SUMMARY
Coxiella burnetii is a gram-negative intracellular bacterium that forms a large, lysosome-like parasitophorous vacuole (PV) essential for bacterial replication. Host membrane lipids are critical for the formation and maintenance of this intracellular niche, yet the mechanisms by which Coxiella manipulates host cell lipid metabolism, trafficking, and signaling are unknown. ORP1L (oxysterol-binding protein (OSBP)-related protein 1, long) is a mammalian lipid-binding protein that plays a dual role in cholesterol-dependent endocytic trafficking as well as interactions between endosomes and the endoplasmic reticulum (ER). We found that ORP1L localized to the Coxiella PV within 12 hours of infection through a process requiring the Coxiella Dot/Icm Type 4B secretion system, which secretes effector proteins into the host cell cytoplasm where they manipulate trafficking and signaling pathways. The ORP1L N-terminal ankyrin repeats were necessary and sufficient for PV localization, indicating ORP1L binds a PV membrane protein. Strikingly, ORP1L simultaneously co-localized with the PV and ER, and electron microscopy revealed membrane contact sites between the PV and ER membranes. In ORP1L-depleted cells, PVs were significantly smaller than PVs from control cells. These data suggest ORP1L is specifically recruited by the bacteria to the Coxiella PV, where it influences PV membrane dynamics and interactions with the ER.
INTRODUCTION
Intracellular bacterial pathogens hijack host cell processes as a mechanism to create or modify their intracellular niche. In the case of pathogens that reside in membrane-bound compartments, membrane and membrane lipids are obtained from the host cell through a variety of mechanisms. One strategy, used by Legionella pneumophila and Brucella spp., is to recruit membrane directly from an organelle such as the endoplasmic reticulum (ER) (Swanson et al., 1995, Pizarro-Cerda et al., 1998). Other pathogens such as Chlamydia trachomatis intercept ceramide and cholesterol trafficking from the ER and Golgi, respectively, using these lipids as building blocks for the bacteria-containing compartment (Hackstadt et al., 1995, Carabeo et al., 2003, Derre et al., 2011, Elwell et al., 2011). A third strategy, used by the Q fever agent Coxiella burnetii, is to create a compartment that interacts with a wide range of host vesicular trafficking pathways (Moffatt et al., 2015). Upon infection, nascent C. burnetii-containing vacuoles mature into acidic phagolysosomes by trafficking through the default endocytic pathway, a process essential for formation of a mature parasitophorous vacuole (PV) that supports growth (Heinzen et al., 1996). Early during infection, C. burnetii acquires the autophagic marker LC3, and early interactions with the host autophagy pathway enhance PV development and bacterial growth (Beron et al., 2002, Gutierrez et al., 2005, Newton et al., 2014). Approximately 24–48 hours after infection, the C. burnetii PV expands, fusing with early and late endosomes, lysosomes, and autophagosomes (Voth et al., 2007). The PV contains the late endosomal markers CD63 and Rab7 (Beron et al., 2002, Ghigo et al., 2002), lysosomal LAMP1 (Heinzen et al., 1996), autophagic markers LC3 and Rab24 (Beron et al., 2002, Gutierrez et al., 2005), and the SNARE proteins Vamp2, Vamp7, Vamp8, and Vti1b (Campoy et al., 2013). Further, both clathrin-mediated vesicular trafficking and retrograde trafficking play a role in PV development (Larson et al., 2013) (McDonough et al., 2013). The mature PV, however, does not appear to fuse directly with early endosomes or the golgi apparatus, based on the lack of markers for these organelles on the PV membrane (Heinzen et al., 1996, Howe et al., 2003).
C. burnetii actively directs PV formation through a Dot/Icm type 4B secretion system (T4BSS), which secretes bacterial effector proteins into the host cell cytoplasm. In the absence of a functional T4BSS, PV expansion and robust bacterial replication does not occur (Carey et al., 2011, Beare et al., 2012). Activation of the T4BSS requires the acidic environment of a late endosome/lysosome, with effector secretion detectable by 8 hours post infection (Newton et al., 2013). Effector proteins have been localized to several major host cell structures, with a subset known as Coxiella vacuolar proteins (Cvp) also localizing to the C. burnetii PV (Larson et al., 2013, Larson et al., 2015). Mutants in any one of the five PV-localized Cvp effectors show reduced intracellular replication, indicating bacterial proteins on the PV membrane are required to maintain an intracellular niche that supports C. burnetii growth (Larson et al., 2015).
In addition to a unique protein profile, the PV membrane contains a high sterol concentration, as evident by staining with the fluorescent sterol-binding compound filipin (Howe et al., 2006, Gilk, 2012). Further, altering cholesterol homeostasis has a dramatic impact on PV formation and bacterial growth (Howe et al., 2006, Czyz et al., 2014), thus indicating cholesterol plays an important, yet unknown, role in Coxiella pathogenesis. In addition to its role in membrane fluidity, cholesterol regulates membrane trafficking and signaling through a large family of sterol sensor and transfer proteins. One member of this family, Oxysterol-binding protein (OSBP)-Related Protein 1 Long, or ORP1L, plays a major role in cholesterol-dependent endosomal trafficking and formation of membrane contact sites (MCS) between late endosomes/lysosomes and the ER. ORP1L undergoes cholesterol-dependent conformational changes that regulate interactions between endosomes and either microtubules or the ER. On cholesterol-rich endosomes, ORP1L adopts a compact conformation with the N-terminal ankyrin repeats binding the Rab7-RILP (Rab-interacting lysosomal protein) complex and the C-terminal ORD (OSBP-related domain) binding cholesterol (Rocha et al., 2009). When ORP1L is in this conformation, RILP is free to bind the homotypic fusion and protein sorting (HOPS)-tethering complex, which in turn binds the motor protein dynein to facilitate minus end-directed transport of endosomes along microtubules (van der Kant et al., 2013a). On low cholesterol membranes, the absence of ORD binding to cholesterol results in an extended ORP1L conformation, allowing the ORP1L FFAT (two phenylalanines (FF) in an acidic tract) motif to interact with the resident ER protein VAP (VAMP-associated ER protein). As a result of binding to both the ER and late endosomes/lysosomes, ORP1L forms MCS between these two organelles and disrupts association between Rab7-RILP and microtubules (Rocha et al., 2009). Related to its role in late endosome trafficking, ORP1L has roles in transport of low density lipoprotein (LDL) cholesterol to the ER (Cianciola et al., 2013), multivesicular body (MVB) formation, and membrane protein degradation (Kobuna et al., 2010). These roles in lipid trafficking may additionally influence transcriptional regulation of cholesterol efflux genes (Johansson et al., 2003).
Given the cholesterol-dependent role of ORP1L in late endosome and lysosome trafficking, we examined ORP1L localization during C. burnetii host cell infection. Here we show that ORP1L localizes to the C. burnetii PV, a process dependent on the Coxiella T4BSS. Despite the high PV sterol content, ORP1L does not primarily bind PV membrane cholesterol, but instead a PV membrane protein through the N-terminal ankyrin repeats. Furthermore, ORP1L simultaneously associates with both the PV and ER, demonstrating for the first time a direct link between these two organelles. Finally, ORP1L depletion with siRNA resulted in smaller PVs compared to PVs from wild type cells, suggesting ORP1L plays a role in PV expansion.
RESULTS
ORP1L localization to the C. burnetii parasitophorous vacuole is T4BSS-dependent
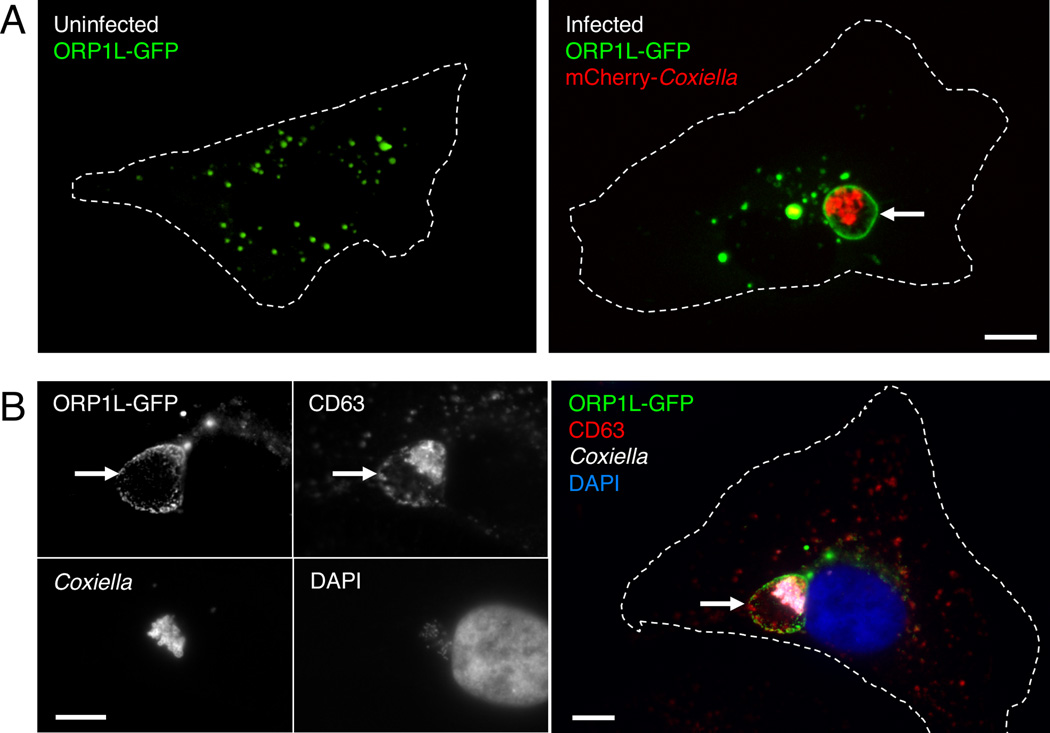
We first examined ORP1L localization in uninfected and infected HeLa cells expressing ORP1L fused to green fluorescent protein (GFP). By live cell microscopy, ORP1L-GFP localized to small vesicular structures in uninfected cells (Figure 1A), consistent with published observations that ORP1L is found on endosomes (Johansson et al., 2003). In C. burnetii-infected cells, ORP1L-GFP also localized to the C. burnetii PV (Figure 1A, arrow). PV localization was confirmed by indirect immunofluorescence, where ORP1L-GFP co-localized with the PV marker CD63 (Figure 1B, arrow). We observed identical localization for both N- and C-terminal GFP fusion proteins (Supplemental Figure 1).
Figure 1. ORP1L localizes to the C. burnetii PV membrane.
Live cell confocal (A) or fixed immunofluorescence (B) microscopy images of uninfected or infected HeLa cells expressing ORP1L-GFP. In uninfected HeLa cells, ORP1L-GFP is found on vesicular structures (A, left). ORP1L-GFP also localizes to the PV membrane (A and B, arrows) in HeLa cells infected for three days with C. burnetii expressing red fluorescent protein mCherry. Cell boundaries are shown with dotted white lines. CD63 is a marker for the PV membrane in B. Scale bars = 10 µm.
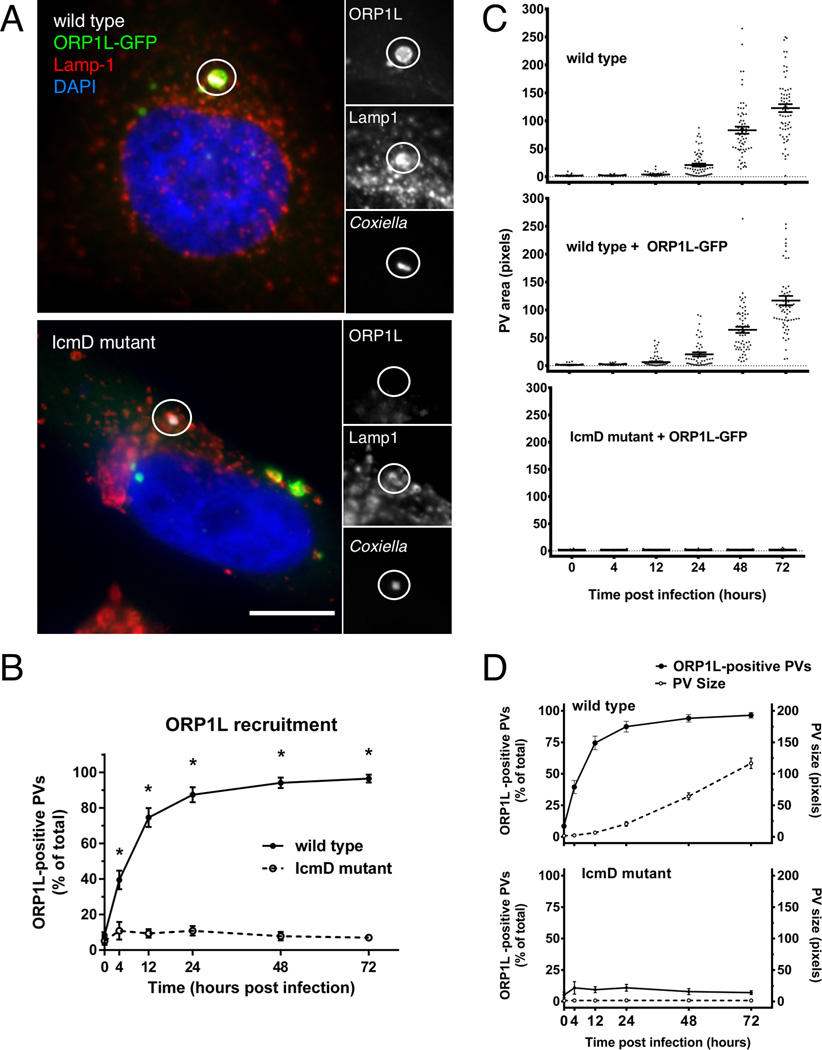
The C. burnetii T4BSS transports bacterial effector proteins across the PV membrane and into the host cell cytoplasm where they manipulate host cell functions such as vesicular trafficking and apoptosis (reviewed in (Moffatt et al., 2015). T4BSS effector proteins recruit several host cell proteins to the PV including clathrin (Larson et al., 2013) and LC3 (Winchell et al., 2014). To determine if the T4BSS is involved in ORP1L PV localization, we examined ORP1L-GFP-expressing cells infected with either wild type C. burnetii or a C. burnetii mutant lacking IcmD, an essential component of the T4BSS (Beare et al., 2011). Cells were fixed and stained for C. burnetii and Lamp1, a lysosomal protein that also serves as a marker for the C. burnetii PV. Unlike PVs harboring wild type bacteria, ORP1L-GFP did not localize to PVs containing the IcmD mutant (Figure 2A). When quantified, over 85% of wild type PVs were ORP1L-positive by 24 hours post infection, with nearly 95% being positive by 48 hours (Figure 2B). In contrast, less than 10% of the IcmD-mutant PVs were positive at any point during a 72 hour infection, demonstrating the T4BSS is required for ORP1L localization to the C. burnetii PV.
Figure 2. T4BSS-dependent recruitment of ORP1L to the PV membrane.
(A) Light microscopy images of HeLa cells transfected with ORP1L-GFP and then infected with C. burnetii for 2 days. ORP1L-GFP is recruited to the PV of wild type (top) but not IcmD mutant (bottom) bacteria. Green = ORP1L-GFP, Red = Lamp1, Gray = C. burnetii, Blue = DAPI. The PV (circled) is shown magnified with individual fluorescent channels. Scale bar = 10 µm. (B) The number of ORP1L-positive PVs was quantified over a 72 hour time course of infection in HeLa cells infected with wild type (solid) or IcmD mutant (dashed) C. burnetii. Cells were transfected with ORP1L-GFP, infected, stained by immunofluorescence for C. burnetii and Lamp1, and visually scored for the presence or absence of ORP1L on the PV. Each data point represents the average of 3–4 experiments, with 20 PVs counted per experiment. Error bars represent standard error of the mean (SEM). Means compared by unpaired Welch’s t-test. * = p <0.01. (C) Measurements of PV size shows that PVs harboring wild type bacteria expand between 24 and 48 hours, while IcmD mutant PVs do not expand. HeLa cells, untransfected or expressing ORP1L-GFP, were infected with either wild type C. burnetii or the IcmD mutant. At the indicated times, the cells were fixed, stained for C. burnetii and CD63, and the PVs measured using ImageJ. PVs harboring wild type bacteria expanded between 24 and 48 hours, while IcmD PVs did not expand. Shown are individual PV measurements from three separate experiments, with at least 20 PVs per timepoint per experiment. Bars represent mean ± SEM. (D) In the comparison of ORP1L localization compared to PV expansion, ORP1L localizes to the PV 12 hours prior to significant expansion of wild type PVs (top). Error bars represent SEM.
ORP1L recruitment occurs prior to PV expansion
T4BSS-dependent localization of ORP1L to the PV could occur either through fusion with endosomes, or through direct recruitment by a PV-associated protein. ORP1L does not contain transmembrane domains, and membrane association requires binding to membrane proteins or lipids. We therefore hypothesized that ORP1L interacts with the PV by protein-protein or protein-lipid interactions, and is not delivered through membrane fusion events during PV expansion. To test this, we assessed the timing of PV expansion in comparison to ORP1L PV localization. To determine when PV expansion occurs in HeLa cells, we measured PVs harboring either wild type or IcmD mutant bacteria during a 72 hour infection time course (Figure 2C). PVs harboring IcmD mutant bacteria never expanded, consistent with the role of the T4BSS in PV fusion and expansion. For wild type bacteria, a small amount of expansion was observed by 24 hours, with the vast majority of PVs expanding between 24 and 48 hours. At 48 hours post infection, there was a small but statistically significant difference in PV size between untransfected and ORP1L-GFP transfected cells (Supplemental Figure 2). However, the timing of expansion was identical, suggesting that ectopic expression of ORP1L-GFP does not affect the overall timing of expansion. When PV size was compared to ORP1L PV localization in wild type PVs, ORP1L was present on the PV prior to significant expansion (Figure 2D). Furthermore, ORP1L recruitment was independent of PV size, as wild type ORP1L-positive PVs were nearly identical in size to IcmD mutant ORP1L-negative PVs at early time points (Figure 2D). Based on these data, we hypothesize ORP1L is directly recruited by a PV membrane protein or lipid prior to PV expansion.
ORP1L ankyrin repeats are necessary and sufficient for PV localization
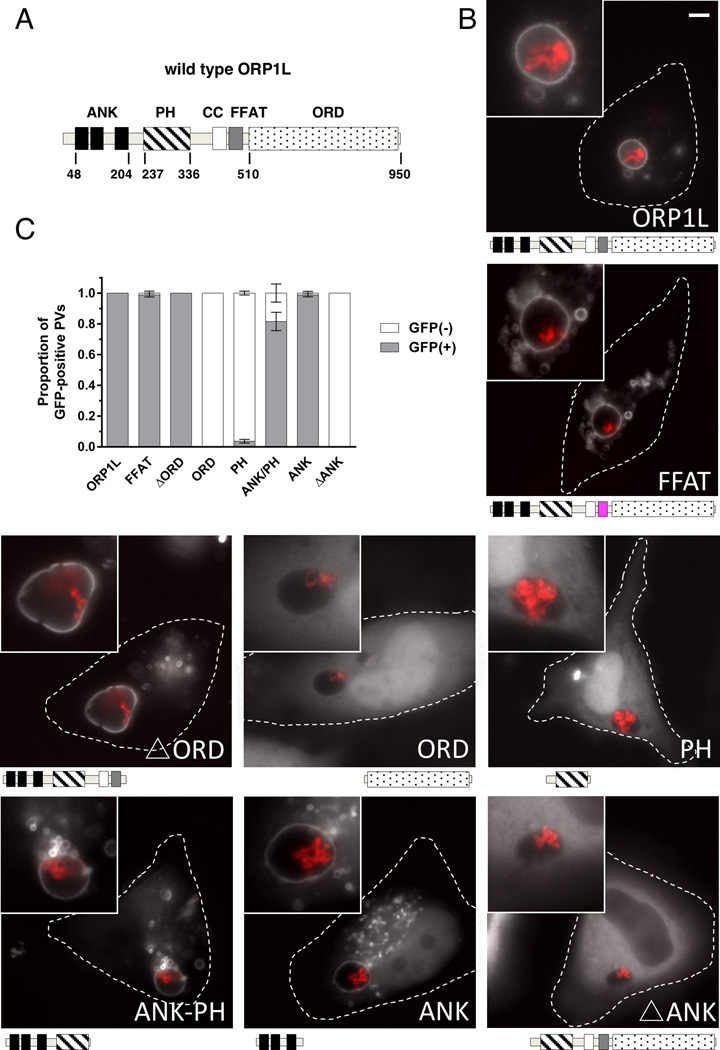
ORP1L contains multiple domains responsible for both protein-protein interactions and protein-lipid interactions (Figure 3A). Three N-terminal ankyrin repeats interact with the small GTPase Rab7 (Johansson et al., 2005b). ORP1L association with the ER is mediated through an FFAT motif that binds the ER protein VAMP-associated protein (VAP) (Rocha et al., 2009). The plekstrin homology (PH) domain binds with low affinity and specificity to the signaling lipids PI(3,4)P2 and PI(3,4,5)P3 (Johansson et al., 2005a), while the C-terminal ORD binds cholesterol and several oxysterol species (Yan et al., 2007, Vihervaara et al., 2011). To identify the ORP1L domain(s) responsible for PV localization, we generated GFP fusions of the ORP1L domains and assessed their localization using live cell imaging of C. burnetii-infected cells (Figure 3B). Like wild type ORP1L, ORP1L with a mutated FFAT motif localized to the PV. Removal of the cholesterol-binding domain (ΔORD) did not alter the PV localization and the ORD domain by itself was cytoplasmic, indicating cholesterol is not significantly involved in ORP1L PV localization. In addition to cholesterol, the signaling lipids PI(3,4)P2 and PI(3,4,5)P3 also appear to not be sufficient for PV localization, as GFP fusions of the phosphoinositide-binding PH domain was also cytoplasmic with no PV association. In contrast, PV localization was observed for both the ORP1L ankyrin repeat domain along with the PH domain (Ank-PH) and the ORP1L ankyrin repeat domain alone (Ank). To determine if the ankyrin repeats are required for PV association, we generated an ORP1L ankyrin truncation mutant (ΔAnk). ΔAnk-GFP did not localize to the PV and was cytoplasmic, demonstrating that the N-terminal ankyrin repeats are necessary and sufficient for ORP1L association with the PV. Further, these data show that protein-protein interactions, and not protein-lipid, are primarily responsible for ORP1L PV localization.
Figure 3. The ankryin repeat domains are necessary and sufficient to target ORP1L to the PV.
(A) Stick diagram of ORP1L protein domains. ANK = ankryin repeats (black), PH = plekstrin homology domain (stripe), CC = coiled-coil domain (white), FFAT = two phenylalanines in an acidic tract (gray), ORD = OSBP-related ligand binding domain (dots). Numbers represents amino acid position. (B) HeLa cells were transfected with C-terminal GFP constructs and infected with mCherry-expressing C. burnetii. Domain constructs are represented below each image. At 3 days post infection, live cells were imaged by wide field fluorescence microscopy. FFAT mutant (D478A) (magenta), Ank-PH, and Ank localize to the PV membrane, while ORD, Δ Ank, and PH remain cytoplasmic without PV membrane localization. Green = ORP1L-GFP, Red = mCherry-C. burnetii. Scale bar = 10 µm. (C) Quantitation of PV localization of ORP1L domain constructs. Shown are the results from two independent experiments, with at least 30 PVs per condition per experiment. Error bars represent SEM.
ORP1L can simultaneously associate with the C. burnetii PV and host endoplasmic reticulum
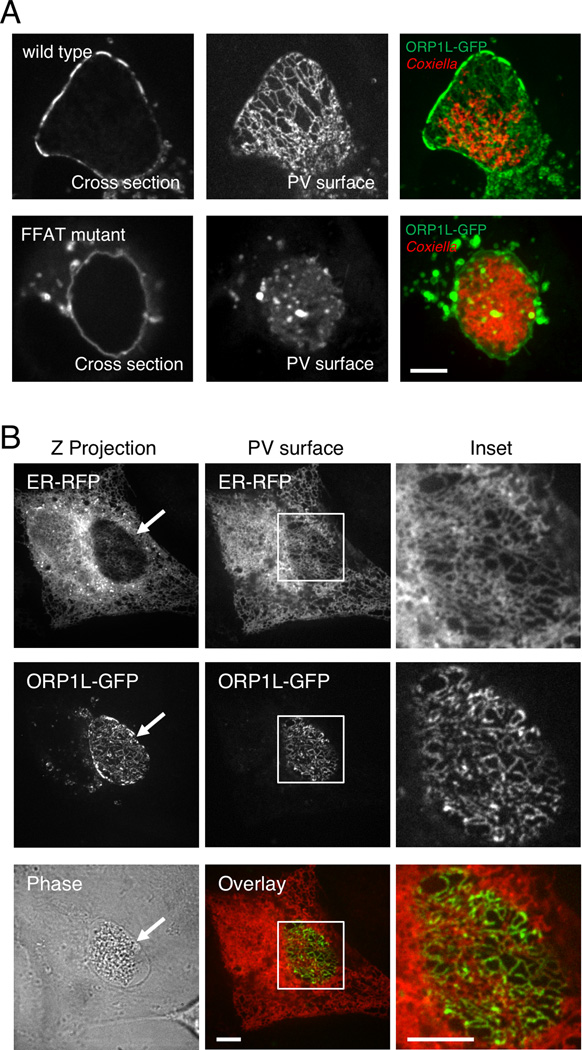
Using confocal microscopy to more precisely define ORP1L localization on the PV membrane, we observed a striking ER-like reticulate pattern most clearly seen when looking at the top surface of the PV (Figure 4A). ORP1L contains an FFAT motif, a conserved protein motif which binds to VAP on the cytosolic surface of the ER (Kaiser et al., 2005). We hypothesized that the PV ORP1L reticulate pattern was due to ORP1L also interacting with the ER via the FFAT motif. To test this, we mutated the FFAT motif such that it could longer bind to VAP and examined the ORP1L pattern on the C. burnetii PV. While the ORP1L FFAT mutant still localized to the PV (Figure 3B and 4A), the reticulate pattern was lost (Figure 4A, bottom panel), suggesting PV-associated ORP1L also binds to VAP on the ER. To futher confirm this observation, we tested for co-localization between PV-associated ORP1L and the ER marker KDEL-RFP (red fluorescent protein). We observed significant overlap between PV-associated ORP1L-GFP and KDEL-RFP (Figure 4B), demonstrating that ORP1L can simultaneously interact with both the PV and the ER. Together with the domain studies, these data suggest that PV-associated ORP1L binds to a PV membrane protein via the ankyrin repeats, while the FFAT motif binds to VAP on the ER cytosolic surface.
Figure 4. ORP1L simultaneously co-localizes with the PV and ER.
(A) Live cell confocal microscopy images of ORP1L-GFP on the C. burnetii PV. HeLa cells were transfected with wild type ORP1L-GFP or an FFAT mutant, ORP1L(D478A)-GFP. Three days after infection with mCherry-expressing C. burnetii, the PVs were identified by phase microscopy and fluorescence imaged by live cell spinning disk confocal microscopy. When the top surface of the PV is examined, wild type ORP1L exhibits a striated pattern. This pattern is disrupted by the FFAT D478A mutation, which prevents binding to the ER protein VAP. (B) Live cell fluorescence microscopy images of C. burnetii-infected HeLa cells expressing ER-localized red fluorescent protein (KDEL-RFP) and ORP1L-GFP. The maximum Z projection (Z projection) shows the flattened confocal stack through the entire cell, while the PV surface is a confocal slice of the top surface of the PV (arrow). As shown in the magnification of the boxed PV, ORP1L-GFP on the PV co-localized with the host ER. Red = ER, Green = ORP1L-GFP. Scale bars = 10 µm.
Membrane contact sites between the PV and ER
The finding that ORP1L simultaneously associates with both the PV and the ER suggests the presence of membrane contact sites (MCS) between these two membranes. MCS are sites where membranes closely interact with an intermembrane distance of ~10–30 nm, or the length of a single protein. Given that this is the first indication of MCS between the C. burnetii PV and ER, we used transmission electron microscopy (TEM) and electron tomography (ET) to more closely define the interactions between these two membranes. By TEM, the ER was found in close apposition to the PV membrane at several sites in HeLa cells (Figure 5). We also observed close interactions between the PV and ER in Vero epithelial cells and THP-1 macrophage-like cells (Supplemental Figure 4), indicating this interaction is not cell type specific. Using ET to generate a higher resolution 3-D structure, we identified numerous PV-ER MCS with less than 3nm distance between the two membranes as well as sites where the PV and ER appear to be in direct contact (Supplemental Figure 4 and Supplemental Movies 1–3). These data demonstrate the presence of membrane contact sites between the C. burnetii PV and the host endoplasmic reticulum.
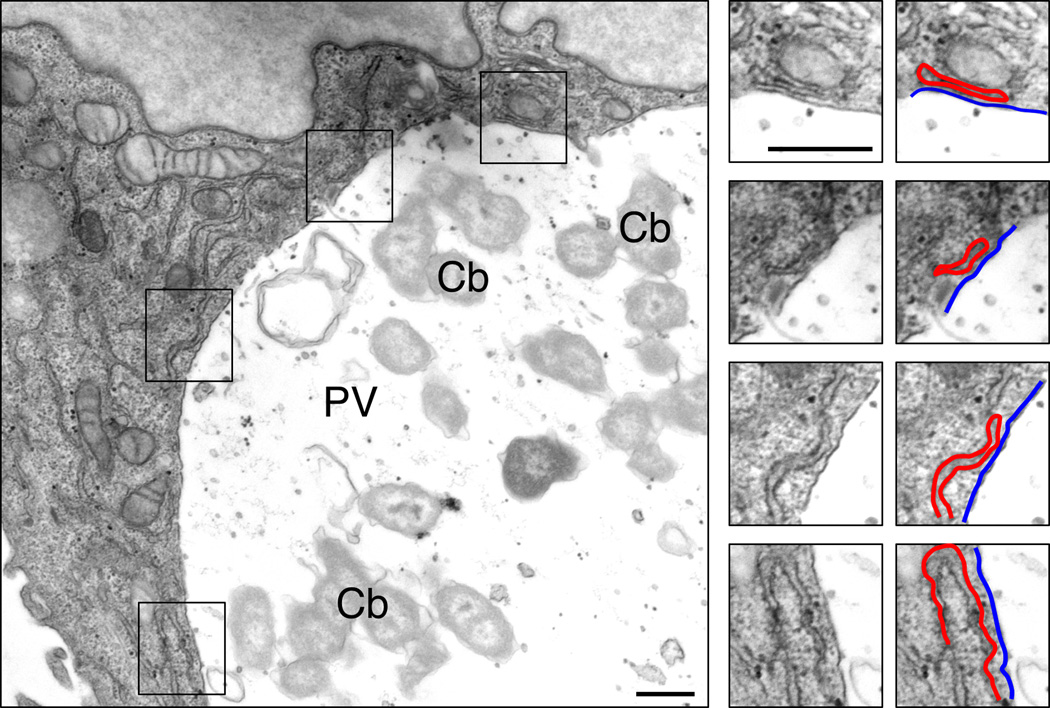
Figure 5. Electron microscopy reveals membrane contact sites between the PV and ER.
Transmission electron micrographs of HeLa cells infected with C. burnetii for 2 days. The boxed areas are further magnified, showing several areas where the PV membrane (outlined in blue) and ER membrane (red) are in close proximity. Cb = C. burnetii; PV = parasitophorous vacuole. Scale bars = 200 nm.
ORP1L depletion leads to smaller PVs
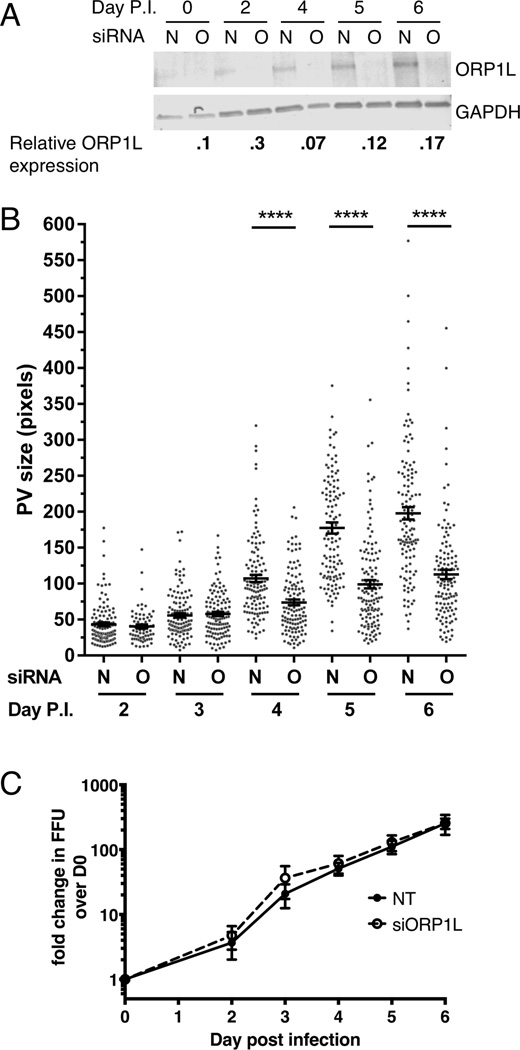
To further understand the role of ORP1L during C. burnetii colonization of host cells, we used an ON-TARGETplus siRNA pool to deplete ORP1L. ORP1L protein levels were decreased by 90% at the time of infection, with levels remaining less than 30% of non-targeting control siRNA for the duration of a 6 day infection time course (Figure 6A). At two and three days post infection, the PV size in ORP1L-depleted cells was similar to control cells (Figure 6B). However, while control PVs continued to expand approximately four-fold between days three and six, PVs in ORP1L-depleted cells had significantly less expansion, and were half the size of non-targeting PVs at day 6. Similarly sized PVs were observed at day 6 post infection in cells treated with a single siRNA duplex (Supplemental Figure 3). Interestingly, C. burnetii growth was similar in cells treated with nontargeting or ORP1L siRNA (Figure 6C). RILP and Rab7 localize to the C. burnetii PV in both normal and ORP1L-depleted cells (Supplemental Figure 6), indicating the smaller PVs in ORP1L-depleted cells are not due to the absence of Rab7 or RILP from the PV. These data suggest ORP1L plays a role in PV size; further, a large, spacious PV is not required for robust bacterial growth.
Figure 6. Depletion of ORP1L results in smaller PVs.
HeLa cells were treated with siRNA and then infected 2 days later (day 0 post infection (p.i.)). Infected cells were re-transfected with siRNA at 0 days p.i., and samples processed for immunoblotting, immunofluorescence, or growth assays. (A) ORP1L protein expression in cells treated with either non-targeting (N) or ORP1L siRNA (O) over a six day C. burnetii infection. Cell lysates were immunoblotted and ORP1L protein levels quantitated by normalizing to the loading control GAPDH. ORP1L protein levels remained less than 30% of the non-targeting control for the duration of the experiment. Shown is a representative blot from 6 experiments. (B) PV measurements in cells with either wild type or depleted ORP1L. At various times post infection, coverslips were fixed with paraformaldehyde and the PV membrane stained with anti-CD63 and anti-C. burnetii. The PV size was determined using ImageJ and standard error of the mean determined by ordinary one-way ANOVA with Sidak’s multiple comparisons test. The scatter plot shows individual PV measurements from three independent experiments, with at least 30 PVs per condition in each experiment. Bars indicate average ± SEM. **** = p<0.0001. (C) C. burnetii growth in ORP1L-depleted cells is similar to control cells. The number of viable bacteria was determined by fluorescent foci unit (FFU) assay at the days indicated, and normalized to day 0 to determine fold change in bacterial growth. The results are expressed as the mean of three experiments done in duplicate. Error bars represent SEM.
DISCUSSION
The endoplasmic reticulum plays essential roles in mammalian cells, including lipid and protein synthesis, calcium storage, and the cellular stress response. Membrane contact sites (MCS) between the ER and other cellular organelles serve as a location for exchange of small molecules and lipids. The formation of MCS relies on multi-protein complexes that tether the two membranes together within a distance of 15 nm (Phillips et al., 2016). Prior studies have implicated ORP1L, a member of the sterol transfer protein family, in the formation of late endosome/lysosome-ER MCS (Rocha et al., 2009, Weber-Boyvat et al., 2015). In this study, we discovered that ORP1L also localizes to the PV harboring the intracellular bacterial pathogen Coxiella burnetii. ORP1L can simultaneously bind to the C. burnetii PV and the ER, suggesting ORP1L is a component of PV-ER MCS. This is the first description of MCS between the C. burnetii PV and an intracellular organelle, and is supported by a recent study showing that the ER marker calnexin is closely associated with the PV during C. burnetii infection (Graham et al., 2015). ORP1L depletion results in smaller PVs, indicating ORP1L plays a role in PV membrane dynamics.
Our data show that ORP1L localization to the C. burnetii PV requires both the N-terminal ankyrin repeats and a functional C. burnetii Type 4B secretion system (T4BSS). The requirement for the ankyrin repeats indicates that protein-protein interactions are primarily responsible for ORP1L association with the PV. This is true for ORP1L localization during adenovirus infection, where the adenovirus protein RIDα binds to ORP1L in the formation of endosome-ER MCS (Cianciola et al., 2013). Several C. burnetii T4BSS effector proteins localize to the PV membrane (Larson et al., 2013), and are potential binding partners for ORP1L. In an analogous system, the C. trachomatis protein IncD recruits the ceramide-transfer protein CERT to the bacterial inclusion, which in turn binds VAP on the ER to form inclusion-ER MCS (Derre et al., 2011, Elwell et al., 2011). Thus, intracellular pathogens may have a common strategy to recruit host cell proteins in the formation of MCS with the ER.
Besides directly binding a T4BSS effector, a T4BSS effector protein might modulate the small GTPase Rab7, the ORP1L binding partner on late endosomes and lysosomes (Johansson et al., 2005b). Rab7 cycles between an active, GTP-bound form and an inactive, GDP-bound form. ORP1L can bind both forms and stabilizes GTP-bound Rab7, although it is not known if the Rab7 activation state affects ORP1L function (Johansson et al., 2005a). A constitutively active mutant of Rab7 is found on the PV (Beron et al., 2002), and Rab7 is required for PV maturation and expansion through an unknown mechanism (Beron et al., 2002, Newton et al., 2013). It is possible that a T4BSS effector protein regulates Rab7 activation, leading to the recruitment of ORP1L. We also cannot rule out the possibility that the lack of ORP1L on IcmD mutant PVs is indirectly due to a lack of PV maturation. However, because PVs harboring the IcmD mutant do support limited bacterial replication (Beare et al., 2011), maturation to an acidic phagolysosome is most likely not the sole requirement for PV ORP1L localization. This is further supported by ORP1L localization to the majority of wild type C. burnetii PVs at 12 hours post infection, shortly after T4BSS secreton is believed to begin around 8 hours post infection. Further studies are needed to identify the ORP1L PV binding partner(s) and understand how ORP1L is recruited to the PV membrane.
ORP1L exhibited a striated pattern on the C. burnetii surface that co-localized with an ER marker; this ER-like pattern was lost when the ORP1L FFAT motif was mutated. FFAT motifs bind to VAP on the ER cytosolic surface, and ORP1L has been shown to directly bind VAP (Weber-Boyvat et al., 2015). Based on these data, we propose a model where PV-associated ORP1L binds to VAP on the ER, forming MCS between the PV and ER (Figure 7). This model is further supported by our observation of PV-ER MCS by electron microscopy. While our data suggest ORP1L and VAP are components of these MCS, other proteins or protein complexes may be involved and ORP1L may not be a component of every PV-ER MCS. Indeed, many lipid transfer proteins with FFAT motifs have been found at MCS between the ER and the plasma membrane, golgi, and mitochondria (reviewed in (Kentala et al., 2016).
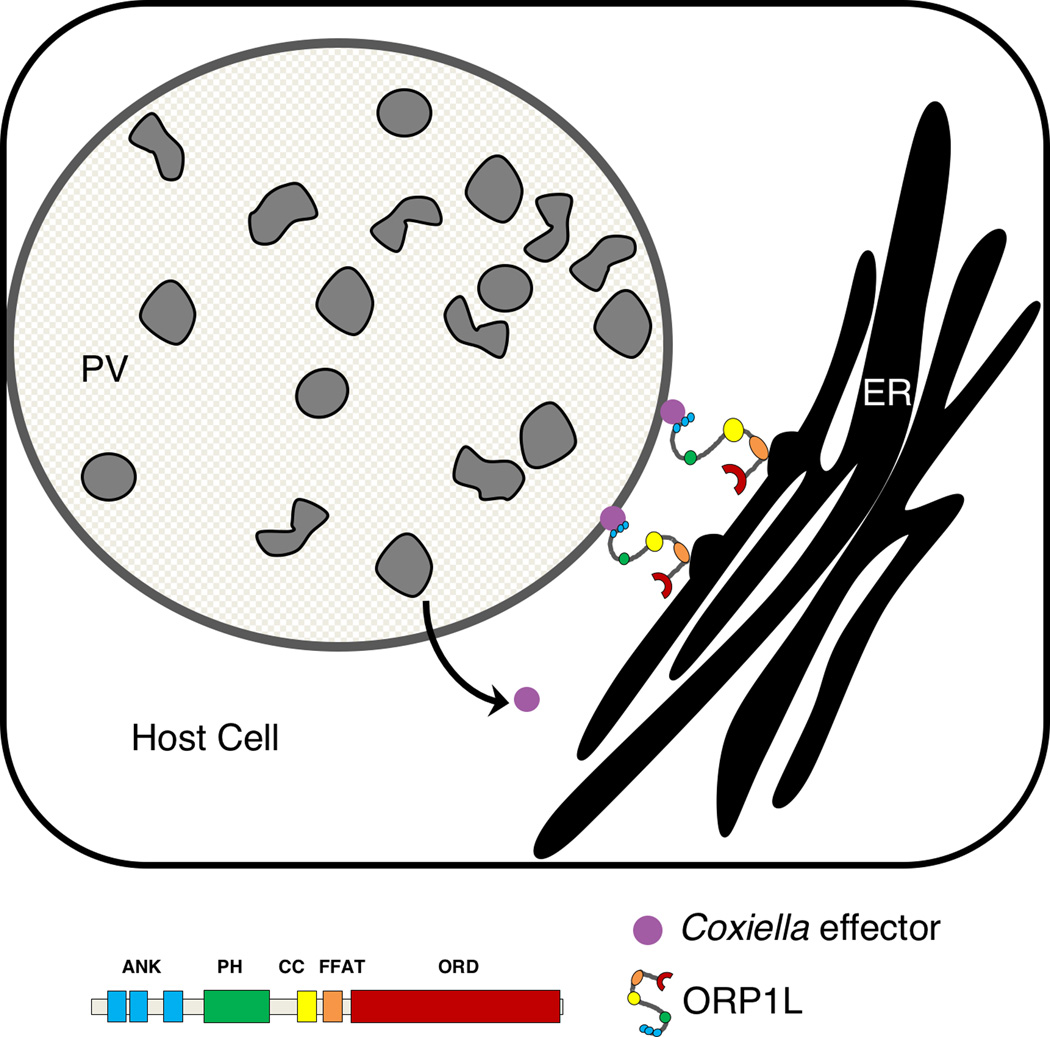
Figure 7. Model for ORP1L in membrane contact sites between the C. burnetii parasitophorous vacuole (PV) and host cell endoplasmic reticulum.
ORP1L is recruited directly or indirectly to the C. burnetii PV by the T4BSS, where it is involved in membrane contact sites between the PV and host ER.
ORP1L depletion led to smaller PVs, yet did not have an effect on bacterial growth. A similar observation was made recently in LAMP 1/2 knockout fibroblasts (Schulze-Luehrmann et al., 2016), suggesting that a large, spacious PV is not essential for C. burnetii growth. Further, a mutant in the Coxiella T4SS effector protein cvpB forms multiple PVs per cell, but does not have a growth defect in vitro (Newton et al., 2014, Martinez et al., 2016). Collectively, Coxiella appears to modulate a complex interplay of different, and possibly redundant, pathways to build the optimal vacuole to support bacterial replication. This is supported by the finding that a cvpB mutant has an attenuated virulence phenotype in the in vivo insect model Galleria mellonella (Martinez et al., 2016). The reason behind smaller PVs in the absence of ORP1L is not clear. Interactions between host vesicular trafficking and the PV membrane are not well understood, but it has been assumed that a significant portion of the PV membrane comes from fusion with host vesicles. It is possible that the ER might serve to stabilize the PV membrane, with the absence or decrease in PV-ER MCS leading to altered PV membrane dynamics and decreased fusion with host trafficking pathways. Alternatively, ORP1L may play a direct role in fusion between the PV and host cell endosomes, and smaller PVs might be the result of decreased fusion. In uninfected cells, ORP1L regulates endosomal tethering and subsequent fusion by mediating interactions between RILP and the HOPS complex (van der Kant et al., 2013a). Both Rab7 and RILP localize to the C. burnetii PV in the absence of ORP1L, suggesting these proteins are not directly responsible for the size defect. Given that ORP1L’s dual role in endosomal fusion and endosome-ER membrane contact sites, further investigation of the molecular machinery involved in endosome-PV fusion is needed.
The function of ORP1L and PV-ER MCS is not clear. ORP1L belongs to a family of sterol-transfer proteins and binds phosphatidylinositol phosphates, oxysterols, and cholesterol. ORP1L may transfer cholesterol or other sterols between two membranes, similar to the proposed function of CERT at the C. trachomatis inclusion (Derre et al., 2011, Elwell et al., 2011). ORP1L is hypothesized to transfer cholesterol from lysosomes to the ER independent of the lysosomal cholesterol transporter NPC1, with the cholesterol then incorporated into lipid droplets (Cianciola et al., 2013, van der Kant et al., 2013b). PV-ER MCS may also participate in the exchange of calcium or other small molecules, or be a mechanism to manipulate ER stress and apoptosis.
In summary, this study demonstrates the presence of membrane contact sites between the C. burnetii PV and the host ER. The sterol-binding protein ORP1L is one component of these MCS, and appears to play a role in PV membrane dynamics. Future experiments identifying the PV binding partner and elucidating the function of PV-ER interactions will give new insight into C. burnetii pathogenesis and manipulation of the host cell.
MATERIALS AND METHODS
Bacteria and mammalian cells
C. burnetti Nine Mile Phase II (NMII; clone 4, RSA439) and mCherry-expressing C. burnetti NMII (Beare et al., 2009) were purified from Vero cells (African green monkey kidney epithelial cells, ATCC CCL-81; American Type Culture Collection, ATCC, Manassas, VA) and stored as previously described (Cockrell et al., 2008). For experiments examining T4BSS-dependent recruitment of ORP1L, NMII and the icmD mutant (Beare et al., 2011) were grown for 4 days in ACCM-2, washed twice with PBS (phosphate buffered saline), and stored as previously described (Omsland et al., 2009). The multiplicity of infection (MOI) for each bacteria stock was optimized for ~1 internalized bacterium per cell. Vero, human cervical epithelial cells (HeLa; ATCC CCL-2), and human monocytes (THP-1; ATTC TIB-202) were maintained in RPMI 1640 medium (Corning, New York, NY) containing 10% fetal bovine serum (FBS) (Atlanta Biologicals, Norcross, GA) at 37°C and 5% CO2. THP-1 cells were differentiated with 200nM phorbol 12-myristate 13-acetate (PMA) for 24 hours. The PMA was removed and the cells rested for 48 hours prior to infection.
Generation of ORP1L constructs
ORP1L-GFP was a generous gift of J. Neefjes (Netherlands Cancer Institute). For construction of ORP1L domain GFP constructs, ORP1L domains (Johansson et al., 2005a) were amplified and cloned into the BsrG1 site of pcDNA6.2/N-EmeraldGFP using In-Fusion (Clontech). Oligonucleotide sequences used in cloning were:
ORP1L-1-F:ATGGACGAGCTGTACAAGATGAACACAGAAGCGGAGCAAC;ORP1L-211-F:ATGGACGAGCTGTACAAGAAACCTCTTGACCTTGCCCAGGGTG;ORP1L-510-F: ATGGACGAGCTGTACAAGAGAAAACACAGAATGTCCGAAGAAAAAGAC;ORP1L-237-R AATATCACTCTGTACATTATCGTTTCAATGCTTTGTAGATGACC;ORP1L-345-R AATATCACTCTGTACATTACAGCTGGTCCTGGGAACAGTAG;ORP1L-408-R AATATCACTCTGTACATTAAGAAGCTTCTGAGACAACTTCAAC;ORP1L-514-R TCAACCACTTTGTACATTACATTCTGTGTTTTCTACTGCCC;
ORP1L-950-R AATATCACTCTGTACATTAATAAATGTCAGGCAAATTGAAG
ORP1L-D478A–F GCGAGGACGAGTTCTATGCTGCGCTGTCAGATTCCGA
ORP1L-D478A–R TCGGAATCTGACAGCGCAGCATAGAACTCGTCCTCGC.
The following primer combinations were used: Full length ORP1L (amino acids (AA) 1–750) = ORP1L-1-F and ORP1L-950-R; ORD (AA 510–950) = ORP1L-510-F and ORP1L-950-R; PH (AA 211–345) = ORP1L-211-F and ORP1L-345-R; Ank-PH (AA 1–408) = ORP1L-1-F and ORP1L-408-R; Ank (AA 1–237) = OFP1L-1-F and ORP1L-237-R; ΔAnk (AA 211–950) = ORP1L-211-F and ORP1L-1-R; ΔORD (AA 514–950) = ORP1L-514-F and ORP1L-950-R. FFAT mutant (D478A) was generated using site directed mutagenesis with primers ORP1L-D478A–F and ORP1L-D478A–R according to the manufactor’s directions (Clontech).
Transfection and immunofluorescence assay (IFA)
HeLa cells (2×104 cells per well of a 24 well plate) were plated on glass coverslips and simultaneously transfected with 0.4 µg of plasmid with Fugene 6 (Promega, Madison, WI) according to the manufacturers reverse transfection protocol. Approximately 24 hours post-transfection, cells were infected with C. burnetii in 0.25 mL RPMI with 10% FBS for two hours at 37°C, 5% CO2, washed extensively with PBS, and incubated in 10% RPMI. At the indicated times post-infection, the coverslips were fixed with 4% paraformaldehyde (PFA) in PBS for 15 minutes. Cells were permeabilized and blocked by treatment with 0.1% saponin/1% bovine serum albumin (BSA)/PBS for 15 minutes, and then incubated with mouse anti-CD63 (556019; BD Biosciences, San Jose, CA) or rabbit anti-LAMP1 (ab24170; Abcam, Cambridge, MA), and guinea pig anti-C. burnetti serum (Coleman et al., 2007) for one hour in saponin/BSA/PBS. Following incubation with AlexFluor secondary antibodies (Invitrogen, Carlsbad, CA) in saponin BSA/PBS, coverslips were mounted using ProLong Gold with DAPI (Invitrogen), and visualized.
Recruitment and PV measurements
Fixed cells were visualized with a Leica inverted DMI6000B microscope (63X oil objective), and CD63-positive or Lamp1-positive PVs were visually scored as GFP-positive or negative to determine ORP1L-GFP recruitment. PVs with co-localization between the PV marker (CD63 or Lamp1) and protein of interest were scored as positive. Images were captured using a DFC365FX camera with Leica Application Suite X software (version 1.5.0). For recruitment of ORP1L to wild type and IcmD mutant PVs, 15–20 PVs were scored for each of four independent experiments. For PV size measurements, each PV was imaged and the cross-sectional area through the middle of the PV was measured with ImageJ software (NIH).
Live cell microscopy
For localization of ORP1L domain GFP constructs, HeLa cells were transfected and infected with mCherry-expressing C. burnetii as described above. At 3 days post infection, the cells were replated onto ibidi u-Dishes (ibidi, Verona, Wisconsin) and visualized live using the Leica inverted microscope described above. For ER localization studies, the cells were transduced with the ER marker KDEL-RFP CellLights BacMam (Invitrogen; MOI of 10) 24 hours prior to imaging on a modified Perkin-Elmer UltraView spinning-disk confocal system connected to a Nikon Eclipse Ti-E inverted microscope with a 100X objective and environmental chamber for temperature and CO2 control (Pathology Devices, Inc., Westminster, MD.
Transmission Electron Microscopy
Infected cells grown in 6 well plates were fixed with 2.5% glutaraldehyde/0.5 M sucrose in 0.1 M sodium cacodylate pH 6.8. Samples were processed and visualized at 80 kV with a model H7500 electron microscope as previously described (Starr et al., 2012).
Electron Tomography
Samples were prepared as described in TEM section. Sections (200 nm) were collected on glow discharged carbon grids, and 10 colloidal gold fiducial markers were applied. Using a linear tilt scheme, single axis tilt series were collected on a Tecnai BioTwin Spirit TEM (FEI) operated at 120 kV. Images captured over a tilt range of +/− 68° (1° increments) at 1µm defocus were recorded on a 2048 × 2048 pixel UltraScan 1000 Gatan CCD camera using the automated tomography acquisition software (Xplore 3D, FEI). The tilt series were aligned using the IMOD software package (version 4.8.40). All measurements were done using the Amira Visualization Package (version 6.0.0, FEI).
siRNA knockdown
ORP1L and non-targeting ON-TARGETplus SMARTpool siRNA duplexes were obtained from GE Dharmacon (Lafayette, CO). HeLa cells (1.2×105 cells) were transfected with 50 nM siRNA in 6 well plates using DharmaFECT 1 according to the manufacturer’s reverse transfection protocol (GE Dharmacon). Two days later, the cells were infected with C. burnetii for one hour as described above, trypsinized, and re-transfected with 50 nM siRNA in 24 well plates (2.5×104 cells per well). At the given time points, wells were processed for either immunoblotting, FFU growth assay, or IFA. Single target ORP1L siRNA (UGCCAGUGCCGGAUUCUGAdTdT, (Johansson et al., 2007) was obtained from Sigma-Aldrich (St. Louis, MO). Cells were treated with siRNA as above using 50nM siRNA. For immunoblotting, cells were harvested in 2% sodium dodecyl sulfate (SDS). Total protein (15 µg) was separated by SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membrane (EMD Millipore, Kankakee, IL), and probed with rabbit anti-ORP1L (Abcam 131165) and mouse anti-GAPDH (ThermoFisher MA5–15738), followed by incubation with LI-COR infrared secondary antibodies (ThermoFisher). Blots were imaged using an Odyssey Imager (LI-COR, Lincoln, NE) and bands quantitated using Image Studio Software (LI-COR). ORP1L knockdown efficiency was calculated by first normalizing to GAPD, and then to the corresponding non-targeting sample.
For Rab7 and RILP localization experiments, 2.5×104 HeLa cells were transfected with 50nM ORP1L or non-targeting ON-TARGETplus SMARTpool siRNA duplexes in 24-well plates. 24 hours later, cells were transfected with EGFP-Rab7A or RILP-GFP plasmids using standard Fugene6 transfection protocol and incubated overnight. The next day, double-transfected cells were infected with mCherry-expressing Coxiella. 3 days post infection, cells were fixed and stained for IFA. EGFP-Rab7A was a gift from Qing Zhong (Addgene plasmid # 28047 (Sun et al., 2010))
Fluorescent Infectious Foci Forming Unit (FFU) assay
To quantitate bacterial growth in HeLa cells, FFU assays were performed as previously described (Coleman et al., 2004). Briefly, bacteria were released from infected HeLa cells by incubating for 5 minutes in sterile water, followed by serial dilutions in 2% RPMI and addition to Vero cell monolayers in 24 well plates. After 5 days, the cells were fixed with methanol and stained with rabbit anti-C. burnetii and Alexa488 secondary antibodies. Fluorescent foci were visualized using an EVOS FL Auto Imaging System (ThermoScientific) and counted using ImageJ. Each experiment was done in duplicate.
Data analysis
Image processing and analysis was done with ImageJ software (W.S. Rasband, National Institutes of Health, Bethesda, MD). Statistical analyses were performed using unpaired Welch’s t-test or ordinary one-way ANOVA with Sidak multiple comparisons tests in Prism (GraphPad Software, Inc., La Jolla, CA).
Supplementary Material
Supplemental Figure 1. GFP-ORP1L localization. Live cell confocal microscopy image of C. burnetii-infected HeLa cell expressing GFP-ORP1L. GFP-ORP1L localizes to the C. burnetii PV (arrow). The cell boundary is outlined. Scale bar = 10 µm.
Supplemental Figure 2. PV size in ORP1L–GFP expressing cells. At 48 hours post infection, wild type PVs in cells ectopically expressing ORP1L–GFP are smaller than untransfected cells. Error bars represent SEM. Means were compared by ordinary one-way ANOVA with Sidak’s multiple comparisons tests. ** = p <0.01.
Supplemental Figure 3. Depletion of ORP1L with single siRNA duplex results in smaller PVs. HeLa cells were treated with a single siRNA duplex at 50nM. At 6 days post infection, coverslips were fixed with paraformaldehyde and the PV membrane stained with anti-CD63 and anti-C. burnetii. The PV size was determined using ImageJ and means compared by unpaired Welch’s t-test. The scatter plot shows individual PV measurements from two experiments, with at least 40 PVs per condition in each experiment. Bars indicate average ± SEM. **** = p<.0001
Supplemental Figure 4. PV-ER membrane contact sites in C. burnetii infected Vero and THP-1 cells. Vero and THP-1 cells were infected with C. burnetii, fixed at 1 day post infection (THP-1) or 2 days post infection (Vero), and analyzed by transmission electron microscopy. Multiple membrane contact sites between the PV and ER are observed (boxed magnification, arrows).
Supplemental Figure 5. Measurement of PV-ER membrane contact sites using electron tomography. HeLa cells were infected with C. burnetii, and examined by electron tomography at 2 days post infection. Shown are slices from supplemental movie 1. The boxed areas are further magnified, and the distances between the PV (blue line) and ER membranes (red lines) shown. In a, the PV and ER membranes are indistinguishable (arrow), while b-d show areas where the two membranes are less than 3 nm apart. Scale bar = 50 nm.
Supplemental Figure 6. Depletion of ORP1L does not alter Rab7 and RILP PV-localization. HeLa cells were treated with siRNA, transfected with plasmid DNA the next day, and then infected 1 day later. At 2 days post infection, cells were fixed and stained for CD63 and Coxiella. Both Rab7-GFP (A) and RILP-GFP (B) localize to the Coxiella PV in cells treated with siRNA targeting ORP1L or non-targeting (NT) negative control. Scale bar = 10µm.
Supplemental Movies 1–3. Electron tomography tilt series of three different C. burnetii PVs. Tilt series for three separate PVs are shown; the arrow denotes areas where the PV and ER membranes are indistinguishable from one another.
Acknowledgments
This research was supported by the Showalter Trust Fund (S.D.G), NIH NRSA Immunology and Infectious Disease Training Program T32AI060519 (A.J.V.), and the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Disease (R.A.H.). We thank Seth Winfree, Minal Mulye, and Dhritiman Samanta for critical reading of the manuscript, and members of the IU Biology of Intracellular Pathogens Group for helpful suggestions.
Footnotes
The authors have no conflict of interest to declare.
References
- Beare PA, Gilk SD, Larson CL, Hill J, Stead CM, Omsland A, et al. Dot/Icm type IVB secretion system requirements for Coxiella burnetii growth in human macrophages. MBio. 2011;2:e00175–e00111. doi: 10.1128/mBio.00175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beare PA, Howe D, Cockrell DC, Omsland A, Hansen B, Heinzen RA. Characterization of a Coxiella burnetii ftsZ mutant generated by Himar1 transposon mutagenesis. J. Bacteriol. 2009;191:1369–1381. doi: 10.1128/JB.01580-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beare PA, Larson CL, Gilk SD, Heinzen RA. Two systems for targeted gene deletion in Coxiella burnetii. Appl. Environ. Microbiol. 2012;78:4580–4589. doi: 10.1128/AEM.00881-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beron W, Gutierrez MG, Rabinovitch M, Colombo MI. Coxiella burnetii localizes in a Rab7-labeled compartment with autophagic characteristics. Infect. Immun. 2002;70:5816–5821. doi: 10.1128/IAI.70.10.5816-5821.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campoy EM, Mansilla ME, Colombo MI. Endocytic SNAREs are involved in optimal Coxiella burnetii vacuole development. Cell Microbiol. 2013;15:922–941. doi: 10.1111/cmi.12087. [DOI] [PubMed] [Google Scholar]
- Carabeo RA, Mead DJ, Hackstadt T. Golgi-dependent transport of cholesterol to the Chlamydia trachomatis inclusion. Proc. Natl. Acad. Sci. USA. 2003;100:6771–6776. doi: 10.1073/pnas.1131289100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey KL, Newton HJ, Luhrmann A, Roy CR. The Coxiella burnetii Dot/Icm system delivers a unique repertoire of type IV effectors into host cells and is required for intracellular replication. PLoS Pathogens. 2011;7:e1002056. doi: 10.1371/journal.ppat.1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianciola NL, Greene DJ, Morton RE, Carlin CR. Adenovirus RIDalpha uncovers a novel pathway requiring ORP1L for lipid droplet formation independent of NPC1. Mol. Biol. Cell. 2013;24:3309–3325. doi: 10.1091/mbc.E12-10-0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockrell DC, Beare PA, Fischer ER, Howe D, Heinzen RA. A method for purifying obligate intracellular Coxiella burnetii that employs digitonin lysis of host cells. J. Microbiol. Methods. 2008;72:321–325. doi: 10.1016/j.mimet.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman SA, Fischer ER, Cockrell DC, Voth DE, Howe D, Mead DJ, et al. Proteome and antigen profiling of Coxiella burnetii developmental forms. Infect. Immun. 2007;75:290–298. doi: 10.1128/IAI.00883-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman SA, Fischer ER, Howe D, Mead DJ, Heinzen RA. Temporal analysis of Coxiella burnetii morphological differentiation. J. Bacteriol. 2004;186:7344–7352. doi: 10.1128/JB.186.21.7344-7352.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czyz DM, Potluri LP, Jain-Gupta N, Riley SP, Martinez JJ, Steck TL, et al. Host-directed antimicrobial drugs with broad-spectrum efficacy against intracellular bacterial pathogens. MBio. 2014;5:e01534–e01514. doi: 10.1128/mBio.01534-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derre I, Swiss R, Agaisse H. The lipid transfer protein CERT interacts with the Chlamydia inclusion protein IncD and participates to ER-Chlamydia inclusion membrane contact sites. PLoS Pathogens. 2011;7:e1002092. doi: 10.1371/journal.ppat.1002092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elwell CA, Jiang S, Kim JH, Lee A, Wittmann T, Hanada K, et al. Chlamydia trachomatis co-opts GBF1 and CERT to acquire host sphingomyelin for distinct roles during intracellular development. PLoS Pathogens. 2011;7:e1002198. doi: 10.1371/journal.ppat.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghigo E, Capo C, Tung CH, Raoult D, Gorvel JP, Mege JL. Coxiella burnetii survival in THP-1 monocytes involves the impairment of phagosome maturation: IFN-gamma mediates its restoration and bacterial killing. J Immunol. 2002;169:4488–4495. doi: 10.4049/jimmunol.169.8.4488. [DOI] [PubMed] [Google Scholar]
- Gilk SD. Role of lipids in Coxiella burnetii infection. Adv. Exp. Med. Biol. 2012;984:199–213. doi: 10.1007/978-94-007-4315-1_10. [DOI] [PubMed] [Google Scholar]
- Graham JG, Winchell CG, Sharma UM, Voth DE. Identification of ElpA, a Coxiella burnetii pathotype-specific Dot/Icm type IV secretion system substrate. Infect. Immun. 2015;83:1190–1198. doi: 10.1128/IAI.02855-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez MG, Vazquez CL, Munafo DB, Zoppino FC, Beron W, Rabinovitch M, Colombo MI. Autophagy induction favours the generation and maturation of the Coxiella-replicative vacuoles. Cell. Microbiol. 2005;7:981–993. doi: 10.1111/j.1462-5822.2005.00527.x. [DOI] [PubMed] [Google Scholar]
- Hackstadt T, Scidmore MA, Rockey DD. Lipid metabolism in Chlamydia trachomatis-infected cells: directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc. Natl. Acad. Sci. USA. 1995;92:4877–4881. doi: 10.1073/pnas.92.11.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzen RA, Scidmore MA, Rockey DD, Hackstadt T. Differential interaction with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetii and Chlamydia trachomatis. Infect. Immun. 1996;64:796–809. doi: 10.1128/iai.64.3.796-809.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe D, Heinzen RA. Coxiella burnetii inhabits a cholesterol-rich vacuole and influences cellular cholesterol metabolism. Cell. Microbiol. 2006;8:496–507. doi: 10.1111/j.1462-5822.2005.00641.x. [DOI] [PubMed] [Google Scholar]
- Howe D, Melnicakova J, Barak I, Heinzen RA. Fusogenicity of the Coxiella burnetii parasitophorous vacuole. Ann. N. Y. Acad. Sci. 2003;990:556–562. doi: 10.1111/j.1749-6632.2003.tb07426.x. [DOI] [PubMed] [Google Scholar]
- Johansson M, Bocher V, Lehto M, Chinetti G, Kuismanen E, Ehnholm C, et al. The two variants of oxysterol binding protein-related protein-1 display different tissue expression patterns, have different intracellular localization, and are functionally distinct. Mol. Biol. Cell. 2003;14:903–915. doi: 10.1091/mbc.E02-08-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M, Lehto M, Tanhuanpaa K, Cover TL, Olkkonen VM. The oxysterol-binding protein homologue ORP1L interacts with Rab7 and alters functional properties of late endocytic compartments. Mol. Biol. Cell. 2005a;16:5480–5492. doi: 10.1091/mbc.E05-03-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M, Olkkonen VM. Assays for interaction between Rab7 and oxysterol binding protein related protein 1L (ORP1L) Methods Enzymol. 2005b;403:743–758. doi: 10.1016/S0076-6879(05)03065-X. [DOI] [PubMed] [Google Scholar]
- Johansson M, Rocha N, Zwart W, Jordens I, Janssen L, Kuijl C, et al. Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor betalll spectrin. J Cell Biol. 2007;176:459–471. doi: 10.1083/jcb.200606077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser SE, Brickner JH, Reilein AR, Fenn TD, Walter P, Brunger AT. Structural basis of FFAT motif-mediated ER targeting. Structure. 2005;13:1035–1045. doi: 10.1016/j.str.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Kentala H, Weber-Boyvat M, Olkkonen VM. OSBP-Related Protein Family: Mediators of Lipid Transport and Signaling at Membrane Contact Sites. Int Rev Cell Mol Biol. 2016;321:299–340. doi: 10.1016/bs.ircmb.2015.09.006. [DOI] [PubMed] [Google Scholar]
- Kobuna H, Inoue T, Shibata M, Gengyo-Ando K, Yamamoto A, Mitani S, Arai H. Multivesicular body formation requires OSBP-related proteins and cholesterol. PLoS Genet. 2010:6. doi: 10.1371/journal.pgen.1001055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson CL, Beare PA, Howe D, Heinzen RA. Coxiella burnetii effector protein subverts clathrin-mediated vesicular trafficking for pathogen vacuole biogenesis. Proc. Natl. Acad. Sci. USA. 2013;110:E4770–E4779. doi: 10.1073/pnas.1309195110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson CL, Beare PA, Voth DE, Howe D, Cockrell DC, Bastidas RJ, et al. Coxiella burnetii Effector Proteins That Localize to the Parasitophorous Vacuole Membrane Promote Intracellular Replication. Infect. Immun. 2015;83:661–670. doi: 10.1128/IAI.02763-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E, Allombert J, Cantet F, Lakhani A, Yandrapalli N, Neyret A, et al. Coxiella burnetii effector CvpB modulates phosphoinositide metabolism for optimal vacuole development. Proc Natl Acad Sci U S A. 2016 doi: 10.1073/pnas.1522811113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough JA, Newton HJ, Klum S, Swiss R, Agaisse H, Roy CR. Host pathways important for Coxiella burnetii infection revealed by genome-wide RNA interference screening. MBio. 2013;4:e00606–e00612. doi: 10.1128/mBio.00606-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt JH, Newton P, Newton HJ. Coxiella burnetii: turning hostility into a home. Cell. Microbiol. 2015;17:621–631. doi: 10.1111/cmi.12432. [DOI] [PubMed] [Google Scholar]
- Newton HJ, Kohler LJ, McDonough JA, Temoche-Diaz M, Crabill E, Hartland EL, Roy CR. A screen of Coxiella burnetii mutants reveals important roles for Dot/Icm effectors and host autophagy in vacuole biogenesis. PLoS Pathogens. 2014;10:e1004286. doi: 10.1371/journal.ppat.1004286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton HJ, McDonough JA, Roy CR. Effector protein translocation by the Coxiella burnetii Dot/Icm type IV secretion system requires endocytic maturation of the pathogen-occupied vacuole. PLoS One. 2013;8:e54566. doi: 10.1371/journal.pone.0054566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omsland A, Cockrell DC, Howe D, Fischer ER, Virtaneva K, Sturdevant DE, et al. Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc. Natl. Acad. Sci. USA. 2009;106:4430–4434. doi: 10.1073/pnas.0812074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips MJ, Voeltz GK. Structure and function of ER membrane contact sites with other organelles. Nat Rev Mol Cell Biol. 2016;17:69–82. doi: 10.1038/nrm.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizarro-Cerda J, Meresse S, Parton RG, van der Goot G, Sola-Landa A, Lopez-Goni I, et al. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect. Immun. 1998;66:5711–5724. doi: 10.1128/iai.66.12.5711-5724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha N, Kuijl C, van der Kant R, Janssen L, Houben D, Janssen H, et al. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. J. Cell. Biol. 2009;185:1209–1225. doi: 10.1083/jcb.200811005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Luehrmann J, Eckart RA, Olke M, Saftig P, Liebler-Tenorio E, Luhrmann A. LAMP proteins account for the maturation delay during the establishment of the Coxiella burnetii-containing vacuole. Cell Microbiol. 2016;18:181–194. doi: 10.1111/cmi.12494. [DOI] [PubMed] [Google Scholar]
- Starr T, Child R, Wehrly TD, Hansen B, Hwang S, Lopez-Otin C, et al. Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe. 2012;11:33–45. doi: 10.1016/j.chom.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Westphal W, Wong KN, Tan I, Zhong Q. Rubicon controls endosome maturation as a Rab7 effector. Proc Natl Acad Sci U S A. 2010;107:19338–19343. doi: 10.1073/pnas.1010554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson MS, Isberg RR. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect. Immun. 1995;63:3609–3620. doi: 10.1128/iai.63.9.3609-3620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kant R, Fish A, Janssen L, Janssen H, Krom S, Ho N, et al. Late endosomal transport and tethering are coupled processes controlled by RILP and the cholesterol sensor ORP1L. J. Cell Sci. 2013a;126:3462–3474. doi: 10.1242/jcs.129270. [DOI] [PubMed] [Google Scholar]
- van der Kant R, Zondervan I, Janssen L, Neefjes J. Cholesterol-binding molecules MLN64 and ORP1L mark distinct late endosomes with transporters ABCA3 and NPC1. J. Lipid Res. 2013b;54:2153–2165. doi: 10.1194/jlr.M037325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihervaara T, Uronen RL, Wohlfahrt G, Bjorkhem I, Ikonen E, Olkkonen VM. Sterol binding by OSBP-related protein 1L regulates late endosome motility and function. Cell Mol. Life Sci. 2011;68:537–551. doi: 10.1007/s00018-010-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voth DE, Heinzen RA. Lounging in a lysosome: the intracellular lifestyle of Coxiella burnetii. Cell. Microbiol. 2007;9:829–840. doi: 10.1111/j.1462-5822.2007.00901.x. [DOI] [PubMed] [Google Scholar]
- Weber-Boyvat M, Kentala H, Peranen J, Olkkonen VM. Ligand-dependent localization and function of ORP-VAP complexes at membrane contact sites. Cell Mol Life Sci. 2015;72:1967–1987. doi: 10.1007/s00018-014-1786-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winchell CG, Graham JG, Kurten RC, Voth DE. Coxiella burnetii type IV secretion-dependent recruitment of macrophage autophagosomes. Infect. Immun. 2014;82:2229–2238. doi: 10.1128/IAI.01236-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Jauhiainen M, Hildebrand RB, Willems van Dijk K, Van Berkel TJ, Ehnholm C, et al. Expression of human OSBP-related protein 1L in macrophages enhances atherosclerotic lesion development in LDL receptor-deficient mice. Arterioscler. Thromb. Vasc. Biol. 2007;27:1618–1624. doi: 10.1161/ATVBAHA.107.144121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. GFP-ORP1L localization. Live cell confocal microscopy image of C. burnetii-infected HeLa cell expressing GFP-ORP1L. GFP-ORP1L localizes to the C. burnetii PV (arrow). The cell boundary is outlined. Scale bar = 10 µm.
Supplemental Figure 2. PV size in ORP1L–GFP expressing cells. At 48 hours post infection, wild type PVs in cells ectopically expressing ORP1L–GFP are smaller than untransfected cells. Error bars represent SEM. Means were compared by ordinary one-way ANOVA with Sidak’s multiple comparisons tests. ** = p <0.01.
Supplemental Figure 3. Depletion of ORP1L with single siRNA duplex results in smaller PVs. HeLa cells were treated with a single siRNA duplex at 50nM. At 6 days post infection, coverslips were fixed with paraformaldehyde and the PV membrane stained with anti-CD63 and anti-C. burnetii. The PV size was determined using ImageJ and means compared by unpaired Welch’s t-test. The scatter plot shows individual PV measurements from two experiments, with at least 40 PVs per condition in each experiment. Bars indicate average ± SEM. **** = p<.0001
Supplemental Figure 4. PV-ER membrane contact sites in C. burnetii infected Vero and THP-1 cells. Vero and THP-1 cells were infected with C. burnetii, fixed at 1 day post infection (THP-1) or 2 days post infection (Vero), and analyzed by transmission electron microscopy. Multiple membrane contact sites between the PV and ER are observed (boxed magnification, arrows).
Supplemental Figure 5. Measurement of PV-ER membrane contact sites using electron tomography. HeLa cells were infected with C. burnetii, and examined by electron tomography at 2 days post infection. Shown are slices from supplemental movie 1. The boxed areas are further magnified, and the distances between the PV (blue line) and ER membranes (red lines) shown. In a, the PV and ER membranes are indistinguishable (arrow), while b-d show areas where the two membranes are less than 3 nm apart. Scale bar = 50 nm.
Supplemental Figure 6. Depletion of ORP1L does not alter Rab7 and RILP PV-localization. HeLa cells were treated with siRNA, transfected with plasmid DNA the next day, and then infected 1 day later. At 2 days post infection, cells were fixed and stained for CD63 and Coxiella. Both Rab7-GFP (A) and RILP-GFP (B) localize to the Coxiella PV in cells treated with siRNA targeting ORP1L or non-targeting (NT) negative control. Scale bar = 10µm.
Supplemental Movies 1–3. Electron tomography tilt series of three different C. burnetii PVs. Tilt series for three separate PVs are shown; the arrow denotes areas where the PV and ER membranes are indistinguishable from one another.