Abstract
The principal challenge of using classical physics to model biomolecular interactions is capturing the nature of short-range interactions that drive biological processes from nucleic acid base stacking to protein-ligand binding. In particular most classical force fields suffer from an error in their electrostatic models that arises from an ability to account for the overlap between charge distributions occurring when molecules get close to each other, known as charge penetration. In this work we present a simple, physically motivated model for including charge penetration in the AMOEBA (Atomic Multipole Optimized Energetics for Biomolecular Applications) force field. With a function derived from the charge distribution of a hydrogen-like atom and a limited number of parameters, our charge penetration model dramatically improves the description of electrostatics at short range. On a database of 101 biomolecular dimers, the charge penetration model brings the error in the electrostatic interaction energy relative to the ab initio SAPT electrostatic interaction energy from 13.4 kcal/mol to 1.3 kcal/mol. The model is shown not only to be robust and transferable for the AMOEBA model, but also physically meaningful as it universally improves the description of the electrostatic potential around a given molecule.
Graphical abstract
1. Introduction
A grand challenge of molecular mechanics (MM) force fields is modeling the physics of molecular interactions with an accuracy and efficiency that allows realistic, tractable simulations of large systems. The goal is not only to correctly capture the physics of molecular interactions, but also to be able to answer important practical questions posed by biology, materials science and a number of other fields. To do this, MM models make classical approximations to the 1st principles quantum mechanics driving the true dynamics of a molecular system. Typically, this is done via a set of classical harmonic potential terms describing the intramolecular interactions of bonded atoms in the system and a separate set of non-bonded terms to describe intermolecular interactions. In particular, the electrostatic nonbonded terms are especially important for accurately modeling both short and long range molecular interactions.1
The AMOEBA force field is unique in its treatment of these important intermolecular electrostatic interactions. Most MM force fields use point charges to approximate the charge distribution around atoms in a system and parameterize these point charges based on thermodynamic measurements. AMOEBA takes a more physically realistic approach. The AMOEBA model approximates the charge distribution around atoms as a point multipole expansion of the charge distribution obtained from ab initio quantum mechanics (QM) calculations.2, 3 Using a multipole expansion derived from ab initio QM calculations provides a much more accurate description of electrostatic interactions at medium-range (~2 to 4 times the vdW radius), and has been shown to yield satisfactory results for simulations of water, proteins, nucleic acids and small molecules.1, 2, 4, 5
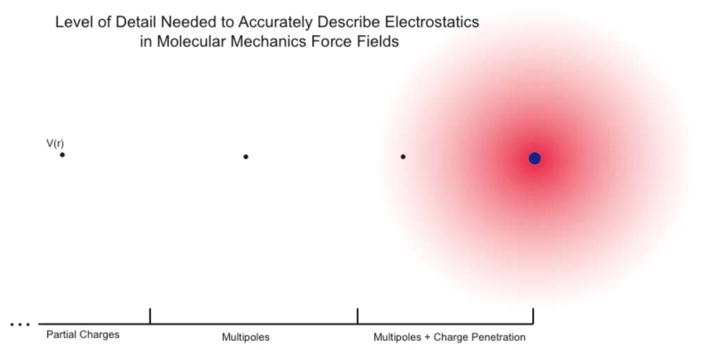
The multipole approximation of electrostatics, however, starts to break down at short-range. While the multipole expansion is rigorously correct for interactions of atoms at sufficient distance, it is no longer strictly valid once the electron clouds of interacting atoms start to overlap. This phenomenon is known as charge penetration. Charge penetration is simply the change in the electrostatic interaction between two atoms due to their electron cloud overlap and the associated loss of nuclear screening. It is a simple accounting for the fact that atoms in a system are not points; they represent finite charge distributions. Accurately modeling electrostatics has been a priority with AMOEBA since its inception. The importance of these interactions was a key motivation for the original AMOEBA multipole model. Qualitatively, accounting for charge penetration is the logical next step in improving this model. As depicted in figure 1, the current model covers the accuracy of long- and medium-range electrostatic interactions. What is needed is a description of charge penetration to accurately model short-range interactions.
Figure 1.
Electrostatic potential as a function of distance. An increasing level of theory is needed as the radial distance from an atom of interest decreases.
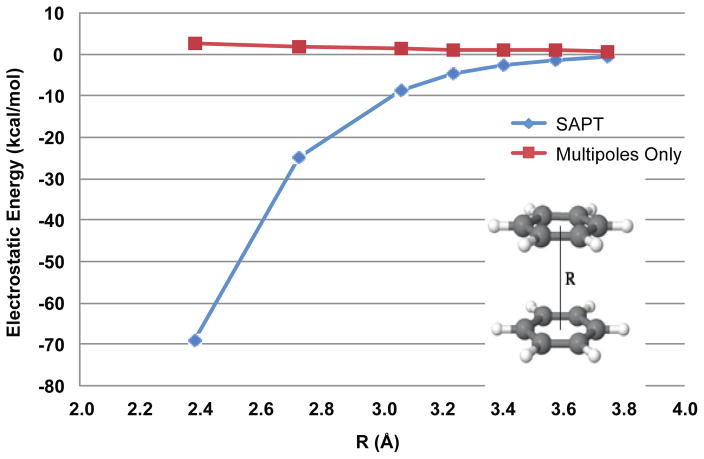
In addition to being physically relevant, charge penetration has been shown to be an important factor in many intermolecular interactions. A particularly instructive set of examples lies with what are commonly called “pi-pi” stacking interactions.6 The benzene sandwich dimer, as illustrated in figure 2, should classically be considered electrostatically repulsive since like charges are lined up across from one another. High level ab initio quantum mechanical calculations, however, show the counterintuitive result that the benzene sandwich dimer is electrostatically attractive.7 This is almost entirely due to charge penetration. Figure 2 shows that the overlap of electron clouds causes the electrostatic energy of the interaction to become more negative as the two monomers get closer together. This same phenomenon is observed with stacking interactions between nucleobases. Parker and Sherrill have recently shown that without charge penetration, it is difficult, if not impossible to accurately capture the electrostatics of interacting nucleobases.8 These considerations show that if AMOEBA is to be successful in accurately modeling biologically relevant interactions such as nucleic acid folding or ligand binding, we must account for the short-range electrostatics of charge penetration.
Figure 2.
Electrostatic energy of the benzene sandwich dimer. AMOEBA overestimates the electrostatic energy of the interaction compared with the benchmark QM calculations. The error gets progressively worse at short-range.
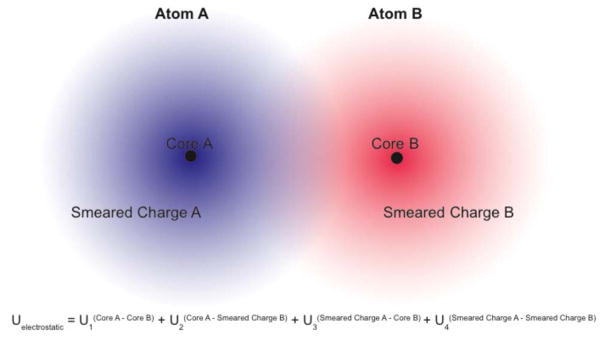
A number of studies have suggested functions for incorporating charge penetration into existing molecular mechanics force fields.9–20 The derivation of most of these functions has followed the same basic strategy. The electrostatic description of each atom in the system is split into two parts. The first is the core charge (often, but not necessarily simply the nuclear charge), treated as a point and second a smeared electron cloud charge representing the remaining charge of the atom. This splits what was a single interaction into four interactions, as illustrated in figure 3. The functions listed in table 1 are four methods suggested for how best to handle this four-part interaction between atoms. Tafipolsky and Engels took a more direct approach and calculated a numerical integral between spherical pro-molecule charge densities.17 This is similar in spirit to the approach of the GEM (Gaussian Electrostatic Model) force field, where hermite gaussians are used to reproduce the ab initio electron density.9, 21, 22 While being physically straightforward, these methods currently lack the efficiency needed for simulating large systems. The other three methods use damping functions to approximate how the electrostatic potential of an atom changes in its electron cloud and use those damping functions to approximate the value of the overlap integral for U4.
Figure 3.
Electrostatic energy of charge penetration-corrected, smeared-charge atomic interactions. The total electrostatic energy is split into four parts. The first term is the energy of the core-core, point-point interaction. The second and third terms are the energies of each core in the electrostatic potential of the opposing smeared charge. The fourth term is the energy of the overlap between smeared charge distributions.
Table 1.
Proposed methods for incorporating charge penetration into molecular mechanics electrostatic energy. For consistency, Z is the nuclear charge, ρ is the total charge density of the electrons, q is the total charge of the electron cloud, V is the number of valence electrons, c is the partial charge, n is the number of “screening electrons”, and r is the internuclear distance. In the first row, the charge density is either a promolecular charge density (Engels) or a density from hermite gaussians in the GEM model (Cisneros).
| Model | Core A –Core B | Core A – Smeared Charge B | Smeared Charge A – Core B | Smeared Charge A – Smeared Charge B | ||||
|---|---|---|---|---|---|---|---|---|
| Engels; Cisneros |
|
|
|
|
||||
| Gordon |
|
|
|
|
||||
| Piquemal |
|
|
|
|
||||
| Truhlar |
|
|
|
|
In a previous proof-of-principle study, we implemented the form of Piquemal and co-workers in the AMOEBA force field.23 The study showed that accounting for charge penetration can start to recover the true nature of short-range electrostatic interactions between molecules. A follow-up study extended the model for use with smooth particle mesh Ewald.24 In the present work we seek to develop a comprehensive model based on the previous work that best captures the physics of electrostatic intermolecular interactions and the aims of the AMOEBA force field. Given the potential improvement our previous work has shown possible in such a model, the question becomes: what features would we like the AMOEBA charge penetration model to have? In the work presented here we aim to implement a charge penetration function that best meets the following criteria:
The model should be physically derived.
The model should be computationally efficient to compute.
The model should be numerically stable.
The model should accurately reproduce ab initio QM measurements for relevant molecular interactions.
The model should be consistent with the AMOEBA multipole model.
In section 2, we present the physical derivation of the models that were considered and derive corresponding damping terms for higher-order multipoles. In section 3, the scheme for parameterizing the models is presented. Section 4 lays out results comparing the performance of the models. Section 5 shows validation that the charge penetration model is capturing physical reality. And lastly, section 6 draws our conclusions.
2. Theory
Stone illustrated the phenomenon of charge penetration with a simple example.25 Consider the interaction of a proton with a hydrogen-like atom with nuclear charge Z. From quantum mechanics we know that the wave function of a hydrogen-like atom is
| (1) |
This gives us the electron density of the atom,
| (2) |
This tells us how dense the electron distribution of the atom is as a function of the radial distance (r) from its nucleus. To get the potential this density generates, we must apply Poisson’s equation,
| (3) |
where ε0 is the permittivity of free space. Applying Eq. (3) to Eq. (2) we obtain
| (4) |
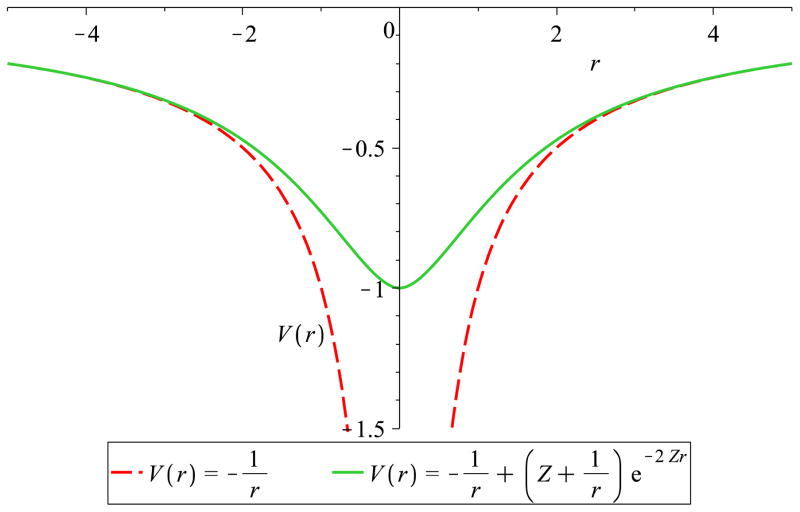
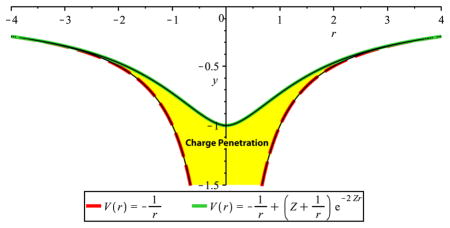
the familiar potential due to the electron density of a hydrogen-like atom. At large distances from the atom, the first term in Eq. 4 dominates the second term due to the second’s exponential decay and we have the classical point charge coulomb approximation of the potential. At closer distances, however, as shown in figure 4, the second term becomes non-negligible. This second term represents the charge penetration.
Figure 4.
Classical coulomb potential vs. Hydrogen-like atom potential. Plotted is the electrostatic potential of a point electron vs. the hydrogen-like electron (Z=2 to emphasize the distinction). The classical potential diverges from the hydrogen-like result at short-range.
We can exploit the fact that V(r) converges to −1/r at large distances and rewrite Eq. 4 as
| (5) |
where,
| (6) |
The potential in this form is represented simply as the point charge coulomb potential multiplied by a damping function. This is convenient because the damping function has the following straightforward properties:
It approaches a value of one as r becomes large.
It approaches a value of zero as r approaches zero.
It is a direct multiplication of the classical point-charge coulomb potential.
It describes charge penetration as a deviation from the classical potential.
To this point there are no approximations made in our derivation. Crucially, however, most atoms in systems of interest for molecular simulation are not strictly hydrogen-like. This means that fdamp for non-hydrogen-like atoms is not exactly given by Eq. 6. The properties and form of Eq. 6 are instructive, however. To capture the physics more generally, we introduce a parameter, α, in place of the 2Z and remove the prefactor in front of the exponential to obtain
| (7) |
This more general construction of fdamp retains all of the relevant damping function properties listed above and allows us to tune the parameter, α, to reproduce ab initio electrostatic energies. This is identical to the damping function proposed separately by both Gordon and co-workers11 and Piquemal and co-workers.10
Using the damping formulation of Eq. 7, we have now effectively changed the potential due to every atom in a given system. The potential at any point in the system is described by,
| (8) |
where the potential due to the nucleus is unchanged, but the potential due to the electrons now accounts for the charge penetration effect. This, however, is not quite enough to get the interaction energy between two atoms. Recall from figure 3 that although the second and third terms of the charge penetration corrected electrostatic interaction energy involve simple point charges interacting with the potential due to smeared charge distributions, the fourth term has two smeared charge distributions interacting with each other. In this unique case, we must derive a second “overlap” damping function to account for this interaction.
For the fourth, overlap term we are attempting to approximate the overlap integral between the two charge distributions,
| (9) |
where VA and VB are the charge penetration corrected potentials due to atoms A and B respectively. Gordon and co-workers approximate this integral using the one-center method given by Coulson26 to yield
| (10a) |
where qA and qb are the total electron charges of atoms A and B, for the charge-charge portion of the interaction. Piquemal and co-workers take a two-center approach to approximating the integral,
| (10b) |
where, as laid out in our previous work (Ref. 20), a second parameter is introduced to describe the overlap. While the derivations of these formulae are slightly different, mathematically these U4 overlap damping functions constitute the only functional difference between the models of Gordon and co-workers and Piquemal and co-workers. For simplicity’s sake, the approach of Eq. 10a will be referred to as model 1 and Eq. 10b as model 2. They can be implemented, however, in an identical manner. These overlap damping functions allow us to calculate the charge penetration corrected charge-charge electrostatic interaction between any two sites:
| (11) |
The AMOEBA model, however, has more than just charges on every atom. It uses a multipole expansion representing the charge distribution at every site. The energy between two AMOEBA multipole sites, i and j, is given by,
| (12) |
where Mi and Mj represent the multipole moments on atoms i and j respectively, and
| (13) |
is the classical point multipole interaction matrix. We can see in Eq. 13 that the interaction matrix, Tij, for AMOEBA without charge penetration is obtained simply by taking repeated derivatives of the classical coulomb potential, 1/r. To account for charge penetration, not just in charge-charge interactions, but in all multipole interactions up to arbitrary order, we simply insert the charge penetration damped potential in place of the classical potential. This yields the charge penetration corrected multipole interaction matrix,
| (14) |
where fdamp is either 1 (for nuclear-nuclear interactions), the damping function from Eq. 7 (for the second and third terms of the interaction energy), or the overlap damping function from Eqs. 10a or 10b (for the fourth term of the interaction energy). Using the charge penetration corrected multipole interaction matrices, we can express the new AMOEBA multipole interaction energy of any two sites as:
| (15) |
Eq. 15 allows us to account for the effects of charge penetration up to arbitrary order multipole expansion. For AMOEBA, which has multipole interactions up to quadrupole-quadrupole, this means that the charge penetration model can be made fully consistent with the multipole model. See Supplementary Material for explicit damping functions for all AMOEBA multipole interaction components.
3. Parameterization
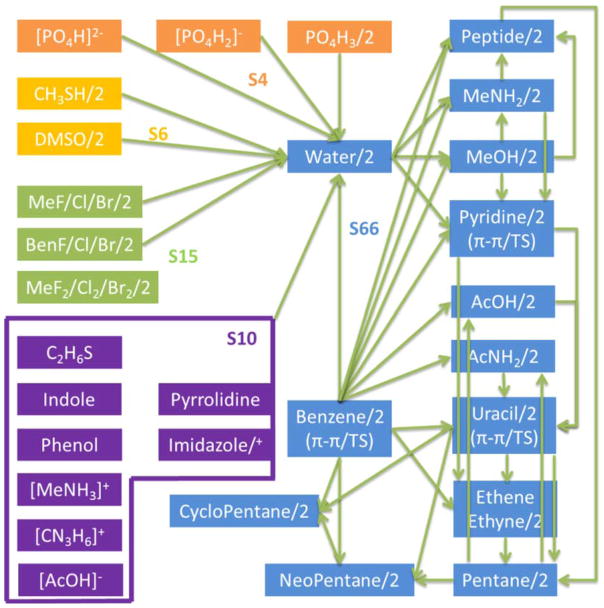
The goal of including charge penetration in the AMOEBA model is to more accurately reproduce the energies of electrostatic interactions between molecules at short range. Because both models 1 and 2 contain empirical parameters, we will seek to optimize them by fitting to a database of relevant intermolecular electrostatic energies. In our previous work, the S101 and S101x7 databases where constructed for this purpose.23 The S101 database contains 101 unique pairs of both homodimers and heterodimers of common organic molecules. It contains the widely used S66 database27 along with some additional relevant biomolecular interactions. The S101x7 database is constructed by placing each dimer pair from the S101 database at 0.70, 0.80, 0.90, 0.95, 1.00, 1.05 and 1.10 times their equilibrium intermolecular distance. A schematic representation of all the dimer pairs included in the S101 database is shown in figure 5. In all of the parameterization that follows, the entire S101x7 database was used with the exception of interactions involving ethyne. The omission of ethyne allows direct comparison with the results from our previous work.
Figure 5.
Dimer pairs in the S101 database. Arrows connect monomers that form dimers. A “/2” designation indicates a homodimer. A “/+” designation indicates both neutral and positively charged forms. Reproduced from Ref. 20.
To parameterize the charge penetration models against the S101x7 database, accurate intermolecular electrostatic energies are needed for all dimer pairs. In the previous work, Symmetry Adapted Perturbation Theory (SAPT)28 calculations where performed to obtain these energies. SAPT calculations decompose intermolecular energies into physically meaningful components; the intermolecular energy between two monomers is broken down into electrostatic, induction, exchange-repulsion and dispersion energies. For the S101x7 database, SAPT2+ calculations29, 30, estimated at the complete basis set (CBS) limit as described in Ref. 22, were carried out to return the ab initio electrostatic interaction energy of each dimer pair.
The parameters of model 1 and model 2 were optimized by performing a nonlinear least squares fit to minimize the difference between the AMOEBA electrostatic energy (with charge penetration), , and the SAPT electrostatic energy, , for each dimer pair. For models 1 and 2, two methods of parameterizing are proposed. In the first method one parameter, α, is assigned per element. In the second, one α is assigned per charge penetration class. These classes, as listed in table 2, are simply chosen to allow for different descriptions of atoms of the same element but different physiochemical classifications. The choice of classes is based on the knowledge that the electronic structure of an sp2 hybridized carbon, for example, will be generally different than that of an aromatic carbon. While it is certainly true that differences in electron distribution exist even amongst atoms of the same charge penetration class (the electronic structure of every sp2 hybridized carbon is not exactly the same), the guiding principle is to include only the minimal level of atomic classification to allow the model to be easily transferable.
Table 2.
Atom classes and fitted parameters for charge penetration models.
| Element | Charge-charge Damping | Charge Penetration Class | Charge-charge Damping | Higher-order Damping | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||
| Model 1 α (Å−1) | Model 2 α (Å−1) | Model 2 γ | Model 3 ζ (Å−1) | Model 1 α (Å−1) | Model 2 α (Å−1) | Model 2 γ | Model 1 α (Å−1) | Model 2 α (Å−1) | Model 2 γ | ||
| Hydrogen (H) | 4.0026 | 10.000 | 0.8720 | 1.2976 | non-polar (H-C) | 3.4345 | 3.5474 | 0.8710 | 3.2484 | 3.2624 | 0.8823 |
|
|
|
||||||||||
| aromatic (H-C) | 3.9419 | 4.0006 | 3.4437 | 3.4080 | |||||||
|
|
|
||||||||||
| polar, water (H-X) | 5.0049 | 10.000 | 3.2632 | 3.4317 | |||||||
|
|
|
|
|||||||||
| Carbon (C) | 3.0957 | 2.9137 | 1.2100 | sp3, tetrahedral | 3.3863 | 3.2136 | 3.5898 | 3.7576 | |||
|
|
|
||||||||||
| sp2, aromatic | 3.1205 | 2.9268 | 3.2057 | 3.2569 | |||||||
|
|
|
||||||||||
| sp2, carbonyl, etc. | 3.1702 | 2.9349 | 3.1286 | 3.1971 | |||||||
|
|
|
|
|||||||||
| Nitrogen (N) | 3.7321 | 3.4066 | 1.4502 | sp3, tetrahedral | 3.2519 | 3.2317 | 4.0135 | 3.9410 | |||
|
|
|
||||||||||
| sp2, aromatic | 3.6979 | 3.4199 | 3.6358 | 3.7534 | |||||||
|
|
|
||||||||||
| sp2, other | 3.4264 | 3.3110 | 3.7071 | 3.8244 | |||||||
|
|
|
|
|||||||||
| Oxygen (O) | 4.1390 | 3.5677 | 1.4114 | sp3, hydroxyl, water | 3.7975 | 3.7038 | 4.1615 | 4.2449 | |||
|
|
|
||||||||||
| sp2, aromatic | 3.7770 | 3.6686 | 4.3778 | 5.0908 | |||||||
|
|
|
||||||||||
| sp2, carbonyl | 3.4938 | 3.4509 | 3.7321 | 3.6146 | |||||||
|
|
|
|
|||||||||
| Phosphorous (P) | 3.0661 | 2.5969 | 1.3369 | phosphate | 3.1539 | 2.6076 | 2.7476 | 3.0668 | |||
|
|
|
|
|||||||||
| Sulfur (S) | 2.9570 | 2.5965 | 1.1156 | sulfide, thiol | 3.2046 | 2.7320 | 3.3112 | 3.3826 | |||
|
|
|
||||||||||
| sulfur (IV) | 3.3824 | 2.6353 | 2.6247 | 2.9057 | |||||||
|
|
|
|
|||||||||
| Fluorine (F) | 4.4875 | 4.2333 | 1.5955 | organofluoride | 4.4314 | 4.2730 | 4.4675 | 10.000 | |||
|
|
|
|
|||||||||
| Chlorine (Cl) | 3.5173 | 2.9092 | 1.2102 | organochloride | 3.5060 | 2.8887 | 3.4749 | 3.5035 | |||
|
|
|
|
|||||||||
| Bromine (Br) | 3.7202 | 2.5924 | 1.2259 | organobromide | 3.7150 | 2.5820 | 3.6696 | 3.7146 | |||
For model 2, the parameter, β, is fixed as a fraction of α,
where the parameter, γ, is taken to be universal to avoid over-fitting. Allowing β to float for every charge penetration class has the potential, of course, to improve the overall fit, but at the cost of losing physical meaningfulness. Recall from Eq. 10b that although the β parameter is specific to the overlap function in model 2, the two electron clouds that are overlapping are supposed to already be described by the parameter α. Allowing both α and β to float in the fit would allow two different parameters to describe essentially the same physics. Instead fitting one universal parameter γ simply describes how β should be generally related to α in approximating the overlap between molecules. It should be noted that the parameterization strategy here for model 2 differs slightly from previous work. It is chosen in this way to best fit the AMOEBA multipole model and provide for a direct comparison with model 1 on the same test set.
The results of fitting model 1 and model 2 are shown in table 2. Three fits were performed for each model. First the S101x7 database of intermolecular electrostatic energies was fit using only charge-charge damping with parameters assigned by element. Next, the same charge-charge damping fit was performed with parameters assigned by class. Then the database was fit using higher-order damping with damping of all AMOEBA multipole interactions (up to and including quadrupole-quadrupole).
In addition to parameterizing models 1 and 2, a third model, due to Wang and Truhlar18–20 has been parameterized as well. This model, developed for application in QM/MM calculations is included as a point of comparison. However, it is not developed any further than charge-charge damping using parameters assigned by element as it has several properties that make it unsuitable for implementation in AMOEBA. First, the model can be unstable with respect to the parameters of interacting atoms. If two closely interacting atoms have parameters that are close, but not identical, the overlap damping functions of the model breaks down. Second, expanding the model to include higher-order damping to make it fully consistent with the AMOEBA multipole model is computationally intractable with this model. The expressions that form the overlap damping functions, as seen in Eqs. 8 and 9 in Ref. 19 are much more complex functions of the radial distance between atoms, r. Taking the successive derivatives necessary for higher-order damping terms would produce expressions too expensive to calculate for our purposes. Third, even if such derivatives were deemed necessary, the model’s framework is incompatible with higher-order damping. The damping functions used in Wang and Truhlar’s model are meant to simulate the outer Slater-type orbitals of atoms. With this being the case, rather than treat all of an atom’s electrons as damped, the model only treats a maximum of 2 as damped. This treatment is acceptable for charge-charge damping since charge is spherically symmetric and one simply treats the remaining electrons as part of the “core”. This is, however, problematic for higher-order damping because there is no such simple partitioning of the electrons that make up an atom’s dipole and quadrupole moment. It would be nonsensical to apply the model’s damping terms meant for two electrons, to an atom’s dipole and quadrupole interactions.
In the following section the fits produced by the parameterization of all three models is presented. The fits of each model to the S101x7 database will be used along with some important validation tests and theoretical arguments to determine which model and which parameterization strategy to implement in AMOEBA.
4. Results
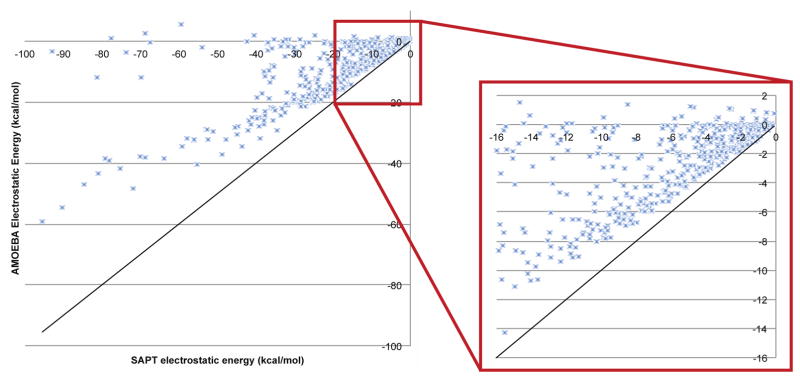
To understand how charge penetration improves the electrostatic model of AMOEBA, we must understand how the current AMOEBA model without a charge penetration correction performs. Figure 6 shows how AMOEBA’s prediction of intermolecular electrostatic energies compares to the SAPT ab initio electrostatic energy values on the S101x7 database. Figure 6 reveals that using only a multipole expansion to describe the electrostatic interactions between molecules systematically overestimates the electrostatic energy at short range. The pervasive gap illustrated in figure 6 illustrates the need for including charge penetration in the electrostatic model of the AMOEBA force field.
Figure 6.
AMOEBA, multipole-only intermolecular electrostatic energy of dimers in S101x7 database. The multipole-only electrostatic energy for each dimer is plotted against the benchmark SAPT electrostatic energy. The diagonal, y=x line indicates what would be perfect agreement. Compared to the benchmark calculations, the multipole-only model systematically overestimates the electrostatic energy.
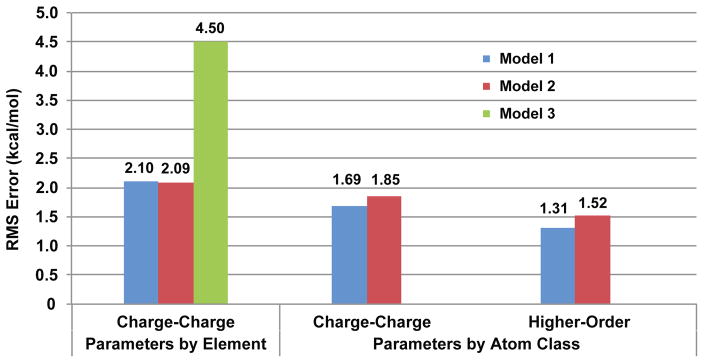
The most naïve method of applying a charge penetration correction is to assign one parameter per element and damp only the charge-charge electrostatic interactions. As a first test of the theory, this strategy was implemented for models 1, 2 and 3. Each model was then parameterized by fitting to the S101x7 database. The overall results of assigning parameters by element and damping only the charge-charge electrostatic interactions are illustrated in the first cluster of columns in figure 7. It is clear that all three models perform much better than the current AMOEBA multipole only model. The RMS error of the multipole-only model for electrostatic energies on the S101x7 database is 13.4 kcal/mol. Models 1, 2 and 3 bring that error down to 2.1 kcal/mol, 2.1 kcal/mol and 4.5 kcal/mol respectively, showing that even a naïve damping strategy starts to capture the missing physics. It is also apparent that models 1 and 2 perform much better, even at this low level of implementation, than model 3. Additionally, note that despite having fewer parameters, model 1 performs nearly identically to model 2 for this implementation. Complete statistics for each of these fits, including a breakdown by intermolecular distance, are available in Supplementary Material.
Figure 7.
Root mean square error of AMOEBA electrostatic energy with charge penetration on S101x7 database. Multiple charge penetration models were tested. The first cluster of columns represents the results of parameters fit by element with charge-charge damping only. The second cluster is the results of having parameters assigned by class and charge-charge damping. The third cluster is the results for including higher-order damping in addition to having parameters assigned by class. (RMS error of AMOEBA with multipoles-only is 13.4 kcal/mol)
While assigning parameters by element produces an improvement over the multipole-only AMOEBA model, it ignores some key physiochemical properties of elements in different bonding environments relevant to interpreting the α parameter. The α parameter with units, Å−1, can be understood as the inverse of the physical extent of the electron cloud of an atom. From ab initio electronic structure calculations we know that in general this property can change substantially based on the bonding environment of an atom. For this reason we fit models 1 and 2 with parameters assigned by class to the S101x7 as described in the preceding section. The overall results of assigning parameters by class and still damping only the charge-charge electrostatic interactions are illustrated in the second cluster of columns in figure 7. The first thing to note is the absence of a fit for model 3. Once the parameter set is expanded to include classes, model 3 becomes highly unstable. As noted before this is due to numerical instability when parameters in the model become close. This is practically unavoidable for class-based parameters, so model 3 is excluded from this point forward. More importantly, however, we notice also that splitting out different parameter classes improves the overall fit to the S101x7 database for models 1 and 2. Assigning parameters by class improves the performance on the RMS error. Again despite having fewer parameters, model 1 outperforms model 2 in this case. This improvement is largely due to allowing different classes for the same element. For example, table 2 shows that for model 1 the parameter for hydrogen in the element based fit splits quite significantly when one allows different classes to vary. The element parameter, 4.0 Å−1 splits into parameters of 3.4 Å−1, 3.9 Å−1 and 5.0 Å−1 for non-polar, aromatic and polar hydrogen respectively. This extra flexibility in the parameterization, rooted in basic physiochemical properties improves our overall description of the electrostatics. Again specific statistics for class-based fits can be found in the Supplementary Information.
Splitting out separate chemical classes for parameters improves the performance of our charge-charge damping charge penetration model, but it unfortunately does not meet the criteria of being fully consistent with the AMOEBA multipole electrostatic model. To test the fully integrated model we implemented charge penetration damping for all multipole interaction terms (up to and including quadrupole-quadrupole) for both models 1 and 2. We will refer to this model as “higher-order” damping. The overall results, illustrated in the third and final cluster of columns in figure 7, show the improvement that this model brings. Implementing a fully integrated higher-order damping model with class-based parameters brings the RMS error on the entire S101x7 database for models 1 and 2 down to 1.31 kcal/mol and 1.52 kcal/mol respectively. Full statistical analysis can be found in Supplementary Information. These numbers represent a dramatic improvement over the current AMOEBA multipole-only RMS error of 13.43 kcal/mol. More importantly they also improve on the errors from our charge-charge damping implementations. A significant portion of the improvement is due to improvement in the performance on the closest dimer pairs in the S101x7 database. Among dimers that are separated by 0.70 and 0.80 of their equilibrium distance, model 1 with higher-order damping reduced that error from 2.75 kcal/mol to 2.27 kcal/mol, and model 2 reduced it from 4.36 kcal/mol to 2.64 kcal/mol. Importantly, this improvement does not sacrifice the fit at more accessible distances. For model 1 the RMS error on dimers with intermolecular separations of 0.90 to 1.10 times their equilibrium distance dropped to under 1 kcal/mol compared with an error of over 4 kcal/mol for the current multipole-only model. Lastly, these fits give a slight edge to the simpler model 1 over model 2. Model 1 performs 16% better than model 2 on overall RMS errors in the S101x7 database when higher-order damping is included. The absolute percent error of model 2 on the electrostatic energies of the S101x7 database is 10%, while model 1 gives 7%.
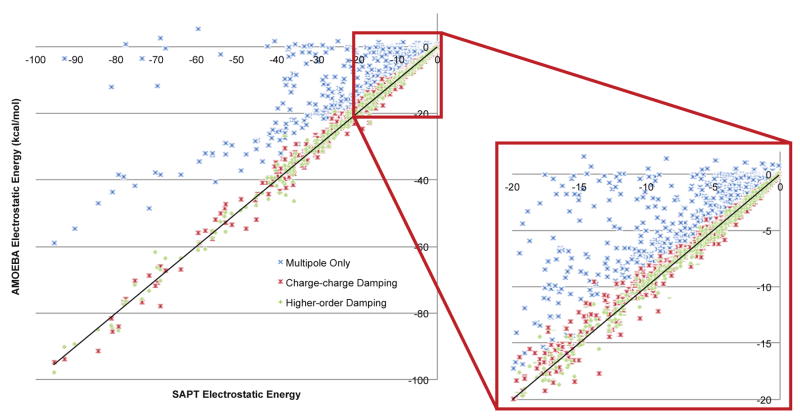
Figure 7 lays out the overall performance of each of the implementations described above. It is clear from this data that model 1 with higher-order damping and parameters assigned by class gives the best fit to the electrostatics of the S101x7 database. The improvement this model gives on each individual dimer pair is shown in figure 8. Figure 8 shows that across the board model 1 with higher-order damping is superior to simple charge-charge damping, and represents a dramatic improvement over the current multipole-only model. This is bourn out in a handful of important and instructive examples. Figure 9 lays out the results for fitting the water dimer, figure 10 shows two important orientations of the benzene dimer and figure 11 shows the model’s performance on phosphate ions. These three examples represent important relevant biomolecular interactions that the current multipole-only model fails to accurately capture. Moreover, all three also show that an integrated higher-order damping model is needed to achieve the highest level of agreement with SAPT electrostatic data. These examples show that not only does the model generally improve the quality of electrostatics across a wide dataset, but it also performs well on individual examples, such as the benzene sandwich dimer, that inspired our investigation of the charge penetration phenomemon.
Figure 8.
AMOEBA intermolecular electrostatic energy with and without charge penetration of S101x7 database dimers. The AMOEBA electrostatic energy both without (mulitpole-only) and with (model 1 with charge-charge or higher-order damping) charge penetration is plotted against benchmark SAPT electrostatic energy calcuations. The diagonal, y=x line indicates what would be perfect agreement. Including higher-order damping in the charge penetration model yields the best agreement with ab initio electrostatic energies.
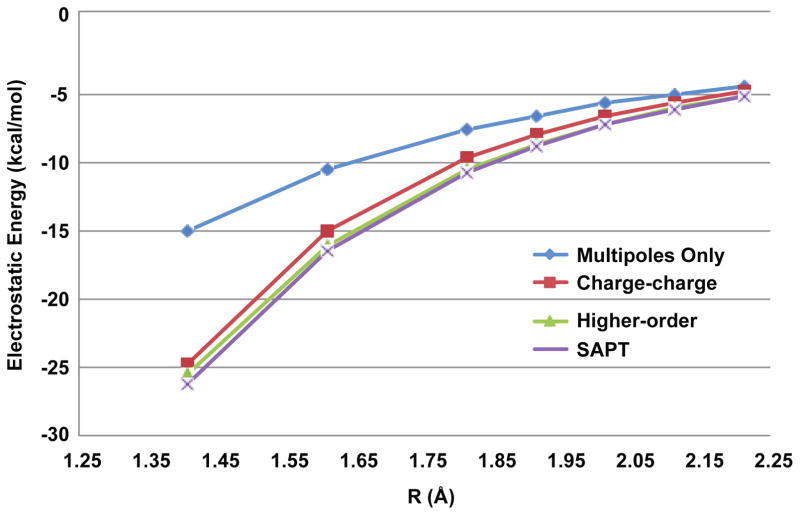
Figure 9.
Water dimer electrostatics. AMOEBA dimer electrostatic energies without (multipoles-only) and with (model 1 with charge-charge and higher-order damping) charge penetration are plotted against benchmark SAPT electrostatic energies.
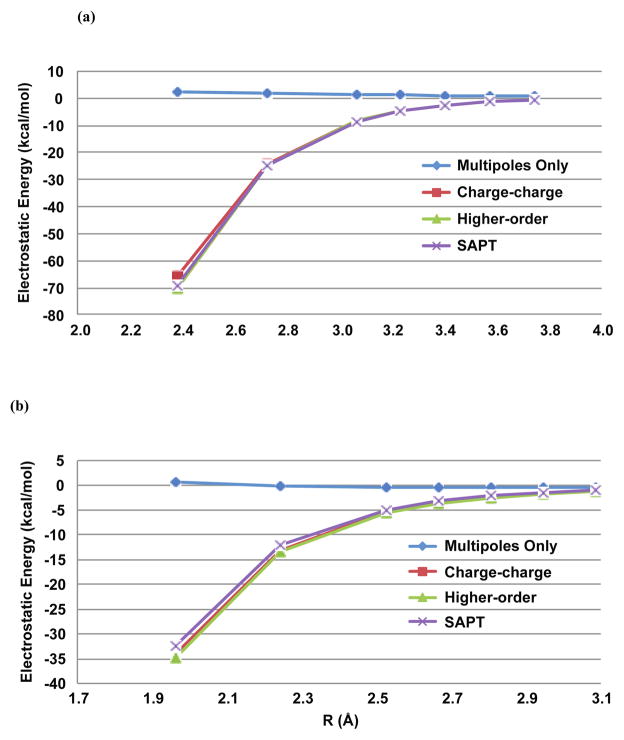
Figure 10.
Benzene (a) Sandwich and (b) T-shape dimer electrostatics. AMOEBA dimer electrostatic energies without (multipoles-only) and with (model 1 with charge-charge and higher-order damping) charge penetration are plotted against benchmark SAPT electrostatic energies.
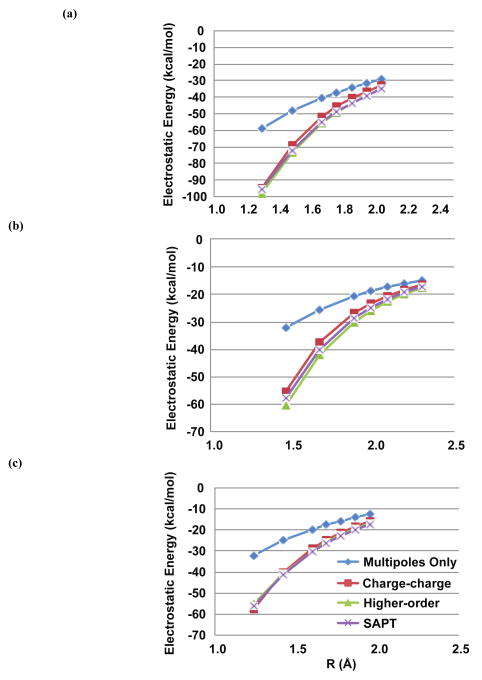
Figure 11.
Phosphate-water dimer electrostatics. AMOEBA dimer electrostatic energies without (multipoles-only) and with (model 1 with charge-charge and higher-order damping) charge penetration are plotted against benchmark SAPT electrostatic energies. Results are shown for PO4H (a), PO4H2 (b) and PO4H3 (c).
5. Validation
The fit to the S101x7 database with model 1 higher-order damping is a welcome result. The model dramatically improves the quality of the electrostatic fit for those electrostatic interactions over AMOEBA’s current multipole-only model and it outperforms all of the other relevant damping models proposed. There are, however, some considerations that need to be addressed to validate model 1 with higher-order damping as the best option for capturing the physics of charge penetration. First, we would like to show that in addition to giving the best fit, model 1 is also the most robust option. Second, we need to know to what extent this charge penetration model is independent of the AMOEBA multipole model. And most importantly, we must validate that this model is capturing a real physical phenomenon.
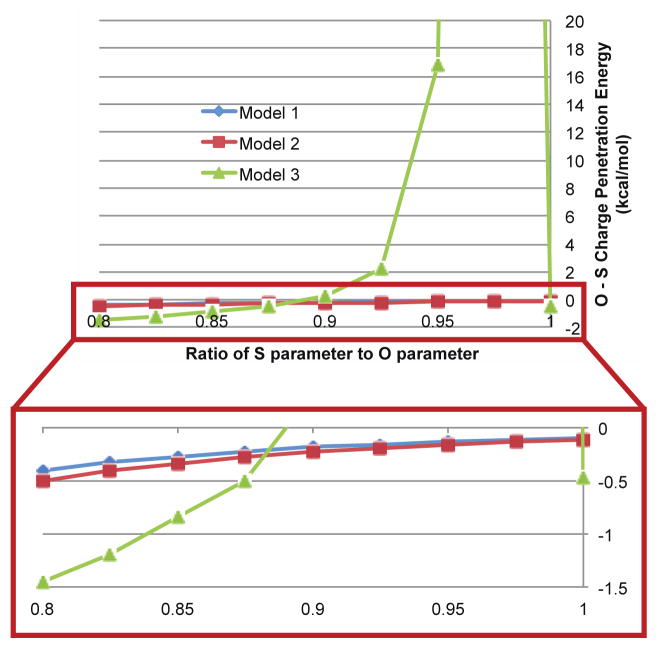
It is important our charge penetration model not only provides a good fit to ab initio electrostatic data, but also that the model is robust. To evaluate robustness we must evaluate the sensitivity of the model to small changes in the parameters. Model 3 does not pass this parameter sensitivity requirement. Figure 12 shows the behavior of the oxygen–sulfur electrostatic interaction in the DMSO–water dimer as the difference between oxygen and sulfur parameters gets smaller. Clearly model 3 breaks down as the two parameters get close to one another. Moreover, the problem is compounded as the intermolecular distance decreases. Since the zeta parameter multiplies the interatomic distance, r, everywhere in the damping function, the problem gets worse as monomers get closer together. Model 2 does not suffer from any such numerical instability, but it is sensitive to the parameter, γ, that determines the overlap damping function. Table 3 shows that if the closest dimers are left out of our fit to the electrostatic data, γ changes from 0.88 to 0.90. Moreover, if we use the γ that comes out of the fit where we leave out the closest points, the RMS error for the full S101x7 database jumps from 1.52 kcal/mol to 1.83 kcal/mol. Model 1 on the other hand does not suffer from any such sensitivity. If we leave out the closest dimer pairs and fit parameters to our model, table 3 shows that those parameters do almost as well as the parameters fit to the full S101x7 database. The RMS error for model 1 in this case goes up by less than 0.1 kcal/mol. By these tests model 1 shows the strength with respect to numerical stability and parameter transferability we expect a robust charge penetration model to have.
Figure 12.
Charge penetration model stablity. The oxygen-sulfur electrostatic interaction energy for the water-DMSO dimer is plotted as a function of the difference between the oxygen and sulfur charge penetration paramters. As the ratio of the paramters approaches unity, model 3 becomes unstable.
Table 3.
Charge penetration model parameter sensitivity. Models 1 and 2 were fit to the S101x7 database excluding the closest points (all dimers except those at 0.7 times the equilibrium distance). The parameters generated from that fit are then tested on the full database. Model 2, particularly the γ parameter, proves to be the more sensitive to this change.
| Model 1 | Model 2 | |
|---|---|---|
| Parameters from fit to full S101x7 database | 1.31 kcal/mol | 1.52 kcal/mol (γ = 0.88) |
| Parameters from fit to S101x7 database excluding the closest points (0.8 – 1.1) | 1.40 kcal/mol | 1.83 kcal/mol (γ = 0.90) |
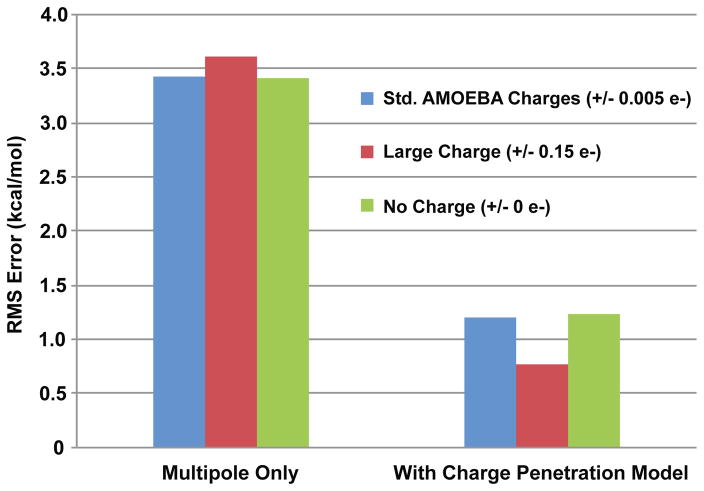
In addition to being the most robust option, model 1 also shows good model independence from the AMOEBA multipole model. AMOEBA follows a defined protocol for determining charge, dipole and quadrupole parameters for each monomer2 and we should expect that our model should, for the most part, be independent of that specific protocol. In other words the multipole model and the charge penetration model should not depend on each other. To test this we use the toy example, benzene. When determining the electrostatic parameters for benzene, multiple values for the opposing charges of the carbons and hydrogens will give nearly identical fits to the electrostatic potential on a grid of points around the molecule. Although the AMOEBA multipole protocol fixes those charge values semi-arbitrarily, we wanted to see if choosing otherwise would break our model 1 charge penetration model. Figure 13 demonstrates that model 1 accurately reproduces the electrostatic potential regardless of which potential-fitted charge-dipole-quadrupole model one chooses. This validates an important feature of the model: that it is independent of the specifics of potential fitting protocol for the AMOEBA multipole model.
Figure 13.
Charge penetration model independence. Three different benzene multipole models were chosen with charges fixed at +/−0.005 e−, +/− 0.15 e−, and 0 e− that give roughly equivalent electrostatic potential fits. The charge penetration model was then applied to all three models. RMS errors of the electrostatic potential on a grid of points around benzene for each model are plotted. The charge penetration significantly lowers the error regardless of multipole model.
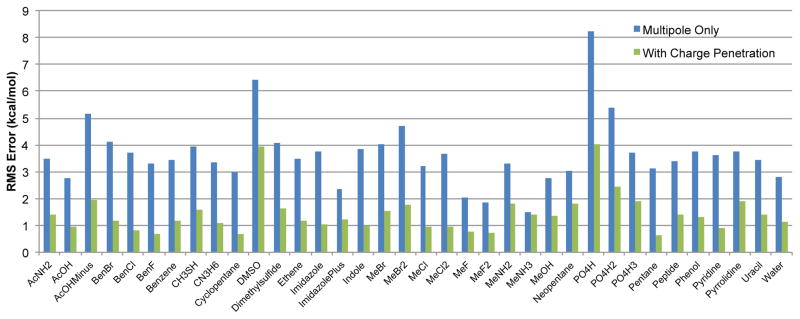
Lastly, but most importantly, for our model to be valid, we must prove that it is capturing a real physical effect. At the heart of the charge penetration phenomenon is the fact that the electrostatic potential around an atom at short range cannot be reproduced by a simple point multipole approximation without accounting for the extent of the atom’s charge density. To validate that the model is describing this physics we tested to see if our charge penetration model, model 1 with higher-order damping, could accurately reproduce the ab initio electrostatic potential around a molecule at short range. Figure 14 shows that without exception the charge penetration model dramatically improves the electrostatic potential fit around every monomer in the S101 database. This is the validation we are looking for. Not only does our model correct the practical problem of bad intermolecular electrostatic energies at close range, but it does so by accurately capturing the physical reality of molecules’ finite charge distributions.
Figure 14.
Charge penetration model performance on electrostatic potential of monomers in S101 database. The RMS error of the electrostatic potential on a grid of points around each monomer is plotted. Including charge penetration improves the fit to the electrostatic potential for every monomer.
6. Test Case: Nucleic Acid Base Stacking
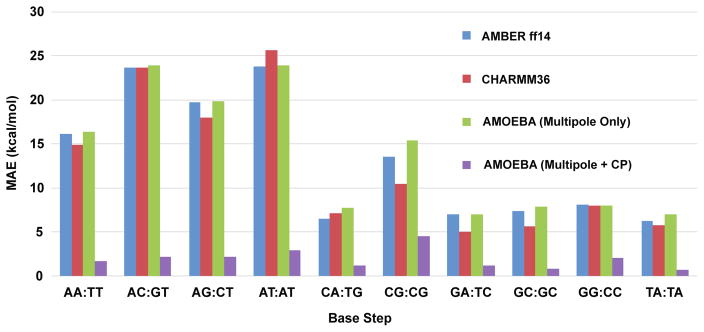
As stated in the introduction, charge penetration effects are important in a broad range of close-contact biomolecular interactions. One essential example is the stacking interactions of nucleobases in DNA and RNA sequences. Parker and Sherrill recently showed that without an explicit accounting for charge penetration, force fields struggle to accurately reproduce the ab initio electrostatic energies of these interactions.8 For instance in an AC:GT base step, the mean absolute errors (MAE) of the AMBER31, 32 and CHARMM33 force fields relative to the SAPT electrostatic energy were over 20 kcal/mol. Likewise, we find that AMOEBA without charge penetration gives an electrostatic energy MAE over 20 kcal/mol as well. However, when we apply our charge penetration function with parameters fixed to their values from the S101x7 fit, the MAE drops dramatically to nearly 2 kcal/mol. This improvement is not unique to the AC:GT base step. As shown in figure 15, the MAE of our AMOEBA model with charge penetration is significantly lower for every base step combination.
Figure 15.
Mean absolute electrostatic interaction energy error relative to SAPT0 for ten stacked base steps. Including charge penetration lowers the MAE in the electrostatic interaction energy for every base step combination.
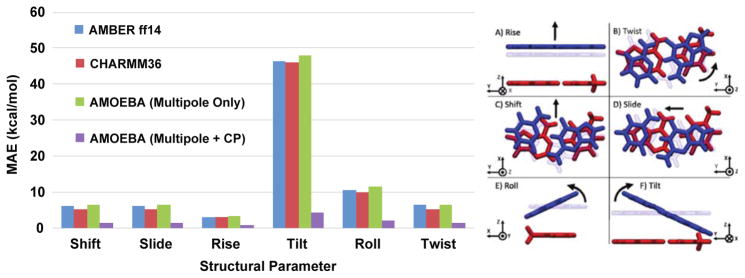
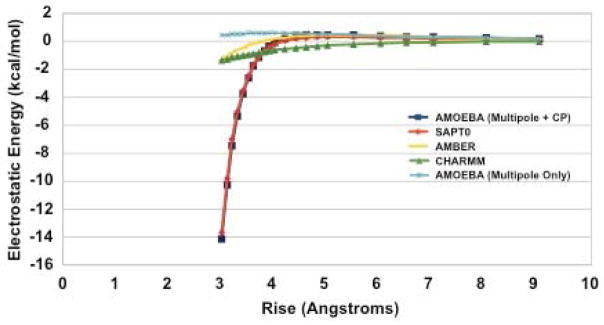
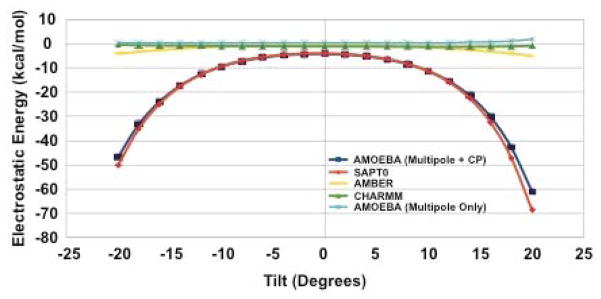
Moreover, this improvement in the electrostatic description of nucleobase stacking holds even for non-equilibrium stacking arrangements. Figure 16 shows that for the six structural parameters that define the stacking interaction, 34 the AMOEBA + charge penetration model does far better than AMBER, CHARMM or the current AMOEBA force field. These data confirm, as asserted by Parker and Sherrill, that including charge penetration is an absolute necessity for a robust nucleic acid force field model. This imperative is highlighted in two standout cases of the TA:TA base step. Figure 17 shows the performance of force field models against SAPT electrostatics versus the nucleobase rise. It is immediately clear that the AMOEBA + charge penetration model put forward here is the only model that accurately reproduces the electrostatic nature of this interaction. The same is seen in figure 18 where we examine the electrostatic energy as a function of the tilt parameter. Again, the model including charge penetration is the only model that agrees with the quantum mechanics. This same improvement persists across all structural parameters of the TA:TA base step. Figures for the other four parameters can be found in the supplementary information. It is worth noting that not only is this an important test case because of its direct relation to biomolecular applications for the force field. It is also important because it shows that the model, parameterized against a particular test set (S101x7) performs well on interactions well outside of that set. These results give us confidence in the transferability of our charge penetration model.
Figure 16.
Mean absolute electrostatic interaction energy error relative to SAPT for six structural parameters. Including charge penetration lowers the MAE for variation along every degree of freedom in the nucleobase stacking interaction. Inset reproduced from Ref. 7.
Figure 17.
Electrostatic energy of a stacked TA:TA interaction vs. Rise. Including charge penetration reproduces the ab initio SAPT electrostatic energy over the range of rise parameters. The behavior is consistent with that of the benzene dimer interaction (see figure 10).
Figure 18.
Electrostatic energy of a stacked TA:TA interaction vs. Tilt. Including charge penetration reproduces the ab initio SAPT electrostatic energy over the range of tilt parameters. Tilt-like interactions are not part of the S101x7 database, so this behavior shows a level of transferability for the model.
7. Conclusions
The goal of the AMOEBA force field is to model the physics of biomolecular interactions using approximations that make calculations on large systems tractable. Our work here shows that to accurately capture the physics of short-range intermolecular interactions, a charge penetration term is absolutely necessary. Without accounting for charge penetration, even an advanced point multipole model cannot accurately reproduce electrostatic interactions at short range. These discrepancies in intermolecular interactions crucial to biomolecular systems are large enough that they cannot be ignored. Fortunately, we have also shown that charge penetration can be corrected for with the implementation of a simple set of damping functions. This is not necessarily a new conclusion. Previous work on AMOEBA as well other classical force field models have demonstrated the efficacy of using damping functions to capture charge penetration. We have demonstrated here that the higher-order damping functions we have developed for model 1 represent the best, most integrated method for implementing charge penetration in the AMOEBA force field.
There are some key reasons why using model 1 with higher-order damping makes the most sense for AMOEBA. The first reason is the most obvious. On an extensive test set of relevant molecular dimers, model 1 with higher-order damping produced the most accurate results. We have shown that including higher-order damping provides a substantial increase in model accuracy and model 1 performs well at this purpose. The practical purpose of including charge penetration in the force field is to accurately describe intermolecular interactions and by this direct measure model 1 with higher-order damping does the best.
The model does more than simply give good numbers, however. Model 1 is derived from the fundamental physics of atomic charge distributions. The damping function that describes the electrostatic potential around an atom in this model comes directly from the charge distribution of a hydrogen-like atom. The overlap damping function comes directly from an approximation of the overlap integral between two hydrogen-like charge densities. The model does contain empirical parameters, but those parameters are given physical meaning by the derived functions they sit in.
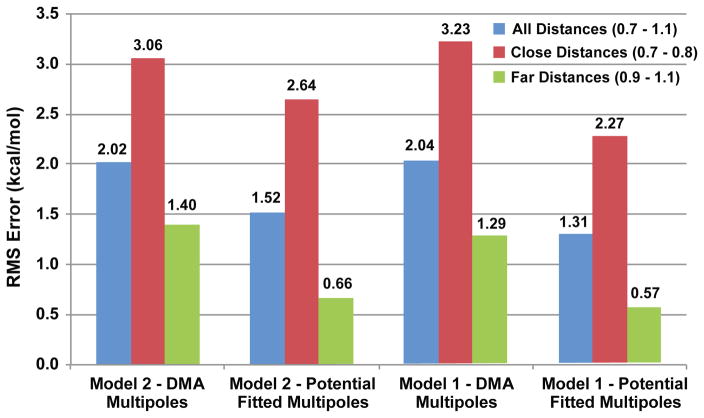
A natural question is why the similar model 2 with one extra parameter does not give better results than model 1. The simple answer is that it appears the two models are intrinsically aligned with different mulitpole models. AMOEBA takes a two step approach to assigning multipole parameters. First distributed multipole analysis (DMA) is performed to obtain initial charge, dipole and quadruople parameters. Then, those parameters are optimized by fitting to the electrostatic potential on a grid of points around the molecule. Because the overlap function in model 1 is constructed starting from a simple one-electron potential, model 1 seems to align nicely with the electrostatic potential fit method for determining AMOEBA multipoles. In contrast it seems that the two-center integral method used by model 2 might perform better with multipoles that are not potential-fitted. This theory is borne out by the results of figure 19. Figure 19 illustrates that model 2 with its extra free parameter, does perform better on the S101x7 database when simple DMA multipoles are used instead of potential fitted ones. Using the AMOEBA potential fitted multipoles however does better overall and much better when paired with model 1. The origin of this difference between models 1 and 2 in instructive. It shows that despite its relative simplicity, model 1 seems to provide a better intrinsic fit for the AMOEBA force field.
Figure 19.
Charge penetration model agreement with AMOEBA potential-fit multipole model. Models 1 and 2 are fit to the S101x7 database using either DMA or potential-fit multipoles. RMS electrostatic energy error is plotted. Model 2 performs slightly better when DMA multipoles are used, but model 1 with potential-fit multpoles gives the best overall fit.
Not only is the model conceptually aligned with the AMOEBA multipole model, but it is fully integrated with it as well. Prior charge penetration models have damped charge-charge interactions or a handful of higher order interactions13, 14, but here we have derived damping functions for multipole interactions up to arbitrary order. This does two important things. First, it improves the overall accuracy of our intermolecular electrostatic energies. And second, it gives us a fully integrated multipole electrostatic–charge penetration model. The charge, dipole, quadrupole moments of a multipole expansion are all functions of the underlying charge density distribution. Thus every interaction of these moments should be damped by the function that describes that charge density. Our higher-order charge penetration model satisfies this requirement and does so in a simple, straightforward way.
Importantly, the charge penetration model doesn’t just fit one set of data. We have demonstrated that it passes multiple validation tests. First, the model proved to be robust. There is no numerical instability and the parameters are not overly sensitive. Second, the model is independent of the multipole model. This means that even if a slightly different set of multipole moments that fit the electrostatic potential are chosen for a given molecule, our charge penetration model will still give the same improvement in the fit. These validation tests indicate not only that our model is viable, but that it is not beholden to the test set or the multipole model. In addition we have shown that our charge penetration model has some measure of predictive power. On the biologically significant test of electrostatics in nucleic acid base stacking, our charge penetration model accurately predicted the electrostatic energies of base stacking over a wide range of non-equilibrium structural parameters. This result displays the promise this model shows in its application to simulations of real biological systems.
Finally, our higher-order charge penetration model captures a real physical effect. The charge penetration phenomenon is a direct result of the fact that atoms have charge distributions representing their electron densities. We have shown that our charge penetration function captures exactly this physics. When we use our model to fit the electrostatic potential on a grid of point surrounding a molecule, the error in the electrostatic fit from the simple point multipole approximation goes down for every tested case. This gives us the highest degree of certainty that we are doing more than just adding in another degree of freedom to our electrostatic function. The damping functions derived for our higher-order damping model accurately describe the electrostatic environment around molecules, and since the effect is necessarily short-range, the computational cost of accounting for charge penetration in this way is minimal. The damping terms can be implemented utilizing a short-range cutoff, or can be computed for every pairwise interaction in the real-space portion of an Ewald summation approach. In either case, the additional cost beyond that of the standard AMOEBA electrostatic model is small. By describing this simple physics in a simple way, our model allows us to more accurately predict intermolecular interactions between biomolecules.
Supplementary Material
Acknowledgments
JWP and PR wish to thank the National Institutes of Health NIGMS for support of AMOEBA development via awards R01 GM106137 and R01 GM114237.
References
- 1.Ren P, Ponder JW. Polarizable Atomic Multipole Water Model for Molecular Mechanics Simulation. J Phys Chem B. 2003;107:5933–47. [Google Scholar]
- 2.Ren P, Wu C, Ponder JW. Polarizable Atomic Multipole-Based Molecular Mechanics for Organic Molecules. J Chem Theory Comput. 2011;7:3143–61. doi: 10.1021/ct200304d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stone AJ, Alderton M. Distributed Multipole Analysis. Mol Phys. 1985;56:1047–64. [Google Scholar]
- 4.Ponder JW, Wu C, Ren P, Pande VS, Chodera JD, Schnieders MJ, Haque I, Mobley DL, Lambrecht DS, DiStasio RA, Head-Gordon M, Clark GNI, Johnson ME, Head-Gordon T. Current Status of the AMOEBA Polarizable Force Field. J Phys Chem B. 2010;114:2549–64. doi: 10.1021/jp910674d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi Y, Xia Z, Zhang J, Best R, Wu C, Ponder JW, Ren P. Polarizable Atomic Multipole-Based AMOEBA Force Field for Proteins. J Chem Theory Comput. 2013;9:4046–63. doi: 10.1021/ct4003702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sinnokrot MO, Sherrill CD. Highly Accurate Coupled Cluster Potential Energy Curves for the Benzene Dimer: Sandwich, T-Shaped, and Parallel-Displaced Configurations. J Phys Chem A. 2004;108:10200–7. [Google Scholar]
- 7.Sherrill CD, Sumpter BG, Sinnokrot MO, Marshall MS, Hohenstein EG, Walker RC, Gould IR. Assessment of Standard Force Field Models Against High-Quality ab Initio Potential Curves for Prototypes of Pi-Pi, CH/Pi, and SH/Pi Interactions. J Comput Chem. 2009;30:2187–93. doi: 10.1002/jcc.21226. [DOI] [PubMed] [Google Scholar]
- 8.Parker TM, Sherrill CD. Assessment of Empirical Models versus High-Accuracy Ab Initio Methods for Nucleobase Stacking: Evaluating the Importance of Charge Penetration. J Chem Theory Comput. 2015;11:4197–204. doi: 10.1021/acs.jctc.5b00588. [DOI] [PubMed] [Google Scholar]
- 9.Piquemal J-P, Cisneros GA, Reinhardt P, Gresh N, Darden TA. Towards a Force Field Based on Density Fitting. J Chem Phys. 2006;124:104101. doi: 10.1063/1.2173256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cisneros GA, Tholander SNI, Parisel O, Darden TA, Elking D, Perera L, Piquemal JP. Simple Formulas for Improved Point-Charge Electrostatics in Classical Force Fields and Hybrid Quantum Mechanical/Molecular Mechanical Embedding. Int J Quantum Chem. 2008;108:1905–12. doi: 10.1002/qua.21675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freitag MA, Gordon MS, Jensen JH, Stevens WJ. Evaluation of Charge Penetration Between Distributed Multipolar Expansions. J Chem Phys. 2000;112:7300–6. [Google Scholar]
- 12.Piquemal J-P, Gresh N, Giessner-Prettre C. Improved Formulas for the Calculation of the Electrostatic Contribution to the Intermolecular Interaction Energy from Multipolar Expansion of the Electronic Distribution. J Phys Chem A. 2003;107:10353–9. doi: 10.1021/jp035748t. [DOI] [PubMed] [Google Scholar]
- 13.Slipchenko LV, Gordon MS. Electrostatic Energy in the Effective Fragment Potential Method: Theory and Application to Benzene Dimer. J Comput Chem. 2007;28:276–91. doi: 10.1002/jcc.20520. [DOI] [PubMed] [Google Scholar]
- 14.Slipchenko LV, Gordon MS. Damping Functions in the Effective Fragment Potential Method. Mol Phys. 2009;107:999–1016. [Google Scholar]
- 15.Spackman MA. The Use of the Promolecular Charge Density to Approximate the Penetration Contribution to Intermolecular Electrostatic Energies. Chem Phys Lett. 2006;418:158–62. [Google Scholar]
- 16.Stone AJ. Electrostatic Damping Functions and the Penetration Energy. J Phys Chem A. 2011;115:7017–27. doi: 10.1021/jp112251z. [DOI] [PubMed] [Google Scholar]
- 17.Tafipolsky M, Engels B. Accurate Intermolecular Potentials with Physically Grounded Electrostatics. J Chem Theory Comput. 2011;7:1791–803. doi: 10.1021/ct200185h. [DOI] [PubMed] [Google Scholar]
- 18.Wang B, Truhlar DG. Including Charge Penetration Effects in Molecular Modeling. J Chem Theory Comput. 2010;6:3330–42. doi: 10.1021/ct1003862. [DOI] [PubMed] [Google Scholar]
- 19.Wang B, Truhlar DG. Partial Atomic Charges and Screened Charge Models of the Electrostatic Potential. J Chem Theory Comput. 2012;8:1989–98. doi: 10.1021/ct2009285. [DOI] [PubMed] [Google Scholar]
- 20.Wang B, Truhlar DG. Screened Electrostatic Interactions in Molecular Mechanics. J Chem Theory Comput. 2014;10:4480–7. doi: 10.1021/ct5005142. [DOI] [PubMed] [Google Scholar]
- 21.Cisneros GA, Piquemal J-P, Darden TA. Generalization of the Gaussian Electrostatic Model: Extension to Arbitrary Angular Momentum, Distributed Multipoles and Speedup with Reciprocal Space Methods. J Chem Phys. 2006;125:184101. doi: 10.1063/1.2363374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duke RE, Starovoytox ON, Piquemal J-P, Cisneros GA. GEM*: A Molecular Electronic Density-Based Force Field for Molecular Dynamics Simulations. J Chem Theory Comput. 2014;10:1361–5. doi: 10.1021/ct500050p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Q, Rackers JA, He C, Qi R, Narth C, Lagardere L, Gresh N, Ponder JW, Piquemal J-P, Ren P. General Model for Treating Short-Range Electrostatic Penetration in a Molecular Mechanics Force Field. J Chem Theory Comput. 2015:2609–18. doi: 10.1021/acs.jctc.5b00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narth C, Lagardere L, Polack E, Gresh N, Wang Q, Bell DR, Rackers JA, Ponder JW, Ren PY, Piquemal J-P. Scalable Improvement of SPME Multipolar Electrostatics in Anisotropic Polarizable Molecular Mechanics Using a General Short-Range Penetration Correction up to Quadrupoles. J Comput Chem. 2016;37:494–506. doi: 10.1002/jcc.24257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stone AJ. The Theory of Intermolecular Forces. New York: Oxford University Press; 1996. [Google Scholar]
- 26.Coulson CA, editor. Mathematical Proceedings of the Cambridge Philosophical Society. Cambridge University Press; 1942. Two-Centre Integrals Occurring in the Theory of Molecular Structure. [Google Scholar]
- 27.Rezac J, Hobza P. Extrapolation and Scaling of the DFT-SAPT Interaction Energies toward the Basis Set Limit. J Chem Theory Comput. 2011;7:685–9. doi: 10.1021/ct200005p. [DOI] [PubMed] [Google Scholar]
- 28.Jeziorski B, Moszynski R, Szalewicz K. Perturbation Theory Approach to Intermolecular Potential Energy Surfaces of van der Waals Complexes. Chem Rev. 1994;94:1887–930. [Google Scholar]
- 29.Hohenstein EG, Sherrill CD. Density Fitting of Intramonomer Correlation Effects in Symmetry-Adapted Perturbation Theory. J Chem Phys. 2010;133:014101. doi: 10.1063/1.3451077. [DOI] [PubMed] [Google Scholar]
- 30.Parker TM, Burns LA, Parrish RM, Ryno AG, Sherrill CD. Levels of Symmetry Adapted Perturbation Theory (SAPT) I Efficiency and Performance for Interaction Energies. J Chem Phys. 2014;140:094106. doi: 10.1063/1.4867135. [DOI] [PubMed] [Google Scholar]
- 31.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. J Am Chem Soc. 1995;117:5179–97. [Google Scholar]
- 32.Wang J, Cieplak P, Kollman PA. How Well Does a Restrained Electrostatic Potential (RESP) Model Perform in Calculating Conformational Energies of Organic and Biological Molecules? J Comput Chem. 2000;21:1049–74. [Google Scholar]
- 33.Foloppe N, MacKerell ADJ. All-Atom Empirical Force Field for Nucleic Acids: I Parameter Optimization Based on Small Molecule and Condensed Phase Macromolecular Target Data. J Comput Chem. 2000;21:86–104. [Google Scholar]
- 34.El Hassan MA, Calladine CR. The Assessment of the Geometry of Dinucleotide Steps in Double-Helical DNA: A New Local Calculation Scheme. J Mol Biol. 1995;251:648–64. doi: 10.1006/jmbi.1995.0462. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.