localisation to lipid rafts correlates with increased function of the Gal/GalNAc lectin in the human protozoan parasite, Entamoeba histolytica. Int J Parasitol. 2011;41(13–14):1409–19. Epub 2011/11/17. S0020-7519(11)00252-9 [pii]. PubMed Central PMCID: PMC3232469. 10.1016/j.ijpara.2011.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loftus B, Anderson I, Davies R, Alsmark UC, Samuelson J, Amedeo P, et al. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433(7028):865–8. Epub 2005/02/25. nature03291 [pii]. 10.1038/nature03291 [DOI] [PubMed] [Google Scholar]
- 7.Mata-Cardenas BD, Vargas-Villarreal J, Martinez-Rodriguez HG, Castro-Garza J, Gonzalez-Garza MT, Said-Fernandez S. Auxotrophy to lipoproteins of Entamoeba histolytica cultivated under axenic conditions. Parasitol Res. 2000;86(12):1018–21. Epub 2001/01/02. [DOI] [PubMed] [Google Scholar]
- 8.Latour NG, Reeves RE, Guidry MA. Steroid Requirement of Entamoeba Histolytica. Exp Parasitol. 1965;16:18–22. Epub 1965/02/01. [DOI] [PubMed] [Google Scholar]
- 9.Gaylor JL, Delwiche CV. Investigation of the multienzymic system of microsomal cholesterol biosynthesis. Ann N Y Acad Sci. 1973;212:122–38. Epub 1973/01/01. [DOI] [PubMed] [Google Scholar]
- 10.Ikonen E. Cellular cholesterol trafficking and compartmentalization. Nat Rev Mol Cell Biol. 2008;9(2):125–38. Epub 2008/01/25. nrm2336 [pii]. 10.1038/nrm2336 [DOI] [PubMed] [Google Scholar]
- 11.Vanier MT, Millat G. Niemann-Pick disease type C. Clin Genet. 2003;64(4):269–81. Epub 2003/09/17. 147 [pii]. [DOI] [PubMed] [Google Scholar]
- 12.Akaboshi S, Ohno K. [Niemann-Pick disease type C]. Nihon Rinsho. 1995;53(12):3036–40. Epub 1995/12/01. [PubMed] [Google Scholar]
- 13.Ko DC, Gordon MD, Jin JY, Scott MP. Dynamic movements of organelles containing Niemann-Pick C1 protein: NPC1 involvement in late endocytic events. Mol Biol Cell. 2001;12(3):601–14. Epub 2001/03/17. PubMed Central PMCID: PMC30967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies JP, Ioannou YA. Topological analysis of Niemann-Pick C1 protein reveals that the membrane orientation of the putative sterol-sensing domain is identical to those of 3-hydroxy-3-methylglutaryl-CoA reductase and sterol regulatory element binding protein cleavage-activating protein. J Biol Chem. 2000;275(32):24367–74. Epub 2000/05/24. M002184200 [pii]. 10.1074/jbc.M002184200 [DOI] [PubMed] [Google Scholar]
- 15.Inohara N, Nunez G. ML—a conserved domain involved in innate immunity and lipid metabolism. Trends Biochem Sci. 2002;27(5):219–21. Epub 2002/06/22. S0968-0004(02)02084-4 [pii]. [DOI] [PubMed] [Google Scholar]
- 16.Kirchhoff C, Osterhoff C, Young L. Molecular cloning and characterization of HE1, a major secretory protein of the human epididymis. Biol Reprod. 1996;54(4):847–56. Epub 1996/04/01. [DOI] [PubMed] [Google Scholar]
- 17.McCauliff LA, Xu Z, Li R, Kodukula S, Ko DC, Scott MP, et al. Multiple Surface Regions on the Niemann-Pick C2 Protein Facilitate Intracellular Cholesterol Transport. J Biol Chem. 2015;290(45):27321–31. Epub 2015/08/25. M115.667469 [pii]. PubMed Central PMCID: PMC4646368. 10.1074/jbc.M115.667469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Storch J, Xu Z. Niemann-Pick C2 (NPC2) and intracellular cholesterol trafficking. Biochim Biophys Acta. 2009;1791(7):671–8. Epub 2009/02/24. S1388-1981(09)00043-2 [pii]. PubMed Central PMCID: PMC4281484. 10.1016/j.bbalip.2009.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Infante RE, Wang ML, Radhakrishnan A, Kwon HJ, Brown MS, Goldstein JL. NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc Natl Acad Sci U S A. 2008;105(40):15287–92. Epub 2008/09/06. 0807328105 [pii]. PubMed Central PMCID: PMC2563079. 10.1073/pnas.0807328105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwon HJ, Abi-Mosleh L, Wang ML, Deisenhofer J, Goldstein JL, Brown MS, et al. Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell. 2009;137(7):1213–24. Epub 2009/07/01. S0092-8674(09)00393-6 [pii]. PubMed Central PMCID: PMC2739658. 10.1016/j.cell.2009.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang ML, Motamed M, Infante RE, Abi-Mosleh L, Kwon HJ, Brown MS, et al. Identification of surface residues on Niemann-Pick C2 essential for hydrophobic handoff of cholesterol to NPC1 in lysosomes. Cell Metab. 2010;12(2):166–73. Epub 2010/08/03. S1550-4131(10)00233-0 [pii]. PubMed Central PMCID: PMC3034247. 10.1016/j.cmet.2010.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chevallier J, Chamoun Z, Jiang G, Prestwich G, Sakai N, Matile S, et al. Lysobisphosphatidic acid controls endosomal cholesterol levels. J Biol Chem. 2008;283(41):27871–80. Epub 2008/07/23. M801463200 [pii]. 10.1074/jbc.M801463200 [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi T, Beuchat MH, Lindsay M, Frias S, Palmiter RD, Sakuraba H, et al. Late endosomal membranes rich in lysobisphosphatidic acid regulate cholesterol transport. Nat Cell Biol. 1999;1(2):113–8. Epub 1999/11/13. 10.1038/10084 [DOI] [PubMed] [Google Scholar]
- 24.Christy NC, Buss SN, Petri WA Jr. Common pathways for receptor-mediated ingestion of Escherichia coli and LDL cholesterol by Entamoeba histolytica regulated in part by transmembrane kinase 39. Int J Parasitol. 2012;42(4):393–400. Epub 2012/05/24. S0020-7519(12)00050-1 [pii]. PubMed Central PMCID: PMC3449034. 10.1016/j.ijpara.2012.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banuelos C, Garcia-Rivera G, Lopez-Reyes I, Orozco E. Functional characterization of EhADH112: an Entamoeba histolytica Bro1 domain-containing protein. Exp Parasitol. 2005;110(3):292–7. Epub 2005/06/16. S0014-4894(05)00080-9 [pii]. 10.1016/j.exppara.2005.03.005 [DOI] [PubMed] [Google Scholar]
- 26.Castellanos-Castro S, Cerda-García-Rojas CM, Javier-Reyna R, Pais-Morales J, Chávez-Munguía B, Orozco E. Identification of the phospholipid lysobisphosphatidic acid in the protozoan Entamoeba histolytica: An active molecule in endocytosis. Biochemistry and Biophysics Reports. 2016;5:224–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perdomo D, Manich M, Syan S, Olivo-Marin JC, Dufour AC, Guillen N. Intracellular traffic of the lysine and glutamic acid rich protein KERP1 reveals features of endomembrane organization in Entamoeba histolytica. Cell Microbiol. 18(8):1134–52. Epub 2016/02/10. 10.1111/cmi.12576 [DOI] [PubMed] [Google Scholar]
- 28.Perdomo D, Ait-Ammar N, Syan S, Sachse M, Jhingan GD, Guillen N. Cellular and proteomics analysis of the endomembrane system from the unicellular Entamoeba histolytica. J Proteomics. 112:125–40. Epub 2014/08/12. S1874-3919(14)00395-9 [pii]. 10.1016/j.jprot.2014.07.034 [DOI] [PubMed] [Google Scholar]
- 29.Ioannou YA. The structure and function of the Niemann-Pick C1 protein. Mol Genet Metab. 2000;71(1–2):175–81. Epub 2000/09/26. S1096-7192(00)93061-0 [pii]. 10.1006/mgme.2000.3061 [DOI] [PubMed] [Google Scholar]
- 30.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. Epub 2001/03/10. 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- 31.Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435(7038):43–57. Epub 2005/05/06. nature03481 [pii]. PubMed Central PMCID: PMC1352341. 10.1038/nature03481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu S, Benoff B, Liou HL, Lobel P, Stock AM. Structural basis of sterol binding by NPC2, a lysosomal protein deficient in Niemann-Pick type C2 disease. J Biol Chem. 2007;282(32):23525–31. Epub 2007/06/19. M703848200 [pii]. PubMed Central PMCID: PMC3645284. 10.1074/jbc.M703848200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez-Higuera A, Salas-Casas A, Calixto-Galvez M, Chavez-Munguia B, Perez-Ishiwara DG, Ximenez C, et al. Identification of calcium-transporting ATPases of Entamoeba histolytica and cellular localization of the putative SERCA. Exp Parasitol. 2013;135(1):79–86. Epub 2013/06/27. S0014-4894(13)00162-8 [pii]. 10.1016/j.exppara.2013.06.004 [DOI] [PubMed] [Google Scholar]
- 34.Phillips MJ, Voeltz GK. Structure and function of ER membrane contact sites with other organelles. Nat Rev Mol Cell Biol. 17(2):69–82. Epub 2015/12/03. nrm.2015.8 [pii]. 10.1038/nrm.2015.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verma K, Saito-Nakano Y, Nozaki T, Datta S. Insights into endosomal maturation of human holo-transferrin in the enteric parasite Entamoeba histolytica: essential roles of Rab7A and Rab5 in biogenesis of giant early endocytic vacuoles. Cell Microbiol. 2015;17(12):1779–96. Epub 2015/06/23. 10.1111/cmi.12470 [DOI] [PubMed] [Google Scholar]
- 36.Nakada-Tsukui K, Saito-Nakano Y, Ali V, Nozaki T. A retromerlike complex is a novel Rab7 effector that is involved in the transport of the virulence factor cysteine protease in the enteric protozoan parasite Entamoeba histolytica. Mol Biol Cell. 2005;16(11):5294–303. Epub 2005/08/27. E05-04-0283 [pii]. PubMed Central PMCID: PMC1266427. 10.1091/mbc.E05-04-0283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X, Wang J, Coutavas E, Shi H, Hao Q, Blobel G. Structure of human Niemann-Pick C1 protein. Proc Natl Acad Sci U S A. 113(29):8212–7. Epub 2016/06/17. 1607795113 [pii]. PubMed Central PMCID: PMC4961162. 10.1073/pnas.1607795113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kissoon-Singh V, Moreau F, Trusevych E, Chadee K. Entamoeba histolytica exacerbates epithelial tight junction permeability and proinflammatory responses in Muc2(-/-) mice. Am J Pathol. 2013;182(3):852–65. Epub 2013/01/30. S0002-9440(13)00003-5 [pii]. 10.1016/j.ajpath.2012.11.035 [DOI] [PubMed] [Google Scholar]
- 39.Lange S, Delbro DS, Jennische E. Evans blue permeation of intestinal mucosa in the rat. Scand J Gastroenterol. 1994;29(1):38–46. Epub 1994/01/01. [DOI] [PubMed] [Google Scholar]
- 40.Bracha R, Nuchamowitz Y, Mirelman D. Transcriptional silencing of an amoebapore gene in Entamoeba histolytica: molecular analysis and effect on pathogenicity. Eukaryot Cell. 2003;2(2):295–305. Epub 2003/04/10. PubMed Central PMCID: PMC154849. 10.1128/EC.2.2.295-305.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lige B, Romano JD, Bandaru VV, Ehrenman K, Levitskaya J, Sampels V, et al. Deficiency of a Niemann-Pick, type C1-related protein in toxoplasma is associated with multiple lipidoses and increased pathogenicity. PLoS Pathog. 2011;7(12):e1002410 Epub 2011/12/17. PPATHOGENS-D-11-00951 [pii]. PubMed Central PMCID: PMC3234224. 10.1371/journal.ppat.1002410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Higaki K, Almanzar-Paramio D, Sturley SL. Metazoan and microbial models of Niemann-Pick Type C disease. Biochim Biophys Acta. 2004;1685(1–3):38–47. Epub 2004/10/07. S1388-1981(04)00133-7 [pii]. 10.1016/j.bbalip.2004.08.010 [DOI] [PubMed] [Google Scholar]
- 43.Lu F, Liang Q, Abi-Mosleh L, Das A, De Brabander JK, Goldstein JL, et al. Identification of NPC1 as the target of U18666A, an inhibitor of lysosomal cholesterol export and Ebola infection. Elife. 2015;4. Epub 2015/12/10. e12177 [pii]. PubMed Central PMCID: PMC4718804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dixit SS, Sleat DE, Stock AM, Lobel P. Do mammalian NPC1 and NPC2 play a role in intestinal cholesterol absorption? Biochem J. 2007;408(1):1–5. Epub 2007/09/21. BJ20071167 [pii]. PubMed Central PMCID: PMC2049080. 10.1042/BJ20071167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orozco E, Guarneros G, Martinez-Palomo A, Sanchez T. Entamoeba histolytica. Phagocytosis as a virulence factor. J Exp Med. 1983;158(5):1511–21. Epub 1983/11/01. PubMed Central PMCID: PMC2187140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diamond LS. Axenic cultivation of Entamoeba hitolytica. Science. 1961;134(3475):336–7. Epub 1961/08/04. [DOI] [PubMed] [Google Scholar]
- 47.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–9. Epub 2011/05/07. msr121 [pii]. PubMed Central PMCID: PMC3203626. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Montano S, Orozco E, Correa-Basurto J, Bello M, Chavez-Munguia B, Betanzos A. Heterodimerization of the Entamoeba histolytica EhCPADH virulence complex through molecular dynamics and protein-protein docking. J Biomol Struct Dyn. 2016:1–18. Epub 2016/02/11. [DOI] [PubMed] [Google Scholar]
- 49.Kettis AA, Lidman K, Fagraeus A. Actin in Entamoeba histolytica trophozoites revealed by human actin antibodies. J Parasitol. 1977;63(3):581–3. Epub 1977/06/01. [PubMed] [Google Scholar]
- 50.Petri WA Jr., Chapman MD, Snodgrass T, Mann BJ, Broman J, Ravdin JI. Subunit structure of the galactose and N-acetyl-D-galactosamine-inhibitable adherence lectin of Entamoeba histolytica. J Biol Chem. 1989;264(5):3007–12. Epub 1989/02/15. [PubMed] [Google Scholar]
- 51.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224(Pt 3):213–32. Epub 2007/01/11. JMI1706 [pii]. 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- 52.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–5. Epub 1970/08/15. [DOI] [PubMed] [Google Scholar]
- 53.Avalos-Padilla Y, Betanzos A, Javier-Reyna R, Garcia-Rivera G, Chavez-Munguia B, Lagunes-Guillen A, et al. EhVps32 Is a Vacuole-Associated Protein Involved in Pinocytosis and Phagocytosis of Entamoeaba histolytica. PLoS Pathog. 2015;11(7):e1005079 Epub 2015/08/01. PPATHOGENS-D-15-00139 [pii]. PubMed Central PMCID: PMC4521941. 10.1371/journal.ppat.1005079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Talamas-Lara D, Chavez-Munguia B, Gonzalez-Robles A, Talamas-Rohana P, Salazar-Villatoro L, Duran-Diaz A, et al. Erythrophagocytosis in Entamoeba histolytica and Entamoeba dispar: a comparative study. Biomed Res Int. 2014;2014:626259 Epub 2014/07/09. PubMed Central PMCID: PMC4066688. 10.1155/2014/626259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Novikoff AB, Novikoff PM, Davis C, Quintana N. Studies on microperoxisomes. II. A cytochemical method for light and electron microscopy. J Histochem Cytochem. 1972;20(12):1006–23. Epub 1972/12/01. [DOI] [PubMed] [Google Scholar]
- 56.Shibayama M, Campos-Rodriguez R, Ramirez-Rosales A, Flores-Romo L, Espinosa-Cantellano M, Martinez-Palomo A, et al. Entamoeba histolytica: liver invasion and abscess production by intraperitoneal inoculation of trophozoites in hamsters, Mesocricetus auratus. Exp Parasitol. 1998;88(1):20–7. 10.1006/expr.1998.4218 [DOI] [PubMed] [Google Scholar]
- 57.Baxt LA, Rastew E, Bracha R, Mirelman D, Singh U. Downregulation of an Entamoeba histolytica rhomboid protease reveals roles in regulating parasite adhesion and phagocytosis. Eukaryot Cell. 9(8):1283–93. Epub 2010/06/29. EC.00015-10 [pii]. PubMed Central PMCID: PMC2918930. 10.1128/EC.00015-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Western blot assays of rEhNPC2a and rEhNPC2b recombinant proteins immunodetected with rat α-EhNPC2a antibodies. Trophozoites lysates (ET) were used as a positive control.
(TIF)
(DOCX)
(DOCX)
BamHI and SalI restriction enzyme sites are underlined in forward and reverser primers, respectively.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.