Abstract
Age-associated changes in lung structure and function are some of the most important predictors of overall health, cognitive activities and longevity. Common to all aging cells is an increase in oxidatively modified DNA bases, primarily 8-oxo-7,8-dihydroguanine (8-oxoG). It is repaired via DNA base excision repair pathway driven by 8-oxoguanine DNA glycosylase-1 (OGG1-BER), whose role in aging has been the focus of many studies. This study hypothesizes that signaling and consequent gene expression during cellular response to OGG1-BER “wires” senescence/aging processes. To test OGG1-BER was mimicked by repeatedly exposing diploid lung fibroblasts cells and airways of mice to 8-oxoG base. Results showed that repeated exposures led to G1 cell cycle arrest and pre-matured senescence of cultured cells in which over 1000 genes were differentially expressed --86% of them been identical to those in naturally senesced cells. Gene ontology analysis of gene expression displayed biological processes driven by small GTPases, phosphoinositide 3-kinase and mitogen activated kinase cascades both in cultured cells and lungs. These results together, points to a new paradigm about the role of DNA damage and repair by OGG1 in aging and age-associated disease processes.
Keywords: OGG1, 8-oxoguanine, senescence, aging
Graphical Abstract
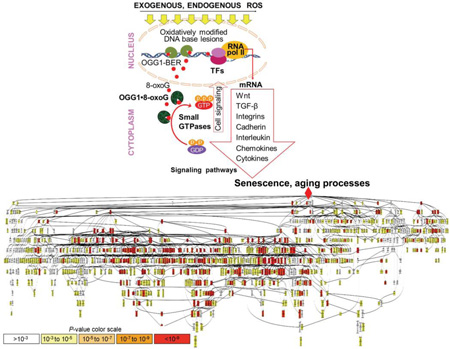
Visual illustration for role of OGG1-BER in senescence/aging processes. In this model OGG1-BER and generation of OGG1•8-oxoG complex (a guanine nucleotide exchange factor) increases levels of activated small GTPases. Downstream from small GTPases transcription factors (TFs) are activated leading to differential expression of mediators and “fueling” senescence/aging processes as shown by gene ontology enrichment and visualization analysis. P values of biological processes are depicted by colors.
1. Introduction
Aging of the respiratory system leads to decrease in lung function (elastic recoil of the lungs, inefficient gas-exchange and respiratory muscle performance) correlating well with poor health conditions and vital functions including e.g., poorer cognitive activities, increased levels of subcortical atrophy, dementia and decline in cardiovascular performance in humans (Carvalhaes-Neto et al., 1995; Janssens, 2005). The physiological processes controlling the rate of aging in mammals, at levels of development, growth, reproduction, metabolism and resistance to oxidative stress, and so on involves the cross-talk among various signaling cascades centered around reactive oxygen species (ROS) (Papaconstantinou, 1994; Papaconstantinou, 2009). Despite the universal nature of aging and age-associated complications the underlying molecular mechanism remains poorly understood (Papaconstantinou, 1994). One of the theories of aging proposes that accumulation of oxidized base lesions- and DNA strand breaks-induced signaling alter gene expression leading to a decline in cellular/tissue function (Akbari and Krokan, 2008; David et al., 2007; Rodier et al., 2009; Sohal et al., 1994; Wilson and Bohr, 2007; Wilson et al., 2008).
The most common and abundant oxidative DNA base lesion in all aged cell types is the 7,8-dihydro-8-oxoguanine (8-oxoG) (Chen et al., 2003; Dianov et al., 2001). A great abundance of this lesion is attributed to guanine’ lowest redox potential among the all nucleobases in DNA and RNA (Dizdaroglu, 1985; Radak and Boldogh, 2010; Steenken, 1997). Repair of 8-oxoG is initiated by the 8-oxoguanine DNA glycosylase1 (OGG1) base excision repair pathway (OGG1-BER) (David et al., 2007; Mitra et al., 2002). Despite large numbers of publications there is a loose etiological association has been established between accumulation of genomic 8-oxoG lesions and aging processes (Bacsi et al., 2007; Chen et al., 1995; David et al., 2007; Hamilton et al., 2001; Lovell and Markesbery, 2007; Szczesny et al., 2003; Weissman et al., 2007). The lack of a strong association could well be correct as the phenotype of OGG1 knock out (Ogg1−/−) mice is mild. Despite the supra-physiological increase in mitochondrial and genomic 8-oxoG levels, Ogg1−/− mice developed normally, are fertile, showed only limited pathological changes, and have a life span similar to that of wild type mice (Klungland et al., 1999; Minowa et al., 2000; Osterod et al., 2001; Sakumi et al., 2003). Under experimental conditions (e.g., high-fat diet) Ogg1−/− mice exhibit altered insulin levels, glucose tolerance, adiposity, hepatic steatosis (Sampath et al., 2012).
It is estimated that several thousands 8-oxoG lesions could be formed in genome per cell daily due to production of endogenous electrophilic molecules (Nakamura et al., 2014), while the number of such guanine lesions can be higher upon exogenous environmental exposures (Lindahl and Barnes, 2000). Estimates on the absolute numbers of genomic 8-oxoG lesions in airways (nasal, bonchial, bronchiolar epithelium, or subepihelial lung tissues) which directly interact with the environment is not available; however, the levels of the OGG1-BER repair products (e.g., 8-oxoG base) in serum or urine correlates well with dose and length of exposure, chemical composition, and physical nature of the inhaled environmental agents (Ba et al., 2014; Ba et al., 2015). Moreover, an increase free 8-oxoG levels in sputum and bronchoalveolar lavage fluid after environmental exposures (Ba et al., 2014; Bacsi et al., 2016; Proklou et al., 2013). In experimental animal models of lung diseases or in age-associated human lung pathologies (e.g., COPD, emphysema, and asthma) showed that one of the most referenced DNA base damage(s) is 8-oxoG (Ba et al., 2014; Ba et al., 2015; Deslee et al., 2009; Igishi et al., 2003).
Studies have also demonstrated that when free 8-oxoG base released from genome or added to cells [which rapidly enter into cells (Hajas et al., 2012)] it is bound by OGG1 with high affinity, and the resulting complex (OGG1•8-oxoG) physically interacts with small GTPases (Boldogh et al., 2012). Importantly, the OGG1•8-oxoG complex caused GDP → GTP exchange in Kirsten (K)-RAS, neuroblastoma RAS viral oncogene homolog (N)-RAS, Harvey (H)-RAS, RHOA and RHO family member RAC1 (Aguilera-Aguirre et al., 2014; Boldogh et al., 2012) and thus it functions as a guanine nucleotide exchange factor (GEF) in a manner similar to that of other GEFs (Mosteller et al., 1994). In airways, OGG1•8-oxoG complex increases KRAS-GTP, which than lead to activation of Raf1 kinase (v-raf-leukemia viral oncogene 1 (Raf1) homolog), phosphatidylinositol-, mitogen-, stress-activated as well as IκB (nuclear factor of kappa light chain gene enhancer in B-cells) kinases. These signaling pathways activate various transcription factors including nuclear factor-κB (NF-κB). The later has the capacity to alter cell proliferation, differentiation, modulate state of the host tissue and of multiple components of the immune system (Brasier, 2006; Vlahopoulos et al., 2015).
In this study, we postulated that supra-physiologically generated levels of OGG1•8-oxoG complexes by OGG1-BER, activates small GTPases leading to changes in gene expression, which eventually results in cellular senescence/aging. To test this hypothesis, we repeatedly challenged human diploid lung fibroblasts and airways to mimic OGG1-BER and utilized molecular biological approaches, high throughput gene expression profiling and gene ontology (GO) analysis. Results showed that 8-oxoG-exposed OGG1-exprssing cells showed cell cycle arrest at G1 phase mediated by increased expression of TP53/p21/p16. Gen ontology analysis of gene expression displayed signaling pathways and biological processes consistent with senescence in cell culture and aging in airways.
2. Materials and Methods
2.1. Reagents
8-OxoG (Cayman Chemical, Ann Arbor, MI), diphenyleneiodonium chloride (DPI), 7,8-dihydro-8-oxo-2'-deoxyguanosine and guanine (8-oxo-dG; Sigma-Aldrich, St. Lois, MO), 8-hydroxyadenine (8-OH-Ade; Biolog Life Science Institute, Bremen, Germany), N-acetyl-cysteine (NAC), (Sigma, St. Louis, MO); AA-861 [2,3,5-trimethyl-6-(12-hydroxy-5,10-dodecadiynyl)-1,4-benzoquinone; Sigma, St. Louis, MO], methyl arachidonyl fluorophosphonate (MAFP) and nordihydroguaiaretic acid (NDGA, Cayman Chemical, Ann Arbor, MI), PD98059 (Calbiochem, Gibbstown, NJ). 8-oxoG was provided as a hydroacetate salt and dissolved in 12 mM NaOH to a final concentration of 4 mM (stock solution). It was prepared freshly, diluted in PBS (w/o Ca2+/Mg2+, pH: 7.4) to attain experimental concentrations (10, 1, and 0.1 µM), filter-sterilized and used within 1 h for experiments. All other intact and oxidized nucleotide bases and nucleosides were dissolved in the same manner.
2.2. Cell cultures and treatment
Human embryonic diploid lung fibroblasts (HDLF = MRC-5; American Type Culture Collection (ATCC, Cat # CCL-171) were grown in MEM (Hyclone) supplemented with 10% fetal bovine serum (FBS; HyClone). To determine population doubling levels (PDLs), cells were subcultured at approximately 80% confluence, counted, and seeded at a density of 4×104 cells. PDL was calculated at each passage, as described above (Bacsi et al., 2007). Mouse embryonic lung fibroblast (MELF) cultures were developed from nine days old embryos of C57Bl/6 mice and maintained in DMEM (HyClone). All media was supplemented with 10% FBS, 100 U penicillin and 100 µg/mL streptomycin and L-glutamine (200 mg per L) (Bacsi et al., 2007).
2.3. Animals and treatment
Animal experiments were performed according to the NIH Guide for Care and Use of Experimental Animals and approved by the UTMB Animal Care and Use Committee (approval no. 0807044A). Eight-week-old female BALB/c mice (The Jackson Laboratory, Bar Harbor, ME, USA) were used for these studies. Mice (n = 5 per group) were challenged intranasally on days 0, 2 and 4 (multiple challenge) with 60 µL of pH-balanced 8-oxoG (Cayman Chemicals, Ann Arbor, MI) solution (pH: 7.4; 0.0005 mg per kg), or saline (mock) under mild anesthesia (Boldogh et al., 2005). The lipopolysaccharide concentration was below detectable levels in all reagents. Animals were sacrificed after the last challenge on day four at times 0, 30, 60 and 120 min.
2.4. Detection of senescent markers
Senescent associated (SA)–galactosidase (SA-β-gal) expression was undertaken as described previously (Dimri et al., 1995). Briefly, cells were fixed in 2% formaldehyde/0.2% glutaradehyde then incubated at 37 °C with β-gal staining solution (1 mg of X-Gal per ml in dimethylformamide) in buffer containing 40 mM citric acid/sodium phosphate; 5 mM potassium ferrocyanide; 5 mM potassium ferricyanide; 150 mM NaCl and 2 mM MgCl2). Cultures were evaluated after 16 h incubation. Lipofuscin: cells on microscope cover-slips were placed in a thermo-controlled microscopic chamber and cellular autofluorescence were visualized at 580–630 nm. Microscopy and co-localization was performed on a NIKON Eclipse Ti System using Nikon NIS Elements Version 3.5 (NIKON Instruments, Tokyo, Japan). Magnification: ×125.
2.5. Immunohistochemistry
Cell on coverslips were fixed in 4% paraformaldehyde at 4°C for 15 min. The air-dried cells were permeabilized with acetone-methanol (ratio: 1:1). The cells were then incubated for 60 min at 37°C with primary antibodies to TP53-serine15, y-pH2A.X, p16INK4a or p21WAF/CIP1. After washing cells (3 times for 15 min) with PBS-Tween 20 (PBS-T), they were incubated with affinity-purified FITC-conjugated secondary antibodies (Santa Cruz Biotechnology) for 1 h at room temperature. Nuclei of cells were stained with 10 ng/ml DAPI (4′6-diamidino-2-phenylindole dihydrochloride) for 15 min and then mounted in antifade medium (Dako Inc. Carpinteria, CA) on microscope slides. Microscopy was performed on a NIKON Eclipse Ti System using Nikon NIS Elements Version 3.5 (NIKON Instruments, Tokyo, Japan). Magnification: ×125.
2.6. Analysis of cell cycle
Cells were synchronized in serum free media and treated with 8-oxoG. Cells were harvested after 0, 3, 6, 12, 18, and 24h treatment and processed for cell cycle analysis (Bresnahan et al., 1996). Briefly, cells were suspended in low salt buffer containing 3% polyethylene glycol, propidium iodine (5 µg/ml), 0.1% Triton X-100, 4 mM Na-citrate, RNase (100 µg/ml) and incubated at 37°C for 30 min. High salt buffer (3% polyethylene glycol, propidium iodine (5 µg/ml), 0.1% Triton X100, and 400 mM NaCl) was added and the cells were kept at 4°C overnight. The cellular DNA content was evaluated by flow cytometry using a FACScan flow cytometer (Becton Dickinson). Histograms were analyzed using ModFit LT cell cycle analysis software (Verity Software House, Inc) to determine the percent of cells in various stages of the cell cycle. 12,000 events were collected for each sample.
2.7. Mutagenicity assay
V79 cells were exposed to 8-oxoG (10 µM twice a day ) for 4 days or same concentrations of solvent, or H2O2 (10 µM) and monolayers were dissociated and subcultured into 100-mm dishes (106 cells per dish) in growth medium containing 6-thioguanine (6-TG; 7 µg/ml). Colonies of 6-TG-resistant cells were allowed to develop for 7 to 8 days and, then fixed in 3.7% formalin and stained with 0.1% methylene blue. The mutation frequency was calculated by determining the number of 6-TG-resistent colonies relative to the number of cells seeded in the 6-TG containing medium and correcting for the plating efficiency in the absence of the selective agent (Albrecht et al., 1997).
2.8. Immunoblotting
Cells were lysed in a lysis buffer containing 25 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 1% NP-40, 1 mM DTT, 5% glycerol, 20 mM NaF, 1 mM, 1 µg/ml leupeptin and 1 µg/ml aprotinin. Protein samples (10 to 40 µg per lane) were separated by 5-20% SDS-PAGE. Proteins were transferred to Hybond-ECL nitrocellulose (Amersham, Biosciences, UK Limited) membrane and then blocked with 3% BSA in Tris-buffered saline (TBS) containing 0.1% Tween (TBS-T) for 3 h, and incubated overnight at 4°C with the primary Ab diluted in 3% BSA in TBS-T. The blots were then washed four times with TBS-T and incubated for 1 h with HRP-conjugated secondary HPR-conjugated Ab (anti mouse IgG, GE Healthcare, UK Ltd) in 5% non-fat dry milk in TBS-T. Immunoreactive bands were detected using an ECL reagent (Amersham Biosciences, UK Limited) and visualized by autoradiography.
2.9. Telomere length analysis
Proliferating and senescent cell were harvested, washed and DNA extracted with a Qiagen DNA isolation kit (Qiagen, Valencia, CA). DNAs were eluted with Qiagen elution buffer (pH 8.0) and stored at −80°C until use. Relative telomere length was determined by real-time qPCR using an ABI PRISM 7000 Sequence Detection System (Applied Biosystems, Foster City, CA) according to the protocol described previously by (Cawthon, 2002). The assay included amplification of the telomere (Tel) and a single-copy gene (36B4 gene) for normalization. Primers for Tel: F, 5'-GGTTTTTGAGGGTGAGGGTGAGGGTGAGGGTG AGGGT-3'; R, (5'- TCCCGACTATCCCTATCCCTATCCCTATCCCTATCCCTA -3'). The 36B4u primers F, 5'- CAGCAAGTGGGAAGGTGTAATCC-3' and 36B4d: 5'-CCCATTCTATCATCAACGGGTACAA-3'. The T/S ratio (Telomere-to-single copy gene ratio) was defined as the difference in Cp between the telomere and single-copy gene, or [2Cp(telomere)/2Cp(BTF3)] = 2−ΔCp (Cawthon, 2002).
2.10. Assessment of DNA strand breaks in telomeres
Parallel coverslip cultures of cells were fixed with 2% formaldehyde, permebilized as we described previously (Bacsi FRBM) and incubated with monoclonal antibody against γ-H2A.X (Ser 139) (Upstate Biotechnology, Lake Placid, NY, USA), diluted 1:2000 for 1h at room temperature and binding was visualized by Alexa Fluor 594-goat anti-mouse IgG (1:4000; Molecular Probes, Eugene, OR, USA). The specimens were then washed to perform immuno-FISH (Ksiazek et al., 2007). In brief, cells were washed with SSC/0.5% Tween, air dried and baked at 60 °C and incubated with 4 ng/µl of Cy3-labeled telomere specific (CCCTAA)3 peptide nucleic acid probe (Dako, Glostrup, Denmark), followed by denaturation at 85°C and hybridization for 2 h. Cells were stained with DAPI and microscopy was performed on a NIKON Eclipse Ti System using Nikon NIS Elements Version 3.5 (NIKON Instruments, Tokyo, Japan). Magnification: ×125. Colocalization was visualized using Nikon NIS Elements Version 3.5 (NIKON Instruments, Tokyo, Japan).
2.11. siRNA-mediated depletion of gene expression
NRAS, HRAS and KRAS expression was down-regulated using a simultaneous siRNA transfection and plating method (Omerovic et al., 2008). In brief, cells were transfected with control siRNA (siGENOME Non-Targeting siRNA) or target-specific siRNAs for Harwey (H)-Ras (# M-004142), Kirsten (K)-Ras (# M-005069), neuroblastome RAS viral oncogene homolog (N)-Ras (#M-003919) and/or OGG1 (#NM-010957) (siGENOME SMARTpools, Thermo Scientific) using INTERFERinTM reagent (Polyplus-transfection Inc., NY), and incubated in growth medium for 72 h. The levels of RNA for the target sequences were determined with melting curve analysis using the ABI PRISM 7000 sequence Detector software (Applied Biosystems, Foster City, CA, USA). A ΔΔCT analysis was performed as we described previously (Aguilera-Aguirre, 2009).
2.12. Assessment of GTP-bound small GTPases
The active form of GTPases was determined using pull-down affinity purification assays (Active Ras Pull-Down and Detection Kit, Thermo Fisher Scientific) as described previously (Boldogh et al., 2012). Briefly, cells (or lungs) were washed with ice-cold PBS and lysed with 1x Lysis/Binding/Washing buffer [25 mM Tris-HCl (pH 7.5), 150 mM NaCl, 50 mM MgCl2, 5 mM EDTA, 1% NP-40, 1 mM DTT, 5% glycerol, 20 mM NaF, 1 mM, 1 µg/ml leupeptin and 1 µg/ml aprotinin]. Lysates were cleared by centrifugation at 12,000×g, protein concentrations were determined, and active-GTPase was captured by the binding domain of Raf1 (Ras-RBD) (Taylor et al., 2001). The proteins were fractionated on 4-20% PAGE, and changes in the levels of activated GTPases were determined by Western immunoblot analysis using type-specific antibodies to HRAS, KRAS, N-RAS or anti-pan-RAS (Aguilera-Aguirre et al., 2014; Boldogh et al., 2012; Hajas et al., 2013).
2.13. RNA isolation
RNA was extracted using an RNeasy kit (Qiagen, Valencia, CA) per the manufacturer’s instructions. Briefly, cells or lung tissue lysates were loaded onto an RNeasy column and RNA was isolated. The RNA concentration was determined spectrophotometrically on an Epoch Take-3™ system (Biotek, Winooski, VT) using Gen5 v2.01 software. The quality of the total RNA was confirmed spectrophotometrically via the 260/280 nm ratio, which varied from 1.9 to 2.0.
2.14. Affimetrix gene array and data analysis
Affymetrix GeneChip® Human Genome Focus Arrays were used to analyze gene expression, which were performed in UTMB’s Next- Generation Sequencing (NGS) Core Facility (Director: Dr. Thomas G. Wood). Gene expression data were analyzed by Microsoft Excel 2003 software (Microsoft, Redmond, CA), the Affymetrix NetAFFX Analysis Center online tools, and the Spotfire Decision Site 9.0 software (TIBCO Spotfire, Somerville, MA). The array data from all samples were normalized by the overall expression level of the given array. As a background corresponding to approximately 200 units of fluorescent intensity level in the raw data file) was used. Genes with increased or decreased expression levels (≥ 1.5-fold) were selected for further analysis. Network-, pathway-, and functional analyses were performed using the Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Inc., Redwood City, CA; www.ingenuity.com). Each gene identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base. These genes, called focus genes, were overlaid onto a global molecular network developed from information contained in the Ingenuity Pathways Knowledge Base. These networks were ranked based on the significance score value calculated by the IPA software. A graphical representation of the molecular relationships between genes/gene products for each of the top ranked networks was generated where genes or gene products are represented as nodes, and the biological relationship between two nodes is represented as an edge (Szaniszlo et al., 2009).
2.15. RNA sequencing
Poly-A+ RNA was selected from total RNA (1 µg) with poly-T oligo-attached magnetic beads. Bound RNA was fragmented by incubation at 94°C for 8 minutes in 19.5 µl of fragmentation buffer (Illumina, Part # 15016648). First- and second-strand synthesis, adapter ligation, and amplification of the library were performed using the Illumina TruSeq RNA Sample Preparation kit per the manufacturer’s instructions. Samples were tracked through the “index tags” incorporated into the adapters. Library quality was evaluated using an Agilent DNA-1000 chip on an Agilent 2100 Bioanalyzer. Library DNA templates were quantitated by qPCR and a known-size reference standard. Sequencing analysis were performed in UTMB’s Next- Generation Sequencing (NGS) Core Facility (Director: Dr. Thomas G. Wood) on an Illumina HiSeq 1000 sequencing system (Illumina Inc., San Diego, CA, USA).
Cluster formation of the library DNA templates was performed using the TruSeq PE Cluster Kit v3 (Illumina) and the Illumina cBot workstation under the conditions recommended by the manufacturer. Template input was adjusted to obtain a cluster density of 700-1000 K/mm2. Paired-end 50-base sequencing-by-synthesis was performed with a TruSeq SBS kit v3 (Illumina) on an Illumina HiSeq 1000 per the manufacturer’s protocols. Base calls were converted to sequence reads using CASAVA-1.8.2. Sequence data were analyzed with the Bowtie2, Tophat and Cufflinks programs using NCBI’s mouse (Mus musculus) genome build reference mm10. RNA-Seq data have been deposited in the NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE65031.
2.16. Gene ontology analysis
A ranked list was generated from GeneCards human gene data base (http://www.genecards.org/) including the key words “lung” and “aging”. The search generated a 4956 genes ranked list. A minimum relevance score of 5 was used to render a top ranked list containing 1551 genes. These genes were overlaid on our RNA-sequencing data set to identify differentially expressed genes induced by 8-oxoG challenge. The identified 1351 differentially expressed genes were submitted to GENE-E online software (version 3.0.204) from the Broad Institute (http://www.broadinstitute.org/cancer/software/GENE-E/) to generate heat-map. Gene ontology (GO) analysis was performed using the Gene Ontology enrichment analysis and visualization (GOrilla) data base (http://cbl-gorilla.cs.technion.ac.il/), which employs the hypergeometric distribution to determine overrepresented GO categories in a ranked set of genes (Eden et al., 2009).
2.17. Statistical analysis
Statistical analysis was performed using Student’s t-test or ANOVA, followed by post hoc tests: Bonferroni’s and Dunnett’s T3 with SPSS 14.0 software. The data are presented as the means ± the standard error of the mean. Differences were considered statistically significant at p < 0.05. (*p < 0.05; **p < 0.01; ***p < 0.001).
3. Results
3.1. Exposure of OGG1-expressing cells to 8-OxoG base accelerate senescence
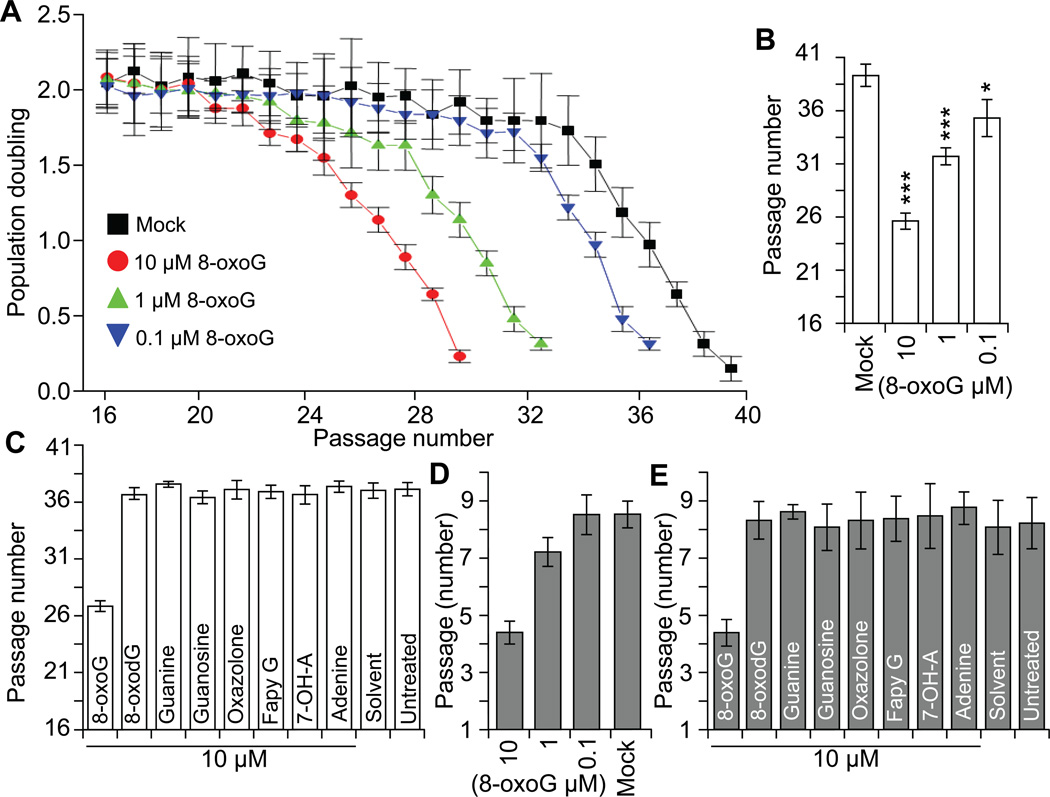
Parallel human diploid lung fibroblast cell (HDLF; between 16 and 20 passages) cultures were treated with increasing concentrations of 0.1, 1, and 10 µM, pH-balanced 8-oxoG solution. At 90% confluence cells were sub-cultured and population doublings (PD) were calculated. Results show that 8-oxoG (10 µM, every 24h) treatment significantly decreased PDs after 4 to 5 passages and after an additional 3-4 passages cells entered into a growth arrest (passage 26-27; population doubling level [PDL]: 29 ±4.4) (Fig. 1A). Treating cells with 1 µM 8-oxoG (every 24h) cell proliferation was not affected for 14 passages, but thereafter a decline in cell replication was significant and cells entered into senescence. Treatment of cells with 0.1 µM 8-oxoG twice a day was required to observe a decrease in life span (Fig. 1AB). Cells’ life-span of those exposed to 8-oxodG, guanine, deoxyguanosine, oxazolone, FapyG, 7-hydroxy-adenine, adenine, deoxy-adenosine were similar to those solvent- or un-treated ones and they reached replicative senescence (RS) at passage 38-39 (PDL: 79 ±4 (Fig. 1C).
Figure 1.
Exposure to 8-oxoG accelerate senescence of diploid cells. A, 8-oxoG base exposure decreases the life-span of HDLF cells. Parallel cultures of HDLF cells were exposed serially passaged and exposed to 10, 1 or 01 uM 8-oxoG (final concentration) or to solvent only. Population doubling (PD) was calculated at passage and when it decreased under 0.5 cells were considered senescence. B, Graphical depiction of HDLF cell’s life-span (as given by number of passage). *** < 0.001; * < 0.05. C, 8-oxoG but not 8-oxodG, guanine, guanosine, FapyG, 7-OH-A or adenine induce pre-matured senescence. Cells were exposed nucleotides/nucleosides daily until PD decreased under 0.5 and total number of passage calculated. D, 8-oxoG-base exposure decreases life-span of MELF cells. Experiments were performed as described to legend B. E, Only 8-oxoG base exposure decreases life-span of MELF cells. Experiments were performed as described in legend C.
To test whether 8-oxoG-accelerated senescence is a general phenomenon, we undertook lifespan assays in which mouse embryonic lung fibroblasts (MELF) were used. Under regular culture conditions the mock- and un-treated cells replicated through 7 to 9 passages (14−16 PDs) (Fig. 1D,E). When treated with 10 µM 8-oxoG solution daily, cells entered into growth arrest at a point between 4 to 5 passages (average of 8.6 PDs). The low concentrations (0.1 or 1.0 µM) of 8-oxoG had no significant effect (Fig. 1D). 8-OxodG, guanine, deoxyguanosine, adenine, deoxy-adenosine, 7-hydroxy-adenine did not affect lifespan (Fig. 1E). Notably, frequency of immortalization in 8-oxoG-treated MELF cultures was under detectable levels, while mock-treated cells immortalized with high frequency. The second wave of proliferation in mock-treated cells is in line with previous observations (Parrinello et al., 2003).
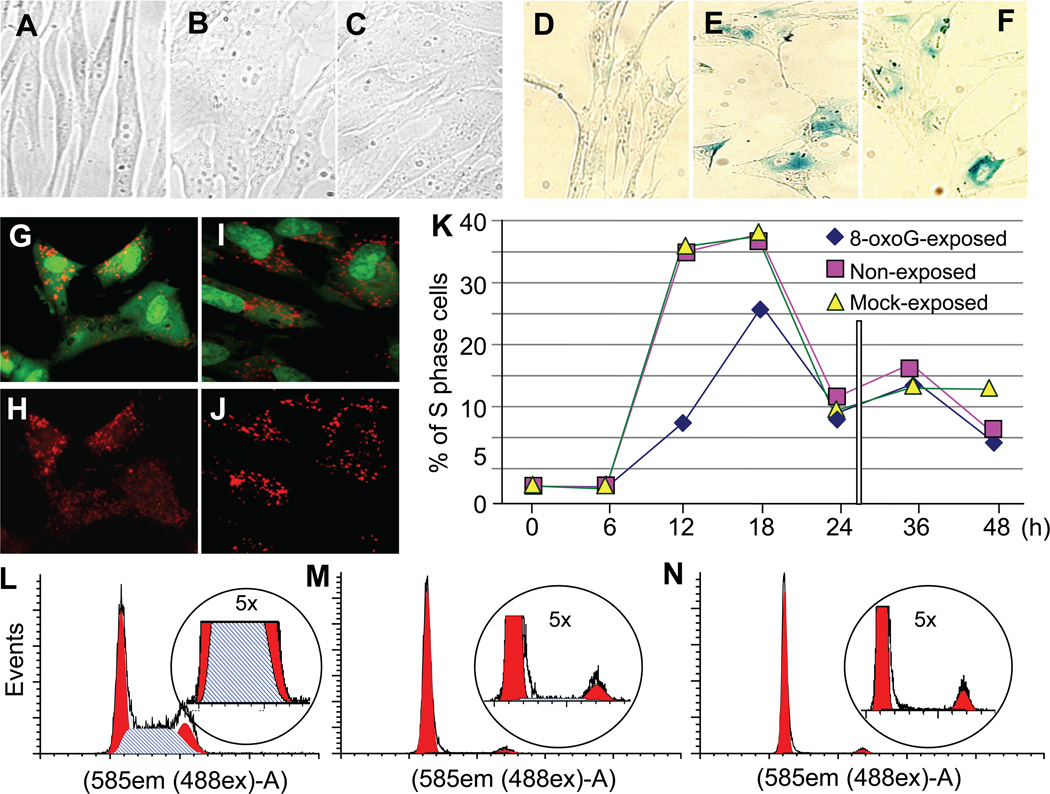
There were no morphological differences between 8-oxoG-exposure-induced pre-mature (passage 26; PDL: 29 ±4.4) and RS cells (passage 38: PDL: 79 ±4) (Fig. 2 ABC). We also showed that 8-oxoG-senesced cells expressed typical senescence markers, including expression of senescence-associated β-galactosidase (SA-βGal; Fig. 2DEF), and accumulation of lipofuscin (Fig. 2FGH). Similar results were obtained for RS cells (data not shown).
Figure 2.
Morphology and senescence markers developed upon 8-oxoG exposure of HDLF cells. A, Morphology of HDLFs at passage 26; B,C Morphological changes in 8-oxoG-exposed HDLF cells at passage 26 is similar to those in cells attained at replicative senescence (C, passage 39). Microphotographs were captured on a NIKON TE system. Magnification 40 X. D-J, Marker of senescence. D,E,F, Expression of β-galactosidase (β-Gal) and G,H,I,J accumulation of lipofuscin (shown by increased autofluorescence) in cells senesced by 8-oxoG exposure (G,H) or replicatively (I,J). Cells were photographed at a magnification of 40 X. K, Kinetic changes in cell cycle progression after repeated 8-oxoG-exposure. Cells were exposed to 8-oxoG (10 µM)- or solvent or were left unexposed. After the 4th passage cells were plated and at 60% confluence were serum starved (but exposed to 8-oxoG or solvent) and the 5th day cell cycle was initiated by the addition of FBS (10%), and DNA content of cells was analyzed by using flow cytometry (Materials and Methods). L, Cell cycle stage distribution is shown in solvent-exposed control cell culture. M, Cell cycle arrest in G-phase is shown in 8-oxoG-senesced and N, RS cultures of cells. DNA contents of cells were analyzed by flow cytometry (Materials and Methods)
To examine if 8-oxoG exposure alters cell cycle progression, HDLF cells were passaged 4 times with or without 8-oxoG exposure (pre-treatment and treatment (total) passage number = 23). Cells were synchronized by serum starvation, and then these cells were either exposed to 10 µM of 8-oxoG or solvent (mock-exposed) or were left unexposed, and cell cycle was initiated by addition of FBS (10%). Cells were harvested at 6-h intervals and subjected to DNA content analysis (Materials and Methods). Results from these experiments show that in non- or solvent-exposed cultures cells enter the S-phase between 12h and 18h after initiation by serum addition. In contrast, 8-oxoG exposure decreased cell cycle progression as shown by lower numbers of cells entering the S-phase of the cell cycle (Fig. 2K) as defined by ModFit LT cell cycle analysis software. We found that 8-oxoG-exposed and RS cells were both arrested at G1 stage of cell cycle (Fig. 2M,N). Fig. 2L shows cell cycle distribution of a replicating HDLF culture (passage 16).
3.2. 8-OxoG exposure-induced senescence is independent from telomere shortening
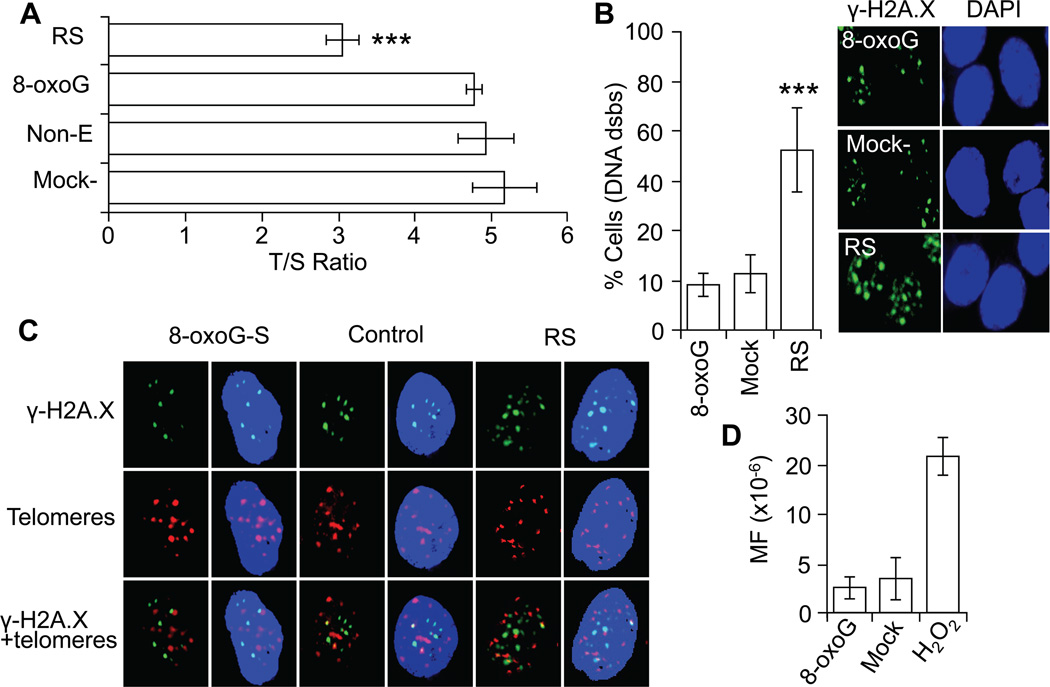
The replicative lifespan of human normal diploid cells is limited by the erosion of the protective caps (telomeres) of chromosomes of which generates signals, leading to senescence (Campisi and d'Adda di Fagagna, 2007). To test if 8-oxoG exposures-generated signals accelerate telomere length shortening we performed qPCR and the results are expressed as telomere-to-single copy gene ratio (T/S ratio; Materials and Methods). Results from qPCR showed that there are no significant differences in telomere length between 8-oxoG-senesced and PDL-matched controls. In contrast, telomere length of RS cells were significantly shorter (Fig 3A) compared to PDL-matched HDLF cells.
Figure 3.
Telomere length and levels dsbs in DNA of 8-oxoG-senesced cells. A, Decreased telomere length in RS, but not 8-oxoG-senesced cells. T/S ratio was determined as described in (Materials and Methods). B, Percentage of cells showing DNA dsbs. Cells at stage of senescence were fixed and immunostained by using antibody to γ-H2A.X. The number of positively stained (3 or more γ -H2A.X foci) cells are expressed as a percentage of 100 counted cells. *** = 0.001. Right panel, representative images of g-H2A.X-stained cells. C, DNA dsbs are in non-telomeric regions of the genome in 8-oxoG-senesced cells. Cells were immunostained by using antibody to γ-H2A.X, and immuno FISH staining was performed by using telomere-specific (CCCTAA)3 peptide nucleic acid probe (Materials and Methods). Images were visualized on a NIKON Eclipse Ti System by using Nikon NIS Elements Version 3.5. Magnification x320. D, Repeated exposure to 8-oxoG does not increase mutation frequency. Low density cultures of V79 cells were exposed to 8-oxoG (10 µM) twice a day for 4 days, and the number of 6-TG-resistant colonies was determined. In A, B, and C, RS, replicative senescence; 8-oxoG-, 8-oxoG-induced senescence; Non-T, non-treated cells; Mock-, solvent treated; MF, mutation frequency; T/S ratio, telomere-to-single copy gene ratio; and Non-E, non-exposed.
The signaling pathways activated by persistent DNA lesions, such as DNA single- (ssbs) and double-strand breaks (dsbs) have been associated with cellular senescence (Rodier et al., 2009). To test whether repeated 8-oxoG exposures lead to the formation of DNA strand lesions, cells were stained by using antibody to phosphorylated histone H2A.X (γ-H2A.X) (Celeste et al., 2003). Results showed 3 or more DNA damage foci only in 3.3±1,4% of 8-oxoG-senesced and 4.5±3.5% of PDL-matched control (mock-treated) cells (Fig. 3B, right panel). On the other hand, RS cells exhibited 3 or more DNA damage foci per cells in >50% of the cell populations (Fig 3B). In Fig 3B, right panels show representative images of γ-H2A.X-stained cells. Although a low percentage of 8-oxoG-senesced cells showed DNA strand lesions, they were examined for telomere localization using co-staining of γ-H2A.X foci with telomere-specific (CCCTAA)3 peptide [using immuno FISH technology (Ksiazek et al., 2007)]. 8-OxoG-senesced cells γ-H2A.X foci were undetectable at telomeric DNA whereas in RS cells (Fig. 3C) 9,5% of γ-H2A.X foci localized to telomeres (data not shown). Together these data and those from qPCR suggest that 8-oxoG-driven senescence is not associated with telomere erosion. Moreover, it is unlikely that multiple treatments with an 8-oxoG increase in mutation frequency (MF) in cells, as MF is similar to those observed in mock-treated V79 cultures (Fig. 3D).
3.3. OGG1 down-regulation increased lifespan of diploid lung fibroblasts
To investigate the physiological relevance of 8-oxoG released from DNA to accelerate senescence processes, OGG1 was depleted by stable expression of specific shRNA (Materials and Methods) and colonies were isolated (antibiotic selection) from cultures of diploid lung fibroblasts. Only colonies with mRNA levels of OGG1 less than 75% of control plasmid transfected cells were utilized. A decrease in protein levels was confirmed by immunoblotting (Fig. 4B). At the first passage, clones were divided and 8-oxoG- (10 µM) or solvent-exposed daily. Cell clones with or without 8-oxoG exposure were maintained and total cell numbers (cells yield) were determined. Cell-yield attained by OGG1 depleted colonies are significantly higher than that of OGG1 expressing ones (control shRNA) (Fig. 4A). Importantly, 8-oxoG exposure had minor impact on total cell numbers in OGG1-depleted clones, while the same dose of 8-oxoG significantly decreased total cell numbers in clones expressing control shRNA similar to those shown in Fig. 1AB. These data together, suggest roles of both OGG1 and 8-oxoG base in senescence processes.
Figure 4.
OGG1 depletion prevented 8-oxoG-induced premature senescence. A, 8-oxoG exposures do effect life-span of OGG1-depleted cells. Cells were transfected with shRNAs to OGG1 and shRNA-expressing stable cell clones were developed and passaged serially ± 8-oxoG (10 µM daily). Total cells numbers were calculated at the end of their life span. B, Characterization of shRNA expressing cell clones. Level of OGG1 mRNA in clones of expressing OGG1-shRNA and control shRNA was determined by qPRC. Inset, OGG1 protein levels in lysates of cells stable expressing target specific and control shRNA (immunoblotting).
3.4. Activation of NRAS in response to 8-oxoG base exposure
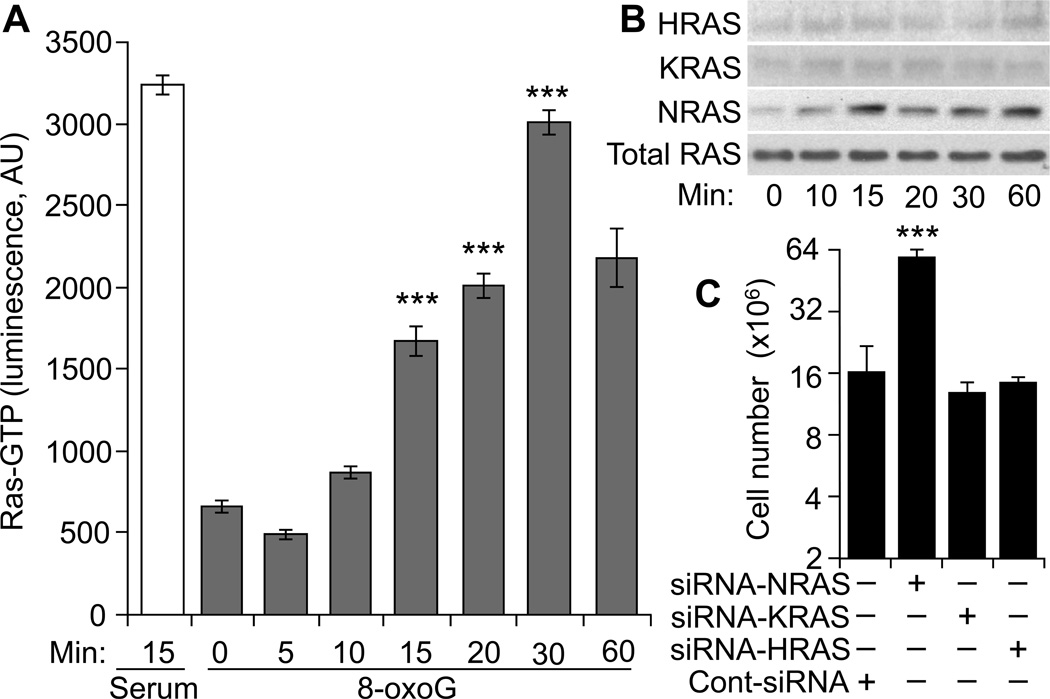
Expression of oncogenic RAS (HRAS-V12; protein lacks GTPase activity, so it is constitutively activated) in primary human and rodent cells results in a permanent G1 arrest and prematured senescence (Benanti and Galloway, 2004; Serrano et al., 1997). Therefore we asked whether and which RAS family member is activated by repeated 8-oxoG exposure of human diploid fibroblast cells. Results in Fig. 5A, show a time-dependent increase in total GTP-bound RAS levels in response to challenge. However, activated RAS pull-down assay show that only NRAS is activated, while GTP-bound HRAS and KRAS were not increased (Fig. 5B). These results are consistent with previous observations showing increase in level primarily of the NRAS-GTP in human (and mouse) lung fibroblast (Aguilera-Aguirre et al., 2014).
Figure 5.
Activation of small GTPases upon 8-oxoG exposure of HDLF cells. A, Changes in total RAS-GTP levels. Cells were exposed to 8-oxoG (10 µM) or serum (20%, for 15 min) lysates were prepared and RAS-GTP levels were assessed by an ELISA (Materials and Methods). B, 8-oxoG exposure activates NRAS in human fibroblasts. Cells exposed to 8-oxoG (10 µM) lysates were prepared at the times indicated. Active RAS was collected by pull-down assays (Materials and Methods). Eluted proteins were subjected to SDS-PAGE, and RAS isoforms were identified by immunoblotting using type-specific antibodies. C, NRAS depletion increased cell numbers. Parallel cultures of cells were NRAS, KRAS or HRAS depleted by using siRNA transfection and 8-oxoG exposed. Results are expressed as total number of cells.
8-OxoG exposure of HDLF cells induced activation of NRAS, and therefore its possible role in senescence was evaluated. NRAS expression was depleted using a simultaneous siRNA transfection and plating method (Omerovic et al., 2008). In controls, HRAS and KRAS expression was down-regulated by using a similar method. Control and target-specific, siRNA-transfected HDLF cells were maintained up to their life-span in the presence of 8-oxoG (10 µM). Results showed that NRAS depletion significantly increased the life-span and the number of cells from 2x106 to 59.4 ± 3.1 x106 (Fig 5C). Compared to cont-siRNA-transfected cells, depletion of HRAS or KRAS did not significantly change total cell numbers (Fig 5C). Together these data imply that NRAS-driven signaling is the primary cause of pre-mature senescence (Fig. 1) and possibly of the G1-phase arrest of the cell cycle at stage of senescence (Fig. 2N,M).
3.5. Gene expression profiles of 8-oxoG- and naturally-senesced cells are similar
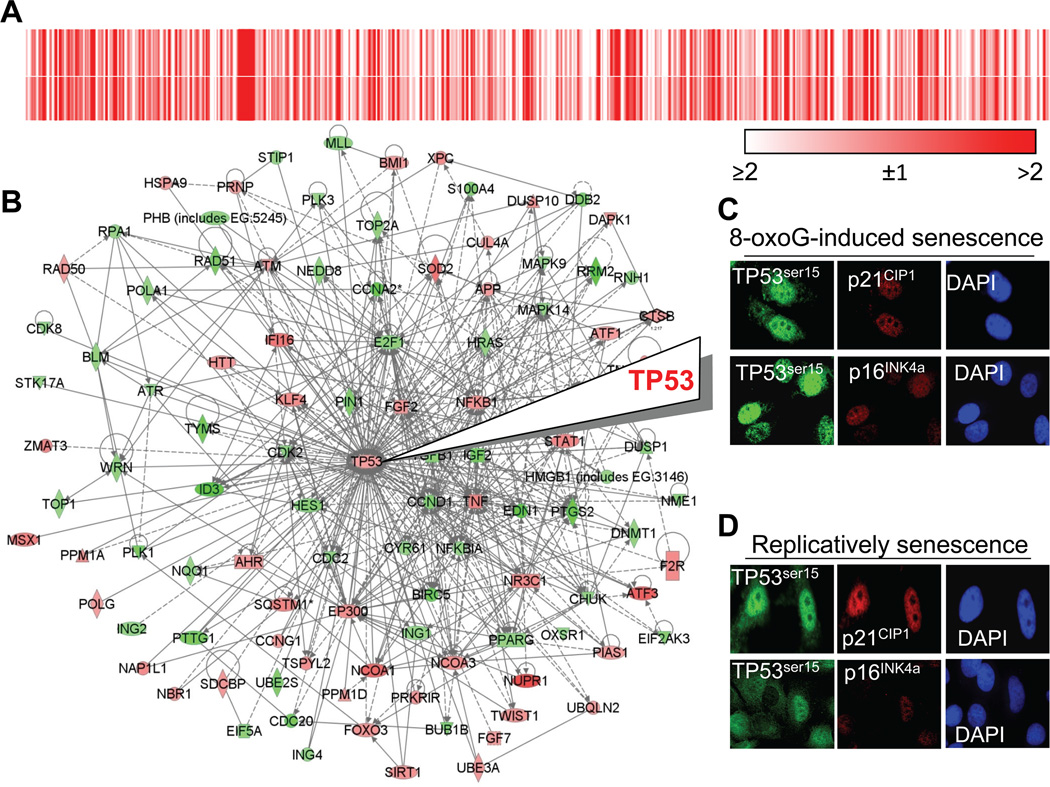
To obtain data on gene expression maintaining senescent phenotype, RNAs were isolated from 8-oxoG- and naturally-senesced cells and subjected to microarray analysis using the Affymetrix gene arrays (Materials and Methods). Genes with increased or decreased expression levels (≥ 2-fold) were identified and selected for analysis. To define genes associated with senescent on the array 1617 genes were selected based on the key word “senescence” in the GeneCards data base. The 1617 genes were then overlaid on significantly modulated gene lists corresponding to those from 8-oxoG- and RS cells and 1116 senescence-associated genes were found. To visualize similarities in gene expression patterns the list were submitted to GEN-E online software and heat maps were generated. The heat-maps show that gene sets and their expression level maintaining terminal cell cycle arrest is similar between 8-oxoG- and RS cells (Fig. 6A). Specifically, out of 1116, genes 959 (86%) showed nearly similar expression patterns.
Figure 6.
Signaling network associated with TP53 in 8-oxoG senesced cells. A, Similar differential gene expression in 8-oxoG-senesced and RS cells. A list of differentially expressed genes (1116) was submitted to GENE-E online software and heat-maps were generated. Upper heat-map, signal intensities of genes expressed in 8-oxoG-senesced cells; lower panel, signal intensities of genes in RS cells. B, Visual depiction of TP53-associated network in 8-oxoG-senesced cells. Expression was analyzed by Affimetrix gene array. Microarray data sets containing differentially expressed genes were overlaid onto molecular network of the Ingenuity Pathways Knowledge Base. The intensity of the node color corresponds to the degree of change in gene expression. Red, up-regulated, green: down-regulated. ——, direct interactions, ----, indirect interactions.
C,D Expression of TP53Ser15 and cyclin kinase inhibitors in 8-oxoG- and RS cells. Senesced cells were subcultured onto coverslips, fixed and immuno-stained using antibodies to TP53Ser15, p16INK4a or p21WAF1. Microscopy was performed on a NIKON Eclipse Ti System using Nikon NIS Elements Version 3.5 (NIKON Instruments, Tokyo, Japan). Magnification: ×125.
Gene symbols: SUPPLEMENTARY MATERIAL
To analyze gene sets expressed in senescent cells, the gene networks were ranked based on the significance score calculated by the IPA software. As immuno-staining showed increased levels of TP53Ser15 (Fig. 6B,C), we selected one of the top ranked signaling networks regulating TP53 expression and its post-translational modifications central to maintaining G1 phase arrest in 8-oxoG-senesced diploid fibroblasts (Fig. 6D). This network includes key transcription factors such as nuclear factor kappa B1 (NFKB1); the Kruppel family of transcription factor 4 (KLF4, regulates TP53 expression); activating transcription factor 1 and 3 (ATF1,3); twist basic helix-loop-helix transcription factor 1 (TWIST1); signal transducer and activator of transcription 1 (STAT1); polycomb group RING finger protein 1 (BMI1); forkhead Box O3 family of transcription factors (FOXO3); and Msh Homeobox 1 (MSX1, a transcriptional repressor). Genes encoding for cytokines upstream from TP53 include interferon gamma family protein 16 (IFI16); tumor necrosis factor ligand superfamily member (TNF); tumor necrosis factor receptor superfamily 10B (TNFRSF10B); growth factors such as fibroblast growth factor 2 (FGF2) and transforming growth factor, beta1 (TGFβ1, regulates proliferation, differentiation, adhesion, migration, and cell tissue remodeling).
DNA damage-activated genes such as ataxia telangiectasia mutated (ATM), xeroderma pigmentosum group C-complementing protein (XPC), S. cerevisiae homolog, double strand break repair protein (RAD50) and others listed in the legend to Fig. 6D were also modulated. The change in gene expression resembles persistent DNA damage response signaling, an unexpected observation as 8-oxoG-mediated, prematurely-senesced cells did not show increased DNA damage. A nearly identical TP53 top-ranked network was identified in RS cells (data not shown). Thus it appears that genes upstream from TP53 not only modulate its function, but encode for multifunctional proteins that alone or in combination with other proteins in this network is associated with the irreversible arrest of cell proliferation and pre-matured senescence by the product of OGG1-BER.
To examine the validity of microarray data, cells were immuno-stained with antibodies to phosphorylated TP53 at serine15 (TP53Ser15), p16INK4a or p21WAF/CIP1 cyclin kinase inhibitors. Results summarized in Fig. 6C,D show that the nuclei of 8-oxoG- and naturally senesced cells accumulate TP53Ser15, p16INK4a and p21WAF/CIP1. These observations were not surprising for RS cells, which have DNA strand damage as shown by γ-H2A.X staining (Fig. 3B). However, it is unexpected that 8-oxoG-senesced cells, which do not have significant levels of DNA damage (Fig. 3A,B,C), to accumulate TP53Ser15, p16INK4a and p21WAF/CIP1. These results could be due to unscheduled signaling by activated NRAS (Fig. 5B) and are in line with the microarray analysis of gene expression (Fig. 6B). In support, constitutively active RASVal12 expressed in diploid fibroblast cells induced the activation of TP53 and p16INK4a tumor suppressor proteins (Hara et al., 1996). In early passage-cells TP53Ser15, p16INK4a or p21WAF1 were at under detectable levels (data not shown).
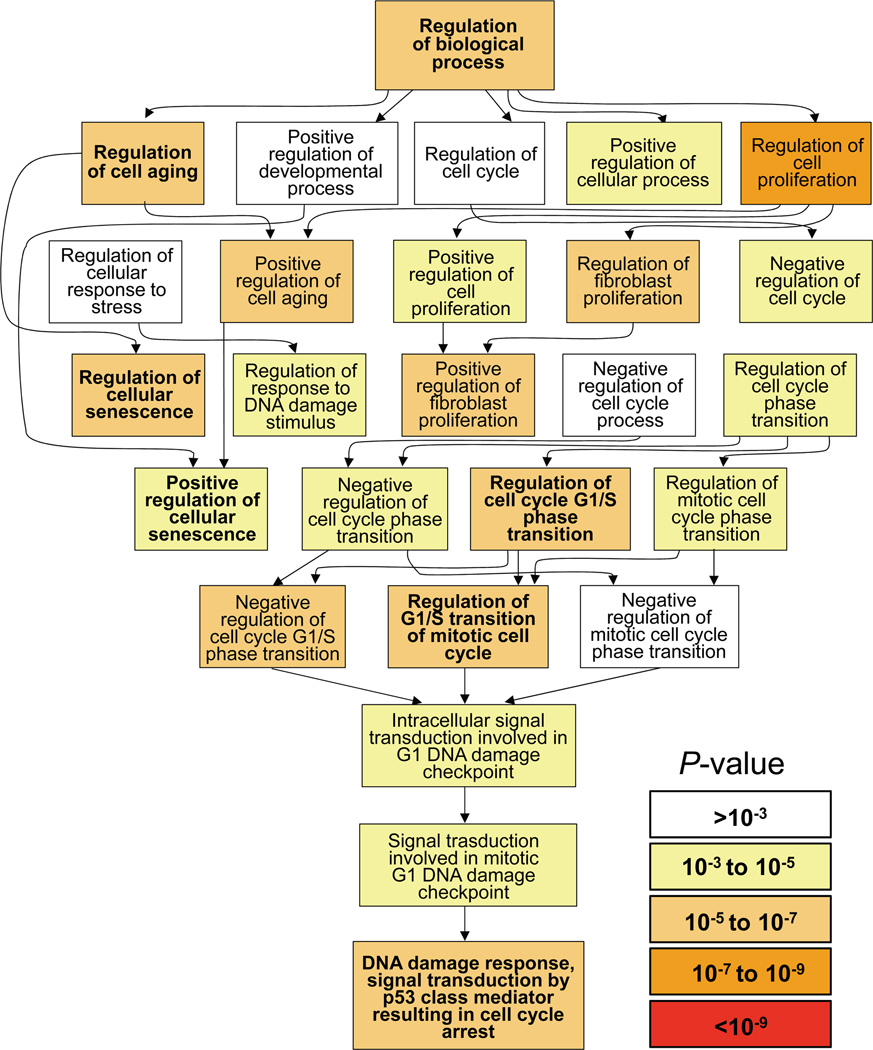
To document the biological processes that are associated with terminal cell cycle arrest in 8-oxoG-senesced cells, the list of differentially expressed genes (600 top-ranked out of 1116 genes based on GeneCard’s significance score >5) were submitted to Gene Ontology enrichment analysis and visualization tool (GOrilla) (Eden et al., 2009). Results show that the gene ontology (GO) categories involve regulation of cell aging (P = 9.11E-08), cell proliferation (P = 1.92E-09), positive regulation of cell aging (P = 8.53E-07) and regulation of cellular senescence (P = 3.98E07). The analysis also identified processes associated with regulation of cell cycle G1/S phase transition (P = 2.11E-06), and negative regulation of cell cycle G1/S phase transition (P = 8.96E-06). Although no significant increase was observed in level of DNA strand breaks (Fig. 3B) GO analysis identified processes that were regulated via intracellular signal transduction modulating G1 DNA damage check points (P = 5.11E-05). Accordingly, DNA damage response signal transduction by TP53 class mediators resulting in significant cell cycle arrest (P = 2.18E-05). Additional biological processes and their P-values are shown in Fig. 7 visually and numerically in Table 1 (SUPPLEMENTARY MATERIAL). GOrilla database-defined biological processes were consistent with 8-oxoG exposures-induced delay in G1-S phase of the cell cycle (Fig. 2K) and arrest of cells in the G1 phase (Fig. 2M). Furthermore, GO analysis supports our observations showing accumulation of TP53Ser15 and p16INK4a as well as p21WAF/CIP1 in nuclei of OGG1-expressing 8-oxoG-senesced cells (Fig. 6B,C,D). These data together strongly suggest that biological processes induced by OGG1-BER (mimicked via 8-oxoG exposures) play a role in terminal cell cycle arrest of cultured HDLF cells.
Figure 7.
Biological processes associated with 8-oxoG-induced senescence. A list of differentially expressed genes was submitted to GOrilla analysis data base for identifying and visualizing enriched GO terms. P-value is the proportion of group of genes modulated under 8-oxoG-induced senescence that are annotated to that GO term. Inset: P-value ranges. P-values of each biological process are numerically shown in Table 1 (SUPPLEMENTARY MATERIAL)
3.6. OGG1-BER → KRAS signaling induces aging-associated gene expression in airways
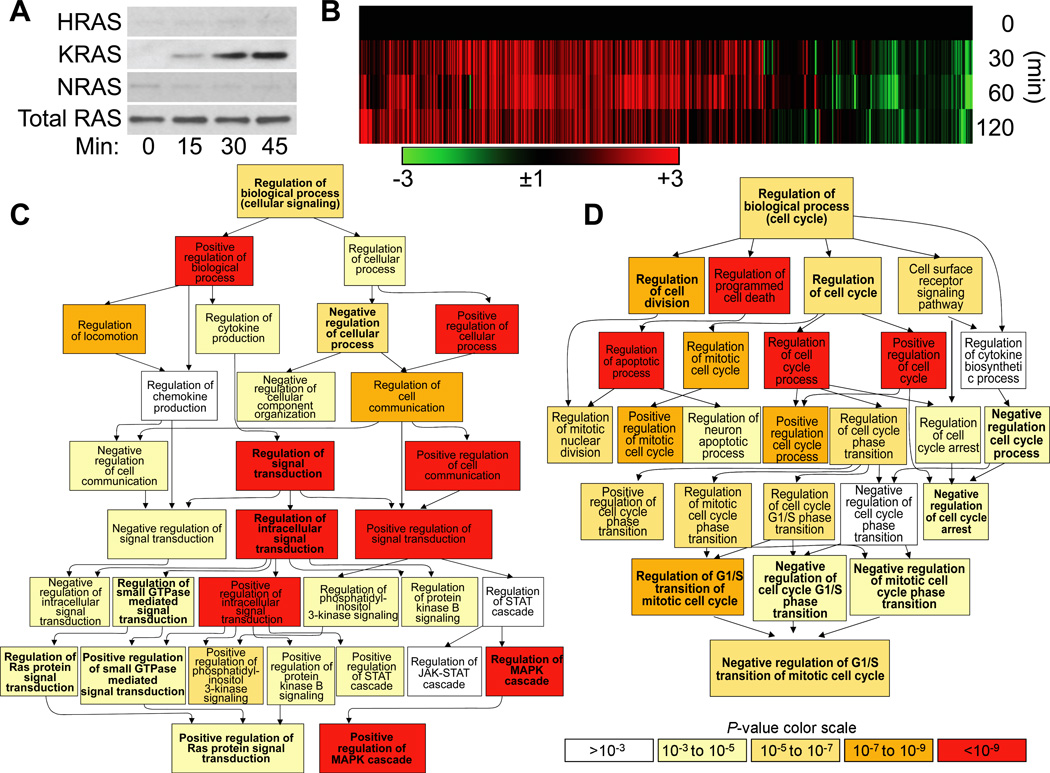
Exposure of lungs to environmental pollutants, changes in osmotic, temperature and oxygen levels, and oxidative burst by immune cells leads to 8-oxoG formation, consequently OGG1-BER. To test whether liberation of genomic 8-oxoG base caused aging-associated gene expression in lungs mouse airways were repeatedly challenged with 8-oxoG base (Materials and Methods). First, changes in activated ras family protein levels were examined. Results summarized in Fig. 7A shows a time-dependent increase in GTP-bound levels of KRAS, while activation of HRAS or NRAS was under detectable level in line with previous observations (Aguilera-Aguirre et al., 2014). Next whether 8-oxoG challenge-induced KRAS signaling in airways resembles that of aging-associated gene expression was also queried. RNAs were isolated at 0, 30, 60 and 120 min after challenge and subjected to RNA-sequencing and gene ontology analysis (Materials and Methods) (Aguilera-Aguirre et al., 2015; Aguilera-Aguirre, 2015).
RNA-sequencing analysis showed a rapid response (from 30 min on) at mRNA levels to 8-oxoG challenge. Utilizing gene identification programs and NCBI’s mouse data base (Materials and Methods) we identified 3252 differentially expressed transcripts (2435 up- and 817 down-regulated) including 2080 protein-coding mRNAs, 144 microRNAs, and 326 non-coding, full-length RNAs as we published previously (Aguilera-Aguirre, 2015). To identify genes associated with lung aging, a ranked list was generated from the GeneCards gene data base based on the key words “lung” and “aging.” The search generated a 4956-gene-ranked list. A minimum relevance score of 5 was used to render a top ranked list containing 1551 genes that had previously been linked to lung aging. This list was overlaid on RNA-sequencing data and fold changes in gene expressions were added. Out of 1551 (score 5 or higher; defined by GeneCards) 1,357 (462 were up-regulated (>3-fold) 825 were unchanged or modulated <3-fold) and 70 genes were down-regulated) were modulated by 8-oxoG challenge of airways containing genes (Fig. 7). To visualize the changes and expression patterns a heat maps was generated (Fig 7) by using GENE-E online software (Broad Institute) (Aguilera-Aguirre et al., 2014).
To examine the integrated roles of 1,357 genes induced by 8-oxoG challenge of airways in biological processes, they were submitted to GOrilla for gene ontology analysis (Eden et al., 2009). The GOrilla database defined top-ranked biological system processes in lungs as summarized in Table 2 (SUPPLEMENTARY MATERIAL). Examples of the most significantly modulated biological processes include negative regulation of the biological process (P= 2.70E-07), negative regulation of the developmental process (P = 1.84E-07), regulation of multicellular organismal process (P = 9.67E-08), negative regulation of cell motility (P = 7.21E-08), regulation of cell differentiation (P = 6.63E-08), negative regulation of cell differentiation (P = 5.42E-06) and others (Table 2, SUPPLEMENTARY MATERIAL). Among GOrilla GO database-defined biological processes for detailed analysis, we selected regulation of cell signaling (P = 1.37E-07) and regulation of cell cycle process (P = 3.96E-10).
According to GOrilla GO database positively/negatively regulated signal transduction pathways were mediated through small GTPases [e.g., regulation of Ras protein signal transduction (P = 4.52E-04); positive regulation of small GTPase mediated signal transduction (P= 1.73E-04)]. Down-stream from GTPases, highly significant changes were identified regarding positive regulation of phosphatidyl-inositol 3-kinase signaling (P = 6.60E-06), regulation of MAPK (P = 3.48E-11) and positive regulation of MAPK cascade (P = 4.95E-13) and others (Fig. 8C and Table 3 (SUPPLEMENTARY MATERIAL). Interestingly, Janus kinase/signal transducers (P = 3.88E-02) and activators of transcription (JAK/STAT) pathways (P = 3.88E-04) were only marginally significant. The signaling cascades generated in lungs by 8-oxoG challenge are in line with 8-oxoG-driven GEF activity of OGG1 and consequent activation of small GTPase KRAS (Aguilera-Aguirre et al., 2014; Boldogh et al., 2012).
Figure 8.
OGG1•8-oxoG-KRAS signaling-induced biological processes in airways. A, Increases in KRAS-GTP levels upon 8-oxoG challenge of lung. Lungs were challenged with 60 µl of 8-oxoG (1 µM) solution for 0, 15, 30 and 45 min, and extracts were prepared. GTP-bound RAS levels were determined by using active RAS pull down assays. Proteins were fractionated by SDS-PAGE and transferred, and RAS was detected by type-specific antibodies (Materials and Methods). B, 8-OxoG challenge induces gene expression consistent with aging in lungs. Lungs were repeatedly challenged on day 0, 2 and 4 with 8-oxoG base (60 µl of 8-oxoG; 10 µM solution). After the last challenge (4th day), RNAs were isolated at 0, 30, 60 and 120 min and subjected to RNA-seq (Materials and Methods). Then, 1357 gene products were identified by overlaying them on gene sets that had previously been associated with lung aging as defined by the GeneCards database. The selected genes along with their associated fold changes were heat mapped and hierarchically clustered using GENE-E online software (Aguilera-Aguirre, 2015). C,D, Regulation of biological processes induced by 8-oxoG exposure of lungs. The list of 1357 genes (up- down-regulated as well as unchanged) was submitted to the GOrilla data base to identify GO categories and their association with biological processes. Significance levels of processes are color-coded (D, lower panel). P-values of each biological process in D and C are numerically shown in Table 3 (SUPPLEMENTARY MATERIAL) and Table 4 (SUPPLEMENTARY MATERIAL)
GOrilla GO database generated images (Fig. 8D and Table 4 (SUPPLEMENTARY MATERIAL) showed biological processes [(regulation of cell cycle process (P = 3.96E-10)] modulated by 8-oxoG challenge of OGG1 expressing lungs. Top-ranked processes include positive regulation cell cycle process (P = 2.22E-08), regulation of cell cycle phase transition (P = 6.87E-06), regulation of mitotic cell cycle phase transition (P = 6.87E-06), regulation of G1/S transition of mitotic cell cycle (P = 2.21E-07), and negative regulation of G1/S transition of mitotic cell cycle (P = 7.53E-06). It is interesting to note that 8-oxoG challenge of airways mediated regulation of cell cycle process similar to that HDLF (shown in Fig. 7).
4. Discussion
It has long been suspected that DNA base and strand lesions have important etiological role in senescence, aging processes and age-associated diseases (Beckman and Ames, 1998; Hart and Setlow, 1974; Sohal et al., 2002; von Zglinicki et al., 2001). Although signaling by DNA strand breaks convincingly mediate senescence, the role of base lesions is however not fully understood. The most commonly observed DNA lesion in all types of aged is the oxidative modified guanine 8-oxoG. It is removed from the genome as a free base by OGG1-BER. Here we show that repeated activation of OGG1’ guanine nucleotide exchange factor activity by 8-oxoG base resulted in pre-matured senescence. Permanent OGG1 down-regulation extended life span of cells upon 8-oxoG exposures. Importantly, 8-oxoG exposed airways displayed differential gene expression, which are consistent with lung aging, These data point to a novel paradigm wherein life-long release of oxidatively-modified base 8-oxoG from DNA mediate differential gene expression, which wires cellular senescence/aging processes.
Repeated 8-oxoG exposure terminally arrested cells after 7-8 passages, while mock-(solvent) exposed cells needed 20-22 passage to reach senescence state. 8-oxoG exposures first decreased proliferative potential and subsequently cells entered a state of permanent growth arrest (at G1 phase of cell cycle), expressed SA β-Gal and accumulated lipofuscin as well as displayed morphological changes. Permanent growth arrest was not associated with accumulation of DNA strand breaks, increased frequency of mutations or shortening of telomeres; however, phenotypic cellular characteristics are fully satisfy the previously described “senescence” or Hayflick limit (Hayflick and Moorhead, 1961).
Studies have shown that DNA-damage signaling mediates arrest of cell proliferation and consequently inducing senescence (Jackson and Bartek, 2009; Sedelnikova et al., 2008; von Zglinicki et al., 2001). Indeed, in RS cells levels of DNA strand lesions were significantly increased and showed excessive telomere shortening. In contrast, 8-oxoG-senescence may not be linked to DNA damage signaling as cells did not show significant accumulation of DNA strand breaks or telomere length shortening. Although it requires additional studies these results could be explained by small GTPases-driven senescence as previously proposed (Benanti and Galloway, 2004; Campisi, 2005). Repeated addition of 8-oxoG to cells persistently increased GTP-bound levels of NRAS in HDLF cells, so it is feasible to propose that senescence is associated with continuous/intermittent unscheduled activation of small GTPases. In support, NRAS-depleted (but not K or HRAS) HDLF cells were partially refractory to 8-oxoG exposures-induced senescence. These results not only imply, but warrant contribution of other small GTPases-signaling in establishment of senescence phenotype. Indeed, 8-oxoG base-induced conformational changes in the OGG1 molecule allow its interaction with and activation of other small GTPases including RHO and RAC family GTPases (Hajas et al., 2013; Luo et al., 2014). There were profound changes observed in morphology of cells at proximity and in the state of permanent growth arrest implying changes in cytoskeleton that required actions of the small GTPase RAC1 and/or RHOA (Kaibuchi et al., 1999). Indeed, independent studies documented that 8-oxoG base exposure of cells increased activity of RAC1 and RHOA-GTPases in association with smooth muscle alpha-actin (α-SMA) polymerization into stress fibers, and thus increased the level of α-SMA in insoluble cellular/tissue fractions (Hajas et al., 2013; Luo et al., 2014).
To address the molecular mechanism by which the senescence state is developed and maintained in 8-oxoG-induced senescent cells unexpected increase were observed in the expressions of DNA damage-regulated genes, including TP53, p21WAF1 and p16INK4a. It is intriguing that expression of these key mediators of cell cycle progression is similar to those in RS cells. Our goal was to compare expression of potential mediators of cell cycle progression between 8-oxoG- and replicatively-senesced cells, therefore their kinetic activation were not examined. In line with expressions of DNA damage-regulated genes, a significant increase in DNA dsbs was observed only in RS cells. 8-OxoG-senesced cells DNA damage was similar to PDL-matched proliferating control cells. Thus one may propose that enhanced levels of phosphorylated TP53 and increased expression of p21WAF1 and p16INK4a in 8-oxoG-senesced cells could be due to activation of small GTPases. Expression of p19ARF, another functional regulator of TP53 was not investigated, but it is most likely to be activated as it has a role in the stabilization of TP53 by interfering with its negative regulator MDM2 (Palmero et al., 1998; Weber et al., 2000). Activated (stabilized) TP53 induced the expression of the p21WAF1, which can inhibit cyclin E- and A, thus promote cell cycle arrest shown previously (Benanti and Galloway, 2004; Bringold and Serrano, 2000; Zhu et al., 1998). In support, studies have shown that cells lacking expression of cyclin kinase inhibitors (p21WAF1 or p16INK4a) fail to undergo senescence in response to activated RAS (Benanti and Galloway, 2004; Zhu et al., 1998). Signaling by activated small GTPases of TP53 and cyclin kinase inhibitors is also well documented (Benanti and Galloway, 2004; Zhu et al., 1998). Thus increased levels of phosphorylated TP53, expression of p21WAF1 and p16INK4a in 8-oxoG-senesced cells can occur via OGG1’ GEF-induced RAS activation in lack of DNA strand damage. .
To address gene expression networks by which the senescence state is maintained as well as define the differences and similarities between 8-oxoG-senesced and RS cells, microarray analysis was performed. Intriguingly, 86% of the differentially expressed genes between 8-oxoG- and RS cells were similar. Sets of the differentially expressed genes were submitted for gene ontology analysis (GOrilla-data base). As examples, the top biological processes indentified were regulation of cell aging (P = 9.11E-8), positive regulation of cell aging (P = 8.53E-7), and negative regulation of cell cycle G1/S phase transition (P = 8.9E-6) in cells pre-maturely senesced by 8-oxoG exposures. Strikingly, nearly similar results were seen for RS-senesced cells. In both 8-oxoG- and RS-senesced cells positive regulation of cells aging and negative modulations of G1/S phase transition are in line with roles of phosphorylated TP53, and expression cyclin kinase inhibitors (p16INK4a p21WAF/CIP1). These results together suggest that signaling and molecular networks maintaining are similar to those in RS after repeated 8-oxoG exposures.
To extend our understanding on senescence/aging OGG1-initiated BER were mimicked by challenging lungs with low doses of 8-oxoG (0.0005 mg/kg body weight). We have chosen lungs as it forms a barrier between environment and vertebrate organisms and its decreased function impacts entire health e.g., correlates with poorer cardiovascular performance, cognitive activities and increased subcortical atrophy, and dementia in humans (Carvalhaes-Neto et al., 1995; Janssens, 2005). In addition, environmental pollutants, osmotic, mechanical stress, and oxidative burst by immune cells in airways leads to 8-oxoG formation, consequently OGG1-BER rev in (Ba et al., 2014; Ba et al., 2015; Bacsi et al., 2016; Radak and Boldogh, 2010). Challenge of lungs with 8-oxoG base robustly increases in GTP-bound levels of KRAS but not activation of HRAS or NRAS in line with previous observations (Aguilera-Aguirre et al., 2014). Studies also showed that KRAS-GTP binds to its effecter protein (Raf proto-oncogene homolog serine/threonine kinase kinase) and activate it, leading to phosphorylation of MAP and PI3 kinases, which are strongly interconnected and play a central role in inflammation, cell growth, differentiation and tumorigenesis. Indeed, analysis by RNA-sequencing showed that 8-oxoG challenge induced differential expression of 1551 genes, out of which 1357 are associated with senescence/aging processes.
Differential gene expression-driven top-ranked processes included regulation of biological processes, negative regulation of developmental processes, cell motility, and cell differentiation (P-values are highly significant). Intriguingly, GOrilla analysis also showed that differential gene expressions are regulated by small GTPase-signaling and down-stream kinases (PI3K, MAPK). To this end, recent studies revealed that OGG1-BER or when it is mimicked by 8-oxoG base challenge resulted in increased phosphorylation levels of MAPK, PI3K and IκB kinases in airways (Aguilera-Aguirre et al., 2014). Activation of these kinases were prerequisite events in induction of biological processes including positive/negative regulation of signal transduction, intracellular signal transduction, cell communication and others (Aguilera-Aguirre et al., 2015). Studies have also shown that OGG1·8-oxoG → small GTPases significantly upregulated signaling pathways including cadherin, integrin, Rho GTPase, transforming growth factor beta (TGFβ), and those modulated by wingless type (Wnt) family growth factors as well as cytokines/chemokines (P values are between 7.61E-04 and P = 7.61E-09) (Aguilera-Aguirre, 2015). These signaling pathways have previously been related to airway remodeling --well characterized changes in aged and diseased lungs (Chilosi et al., 2013; Ijaz et al., 2014). It has also been shown that Wnt-, TGFβ- and catenin-signaling pathways are key to lung parenchyma damage-repair, development, and control of cellular senescence as well as tissue remodeling (Cabrera-Benitez et al., 2012; Halder et al., 2012; Konigshoff et al., 2009).
It has been documented that OGG1·8-oxoG → small GTPases-induced signaling involved gene expression linked to regulation of immune-system, immunohomeostasis and macrophage functions, which were mediated by chemokines, cytokines, and interleukin signaling after 8-oxoG challenge (Aguilera-Aguirre et al., 2015). From these data one may propose that OGG1-BER signaling not only induce and maintain a senescence phenotype directly via activated small GTPases → TP53 and cyclin kinase inhibitors, but also through supra-physiological expression pro-inflammatory mediators. If it is proven to be correct maintenance of OGG1-BER-induced senescence is even more complex involving gene expression similar to that of senescence-associated (SA) “secretome” described previously (Campisi, 2005; Coppe et al., 2010). Because OGG1-BER is continuous process may maintain inflammatory state and contribute to progressive aging processes. Indeed, discrete clusters of senescence cells (SA secretomes) are documented in various pulmonary diseases, including asthma, COPD, emphysema and IPF (Campisi, 2005; Chilosi et al., 2013; Minagawa et al., 2011).
Attempt were made to compare senescence/aging-associated gene expression networks between HDLF and mouse lungs; although previous cross-species comparisons suggests only a potential relationship between senescence of cultured cells and organ aging or organismal life span, and the connection was neither quantitative nor direct (Campisi, 2001). However, we show that GO analysis of differentially expressed genes in both HDLF and lungs were related to regulation of cell cycle. The common, top-ranked biological processes in both were regulation of cell cycle G1/S phase transition, regulation of G1/S transition of mitotic cell cycle, and negative regulation of cell cycle G1/S phase transition. The difference between lung and cultured cells was that in HDLF the cell cycle, G1/S phase transition/arrest was primarily linked to signal transduction by TP53, while in lungs, GO analysis linked cell cycle and G1/S phase transition to signaling by RAS-PI3K and MAPK cascades. However, these results are not controversial as RAS-PI3K-MAPK pathways up regulate TP53 signal transduction. Based on these data one may propose that OGG1-BER could indeed be linked to aging processes at cellular and organ level. We note that increased OGG1 activity and release of 8-oxoG base from mitochondrial DNA of Oxys rats was previously associated with accelerated aging and progression of age-related diseases such as cataract, macular dystrophy, hypertension, osteoporosis, cognitive behavioral dysfunctions, lungs and liver pathologies (Kemeleva et al., 2006a; Kemeleva et al., 2006b).
In conclusion, efficient OGG1-driven DNA BER conventionally thought to maintain genome integrity, prevent mutagenesis, and increase heath-span via releasing the mutagenic 8-oxoG from the genome. However, current data suggest that generation of 8-oxoG base, unscheduled activation of small GTPases via OGG1’s GEF activity and downstream signaling, led to differential gene expression, consequently biological processes resulting in senescence and aging. It is well recognized that he senescence response is highly efficacious tumor suppressive mechanism. So it is feasible to propose that upon excessive stress the DNA repair protein OGG1 is also utilized to induce senescence in order to protect cells at risk from unwanted proliferation, malignant transformation and potential tumorigenesis.
Supplementary Material
HIGHLIGHTS.
-
□
Aging is genetically regulated, and is accelerated by stress from environment
-
□
Generation of genomic 8-oxoguanine and aging are parallel processes
-
□
OGG1-BER of 8-oxoG, fuels activation of small GTPases and gene expression
-
□
GO analyses of OGG1-BER-associated gene expression are relevant to senescence-aging
-
□
Pharmaceutical modulation of guanine oxidation in DNA should decelerate aging processes
Acknowledgments
This work was supported by grants NIA, AG21830 (IB,JP, SM), NIEHS RO1 ES018948 (IB), NIAID/AI062885 (IB), NINDS R01 NS088645 (MLH); NIEHS Center Grant P30 ES006676 (IB); National Science Foundation of China (No. 31571339 (XB), the European Union and the European Social Fund, TAMOP 4.2.2.A-11/1/KONV-2012-2023 (AB). L. Aguilera-Aguirre is an Environmental Toxicology Research Training Fellow (NIEHS T32 ES007254). We thank Dr. Linsey Jaeger (Department of Microbiology and Immunology, Institute of Human Infections and Immunity) for both for their scientific input and for critically editing the manuscript.
Abbreviations
- 8-oxoG
7,8-dihydro-8-oxoguanine
- 8-oxodG
7,8-dihydro-8-oxo-2′-deoxyguanosine
- BER
base excision repair
- DNA dsbs
double strand breaks
- DNA ssbs
single strand breaks
- GEF
guanine nucleotide exchange factor
- GOrilla
Gene Ontology enrichment analysis and visualization data base
- HDLF
human diploid lung fibroblast cells
- HRAS
mammalian homolog of Harvey rat sarcoma viral oncogene
- KRAS
mammalian homolog of Kirsten rat sarcoma virus oncogene
- MAPK
mitogen activated kinase
- MEF
murine embryonic diploid fibroblast
- NRAS
neuroblastoma RAS viral oncogene homolog
- OGG1
8-oxoguanine-DNA glycosylase1
- OGG1-BER
OGG1-initiated DNA base excision repair
- PDL
population doubling levels
- PI3K
phosphoinositide 3-kinase
- ROS
reactive oxygen species
- RS
replicative senescence
- CASAVA
consensus assessment of sequence and variation
- GENE-E
is a matrix visualization and analysis platform
- GEO
gene expression omnibus
- GO
gene ontology
- PANTHER
protein analysis through evolutionary relationships
- RPKM
reads per kilobase of transcript per million
- RNA-Seq
RNA sequencing
REFERENCES
- Aguilera-Aguirre L, Bacsi A, Radak Z, Hazra TK, Mitra S, Sur S, Brasier AR, Ba X, Boldogh I. Innate inflammation induced by the 8-oxoguanine DNA glycosylase-1-KRAS-NF-kappaB pathway. J Immunol. 2014;193:4643–4653. doi: 10.4049/jimmunol.1401625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera-Aguirre L, Bacsi A, Saavedra-Molina A, Kurosky A, Sur S, Boldogh I. Mitochondrial dysfunction increases allergic airway inflammation. Journal of Immunology. 2009;183:5379–5387. doi: 10.4049/jimmunol.0900228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera-Aguirre L, Hosoki K, Bacsi A, Radak Z, Wood TG, Widen SG, Sur S, Ameredes BT, Saavedra-Molina A, Brasier AR, Ba X, Boldogh I. Whole transcriptome analysis reveals an 8-oxoguanine DNA glycosylase-1-driven DNA repair-dependent gene expression linked to essential biological processes. Free Radic Biol Med. 2015;81:107–118. doi: 10.1016/j.freeradbiomed.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera-Aguirre L, Hosoki K, Bacsi A, Radák Zs, Sur S, Hegde ML, Tian B, Saavedra-Molina A, Brasier AR, Ba X, Boldogh I. Whole transcriptome analysis reveals the implications of OGG1-intitiated DNA repair signaling in airway remodeling. Free Rad. Biol. Med. 2015 doi: 10.1016/j.freeradbiomed.2015.07.007. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari M, Krokan HE. Cytotoxicity and mutagenicity of endogenous DNA base lesions as potential cause of human aging. Mech Ageing Dev. 2008;129:353–365. doi: 10.1016/j.mad.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Albrecht T, Fons MP, Deng CZ, Boldogh I. Increased frequency of specific locus mutation following human cytomegalovirus infection. Virology. 1997;230:48–61. doi: 10.1006/viro.1997.8467. [DOI] [PubMed] [Google Scholar]
- Ba X, Aguilera-Aguirre L, Rashid QT, Bacsi A, Radak Z, Sur S, Hosoki K, Hegde ML, Boldogh I. The role of 8-oxoguanine DNA glycosylase-1 in inflammation. Int J Mol Sci. 2014;15:16975–16997. doi: 10.3390/ijms150916975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ba X, Aguilera-Aguirre L, Sur S, Boldogh I. 8-Oxoguanine DNA glycosylase-1-driven DNA base excision repair: role in asthma pathogenesis. Curr Opin Allergy Clin Immunol. 2015;15:89–97. doi: 10.1097/ACI.0000000000000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacsi A, Chodaczek G, Hazra TK, Konkel D, Boldogh I. Increased ROS generation in subsets of OGG1 knockout fibroblast cells. Mech Ageing Dev. 2007;128:637–649. doi: 10.1016/j.mad.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacsi A, Pan L, Ba X, Boldogh I. Pathophysiology of bronchoconstriction: role of oxidatively damaged DNA repair. Curr Opin Allergy Clin Immunol. 2016;16:59–67. doi: 10.1097/ACI.0000000000000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman KB, Ames BN. The free radical theory of aging matures. Physiol Rev. 1998;78:547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- Benanti JA, Galloway DA. The normal response to RAS: senescence or transformation? Cell Cycle. 2004;3:715–717. [PubMed] [Google Scholar]
- Boldogh I, Bacsi A, Choudhury BK, Dharajiya N, Alam R, Hazra TK, Mitra S, Goldblum RM, Sur S. ROS generated by pollen NADPH oxidase provide a signal that augments antigen-induced allergic airway inflammation. J Clin Invest. 2005;115:2169–2179. doi: 10.1172/JCI24422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh I, Hajas G, Aguilera-Aguirre L, Hegde ML, Radak Z, Bacsi A, Sur S, Hazra TK, Mitra S. Activation of ras signaling pathway by 8-oxoguanine DNA glycosylase bound to its excision product-8-oxoguanine. J Biol Chem. 2012;287:20769–20773. doi: 10.1074/jbc.C112.364620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasier AR. The NF-kappaB regulatory network. Cardiovasc Toxicol. 2006;6:111–130. doi: 10.1385/ct:6:2:111. [DOI] [PubMed] [Google Scholar]
- Bresnahan WA, Boldogh I, Thompson EA, Albrecht T. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology. 1996;224:150–160. doi: 10.1006/viro.1996.0516. [DOI] [PubMed] [Google Scholar]
- Bringold F, Serrano M. Tumor suppressors and oncogenes in cellular senescence. Exp Gerontol. 2000;35:317–329. doi: 10.1016/s0531-5565(00)00083-8. [DOI] [PubMed] [Google Scholar]
- Cabrera-Benitez NE, Parotto M, Post M, Han B, Spieth PM, Cheng WE, Valladares F, Villar J, Liu M, Sato M, Zhang H, Slutsky AS. Mechanical stress induces lung fibrosis by epithelial-mesenchymal transition. Crit Care Med. 2012;40:510–517. doi: 10.1097/CCM.0b013e31822f09d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J. From cells to organisms: can we learn about aging from cells in culture? Exp Gerontol. 2001;36:607–618. doi: 10.1016/s0531-5565(00)00230-8. [DOI] [PubMed] [Google Scholar]
- Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- Carvalhaes-Neto N, Lorino H, Gallinari C, Escolano S, Mallet A, Zerah F, Harf A, Macquin-Mavier I. Cognitive function and assessment of lung function in the elderly. Am J Respir Crit Care Med. 1995;152:1611–1615. doi: 10.1164/ajrccm.152.5.7582303. [DOI] [PubMed] [Google Scholar]
- Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30:e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- Chen Q, Fischer A, Reagan JD, Yan LJ, Ames BN. Oxidative DNA damage and senescence of human diploid fibroblast cells. Proc Natl Acad Sci U S A. 1995;92:4337–4341. doi: 10.1073/pnas.92.10.4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SK, Hsieh WA, Tsai MH, Chen CC, Hong AI, Wei YH, Chang WP. Age-associated decrease of oxidative repair enzymes, human 8-oxoguanine DNA glycosylases (hOgg1), in human aging. J Radiat Res (Tokyo) 2003;44:31–35. doi: 10.1269/jrr.44.31. [DOI] [PubMed] [Google Scholar]
- Chilosi M, Carloni A, Rossi A, Poletti V. Premature lung aging and cellular senescence in the pathogenesis of idiopathic pulmonary fibrosis and COPD/emphysema. Transl Res. 2013;162:156–173. doi: 10.1016/j.trsl.2013.06.004. [DOI] [PubMed] [Google Scholar]
- David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslee G, Woods JC, Moore C, Conradi SH, Gierada DS, Atkinson JJ, Battaile JT, Liu L, Patterson GA, Adair-Kirk TL, Holtzman MJ, Pierce RA. Oxidative damage to nucleic acids in severe emphysema. Chest. 2009;135:965–974. doi: 10.1378/chest.08-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianov GL, Souza-Pinto N, Nyaga SG, Thybo T, Stevnsner T, Bohr VA. Base excision repair in nuclear and mitochondrial DNA. Prog Nucleic Acid Res Mol Biol. 2001;68:285–297. doi: 10.1016/s0079-6603(01)68107-8. [DOI] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dizdaroglu M. Formation of an 8-hydroxyguanine moiety in deoxyribonucleic acid on gamma-irradiation in aqueous solution. Biochemistry. 1985;24:4476–4481. doi: 10.1021/bi00337a032. [DOI] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajas G, Bacsi A, Aguilera-Aguirre L, Hegde ML, Tapas KH, Sur S, Radak Z, Ba X, Boldogh I. 8-Oxoguanine DNA glycosylase-1 links DNA repair to cellular signaling via the activation of the small GTPase Rac1. Free Radic Biol Med. 2013;61:384–394. doi: 10.1016/j.freeradbiomed.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajas G, Bacsi A, Aguilerra-Aguirre L, German P, Radak Z, Sur S, Hazra TK, Boldogh I. Biochemical identification of a hydroperoxide derivative of the free 8-oxo-7,8-dihydroguanine base. Free Radic Biol Med. 2012;52:749–756. doi: 10.1016/j.freeradbiomed.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, Dupont S, Piccolo S. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat Rev Mol Cell Biol. 2012;13:591–600. doi: 10.1038/nrm3416. [DOI] [PubMed] [Google Scholar]
- Hamilton ML, Van Remmen H, Drake JA, Yang H, Guo ZM, Kewitt K, Walter CA, Richardson A. Does oxidative damage to DNA increase with age? Proc Natl Acad Sci U S A. 2001;98:10469–10474. doi: 10.1073/pnas.171202698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara MR, Kovacs JJ, Whalen EJ, Rajagopal S, Strachan RT, Grant W, Towers AJ, Williams B, Lam CM, Xiao K, Shenoy SK, Gregory SG, Ahn S, Duckett DR, Lefkowitz RJ. A stress response pathway regulates DNA damage through beta2-adrenoreceptors and beta-arrestin-1. Nature. 1996;477:349–353. doi: 10.1038/nature10368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart RW, Setlow RB. Correlation between deoxyribonucleic acid excision-repair and life-span in a number of mammalian species. Proc Natl Acad Sci U S A. 1974;71:2169–2173. doi: 10.1073/pnas.71.6.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- Igishi T, Hitsuda Y, Kato K, Sako T, Burioka N, Yasuda K, Sano H, Shigeoka Y, Nakanishi H, Shimizu E. Elevated urinary 8-hydroxydeoxyguanosine, a biomarker of oxidative stress, and lack of association with antioxidant vitamins in chronic obstructive pulmonary disease. Respirology. 2003;8:455–460. doi: 10.1046/j.1440-1843.2003.00490.x. [DOI] [PubMed] [Google Scholar]
- Ijaz T, Pazdrak K, Kalita M, Konig R, Choudhary S, Tian B, Boldogh I, Brasier AR. Systems biology approaches to understanding Epithelial Mesenchymal Transition (EMT) in mucosal remodeling and signaling in asthma. World Allergy Organ J. 2014;7:13. doi: 10.1186/1939-4551-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens JP. Aging of the respiratory system: impact on pulmonary function tests and adaptation to exertion. Clin Chest Med. 2005;26:469–484. doi: 10.1016/j.ccm.2005.05.004. vi-vii. [DOI] [PubMed] [Google Scholar]
- Kaibuchi K, Kuroda S, Amano M. Regulation of the cytoskeleton and cell adhesion by the Rho family GTPases in mammalian cells. Annu Rev Biochem. 1999;68:459–486. doi: 10.1146/annurev.biochem.68.1.459. [DOI] [PubMed] [Google Scholar]
- Kemeleva EA, Sinitsyna OI, Conlon KA, Berrios M, Kolosova NG, Zharkov DO, Vasyunina EA, Nevinsky GA. Oxidation of guanine in liver and lung DNA of prematurely aging OXYS rats. Biochemistry (Mosc) 2006a;71:612–618. doi: 10.1134/s0006297906060046. [DOI] [PubMed] [Google Scholar]
- Kemeleva EA, Sinitsyna OI, Kolosova NG, Vasyunina EA, Zharkov DO, Conlon KA, Berrios M, Nevinsky GA. Immunofluorescent detection of 8-oxoguanine DNA lesions in liver cells from aging OXYS rats, a strain prone to overproduction of free radicals. Mutat Res. 2006b;599:88–97. doi: 10.1016/j.mrfmmm.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, Barnes DE. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc Natl Acad Sci U S A. 1999;96:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konigshoff M, Kramer M, Balsara N, Wilhelm J, Amarie OV, Jahn A, Rose F, Fink L, Seeger W, Schaefer L, Gunther A, Eickelberg O. WNT1-inducible signaling protein-1 mediates pulmonary fibrosis in mice and is upregulated in humans with idiopathic pulmonary fibrosis. J Clin Invest. 2009;119:772–787. doi: 10.1172/JCI33950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek K, Passos JF, Olijslagers S, Saretzki G, Martin-Ruiz C, von Zglinicki T. Premature senescence of mesothelial cells is associated with non-telomeric DNA damage. Biochem Biophys Res Commun. 2007;362:707–711. doi: 10.1016/j.bbrc.2007.08.047. [DOI] [PubMed] [Google Scholar]
- Lindahl T, Barnes DE. Repair of endogenous DNA damage. Cold Spring Harb Symp Quant Biol. 2000;65:127–133. doi: 10.1101/sqb.2000.65.127. [DOI] [PubMed] [Google Scholar]
- Lovell MA, Markesbery WR. Oxidative damage in mild cognitive impairment and early Alzheimer's disease. J Neurosci Res. 2007;85:3036–3040. doi: 10.1002/jnr.21346. [DOI] [PubMed] [Google Scholar]
- Luo J, Hosoki K, Bacsi A, Radak Z, Hegde ML, Sur S, Hazra TK, Brasier AR, Ba X, Boldogh I. 8-Oxoguanine DNA glycosylase-1-mediated DNA repair is associated with Rho GTPase activation and alpha-smooth muscle actin polymerization. Free Radic Biol Med. 2014;73:430–438. doi: 10.1016/j.freeradbiomed.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minagawa S, Araya J, Numata T, Nojiri S, Hara H, Yumino Y, Kawaishi M, Odaka M, Morikawa T, Nishimura SL, Nakayama K, Kuwano K. Accelerated epithelial cell senescence in IPF and the inhibitory role of SIRT6 in TGF-beta-induced senescence of human bronchial epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2011;300:L391–L401. doi: 10.1152/ajplung.00097.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minowa O, Arai T, Hirano M, Monden Y, Nakai S, Fukuda M, Itoh M, Takano H, Hippou Y, Aburatani H, Masumura K, Nohmi T, Nishimura S, Noda T. Mmh/Ogg1 gene inactivation results in accumulation of 8-hydroxyguanine in mice. Proc Natl Acad Sci U S A. 2000;97:4156–4161. doi: 10.1073/pnas.050404497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S, Izumi T, Boldogh I, Bhakat KK, Hill JW, Hazra TK. Choreography of oxidative damage repair in mammalian genomes. Free Radic Biol Med. 2002;33:15–28. doi: 10.1016/s0891-5849(02)00819-5. [DOI] [PubMed] [Google Scholar]
- Mosteller RD, Han J, Broek D. Identification of residues of the H-ras protein critical for functional interaction with guanine nucleotide exchange factors. Mol Cell Biol. 1994;14:1104–1112. doi: 10.1128/mcb.14.2.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura J, Mutlu E, Sharma V, Collins L, Bodnar W, Yu R, Lai Y, Moeller B, Lu K, Swenberg J. The endogenous exposome. DNA Repair (Amst) 2014;19:3–13. doi: 10.1016/j.dnarep.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omerovic J, Hammond DE, Clague MJ, Prior IA. Ras isoform abundance and signalling in human cancer cell lines. Oncogene. 2008;27:2754–2762. doi: 10.1038/sj.onc.1210925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterod M, Hollenbach S, Hengstler JG, Barnes DE, Lindahl T, Epe B. Age-related and tissue-specific accumulation of oxidative DNA base damage in 7,8-dihydro-8-oxoguanine-DNA glycosylase (Ogg1) deficient mice. Carcinogenesis. 2001;22:1459–1463. doi: 10.1093/carcin/22.9.1459. [DOI] [PubMed] [Google Scholar]
- Palmero I, Pantoja C, Serrano M. p19ARF links the tumour suppressor p53 to Ras. Nature. 1998;395:125–126. doi: 10.1038/25870. [DOI] [PubMed] [Google Scholar]
- Papaconstantinou J. Unifying model of the programmed (intrinsic) and stochastic (extrinsic) theories of aging. The stress response genes, signal transduction-redox pathways and aging. Ann N Y Acad Sci. 1994;719:195–211. doi: 10.1111/j.1749-6632.1994.tb56829.x. [DOI] [PubMed] [Google Scholar]
- Papaconstantinou J. Insulin/IGF-1 and ROS signaling pathway cross-talk in aging and longevity determination. Mol Cell Endocrinol. 2009;299:89–100. doi: 10.1016/j.mce.2008.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello S, Samper E, Krtolica A, Goldstein J, Melov S, Campisi J. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat Cell Biol. 2003;5:741–747. doi: 10.1038/ncb1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proklou A, Soulitzis N, Neofytou E, Rovina N, Zervas E, Gaga M, Siafakas NM, Tzortzaki EG. Granule cytotoxic activity and oxidative DNA damage in smoking and nonsmoking patients with asthma. Chest. 2013;144:1230–1237. doi: 10.1378/chest.13-0367. [DOI] [PubMed] [Google Scholar]
- Radak Z, Boldogh I. 8-Oxo-7,8-dihydroguanine: links to gene expression, aging, and defense against oxidative stress. Free Radic Biol Med. 2010;49:587–596. doi: 10.1016/j.freeradbiomed.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodier F, Coppe JP, Patil CK, Hoeijmakers WA, Munoz DP, Raza SR, Freund A, Campeau E, Davalos AR, Campisi J. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol. 2009;11:973–979. doi: 10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakumi K, Tominaga Y, Furuichi M, Xu P, Tsuzuki T, Sekiguchi M, Nakabeppu Y. Ogg1 knockout-associated lung tumorigenesis and its suppression by Mth1 gene disruption. Cancer Res. 2003;63:902–905. [PubMed] [Google Scholar]
- Sampath H, Vartanian V, Rollins MR, Sakumi K, Nakabeppu Y, Lloyd RS. 8-Oxoguanine DNA glycosylase (OGG1) deficiency increases susceptibility to obesity and metabolic dysfunction. PLoS One. 2012;7:e51697. doi: 10.1371/journal.pone.0051697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedelnikova OA, Horikawa I, Redon C, Nakamura A, Zimonjic DB, Popescu NC, Bonner WM. Delayed kinetics of DNA double-strand break processing in normal and pathological aging. Aging Cell. 2008;7:89–100. doi: 10.1111/j.1474-9726.2007.00354.x. [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- Sohal RS, Agarwal S, Candas M, Forster MJ, Lal H. Effect of age and caloric restriction on DNA oxidative damage in different tissues of C57BL/6 mice. Mech Ageing Dev. 1994;76:215–224. doi: 10.1016/0047-6374(94)91595-4. [DOI] [PubMed] [Google Scholar]
- Sohal RS, Mockett RJ, Orr WC. Mechanisms of aging: an appraisal of the oxidative stress hypothesis. Free Radic Biol Med. 2002;33:575–586. doi: 10.1016/s0891-5849(02)00886-9. [DOI] [PubMed] [Google Scholar]
- Steenken S, Jovanovic SV. How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J. Am. Chem. Soc. 1997;119:617–618. [Google Scholar]
- Szaniszlo P, German P, Hajas G, Saenz DN, Woodberry MW, Kruzel ML, Boldogh I. Effects of Colostrinin on gene expression-transcriptomal network analysis. Int Immunopharmacol. 2009;9:181–193. doi: 10.1016/j.intimp.2008.10.022. [DOI] [PubMed] [Google Scholar]
- Szczesny B, Hazra TK, Papaconstantinou J, Mitra S, Boldogh I. Age-dependent deficiency in import of mitochondrial DNA glycosylases required for repair of oxidatively damaged bases. Proc Natl Acad Sci U S A. 2003;100:10670–10675. doi: 10.1073/pnas.1932854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SJ, Resnick RJ, Shalloway D. Nonradioactive determination of Ras-GTP levels using activated ras interaction assay. Methods Enzymol. 2001;333:333–342. doi: 10.1016/s0076-6879(01)33067-7. [DOI] [PubMed] [Google Scholar]
- Vlahopoulos SA, Cen O, Hengen N, Agan J, Moschovi M, Critselis E, Adamaki M, Bacopoulou F, Copland JA, Boldogh I, Karin M, Chrousos GP. Dynamic aberrant NF-kappaB spurs tumorigenesis: a new model encompassing the microenvironment. Cytokine Growth Factor Rev. 2015;26:389–403. doi: 10.1016/j.cytogfr.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Zglinicki T, Burkle A, Kirkwood TB. Stress, DNA damage and ageing -- an integrative approach. Exp Gerontol. 2001;36:1049–1062. doi: 10.1016/s0531-5565(01)00111-5. [DOI] [PubMed] [Google Scholar]
- Weber JD, Jeffers JR, Rehg JE, Randle DH, Lozano G, Roussel MF, Sherr CJ, Zambetti GP. p53-independent functions of the p19(ARF) tumor suppressor. Genes Dev. 2000;14:2358–2365. doi: 10.1101/gad.827300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman L, Jo DG, Sorensen MM, de Souza-Pinto NC, Markesbery WR, Mattson MP, Bohr VA. Defective DNA base excision repair in brain from individuals with Alzheimer's disease and amnestic mild cognitive impairment. Nucleic Acids Res. 2007;35:5545–5555. doi: 10.1093/nar/gkm605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DM, 3rd, Bohr VA. The mechanics of base excision repair, and its relationship to aging and disease. DNA Repair (Amst) 2007;6:544–559. doi: 10.1016/j.dnarep.2006.10.017. [DOI] [PubMed] [Google Scholar]
- Wilson DM, 3rd, Bohr VA, McKinnon PJ. DNA damage, DNA repair, ageing and age-related disease. Mech Ageing Dev. 2008;129:349–352. doi: 10.1016/j.mad.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Woods D, McMahon M, Bishop JM. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev. 1998;12:2997–3007. doi: 10.1101/gad.12.19.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.