Abstract
Background
Recent genome-wide association studies have identified genetic loci that jointly make a considerable contribution to risk of developing Alzheimer’s disease (AD). Because neuropathological features of AD can be present several decades before disease onset, we investigated whether effects of polygenic risk are detectable by neuroimaging in young adults. We hypothesized that higher polygenic risk scores (PRSs) for AD would be associated with reduced volume of the hippocampus and other limbic and paralimbic areas. We further hypothesized that AD PRSs would affect the microstructure of fiber tracts connecting the hippocampus with other brain areas.
Methods
We analyzed the association between AD PRSs and brain imaging parameters using T1-weighted structural (n = 272) and diffusion-weighted scans (n = 197).
Results
We found a significant association between AD PRSs and left hippocampal volume, with higher risk associated with lower left hippocampal volume (p = .001). This effect remained when the APOE gene was excluded (p = .031), suggesting that the relationship between hippocampal volume and AD is the result of multiple genetic factors and not exclusively variability in the APOE gene. The diffusion tensor imaging analysis revealed that fractional anisotropy of the right cingulum was inversely correlated with AD PRSs (p = .009). We thus show that polygenic effects of AD risk variants on brain structure can already be detected in young adults.
Conclusions
This finding paves the way for further investigation of the effects of AD risk variants and may become useful for efforts to combine genotypic and phenotypic data for risk prediction and to enrich future prevention trials of AD.
Keywords: Alzheimer’s disease, Cingulum, Fornix, Hippocampus, Imaging, Polygenic risk
Alzheimer’s disease (AD) is the most common neurodegenerative disease, affecting about 5% to 7% of the population over 60 years of age (1). Although a small proportion of cases, often with a younger onset, are caused by autosomal dominant mutations, the vast majority of cases do not follow Mendelian heritability. Such sporadic AD is mediated by both environmental and genetic factors, with many genes contributing different degrees of risk (2). The most highly penetrant common genetic risk factor for AD is the apolipoprotein E(APOE) ε4 allele, each copy of which increases AD risk by a factor of about 3 (3). However, recent genome-wide association studies (GWASs) have identified a further 19 genome-wide significant loci for AD (4, 5, 6), which provide new insights into possible biological mechanisms underlying the neurodegenerative process (7). Individually, the most powerful of these variants only marginally increase an individual’s risk for developing AD (~1% to 8%) (4). Polygenic risk scores (PRSs), which are based on the additive effect of multiple loci across the genome, may be better suited to capture the variance explained by common alleles (8). PRSs based on the most recent GWASs have considerable predictive utility for AD risk (9).
Structural brain imaging has consistently revealed both global and local atrophic changes in patients with AD (10) and is a useful biomarker for preclinical disease (11). Local atrophy in medial temporal areas, including the hippocampus, is observed early in the course of the disorder (12, 13) and in patients with mild cognitive impairment (MCI) (14), a clinical state that may be a precursor to AD (15). Hippocampal atrophy predicts conversion from MCI to AD (16) and has also been reported in carriers of rare dominant AD risk variants in the genes coding for amyloid precursor protein and presenilin 1 (17), as well as carriers of highly penetrant common variants such as APOE ε4 (18). Indeed, hippocampal volume is already a key imaging phenotype to identify preclinical stages of AD (19). Other brain structures showing significant atrophy early in disease progression include other medial temporal lobe regions including the entorhinal cortex (ERC) (20, 21), parahippocampal gyrus (PHG) (22), and posterior cingulate gyrus (PCG) (23). We therefore focused on early changes in these structures.
The combination of PRSs and neuroimaging data is likely to be particularly informative in identifying markers of early risk for AD (24, 25), even before the putative onset of amyloid accumulation. In the current study, we sought to investigate the correlation between polygenic risk for AD based on the largest genetic training dataset available (4) and gray and white matter structural differences in a healthy young population without any signs of cognitive impairment. So far, the only studies conducted with AD PRSs have used 24 risk loci (26) or PRSs derived from a smaller AD GWASs (23, 27), making ours the genetically most powerful study conducted on this topic to date.
We predicted that the PRSs for AD would be negatively correlated with hippocampal volume. We were also interested in exploring the association of cortical thickness of ERC, PHG, and PCG, due to the involvement of these areas in early AD (28, 29, 30), where we would expect to see a decrease in thickness. In addition to gray matter parameters, we also investigated the microstructure of the main white matter pathways connecting our candidate areas to ascertain whether any gray matter loss would already have impacted on the fiber tracts by way of anterograde or retrograde degeneration. We measured fractional anisotropy (FA), a measure of white matter microstructure derived from diffusion tensor imaging (DTI) (31) in the main connecting tracts of the hippocampus, the cingulum, and the fornix. We expected FA to be lower in participants with higher AD PRSs.
Methods and Materials
Participants
Brain scans used in this study were obtained from a repository of neuroimaging and genetic data obtained between 2009 and 2014 from healthy subjects recruited through a range of research projects at Cardiff University Brain Research Imaging Centre, which has received ethical approval from the Cardiff University School of Psychology. All subjects were screened for the exclusion of any neuropsychiatric disorders either by interview or by questionnaires. Participants provided informed consent for genotyping and use of their imaging data for genetic imaging analysis. After genotyping and data quality control standards, 272 individuals with structural T1 data remained (195 female, 77 male) with an average age at time of inclusion of 24.8 years (SD 6.9). Tractography data were available for a subset of 197 participants (138 female, 59 male) with an average age at time of inclusion of 23.9 years (SD 5.1). For a subgroup of 87 participants (53 female, 34 male; mean age 23.9 years [SD 4.4]), data on the Hopkins Verbal Learning Task were available. This task measures declarative verbal learning capacity (32) and forms part of the MATRICS Consensus Cognitive Battery.
Genotyping
Genomic DNA was obtained from saliva using Oragene OG-500 saliva kits (DNA Genotek, Inc., Ontario, Canada). Genotyping was performed using custom Illumina HumanCoreExome-24 BeadChip genotyping arrays, which contain 570,038 genetic variants (Illumina, Inc., San Diego, CA). Quality control was implemented in PLINK (33). Individuals were excluded for any of the following reasons: 1) ambiguous sex (genotypic sex and phenotypic sex not aligning); 2) cryptic relatedness up to third-degree relatives as ascertained using identity by descent; 3) genotyping completeness less than 97%; and 4) non-European ethnicity admixture. The latter was detected as outliers in an iterative EIGENSTRAT analysis of a linkage-disequilibrium-pruned dataset (34). Single nucleotide polymorphisms (SNPs) were excluded where the minor allele frequency was less than 1%, if the call rate was less than 98%, or if the χ2 test for Hardy-Weinberg equilibrium had a p value less than 1 × 10−4. Individuals’ genotypes were imputed using the prephasing/imputation stepwise approach implemented in IMPUTE2/SHAPEIT (35, 36) and 1000 Genomes (December 2013, release 1000 Genomes haplotypes Phase I integrated variant set) (37) as the reference dataset. This resulted in a dataset of 274 individuals with information for 7,413,342 SNPs.
Polygenic Scoring Method
Polygenic score calculations were performed according to the procedure described by the International Schizophrenia Consortium (38). Training data were from the International Genomics of Alzheimer’s Project consortium that comprises 17,008 AD cases and 37,154 control subjects (4). These data are publicly available from http://www.pasteur-lille.fr/en/recherche/u744/igap/igap_download.php. SNPs were removed from all analyses if they had a low minor allele frequency (< .01). Subsequently, the data were pruned for linkage disequilibrium using the clumping function (--clump) in PLINK (33) removing SNPs within 500 kilobase (--clump-kb) and r2 > .25 (--clump-r2) with a more significantly associated SNP. We used the --score command in PLINK to calculate polygenic scores (33). Nine different progressive training p value thresholds (39) were investigated (polygenic threshold [PT] < 1 × 10−8, 1 × 10−7, 1 × 10−6, 1 × 10−5, 1 × 10−4, .01, .1, .3, and .5). Lower PT indicates that SNPs are more significantly associated with AD case status in the training dataset (AD case-control study) (4).
These polygenic risk scores include the APOE loci on chromosome 19, the greatest common genetic risk factor for AD. If a significant association was observed between AD PRSs and brain imaging phenotypes, the data were reanalyzed with polygenic scores excluding any SNPs within the APOE locus (chromosome 19: 45.053–45.73 Mb), to assess if the association was purely due to variance in APOE.
Magnetic Resonance Imaging Data
Data Acquisition
Magnetic resonance imaging was carried out in Cardiff University Brain Research Imaging Centre on a GE Signa HDx 3T scanner (GE Healthcare, Milwaukee, WI). T1-weighted structural data were acquired using an axial three-dimensional fast, spoiled gradient recalled sequence with the following parameters: repetition time/echo time/inversion time = 8/3/450 ms; flip angle = 20°; 1 mm resolution; field of view ranging from 256 × 192 × 160 mm3 to 256 × 256 × 256 mm3 (anterior–posterior/left–right/superior–inferior), with acquisition time ranging from approximately 6 minutes to 10 minutes.
DTI data were acquired using a cardiac-gated sequence with the following parameters; b-values 0 and 1200, repetition time ~20 seconds (dependent on heart rate); echo time = 90 ms; 60 2.4-mm slices aligned with the anterior commissure-posterior commissure, zero slice gap; acquisition matrix 96 × 96; field of view = 230 mm; 2.4-mm isotropic resolution. Data were either acquired from 30 unique diffusion directions plus 3 b0 images or from a subsample of 30 optimal directions from an acquired set of 60 directions with the first 3 b0 images.
Data Processing
Hippocampal volume, ERC, PHG, PCG thickness, and intracranial volume (ICV) were determined through analysis with FreeSurfer (surfer.nmr.mgh.harvard.edu), which has been validated as a suitable method for hippocampal segmentation in large samples (40). The resulting output was quality controlled following a publically available protocol from ENIGMA (http://enigma.ini.usc.edu/) (41). Whenever a region of interest was detected as inadequately segmented by XC or SFF, its metric was declared missing and excluded from analysis. The numbers of brain regions included in the final analysis were hippocampus right and left (n = 270), entorhinal cortex left (n = 257) and right (n = 268), posterior cingulate gyrus left (n = 272) and right (n = 271), and parahippocampal gyrus left (n = 259) and right (n = 271).
DTI data were analyzed using ExploreDTI (42) version 4.8.3 and were corrected for eddy current distortions and subject motion using an affine registration to the nondiffusion-weighted images, with appropriate reorienting of the encoding vectors (43). An echo planar imaging (4) correction was applied, warping the DTI data to the fast, spoiled gradient recalled images, resulting in a 1 × 1 × 1 mm3 resolution in the resulting output. A single diffusion tensor model was fitted to the DTI data (44) to compute quantitative parameters such as FA.
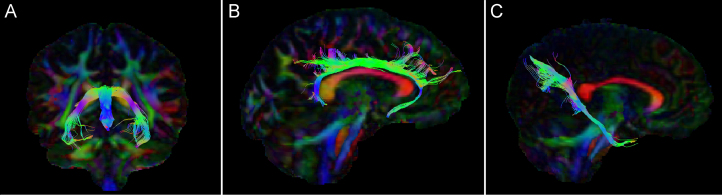
Subsequently, the damped Richardson Lucy pipeline (45) was used to perform whole-brain tractography. Termination criteria were an angle threshold greater than 45° or a drop in the magnitude of the minimally subtending fiber orientation density function peak below 0.05. Tracts were obtained using in-house automated tractography software (46). The automated tractography models for the fornix, cingulum, and parahippocampal cingulum (PHC) were based on manual tractography performed by SFF. Each automated tract underwent quality control through visual inspection and was brought to manual tractography standards by post hoc removal of any fiber bundles considered spurious, where necessary. Final numbers of tracts included were fornix n = 157 and cingulum and PHC right and left n = 197. The fornix was defined according to Metzler-Baddeley et al. (47); the high levels of dropout were most probably due to the high curvature of the tract and proximity to the ventricles. To segment the PHC, the restricted method was used, which incorporates a NOT gate-blocking inclusion of all tracts projecting toward the frontal cortex (48). For an example of the tracts, see Figure 1. Free water correction was applied (49, 50) before extracting FA values for further analysis. FA values were extracted using customized MATLAB scripts (The MathWorks, Inc., Natick, MA).
Figure 1.
(A) Coronal view of fornix fibers. (B) Sagittal view of right cingulum fibers. (C) Sagittal view of right parahippocampal cingulum fibers.
Analysis
Regional thickness data determined from T1 scans were analyzed using hierarchical linear multiple regression in IBM SPSS statistics 20 (IBM Corp., Armonk, NY), covarying for age, gender, and ICV. The hippocampal volumes were adjusted for ICV of each participant with the formula: hippocampal volume corrected = hippocampal volume − [beta * (ICV − mean ICV across the group studied)] (51). Subsequently, they were analyzed using hierarchical linear multiple regression covarying for age and gender.
The DTI data were also analyzed using hierarchical linear multiple regression, including FA as the dependent variable and controlling for independent variables age, gender, and scan type (30 or downsampled 60 directions).
The p values were then corrected for multiple comparisons using false discovery rate in statistics package R (52), resulting in q values. False discovery rate was applied over the regions of interest studied. Where significant results remained after false discovery rate correction, further analysis was performed using the PRSs without APOE SNPs.
It is standard practice to compare PRSs over multiple thresholds (38), and it is difficult to correct for multiple comparisons due to the highly correlated nature of the thresholds. Permutation testing is a robust way to correct for multiple comparisons in a dependent sample (53). The Supplement contains an outline of permutation tests performed on all the nominally significant results.
Results
Hippocampal Volume
PRSs correlated negatively with left hippocampal volume corrected for intracranial volume (R2 = .039; p = .001; q = 0.008; PT < 1 × 10−4) as demonstrated in Table 1. PRSs calculated at all PTs were nominally associated with decreased left hippocampal volume, and four of these survived correction for multiple testing (Table 1). Analysis of the R2 change showed that the PRSs accounted for an additional 1.9% to 3.9% of the variance in left hippocampal volume over the nine thresholds, after removing variance explained by age and gender, which accounted for 0.5%. No such effect was seen in the right hippocampus, where R2 changed ≤.001 across the whole range of PRS thresholds.
Table 1.
The Influence of Polygenic AD Risk Scores on Brain Structure
| Training PT Value | Hippo-campus L R2 (p Value) | Hippo-campus L No APOE R2 (p Value) | Hippo-campus R R2 (p Value) | ERC L R2 (p Value) | ERC R R2 (p Value) | PCG L R2 (p Value) | PCG L No APOE R2 (p Value) | PCG R R2 (p Value) | PHG L R2 (p Value) | PHG R R2 (p Value) |
|---|---|---|---|---|---|---|---|---|---|---|
| PT < 1 × 10−8 | .019 (.024)a | <.001 (.966) | <.001 (.795) | <.001 (.742) | .009 (.118) | .005 (.224) | .003 (.391) | .002 (.500) | .003 (.386) | .003 (.393) |
| PT < 1 × 10−7 | .020 (.021)a | <.001 (.991) | .001 (.683) | <.001 (.869) | .009 (.127) | .005 (.213) | .001 (.550) | .002 (.486) | .002 (.433) | .004 (.325) |
| PT < 1 × 10−6 | .023 (.012)a | .003 (.409) | <.001 (.814) | <.001 (.833) | .006 (.215) | .005 (.215) | <.001 (.788) | .003 (.362) | .002 (.522) | .005 (.227) |
| PT < 1 × 10−5 | .034 (.002)a,b | .019 (.023)a | <.001 (.958) | <.001 (.873) | .007 (.173) | .006 (.201) | .001 (.635) | .004 (.287) | .001 (.592) | .005 (.256) |
| PT < 1 × 10−4 | .039 (.001)a,b | .017 (.031)a | <.001 (.932) | .001 (.620) | .009 (.128) | .011 (.073) | .009 (.112) | .007 (.156) | .002 (.481) | .006 (.212) |
| PT < .01 | .035 (.002)a,b | .022 (.014)a | <.001 (.933) | .005 (.257) | .007 (.181) | .015 (.034)a | .012 (.060) | .001 (.554) | .003 (.406) | .012 (.076) |
| PT < .1 | .023 (.012)a,b | .017 (.032)a | <.001 (.863) | .004 (.289) | .005 (.253) | .025 (.006)a,b | .023 (.009)a | .001 (.645) | .010 (.109) | .010 (.099) |
| PT < .3 | .022 (.015)a | .017 (.033)a | <.001 (.850) | .002 (.512) | .006 (.206) | .019 (.019)a | .017 (.025)a | .002 (.488) | .009 (.123) | .010 (.094) |
| PT < .5 | .021 (.017)a | .016 (.037)a | <.001 (.748) | .002 (.530) | .009 (.123) | .021 (.014)a | .019 (.019)a | .002 (.483) | .012 (.082) | .009 (.128) |
R2 change and p values for hippocampal volume (left n = 270; right n = 270), entorhinal cortex thickness (left n = 257; right n = 268), posterior cingulate gyrus thickness (left n = 272; right n = 271), and parahippocampal gyrus thickness (left n = 259; right n = 271). The top axis shows the region of interest, while the vertical axis shows the polygenic threshold.
AD, Alzheimer’s disease; ERC, entorhinal cortex; FDR, false discovery rate; L, left; PCG, posterior cingulate gyrus; PHG, parahippocampal gyrus; PT, polygenic threshold; R, right.
Nominally significant associations (p value < .05).
Denotes q-FDR corrected <0.05.
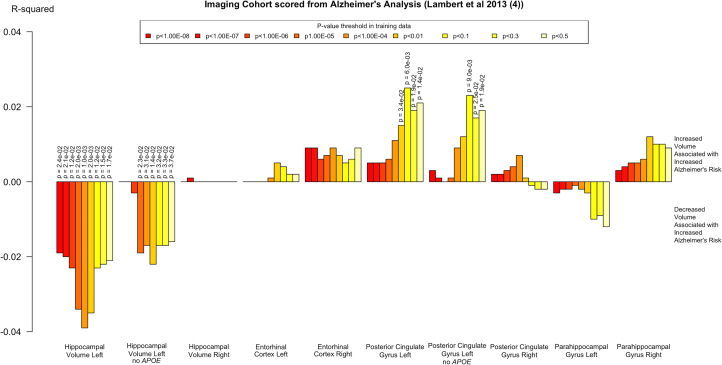
Subsequent analysis of hippocampal volume with the APOE locus removed from PRSs showed that the significant association between AD PRSs and decreased left hippocampal volume persisted (R2 = .022; p = .014; PT < .01), particularly with the more inclusive pTs (Figure 2).
Figure 2.
Effects of polygenic risk scores on regions of interest before false discovery rate correction. Vertical axis denotes the R2 change, with upward indicating an increase and downward indicating a decrease of volume or thickness of the region of interest.
Entorhinal Cortex, Posterior Cingulate Gyrus, and Parahippocampal Gyrus Thickness
No significant effects of AD PRSs were seen on the volume of the ERC or PHG. A small increase in volume was seen in the left PCG, which was conserved at one threshold (R2 = .025; p = .006; q = 0.048; PT < .1), after correction for multiple comparisons. This effect persisted after removal of the APOE locus from the PRSs (Table 1).
Tractography Fornix, Cingulum, and PHC
No effect of AD PRSs on FA was found in the fornix or PHC (Table 2). However, a significant effect of AD PRSs on FA of the right cingulum was found (R2 = .032; p = .009; q = 0.045; PT < 1 × 10−4), with a negative association between PRS and FA at five thresholds. This effect was conserved for one of these thresholds (p ≤ .05) when correcting for multiple comparisons over all five regions of interest. Subsequent analysis of cingulum FA, excluding the APOE locus from the AD PRSs, only showed a significant association at one threshold (R2 = .019; p = .044; PT < 1 × 10−6).
Table 2.
R2 Change and p Values for the Change in FA Correlating With AD PRS for Each Threshold
| Training PT Value | Fornix R2 (p Value) | Cingulum L R2 (p Value) | Cingulum R R2 (p Value) | Cingulum R No APOER2 (p Value) | PHC L R2 (p Value) | PHC R R2 (p Value) |
|---|---|---|---|---|---|---|
| PT < 1 × 10−8 | <.001 (.791) | .009 (.164) | .030 (.011)a | .006 (.254) | .008 (.194) | .002 (.534) |
| PT < 1 × 10−7 | <.001 (.813) | .009 (.162) | .029 (.013)a | .006 (.268) | .006 (.233) | .002 (.515) |
| PT < 1 × 10−6 | <.001 (.813) | .005 (.270) | .023 (.026)a | .019 (.044)a | .006 (.255) | .002 (.538) |
| PT < 1 × 10−5 | <.001 (.846) | .004 (.344) | .025 (.020)a | .006 (.270) | .005 (.292) | .001 (.655) |
| PT < 1 × 10−4 | <.001 (.780) | .009 (.165) | .032 (.009)a,b | <.001 (.885) | .002 (.547) | <.001 (.949) |
| PT < .01 | <.001 (.952) | .001 (.610) | .006 (.273) | <.001 (.858) | .003 (.432) | .001 (.727) |
| PT < .1 | <.001 (.886) | .001 (.626) | .005 (.305) | .001 (.588) | .005 (.289) | .006 (.268) |
| PT < .3 | <.001 (.840) | .003 (.440) | .002 (.493) | <.001 (.777) | .003 (.388) | .004 (.357) |
| PT < .5 | <.001 (.806) | .002 (.486) | .002 (.471) | .001 (.731) | .003 (.387) | .007 (.240) |
Fornix (n = 157), cingulum left (n = 197) and right (n = 197) (with and without APOE SNPs), and parahippocampal cingulum left (n = 197) and right (n = 197) are shown here.
AD, Alzheimer’s disease; FA, fractional anisotropy; FDR, false discovery rate; L, left; PRS, polygenic risk score; PHC, parahippocampal cingulum; PT, polygenic threshold; R, right; SNP, single nucleotide polymorphism.
Nominally significant associations (p value < .05).
Associations that survive FDR correction.
Cognitive Effects
We found no correlation between scores on the Hopkins Verbal Learning Task and polygenic risk for AD (Supplemental Table S1).
Discussion
As predicted, PRS for AD was associated with lower hippocampal volume. While hippocampal volume reductions in patients are a robust finding in AD (10, 13) and MCI patients converting to AD undergo greater hippocampal atrophy (16), previous reports of hippocampal volume effects of genetic risk variants in healthy subjects have been varied. Some studies (54, 55, 56) found no effect of APOE risk alleles on hippocampal volume in young participants, while others found a significant difference between young ε4 and ε2 carriers (18). Previously, right, but not left, hippocampal volume reductions were found in APOE ε4 risk allele carriers in comparison with ε3 carriers in a middle-aged population (57), whereas we found effects in the left, but not right, hippocampus. However, most of the previous literature has reported a slight preponderance of left compared with right hippocampal changes, particularly in the preclinical and early stages of AD progression (58), which would be in line with our findings.
We may see significant effects where others have obtained mixed results with single locus studies because of the benefits of using polygenic risk scores. Although the SNPs contributing to the PRSs have much smaller individual effect sizes than the APOE locus, cumulatively they explain a large amount of variance (9).
Although previous reports have implicated the ERC in AD pathology (20, 21), we do not see an effect of AD genetic risk on ERC thickness. This is consistent with previous studies (39) that failed to find an effect of APOE on ERC thickness in a healthy middle-aged and older adult population. Conversely, the small increase in PCG thickness was unexpected because both manifest AD and genetic risk status have been associated with thinning of this area (59, 60). Replication of this finding in future studies would be needed before strong conclusions can be drawn.
It has been proposed that AD, like many other neuropsychiatric disorders, is a disconnection syndrome (61). Connectivity models of pathophysiology can be supported by the investigation of the microstructural properties of white matter. Changes in white matter parameters have indeed been observed in many DTI studies of AD, for example, decreased FA in the right fornix (10) and the left PHC (62), superior longitudinal fasciculus, temporal lobe (63, 64), and cingulum (65). A significant decrease of FA was also found in the PHC in MCI patients (66). Differences in FA relating to AD risk genes have previously been observed in healthy participants as well; APOE ε4 was linked to alterations in FA in the left medial temporal lobe and in the corpus callosum in healthy individuals (67). One study found widespread FA decreases in a young healthy population (age 20–35) with APOE risk variants (68), while another found widespread decreases in FA, including in the fornix and cingulum, in young individuals carrying the clusterin risk allele (69). A population of preclinical subjects with an autosomal dominant AD variant also had significantly lower FA in the columns of the fornix (70).
Our finding of reduced FA in the right cingulum, which was not exclusively driven by the APOE locus, is in line with models that assume early white matter changes in the course of the development of AD pathology. The fact that this correlation between cingulum FA and AD PRSs was not driven by the PHC, which connects the medial temporal lobe with areas in the parietal and occipital lobes such as the posterior cingulate cortex (47), was contrary to expectations. In our sample, the white matter findings were not directly related to the gray matter structural differences that occurred in the opposite hemisphere.
We used the results from the largest AD GWASs undertaken to date as our training data (4). Therefore, the estimates of SNP effects on AD risk utilized in this study are the best reported to date with better power than previous estimates, as it is known that polygenic risk score and R2 values are highly affected by the sample size of the training dataset (71).
Hippocampal volume and cortical thickness are highly heritable, which makes them appropriate parameters for genetic imaging analysis (72, 73). A limitation of our study is that, by the nature of the polygenic analysis, which pools risk variants across the whole genome, no inferences can be made on the specific molecular mechanisms contributing to the structural brain differences. Although neurofibrillary tangles can be present in the hippocampus in young adults, the Braak staging model would suggest that entorhinal/transentorhinal cortex is affected even earlier by this process (74, 75). Although amyloid pathology can be detected in young carriers of Mendelian variants affecting the amyloid pathway (76), it would be unlikely to be confined to the hippocampus (75), and our sample was probably too young to have a significant amyloid load (77). GWASs for AD have revealed novel pathways associated with AD, including in lipid metabolism, immune responses, and endocytosis, while also finding no enrichment of genes associated with either tau or amyloid pathways, suggesting other factors may play a role in risk and development of AD (7). Furthermore, it is possible that the cumulative effect of common AD risk variants affects the development of the hippocampus in a similar way, as they have been shown to affect cognition across the life course of an individual (78). Longitudinal studies of genetic imaging, involving even younger populations than that of the present study, and pathway-based analysis (79, 80, 81) will be needed to address the biological significance of our findings. However, pathway-based analyses will likely require larger samples and new approaches to the multiple testing problem.
In conclusion, AD polygenic risk was associated with smaller volume of the left hippocampus, increased volume of left PCG, and lower FA in the right cingulum bundle in healthy young adults. Thus, AD genetic risk can be linked to structural differences in brain areas that have been implicated in the early stages of AD pathology many decades before potential illness onset. This effect was not driven exclusively by contributions from APOE, as the associations persisted after removal of the APOE locus. Overall, this work suggests that genetic risk for AD is mediated, in part, through brain morphological differences, mainly in the hippocampus, confirming hippocampal volume changes as an important early biomarker of risk for AD.
Acknowledgments and Disclosures
SFF was funded on an Institutional Strategic Support Fund Grant No. 504182 awarded to Cardiff University by the Wellcome Trust. DL, KET, and JH were supported by Medical Research Council Grant No. MR/K004360/1. TB was funded by the Swiss National Science Foundation, Switzerland (Grant No. PBBEP3_144797). The genetic imaging study was funded by the Neuroscience and Mental Health Research Institute at Cardiff University and by the National Centre for Mental Health at Cardiff University with funds from the National Institute for Social Care and Health Research and Welsh Government, Wales (Grant No. BR09). We would like to acknowledge the MRC Centre for Neuropsychiatric Genetics and Genomics (Grant No. G0800509).
We acknowledge Dr. Claudia Metzler-Baddeley for demonstrating the manual tractography methodology and Jilu Mole and Sonya Bells for allowing use of their MATLAB scripts. We thank all the researchers in the Cardiff University Brain Research Imaging Centre who have contributed to the genetic neuroimaging project. We are grateful to all professionals, patients, and volunteers involved with the National Centre for Mental Health.
The authors report no biomedical financial interests or potential conflicts of interest.
Footnotes
Supplementary material cited in this article is available online at 10.1016/j.biopsych.2016.02.033.
Appendix A. Supplementary material
Supplementary material
References
- 1.Prince M., Bryce R., Albanese E., Wimo A., Ribeiro W., Ferri C.P. The global prevalence of dementia: A systematic review and metaanalysis. Alzheimers Dement. 2013;9:63–75. doi: 10.1016/j.jalz.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Tanzi R.E., Bertram L. New frontiers in Alzheimer’s disease genetics. Neuron. 2001;32:181–184. doi: 10.1016/s0896-6273(01)00476-7. [DOI] [PubMed] [Google Scholar]
- 3.Medland S.E., Jahanshad N., Neale B.M., Thompson P.M. Whole-genome analyses of whole-brain data: Working within an expanded search space. Nat Neurosci. 2014;17:791–800. doi: 10.1038/nn.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambert J.-C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Escott-Price V., Bellenguez Cl, Wang L.-S., Choi S.-H., Harold D., Jones L. Gene-wide analysis detects two new susceptibility genes for Alzheimer’s disease. PLoS One. 2014;9:e94661. doi: 10.1371/journal.pone.0094661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harold D., Abraham R., Hollingworth P., Sims R., Gerrish A., Hamshere M.L. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones L., Holmans P.A., Hamshere M.L., Harold D., Moskvina V., Ivanov D. Genetic evidence implicates the immune system and cholesterol metabolism in the aetiology of Alzheimer’s disease. PLoS One. 2010;5:e13950. doi: 10.1371/journal.pone.0013950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 2013;9:e1003348. doi: 10.1371/journal.pgen.1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Escott-Price V., Sims R., Bannister C., Harold D., Vronskaya M., Majounie E. Common polygenic variation enhances risk prediction for Alzheimer’s disease. Brain. 2015;138:3673–3684. doi: 10.1093/brain/awv268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Serra L., Cercignani M., Lenzi D., Perri R., Fadda L., Caltagirone C. Grey and white matter changes at different stages of Alzheimer’s disease. J Alzheimers Dis. 2010;19:147–159. [Google Scholar]
- 11.Jack C.R., Holtzman D.M. Biomarker modeling of Alzheimer’s disease. Neuron. 2013;80:1347–1358. doi: 10.1016/j.neuron.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braskie M.N., Toga A.W., Thompson P.M. Recent advances in imaging Alzheimer’s disease. J Alzheimers Dis. 2013;33:S313–S327. doi: 10.3233/JAD-2012-129016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hampel H., Lista S., Teipel S.J., Garaci F., Nisticò R., Blennow K. Perspective on future role of biological markers in clinical therapy trials of Alzheimer’s disease: A long-range point of view beyond 2020. Biochem Pharmacol. 2014;88:426–449. doi: 10.1016/j.bcp.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 14.Convit A., De Leon M.J., Tarshish C., De Santi S., Tsui W., Rusinek H., George A. Specific hippocampal volume reductions in individuals at risk for Alzheimer’s disease. Neurobiol Aging. 1997;18:131–138. doi: 10.1016/s0197-4580(97)00001-8. [DOI] [PubMed] [Google Scholar]
- 15.Petersen R.C., Morris J.C. Mild cognitive impairment as a clinical entity and treatment target. Arch Neurol. 2005;62:1160–1163. doi: 10.1001/archneur.62.7.1160. [DOI] [PubMed] [Google Scholar]
- 16.Macdonald K.E., Bartlett J.W., Leung K.K., Ourselin S., Barnes J. The value of hippocampal and temporal horn volumes and rates of change in predicting future conversion to AD. Alzheimer Dis Assoc Disord. 2013;27:168–173. doi: 10.1097/WAD.0b013e318260a79a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ridha B.H., Barnes J., Bartlett J.W., Godbolt A., Pepple T., Rossor M.N., Fox N.C. Tracking atrophy progression in familial Alzheimer’s disease: A serial MRI study. Lancet Neurol. 2006;5:828–834. doi: 10.1016/S1474-4422(06)70550-6. [DOI] [PubMed] [Google Scholar]
- 18.Alexopoulos P., Richter-Schmidinger T., Horn M., Maus S., Reichel M., Sidiropoulos C. Hippocampal volume differences between healthy young apolipoprotein E ε2 and ε4 carriers. J Alzheimers Dis. 2011;26:207–210. doi: 10.3233/JAD-2011-110356. [DOI] [PubMed] [Google Scholar]
- 19.Sperling R.A., Aisen P.S., Beckett L.A., Bennett D.A., Craft S., Fagan A.M. Toward defining the preclinical stages of Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:280–292. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gómez-Isla T., Price J.L., McKeel D.W., Jr, Morris J.C., Growdon J.H., Hyman B.T. Profound loss of layer II entorhinal cortex neurons occurs in very mild Alzheimer’s disease. J Neurosci. 1996;16:4491–4500. doi: 10.1523/JNEUROSCI.16-14-04491.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Killiany R.J., Hyman B.T., Gomez-Isla T., Moss M.B., Kikinis R., Jolesz F. MRI measures of entorhinal cortex vs hippocampus in preclinical AD. Neurology. 2002;58:1188–1196. doi: 10.1212/wnl.58.8.1188. [DOI] [PubMed] [Google Scholar]
- 22.Köhler S., Black S.E., Sinden M., Szekely C., Kidron D., Parker J.L. Memory impairments associated with hippocampal versus parahippocampal-gyrus atrophy: An MR volumetry study in Alzheimer’s disease. Neuropsychologia. 1998;36:901–914. doi: 10.1016/s0028-3932(98)00017-7. [DOI] [PubMed] [Google Scholar]
- 23.Sabuncu M.R., Buckner R.L., Smoller J.W., Lee P.H., Fischl B., Sperling R.A. The association between a polygenic Alzheimer score and cortical thickness in clinically normal subjects. Cereb Cortex. 2011;22:2653–2661. doi: 10.1093/cercor/bhr348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whalley H.C., Papmeyer M., Sprooten E., Romaniuk L., Blackwood D.H., Glahn D.C. The influence of polygenic risk for bipolar disorder on neural activation assessed using fMRI. Transl Psychiatry. 2012;2:e130. doi: 10.1038/tp.2012.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braskie M.N., Ringman J.M., Thompson P.M. Neuroimaging measures as endophenotypes in Alzheimer’s disease. Int J Alzheimers Dis. 2011;2011:490140. doi: 10.4061/2011/490140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chauhan G., Adams H.H.H., Bis J.C., Weinstein G., Yu L., Töglhofer A.M. Association of Alzheimer’s disease GWAS loci with MRI markers of brain aging. Neurobiol Aging. 2015;36(1765):e7–e16. doi: 10.1016/j.neurobiolaging.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biffi A., Anderson C.D., Desikan R.S., Sabuncu M., Cortellini L., Schmansky N. Genetic variation and neuroimaging measures in Alzheimer disease. Arch Neurol. 2010;67:677–685. doi: 10.1001/archneurol.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lampert E.J., Roy Choudhury K., Hostage C.A., Rathakrishnan B., Weiner M., Petrella J.R. Vol. 6. 2014. Brain atrophy rates in first degree relatives at risk for Alzheimer’s; pp. 340–346. (Neuroimage Clin). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raji C.A., Lopez O.L., Kuller L.H., Carmichael O.T., Becker J.T. Age, Alzheimer disease, and brain structure. Neurology. 2009;73:1899–1905. doi: 10.1212/WNL.0b013e3181c3f293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choo I.H., Lee D.Y., Oh J.S., Lee J.S., Lee D.S., Song I.C. Posterior cingulate cortex atrophy and regional cingulum disruption in mild cognitive impairment and Alzheimer’s disease. Neurobiol Aging. 2010;31:772–779. doi: 10.1016/j.neurobiolaging.2008.06.015. [DOI] [PubMed] [Google Scholar]
- 31.Horsfield M.A., Jones D.K. Applications of diffusion-weighted and diffusion tensor MRI to white matter diseases – a review. NMR Biomed. 2002;15:570–577. doi: 10.1002/nbm.787. [DOI] [PubMed] [Google Scholar]
- 32.Brandt J. The Hopkins Verbal Learning Test: Development of a new memory test with six equivalent forms. Clin Neuropsychol. 1991;5:125–142. [Google Scholar]
- 33.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 35.Delaneau O., Marchini J., Zagury J-Fo A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 36.Howie B., Marchini J., Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1:457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.1000 Genomes Project Consortium. Auton A., Brooks L.D., Durbin R.M., Garrision E.P., Kang H.M. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.International Schizophrenia Consortium, Purcell S.M., Wray N.R., Stone J.L., Visscher P.M., O’Donovan M.C. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bunce D., Anstey K.J., Cherbuin N., Gautam P., Sachdev P., Easteal S. APOE genotype and entorhinal cortex volume in non-demented community-dwelling adults in midlife and early old age. J Alzheimers Dis. 2012;30:935–942. doi: 10.3233/JAD-2012-112126. [DOI] [PubMed] [Google Scholar]
- 40.Cherbuin N., Anstey K.J., Réglade-Meslin C., Sachdev P.S. In vivo hippocampal measurement and memory: A comparison of manual tracing and automated segmentation in a large community-based sample. PLoS One. 2009;4:e5265. doi: 10.1371/journal.pone.0005265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stein J.L., Medland S.E., Vasquez A.A., Hibar D.P., Senstad R.E., Winkler A.M. Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet. 2012;44:552–561. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leemans A, Jeurissen B, Sijbers J, Jones DK (2009): ExploreDTI: A graphical toolbox for processing, analyzing, and visualizing diffusion MR data. Presented at the 17th Annual Meeting and Exhibition of the International Society for Magnetic Resonance in Medicine, April 18–24, Honolulu, Hawaii. In: ISMRM 17th Annual Scientific Meeting & Exhibition, Honolulu, HI, 3537.
- 43.Leemans A., Jones D.K. The B-matrix must be rotated when correcting for subject motion in DTI data. Magn Reson Med. 2009;61:1336–1349. doi: 10.1002/mrm.21890. [DOI] [PubMed] [Google Scholar]
- 44.Basser P.J., Mattiello J., LeBihan D. Estimation of the effective self-diffusion tensor from the NMR spin echo. J Magn Reson B. 1994;103:247–254. doi: 10.1006/jmrb.1994.1037. [DOI] [PubMed] [Google Scholar]
- 45.Dell’Acqua F., Scifo P., Rizzo G., Catani M., Simmons A., Scotti G., Fazio F. A modified damped Richardson-Lucy algorithm to reduce isotropic background effects in spherical deconvolution. Neuroimage. 2010;49:1446–1458. doi: 10.1016/j.neuroimage.2009.09.033. [DOI] [PubMed] [Google Scholar]
- 46.Parker G, Rosin PL, Marshall D (2012): Automated Segmentation of Diffusion Weighted MRI Tractography. Presented at the AVA, AVA/BMVA Meeting on Biological and Computer Vision, Spring (AGM) Meeting 2012, May 22, Cambridge, United Kingdom.
- 47.Metzler-Baddeley C., Jones D.K., Belaroussi B., Aggleton J.P., O’Sullivan M.J. Frontotemporal connections in episodic memory and aging: A diffusion MRI tractography study. J Neurosci. 2011;31:13236–13245. doi: 10.1523/JNEUROSCI.2317-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones D.K., Christiansen K.F., Chapman R.J., Aggleton J.P. Distinct subdivisions of the cingulum bundle revealed by diffusion MRI fibre tracking: Implications for neuropsychological investigations. Neuropsychologia. 2013;51:67–78. doi: 10.1016/j.neuropsychologia.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Metzler-Baddeley C., O’Sullivan M.J., Bells S., Pasternak O., Jones D.K. How and how not to correct for CSF-contamination in diffusion MRI. Neuroimage. 2012;59:1394–1403. doi: 10.1016/j.neuroimage.2011.08.043. [DOI] [PubMed] [Google Scholar]
- 50.Pasternak O., Sochen N., Gur Y., Intrator N., Assaf Y. Free water elimination and mapping from diffusion MRI. Magn Reson Med. 2009;62:717–730. doi: 10.1002/mrm.22055. [DOI] [PubMed] [Google Scholar]
- 51.Jack C.R., Jr, Twomey C.K., Zinsmeister A.R., Sharbrough F.W., Petersen R.C., Cascino G.D. Anterior temporal lobes and hippocampal formations: Normative volumetric measurements from MR images in young adults. Radiology. 1989;172:549–554. doi: 10.1148/radiology.172.2.2748838. [DOI] [PubMed] [Google Scholar]
- 52.Core Team R. R Foundation for Statistical Computing; Vienna: 2012. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 53.Sham P.C., Purcell S.M. Statistical power and significance testing in large-scale genetic studies. Nat Rev Genet. 2014;15:335–346. doi: 10.1038/nrg3706. [DOI] [PubMed] [Google Scholar]
- 54.Filippini N., MacIntosh B.J., Hough M.G., Goodwin G.M., Frisoni G.B., Smith S.M. Distinct patterns of brain activity in young carriers of the APOE-ε4 allele. Proc Natl Acad Sci U S A. 2009;106:7209–7214. doi: 10.1073/pnas.0811879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khan W., Giampietro V., Ginestet C., Dell’Acqua F., Bouls D., Newhouse S. No differences in hippocampal volume between carriers and non-carriers of the ApoE ε4 and ε2 alleles in young healthy adolescents. J Alzheimers Dis. 2014;40:37–43. doi: 10.3233/JAD-131841. [DOI] [PubMed] [Google Scholar]
- 56.Prvulovic D., Matura S., Miller J., Scheibe M., O’Dwyer L., Fusser F. APOE-ε4 genotype affects brain function without apparent micro- and macrostructural changes in young adults: A multimodal fMRI, DTI and VBM study. Alzheimers Dement. 2012;8(suppl 4):P619–P620. [Google Scholar]
- 57.Lind J., Larsson A., Persson J., Ingvar M., Nilsson L.-G., Bäckman L. Reduced hippocampal volume in non-demented carriers of the apolipoprotein E ε4: Relation to chronological age and recognition memory. Neurosci Lett. 2006;396:23–27. doi: 10.1016/j.neulet.2005.11.070. [DOI] [PubMed] [Google Scholar]
- 58.Shi F., Liu B., Zhou Y., Yu C., Jiang T. Hippocampal volume and asymmetry in mild cognitive impairment and Alzheimer’s disease: Meta-analyses of MRI studies. Hippocampus. 2009;19:1055–1064. doi: 10.1002/hipo.20573. [DOI] [PubMed] [Google Scholar]
- 59.Lehmann M., Rohrer J.D., Clarkson M.J., Ridgway G.R., Scahill R.I., Modat M. Reduced cortical thickness in the posterior cingulate gyrus is characteristic of both typical and atypical Alzheimer’s disease. J Alzheimers Dis. 2010;20:587–598. doi: 10.3233/JAD-2010-1401. [DOI] [PubMed] [Google Scholar]
- 60.Knight W.D., Kim L.G., Douiri A., Frost C., Rossor M.N., Fox N.C. Acceleration of cortical thinning in familial Alzheimer’s disease. Neurobiol Aging. 2011;32:1765–1773. doi: 10.1016/j.neurobiolaging.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 61.Reid A.T., Evans A.C. Structural networks in Alzheimer’s disease. Eur Neuropsychopharmacol. 2013;23:63–77. doi: 10.1016/j.euroneuro.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 62.Honea R.A., Vidoni E., Harsha A., Burns J.M. Impact of APOE on the healthy aging brain: A voxel-based MRI and DTI study. J Alzheimers Dis. 2009;18:553–564. doi: 10.3233/JAD-2009-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Teipel S.J., Meindl T., Grinberg L., Heinsen H., Hampel H. Vol. 35. 2008. Novel MRI techniques in the assessment of dementia; pp. 58–69. (Eur J Nucl Med Mol Imaging). [DOI] [PubMed] [Google Scholar]
- 64.Xie S., Xiao J.X., Gong G.L., Zang Y.F., Wang Y.H., Wu H.K., Jiang X.X. Voxel-based detection of white matter abnormalities in mild Alzheimer disease. Neurology. 2006;66:1845–1849. doi: 10.1212/01.wnl.0000219625.77625.aa. [DOI] [PubMed] [Google Scholar]
- 65.Douaud G.L., Jbabdi S.D., Behrens T.E.J., Menke R.A., Gass A., Monsch A.U. DTI measures in crossing-fibre areas: Increased diffusion anisotropy reveals early white matter alteration in MCI and mild Alzheimer’s disease. Neuroimage. 2011;55:880–890. doi: 10.1016/j.neuroimage.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rose S.E., McMahon K.L., Janke A.L., O’Dowd B., de Zubicaray G., Strudwick M.W., Chalk J.B. Diffusion indices on magnetic resonance imaging and neuropsychological performance in amnestic mild cognitive impairment. J Neurol Neurosurg Psychiatry. 2006;77:1122–1128. doi: 10.1136/jnnp.2005.074336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Persson J., Lind J., Larsson A., Ingvar M., Cruts M., Van Broeckhoven C. Altered brain white matter integrity in healthy carriers of the APOE ε4 allele A risk for AD? Neurology. 2006;66:1029–1033. doi: 10.1212/01.wnl.0000204180.25361.48. [DOI] [PubMed] [Google Scholar]
- 68.Heise V., Filippini N., Ebmeier K.P., Mackay C.E. The APOE ε4 allele modulates brain white matter integrity in healthy adults. Mol Psychiatry. 2011;16:908–916. doi: 10.1038/mp.2010.90. [DOI] [PubMed] [Google Scholar]
- 69.Braskie M.N., Jahanshad N., Stein J.L., Barysheva M., McMahon K.L., de Zubicaray G.I. Common Alzheimer’s disease risk variant within the CLU gene affects white matter microstructure in young adults. J Neurosci. 2011;31:6764–6770. doi: 10.1523/JNEUROSCI.5794-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ringman J.M., O’Neill J., Geschwind D., Medina L., Apostolova L.G., Rodriguez Y. Diffusion tensor imaging in preclinical and presymptomatic carriers of familial Alzheimer’s disease mutations. Brain. 2007;130:1767–1776. doi: 10.1093/brain/awm102. [DOI] [PubMed] [Google Scholar]
- 71.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Winkler A.M., Kochunov P., Fox P.T., Duggirala R., Almasy L., Blangero J. Heritability of volume, surface area and thickness for anatomically defined cortical brain regions estimated in a large extended pedigree. Neuroimage. 2009;47:S162. [Google Scholar]
- 73.Winkler A.M., Kochunov P., Blangero J., Almasy L., Zilles K., Fox P.T. Cortical thickness or grey matter volume? The importance of selecting the phenotype for imaging genetics studies. Neuroimage. 2010;53:1135–1146. doi: 10.1016/j.neuroimage.2009.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Braak H., Del Tredici K. The pathological process underlying Alzheimer’s disease in individuals under thirty. Acta Neuropathol. 2011;121:171–181. doi: 10.1007/s00401-010-0789-4. [DOI] [PubMed] [Google Scholar]
- 75.Braak H., Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 76.Fleisher A.S., Chen K., Quiroz Y.T., Jakimovich L.J., Gomez M.G., Langois C.M. Florbetapir PET analysis of amyloid-β deposition in the presenilin 1 E280A autosomal dominant Alzheimer’s disease kindred: A cross-sectional study. Lancet Neurol. 2012;11:1057–1065. doi: 10.1016/S1474-4422(12)70227-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Duyckaerts C. Vol. 121. 2011. Tau pathology in children and young adults: Can you still be unconditionally baptist? pp. 145–147. (Acta Neuropathol). [DOI] [PubMed] [Google Scholar]
- 78.Hill W.D., Davies G., CHARGE Cognitive Working Group. Liewald D.C., McIntosh A.M., Deary I.J. Age-dependent pleiotropy between general cognitive function and major psychiatric disorders. Biol Psychiatry. 2016;80:266–273. doi: 10.1016/j.biopsych.2015.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Desikan R.S., Schork A.J., Wang Y., Thompson W.K., Dehghan A., Ridker P.M. Polygenic overlap between c-reactive protein, plasma lipids, and Alzheimer disease. Circulation. 2015;131:2061–2069. doi: 10.1161/CIRCULATIONAHA.115.015489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jones L., Harold D., Williams J. Genetic evidence for the involvement of lipid metabolism in Alzheimer’s disease. Biochim Biophys Acta. 2010;1801:754–761. doi: 10.1016/j.bbalip.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 81.International Genomics of Alzheimer’s Disease Consortium (IGAP) Convergent genetic and expression data implicate immunity in Alzheimer’s disease. Alzheimers Dement. 2015;11:658–671. doi: 10.1016/j.jalz.2014.05.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material