Abstract
Objective
Repeat associated non-AUG (RAN) translation drives production of toxic proteins from pathogenic repeat sequences in multiple untreatable neurodegenerative disorders. Fragile X-associated tremor/ataxia syndrome (FXTAS) is one such condition, resulting from a CGG trinucleotide repeat expansion in the 5′ leader sequence of the FMR1 gene. RAN proteins from the CGG repeat accumulate in ubiquitinated inclusions in FXTAS patient brains and elicit toxicity. In addition to the CGG repeat, an antisense mRNA containing a CCG repeat is also transcribed from the FMR1 locus. We evaluated whether this antisense CCG repeat supports RAN translation and contributes to pathology in FXTAS patients.
Methods
We generated a series of CCG RAN translation specific reporters and utilized them to measure RAN translation from CCG repeats in multiple reading frames in transfected cells. We also developed antibodies against predicted CCG RAN proteins and used immunohistochemistry and immunofluorescence on FXTAS patient tissues to measure their accumulation and distribution.
Results
RAN translation from CCG repeats is supported in all three potential reading frames, generating polyproline, polyarginine, and polyalanine proteins, respectively. Their production occurs whether or not the natural AUG start upstream of the repeat in the proline reading frame is present. All three frames show greater translation at larger repeat sizes. Antibodies targeted to the antisense FMR polyproline and polyalanine proteins selectively stain nuclear and cytoplasmic aggregates in FXTAS patients and colocalize with ubiquitinated neuronal inclusions.
Interpretation
RAN translation from antisense CCG repeats generates novel proteins that accumulate in ubiquitinated inclusions in FXTAS patients.
INTRODUCTION
Nucleotide repeat expansions are among the most common inherited causes of neurodegeneration and neurological disease1. These progressive disorders are currently without any effective treatment. Repeat expansions elicit toxicity through a range of different pathogenic mechanisms, including polyglutamine mediated aggregation and associated impairment of protein quality control pathways, RNA repeat mediated sequestration of protein bound factors, and DNA mediated transcriptional silencing of the genes in which they reside2–4.
Work by a number of groups recently demonstrated that expanded nucleotide repeats can also support an unusual form of protein translational initiation known as repeat-associated non-AUG (RAN) translation5, 6. RAN translation allows for production of aberrant homopolymeric or dipeptide repeat proteins in the absence of an AUG start codon. It has now been described at CAG, CUG, GGGGCC, CCCCGG, and CGG repeats5, 7–13. RAN proteins can be generated from multiple reading frames of the same repeat, and in disorders where bidirectional transcription through the repeat is present, RAN translation can also on the antisense transcript, leading to a series of potentially toxic products that accumulate in patient neurons5, 6, 9, 10, 12.
Fragile X-associated tremor/ataxia syndrome (FXTAS) is an age-related neurodegenerative disease caused by a CGG trinucleotide repeat expansion in the 5′ UTR (hereafter referred to as the 5′ leader sequence) of the FMR1 gene14. FXTAS is under-recognized clinically, with a predicted incidence of 1:3000 men over the age of 5015. Clinical characteristics include progressive gait ataxia, intention tremor, Parkinsonism, and dementia16. FXTAS patients and animals models of disease are characterized by elevated FMR1 mRNA levels, decreased levels of the Fragile X protein, FMRP, and intranuclear ubiquitinated neuronal inclusions in the cerebellum and other brain regions14, 17, 18.
Our group previously demonstrated that RAN translation occurs on CGG repeats in FXTAS to produce homopolymeric polyglycine (FMRpolyG) and polyalanine (FMRpolyA) proteins7. FMRpolyG proteins aggregate in cellular, fly, and mouse models of FXTAS and in ubiquitinated inclusions in patient brain tissue, and FMRpolyG production drives CGG repeat toxicity in simple model systems7, 19. The FMR1 locus also produces an antisense transcript, ASFMR1, that contains an antisense CCG repeat20, 21. This antisense repeat lies within a putative open reading frame, that if utilized would generate a short polyproline containing protein, ASFMRP20. ASFMR1 mRNA is ubiquitously expressed in human tissue, with highest expression in the brain, and its expression is elevated in FXTAS patients and models20. Because the CCG repeat in the ASFMR1 transcript is predicted to form a stable secondary structure22, we hypothesized that it might support RAN translation and potentially contribute to disease pathogenesis in FXTAS, similar to the CGG repeat in the sense transcript23. Here we provide evidence from cellular models that the CCG repeat can support RAN translation in all three reading frames to produce homopolymeric proteins. We further demonstrate that antibodies generated against the predicted proline (ASFMRpolyP) and alanine (ASFMRpolyA) RAN products stain ubiquitinated neuronal inclusions in FXTAS patients. These data suggest a role for CCG repeat RAN translation in neuronal inclusion formation in FXTAS.
MATERIALS AND METHODS
Constructs
Base constructs and cloning strategies used were previously described24. Briefly, each frame of ASFMR1 were cloned into GGG-NL-3xFLAG pcDNA by two rounds of Q5 site directed mutagenesis (New England BioLabs) followed by two rounds of annealing primer ligation using XhoI and EcoRV and NarI and EcoRV restriction sites, respectively. All constructs were verified by Sanger sequencing. Expanded repeats were inserted using XhoI and NarI from FMRpolyG100 GFP7. Repeat sizes were determined by restriction digest. Primer and construct sequences are available upon request.
Western blotting
COS-7 cells were transfected with reporter constructs using Lipofectamine LTX with Plus reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol. SDS-PAGE and western blotting were done at 24hrs post transfection as described previously7, 24. For analysis of tagged constructs, membranes were incubated with mouse monoclonal FLAG-M2 (1:1000, Sigma, F1804) and mouse monoclonal GAPDH (1:1000, Santa Cruz Biotechnology, sc32233) overnight at 4°C. Detection was performed using Western Lightning ECL-Plus (Perkin Elmer) on autoradiography film. AUG-nanoLuciferase-3xFLAG (AUG-NL) was loaded at 1/10th the amount of the other reporters to avoid overexposure on the film.
Luciferase assay
COS-7 cells were plated on a 96-well tissue culture plate and were co-transfected with ASFMR1 reporter constructs and pGL4.13 (Firefly luciferase, Promega) using Viafect (Promega) according to the manufacturer’s protocol. Luciferase assays were performed at 24hrs post transfection as previously described24. NanoLuciferase signal was normalized to the internal Firefly luciferase control. Each experiment included three technical replicates averaged to generate a single “n”. All experiments were carried out at least in triplicate. Experimental results were analyzed by one-way ANOVA using GraphPad Prism. Significance of the difference between individual constructs was determined by Fisher’s LSD with Bonferroni correction for multiple comparisons.
Generation of polyclonal antibodies
Rabbit polyclonal antibodies were generated by NeoScientific to synthetic peptides corresponding to the repeat and C-terminal sequence of the predicted proteins (exact epitopes shown in Fig 1). Antisera were affinity purified using the respective peptide immunogens. Pre-immune sera were used as a negative control.
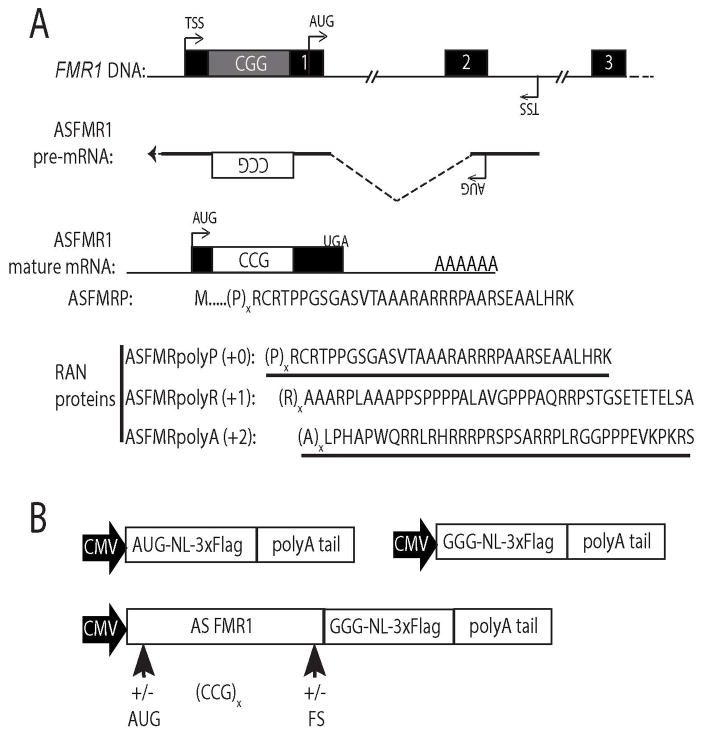
Figure 1. ASFMR1 transcript and putative RAN translation products.
A) The FMR1 locus is bidirectionally transcribed with a start site (TSS) between exons 2 and 3 in the antisense orientation. ASFMR1a mRNA includes an AUG start codon upstream of the CCG repeat, creating a polyproline containing open reading frame (ASFMRP). RAN translation of ASFMR1a mRNA could result in proline (+0 reading frame, ASFMRpolyP), arginine (+1, ASFMRpolyR), and alanine (+2, ASFMRpolyA) repeat proteins. Underlined regions are epitopes used for antibody generation. B) ASFMR1 RAN specific reporters contain the ASFMR1 mRNA sequence including the repeat upstream of a C-terminally 3xFLAG tagged nanoLuciferase lacking a start codon (GGG-NL). Expression constructs were generated for each reading frame by addition of nucleotide frameshifts (+/−FS) at different CCG repeat lengths and with or without the AUG start codon for ASFMRP (+/−AUG).
Immunocytochemistry
COS-7 cells were grown on 4-well chamber slides and transfected with Lipofectamine LTX with Plus reagent. Twenty four hours post transfection, cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton-X in phosphate buffered saline, 1mM MgCl2, 0.1mM CaCl2 (PBS-MC) and then blocked in 5% normal goat serum (NGS) in PBS-MC for 1hr at room temperature. Incubation with primary antibodies (FLAG-M2, 1:100, Sigma; ASpolyP, 1:50, NeoScientific; ASpolyA, 1:10, NeoScientific; Nucleolin, 1:500, Abcam, ab22758) diluted in 5% NGS in PBS-MC was done overnight at 4°C. After rinsing cells with PBS-MC, slides were incubated with goat anti-mouse and goat anti-rabbit IgG antibodies conjugated with Alexa488 and Alexa555, respectively (1:500 each, Thermo Fisher Scientific, A11029 and A21428, respectively) for 1hr at room temperature. Slides were washed and coverslips were mounted with Prolong Gold with DAPI (Thermo Fisher Scientific). Images were captured on an inverted Olympus IX71 microscope at the same exposure and processed using SlideBook 5.5 software, with changes in brightness and contrast applied to the whole image and identically to all images used in a given figure according to published standards25.
Immunohistochemistry and Co-immunofluorescence
Control and FXTAS autopsy tissue was obtained from the University of Michigan Brain Bank and the New York Brain Bank with informed consent of patients or their relatives and approval of local institutional review boards. Two of the FXTAS cases were previously described26. The third case had parkinsonism, progressive gait difficulties and dementia with onset in his 70s. Autopsy showed ubiquitinated inclusions in his brainstem, hippocampus, and cortex with rare Lewy bodies in the substantia nigra (data not shown). Tissue sections were processed as previously described7. For immunohistochemistry, primary antibodies (ASpolyA, 1:10; ASpolyP, 1:100; Ubiquitin, 1:250, Dako Z0458) were diluted in 5% NGS in Tris, pH7.6, 0.1% Triton X-100, 0.5% bovine serum albumin (Tris-B) and incubated overnight at 4°C. Antigen retrieval was required for some antibodies (Ubiquitin: 0.1mM sodium citrate, pH8, 10min at 80°C; ASpolyA: 0.1mM sodium citrate, pH6, 5min at 80°C). For co-immunofluorescence studies with ubiquitin, primary antibodies (Ubiquitin, 1:250, Millipore, MAB1510 with ASpolyA, 1:10 or ASpolyP 1:50) were incubated overnight at 4°C with 5% NGS in PBS-MC, and incubated with goat anti-rabbit and goat anti-mouse IgG antibodies conjugated with Alexa488 and Alexa635, respectively (1:500, Thermo Fisher Scientific, A11008 and A31574, respectively). Images were captured on an Olympus confocal microscope, compiled using ImageJ, and analyzed as previously described7.
RESULTS
The FMR1 locus is bidirectionally transcribed under both normal and pathological conditions20, 21 (Fig 1). The antisense transcript, ASFMR1, has multiple promoters and alternative splice isoforms. One isoform, ASFMR1a, includes a region of the second intron and exon of FMR1 fused by alternative splicing to ~500 nucleotides of exon 1 extending past the annotated transcription start site of FMR120. ASFMR1a transcripts include the 5′ leader sequence of FMR1 and the repeat in a CCG orientation. This transcript has a predicted open reading frame with an AUG start codon that includes the repeat in the polyproline reading frame (ASFMRP) (Fig 1A)20. If RAN translation of ASFMR1a mRNA were to occur upstream of the repeat, it would produce three different repeat proteins: ASFMRpolyP, a polyproline protein derived from the same CCG (+0) reading frame as ASFMRP; ASFMRpolyR, a polyarginine protein from the CGC (+1) reading frame; and ASFMRpolyA, a polyalanine protein from the GCC (+2) reading frame.
To evaluate whether CCG repeats in the context of the ASFMR1a transcript can support RAN translation, we generated a series of CCG RAN translation-specific nanoLuciferase (NL) reporters tagged with a carboxy-terminal 3xFLAG epitope to allow for easy detection by western blot and immunocytochemistry and quantification by luciferase activity24 (Fig 1B). To assure that the reporter only provided signal if initiation occurred within the ASFMR1 sequence, we mutated the AUG start codon of NL to GGG, which eliminated most of its translation and luciferase activity24. We then cloned the ASFMR1a sequence upstream of the modified nanoLuciferase coding sequence. Two sets of constructs were generated. One set retained the native AUG start codon naturally present in ASFMR1 and a second set where this AUG was removed (Fig 1B). In addition, frameshifts were introduced below the repeat such that the NL coding sequence would be in frame each of the three different potential RAN products, creating repeat protein-luciferase fusions. We also generated constructs with a range of repeat sizes, from the normal range in humans (~25 repeats) up into the pathologic repeat size in FXTAS patients (>55 repeats) (Fig 1B).
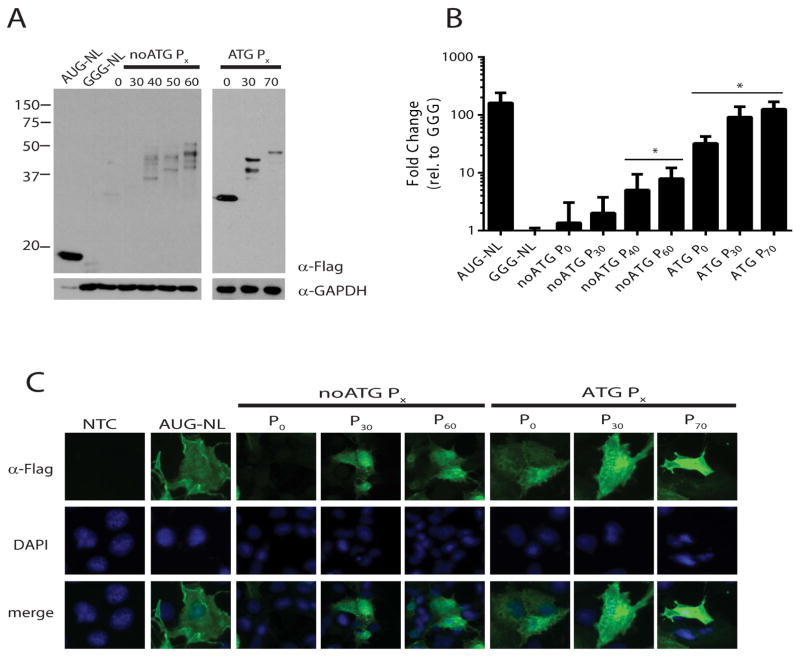
Transient transfection of COS-7 cells expression of AUG-NL ran as a single band of ~18kD by western blot analysis of the 3X-FLAG tag (Fig 2A). Mutation of this start codon to GGG led to a marked reduction in NL production and luciferase signal (Fig 2A and B)24. When the ASFMR1 sequence was introduced above GGG NL in the +0 (proline) reading frame, higher molecular weight products were detected that increased in size with increasing CCG repeat length, consistent with translation initiating in ASFMR1 above the repeat (Fig 2A). To determine if RAN translation could occur, the natural upstream AUG start codon initiating ASFMRP was removed. This change markedly decreased the higher molecular weight protein products in constructs lacking the CCG repeat or with a normal repeat size (30 CGG repeats) (Fig 2A and B). However, at larger repeat sizes (>40 CGG repeats) these higher molecular weight species persisted in the absence of the AUG codon, with enhanced abundance with increasing repeat size as measured by luciferase activity (Fig 2A and B), consistent with RAN translational initiation.
Figure 2. RAN translation from CCG repeats in the proline reading frame of ASFMR1.
A) Anti-FLAG western blot of whole cell lysates from COS-7 cells transfected with the indicated reporters for ASFMRP and ASFMRpolyP. Molecular weight of ASFMR1 derived proteins increased with expanded repeats (ATG Px). Removal of the AUG start codon for ASFMRP (noATG Px, left) eliminated these proteins in the absence of a repeat, but did not prevent their generation at larger repeat sizes. GAPDH served as a loading control and AUG-NL and GGG-NL served as positive and negative controls, respectively. B) NanoLuciferase activity from indicated constructs. For all figures, the Y axis is mean ± standard deviation expressed as fold change above GGG-NL (n>3). *=p<0.05 by Fisher’s LSD with Bonferroni correction for individual comparisons to GGG-NL and by ANOVA for repeat length dependent differences among RAN reporters. C) Localization of ASFMRP and ASFMRpolyP in transfected COS-7 cells stained for FLAG (green). There was no change in distribution with increasing repeat size and FLAG positive inclusions were not observed. 4′,6-Diamidino-2-phenylindole (DAPI, blue) was used to counterstain nuclei.
To determine the subcellular distribution of these novel polyproline containing proteins, we performed immunofluorescence against the FLAG tag epitope in transfected cells (Fig 2C). AUG initiated ASFMRP fused to NL was distributed throughout the cytoplasm and nucleus in a pattern that was similar to AUG-NL alone. Neither increasing repeat size nor the removal of the AUG start codon significantly altered this distribution.
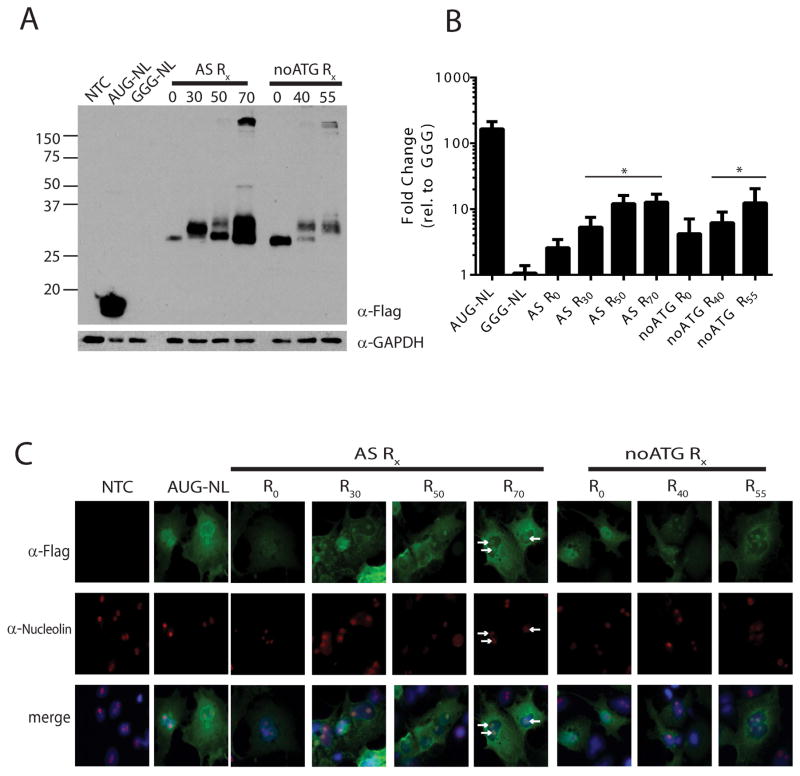
We next tested whether RAN translation could occur in the other two potential repeat reading frames (CGC, Arginine and GCC, Alanine). When the GGG-NL reporter is placed in the +1 (Arginine) reading frame, we observed a higher molecular weight species by western blot that increased with increasing repeat number and formed large(>150kD) complexes at higher repeat sizes (Fig 3A), consistent with published reports of arginine containing RAN proteins8, 11. This product was generated and present regardless of whether the AUG codon normally in the proline reading frame was present or removed (Fig 3A, noATG). As with ASFMRpolyP, translation of ASFMRpolyR was enhanced at increasing repeat sizes (Fig 3B). Unlike the staining pattern of ASFMRpolyP, ASFMRpolyR exhibited a clear redistribution in its intracellular staining pattern that was dependent on the arginine repeat, such that at normal and expanded repeat numbers, the protein localized to the nucleolus (Fig 3C). This change in localization was confirmed by co-immunofluorescence with the nucleolar marker nucleolin and is consistent with findings reported for dipeptide repeat containing RAN products generated in C9orf7227, 28.
Figure 3. RAN translation from CGC repeats in the arginine reading frame of ASFMR1.
A) western blot against FLAG in cells expressing the indicated ASMFRpolyR reporters. GAPDH was used as a loading control. “noATG” indicates the ATG codon in the proline reading frame was removed. B) NanoLuciferase activity derived from the indicated constructs 24hrs post transfection. *=p<0.05 by Fisher’s LSD with Bonferroni correction for individual comparisons to GGG-NL and by ANOVA for repeat length dependent differences among RAN reporters. C) FLAG staining of ASFMRpolyR constructs transfected into COS-7 cells. ASFMRpolyR staining (green) shifted to the nucleus in the presence of the expanded CCG repeats and co-localized (arrows) with the nucleolar marker nucleolin (red). DAPI (blue) was used to counter stain nuclei.
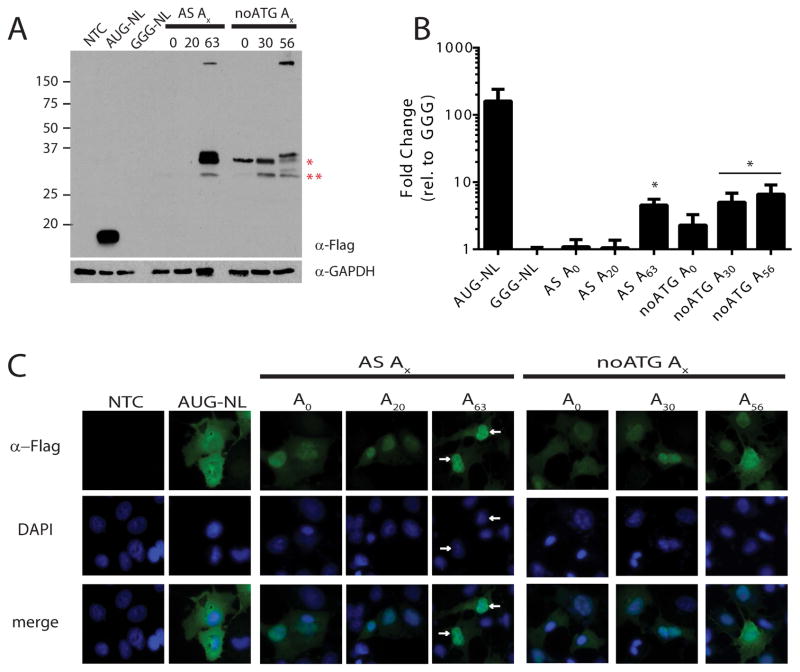
In the third (+2, Alanine, GCC) potential reading frame, a similar pattern was observed (Fig 4). Translation of higher molecular weight species were observed in this reading frame in both the presence and absence of the repeat. Mutational analysis demonstrated that in the absence of any repeat, initiation occurred predominantly below the repeat at two different near-AUG codons found in the human sequence (Fig 4A and data not shown). Initiation at these sites did not significantly increase the luciferase signal above that of our negative control construct (Fig 4B). However at increasing repeat sizes, an additional higher molecular weight product appeared that increased in size in line with repeat number (Fig 4A). At greater than 50 CCG repeats, products accumulated as large complex (>150kD) near the top of the gel. ASFMRpolyA abundance increased with addition of repeats and was greatest at the largest repeat sizes (Fig. 4B). ASFMRpolyA production was significantly suppressed by inclusion of the upstream AUG codon in the proline reading frame in the absence of any repeat or at normal repeat sizes, but not at expanded repeats (Fig 4A and B). This alanine translation product, ASFMRpolyA, exhibited a clear change in its cellular distribution in transfected cells compared to AUG-NL alone, with a repeat length dependent redistribution into the nucleus (Fig 4C).
Figure 4. RAN translation from GCC repeats in the alanine reading frame of ASFMR1.
A) western blot against FLAG on lysates from COS-7 cells transfected with indicated ASFMRpolyA reporters. Red asterisks indicate bands generated from initiation 3′ to the repeat site but in the human sequence at a non-canonical start codon. “noATG” indicates the ATG codon in the proline reading frame was absent. B) NanoLuciferase activity from ASFMRpolyA constructs compared to GGG-NL. *=p<0.05 by Fisher’s LSD with Bonferroni correction for individual comparisons to GGG-NL and by ANOVA for repeat length dependent differences among RAN reporters. C) Localization of ASFMRpolyA (green) was primarily nuclear (arrows) compared to AUG-NL which was cytoplasmic. DAPI (blue) was used to counterstain nuclei.
If RAN translation products from ASFMR1a RNA are generated in vivo, then we would predict that we should be able to identify them in FXTAS patients. To test this hypothesis, we generated a series of polyclonal antibodies against short stretches of the predicted repeats and the C-terminal portion of the predicted proteins (Fig 1A, underlined region of protein sequences). To assess the specificity of these antibodies, we performed western blotting and immunofluorescence on cells expressing the appropriate RAN translation product or control constructs lacking the 5′ leader sequence. Both the ASFMRpolyP and ASFMRpolyA specific antibodies exhibited specificity for their cognate RAN protein by western blot (Fig 5A and C) and by immunofluorescence (Fig 5B and D), respectively. Unfortunately, despite multiple attempts and use of different epitopes, antibodies generated against the predicted ASFMRpolyR protein failed to exhibit sufficient specificity in these validation assays to support further testing in human tissues (data not shown).
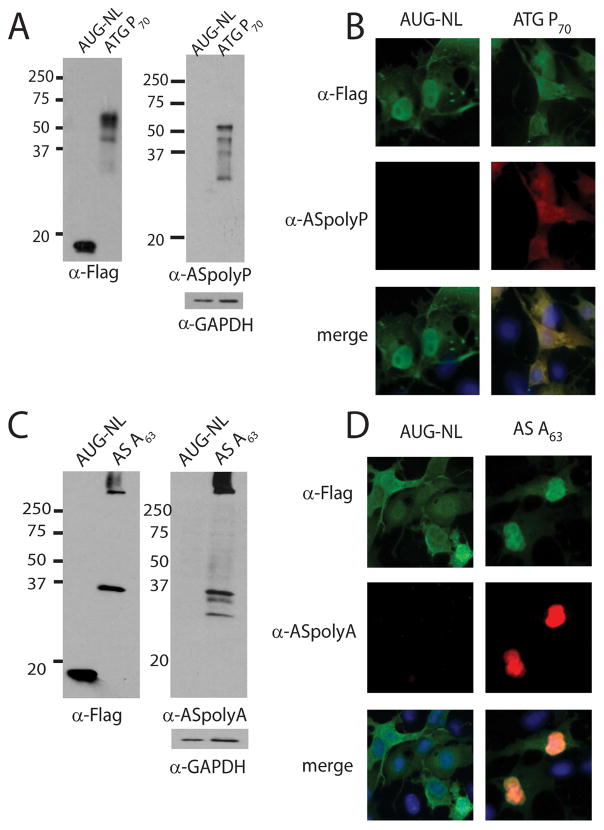
Figure 5. ASFMRpolyP and ASFMRpolyA antibody validation.
A and C) Western blots of constructs probed with FLAG or antibody generated against ASFMRpolyP (α–ASpolyP, panel A), or against ASFMRpolyA (α–ASpolyA, panel C). GAPDH was used as a loading control. B and D) α–ASpolyP (red, panel B) recognized ASFMRPolyP but not AUG-NL by co-immunofluorescence. Similarly, α–ASpolyA (red, panel D) specifically recognized ASFMRpolyA but not AUG-NL.
With the validated antibodies, we looked for the presence of the ASFMR1 protein products in human brain tissue from FXTAS patients. Pre-immune sera for both antibodies showed minimal background staining in both controls and FXTAS cases (Fig 6C and 7C). When FXTAS tissues were stained with ASFMRpolyP antibodies, we observed staining in both FXTAS and control tissues in the hippocampus, cortex, and cerebellum (Fig 6A). Staining was most intense in the perinuclear region of neurons and was more robust in most tissues in FXTAS cases than in controls. In addition, ASFMRpolyP antibodies reliably stained intranuclear neuronal aggregates in FXTAS tissues that were not observed in control tissues (Fig 6B). These aggregates were present in multiple brain regions and were primarily neuronal. To better characterize these aggregates, we performed co-immunofluorescence with ASFMRpolyP and ubiquitin followed by confocal microscopy. We observed numerous aggregates in neurons in FXTAS cases that were both ubiquitin and ASFMRpolyP positive (Fig 6D).
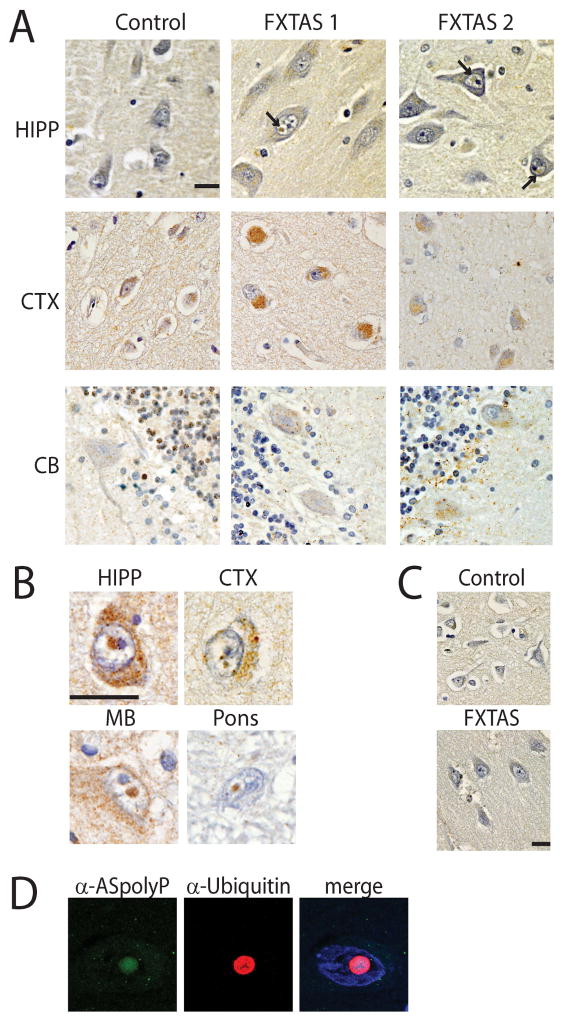
Figure 6. ASFMRpolyP accumulates in FXTAS brain tissue and intranuclear inclusions.
A) Control and FXTAS tissue from the indicated brain regions stained with α–ASpolyP. Hematoxylin (blue) was used as a counterstain to identify nuclei. In addition to strong perinuclear staining, nuclear aggregates (arrows) were observed in FXTAS cases that were not seen in control tissue. B) Higher magnification of intranuclear neuronal inclusions from the indicated brain regions from FXTAS cases probed with α–ASpolyP. C) Pre-immune sera for α–ASpolyP did not show staining in control or FXTAS patient cortex. D) Immunofluorescence of FXTAS hippocampus showed colocalization of ASFMRpolyP (green) with ubiquitin (red). DAPI (blue) was used to identify nuclei. Scale bar is 20μm. HIPP: hippocampus; CTX: frontal cortex; CB: cerebellum; MB: midbrain.
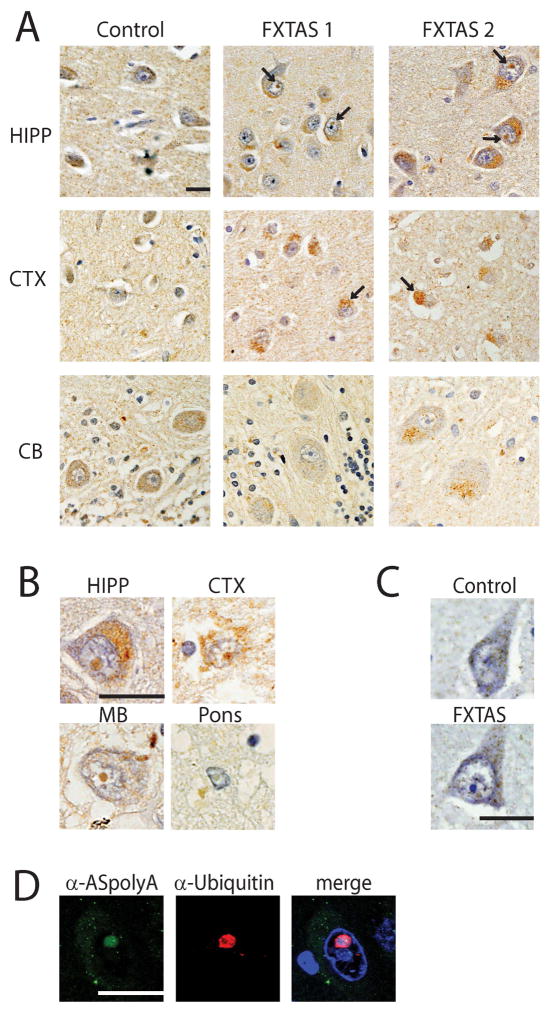
Figure 7. ASFMRpolyA RAN proteins aggregate in FXTAS brain tissue.
A) Control and FXTAS tissue from the indicated brain regions stained with α–ASpolyA. Hematoxylin (blue) was used to identify nuclei. In addition to strong perinuclear staining, nuclear aggregates (arrows) were observed in FXTAS cases that were not seen in control tissue. B) Higher magnification of intranuclear neuronal inclusions from the indicated brain regions from FXTAS cases probed with α–ASpolyA. C) Pre-immune sera for α–ASpolyA did not show specific staining in control or FXTAS patient cortex. D) Co-immunofluorescence on FXTAS hippocampus showed colocalization of ASFMRpolyA (green) with ubiquitin (red). DAPI (blue) was used to identify nuclei. Scale bar is 20μm. HIPP: hippocampus; CTX: frontal cortex; CB: cerebellum; MB: midbrain.
To determine if ASFMRpolyA also accumulated in FXTAS cases, we performed similar immunohistochemical and co-immunoflourescence studies. Like ASFMRpolyP, ASFMRpolyA was found extensively in FXTAS, with the greatest staining in the perinuclear regions of neurons in the hippocampus and cortex (Fig 7A). ASFMRpolyA stained numerous intranuclear neuronal inclusions in multiple tissues and these inclusions were ubiquitin positive by co-immunoflourescence (Fig 7B, D).
DISCUSSION
Aggregation of misfolded proteins is a hallmark in neurodegenerative disorders across a spectrum of etiologies2, 29. Identification of the misfolded protein species in each disorder is a critical first step in elucidating the pathogenic cascades responsible for neurodegeneration in that condition. Here we demonstrate at least two new proteins that accumulate in the neurodegenerative disorder FXTAS: a polyproline containing protein, ASFMRpolyP, and a polyalanine containing protein, ASFMRpolyA. Using a series of reporter constructs, we further demonstrate that both of these proteins can be generated through RAN translation: a recently described unconventional form of initiation that occurs at multiple repeat expansions, including the CGG repeat in the sense strand of FMR1 in FXTAS7.
RAN translation has now been reported for six different repeats, with three derived from sense strand mRNA transcripts (CAG, GGGGCC, and CGG repeats) and three derived primarily from antisense RNAs (CAG, CUG, CCCCGG, and CCG repeats)5, 7–13. All of these transcripts are capable of forming strong secondary structures in vitro, either RNA hairpins or G-quadruplexes, and the ability to form these secondary structures appears important to the process underlying RAN translation5, 27, 30, 31. The fact that this CCG repeat resides within an open reading frame is also consistent with previous data on CAG repeats in Huntington disease and spinocerebellar ataxia type 8, both of which allow RAN translation to occur in all three potential reading frames within an open reading frame13. However, recent work on RAN translation at CGG repeats supports a model for initiation that retains a requirement for a 7-methylguanosine 5′-cap on the mRNA and ribosomal scanning24, both of which superficially do not fit with a location of the repeat downstream within an open reading frame and would require RAN translation to bypass the canonical AUG start codon. Thus, studies of initiation mechanisms at CCG repeats in its native sequence context will be needed to delineate how this atypical process occurs in human cells and how it agrees or disagrees with findings at other repeats.
What role ASFMR1 mRNA derived protein products have in FXTAS disease pathogenesis is also unclear. Expression of CCG repeats in isolation in the 5′ leader sequence of GFP was sufficient to elicit toxicity in a Drosophila model system23. However, whether this toxicity was driven by the repeats as RNA or as RAN translated proteins is not known. Polyalanine-containing proteins and RAN products have previously been shown to be toxic in isolation5, 32, and oculopharyngeal muscular dystrophy results from a polyalanine expansion in the polyadenylate binding protein 233, 34. In contrast, little is known about the potential for polyproline or polyarginine proteins to cause toxicity. Expression of dipeptide-repeat containing proteins generated from GGGGCC repeats in C9orf72 containing arginine and/or proline (glycine-arginine repeats, proline-alanine, and proline-arginine repeats) are toxic in cells and simple model systems when expressed at high levels in the absence of a structured RNA repeat 35–38. Moreover, the repetitive arginine elements target these proteins to the nucleolus38, which we also observe with ASFMRpolyR in cell transfection studies. Thus, defining both the potential for each of these ASFMR1 RAN proteins to elicit toxicity in isolation in model systems and their relative abundance in FXTAS tissues will be important next steps in determining their potential roles in disease pathogenesis.
Our current study and antibodies cannot differentiate between polyproline products generated by AUG initiated translation and RAN translation. Our cell-based luciferase reporter assays suggests that removal of the AUG start codon from the ASFMRP open reading frame decreases the signal by approximately 10 fold (Fig 2B). In addition, we observe staining in control tissues with this antibody (although no staining of intranuclear aggregates) (Fig 6A), but limited RAN products were detected based on reporter assays at normal repeat sizes (Fig. 2A, B). Thus, our antisense proline antibody is likely staining both the AUG initiation derived ASFMRP protein and the RAN derived ASFMRpolyP protein, and both are potentially contributing to aggregation formation and toxicity.
In summary, we provide evidence for RAN translation at CCG repeats derived from an antisense FMR1 transcript. This work expands the list of potential pathogenic species at play in Fragile X-associated tremor/ataxia syndrome and provides further support for non-canonical translation of microsatellite repeat expansions in the pathology of human neurodegenerative disease.
Acknowledgments
This work was funded by grants from the NIH (R01NS086810) and the Veterans Health Administration (1I01BX001689) to P.K.T. M.G.K. was supported by NIH F3211541507.
The authors would like to thank members of the Todd lab for technical suggestions and contributions. We thank Hank Paulson and Andy Lieberman for comments on the manuscript. Histological samples from FXTAS cases were provided by the New York Brain Bank at Columbia University and University of Michigan Brain Bank MADC P30 grant (supported by P30 AG053760). Dr. Laura Ranum supplied unpublished reagents which were used in preliminary work not included in this manuscript.
Footnotes
AUTHOR CONTRIBUTIONS
A.K., M.G.K, and P.K.T. contributed to the conception, design of the study, and data analysis; A.K. performed the experiments; A.K. and P.K.T. wrote the manuscript with editorial input from M.G.K.
POTENTIAL CONFLICTS OF INTEREST
All authors declare no conflicts of interest.
References
- 1.Mason AR, Ziemann A, Finkbeiner S. Targeting the low-hanging fruit of neurodegeneration. Neurology. 2014;83:1470–1473. doi: 10.1212/WNL.0000000000000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williams AJ, Paulson HL. Polyglutamine neurodegeneration: protein misfolding revisited. Trends Neurosci. 2008;31:521–528. doi: 10.1016/j.tins.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Todd PK, Paulson HL. RNA-mediated neurodegeneration in repeat expansion disorders. Ann Neurol. 2010;67:291–300. doi: 10.1002/ana.21948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson DL, Orr HT, Warren ST. The unstable repeats--three evolving faces of neurological disease. Neuron. 2013;77:825–843. doi: 10.1016/j.neuron.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zu T, Gibbens B, Doty NS, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green KM, Linsalata AE, Todd PK. RAN translation-What makes it run? Brain research. 2016 doi: 10.1016/j.brainres.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Todd PK, Oh SY, Krans A, et al. CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron. 2013;78:440–455. doi: 10.1016/j.neuron.2013.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mori K, Weng SM, Arzberger T, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 9.Mori K, Arzberger T, Grasser FA, et al. Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 2013;126:881–893. doi: 10.1007/s00401-013-1189-3. [DOI] [PubMed] [Google Scholar]
- 10.Zu T, Liu Y, Banez-Coronel M, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci U S A. 2013;110:E4968–4977. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ash PE, Bieniek KF, Gendron TF, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gendron TF, Bieniek KF, Zhang YJ, et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 2013;126:829–844. doi: 10.1007/s00401-013-1192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banez-Coronel M, Ayhan F, Tarabochia AD, et al. RAN Translation in Huntington Disease. Neuron. 2015;88:667–677. doi: 10.1016/j.neuron.2015.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagerman RJ, Leehey M, Heinrichs W, et al. Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology. 2001;57:127–130. doi: 10.1212/wnl.57.1.127. [DOI] [PubMed] [Google Scholar]
- 15.Jacquemont S, Hagerman RJ, Leehey MA, et al. Penetrance of the fragile X-associated tremor/ataxia syndrome in a premutation carrier population. JAMA. 2004;291:460–469. doi: 10.1001/jama.291.4.460. [DOI] [PubMed] [Google Scholar]
- 16.Berry-Kravis E, Abrams L, Coffey SM, et al. Fragile X-associated tremor/ataxia syndrome: clinical features, genetics, and testing guidelines. Movement disorders : official journal of the Movement Disorder Society. 2007;22:2018–2030. doi: 10.1002/mds.21493. quiz 2140. [DOI] [PubMed] [Google Scholar]
- 17.Hagerman P. Fragile X-associated tremor/ataxia syndrome (FXTAS): pathology and mechanisms. Acta Neuropathol. 2013;126:1–19. doi: 10.1007/s00401-013-1138-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greco CM, Hagerman RJ, Tassone F, et al. Neuronal intranuclear inclusions in a new cerebellar tremor/ataxia syndrome among fragile X carriers. Brain : a journal of neurology. 2002;125:1760–1771. doi: 10.1093/brain/awf184. [DOI] [PubMed] [Google Scholar]
- 19.Oh SY, He F, Krans A, et al. RAN translation at CGG repeats induces ubiquitin proteasome system impairment in models of fragile X-associated tremor ataxia syndrome. Human molecular genetics. 2015;24:4317–4326. doi: 10.1093/hmg/ddv165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ladd PD, Smith LE, Rabaia NA, et al. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum Mol Genet. 2007;16:3174–3187. doi: 10.1093/hmg/ddm293. [DOI] [PubMed] [Google Scholar]
- 21.Khalil AM, Faghihi MA, Modarresi F, Brothers SP, Wahlestedt C. A novel RNA transcript with antiapoptotic function is silenced in fragile X syndrome. PLoS One. 2008;3:e1486. doi: 10.1371/journal.pone.0001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiliszek A, Kierzek R, Krzyzosiak WJ, Rypniewski W. Crystallographic characterization of CCG repeats. Nucleic Acids Res. 2012;40:8155–8162. doi: 10.1093/nar/gks557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sofola OA, Jin P, Botas J, Nelson DL. Argonaute-2-dependent rescue of a Drosophila model of FXTAS by FRAXE premutation repeat. Hum Mol Genet. 2007;16:2326–2332. doi: 10.1093/hmg/ddm186. [DOI] [PubMed] [Google Scholar]
- 24.Kearse MG, Green KM, Krans A, et al. CGG Repeat-Associated Non-AUG Translation Utilizes a Cap-Dependent Scanning Mechanism of Initiation to Produce Toxic Proteins. Molecular cell. 2016;62:314–322. doi: 10.1016/j.molcel.2016.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossner M, Yamada KM. What’s in a picture? The temptation of image manipulation. J Cell Biol. 2004;166:11–15. doi: 10.1083/jcb.200406019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Louis E, Moskowitz C, Friez M, Amaya M, Vonsattel JP. Parkinsonism, dysautonomia, and intranuclear inclusions in a fragile X carrier: a clinical-pathological study. Mov Disord. 2006;21:420–425. doi: 10.1002/mds.20753. [DOI] [PubMed] [Google Scholar]
- 27.Haeusler AR, Donnelly CJ, Periz G, et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schludi MH, May S, Grasser FA, et al. Distribution of dipeptide repeat proteins in cellular models and C9orf72 mutation cases suggests link to transcriptional silencing. Acta neuropathologica. 2015;130:537–555. doi: 10.1007/s00401-015-1450-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forman MS, Trojanowski JQ, Lee VM. Neurodegenerative diseases: a decade of discoveries paves the way for therapeutic breakthroughs. Nat Med. 2004;10:1055–1063. doi: 10.1038/nm1113. [DOI] [PubMed] [Google Scholar]
- 30.Reddy K, Zamiri B, Stanley SY, Macgregor RB, Jr, Pearson CE. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J Biol Chem. 2013;288:9860–9866. doi: 10.1074/jbc.C113.452532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sobczak K, de Mezer M, Michlewski G, Krol J, Krzyzosiak WJ. RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Res. 2003;31:5469–5482. doi: 10.1093/nar/gkg766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Eyk CL, McLeod CJ, O’Keefe LV, Richards RI. Comparative toxicity of polyglutamine, polyalanine and polyleucine tracts in Drosophila models of expanded repeat disease. Human molecular genetics. 2012;21:536–547. doi: 10.1093/hmg/ddr487. [DOI] [PubMed] [Google Scholar]
- 33.Brais B, Bouchard JP, Xie YG, et al. Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nature genetics. 1998;18:164–167. doi: 10.1038/ng0298-164. [DOI] [PubMed] [Google Scholar]
- 34.Becher MW, Kotzuk JA, Davis LE, Bear DG. Intranuclear inclusions in oculopharyngeal muscular dystrophy contain poly(A) binding protein 2. Annals of neurology. 2000;48:812–815. [PubMed] [Google Scholar]
- 35.Yamakawa M, Ito D, Honda T, et al. Characterization of the dipeptide repeat protein in the molecular pathogenesis of c9FTD/ALS. Hum Mol Genet. 2015;24:1630–1645. doi: 10.1093/hmg/ddu576. [DOI] [PubMed] [Google Scholar]
- 36.Mizielinska S, Lashley T, Norona FE, et al. C9orf72 frontotemporal lobar degeneration is characterised by frequent neuronal sense and antisense RNA foci. Acta Neuropathol. 2013;126:845–857. doi: 10.1007/s00401-013-1200-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wen X, Tan W, Westergard T, et al. Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron. 2014;84:1213–1225. doi: 10.1016/j.neuron.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tao Z, Wang H, Xia Q, et al. Nucleolar stress and impaired stress granule formation contribute to C9orf72 RAN translation-induced cytotoxicity. Hum Mol Genet. 2015;24:2426–2441. doi: 10.1093/hmg/ddv005. [DOI] [PubMed] [Google Scholar]