Abstract
Purpose
To develop and validate a method for the simultaneous measurement of adenosine, guanosine, and inosine derived from mono (MP) and triphosphate (TP) forms in peripheral blood mononuclear cells (PBMCs), red blood cells (RBCs) and dried blood spots (DBS).
Methods
Solid phase extraction of cell lysates followed by dephosphorylation to molar equivalent nucleoside and LC-MS/MS quantification.
Results
The assay was linear for each of the three quantification ranges: 10–2000, 1.0–200 and 0.25–50 pmol/sample for adenosine, guanosine, and inosine, respectively. Intraassay (n=6) and interassay (n=18) precision (%CV) were within 1.7% to 16% while accuracy (%deviation) was within −11.5 % to 14.7 % for all three analytes. Nucleotide monophosphates were less concentrated than triphosphates (except for inosine) and levels in PBMCs were higher than RBCs for all three nucleotides (10, 55, and 5.6 fold for ATP, GTP and ITP, respectively). DBS samples had an average (SD) of −26% (22.6%) lower TP and 184% (173%) higher MP levels compared to paired RBC lysates, suggesting hydrolysis of the TP in DBS.
Conclusion
This method was accurate and precise for physiologically relevant concentrations of adenosine, guanosine and inosine nucleotides in mono- and triphosphate forms, providing a bioanalytical tool for quantitation of nucleotides for clinical studies.
Introduction
The study of purines has been evolving since Scheele discovered uric acid in the renal calculus in 1776 (1). It was a quarter of a century later that an assay to measure uric acid was developed by Garrod who correlated high levels of this byproduct with the occurrence of gout (2). Gout, however, is only one of many diseases caused by complications from the misbalance of purine levels in the body. There are 14 different disorders resulting from inborn errors in purine and pyrimidine metabolism (3, 4). Low expression of adenosine deaminase and purine nucleoside phosphorylase, for example, results in immunodeficiency caused by raised concentrations of deoxyadenosine and deoxyguanosine (5–7). Recent studies have shown that adenosine and guanosine play important roles in the protection of the nervous system (8) and may also be involved in the regulation of cortisol or other hormones (9). Anti-viral, anti-cancer and immunosuppressive therapies are commonly based on analogs of endogenous nucleobases. Because of this, many of the associated toxicities (i.e. anemia, weakened immune system) and efficacies (i.e. inhibition of cancerous and/or viral DNA) of these drugs occur by competing with, and in some cases altering, endogenous nucleotide pools in the body (10, 11).
In order to measure endogenous nucleotides in vivo, it is necessary to develop sophisticated techniques for quantification of these bases in different cellular matrices. Several methods were previously developed to specifically measure adenosine and guanosine related nucleotides in red blood cells (RBCs) with HPLC coupled to ultraviolet-visible and/or diode array detection (10, 12–17). The range of concentrations detected specifically for ATP and GTP was similar for all of these methods (~114 to 213 and ~3.3 to 8.6 pmol/106 cells (10, 13, 16, 17)). Methods that assayed other cell types, like peripheral blood mononuclear cells (PBMCs), found adenosine and guanosine nucleotide concentrations that were roughly 10 fold higher than RBCs (10, 13). Newer methods utilize mass spectrometry as a more selective and sensitive detection method. Quantification has been performed in multiple matrices and for multiple nucleotides in these methods, (18–21) however, most utilize direct analysis techniques where phosphate fractions are separated on an HPLC column using ion pairing based mobile phases. This separation method may cause ion suppression from the mobile phase and also affects the column chromatography for measuring other molecules with HPLC. The indirect method described in this work is advantageous for preventing ion suppression caused by ion pairing agents and allows the detection of monophosphate (MP), diphosphate (DP) and triphosphate (TP) fractions in the free base form. This is valuable because one calibration curve can be utilized and does not need to be made with the phosphorylated moieties for the measurement of clinical samples. Additionally, the chromatography is simplified and use of non-phosphorylated samples allows more accurate mass spectrometry detection since the analytes do not carry extra negative charge.
A sensitive method was developed and validated to measure MP and TP fractions of adenosine, guanosine and inosine in human cells. This is the first method to measure MP and TP forms of adenosine, guanosine and inosine simultaneously in several cell types. In particular, this method was used for PBMCs, RBCs and explores the possibility of dried blood spot (DBS) measurement as a more clinically affordable and simple sample type for future analysis. DBS measurement is useful in a clinical setting because of its ease and affordability and has been used recently for the quantification of adenosine and 2′deoxyadenosine for the purpose of identifying adenosine deaminase deficiency in infants (22). As such, it is important to determine the feasibility of DBS as a matrix for the accurate measurement of endogenous nucleotides. Stability of these nucleotides at varying conditions as well as specificity and selectivity was assessed in addition to the accuracy and precision of this method.
Materials and methods
2.1 Chemicals and supplies
Adenosine, Adenosine Monophosphate (AMP), Adenosine triphosphate (ATP), Guanosine, Guanosine monophosphate (GMP), guanosine triphosphate (GTP), Inosine, Inosine monophosphate (IMP), and Inosine triphosphate (ITP) were purchased from Sigma Aldrich, St. Louis, MO; isotopic internal standards for each, (Ribose 13C5) were from Cambridge Isotope Laboratories, Andover, MA.
Analytical grade reagents were purchased from Fisher Scientific, Fairlawn, NJ, (acetonitrile, methanol, formic acid, potassium chloride, phosphoric acid, and ammonium hydroxide) as well as Whatman® 903 DBS cards, bags and desiccant for DBS preparation and storage. Ammonium acetate 5M solution was purchased from Ambion® and alkaline phosphatase was purchased from Sigma Aldrich, St. Louis. Ultrapure (UP) water was prepared in house from deionized water with a Barnstead Nanopure System (Thermo Fisher Scientific, Waltham, MA). Other supplies included Waters Sep-Pak Accell Plus QMA Cartridge, 3cc (500mg) (Waters Corporation, Milford, MA) and Varian Bond-Elut LRC Phenylboronic Acid (PBA) Cartridge 100mg/10mL (Agilent, Santa Clara, CA). Blood products for lysed cellular matrix were obtained from Bonfils, Denver CO. Cell preparation tubes (CPT) and dipotassium EDTA tubes were purchased from Becton, Dickinson and Company, Franklin Lakes, NJ.
2.2 Sample collection and matrix preparation
Clinical samples (PBMCs, RBCs and DBS) were obtained from 36 hepatitis C-infected individuals participating in an IRB-approved study of ribavirin (RBV) pharmacokinetics (23). All patients provided written informed consent. Whole blood was drawn into EDTA or cell preparation tubes prior to centrifugation and separation of the different cell types. Cells (RBCs and PBMCs) were counted using the Countess™ automated cell counter (Thermo Fisher Scientific). DBS were spotted on Whatman® 903 cards (followed by 2 hour drying prior to storage) from EDTA whole blood before separation of RBCs and PBMCs. Cells were lysed and DBS extracted in 70:30 methanol:water and stored at -70°C prior to analysis.
2.3 Sample extraction
For extraction, 70:30 lysates are added to a QMA anion exchange cartridge to separate MP (5 mL of 75mM KCl), DP (7 mL of 90mM KCl) and TP (2 mL of 1M KCl) fractions using a potassium chloride salt gradient. The resulting separated fractions were then dephosphorylated to molar equivalent parent adenosine, guanosine and inosine with excess alkaline phosphatase for 10 minutes at 37°C followed by addition of internal standard (20 μL), de-salting and concentration with PBA solid phase extraction. The latter consisted of several steps. First, the PBA cartridge was activated according to product specifications. Second, the QMA fractions were added to the cartridge followed by 100xg centrifugation. Washes consisted of 2 × 1 mL of 250 mM ammonium acetate followed by 2 × 1 mL of methanol. Analytes were then eluted with 20% formic acid in methanol solution, dried down and reconstituted in UP-water followed by injection on the LC-MS/MS system. Additional details on the sample preparation and extraction can be found in previous publications (24, 25).
2.4 Instrumentation
MS-MS detection of the analytes and their respective internal standards was accomplished on a Thermo Scientific TSQ Vantage® triple quadrupole mass spectrometer coupled with a Thermo Scientific Accela 1250® pump and CTC Analytics HTC PAL® autosampler. The mobile phase was aqueous with 4% acetonitrile and 0.1% formic acid pumped at a flow rate of 200 μL/min. Chromatographic separation was accomplished with Develosil C30 Reversed-Phase-Aqueous, 140Å, 150–2.0mm, 3μm particle size column purchased from Phenomenex (Torrance, CA). Adenosine, guanosine and inosine analysis was operated in ESI + and the analytes SRM [precursor/product] + transitions (m/z) were detected as described in Table I. The adenosine+1 m/z transition was used for detection of adenosine to avoid signal saturation at the upper range of quantification.
Table I.
Parent and product m/z with instrument settings in ESI+ mode. Analytea
| Parent | Product | Scan width | Time | Collision energy | Q1 | Q3 | S-Lens | |
|---|---|---|---|---|---|---|---|---|
| A+1 | 269.3 | 137.2 | 0.002 | 0.05 | 13 | 0.7 | 0.7 | 74 |
| A+1-IS | 274.3 | 137.2 | 0.002 | 0.05 | 13 | 0.7 | 0.7 | 73 |
| G | 284.3 | 152.2 | 0.002 | 0.05 | 13 | 0.7 | 0.7 | 47 |
| G-IS | 289.3 | 157.2 | 0.002 | 0.05 | 13 | 0.7 | 0.7 | 47 |
| I | 269.2 | 137 | 0.002 | 0.05 | 13 | 0.7 | 0.7 | 47 |
| I-IS | 273.2 | 141.0 | 0.002 | 0.05 | 13 | 0.7 | 0.7 | 47 |
A=adenosine, G=guanosine and I=inosine.
2.5 Standard, quality control and internal standard (IS) preparation
Triphosphate stocks from reference powder were prepared in UP-water to approximately 1 mg/mL for ATP, GTP and ITP. Prior to dilution and combination into a working stock for preparing standards (STDs) and quality controls (QCs), purity and potency was determined for each analyte according to a previously presented method (26). The correction factor and resulting pmol/μL concentration of each preparation stock were 0.83 and 1900 pmol/μL for ATP, 0.91 and 1459 pmol/μL for GTP and 0.88 and 840 pmol/μL for ITP. The second preparation stock (PS2), made in 70:30 methanol:water, combined all three analytes at respective concentrations of 100, 10 and 2.5 pmol/μL for ATP, GTP and ITP. PS2 was diluted in 70:30 to make standard solutions at eight levels for each analyte. Methanol:water (70:30) was used because nucleotides are present in blank lysed cells which would cause inaccurate concentrations. The range was 10.0–2000 pmol/sample for ATP, 1.0–200 pmol/sample for GTP and 0.25–50.0 pmol/sample for ITP where sample=200 μL of solution. These ranges were chosen to accommodate a varying number of cells/sample and because of the concentration differences for each of the analytes (i.e. ATP>GTP>ITP concentration for all cell types).
QCs were also prepared from PS2 at four levels: lower limit of quantitation: LLOQ, low: QL, medium: QM and high: QH. For each analyte, the concentration for QL is three times the LLOQ, while QH is 80% of the upper limit of quantification. Specific concentrations of QCs for ATP, GTP and ITP are detailed in Table II. Isotopic, internal standard (IS) was prepared at a combined concentration of 1.25 pmol/μL from 1mg/mL stocks and stored at 4°C.
Table II.
Inter-and Intraassay accuracy and precison for quantification of ATP, GTP and ITP. Top portion of table shows back-calculated standard statistics for each analyte. Bottom half of table shows the QC response for each analyte at all four QC levels for the three accuracy and precision analytical runs.
| Back Calculated Calibration Standards | ATP | GTP | ITP |
|---|---|---|---|
| Calibration Standard Range | 10.0–2000 pmol/spl | 1.0–200 pmol/spl | 0.25–50.0 pmol/spl |
| Interassay Accuracy (% Dev ) | −2.8 to 4.0 | −4.4 to 5.4 | −1.8 to 8.4 |
| Interassay Precision (%CV ) | 0.5 to 4.2 | 0.3 to 4.2 | 0.6 to 9.4 |
| Mean Ratio of Blank IS to Std H | 0.016 | 0.107 | 0.273a |
| Coefficient of Determination (r2) Mean | 0.9995 | 0.9998 | 0.9995 |
|
| |||
| Quality Control Accuracy and Precision | ATP | GTP | ITP |
| Quality Control Level 1 (LLOQ) | 10.0 pmol/spl | 1.0 pmol/spl | 0.25 pmol/spl |
| Interassay Accuracy (%Dev) (n=18) | 5.9 | 10.4 | 13.2 |
| Interassay Precision (%CV) (n=18) | 4.7 | 7.9 | 15.0 |
| Intraassay Accuracy (%Dev) (n=6) | 0.2 to 7.5 | 2.8 to 14.7 | 3.5 to 13.2 |
| Intraassay Precision (%CV) (n=6) | 3.9 to 7.1 | 3.4 to 16.0 | 5.8 to 13.9 |
| Quality Control (Low) | 30.0 pmol/spl | 3.0 pmol/spl | 0.75 pmol/spl |
| Interassay Accuracy (%Dev) (n=18) | −1.5 | 0.5 | −8.6 |
| Interassay Precision (%CV) (n=18) | 3.2 | 8.6 | 9.5 |
| Intraassay Accuracy (%Dev) (n=6) | −3.2 to 0.5 | −3.6 to 3.6 | −11.5 to −2.9 |
| Intraassay Precision (%CV) (n=6) | 2.2 to 3.0 | 1.8 to 13.7 | 4.3 to 14.7 |
| Quality Control (Medium) | 160.0 pmol/spl | 16.0 pmol/spl | 4.0 pmol/spl |
| Interassay Accuracy (%Dev) (n=18) | −0.4 | −0.9 | 0.7 |
| Interassay Precision (%CV) (n=18) | 2.0 | 2.6 | 3.4 |
| Intraassay Accuracy (%Dev) (n=6) | −1.4 to 0.8 | −2.9 to 1.4 | −0.9 to 2.4 |
| Intraassay Precision (%CV) (n=6) | 1.8 to 2.1 | 1.2 to 2.3 | 2.8 to 3.8 |
| Quality Control (High) | 1600 pmol/spl | 160.0 pmol/spl | 40.0 pmol/spl |
| Interassay Accuracy (%Dev) (n=18) | 1.0 | −1.3 | −1.5 |
| Interassay Precision (%CV) (n=18) | 2.5 | 2.1 | 1.7 |
| Intraassay Accuracy (%Dev) (n=6) | −0.4 to 3.2 | −2.2 to 0.6 | −2.0 to −1.0 |
| Intraassay Precision (%CV) (n=6) | 0.9 to 3.3 | 1.3 to 2.1 | 1.1 to 2.4 |
Highlight: The mean ratio of Blk IS to lowest STD for ITP was consistently above acceptance and so the next calibration level (Std G, 0.625 pmol/10^6 cells) was considered the true LLOQ for unknown samples.
2.6 Determination of Accuracy and Precision
Accuracy was determined by comparing the mean response from each validation sample (n=6 per QC level, per analyte) to the theoretical concentration for each level and is expressed as % deviation (%dev). Precision was calculated using the coefficient of variance (%CV) for the replicate analysis. Both accuracy and precision were determined for three separate analytical runs and within run (intraassay, n=6) and between run (interassay, n=18) calculations were assessed. Passing criteria were ±15% of nominal concentration for QL, QM and QH and ±20% for LLOQ for both accuracy and precision assessments.
2.7 Matrix Effect, Recovery and Process Efficiency
Matrix effect (ME), recovery and process efficiency (PE) were assessed for TP samples only. Since ATP, GTP and ITP constitute the matrix itself, it was necessary to utilize stable labeled IS as the “analyte” to determine these parameters. Five different lots of blank RBC were subjected to QMA extraction for each of the spiked (set 2) and extracted (set 3) samples for low (5.0 pmol/sample) and high (1600 pmol/sample) concentrations. Spiked samples had IS solution added after final elution from the PBA step while extracted samples had IS added prior to PBA extraction. Neat samples (set 1) were prepared by adding 20 μL of the respective IS concentration into 80 μL of UP-water followed by injection onto the MS system. Peak area responses were used to determine each parameter. ME was calculated by comparing mean peak area response of set 2 to set 1 (set 2/set 1 × 100), recovery (from the PBA extraction) was determined by comparing mean peak area response of set 3 to set 2 (set 3/set 2 × 100), and PE was determined by comparing mean peak area response of set 3 to set 1 (set 3/set 1 × 100).
2.8 Conditional Stability assessment
Several conditional stability experiments were performed: freeze/thaw, room temperature and extracted sample stability. Accuracy and precision were determined for triplicate treated and control samples and acceptance criteria were ±15% for both accuracy and precision determinations for all stability experiments.
Freeze/Thaw stability in lysed intracellular matrix was assessed by allowing QL and QH QC samples to undergo three freeze/thaw cycles from −70°C. Thawing occurred for up to two hours prior to extraction for each cycle. Samples were run in triplicate, and the mean response was compared to the nominal concentration for QL and QH and also compared to fresh triplicate QL and QH controls that did not undergo treatment conditions.
Room temperature (RT) stability in 70% methanol was assessed by removing QL and QH samples from −70°C and allowing a thaw time of 60 and 144 hours at room temperature. The QL and QH were analyzed in triplicate and mean response was compared to both nominal and mean response from freshly prepared QL and QH that were not subjected to the test condition.
Extracted sample stability was assessed by retaining six replicate QL and QH samples at RT after extraction and analysis for re-injection 144 hours later. Mean response of the reinjected samples was compared to both nominal concentration and mean response from their initial injection.
3 Validation Results
3.1 Accuracy and precision
The STD concentrations were back calculated using a weighted (1/concentration) quadratic regression curve. The experimental mean of the three runs was compared against the theoretical concentration for accuracy and was within ±8.4% for all STDs passing acceptance criteria. Overall, the %CV for each validation run was ≤9.4% and the average r2 values were ≥0.9995 for all three analytes. A summary for individual interassay accuracy and precision ranges for each analyte is shown in Table II. It should be noted that a contamination peak was present in the blanks that interfered with quantification of ITP. The lowest ITP STD (0.25 pmol/sample) for each of the validation runs was therefore invalid indicating that the next lowest STD (0.625 pmol/sample) is the method LLOQ. Because of this, the reported accuracy and precision values of the calibration curve do not include the ITP STD at 0.25 pmol/sample, though the curve remained linear when it was included.
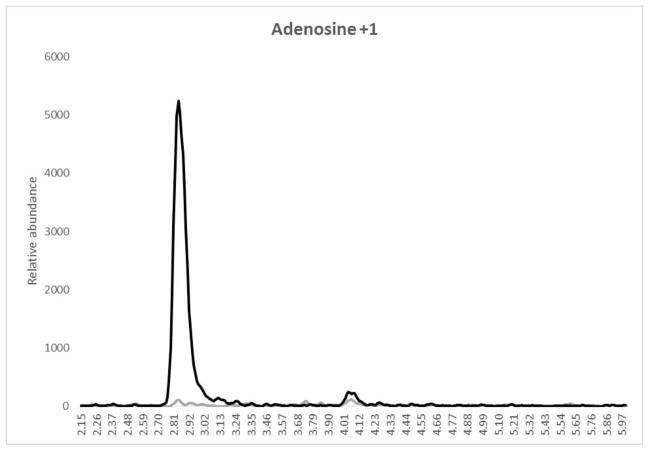
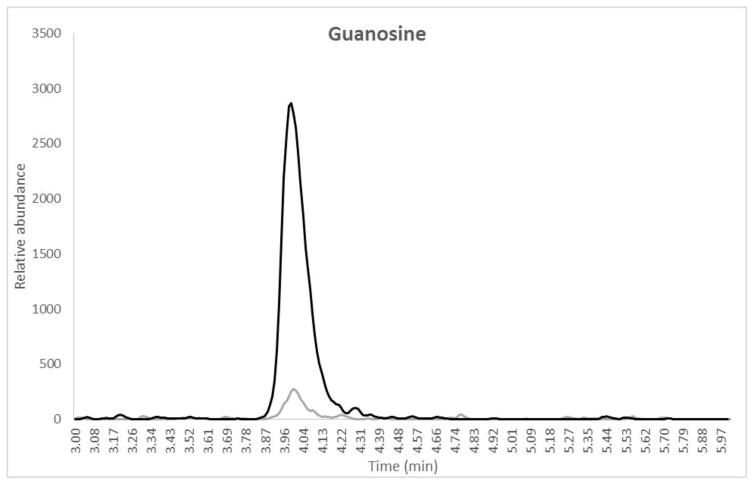
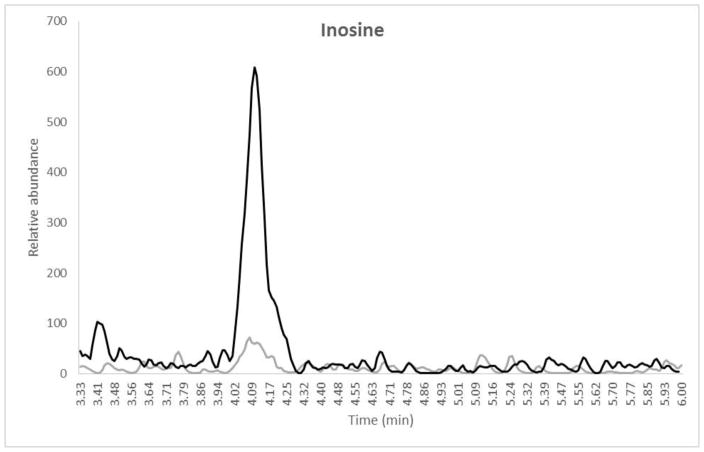
Mean intraassay and interssay data for ATP, GTP and ITP are presented in Table II. Intraassay accuracy for the QCs was −3.2% to 7.5%, −3.6% to 14.7% and −11.5% to 13.2% for ATP, GTP and ITP, respectively. Intraassay precision for all QC levels including the LLOQ was 0.9% to 7.1%, 1.2% to 16.0%, and 1.1% to 14.7% for ATP, GTP and ITP, respectively. Overall interassay precision for all analytes and QC levels was ≤15.0% and interassay accuracy was between −8.6 and 13.2%. The second validation run had 2/6 LLOQ fail high for GTP with one testing as a statistically significant outlier according to Grubb’s outlier test (GraphPad QuickCalcs). These failures were most likely due to sample contamination from environmental factors as is described in the discussion section and so the outlier was not included in the accuracy and precision calculations. The average blank-IS response compared to the lowest STD (representative of the LLOQ for the extraction) are also reported in Table II. A typical chromatogram at the LLOQ level with a blank-IS sample overlay for each analyte is shown in Figure 1 (a, b and c).
Figure 1.
Overlay chromatograms with blank IS (gray) and LLOQ (black) for (a) Adenosine, retention time 3.12 min, (b) Guanosine, retention time 4.05 min and (c) Inosine, retention time 4.16 min. X-axis represents the retention time in minutes and the Y-axis is the relative abundance normalized to the LLOQ response. The total run time for the method was 6.0 minutes. Note that the sample was dephosphorylated from the TP fraction prior to analysis.
3.2 Matrix Effect (ME), Recovery, Process Efficiency (PE)
Adenosine, guanosine and inosine IS were prepared at high (1600 pmol/sample) and low (5.0 pmol/sample) concentrations in lysed cellular matrix that underwent the QMA isolation process. Compared to guanosine and inosine, adenosine IS had a larger suppression of signal due to matrix with a 78%/75% response for low/high concentrations. Guanosine and inosine IS had similar ME to each other at low/high of 88%/84% and 88%/87%, respectively. These data suggest a minimal suppressive effect of the matrix on the stable labeled IS version of the analytes. When comparing the slopes of each lot (peak area over expected concentration) for the low and high sample, the %difference was −10.7 to 9.7% for all analytes. Recovery for stable labeled adenosine, guanosine and inosine for low/high IS levels was 73%/76%, 78%/74% and 66%/70%, respectively. This experiment shows that recoveries were similar between analytes and consistent between the low/high concentration levels. The mean PE for each analyte at low/high levels was 57%/57%, 69%/63%, and 58%/60% for adenosine, guanosine and inosine stable labeled IS, respectively. In general, the method provides process efficiencies that are all above 50% and have overall consistent PE between concentration levels.
3.3.1 Conditional Stability: Freeze/Thaw
All analytes were stable throughout the freeze/thaw test conditions. The %CV for all treated samples was ≤8.8% for QL and ≤0.8% for QH for all analytes. The %dev from control and nominal were within ±9.3% for QL and ±3.0% for QH showing all data are well within acceptance criteria for stability.
3.3.2 Conditional Stability: Room Temperature (RT)
ATP, GTP and ITP were stable in 70% methanol for 144 hours for QH samples. The %CV was ≤5.1 for QL and ≤4.4 for QH while the % difference compared to untreated and nominal were within ±20.6 (GTP failed low compared to control while others remained within acceptance criteria) for QL and ±3.7 for QH. This suggested that 144 hours may be too long at RT for lower concentrations, specifically for GTP. Because of this, another experiment was performed with the treatment at RT being only 60 hours. This experiment gave a %dev at −16.0 for GTP at the QL level compared to control, which is still not within the acceptable criteria. These data suggest that samples should not remain at RT for longer than 2 hours (based on the freeze/thaw experiments) and should be analyzed as soon as possible after removing from storage.
3.3.3 Conditional Stability: Extracted Stability
Extracted QL and QH (n=6) samples (parent nucleosides re-constituted in UP-water after final PBA concentration) were left at RT for 144 hours after analysis followed by re-injection and comparison to the initial response as well as nominal values. The %CV for all treated samples was ≤2.8% for QL and ≤2.4% for QH. The %dev compared to initial and nominal values was within ±4.0% for QL and ±2.1% for QH showing that samples are stable for up to six days post extraction. Although samples should not remain in 70:30 for longer than two hours at room temperature prior to extraction (section 3.3.2), the UP-water that samples are retained in prior to injection onto the LC-MS/MS is a different matrix and therefore may not have the same effect on stability. All conditional stability results are shown in Table III for each individual analyte.
Table III.
Conditional stability results for 3 freeze/thaw cycles, time at room temperature (RT) and time post sample extraction. Top row numbers are precision values while the second row is the %difference compared to control samples.
| Analyte | Condition | QL | QH |
|---|---|---|---|
| ATP | Freeze/thaw (3×) | 2.3 | 1.8 |
| −9.2 | −3.0 | ||
| RT 6 days | 2.6 | 4.4 | |
| −12.8 | −3.7 | ||
| RT 2.5 days | 1.4 | 4.4 | |
| −13.6 | −3.7 | ||
| Extracted 6 days | 1.6 | 1.1 | |
| 0.11 | 0.01 | ||
|
| |||
| GTP | Freeze/thaw (3×) | 1.3 | 0.80 |
| −9.3 | −1.1 | ||
| RT 6 days | 2.4 | 3.7 | |
| −20.6 | −0.35 | ||
| RT 2.5 days | 2.3 | 3.7 | |
| −16 | −0.35 | ||
| Extracted 6 days | 1.7 | 0.80 | |
| 0.7 | −2.1 | ||
|
| |||
| ITP | Freeze/thaw (3×) | 6.7 | 1.8 |
| −7.6 | −1.9 | ||
| RT 6 days | 5.1 | 3.3 | |
| −9.9 | −0.76 | ||
| RT 2.5 days | 15.3 | 3.3 | |
| −10.4 | −0.76 | ||
| Extracted 6 days | 2.8 | 2.4 | |
| −4.0 | −1.5 | ||
3.4 Carryover and Crosstalk Evaluation
Adenosine, guanosine and inosine cross talk evaluation showed no response from the respective IS when STD A was injected without IS present. Additionally, no significant cross talk response in the parent window was evident after the IS-only sample was injected. Carryover was assessed by injecting a blank water sample after STD A. There was no significant detection of adenosine, guanosine or inosine from this water injection signifying no significant carryover effect.
4 Application to Multiple Matrices
4.1 Monophosphate vs Triphosphates
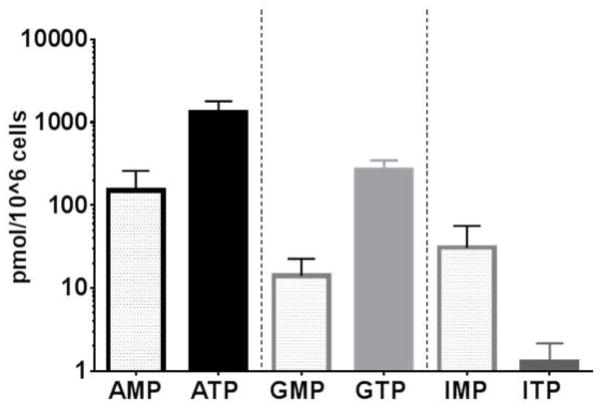
In addition to analyzing the TP fraction (2 mL) obtained from the QMA procedure, it is also possible to quantify the MP fraction (5 mL) as well as the DP fraction (7 mL). This is simple for this particular assay because the Bond-Elut PBA cartridge can hold up to 10 mL of liquid and use of stable labeled IS should account for any difference in recovery of these samples compared to the TP fraction. Because the DP fraction is fleeting in the phosphorylation process (i.e. not the rate limiting step for equilibration to the TP form) and is therefore physiologically less relevant, this work focused on measurement of the MP and TP fractions only. To compare MP vs TP in vivo, 36 patient samples collected at day 1 of RBV treatment (2 hours post dose) were used to quantify paired MP and TP from PBMC fractions. Figure 2 shows the difference in levels of MP and TP for adenosine, guanosine and inosine, respectively. PBMC mean (SD) MP fractions were 150 (108), 14.1 (8.5) and 30.6 (25.9) while TP fractions were 1303 (495), 264 (83) and 1.5 (0.10) pmol/106 cells for adenosine, guanosine and inosine, respectively. For both ATP and GTP these levels were comparable to literature values (10, 13), but AMP and GMP levels were significantly higher (10 to 25 fold) which may be due either to the differing sensitivity of the methods (UV detection vs MS/MS) or a difference in the processing or extracting of cells.
Figure 2.
Mean (SD) concentrations of MP and TP for adenosine, guanosine and inosine at day 1 of RBV treatment. The Y-axis is the back transformed value of each analyte in pmol/106 cells. Black=Adenosine nucleotide, light gray=Guanosine nucleotide and dark gray=Inosine nucleotide. Clear bars are MP and solid bars are the TP fractions measured.
4.2 RBC, PBMC and DBS
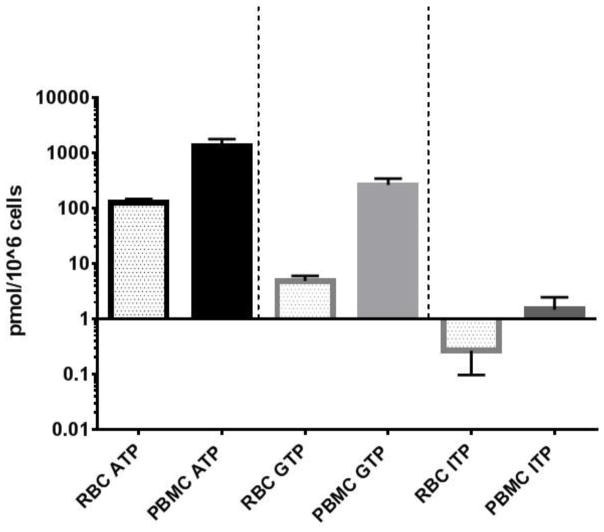
Some of the most common sample types collected for pharmacokinetic studies include RBCs, PBMCs, and more recently, DBS. To assess differences in these matrices for quantification of ATP, GTP and ITP, the same 36 patient samples were utilized (Figure 3). The mean (SD) RBC concentrations were 128 (20.6), 4.8 (1.2) and 0.27 (0.17) compared to PBMCs corresponding to 10, 55, and 5.6 fold lower concentrations for ATP, GTP and ITP, respectively. Although the levels differ in these two matrices, the assay range allows simultaneous quantification of all analytes in both cell types.
Figure 3.
Comparison of mean (SD) adenosine, guanosine and inosine levels in RBCs and PBMCs at day 1 of RBV treatment. The Y-axis is back transformed and denotes analytes in pmol/106 cells. Black=ATP, light gray=GTP and dark gray=ITP with clear bars representing RBCs and solid bars representing PBMCs.
For DBS, paired analysis of RBC lysate and DBS samples were analyzed in five subjects at six different treatment time points at either day 1 or steady state. Both the DBS and RBC lysates were stored at −70°C for 815–929 days prior to this analysis. The pmol/106 cells results for MP and TP for both matrices are reported in Table IV (a, b). DBS pmol/106 cells values were calculated per individual sample by the equation described in Jimmerson et al. and Zheng et al. (25, 27). Briefly, the pmol/punch result is divided by the paired pmol/106 cells result from RBC lysate to yield the approximate (within 20%) amount of cells in a DBS punch (25). Average (SD) TP DBS samples for all analytes were −26% (22.6) lower in response compared to RBC lysates, while MP were 184% (173) higher compared to RBC. This result was unexpected considering nucleoside analogs have been reported to have long term stability in DBS in −70°C storage conditions (28, 29). There is likely a stability issue with endogenous nucleotides in DBS samples during the drying and storing process that needs to be evaluated if this matrix is to be useful for accurate quantification of endogenous nucleotides.
Table IV.
Difference in TP and MP concentrations in paired RBC vs. DBS lysate. (a) Triphosphate results in pmol/106 cells. (b) Monophosphate results in pmol/106 cells.
| (a)
| |||||||||
|---|---|---|---|---|---|---|---|---|---|
| PID | ATP | GTP | ITP | ||||||
|
| |||||||||
| DBS pmol/10^6 cells | RBC pmol/10^6 cells | % difference | DBS pmol/10^6 cells | RBC pmol/10^6 cells | % difference | DBS pmol/10^6 cells | RBC pmol/10^6 cells | % difference | |
| 2629 | 51.1 | 102 | −50.0 | 2.8 | 4.5 | −38.1 | 0.036 | 0.06 | −42.8 |
| 2635 | 47.8 | 124 | −61.3 | 2.1 | 4.5 | −53.9 | 0.031 | 0.08 | −62.9 |
| 2635 | 76.4 | 116 | −33.9 | 3.4 | 4.1 | −17.9 | 0.059 | 0.09 | −36.7 |
| 2632 | 74.2 | 85.8 | −13.5 | 2.7 | 2.7 | 0.3 | 0.049 | 0.07 | −29.3 |
| 2640 | 81.8 | 93.4 | −12.3 | 3.2 | 2.9 | 11.7 | 0.058 | 0.06 | −2.7 |
| 2634 | 62.7 | 81.7 | −23.3 | 3.6 | 4.0 | −11.4 | 0.050 | 0.05 | 2.1 |
| (b)
| |||||||||
|---|---|---|---|---|---|---|---|---|---|
| PID | AMP | GMP | IMP | ||||||
|
| |||||||||
| DBS pmol/10^6 cells | RBC pmol/10^6 cells | % difference | DBS pmol/10^6 cells | RBC pmol/10^6 cells | % difference | DBS pmol/10^6 cells | RBC pmol/10^6 cells | % difference | |
| 2629 | 3.7 | 3.8 | −2.1 | 0.27 | 0.23 | 15.7 | 0.89 | 0.78 | 13.8 |
| 2635 | 2.4 | 0.92 | 164 | 0.16 | 0.10 | 60.7 | 0.37 | 0.24 | 53.7 |
| 2635 | 4.8 | 0.99 | 388 | 0.31 | 0.10 | 215 | 0.87 | 0.22 | 289 |
| 2632 | 2.8 | 1.6 | 79 | 0.20 | 0.14 | 49.6 | 0.46 | 0.23 | 102 |
| 2640 | 5.5 | 0.77 | 616 | 0.30 | 0.09 | 250 | 1.4 | 0.24 | 497 |
| 2634 | 3.4 | 1.0 | 227 | 0.27 | 0.11 | 154 | 0.86 | 0.37 | 133 |
Discussion
An assay for the measurement of TP and MP fractions of adenosine, guanosine and inosine was developed and validated. The method utilizes cellular lysate from different cell types, which increases applicability in clinical studies. The different levels seen in PBMCs vs RBCs are to be expected since PBMCs are nucleated cells and have enzymes that conduct de-novo nucleotide synthesis whereas RBCs only use salvage pathways. Also, RBC do not produce DNA or RNA but use nucleotides as an energy source for the cell itself. As such, there is less need for high concentrations of these molecules and so the quantities are lower compared to PBMCs. Additionally, the MP is the rate limiting step to become the TP, and higher TP is favored when nucleotide pools are at equilibrium (23, 30). The exception to this would be IMP in PBMCs since it is an important molecule for the production of other nucleotides via the de novo synthesis pathway in nucleated cells.
It is important to note that these nucleotides are found primarily in cells because plasma contains phosphatases that convert nucleotides back into the parent form. Additionally, nucleotides are typically dephosphorylated prior to being able to exit the cell and so most extra-cellular purines are in the parent form. This assay eliminated any possible contamination from plasma concentrations of these parent purines because the initial strong ion exchange process does not allow parent forms to interact with the column packing. Therefore, even if there were some plasma left in intracellular samples, particularly for DBS where plasma is a component of the matrix, this would not contribute to the final analysis since it would be washed out prior to desalting/concentration and LC-MS/MS injection.
The application of this method to the DBS matrix provided useful information on the stability of these nucleotides. As mentioned earlier, DBS has been used for the measurement of many endogenous biomarkers including nucleotides, but stability of this matrix has not been thoroughly evaluated (22, 31, 32). Other studies that have measured nucleotide pools in RBC samples have reported that immediate processing is a necessity for accurate results (17, 33). Both the RBCs and DBS samples reported here were stored at −70°C for 815–929 days prior to this analysis, but DBS appeared to have a breakdown of TP to the MP compared to RBCs. One contribution to this instability may be that blood is not immediately dried on the paper prior to storage and remains at RT to dry for up to four hours prior to storage. Therefore, it may be possible that enzymes are continuing to act in cells during the drying period where the enzymes are stopped immediately in RBCs that are lysed directly upon processing.
There are some limitations that should be considered for this method. First, it is important to note that ATP, GTP and ITP are endogenous substances that may be found in the general environment in bacteria, dust, or hair and skin. Because of this, the environment must be more thoroughly regulated for this assay. Several steps were taken to address this issue. 1) Glassware was washed thoroughly with UP-water prior to making solutions or prep stocks 2) Solutions of KCl and 250mM ammonium acetate were prepared fresh within one week for all validation runs while enzyme solution was mixed immediately prior to each extraction 3) Enzyme diluent was prepared and frozen in aliquots prior to use 4) All pipets and secondary storage containers for buffers were sterile, disposable and new for each extraction to avoid any further sources for contamination. Additionally, a blank and blank plus IS sample were extracted through both QMA and PBA procedures in order to determine baseline contamination resulting from the assay process itself. The LLOQ was adjusted to the next calibration level according to whether the blank-IS response was >20% of the LLOQ response. This resulted in some analytes (specifically IMP/ITP) having a calibration curve with seven levels instead of eight as explained in the results.
This assay is sensitive, but the issue of potential contamination at the lower end of quantification may not allow consistent results at the LLOQ level of GMP/GTP or IMP/ITP. Specifically, for IMP/ITP the method LLOQ level was determined to be 0.625 pmol/106 cells based on the low signal of this analyte resulting in contamination issues at the LLOQ (0.25 pmol/106 cells) level. Because ITP concentrations are relatively low in RBCs, the issue of inconsistent quantification at the LLOQ could be more pronounced. More RBCs could be analyzed to increase sensitivity, but results would then be above the limit of quantification for ATP and GTP when assaying more than 10×106 cells. However, for all validation runs the adjusted LLOQ was still relatively sensitive at 0.625 pmol/sample; a lower amount than other methods published for the measurement of inosine or its IMP metabolite (17, 34). Additionally, because ITP is not incorporated into RNA or DNA, previous methods have not, to our knowledge, attempted to quantify this nucleotide. Nevertheless, it is an important anabolite for biological and pharmacological studies such as RBV-induced anemia (35). The lowest STD passed for all GMP/GTP validation runs, but there was some potential contamination noted in the second validation run with the LLOQ samples. However, the LLOQ level did not appear to be a physiologically relevant concentration. All clinical samples analyzed for the guanosine component were well-above the LLOQ of 1.0 pmol/sample in both RBCs (range 5.96 to 44.6 pmol/sample) and PBMCs (range 2.16 to 101 pmol/sample).
The experiments for understanding the effect of the matrix on the isotopically labeled versions of these analytes were important for demonstrating the reproducibility of the method. However, ME is less relevant for this method because this assay utilized stable-labeled IS for quantification. Therefore, the matrix would equally affect the analyte and the IS and would be accounted for by using the ratio (analyte/IS) for calculation of concentrations.
Conclusion
An accurate and precise method was developed to measure the TP and MP moieties of adenosine, guanosine and inosine in RBCs and PBMCs at physiologically relevant concentrations of all analytes. The method was selective and specific and was reasonably stable in regards to freeze/thaw cycles and extracted stability. It is of note that samples should not remain at room temperature for longer than 2 hours prior to extraction, that environmental contamination must be taken into account for accurate results and that DBS needs further stability evaluation for use in measuring these nucleotides.
Acknowledgments
This work was supported by R03 DK096121 (JJK), the International Maternal Pediatric Adolescent AIDS Clinical Trials Network (UM1AI068632 to JJK), and the Colorado Clinical Translational Sciences Institute (1UL1 RR025780).
Abbreviations
- MP
Monophosphate
- DP
diphosphate
- TP
triphosphate
- AMP
adenosine monophosphate
- ATP
adenosine triphosphate
- GMP
guanosine monophosphate
- GTP
guanosine triphosphate
- IMP
inosine monophosphate
- ITP
inosine triphosphate
- RBCs
red blood cells
- PBMCs
peripheral blood mononuclear cells
- DBS
dried blood spots
- NA
nucleoside analog
- RBV
ribavirin
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- HPLC
high performance liquid chromatography
- STD
standard
- IS
stable labeled internal standard
- QC
quality control
- LLOQ
lower limit of quantification
- QL
low control
- QM
medium control
- QH
high control
- UP-water
ultrapure water
References
- 1.Scheele KW. Examen chemicum calculi urinarii. Opuscula. 1776;73(2) [Google Scholar]
- 2.Garrod AB. On the Blood and Effused Fluids in Gout, Rheumatism, and Bright’s Disease. Med Chir Trans. 1854;37:49–601. doi: 10.1177/095952875403700107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nyhan WL. Disorders of purine and pyrimidine metabolism. Mol Genet Metab. 2005;86(1–2):25–33. doi: 10.1016/j.ymgme.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 4.Edwards NL, Fox IH. Disorders associated with purine and pyrimidine metabolism. Spec Top Endocrinol Metab. 1984;6:95–140. [PubMed] [Google Scholar]
- 5.Nuki G. Human purine metabolism: some recent advances and relationships with immunodeficiency. Ann Rheum Dis. 1983;42(Suppl 1):8–11. doi: 10.1136/ard.42.suppl_1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen A, Doyle D, Martin DW, Jr, Ammann AJ. Abnormal purine metabolism and purine overproduction in a patient deficient in purine nucleoside phosphorylase. The New England journal of medicine. 1976;295(26):1449–54. doi: 10.1056/NEJM197612232952603. [DOI] [PubMed] [Google Scholar]
- 7.Simmonds HA, Panayi GS, Corrigall V. A role for purine metabolism in the immune response: Adenosine-deaminase activity and deoxyadenosine catabolism. Lancet. 1978;1(8055):60–3. doi: 10.1016/s0140-6736(78)90002-8. [DOI] [PubMed] [Google Scholar]
- 8.Ribeiro FF, Xapelli S, Miranda-Lourenco C, Tanqueiro SR, Fonseca-Gomes J, Diogenes MJ, et al. Purine nucleosides in neuroregeneration and neuroprotection. Neuropharmacology. 2015 doi: 10.1016/j.neuropharm.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Nishikawa T, Suematsu S, Matsuzawa Y, Saito J, Omura M. Guanosine triphosphate can directly regulate cortisol production by activating Ca(2+)-messenger systems in bovine adrenal fasciculata cells. Endocr J. 2016;63(1):77–85. doi: 10.1507/endocrj.EJ15-0393. [DOI] [PubMed] [Google Scholar]
- 10.Simmonds HA, Fairbanks LD, Morris GS, Webster DR, Harley EH. Altered erythrocyte nucleotide patterns are characteristic of inherited disorders of purine or pyrimidine metabolism. Clinica chimica acta; international journal of clinical chemistry. 1988;171(2–3):197–210. doi: 10.1016/0009-8981(88)90145-3. [DOI] [PubMed] [Google Scholar]
- 11.De Franceschi L, Fattovich G, Turrini F, Ayi K, Brugnara C, Manzato F, et al. Hemolytic anemia induced by ribavirin therapy in patients with chronic hepatitis C virus infection: role of membrane oxidative damage. Hepatology. 2000;31(4):997–1004. doi: 10.1053/he.2000.5789. [DOI] [PubMed] [Google Scholar]
- 12.Dean BM, Perrett D, Sensi M. Changes in nucleotide concentrations in the erythrocytes of man, rabbit and rat during short-term storage. Biochem Biophys Res Commun. 1978;80(1):147–54. doi: 10.1016/0006-291x(78)91116-6. [DOI] [PubMed] [Google Scholar]
- 13.de Korte D, Haverkort WA, van Gennip AH, Roos D. Nucleotide profiles of normal human blood cells determined by high-performance liquid chromatography. Anal Biochem. 1985;147(1):197–209. doi: 10.1016/0003-2697(85)90028-4. [DOI] [PubMed] [Google Scholar]
- 14.Stocchi V, Cucchiarini L, Magnani M, Chiarantini L, Palma P, Crescentini G. Simultaneous extraction and reverse-phase high-performance liquid chromatographic determination of adenine and pyridine nucleotides in human red blood cells. Anal Biochem. 1985;146(1):118–24. doi: 10.1016/0003-2697(85)90405-1. [DOI] [PubMed] [Google Scholar]
- 15.Smolenski RT, Montero C, Duley JA, Simmonds HA. Effects of adenosine analogues on ATP concentrations in human erythrocytes. Further evidence for a route independent of adenosine kinase. Biochemical pharmacology. 1991;42(9):1767–73. doi: 10.1016/0006-2952(91)90514-6. [DOI] [PubMed] [Google Scholar]
- 16.Yeung P, Ding L, Casley WL. HPLC assay with UV detection for determination of RBC purine nucleotide concentrations and application for biomarker study in vivo. Journal of pharmaceutical and biomedical analysis. 2008;47(2):377–82. doi: 10.1016/j.jpba.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 17.Aragon-Martinez OH, Galicia O, Isiordia-Espinoza MA, Martinez-Morales F. A novel method for measuring the ATP-related compounds in human erythrocytes. Tohoku J Exp Med. 2014;233(3):205–14. doi: 10.1620/tjem.233.205. [DOI] [PubMed] [Google Scholar]
- 18.Chen P, Liu Z, Liu S, Xie Z, Aimiuwu J, Pang J, et al. A LC-MS/MS method for the analysis of intracellular nucleoside triphosphate levels. Pharmaceutical research. 2009;26(6):1504–15. doi: 10.1007/s11095-009-9863-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Machon C, Jordheim LP, Puy JY, Lefebvre I, Dumontet C, Guitton J. Fully validated assay for the quantification of endogenous nucleoside mono- and triphosphates using online extraction coupled with liquid chromatography-tandem mass spectrometry. Anal Bioanal Chem. 2014;406(12):2925–41. doi: 10.1007/s00216-014-7711-1. [DOI] [PubMed] [Google Scholar]
- 20.Dudley E, Bond L. Mass spectrometry analysis of nucleosides and nucleotides. Mass Spectrom Rev. 2014;33(4):302–31. doi: 10.1002/mas.21388. [DOI] [PubMed] [Google Scholar]
- 21.Thomas D, Herold N, Keppler OT, Geisslinger G, Ferreiros N. Quantitation of endogenous nucleoside triphosphates and nucleosides in human cells by liquid chromatography tandem mass spectrometry. Anal Bioanal Chem. 2015;407(13):3693–704. doi: 10.1007/s00216-015-8588-3. [DOI] [PubMed] [Google Scholar]
- 22.Azzari C, la Marca G, Resti M. Neonatal screening for severe combined immunodeficiency caused by an adenosine deaminase defect: a reliable and inexpensive method using tandem mass spectrometry. J Allergy Clin Immunol. 2011;127(6):1394–9. doi: 10.1016/j.jaci.2011.03.040. [DOI] [PubMed] [Google Scholar]
- 23.Wu LS, Rower JE, Burton JR, Jr, Anderson PL, Hammond KP, Baouchi-Mokrane F, et al. Population pharmacokinetic modeling of plasma and intracellular ribavirin concentrations in patients with chronic hepatitis C virus infection. Antimicrobial agents and chemotherapy. 2015;59(4):2179–88. doi: 10.1128/AAC.04618-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bushman LR, Kiser JJ, Rower JE, Klein B, Zheng JH, Ray ML, et al. Determination of nucleoside analog mono-, di-, and tri-phosphates in cellular matrix by solid phase extraction and ultra-sensitive LC-MS/MS detection. Journal of pharmaceutical and biomedical analysis. 2011;56(2):390–401. doi: 10.1016/j.jpba.2011.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jimmerson LC, Ray ML, Bushman LR, Anderson PL, Klein B, Rower JE, et al. Measurement of intracellular ribavirin mono-, di- and triphosphate using solid phase extraction and LC-MS/MS quantification. Journal of chromatography B, Analytical technologies in the biomedical and life sciences. 2015;978–979:163–72. doi: 10.1016/j.jchromb.2014.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agarwala S, Eley T, Villegas C, Wang Y, Hughes E, Xie J, Grasela D, editors. Pharmacokinetic interaction between tenofovir and atazanavir coadministered with ritonavir in healthy subjects. 6th International Workshop on the Clinical Pharmacology of HIV Therapy; 2005 April 28–30; Quebec City, Canada. [Google Scholar]
- 27.Zheng JH, Rower C, McAllister K, Castillo-Mancilla J, Klein B, Meditz A, et al. Application of an intracellular assay for determination of tenofovir-diphosphate and emtricitabine-triphosphate from erythrocytes using dried blood spots. Journal of pharmaceutical and biomedical analysis. 2016;122:16–20. doi: 10.1016/j.jpba.2016.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jimmerson LC, Zheng JH, Bushman LR, MacBrayne CE, Anderson PL, Kiser JJ. Development and validation of a dried blood spot assay for the quantification of ribavirin using liquid chromatography coupled to mass spectrometry. Journal of chromatography B, Analytical technologies in the biomedical and life sciences. 2014;944:18–24. doi: 10.1016/j.jchromb.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng JH, Guida LA, Rower C, Castillo-Mancilla J, Meditz A, Klein B, et al. Quantitation of tenofovir and emtricitabine in dried blood spots (DBS) with LC-MS/MS. Journal of pharmaceutical and biomedical analysis. 2014;88:144–51. doi: 10.1016/j.jpba.2013.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Z, Rodman JH, Flynn PM, Robbins BL, Wilcox CK, D’Argenio DZ. Model for intracellular Lamivudine metabolism in peripheral blood mononuclear cells ex vivo and in human immunodeficiency virus type 1-infected adolescents. Antimicrobial agents and chemotherapy. 2006;50(8):2686–94. doi: 10.1128/AAC.01637-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li W, Tse FL. Dried blood spot sampling in combination with LC-MS/MS for quantitative analysis of small molecules. Biomedical chromatography: BMC. 2010;24(1):49–65. doi: 10.1002/bmc.1367. [DOI] [PubMed] [Google Scholar]
- 32.Ji QC, Liu G, D’Arienzo CJ, Olah TV, Arnold ME. What is next for dried blood spots? Bioanalysis. 2012;4(16):2059–65. doi: 10.4155/bio.12.168. [DOI] [PubMed] [Google Scholar]
- 33.Stocchi V, Cucchiarini L, Canestrari F, Piacentini MP, Fornaini G. A very fast ion-pair reversed-phase HPLC method for the separation of the most significant nucleotides and their degradation products in human red blood cells. Anal Biochem. 1987;167(1):181–90. doi: 10.1016/0003-2697(87)90150-3. [DOI] [PubMed] [Google Scholar]
- 34.Coolen EJ, Arts IC, Swennen EL, Bast A, Stuart MA, Dagnelie PC. Simultaneous determination of adenosine triphosphate and its metabolites in human whole blood by RP-HPLC and UV-detection. Journal of chromatography B, Analytical technologies in the biomedical and life sciences. 2008;864(1–2):43–51. doi: 10.1016/j.jchromb.2008.01.033. [DOI] [PubMed] [Google Scholar]
- 35.Hitomi Y, Cirulli ET, Fellay J, McHutchison JG, Thompson AJ, Gumbs CE, et al. Inosine triphosphate protects against ribavirin-induced adenosine triphosphate loss by adenylosuccinate synthase function. Gastroenterology. 2011;140(4):1314–21. doi: 10.1053/j.gastro.2010.12.038. [DOI] [PubMed] [Google Scholar]