Abstract
Background
Targeted therapy with anti-human epidermal growth factor receptor-2 (HER2) monoclonal antibody in patients with HER2 overexpressed esophagogastric adenocarcinoma (EGA) improves survival; however, the effect is transient due to the development of resistance. Some studies suggest that cMet overexpression provides cross talk for epidermal growth factor receptor (EGFR) and HER2 inhibition. We sought to characterize the expression profile of the EGFR family and cMet receptors in untreated, resected EGA.
Methods
This retrospective analysis included all sequential patients with esophageal or gastroesophageal junction (GEJ) adenocarcinoma who underwent primary resection, without neoadjuvant therapy or HER2 inhibition, with adequate tissue, at the University of Florida from 2001 to 2011. Central blinded immunohistochemistry (IHC) was performed on tumor specimens with EGFR, HER2, HER3, HER4 and cMet expression scored as low (0, 1+) or high (2+, 3+). Demographic and tumor characteristics were compared using Fisher exact test. Kaplan-Meier curves and univariate analysis compared survival among different receptors.
Results
Total 52 patients were included in the study with median age 66 years. High expression of EGFR (73%), HER2 (40%), HER3 (75%), HER4 (35%) and cMet (69%) was detected among the study group. HER3 and HER4 co-expression was found in 18 (35%) cases. Pan expression of all four EGFR family members with cMet was noted in only 17% of cases. On univariate analysis, tumor stage and depth correlated with survival, while cMet + HER3 +/– EGFR receptor co-expression trended towards a worse survival.
Conclusions
EGFR family and cMet are frequently co-expressed in treatment naïve resected EGA or GEJ tumors. Although our data do not significantly show receptor status as a prognostic factor, the co-expression profiles support for further investigation to improve targeting of this signal transduction axis.
Keywords: Esophageal cancer, gastric cancer, esophagogastric adenocarcinoma (EGA), epidermal growth factor receptor (EGFR), hepatocyte growth factor receptor (cMet), human epidermal growth factor receptor-2 (HER2), HER3, HER4
Introduction
An estimated 16,980 new cases of and 5,590 deaths from esophageal cancer are in the United States in 2016 (1). At the time of diagnosis, only 30–40% of patients have potentially resectable tumor while most patients have locally advanced or metastatic disease (2-4). Surgical resection following neoadjuvant chemoradiotherapy has significantly improved the outcomes of resectable esophagogastric adenocarcinoma (EGA) resulting in a 5-year survival rate of about 40% compared to surgical resection alone, where the 5-year survival rate was reported to be only 20% (5-7).
The epidermal growth factor receptor (EGFR) pathway has been widely studied in EGA (8-10) with a family consisting of four receptors (EGFR, HER2, HER3 and HER4). Survival is improved when targeting the human epidermal growth factor receptor-2 (HER2) with the monoclonal antibody trastuzumab in combination with chemotherapy in metastatic EGA overexpressing HER2 (11). However, the treatment benefit is limited to only a minority whose tumors overexpress HER2 and subsequent acquired resistance to this therapy follows. Recent data suggested that a prominent mechanism of resistance to EGFR inhibition is through down-stream crosstalk and signaling via the hepatocyte growth factor receptor (cMet) or other EGFR family member (12-15). The cMet proto-oncogene encodes a receptor tyrosine kinase which signals through the mitogen-activated protein kinase (MAPK) pathway (16). Studies have demonstrated cMet overexpression in some digestive cancers as being associated with a poor prognosis, inferior outcomes and more advanced disease (17-19). Moreover, studies have shown that EGFR enhances cMet mediated proliferation and invasion of epithelial cells and cMet can synergize with HER2 to promote a malignant phenotype (20). Therapeutic inhibition of cMet is currently an area of active clinical investigation. Here in, we characterize the co-expression profiles of the EGFR family members and cMet receptor in a cohort of untreated EGA.
Methods
We conducted a retrospective analysis of all sequential patients with distal esophageal or gastroesophageal junction (GEJ) adenocarcinoma who underwent primary tumor resection at the University of Florida from 2001 to 2011 without prior neoadjuvant therapy or HER2 inhibition and with adequate remnant tissue for analysis. Clinical and pathological data were obtained from the medical records of the patients under the approval of UF Institutional Review Board (IRB). Clinical characteristics were correlated with cellular proliferation and expression profiles.
Patient specimens and IHC staining of HER family, cMet and Ki67
All available H&E-stained slides of surgical specimens were reviewed by gastrointestinal pathologists and representative paraffin blocks with adequate tumor for each case were selected for IHC processing. All IHC staining was performed using standard HRP polymer procedure with positive and negative controls. Details of the reagents, suppliers and processing specifications are summarized in Table 1. Importantly, given the current lack of clinical standard established for cMet expression, both Leica and Ventana IHC analyses were performed.
Table 1. Specifications of the reagents and antibodies used in this study.
| Antibody | Clone/code | Company | Dilution | Antigen retrieval |
|---|---|---|---|---|
| EGFR | D3881 | Cell Signaling Technology Danvers, MA | 1:100 | Trilogy; water bath (95 °C 25 min) |
| HER2 | E2-4001/Cat#AH01011 | Invitrogen Camarillo, CA | 1:400 | Citra; steamer 30 min |
| HER3 | 2F12/Cat#05-390 | EMD Millipore Billerica, MA | 1:100 | CC1 30 min |
| HER4 | Cat#PA5-16789 | Thermo Fisher Scientific Rockford, IL | 1:100 | Citra; steamer 30 min |
| cMet (Ventana) | SP44/Cat#790-4430 | Ventana Medical Systems Tucson, AZ | Prediluted | CC1 30 min |
| cMet (Leica) | 8F11/NCL-cMet | Leica Biosystems Newcastle, United Kingdom | 1:80 | Citra; steamer 30 min |
| Ki67 | M1B-1/Cat#M7240 | DAKO Carpinteria, CA | 1:250 | Trilogy; water bath (95 °C 25 min) |
EGFR, epidermal growth factor receptor; HER, human epidermal growth factor receptor; cMet, hepatocyte growth factor receptor.
IHC Scoring and interpretation
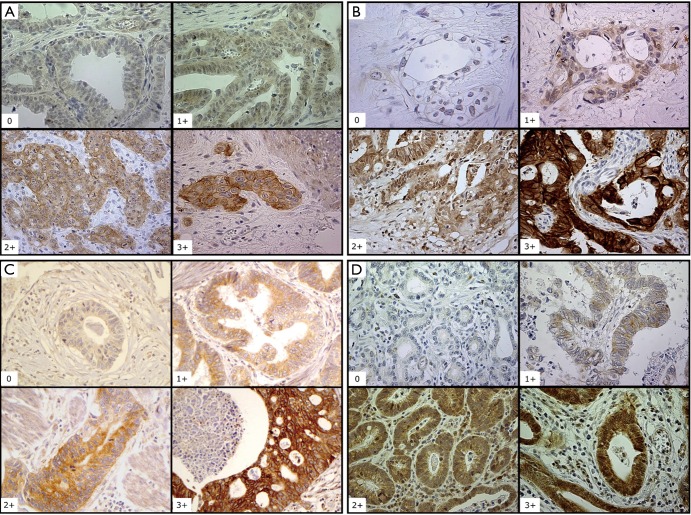
A weighted histoscore method, the Hscore system, was used for EGFR, HER3 and HER4 IHC expression scoring. Histoscores were calculated from the sum of (1 × % cells staining weakly positive) + (2 × % cells staining moderately positive) + (3 × % cells staining strongly positive) with a maximum score of 300. Scores were classified into low or negative (histoscore 0–100), intermediate (histoscore 101–200) or high (histoscore 201–300) (21). Representative histochemical staining of EGFR, HER3 and HER4 are shown in Figure 1.
Figure 1.
Immunohistochemistry analysis and expression of EGFR, HER2, HER3, HER4 in esophagogastric adenocarcinoma (EGA). (A) EGFR, epidermal growth factor receptor staining (100×); (B) HER2, human epidermal growth factor receptor-2 (100× and 400×); (C) HER3, human epidermal growth factor receptor-3 (100×); (D) HER4, human epidermal growth factor receptor-4 (100×); Staining: 0, negative; 1+, weak; 2+, moderate; 3+, strong positivity. The first two photos in (B) (0 and 1+) are 400×, the second two (2+ and 3+) are 100×.
For HER2, the standard clinical scoring system validated for gastric cancer was used (22). Negative (score 0, 1+) meant no reactivity or membranous reactivity (staining) in <10% of invasive tumor cells or faint/barely perceptible membranous reactivity (staining) in ≥10% of invasive tumor cells; cells are reactive (stained) only in part of their membrane (Figure 1). Equivocal (score of 2+) meant weak to moderate complete, basolateral or lateral membranous reactivity (staining) in ≥10% of invasive tumor cells. Positive (score of 3+) meant strong complete, basolateral or lateral membranous reactivity (staining) in ≥10% of invasive tumor cells.
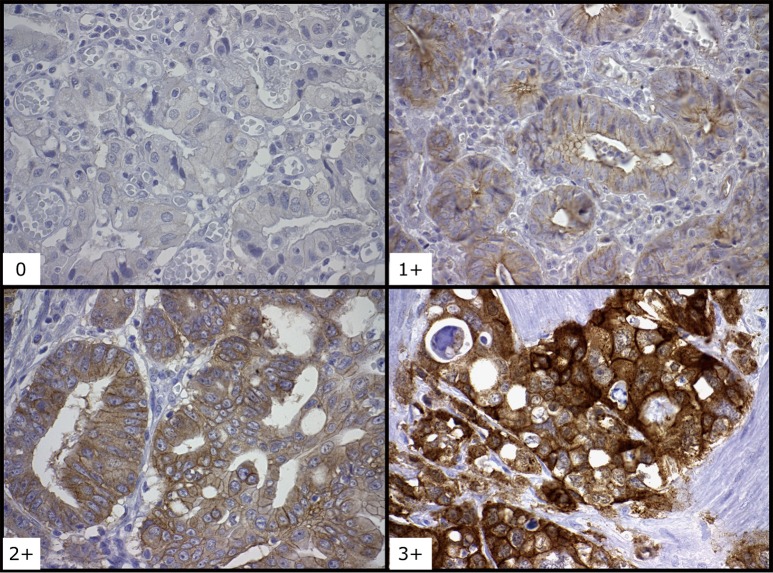
Currently no clinically validated guideline exists for the interpretation and scoring of cMet immunohistochemistry (IHC) in EGA. For our study, we used two different commercially available IHC antibodies (cMet Leica and cMet Ventana). For both antibodies, cMet staining was scored using a four step scale: negative (0), weak (1+), moderate (2+) or strong (3+) positivity (Figure 2). Consistent with published data, if tumor cells stained moderately or strongly positive in more than 30% of tumor cells were considered to be positive (23). The analysis and interpretation of all tissue stains were independently reviewed by three different GI pathologists (TZT, LVD, AA).
Figure 2.
Immunohistochemistry analysis of hepatocyte growth factor receptor (cMet) expression in esophagogastric adenocarcinoma (EGA). Staining: 0, negative; 1+, weak; 2+, moderate; 3+, strong positivity. Magnification is 400×.
Statistical analysis
Descriptive statistics (mean ± SD for continuous variables and proportion for categorical variables) were used for summarizing demographic clinical and pathological characteristics as well as molecular expression profiles. Molecular receptor expressions were coded into dichotomous variables by combining categories into no or weak staining (low) vs. moderate to strong staining (high). Chi-square exact test was used to test the association of demographic and clinicopathological features with receptor expression. Fisher’s exact test was used for measuring the association between receptor (or combined receptor) expressions. Overall survival (OS) curves were compared by log-rank test and the corresponding cumulative survival rates were estimated using Kaplan-Meier method. Univariate analyses were performed to investigate the unadjusted and adjusted association, respectively, of each demographic and clinicopathological factor, and each receptor (or combined receptor) expression with the OS. All statistical analyses were performed using SAS 9.4.
Results
A total of 52 patients were included in this analysis (Table 2) with mean age of 66±10.0 years and BMI of 29.8±6.8. Of these patients, 42 (81%) were men, 38 (79%) were current or former smokers, 41 (93%) had associated Barrett’s esophagus, 30 (58%) had stage I disease and 41 (79%) of patients remained disease free at a median follow up of 3 years.
Table 2. Clinical and pathological characteristics of patients.
| Characteristics | Frequency (N=52) |
|---|---|
| Age (mean ± SD) | 66±10.0 |
| BMI (mean ± SD) | 30±6.8 |
| Male [%] | 42 [81] |
| Stage [%] | |
| I | 30 [58] |
| IIA | 9 [17] |
| IIB | 6 [12] |
| III | 7 [13] |
| Depth of invasion (T) [%] | |
| T1 | 31 [60] |
| T2 | 8 [15] |
| T3 | 10 [19] |
| T4 | 3 [6] |
| Nodal involvement [%] | 11 [21] |
| Tumor differentiation [%] | |
| Well | 4 [8] |
| Moderate | 26 [50] |
| Poor | 13 [25] |
| Unknown | 9 [17] |
| Barrett’s esophagus [%] | 41 [93] |
| Smoking [%] | |
| Current | 9 [19] |
| Former | 29 [60] |
| Never | 10 [21] |
| Recurrence [%] | |
| Yes | 6 [11] |
| No | 41[79] |
| Unknown | 5 [10] |
Receptor expression profiles of EGFR family and cMet
Receptor expression and co-expression profiles are described in Table 3 and Figure 3. High expression of EGFR (73%), HER2 (40%), HER3 (75%), HER4 (35%) and cMet (69%) were detected in this patient sample cohort. Co-expression profiles also demonstrated several common combinations including individual EGFR family members with each other or cMet. Pan expression of all four EGFR family members with cMet was noted in 17% of samples. Among EGFR family member, HER3 and HER4 co-expression was found in 35% of cases.
Table 3. Expression (co)profiles of the EGFR family and cMet receptors in esophagogastric adenocarcinoma.
| Receptor(s) | Frequency (N) | Positivity (%) |
|---|---|---|
| EGFR | 38 | 73 |
| HER2 | 21 | 40 |
| HER3 | 39 | 75 |
| HER4 | 18 | 35 |
| cMet | 36 | 69 |
| EGFR + HER2 | 17 | 33 |
| EGFR + HER3 | 29 | 56 |
| EGFR + HER4 | 15 | 29 |
| EGFR + cMet | 28 | 54 |
| HER2 + HER3 | 18 | 35 |
| HER2 + HER4 | 10 | 19 |
| HER2 + cMet | 15 | 29 |
| HER3 + HER4 | 18 | 35 |
| HER3 + cMet | 27 | 52 |
| HER4 + cMet | 14 | 27 |
| EGFR + HER2 + HER3 + HER4 + cMet | 9 | 17 |
EGFR, epidermal growth factor receptor; HER, human epidermal growth factor receptor; cMet, hepatocyte growth factor receptor.
Figure 3.
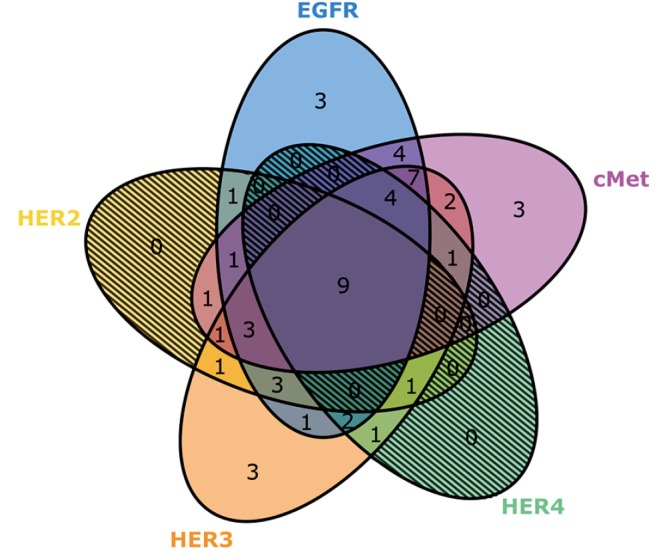
High expression relationships among all EGFR family members and cMet. Numbers reflect incidence of relative expression profiles from entire cohort (n=52). EGFR, epidermal growth factor receptor; HER, human epidermal growth factor receptor; cMet, hepatocyte growth factor receptor.
Comparison of cMet Leica vs. cMet Ventana antibody
While 27 tumors stained positive for cMet using either antibody, concordance of positive staining using both antibodies was present in only 16 cases (59% concordance rate). This inconsistency was despite appropriate positive and negative control verification and repeat testing. Therefore, all results regarding cMet expression throughout this report are based upon the cMet Ventana antibody, unless otherwise specified.
Receptor expression and clinicopathological factors
No significant association was detected between any demographic or clinicopathological factors with receptor expressions or co-expressions, except for, smoking status, depth of invasion and tumor differentiation (data not shown). Lower T stage had a non-significant trend associated with higher expressions of all receptors.
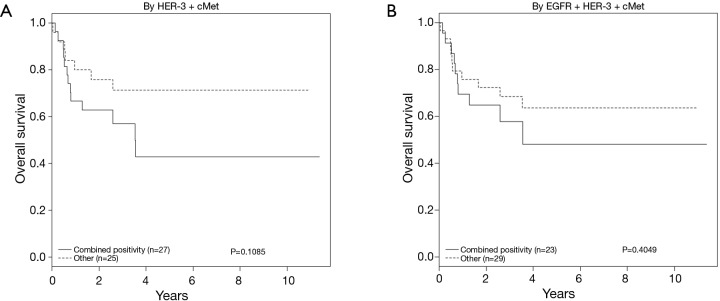
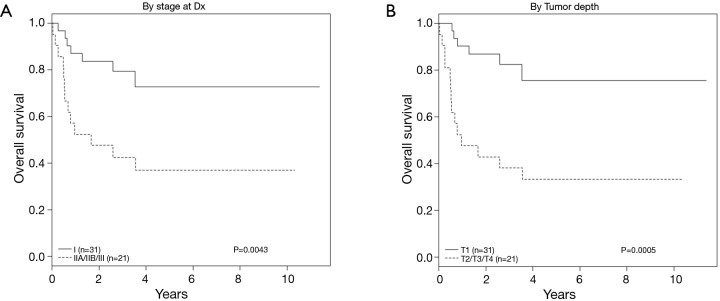
As expected, univariate analysis confirmed survival was independently worse for patients with higher cancer stage (HR =3.52; 95% CI, 1.40–8.85; P=0.004) and depth of invasion (HR =4.71; 95% CI, 1.80–12.03; P≤0.001) (Figure 4, Table 4). Tumors that co-expressed cMet + HER3 with or without EGFR had a non-statistically significant trend towards a worse survival (Figure 5).
Figure 4.
HER3, EGFR and cMet receptor pairs with trend towards worse overall survival (OS). HER, human epidermal growth factor receptor; EGFR, epidermal growth factor receptor; cMet, hepatocyte growth factor receptor.
Table 4. Univariate analysis of clinicopathologic and molecular marker expression on survival.
| Variable | HR | 95% CI of HR | P value |
|---|---|---|---|
| Age | 1.02 | 0.97–1.06 | 0.516 |
| BMI | 0.98 | 0.89–1.06 | 0.566 |
| Male gender | 0.97 | 0.32–2.92 | 0.954 |
| Stage at Dx | 0.004* | ||
| Stage I | Reference | – | |
| Stage IIA/IIB/III | 3.52 | 1.40–8.85 | |
| Tumor depth | <0.001* | ||
| T1 | Reference | – | |
| T2/T3/T4 | 4.71 | 1.80–12.03 | |
| Smoking status | 0.421 | ||
| Never | Reference | – | |
| Current | 2.14 | 0.51–9.02 | |
| Former | 1.11 | 0.30–4.13 | |
| Barrett esophagus | 1.14 | 0.15–8.73 | 0.900 |
| EGFR | 0.86 | 0.33–2.26 | 0.766 |
| HER2 | 0.84 | 0.33–2.13 | 0.718 |
| HER3 | 1.13 | 0.41–3.10 | 0.818 |
| HER4 | 1.14 | 0.46–2.87 | 0.776 |
| cMet | 1.44 | 0.52–3.96 | 0.484 |
| HER3+cMet | 2.01 | 0.83–5.28 | 0.108 |
| EGFR+HER3+cMet | 1.45 | 0.60–3.50 | 0.404 |
*, statistically significant (P<0.05). HR, hazard ratio; EGFR, epidermal growth factor receptor; HER, human epidermal growth factor receptor; cMet, hepatocyte growth factor receptor.
Figure 5.
Overall survival (OS) by tumor stage and depth of invasion.
Discussion
Members of the EGFR family are overexpressed in EGA and represent druggable targets (8-11). Signal transduction pathways are activated through ligand binding and dimerization with the same (homodimerization) or a different (heterodimerization) EGFR family member (23). Thus, the expression pattern of EGFR family members has biologic and treatment resistance implications. Our study importantly focused on tumors that were surgically resected and that were previously untreated. This offers both a strength of ensuring adequate tissue was present in the specimen to avoid sampling errors associated with tumor heterogeneity, but also limited our study to those with an earlier stage of disease. It is unclear if similar results would have been seen in patients with more advanced disease or in those whom an endoscopic biopsy or FNA was used to assess these biomarkers.
In our study of previously untreated tumors, we noted a relatively high expression of EGFR receptor (73%). Previous studies have reported EGFR overexpression in 27–55% of esophageal and GEJ adenocarcinomas (24-27). Wang et al. demonstrated EGFR overexpression in 32% of patients and was correlated with higher tumor stage, lymph node metastasis, but failed to show correlation with shorter disease-free (DFS) or OS, which is in accordance with the results of our study (28). Thus, our high EGFR expression profile may simply represent the relative earlier stages of cancers that underwent primary surgical resection as their initial treatment in our cohort. HER2 was overexpressed in 40% of cases in our study which is similar to that reported in the literature (10–44%) (29). Although the prognostic significance of HER2 overexpression in EGA is controversial, our study failed to demonstrate an association between HER2 overexpression and survival, potentially as a result of sample size (30).
HER3 was found to be overexpressed in 75% of cases which is slightly higher than that shown by Ocana and colleagues (34–59%) (31). Although not demonstrated in our study, prior association of HER3 expression with poor prognosis is an interesting finding since HER3 does not have an intracellular tyrosine kinase domain. Thus, the biologic impact of HER3 co-expression may be related more to its ability to heterodimerize with other EGFR family members thus stimulating downstream growth and signaling pathways (31,32). In fact, our study showed that HER3 was frequently co-expressed with other EGFR family members. In a retrospective analysis by Jácome et al. of 201 patients with esophageal junction and gastric cancer who underwent primary resection, HER2 and HER3 expression were significantly correlated (33). The findings corroborated by our study may have therapeutic importance in EGA given that the addition of pertuzumab—a drug that inhibits HER2-HER3 heterodimerization—to trastuzumab in the treatment of HER2-positive breast cancer improved outcomes (34,35). This suggests that the concept of inhibiting HER3 dimerization may be a potential therapeutic target in the treatment of HER2 overexpressing EGA.
Overexpression of HER4 was noted to be 35% in our study. This receptor was previously suggested to be non-prognostic but knowledge on HER4 expression in EGA is very limited. Begnami et al. found that although HER4 expression was observed in 41% of gastric cancers, only expression of HER2 and HER3 were associated with poor survival (36). We demonstrated that the co-expression of HER3 and HER4 at 35% was statistically significant (P=0.019), suggesting that again, HER3 may be acting as a preferred dimerization partner for EGFR family members. Indeed, Hayashi and colleagues demonstrated that HER3 membranous expression was also significantly correlated with HER4 expression and associated with tumor progression, higher depth of tumor invasion (T1 vs. T2–T4) involved lymph nodes, distant metastasis tumor stage, recurrences and worse survival (37). We noted overexpression of all EGFR family members in only 17% of the cases. These results highlight the potential inherent complex interactions among different receptors in tumors without prior exposure to chemotherapy or radiation.
In addition to EGFR family members, we evaluated cMet expression profiles. Overexpression of cMet detected by IHC is seen in 40–70% of gastric adenocarcinoma and in 50–80% of EGA. Inhibition of cMet in EGA preclinical models resulted in decreased cell motility, viability and invasion of tumor cells (38,39). Our study showed that cMet was overexpressed in 69% of patients but was not associated with a poor prognosis. Determination of cMet expression was challenging and discordance was noted between the different commercially available antibodies for use in IHC. This inconsistency was despite rigorous quality control, usage of CAP-certified processes and facilities and repeated verification. Given that there is no consistent commercial standard for cMet quantification, our findings suggest that great care should be taken in the use of cMet IHC for purposes of predictive biomarker determination.
Despite these limitations, cMet demonstrated a trend towards worse survival when co-expressed with HER3, which is in alignment with the theory posed of HER3 serving as an important biologic partner for aberrant signal transduction and cellular growth. HER3 has been purported to serve an essential role in gastric cancer proliferation, particularly when heterodimerized with HER2 and/or cMet (40). Clinically, co-amplification of cMet with HER2 was associated with poor prognosis and treatment resistance (41). Tuynman and colleagues showed that cMet expression was found in 54% of EGA which was associated with a significantly shorter 5-year disease specific and OS with an increased likelihood to develop both local recurrences and distant metastases (42). Similarly, Lennerz et al. demonstrated that EGA tumors harboring cMet overexpression were associated with high-grade histology, advanced stages and had shorter OS (7.1 vs. 16.2 months) (43). Importantly, they reported 2 patients with cMet-amplified tumors who experienced encouraging tumor shrinkage and improvement in symptoms when treated with the cMet inhibitor crizotinib.
Although HER2 inhibition provides a survival benefit in advanced EGA, the benefits can be short lived for many patients, suggesting that inhibition of one target may not be sufficient for durable benefit (44,45). Indeed, monotherapy with anti-EGFR monoclonal antibodies have failed to prove clinically beneficial. Our study adds to the growing literature regarding the complexity of the EGFR family members and associated signaling pathways which may support treatment resistance, but also offer clues into the rational development of multi targeted therapy.
Conclusions
Our comprehensive expression profile of EGFR family members demonstrate that they are commonly co-expressed with or without cMet. HER3, in particular, appears to play a prominent role in co-expression patterns suggesting an important biologic function in EGA. Our study, taken together with other published reports detailing the biologic function of the EGFR and cMet pathways in EGAs, further supports the need for the rational development of clinical trials to test multi-targeted therapeutic strategies in this disease.
Acknowledgements
Funding: This work was supported by the Gatorade Trust through funds distributed by the University of Florida, Department of Medicine and by NIH (NCATS) CTSA grant UL1TR000064.
Ethical Statement: Clinical and pathological data were obtained from the medical records of the patients under the approval of UF Institutional Review Board (IRB).
Footnotes
Conflicts of Interest: Data was presented in part at the 2013 American Society of Clinical Oncology and 2014 Gastrointestinal Cancers Symposia Annual Meetings.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015;65:5-29. 10.3322/caac.21254 [DOI] [PubMed] [Google Scholar]
- 2.Brown LM, Devesa SS, Chow WH. Incidence of adenocarcinoma of the esophagus among white Americans by sex, stage, and age. J Natl Cancer Inst 2008;100:1184-7. 10.1093/jnci/djn211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corley DA, Kubo A, Zhao W. Abdominal obesity and the risk of esophageal and gastric cardia carcinomas. Cancer Epidemiol Biomarkers Prev 2008;17:352-8. 10.1158/1055-9965.EPI-07-0748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kubo A, Corley DA. Body mass index and adenocarcinomas of the esophagus or gastric cardia: a systematic review and meta-analysis. Cancer Epidemiol Biomarkers Prev 2006;15:872-8. 10.1158/1055-9965.EPI-05-0860 [DOI] [PubMed] [Google Scholar]
- 5.Chang AC, Ji H, Birkmeyer NJ, et al. Outcomes after transhiatal and transthoracic esophagectomy for cancer. Ann Thorac Surg 2008;85:424-9. 10.1016/j.athoracsur.2007.10.007 [DOI] [PubMed] [Google Scholar]
- 6.Tepper J, Krasna MJ, Niedzwiecki D, et al. Phase III trial of trimodality therapy with cisplatin, fluorouracil, radiotherapy, and surgery compared with surgery alone for esophageal cancer: CALGB 9781. J Clin Oncol 2008;26:1086-92. 10.1200/JCO.2007.12.9593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Hagen P, Hulshof MCCM, van Lanschot JJ, et al. Preoperative chemoradiotherapy for esophageal or junctional cancer. N Engl J Med 2012;366:2074-84. 10.1056/NEJMoa1112088 [DOI] [PubMed] [Google Scholar]
- 8.Dragovich T, McCoy S, Fenoglio-Preiser CM, et al. Phase II trial of erlotinib in gastroesophageal junction and gastric adenocarcinomas: SWOG 0127. J Clin Oncol 2006;24:4922-7. 10.1200/JCO.2006.07.1316 [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez CP, Adelstein DJ, Rice TW, et al. A phase II study of perioperative concurrent chemotherapy, gefitinib, and hyperfractionated radiation followed by maintenance gefitinib in locoregionally advanced esophagus and gastroesophageal junction cancer. J Thorac Oncol 2010;5:229-35. 10.1097/JTO.0b013e3181c5e334 [DOI] [PubMed] [Google Scholar]
- 10.Crosby T, Hurt CN, Falk S, et al. Chemoradiotherapy with or without cetuximab in patients with oesophageal cancer (SCOPE1): a multicentre, phase 2/3 randomised trial. Lancet Oncol 2013;14:627-37. 10.1016/S1470-2045(13)70136-0 [DOI] [PubMed] [Google Scholar]
- 11.Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet 2010;376:687-97. 10.1016/S0140-6736(10)61121-X [DOI] [PubMed] [Google Scholar]
- 12.Lai AZ, Abella JV., Park M. Crosstalk in Met receptor oncogenesis. Trends Cell Biol 2009;19:542-51. 10.1016/j.tcb.2009.07.002 [DOI] [PubMed] [Google Scholar]
- 13.Shattuck DL, Miller JK, Carraway KL, 3rd, et al. Met receptor contributes to trastuzumab resistance of Her2-overexpressing breast cancer cells. Cancer Res 2008;68:1471-7. 10.1158/0008-5472.CAN-07-5962 [DOI] [PubMed] [Google Scholar]
- 14.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007;316:1039-43. 10.1126/science.1141478 [DOI] [PubMed] [Google Scholar]
- 15.Leto SM, Sassi F, Catalano I, et al. Sustained Inhibition of HER3 and EGFR Is Necessary to Induce Regression of HER2-Amplified Gastrointestinal Carcinomas. Clin Cancer Res 2015;21:5519-31 10.1158/1078-0432.CCR-14-3066 [DOI] [PubMed] [Google Scholar]
- 16.Karamouzis MV., Konstantinopoulos PA, Papavassiliou AG. Targeting MET as a strategy to overcome crosstalk-related resistance to EGFR inhibitors. Lancet Oncol 2009;10:709-17. 10.1016/S1470-2045(09)70137-8 [DOI] [PubMed] [Google Scholar]
- 17.Kammula US, Kuntz EJ, Francone TD, et al. Molecular co-expression of the c-Met oncogene and hepatocyte growth factor in primary colon cancer predicts tumor stage and clinical outcome. Cancer Lett 2007;248:219-28. 10.1016/j.canlet.2006.07.007 [DOI] [PubMed] [Google Scholar]
- 18.Lagarde SM, ten Kate FJ, Reitsma JB, et al. Prognostic factors in adenocarcinoma of the esophagus or gastroesophageal junction. J Clin Oncol 2006;24:4347-55. 10.1200/JCO.2005.04.9445 [DOI] [PubMed] [Google Scholar]
- 19.Miyamoto M, Ojima H, Iwasaki M, et al. Prognostic significance of overexpression of c-Met oncoprotein in cholangiocarcinoma. Br J Cancer 2011;105:131-8. 10.1038/bjc.2011.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khoury H, Naujokas MA, Zuo D, et al. HGF converts ErbB2/Neu epithelial morphogenesis to cell invasion. Mol Biol Cell 2005;16:550-61. 10.1091/mbc.E04-07-0567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirkegaard T, Edwards J, Tovey S, et al. Observer variation in immunohistochemical analysis of protein expression, time for a change? Histopathology 2006;48:787-94. 10.1111/j.1365-2559.2006.02412.x [DOI] [PubMed] [Google Scholar]
- 22.Hofmann M, Stoss O, Shi D, et al. Assessment of a HER2 scoring system for gastric cancer: results from a validation study. Histopathology 2008;52:797-805. 10.1111/j.1365-2559.2008.03028.x [DOI] [PubMed] [Google Scholar]
- 23.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer 2005;5:341-54. 10.1038/nrc1609 [DOI] [PubMed] [Google Scholar]
- 24.Wilkinson NW, Black JD, Roukhadze E, et al. Epidermal growth factor receptor expression correlates with histologic grade in resected esophageal adenocarcinoma. J Gastrointest Surg 2004;8:448-53. 10.1016/j.gassur.2004.01.006 [DOI] [PubMed] [Google Scholar]
- 25.Kim MA, Lee HS, Lee HE, et al. EGFR in gastric carcinomas: prognostic significance of protein overexpression and high gene copy number. Histopathology 2008;52:738-46. 10.1111/j.1365-2559.2008.03021.x [DOI] [PubMed] [Google Scholar]
- 26.Langer R, Von Rahden BH, Nahrig J, et al. Prognostic significance of expression patterns of c-erbB-2, p53, p16INK4A, p27KIP1, cyclin D1 and epidermal growth factor receptor in oesophageal adenocarcinoma: a tissue microarray study. J Clin Pathol 2006;59:631-4. 10.1136/jcp.2005.034298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L, Sun J, Zhang JQ, et al. Expression and significance of molecular biomarkers in esophageal carcinoma in different nationalities patients in Xinjiang. Genet Mol Res 2014;13:5413-25. 10.4238/2014.July.24.21 [DOI] [PubMed] [Google Scholar]
- 28.Wang KL, Wu T-T, Choi IS, et al. Expression of epidermal growth factor receptor in esophageal and esophagogastric junction adenocarcinomas: association with poor outcome. Cancer 2007;109:658-67. 10.1002/cncr.22445 [DOI] [PubMed] [Google Scholar]
- 29.Okines A, Cunningham D, Chau I. Targeting the human EGFR family in esophagogastric cancer. Nat Rev Clin Oncol 2011;8:492-503. 10.1038/nrclinonc.2011.45 [DOI] [PubMed] [Google Scholar]
- 30.Chan E, Duckworth LV, Alkhasawneh A, et al. HER2 expression and response to neoadjuvant chemoradiotherapy in esophagogastric adenocarcinoma. J Gastrointest Oncol 2016;7:173-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ocana A, Vera-Badillo F, Seruga B, et al. HER3 overexpression and survival in solid tumors: A meta-analysis. J Natl Cancer Inst 2013;105:266-73. 10.1093/jnci/djs501 [DOI] [PubMed] [Google Scholar]
- 32.Holbro T, Beerli RR, Maurer F, et al. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc Natl Acad Sci U S A 2003;100:8933-8. 10.1073/pnas.1537685100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jácome AA, Wohnrath DR, Scapulatempo Neto C, et al. Prognostic value of epidermal growth factor receptors in gastric cancer: a survival analysis by Weibull model incorporating long-term survivors. Gastric Cancer 2014;17:76-86. 10.1007/s10120-013-0236-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baselga J, Cortés J, Kim SB, et al. Pertuzumab plus Trastuzumab plus Docetaxel for Metstatic Breast Cancer. N Engl J Med 2012;366:109-19. 10.1056/NEJMoa1113216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamashita-Kashima Y, Iijima S, Yorozu K, et al. Pertuzumab in combination with trastuzumab shows significantly enhanced antitumor activity in HER2-positive human gastric cancer xenograft models. Clin Cancer Res 2011;17:5060-70. 10.1158/1078-0432.CCR-10-2927 [DOI] [PubMed] [Google Scholar]
- 36.Begnami MD, Fukuda E, Fregnani JH, et al. Prognostic implications of altered human epidermal growth factor receptors (HERs) in gastric carcinomas: HER2 and HER3 are predictors of poor outcome. J Clin Oncol 2011;29:3030-6. 10.1200/JCO.2010.33.6313 [DOI] [PubMed] [Google Scholar]
- 37.Hayashi M, Inokuchi M, Takagi Y, et al. High expression of HER3 is associated with a decreased survival in gastric cancer. Clin Cancer Res 2008;14:7843-9. 10.1158/1078-0432.CCR-08-1064 [DOI] [PubMed] [Google Scholar]
- 38.Herrera LJ, El-Hefnawy T, Queiroz de Oliveira PE, et al. The HGF receptor c-Met is overexpressed in esophageal adenocarcinoma. Neoplasia 2005;7:75-84. 10.1593/neo.04367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watson GA, Zhang X, Stang MT, et al. Inhibition of c-Met as a therapeutic strategy for esophageal adenocarcinoma. Neoplasia 2006;8:949-55. 10.1593/neo.06499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yun C, Gang L, Rongmin G, et al. Essential role of Her3 in two signaling transduction patterns: Her2/Her3 and MET/Her3 in proliferation of human gastric cancer. Mol Carcinog 2015;54:1700-9. 10.1002/mc.22241 [DOI] [PubMed] [Google Scholar]
- 41.Kwak EL, Ahronian LG, Siravegna G, et al. Molecular Heterogeneity and Receptor Coamplification Drive Resistance to Targeted Therapy in MET-Amplified Esophagogastric Cancer. Cancer Discov 2015;5:1271-81. 10.1158/2159-8290.CD-15-0748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tuynman JB, Lagarde SM, Ten Kate FJ, et al. Met expression is an independent prognostic risk factor in patients with oesophageal adenocarcinoma. Br J Cancer 2008;98:1102-8. 10.1038/sj.bjc.6604251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lennerz JK, Kwak EL, Ackerman A, et al. MET amplification identifies a small and aggressive subgroup of esophagogastric adenocarcinoma with evidence of responsiveness to crizotinib. J Clin Oncol 2011;29:4803-10. 10.1200/JCO.2011.35.4928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lordick F, Kang YK, Chung HC, et al. Capecitabine and cisplatin with or without cetuximab for patients with previously untreated advanced gastric cancer (EXPAND): A randomised, open-label phase 3 trial. Lancet Oncol 2013;14:490-9. 10.1016/S1470-2045(13)70102-5 [DOI] [PubMed] [Google Scholar]
- 45.Waddell T, Chau I, Cunningham D, et al. Epirubicin, oxaliplatin, and capecitabine with or without panitumumab for patients with previously untreated advanced oesophagogastric cancer (REAL3): a randomised, open-label phase 3 trial. Lancet Oncol 2013;14:481-9. 10.1016/S1470-2045(13)70096-2 [DOI] [PMC free article] [PubMed] [Google Scholar]