Abstract
Mass spectrometry has traditionally been the technology of choice for small molecule analysis, making significant inroads into metabolism, clinical diagnostics and pharmacodynamics since the 1960s. In the mid 1980s, with the discovery of electrospray ionization (ESI) for biomolecule analysis, a new door opened for applications beyond small molecules. Initially proteins were widely examined, followed by oligonucleotides and other nonvolatile molecules. Then in 1991, three intriguing studies reported using mass spectrometry to examine noncovalent protein complexes, results that have been expanded on for the last 25 years. Those experiments also raised the question, how soft is ESI and can it be used to examine even more complex interactions. Our lab addressed these questions with the analyses of viruses, which were initially tested for viability following electrospray ionization and their passage through a quadrupole mass analyzer by placing them on an active medium that would allow them to propagate. This observation has been replicated on multiple different systems including experiments on an even bigger microbe, a spore. The question of analysis was also addressed in the early 2000’s with charge detection mass spectrometry. This unique technology could simultaneously measure mass-to-charge and charge, allowing for the direct determination of the mass of a virus. More recent experiments on spores and enveloped viruses have given us insight into the range of mass spectrometry’s capabilities (reaching 100 trillion daltons), beginning to answer fundamental questions regarding the complexity of these organisms beyond proteins and genes, and how small molecules are integral to these supramolecular living structures.
Graphical Abstract
Introduction
The origins of mass spectrometry (MS) being applied to biological noncovalent interactions can largely be traced to 1991 when three seminal papers reported the observation of known protein complexes in the gas phase using electrospray ionization (ESI). These included the immunosuppressant ligand-binding to human immunophilin FKBP (Figure 1a) [1], the heme-myoglobin complex [2], and the ternary complex of dimeric HIV-1 protease binding to an inhibitor [3]. This work was quickly followed by numerous other reports on noncovalent structures such as the duplex DNA-drug complex [4], calcium mediated cell-surface carbohydrate association [5] (Figure 1b), and catalytic antibody-hapten/substrate interactions [6, 7]. As these applications further developed, it became increasingly clear that the noncovalent analysis capabilities that ESI-MS was capable of could be used for a variety of purposes. For example, monitoring binding site kinetics by titrating increasing concentrations of ligand [1], and even binding site position by using collision induced dissociation (CID) to disrupt both covalent bonds [8] and noncovalent interactions [3, 9]. These results motivated new instrumentation designs of large complexes, such as DNA (Figure 1c) [10], which were otherwise difficult or impossible to analyze.
Figure 1.
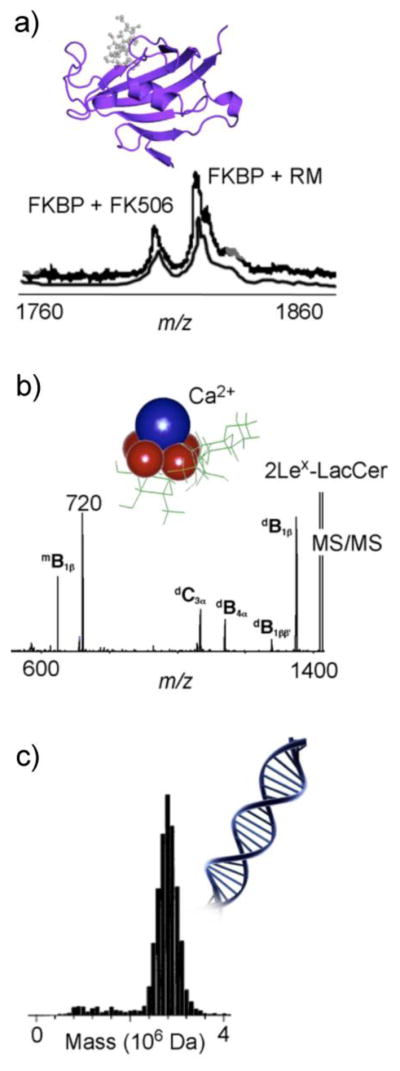
Mass spectrometric analysis of biomolecular complexes (a) the binding of immunosuppressants FK506 and rapamycin (RM) to human immunophilin FKBP [1], (b) Ca2+ mediated dimerization of the glycosphingolipid Lex – LacCer with MS/MS providing site specific information of Ca2+ binding and (c) the analysis of DNA using charge detection mass spectrometry [10]. Images reprinted with permission from [1] and [5], copyright 1991, 1993 American Chemical Society, and from [10], copyright 1998 Springer Publishing.
These breakthroughs were compelling for both chemists and biologists, and for our lab they begged a bigger question: can whole organisms be examined with mass spectrometry? By virtue of their homogeneity and relatively small size (megadaltons), the logical organism to test to answer this question was a virus. An intact virus, or virion, consists of genetic material in the form of RNA or DNA surrounded by a protein shell known as a capsid and may be helical or icosahedral in nature. Some viruses are further encased by a lipid membrane called a viral envelope, which helps in evading the immune system. While advances in viral research have been largely associated with the development of physical techniques such as X-ray crystallography [11, 12] and electron microscopy [13, 14], the development of mass spectrometry to mass measure viral ions was recognized as having the potential to further facilitate their characterization. Another motivation for analyzing viruses was that while convincing evidence existed regarding the observation of noncovalent interactions with mass spectrometry [1–7, 10], a common question was (and still is) whether native conformations are preserved throughout the vaporization, ionization and mass analysis within the vacuum of the mass spectrometer [15]. A third question, reminiscent of the Manhattan Project where Calutron mass spectrometers were used to separate uranium isotopes [16], is whether this technology can be used as a viable separation and collection device for biomolecules. Our work on viruses attempted to address these issues with the analysis 40 MDa tobacco mosaic virus.
Instrumentation for Viral Analysis
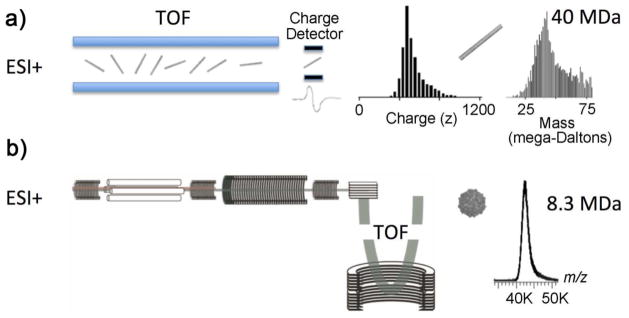
The first breakthrough in accurately measuring the mass of a complete virus particle required the development of a novel detector capable of simultaneously measuring charge, m/z and velocity of an ion. Dr. Henry Benner created this technology by modifying an ESI-TOF mass spectrometer with an additional metal flight tube attached to a charge-sensitive preamplifier [10, 17, 18]. The charge-sensitive preamplifier creates an image current for single ions as they pass through this device and have the added benefit of a signal whose magnitude is proportional to the number of charges they are carrying. To demonstrate the capability of this charge detector design, an entire intact genome was initially analyzed [19]. This represented, at that time, the single largest molecule to be successfully mass analyzed. Charge detection mass analysis circumvents the problem of detecting large ions by making a simultaneous measurement of charge (z) and m/z ratio for individual ions, therefore it enables the direct measurement of mass (as opposed to m/z). Strictly speaking, one could say that this was the first true “mass spectrometer”.
The charge detector instrumentation was successfully used to measure the mass of two intact viruses: rice yellow mottle virus (RYMV) and tobacco mosaic virus (TMV) [18]. The signal from thousands of individual ions was averaged over 30 minutes (Figure 2a) with both RYMV and TMV possessing a charge distribution between 300 and 1,000 positive charges resulting in measured masses between 6 x 106 and 7 x 106 Da for RYMV and between 39 x 106 and 42 x 106 Da for TMV. These masses agreed well with the calcuated masses of 6.5 x 106 Da and 40 x 106 Da for RYMV and TMV respectively. The error associated with these signals could be attributed to the charge detection signal amplification process and the system having a relatively short linear flight path, while the large tailing in the mass distribution is most likely the result of adduct formation and incomplete desolvation [9, 20, 21]. Previous studies have shown that desolvation becomes less efficient with increasing size of the biomolecules and a similar distribution was observed in Benner’s original experiments on DNA [10]. More recent commercial instrumentation with traditional electron multiplier detectors have been especially successful at performing analyses of viruses and virus capsids. The addition of an inert countercurrent gas to time-of-flight (TOF) mass spectrometers, such as nitrogen [18] or xenon [22] to assist with droplet desolvation of large biomolecules, can produce charge-resolved spectra of masses up to approximately 5 MDa [22] (Figure 2b).
Figure 2.
Intact nonenveloped tobacco mosaic and Norwalk viruses were electrosprayed into a time-of-flight (TOF) mass spectrometer with Benner’s charge detection MS (a) and, more recently (b) a modified commercial TOF instrument.
While the initial observations of a single virus ion may seem trivial given today’s mass spectrometry technology, back in the 1990s in the Lawrence Livermore lab, monitoring the data being generated by the instrument as each single ion passed through the detector was thrilling. At the time, this was a completely unique mass spectrometer at the forefront of sensitivity and mass measurement capability. These instrument alterations were highly influential in studying supramolecular living structures by mass spectrometry. Since the 1990s even larger biomolecules have been analyzed using charge detection devices with improvements made by the Jarrold lab on the additional flight tube with optmized dimensions, alignment and programming parameters. In this system, peaks can be rejected if they have baseline fluctuations, signals from more than one droplet or when signficant differences between the entrance and exit peaks are observed [23]. This improved setup results in a tighter distribution and detectable mass limits reaching 1014 Da (100 trillion daltons).
Another interesting offshoot of ESI-MS has been the coupling of ESI to ion mobility spectrometry (IMS) and differential mobility analysis better known as Gas-phase Electrophoretic Molecular Mobility Analyzers (GEMMA) [24]. These devices provide confident measure of nanometer-sized biomolecules and noncovalent complexes with sub-nanometer resolution, and additional validation that these complexes survive the analysis process. IMS allows for the analysis of the cross-sectional area of an ion to be measured and have been widely applied to intact protein complexes [21, 25–29]. This technique has been used to detect intact MS2 bacteriophage at 24 ± 2 nm [30], and human rhinovirus with a diameter of 29.8 ± 0.3 nm [28]. Interestingly, since the measurements are based on the electrophoretic mobility (EM) measurements that are independent of known virus features, these results provide strong evidence that no large-scale disruption of tertiary or quaternary structure of the capsid occurred during ESI desolvation and ionization [31].
Although the ability to detect megadalton sized particles is at the heart of investigating microbe behavior, it is not only size that matters. In native microbe analysis, the resolving power, sensitivity and sampling rate are crucial for deciphering relevant and specific information about the intact particles and how they interact with modulators of function. Consequently, these aspects have been continually improved in both CDMS and GEMMA over the years. Today even conventional mass spectrometers can be harnessed to obtain high resolution mass spectra of megadalton sized complexes. QTOFs are a particularly good choice for simple modifications to aid in the transmission of large ions. For example, analysis of an 18 MDa intact capsid from bacteriophage HK97 was performed on a QTOF with xenon collision gas at higher pressures and voltages. The instrument is capable of resolving masses up to 40 GDa, but the practical limitation lies within the ability to desolvate the ions, thus reducing the limit to ~20 MDa [32]. An Orbitrap mass analyzer has also been configured to accommodate the analysis of megadalton sized capsids. With an additional gas flow to the HCD cell, optimized RF voltages and the removal of analogue filters, an upper mass limit of ~5 MDa was achieved, but restricted by the low pressures required in the C-trap [33]. The benefit of using a modified QTOF or Orbitrap mass analyzer to measure megadalton particles is the capability of generating high resolution spectra without the need for a completely different system.
Virus and Spore Viability
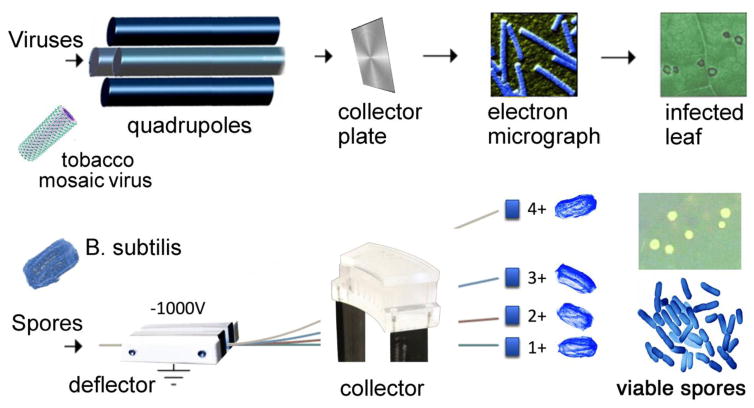
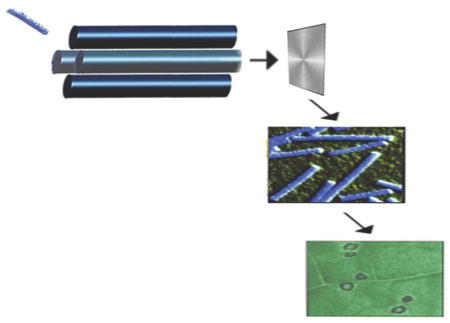
Beyond mass analysis, viruses represent an interesting analytical challenge and would make a good model system for testing the limits of ESI and its ability to examine noncovalent complexes. The primary goal of these first attempts to analyze an intact virus was to determine if viruses could be transferred into the gas phase intact, and at the same time perform mass selection [26]. The upper mass limit of our instrument was 2,400 m/z, a limit that was overcome by operating the quadrupole mass analyzer in radio frequency (RF) mode where only ions of high m/z were allowed to pass. The next step in these experiments was to determine whether the viral ions were intact and finally, whether they were still alive and capable of their native biological function (i.e. infection). To accomplish this, we collected the viral ions from a glycerol-coated brass plate placed in front of the detector (Figure 3a). The separation and collection of ions within a mass spectrometer for purification was inspired by early Calutron mass spectrometers used to separate uranium isotopes [16]. The isolated virus sample was then directly analyzed by negative-stain transmission electron microscopy (TEM). Both RYMV and TMV (Figure 3a) particles remained intact based on the electron microscopy images, indicating the native structure had been preserved. Even with the success of these experiments, it was clear that damage to the protein capsid or the packaged RNA could go undetected in the TEM images. Therefore, the viability of the collected virus following ESI mass spectrometry was tested to provide definitive evidence of whether the native state was retained. This experiment was conducted by inoculating tobacco cultivar Xanthi plants with TMV collected in the mass spectrometer. The tobacco plants developed lesions characteristic of infection (Figure 3a) demonstrating that the viruses were viable. A similar viability experiment with RYMV was not successful indicating it was more labile in its symmetrical icosahedral structure than TMV during mass spectrometry.
Figure 3.
(Top) Intact viruses electrosprayed into the mass spectrometer, collected, detected with electron microscopy and inducing an infection on a leaf. (Bottom) Intact spores are electrosprayed into a Bug Trap where they are electrostatically deflected and collected. Bug collector and B. subtilis images courtesy of Sarah Pratt.
There have since been other reports of testing viability after the electrospray ionization of a virus. Hogan et al. used EM to analyze bacteriophage MS2, λ, T2 and T4 with a highly monodisperse electrospray source operated in cone jet mode to produce a near steady state diffusion charge distribution[34]. An airborne sampler was teed off from the electrospray source to collect and test the viability of the aerosolized virus particles. Bacteriophage MS2, a small, single-unit virus, was found to be viable while the λ bacteriophage had low amounts of viability and the large, multi-subunit viruses T2 and T4 bacteriophages had no viability. The length of bacteriophages T2 and T4 exceeded the mean diameter of the droplets (171 nm), yet both T2 and T4 had mean diameters of 87.03 and 88.32 nm respectively. It was surmised that the T2 and T4 bacteriophages were not the whole viruses, but only the protein capsid heads that remained intact through the electrospray process. Viruses with sizes exceeding the droplet diameter would be exposed to more mechanical and electrical stress, leaving them more susceptible to noncovalent bond disruption.
Another interesting study involving much larger bacterial cells was recently reported by Pratt and Austin [35]. Two different species, E. coli and B. subtilis, were electrosprayed ensuring complete deagglomeration and desolvation, electrostatically deflected and collected to assess whether individual cells in various charge states could survive. Bacterial particles with higher charge or lower mass were deflected more strongly as shown in Figure 3. This device, called a “Bug Trap”, consists of two parallel stainless steel plates with an applied electric field acting as a particle deflector, and a slotted collection plate to capture the charged particles. After electrospraying a bacteria sample, the collector channels were washed with sterile water and transferred to petri dishes and incubated overnight at 39°C. The E. coli did not survive complete desolvation and charging under the applied electric field while B. subtilis did survive. The major difference between these two strains is the ability for B. subtilis to form spore while E. coli does not. This protective spore coat helped this bacterium survive the electrospray process in both atmospheric (~20% recovery) and vacuum (<0.1% recovery) conditions, albeit at low charge states. To keep microorganisms viable as they traverse the harsh conditions of electrospraying and acceleration through electric fields, the size of the microorganism with respect to the droplet diameter becomes important as does the strength of the noncovalent interactions maintaining the native structure. Reducing mechanical and electrical stress by controlling droplet size can be balanced with the ability to completely desolvate the particle to effectively separate it based on electrophoretic mobility and m/z, which is necessary to give accurate and well-resolved measurements. Taken together, these results show the viability of the microorganism was strain dependent, as determined by by their size and structural stability.
Conclusion: Revolution to Evolution
The mass spectrometer has seen dramatic improvements in the past three decades and the revolution of ESI has generated an evolution in the entire mass spectrometer from front to back. ESI has clearly been the method of choice for examining noncovalent interactions in the gas phase, especially intact viruses, yet it is interesting that desorption/ionization techniques (e.g., MALDI) have not been used extensively, despite both of these methods also being relatively soft. This may be because analyzing macromolecular complexes by MALDI is confounded by the need for a denaturing sample preparation involving matrix application, laser optimization and reduced detector sensitivity in the ultrahigh m/z range [24]. However, gentle ablation methods, such as DESI [36] or nanostructure-based desorption and ionization [37] coupled with CDMS or modified TOF-MS may provide possible alternatives for soft desorption/ionization. For the ESI itself, nebulization techniques could also be improved to increase the viability of the microorganisms. As discussed above, previous studies observed that bacteria or viruses with a more spherical shape and smaller diameters were more likely to survive the journey through the MS. Further investigation on the size of nebulizer droplets, desolvation methods and temperature/voltage during ion transfer would be useful in keeping larger microorganisms alive through the MS process.
Advances in analyzer technology have also greatly enhanced our ability to simultaneously and accurately analyze m/z and z of individual ions. With improved mass accuracy, limits of detection, and the ability to decipher charge states from heterogeneous mixtures, it has become possible to detect intermediates in the capsid assembly process (Figure 4) [39] and to examine how protein envelopes form on capsid surfaces [40]. Detector evolution has also played a significant role in the ability to detect large megadalton species as limitations exist in both electron multiplier detectors and condensation particle counters (GEMMA). For example, the recently developed nanoelectromechanical mass sensors (NEMS) showed advantages over both of the currently developed detectors. This cantilever-based mass sensor operates at an ultra-high vibrational frequency, with the adsorption of analytes causing measurable alterations in that frequency that span a very large dynamic range [41]. In contrast to GEMMA, NEMS successfully avoids the drawback of charge neutralization, which makes most particles become uncharged and therefore not separated in the electric field, resulting in decreased sensitivity [34, 42]. Compared with CDMS, interpretation of NEMS spectra are simplified as there are no charge states associated with particles [39]. However, this technique suffers from relatively low mass resolution [41] compared to GEMMA and CDMS and requires experimental run times up to several hours [42]. Although this technique has great potential for analyzing supramolecular living structures, the NEMS-MS technology is still in its early stages of development.
Figure 4.
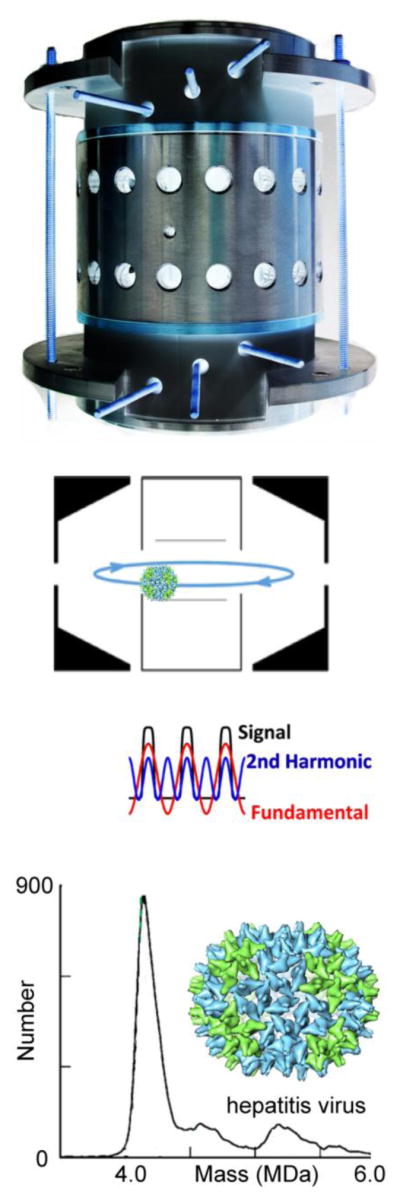
. A “Dalek” Charge Detector designed to investigate heterogeneous organisms. Artist rendition of the Dalek Charge Detector courtesy of Martin Jarrold and reprinted with permission from [38] copyright 2015 American Chemical Society.
Some would say that mass spectrometry is now a mature technology, however a truly comprehensive understanding of a biological system requires the ability to examine all of its components: the genetic material, proteins, macromolecular structures, and small molecules. While there have been many advances in MS instrumentation for analyzing these systems, a complete solution remains tantalizingly out of reach. What are the next mass spectrometry technologies that will allow us to simultaneously observe genes, proteins, metabolites and their interactions within the complexity that is a living biological entity? Perhaps we need to turn to our students for the next revolution.
References
- 1.Ganem B, Li YT, Henion JD. Detection of noncovalent receptor ligand complexes by mass-spectrometry. Journal of the American Chemical Society. 1991;113:6294–6296. [Google Scholar]
- 2.Katta V, Chait BT. Observation of the hemeglobin complex in native myoglobin by electrospray-ionization mass spectrometry. Journal of the American Chemical Society. 1991;113:8534–8535. [Google Scholar]
- 3.Baca M, Kent SBH. Direct observation of a ternary Complex Between the Dimeric Enzyme HIV-1 Protease and a Substrate-Based Inhibitor. Journal of the American Chemical Society. 1992;114:3992–3993. [Google Scholar]
- 4.Gale DC, Goodlett DR, Lightwahl KJ, Smith RD. Observation of duplex DNA-drug noncovalent complexes by electrospray-ionization mass-spectrometry. Journal of the American Chemical Society. 1994;116:6027–6028. [Google Scholar]
- 5.Siuzdak G, Ichikawa Y, Caulfield TJ, Munoz B, Wong CH, Nicolaou KC. Evidence of a Ca2+-dependent carobhydrate association through ion spray mass-spectrometry. Journal of the American Chemical Society. 1993;115:2877–2881. [Google Scholar]
- 6.Siuzdak G, Krebs JF, Benkovic SJ, Dyson HJ. Binding of hapten to a single-chain catalytic antibody demonstrated by electrospray mass-spectrometry. Journal of the American Chemical Society. 1994;116:7937–7938. [Google Scholar]
- 7.Krebs JF, Siuzdak G, Dyson HJ, Stewart JD, Benkovic SJ. Detection of a catalytic antibody species acylated at the active-site by electrospray mass-spectrometry. Bio chemistry. 1995;34:720–723. doi: 10.1021/bi00003a002. [DOI] [PubMed] [Google Scholar]
- 8.Loo JA, Edmonds CG, Smith RD. Tandem mass-spectrometry of very large molecules .2. Dissociation of multiply charged proline-containing proteins from electrospray ionization. Anal Chem. 1993;65:425–438. doi: 10.1021/ac00052a020. [DOI] [PubMed] [Google Scholar]
- 9.Nettleton EJ, Sunde M, Lai ZH, Kelly JW, Dobson CM, Robinson CV. Protein subunit interactions and structural integrity of amyloidogenic transthyretins. Evidence from electrospray mass spectrometry. J Mol Biol. 1998;281:553–564. doi: 10.1006/jmbi.1998.1937. [DOI] [PubMed] [Google Scholar]
- 10.Schultz JC, Hack CA, Benner WH. Mass determination of megadalton-DNA electrospray ions using charge detection mass spectrometry. J Am Soc Mass Spectrom. 1998;9:305–313. doi: 10.1016/S1044-0305(97)00290-0. [DOI] [PubMed] [Google Scholar]
- 11.Stern LJ, Brown JH, Jardetzky TS, Gorga JC, Urban RG, Strominger JL, et al. Crystal-structure of the human class-II MHC protein HLA-DR1 complexed with an influenza-virus peptide. Nature. 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- 12.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 14.Yeager M, Berriman JA, Baker TS, Bellamy AR. 3-Dimensional structure of the rotavirus hemagglutinin VP4 by cryoelectron microscopy and difference map analysis. Embo J. 1994;13:1011–1018. doi: 10.1002/j.1460-2075.1994.tb06349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aplin RT, Robinson CV, Schofield CJ, Westwood NJ. Does the observation of noncovalent complexes between biomolecules by electrospray-ionization mass-spectrometry necessarily reflect specific solution interactions. J Chem Soc-Chem Commun. 1994:2415–2417. [Google Scholar]
- 16.Smith LP, Parkins WE, Forrester AT. On the separation of isotopes in quantity by electromagnetic means. Physical Review. 1947;72:989–1002. [Google Scholar]
- 17.Schultz JC, Hack CA, Benner WH. Polymerase chain reaction products analyzed by charge detection mass spectrometry. Rapid Commun Mass Spectrom. 1999;13:15–20. [Google Scholar]
- 18.Fuerstenau SD, Benner WH, Thomas JJ, Brugidou C, Bothner B, Siuzdak G. Mass spectrometry of an intact virus. Angew Chem-Int Edit. 2001;40:542–544. doi: 10.1002/1521-3773(20010202)40:3<541::AID-ANIE541>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 19.Fuerstenau SD, Benner WH. Molecular weight determination of megadalton DNA electrospray ions using charge detection time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 1995;9:1528–1538. doi: 10.1002/rcm.1290091513. [DOI] [PubMed] [Google Scholar]
- 20.Loo JA, Loo RRO. Applying charge discrimination with electrospray-ionization mass-spectrometry to protein analyses. J Am Soc Mass Spectrom. 1995;6:1098–1104. doi: 10.1016/1044-0305(95)00531-5. [DOI] [PubMed] [Google Scholar]
- 21.Kaufman SL, Skogen JW, Dorman FD, Zarrin F, Lewis KC. Macromolecule analysis based on electrophoretic mobility in air. Globular proteins. Anal Chem. 1996;68:1895–1904. doi: 10.1021/ac951128f. [DOI] [PubMed] [Google Scholar]
- 22.Shoemaker GK, van Duijn E, Crawford SE, Uetrecht C, Baclayon M, Roos WH, et al. Norwalk Virus Assembly and Stability Monitored by Mass Spectrometry. Mol Cell Proteomics. 2010;9:1742–1751. doi: 10.1074/mcp.M900620-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mabbett SR, Zilch LW, Maze JT, Smith JW, Jarrold MF. Pulsed acceleration charge detection mass spectrometry. Application-to weighing electrosprayed droplets. Anal Chem. 2007;79:8431–8439. doi: 10.1021/ac071513s. [DOI] [PubMed] [Google Scholar]
- 24.Laschobert C, Wruss J, Blaas D, Szymanski WW, Allmaier G. Gas-phase electrophoretic molecular mobility analysis of size and stoichiometry of complexes of a common cold virus with antibody and soluble receptor molecules. Anal Chem. 2008;80:2261–2264. doi: 10.1021/ac702463z. [DOI] [PubMed] [Google Scholar]
- 25.Cox KA, Julian RK, Cooks RG, Kaiser RE. Conformer selection of protein ions by ion mobility in a triple quadrupole mass-spectrometer. J Am Soc Mass Spectrom. 1994;5:127–136. doi: 10.1016/1044-0305(94)85025-9. [DOI] [PubMed] [Google Scholar]
- 26.Siuzdak G, Bothner B, Yeager M, Brugidou C, Fauquet CM, Hoey K, et al. Mass spectrometry and viral analysis. Chem Biol. 1996;3:45–48. doi: 10.1016/s1074-5521(96)90083-6. [DOI] [PubMed] [Google Scholar]
- 27.Clemmer DE, Hudgins RR, Jarrold MF. Naked protein nconformations - cytochrome c in the gas-phase. Journal of the American Chemical Society. 1995;117:10141–10142. [Google Scholar]
- 28.Bacher G, Szymanski WW, Kaufman SL, Zollner P, Blaas D, Allmaier G. Charge-reduced nano electrospray ionization combined with differential mobility analysis of peptides, proteins, glycoproteins, noncovalent protein complexes and viruses. J Mass Spectrom. 2001;36:1038–1052. doi: 10.1002/jms.208. [DOI] [PubMed] [Google Scholar]
- 29.Pease LF. Physical analysis of virus particles using electrospray differential mobility analysis. Trends Biotechnol. 2012;30:216–224. doi: 10.1016/j.tibtech.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 30.Wick CH, McCubbin PE. Characterization of purified MS2 bacteriophage by the physical counting methodology used in the integrated virus detection system (IVDS) Toxicol Method. 1999;9:245–252. [Google Scholar]
- 31.Weiss VU, Bereszcazk JZ, Havlik M, Kallinger P, Gosler I, Kumar M, et al. Analysis of a Common Cold Virus and Its Subviral Particles by Gas-Phase Electrophoretic Mobility Molecular Analysis and Native Mass Spectrometry. Anal Chem. 2015;87:8709–8717. doi: 10.1021/acs.analchem.5b01450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snijder J, Rose RJ, Veesler D, Johnson JE, Heck AJR. Studying 18 MDa Virus Assemblies with Native Mass Spectrometry. Angewandte Chemie International Edition. 2013;52:4020–4023. doi: 10.1002/anie.201210197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snijder J, van de Waterbeemd M, Damoc E, Denisov E, Grinfeld D, Bennett A, et al. Defining the Stoichiometry and Cargo Load of Viral and Bacterial Nanoparticles by Orbitrap Mass Spectrometry. Journal of the American Chemical Society. 2014;136:7295–7299. doi: 10.1021/ja502616y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hogan CJ, Kettleson EM, Ramaswami B, Chen DR, Biswas P. Charge reduced electrospray size spectrometry of mega- and gigadalton complexes. Whole viruses and virus fragments. Anal Chem. 2006;78:844–852. doi: 10.1021/ac051571i. [DOI] [PubMed] [Google Scholar]
- 35.Pratt SN, Austin DE. Bacterial Spores Survive Electrospray Charging and Desolvation. J Am Soc Mass Spectrom. 2014;25:712–721. doi: 10.1007/s13361-014-0827-x. [DOI] [PubMed] [Google Scholar]
- 36.Ouyang Z, Takats Z, Blake TA, Gologan B, Guymon AJ, Wiseman JM, et al. Preparing protein microarrays by soft-landing of mass-selected ions. Science. 2003;301:1351–1354. doi: 10.1126/science.1088776. [DOI] [PubMed] [Google Scholar]
- 37.Siuzdak GE, Buriak J, Wei J. Desorption/ionization of analytes from porous light-absorbing semiconductor. 6288390 B1. Patent US. 2001
- 38.Keifer DZ, Shinholt DL, Jarrold MF. Charge Detection Mass Spectrometry with Almost Perfect Charge Accuracy. Anal Chem. 2015;87:10330–10337. doi: 10.1021/acs.analchem.5b02324. [DOI] [PubMed] [Google Scholar]
- 39.Pierson EE, Keifer DZ, Selzer L, Lee LS, Contino NC, Wang JCY, et al. Detection of Late Intermediates in Virus Capsid Assembly by Charge Detection Mass Spectrometry. Journal of the American Chemical Society. 2014;136:3536–3541. doi: 10.1021/ja411460w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keifer DZ, Motwani T, Teschke CM, Jarrold MF. Acquiring Structural Information on Virus Particles with Charge Detection Mass Spectrometry. J Am Soc Mass Spectrom. 2016:1–9. doi: 10.1007/s13361-016-1362-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snijder J, Heck AJR. Analytical Approaches for Size and Mass Analysis of Large Protein Assemblies. In: Cooks RG, Pemberton JE, editors. Annual Reviews. Palo Alto: 2014. [DOI] [PubMed] [Google Scholar]
- 42.Sage E, Brenac A, Alava T, Morel R, Dupre C, Hanay MS, et al. Neutral particle mass spectrometry with nanomechanical systems. Nat Commun. 2015;6:5. doi: 10.1038/ncomms7482. [DOI] [PMC free article] [PubMed] [Google Scholar]