Abstract
Objective
HIV-1 escape from cytotoxic T-lymphocytes (CTL) results in the accumulation of HLA-associated mutations in the viral genome. To understand the contribution of early escape to disease progression, this study investigated the evolution and pathogenic implications of CTL escape in a cohort followed from infection for five years.
Methods
Viral loads and CD4+ counts were monitored in 78 subtype C infected individuals from onset of infection until CD4+ decline to <350 cells/μl or five years post-infection. The gag gene was sequenced and HLA-associated changes between enrolment and 12 months post-infection were mapped.
Results
HLA-associated escape mutations were identified in 48 (62%) of the participants and were associated with CD4+ decline to <350 copies/ml (p=0.05). Escape mutations in variable Gag proteins (p17 and p7p6) had a greater impact on disease progression than escape in more conserved regions (p24) (p=0.03). The association between HLA-associated escape mutations and CD4+ decline was independent of protective HLA allele (B*57, B*58:01, B*81) expression.
Conclusion
The high frequency of escape contributed to rapid disease progression in this cohort. While HLA-adaption in both conserved and variable Gag domains in the first year of infection was detrimental to long term clinical outcome, escape in variable domains had greater impact.
Keywords: HLA-associated, CTL escape, CD4+ decline, Gag, Disease progression
Introduction
HIV-1 subtype C-infected women in South Africa generally have rapid disease progression with more than 31 % having CD4+ decline to <350 cell/μl in the first year of infection [1]. Under the current WHO treatment guidelines (CD4+ count of <500 cell/μl), about 70% of these women would need to go on therapy in the first year of infection [1]. Cytotoxic T lymphocyte (CTL) responses, in particular Gag-targeted responses, are known to contribute to viral control in HIV-1 infections and drive the virus to escape through mutations [2]. The detrimental impact of immune evasion on pathogenesis can be alleviated in certain cases where escape has a fitness cost on the virus [3-7]. While is it known that clinical parameters in early infection are predictive of subsequent disease progression [1], and it is well established that specific escape mutations restricted by protective HLA-alleles [5, 7-11] can have a beneficial impact, less is understood at a population level how immune escape in early infection impacts long term clinical outcome.
Protective HLA alleles, which generally target conserved epitopes, drive the evolution of CTL escape mutations which can attenuate viral replication resulting in slower disease progression [3, 5, 10, 12]. An HLA-independent study of 17 individuals, monitored from acute infection, found CTL escape occurred in all individuals within the first 50 days of infection [13]. The likelihood of escape was determined by a combination of the most dominant CTL response in an individual, together with the entropy of the target [13]. In this cohort of 17, escape in Gag was demonstrated in approximately 30% (n=5) of individuals [13].
Several large cohort studies have demonstrated that CTL escape can be predicted based on the HLA alleles expressed by the host [14-19]. Furthermore, there is an inverse correlation between the potential breadth of HLA-mediated anti-Gag selection pressure, identified as the total number of potential HLA-associated sites, and viral loads [19, 20]. These studies have largely been done in chronic infection, and while it is known that events in the acute/early phase of infection are important predictors of disease progression [21], and there is limited information on the role of evolving HLA-associated polymorphisms during this critical period and their long-term impact on disease.
In this paper, we report on a study of 78 individuals recruited within the first three months of infection, and followed for at least five years, or until CD4+ decline to <350 cells/μl. Taking advantage of well documented databases that have mapped HLA-associated polymorphisms in subtype C infection, we identified putative CTL escape mutations across the Gag protein in the first year of infection and investigated the impact on CD4+ decline. The information reported here augments our understanding of mechanisms associated with rapid disease progression in this cohort. Understanding the extent of escape in early infection is also important to HIV-1 cure studies as it has been shown that these escape variants get archived and could potentially limit vaccine associated interventions aimed at reducing the viral reservoir [22].
Methods
Study Participants
Participants in this study (n=78) were HIV-1 infected women from the CAPRISA 002 acute infection cohort [23] and the placebo arm of the CAPRISA 004 1% tenofovir gel microbicide trial [24]. We excluded participants from the treatment arm as there are ongoing studies to determine whether the 1% tenofovir gel resulted in selection of viruses with altered characteristics. We also excluded individuals who were dually-infected (co- and super-infected) as well as those who were non-subtype C infected. The CAPRISA 002 Acute Infection study was reviewed and approved by the research ethics committees of the University of KwaZulu-Natal (E013/04) and the University of Cape Town (025/2004). Written consent was obtained from all participants.
RNA isolation, RT-PCR and viral sequencing
The gag region was amplified and sequenced from plasma samples collected at enrolment (earliest available sample post-infection) and 12 months post-infection by nested RT-PCR as described previously [25]. RNA isolated from plasma samples using the Magna-Pure Compact Nucleic Acid Extractor (Roche) was reverse transcribed using the Invitrogen Thermoscript Reverse transcription kit (Invitrogen) and the primer Gag D reverse (5′-AAT TCC TCC TAT CAT TTT TGG-3′; HXB2 position 2382-2402). Limiting dilution nested PCR was carried out by serial end-point dilution of the cDNA [26]. The first round PCR primers for amplification were Gag D forward (5′-TCT CTA GCA GTG GCG CCC G-3′; HXB position 626-644) and Gag D reverse. The second round PCR primers were Gag A forward (5′-CTC TCG ACG CAG GAC TCG GCT T-3′; HXB2 position 683-704) and Gag C reverse (5′-TCT TCT AAT ACT GTA TCA TCT GC-3′; HXB2 position 2334-2356). PCR products were sequenced using an ABI PRISM dye terminator cycle-sequencing kit (Applied Biosysytems) and the primers Gag A forward, Gag A reverse (5′-ACA TGG GTA TCA CTT CTG GGC T-3′; HXB2 position 1282-1303), Gag B forward (5′-CCA TAT CAC CTA GAA CTT TGA AT-3′; HXB2 position 1226-1246), Gag B reverse (5′-CTC CCT GAC ATG CTG TCA TCA T-3′; HXB2 position 1825-1846), Gag C forward (5′-CCT TGT TGG TCC AAA ATG CGA-3′; HXB2 position 1748-1768) and Gag C reverse. Sequences were assembled using ChromasPro (www.technelysium.com.au/chromas.html).
For participants included in this study, the gag sequences from the enrolment and 12 months post-infection visits from each participant grouped together on a phylogenetic tree (data not shown).
HLA typing
High resolution (four digit) HLA typing was performed on all participants. DNA was extracted from either PBMCs or granulocytes using the Pel-Freez DNA Isolation kit (Pel-Freez). HLA-A, -B and -C typing was performed by sequencing of exons 2, 3 and 4 using Atria HLA-AlleleSeqr kits (Abbott) or HLA-SBTexcellerator kits (Qiagen) and Assign-SBT v3.5 software (Conexio Genomics). Any ambiguities resulting from either polymorphisms outside the sequenced exons or identical heterozygote combinations were resolved using sequence-specific primers, or resolved statistically using the algorithm developed for this purpose [27].
Identification of CTL escape
HLA-associated CTL escape was determined by comparison of HLA-associated sites, derived from a large subtype C chronic infection dataset [16], between enrolment and 12 months Gag amino acid sequences according to each participant's HLA genotype. The list of HLA-associated polymorphisms resulted from analysis of 2,126 chronically HIV-1 subtype C-infected anti-retroviral-naïve individuals from Southern Africa and found 301 polymorphisms in 90 epitopes [16]. Escape was defined as a change towards an HLA-adapted amino acid or away from a non-adapted amino acid [16]. In addition, mutations at HLA-associated sites which were neither going towards or away from adapted or non-adapted residues were also classified as escape. Plasma viral loads and CD4+ counts at 12 months post-infection were calculated by taking the mean of three time-points closest to 12 months post-infection.
Statistical analyses
All statistical analyses were carried out in Graphpad Prism 5.0 (GraphPad Software, Inc.).
Results
A total of 78 HIV-1 subtype C infected women were studied over a five year period, or until CD4+ decline to <350 cells/μl. The median (IQR) time post-infection for the earliest samples was 40 (26-54) days post-infection. At 12 months post-infection (set-point), the median log plasma viral load and CD4+ count were 4.20 copies/ml and 432 cells/μl, respectively.
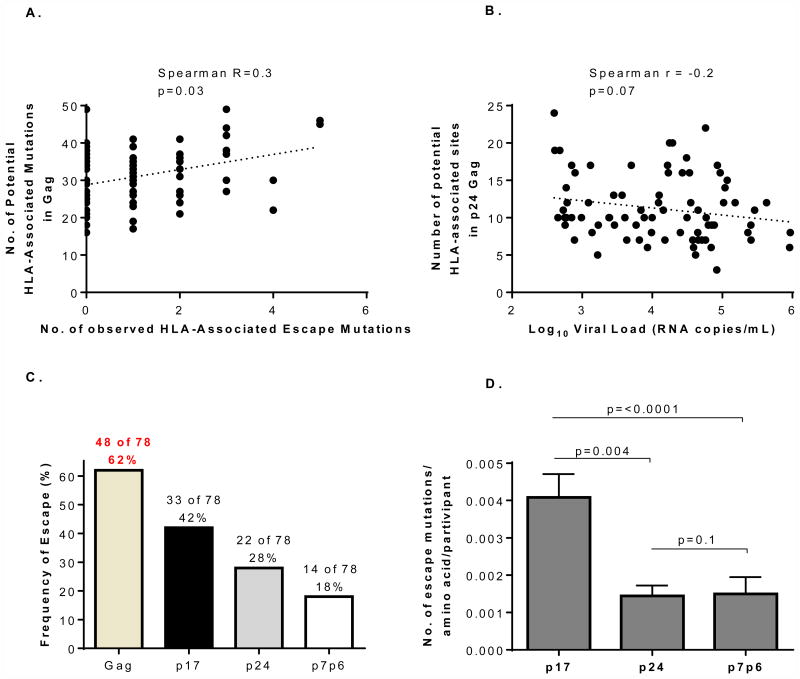
Escape was identified as changes in Gag amino acids sites between enrolment and twelve months post-infection at positions that have previously been shown to positively correlate with the participants HLA-alleles [16]. This list of HLA-associated polymorphism was derived from a chronic database, and it likely reflects both early and late escaping epitopes. We thus evaluated whether there was an association between the observed mutations and the total number expected based on the participants' HLA genotype. We found a significant association between the observed number of HLA-associated escape mutations and the HLA-associated polymorphisms over 12 months post-infection (Spearman r=0.3; p=0.03) (Figure 1A), supporting the use of this list for evaluation sequences from early infection.
Figure 1.
Panel A) Association between number of HLA-associated escape sites and observed HLA-associated escape mutations in Gag. Panel B) Relationship between number of HLA-associated escape sites and log10 plasma viral load at 12 months post-infection. Panel C) Frequency of putative CTL escape at HLA-associated escape sites in the first year of infection in Gag, p17, p24 and p7p6. Panel D) Number of escape mutations per Gag protein normalised to protein size.
Potential CTL breadth in p24 associated with 12 months viral load set-point
To determine if there was a relationship between the total potential CTL pressure that an individual exerts on the virus, and disease outcome, we calculated the total number of potential HLA-associated sites restricted by each participants HLA allele. We found no association between number of potential HLA-associated sites and either viral load or CD4+ count for the entire Gag region or each of the Gag regions (p17, p24 and p7p6) (data not shown). However, there was a strong trend towards a negative association between the number of potential HLA-associated sites in p24 Gag and plasma viral load (Spearman r=-0.2; p=0.07) (Figure 1B). This supports previous findings from acute and chronic infection [12, 28-30] and suggests that the CTL pressure on the conserved p24 Gag region contributed to early viral control.
Higher frequency of escape in more variable p17 Gag
We then investigated actual escape across different Gag proteins (p17, p24 and p7p6). We identified HLA-associated escape mutations in the first year of infection in 48 (62%) study participants, with escape occurring in 1-5 sites per participant (Figure 1C). HLA-associated mutations occurred most frequently in p17 in 33 (42%) individuals, followed by escape in p24 in 22 (28%) individuals, and lastly in p7p6 in 14 (18%) individuals (Figure 1C). Twenty-three percent of the study participants harboured viruses that developed HLA-associated mutations only in p17, 16% only in p24, and 1% only in p7p6 (Supplementary Figure 1). Normalisation of the number of HLA-associated mutations relative to the protein size further accentuated the differences in frequency between proteins with HLA-associated escape mutations per amino acid significantly higher in p17compared to p24 (p=0.004) and p7p6 (p<0.0001) (Figure 1D).
Early HLA-associated escape mutations predict CD4+ decline to < 350 μl/ml
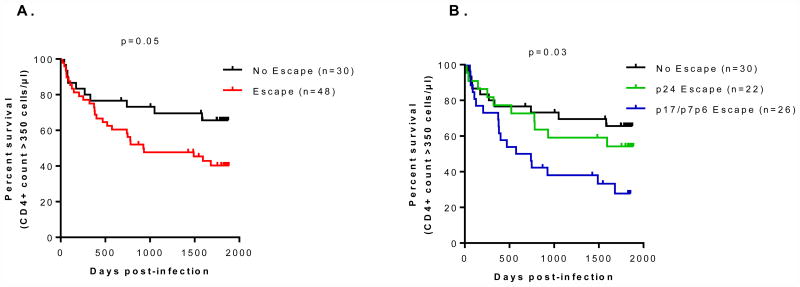
CTL escape in p24 has previously been reported to be beneficial in terms of lowering viral loads in early infection [5, 7]. We were interested in evaluating the relative impact of escape across all Gag proteins. We first compared the time to CD4+ counts of <350 cells/μl in the first five years of infection for participants whose infecting viruses developed CTL escape and those where escape was not observed. The time to CD4+ decline <350 cells/μl was significantly shorter for participants whose infecting viruses developed CTL escape (p=0.05, log rank test) (Figure 2A). As escape in p24 tends to be associated with reduced viral load [5-7], we stratified the HLA-associated escape mutations into p24 escape and p17/p7p6 escape (individuals where HLA-associated escape mutations were observed in both p24 and variable regions were classified as p24 escape). Individuals whose viruses developed HLA-associated escape mutations in variable proteins (p17 and p7p6) progressed more rapidly to CD4+ decline to <350 cells/μl over 5 years, compared to those whose viruses developed p24 HLA-associated escape mutations [the order of progression; no escape > p24 escape > p17/p7p6 escape (p=0.03, log-rank test)] (Figure 2B). Interestingly, of the 22 participants whose infecting viruses developed HLA-associated escape mutations in p24, only 11 (41%) had mutations at sites previously described to have a fitness cost to virus replication (HXB2 positions 146[31], 182[32], and 242[33]) (Supplementary Table 1). This suggests that while CTL escape is associated with rapid CD4+ decline, escape in escape in the variable p17 and p7p6 proteins was more detrimental to disease progression than escape in the conserved p24 region.
Figure 2.
Panel A) Association between HLA-associated escape mutations and CD4+ decline to <350 cells/μl. Panel B) Association between no escape, HLA-associated escape mutations in conserved (p24) and in variable (p17 and p7p6) Gag proteins, and CD4+ decline to <350 cells/μl.
HLA-associated escape in early infection is independent of plasma viral load
As both viral load and CD4+ count are predictors of disease progression, viral load is a potential confounder if the emergence of CTL escape mutations was driven by high viral loads. To interrogate this, we compared the plasma viral loads at 12 months post-infection between participants whose infecting viruses developed HLA-associated escape mutations (escape group) and those who did not (no-escape group). We found no differences in viral loads between the two groups (p=0.4) (data not shown). We also compared viral loads between the escape and no-escape groups at enrolment, three months and six months post-infection and found no difference (p=0.3, p=0.6 and p=0.5, respectively) (data not shown). Repeating these analyses after removal of individuals with protective HLA alleles (HLA B*57, B*58:01 and B*81; n=18) also showed no significant difference (p>0.5) (data not shown). This suggests that the emergence of HLA-associated escape mutations in early infection was independent of viral load.
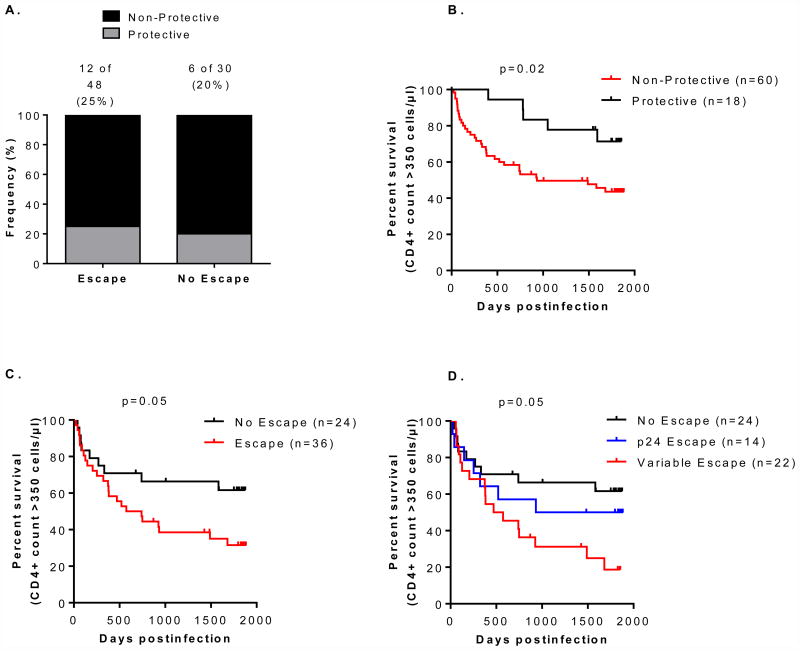
Impact of HLA-associated escape mutations on CD4+ decline is independent of protective HLA alleles
As some HLA alleles are known to be protective and associated with slower disease progression, we investigated whether there was enrichment of protective HLA alleles (HLA B*57, B*58:01 and B*81) in either group (escape and no escape). There was no significant difference in HLA alleles between the two groups [12/48 (25%) vs. 6/30 (20%), respectively; p=0.5; Fisher's exact test] (Figure 3A), and we found that protective HLA alleles were associated with a slower decline to CD4+ counts of <350 cells/μl (p=0.02) (Figure 3B). Assessment of CD4+ decline after excluding individuals with protective HLA alleles showed that individuals whose viruses developed HLA-associated escape mutations had rapid CD4+ decline (p=0.05) (Figure 3C). We also observed that individuals whose viruses developed HLA-associated escape mutations in variable proteins (p17 and p7p6) progressed more rapidly to CD4+ decline to <350 cells/μl over 5 years, compared to those whose viruses developed p24 HLA-associated escape mutations [the order of progression; no escape > p24 escape > p17/p7p6 escape (p=0.05, log-rank test)] (Figure 3D).These results suggest that the effect of HLA-associated escape mutations on CD4+ decline is independent of protective HLA alleles.
Figure 3.
Panel A) Distribution of protective HLA alleles (HLA B*57, B*58:01 and B*81) between the two groups (escape and no escape). Panel B) Association between protective HLA alleles and CD4+ decline to <350 cells/μl. Panel C) Association between HLA-associated escape mutations and CD4+ decline to <350 cells/μl in individuals with non-protective HLA alleles. Panel D) Association between no escape, HLA-associated escape mutations in conserved (p24) and in variable (p17 and p7p6) Gag proteins, and CD4+ decline to <350 cells/μl in individuals with non-protective HLA alleles.
Discussion
To understand factors contributing to rapid disease progression observed in a subtype C cohort in South Africa [1], we examined the impact of early CTL escape in Gag on disease progression. We detected a high frequency of HLA-associated escape mutations (48% of the study participants) which occurred predominantly in the p17 region (42% of participants). The development of HLA-associated escape mutations in the first year of infection was associated with a more rapid decline to CD4+ count of <350 cells/μl, and the effect of escape was more pronounced for mutations occurring in the variable Gag proteins.
The breadth of CTL responses as well as targeting of conserved regions has been shown to be associated with viral control [12, 28, 29]. We reasoned that the number of HLA-associated escape sites for each individual is a measure of the potential CTL pressure that the individual exerts on the virus. We found a trend towards a negative association between the total potential number of HLA-associated escape sites and plasma viral load at 12 months post-infection (Spearman r=-0.2; p=0.07) for p24 Gag. This underscores the importance of CTL responses targeting the functionally and structurally conserved p24 region and supports previous findings from chronic subtype B infections [20].
Previous studies have reported specific immune escape mutations in p24 Gag that are associated with partial attenuation of viral replicative fitness and slower disease progression [5, 7, 8]. In this study participants in whom HLA-associated escape mutations developed in the first year of infection progressed faster to CD4+ count <350 cells/μl compared to those where HLA-associated escape mutations were not detected (p=0.05). However, individuals whose viruses escaped in the more variable p17/p7p6 regions progressed faster to CD4+ count <350 cells/μl compared to those where HLA-associated escape mutations were observed in the more conserved p24. These results suggest that while overall CTL escape is detrimental to the host, escape in the variable regions of the virus (p17 and p7p6) is associated with a more rapid CD4+ decline.
We found protective HLA alleles to be evenly distributed between individuals whose viruses developed escape mutations and those who did not (p=0.5, Fisher's exact test), and participants with protective HLA alleles had slower CD4+ decline. Interestingly, the association between HLA-associated escape mutations and CD4+ decline was robust to the exclusion of participants with protective HLA alleles. These data suggest that protective HLA alleles were not a major driver of the observed impact of HLA-associated escape mutations on CD4+ decline. Interestingly, while viral load is also correlated with CD4+ decline, the emergence of HLA-associated escape mutations in the first year of infection was not associated with plasma viral load, suggesting that CTL escape, independent of other disease progression makers, can predict CD4+ decline.
In this study, we have assessed the impact of HLA-associated escape mutations in the first year on CD4+ decline in the first five years of infection and showed that putative HLA-associated escape mutations are associated with a shorter time to CD4+ decline to <350 cells/μl, which is more pronounced in the variable Gag regions (p17 and p7p6) compared to the conserved p24. The high frequency of early putative escape may be a contributing factor disease progression in HIV-infected South Africans.
Supplementary Material
Acknowledgments
We thank the CAPRISA 002 study participants and the CAPRISA clinical and laboratory personnel.
DRC, NG, CM, QA, SA, CG and CW conceived and designed the study; DRC RN NN performed the experiments; DRC and DM analyzed the data; DRC and CW wrote the paper.
This work was supported by a National Research Foundation (NRF) Research Career Advancement Fellowship and a Poliomyelitis Research Foundation (PRF) research grant to DRC, and the National Institutes of Health (NIH) (Grant no. U19 A151794) to CW. DRC is a recipient of the South African National Research Foundation (NRF) Career Advancement Fellowship. The views expressed by the authors are entirely their own and do not necessarily reflect those of their funding sources.
Footnotes
Conflicts of interest: The authors declare no conflicts of interest.
References
- 1.Mlisana K, Werner L, Garrett NJ, McKinnon LR, van Loggerenberg F, Passmore JA, et al. Rapid disease progression in HIV-1 subtype C-infected South African women. Clin Infect Dis. 2014;59:1322–1331. doi: 10.1093/cid/ciu573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borrow P, Lewicki H, Wei X, Horwitz MS, Peffer N, Meyers H, et al. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med. 1997;3:205–211. doi: 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- 3.Brockman MA, Brumme ZL, Brumme CJ, Miura T, Sela J, Rosato PC, et al. Early selection in Gag by protective HLA alleles contributes to reduced HIV-1 replication capacity that may be largely compensated for in chronic infection. J Virol. 2010;84:11937–11949. doi: 10.1128/JVI.01086-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brockman MA, Chopera DR, Olvera A, Brumme CJ, Sela J, Markle TJ, et al. Uncommon pathways of immune escape attenuate HIV-1 integrase replication capacity. J Virol. 2012;86:6913–6923. doi: 10.1128/JVI.07133-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brockman MA, Schneidewind A, Lahaie M, Schmidt A, Miura T, Desouza I, et al. Escape and compensation from early HLA-B57-mediated cytotoxic T-lymphocyte pressure on human immunodeficiency virus type 1 Gag alter capsid interactions with cyclophilin A. J Virol. 2007;81:12608–12618. doi: 10.1128/JVI.01369-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miura T, Brockman MA, Schneidewind A, Lobritz M, Pereyra F, Rathod A, et al. HLA-B57/B*5801 human immunodeficiency virus type 1 elite controllers select for rare gag variants associated with reduced viral replication capacity and strong cytotoxic T-lymphocyte [corrected] recognition. Journal of virology. 2009;83:2743–2755. doi: 10.1128/JVI.02265-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schneidewind A, Brockman MA, Yang R, Adam RI, Li B, Le Gall S, et al. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J Virol. 2007;81:12382–12393. doi: 10.1128/JVI.01543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crawford H, Lumm W, Leslie A, Schaefer M, Boeras D, Prado JG, et al. Evolution of HLA-B*5703 HIV-1 escape mutations in HLA-B*5703-positive individuals and their transmission recipients. J Exp Med. 2009;206:909–921. doi: 10.1084/jem.20081984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chopera DR, Mlotshwa M, Woodman Z, Mlisana K, de Assis Rosa D, Martin DP, et al. Virological and immunological factors associated with HIV-1 differential disease progression in HLA-B 58:01-positive individuals. J Virol. 2011;85:7070–7080. doi: 10.1128/JVI.02543-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leslie AJ, Pfafferott KJ, Chetty P, Draenert R, Addo MM, Feeney M, et al. HIV evolution: CTL escape mutation and reversion after transmission. Nat Med. 2004;10:282–289. doi: 10.1038/nm992. [DOI] [PubMed] [Google Scholar]
- 11.Ntale RS, Chopera DR, Ngandu NK, Assis de Rosa D, Zembe L, Gamieldien H, et al. Temporal association of HLA-B*81:01- and HLA-B*39:10-mediated HIV-1 p24 sequence evolution with disease progression. J Virol. 2012;86:12013–12024. doi: 10.1128/JVI.00539-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang YE, Li B, Carlson JM, Streeck H, Gladden AD, Goodman R, et al. Protective HLA class I alleles that restrict acute-phase CD8+ T-cell responses are associated with viral escape mutations located in highly conserved regions of human immunodeficiency virus type 1. J Virol. 2009;83:1845–1855. doi: 10.1128/JVI.01061-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu MK, Hawkins N, Ritchie AJ, Ganusov VV, Whale V, Brackenridge S, et al. Vertical T cell immunodominance and epitope entropy determine HIV-1 escape. J Clin Invest. 2013;123:380–393. doi: 10.1172/JCI65330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brumme ZL, John M, Carlson JM, Brumme CJ, Chan D, Brockman MA, et al. HLA-associated immune escape pathways in HIV-1 subtype B Gag, Pol and Nef proteins. PLoS One. 2009;4:e6687. doi: 10.1371/journal.pone.0006687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang KH, Goedhals D, Carlson JM, Brockman MA, Mishra S, Brumme ZL, et al. Progression to AIDS in South Africa is associated with both reverting and compensatory viral mutations. PLoS One. 2011;6:e19018. doi: 10.1371/journal.pone.0019018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlson JM, Listgarten J, Pfeifer N, Tan V, Kadie C, Walker BD, et al. Widespread impact of HLA restriction on immune control and escape pathways of HIV-1. J Virol. 2012;86:5230–5243. doi: 10.1128/JVI.06728-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rousseau CM, Daniels MG, Carlson JM, Kadie C, Crawford H, Prendergast A, et al. HLA class I-driven evolution of human immunodeficiency virus type 1 subtype c proteome: immune escape and viral load. J Virol. 2008;82:6434–6446. doi: 10.1128/JVI.02455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlson JM, Schaefer M, Monaco DC, Batorsky R, Claiborne DT, Prince J, et al. HIV transmission. Selection bias at the heterosexual HIV-1 transmission bottleneck. Science. 2014;345:1254031. doi: 10.1126/science.1254031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Tran G, Chikata T, Carlson JM, Murakoshi H, Nguyen DH, Tamura Y, et al. A strong association of HLA-associated Pol and Gag mutations with clinical parameters in HIV-1 subtype A/E infection. AIDS. 2015 doi: 10.1097/QAD.0000000000000969. [DOI] [PubMed] [Google Scholar]
- 20.Brumme ZL, Tao I, Szeto S, Brumme CJ, Carlson JM, Chan D, et al. Human leukocyte antigen-specific polymorphisms in HIV-1 Gag and their association with viral load in chronic untreated infection. AIDS. 2008;22:1277–1286. doi: 10.1097/QAD.0b013e3283021a8c. [DOI] [PubMed] [Google Scholar]
- 21.Vanhems P, Hirschel B, Phillips AN, Cooper DA, Vizzard J, Brassard J, et al. Incubation time of acute human immunodeficiency virus (HIV) infection and duration of acute HIV infection are independent prognostic factors of progression to AIDS. J Infect Dis. 2000;182:334–337. doi: 10.1086/315687. [DOI] [PubMed] [Google Scholar]
- 22.Deng K, Pertea M, Rongvaux A, Wang L, Durand CM, Ghiaur G, et al. Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature. 2015;517:381–385. doi: 10.1038/nature14053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Loggerenberg F, Mlisana K, Williamson C, Auld SC, Morris L, Gray CM, et al. Establishing a cohort at high risk of HIV infection in South Africa: challenges and experiences of the CAPRISA 002 acute infection study. PLoS One. 2008;3:e1954. doi: 10.1371/journal.pone.0001954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abdool Karim Q, Abdool Karim SS, Frohlich JA, Grobler AC, Baxter C, Mansoor LE, et al. Effectiveness and safety of tenofovir gel, an antiretroviral microbicide, for the prevention of HIV infection in women. Science. 2010;329:1168–1174. doi: 10.1126/science.1193748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chopera DR, Woodman Z, Mlisana K, Mlotshwa M, Martin DP, Seoighe C, et al. Transmission of HIV-1 CTL escape variants provides HLA-mismatched recipients with a survival advantage. PLoS Pathog. 2008;4:e1000033. doi: 10.1371/journal.ppat.1000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodrigo AG, Goracke PC, Rowhanian K, Mullins JI. Quantitation of target molecules from polymerase chain reaction-based limiting dilution assays. AIDS Res Hum Retroviruses. 1997;13:737–742. doi: 10.1089/aid.1997.13.737. [DOI] [PubMed] [Google Scholar]
- 27.Listgarten J, Brumme Z, Kadie C, Xiaojiang G, Walker B, Carrington M, et al. Statistical resolution of ambiguous HLA typing data. PLoS Comput Biol. 2008;4:e1000016. doi: 10.1371/journal.pcbi.1000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007;13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 29.Borghans JA, Molgaard A, de Boer RJ, Kesmir C. HLA alleles associated with slow progression to AIDS truly prefer to present HIV-1 p24. PLoS One. 2007;2:e920. doi: 10.1371/journal.pone.0000920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ndhlovu ZM, Kamya P, Mewalal N, Kloverpris HN, Nkosi T, Pretorius K, et al. Magnitude and Kinetics of CD8+ T Cell Activation during Hyperacute HIV Infection Impact Viral Set Point. Immunity. 2015;43:591–604. doi: 10.1016/j.immuni.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Troyer RM, McNevin J, Liu Y, Zhang SC, Krizan RW, Abraha A, et al. Variable fitness impact of HIV-1 escape mutations to cytotoxic T lymphocyte (CTL) response. PLoS Pathog. 2009;5:e1000365. doi: 10.1371/journal.ppat.1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wright JK, Naidoo VL, Brumme ZL, Prince JL, Claiborne DT, Goulder PJ, et al. Impact of HLA-B*81-associated mutations in HIV-1 Gag on viral replication capacity. J Virol. 2012;86:3193–3199. doi: 10.1128/JVI.06682-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez-Picado J, Prado JG, Fry EE, Pfafferott K, Leslie A, Chetty S, et al. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. Journal of virology. 2006;80:3617–3623. doi: 10.1128/JVI.80.7.3617-3623.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.