Abstract
Background: The gut microbiome has been implicated in various metabolic and neurocognitive disorders and is heavily influenced by dietary factors, but there is a paucity of research on the effects of added sugars on the gut microbiome.
Objective: With the use of a rodent model, our goal was to determine how added-sugar consumption during the juvenile and adolescent phase of development affects the gut microbiome.
Methods: Forty-two juvenile male Sprague-Dawley rats [postnatal day (PND) 26; 50–70 g] were given access to 1 of 3 different 11%-carbohydrate solutions designed to model a range of monosaccharide ratios commonly consumed in sugar-sweetened beverages: 1) 35% fructose:65% glucose, 2) 50% fructose:50% glucose, 3) 65% fructose:35% glucose, and 4) control (no sugar). After ad libitum access to the respective solutions for the juvenile and adolescent period (PND 26–80), fecal samples were collected for next-generation 16S ribosomal RNA sequencing and multivariate microbial composition analyses. Energy intake, weight change, and adiposity index were analyzed in relation to sugar consumption and the microbiota.
Results: Body weight, adiposity index, and total caloric intake did not differ as a result of sugar consumption. However, sugar consumption altered the gut microbiome independently of anthropometric measures and caloric intake. At the genus level, Prevotella [linear discriminant analysis (LDA) score = −4.62; P < 0.001] and Lachnospiraceae incertae sedis (LDA score = −3.01; P = 0.03) were reduced, whereas Bacteroides (LDA score = 4.19; P < 0.001), Alistipes (LDA score = 3.88; P < 0.001), Lactobacillus (LDA score = 3.78; P < 0.001), Clostridium sensu stricto (LDA score = 3.77; P < 0.001), Bifidobacteriaceae (LDA score = 3.59; P = 0.001), and Parasutterella (LDA score = 3.79; P = 0.004) were elevated by sugar consumption. No overall pattern could be attributable to monosaccharide ratio.
Conclusions: Early-life sugar consumption affects the gut microbiome in rats independently of caloric intake, body weight, or adiposity index; these effects are robust across a range of fructose-to-glucose ratios.
Keywords: glucose, fructose, gut microbiota, juvenile, adolescence
Introduction
Colonization of the gut microbiome, which consists of an estimated 100 trillion microorganisms (1), begins during birth and continues into early childhood (2). Early-life gut microbial populations play a critical role in the development of the nervous system and the immune response and have been shown to affect behaviors such as anxiety and motor control into adulthood (3–6). Current findings also identified a role for the gut microbiome in the development of gastrointestinal diseases, such as ulcerative colitis and irritable bowel syndrome (7, 8), in metabolic pathologies such as insulin resistance (9) and obesity (10, 11) and in neurological disorders such as autism (12), Parkinson disease (13), and Alzheimer disease (14). Due to evidence linking the gut microbiome with human health and disease, it has been suggested that nurturing the development of a healthy patient/microbial “superorganism” is a cornerstone in the future of medicine (15, 16). Thus, an understanding of how modifiable environmental factors affect the gut microbiome, particularly during early-life developmental periods in which there is rapid microorganism colonization of the gut, is of critical importance for human health and disease prevention.
Recent experimental rodent studies revealed that dietary factors robustly affect the gut microbiome (17). In particular, an obesogenic high-fat diet (HFD)9, typically composed of 45–60% fat with sucrose as the primary carbohydrate source, has been studied in rodents with regard to changes in bacterial populations. For example, an HFD reduces populations in the phylum Bacteroidetes and increases both Firmicutes and Proteobacteria relative to control diets that are 10–15% kilocalories from fat with complex carbohydrates (starch) as the primary carbohydrate source (18, 19). Bacteroidetes aid in promoting T cell–mediated immune responses in the host and prevent the overgrowth of more harmful pathogens (20–22), whereas Proteobacteria and Firmicutes are generally associated with gut dysbiosis (23) and obesity (24), respectively. However, given that rodent HFD models produce obesity and metabolic syndrome, it is unclear whether HFD-mediated gut microbiome alterations are based directly on dietary factors or, rather, are secondary to increased adiposity and associated metabolic derangements.
Although many experimental rodent models showed that a diet that is high in both FAs and simple sugars (e.g., sucrose) affects the gut microbiome, there is a paucity of research on the contribution of added sugars independent of elevated dietary fat. In humans, over the past half-century, a large increase in the consumption of added sugars has occurred, particularly from sugar-sweetened beverages (SSBs) (25). For example, in children [the highest sugar consumers of any age group (26, 27)], 40% of added sugars come from SSBs (26), which is associated with increased risk of cardiovascular and metabolic disease (27–29) and weight gain (30). Moreover, changes in the food industry in the past decades have created a shift in the biochemical form in which sugars are frequently consumed (25, 31). For example, in the United States, the natural disaccharide sucrose (chemically bound fructose and glucose molecules, 50:50 ratio) has been largely replaced with sweeteners containing unbound fructose and glucose monosaccharide molecules, typically comprising an overall elevation in the fructose-to-glucose ratio compared with sucrose (e.g., high-fructose corn syrup) (32). A current study that used HPLC revealed that the percentage of fructose in high-fructose corn syrup in popular, commercially available SSBs ranged from 47% to 65%, with a mean fructose content of 59% (33). Thus, due to the excessive use of added sugars in the modern food environment, particularly in the form of SSBs, and the industrial development of sweeteners with monosaccharide ratios that diverge from those present in foods in their native form (typically with a higher fructose content than sugar), the current study elucidates the impact of SSBs that vary in the glucose-to-fructose ratio on gut microbial populations.
Evidence from human studies suggests that the gut microbiota that is present during early postnatal and adolescent periods likely plays a major role in subsequent health and disease (34, 35). Rodent studies in which the gut microbiota was manipulated during this critical period confirmed that developmental abnormalities present in germ-free mice are reversible when these mice were colonized with intestinal bacteria during early life but not during adulthood (36, 37). Because added sugars make up an increasing proportion of the diet during early-life periods of development in humans, and the gut microbiota during early life may have profound implications for health during the life span, our goal was to determine (by using a rodent model) how the consumption of added sugars (with varying fructose-to-glucose ratios, and in the form of SSBs) during the juvenile and adolescent period of development affects the gut microbiome, and whether these sugar-induced microbiome alterations are related to caloric intake and body weight gain.
Methods
Experimental design
Effect of different monosaccharide ratios of sugar on the fecal microbiome.
Forty-two juvenile male Sprague-Dawley rats [Envigo; postnatal day (PND) 26; 50–70 g] were housed individually in standard conditions with a 12:12 light-dark cycle and were classified into 4 groups on the basis of solution feeding of the following: 1) 35% fructose and 65% glucose (n = 11), 2) 65% fructose and 35% glucose (n = 11), 3) 50% fructose and 50% glucose (n = 10), and 4) control (no sugar; n = 10). For each of the sugar groups, the concentration of total sugar in solution was 11% wt:vol (comparable to SSBs typically consumed by humans) in reverse osmosis–filtered water. In addition to sugar solutions (or an extra water bottle for the control group), rats were given access to Lab Diet 5001 (29.8% kilocalories from protein, 13.4% kilocalories from fat, 56.7% kilocalories from carbohydrate; PMI Nutrition International) and water ad libitum. Food intake, solution intake, and body weights were monitored 3 times/wk from PNDs 26 to 61, with additional recordings taken at PND 80 (fecal collection) and a terminal recording at PND 92. The percentage of kilocalories consumed from each macronutrient was estimated by first multiplying the grams of feed pellets consumed by the gram percentage of each macronutrient in the feed pellets. The gram contribution of each macronutrient was converted to kilocalories by using the 4-, 4-, and 9-kcal/g conversion factors for carbohydrate, protein, and fat, respectively. This number was then divided by the total number of kilocalories consumed for a percentage contribution of each macronutrient. Feces were collected from the rats according to the following methods: each rat was placed in a sterile cage and gently restrained while lifting its tail until defecation occurred. Feces were immediately placed into dry ice and stored at −80°C until time of processing for RNA sequencing. All of the experiments were performed in accordance with the approval of the Animal Care and Use Committee at the University of Southern California.
A separate group of male Sprague-Dawley rats (n = 42; PND 26; 50–70 g) were housed individually in standard conditions with a 12:12 light-dark cycle and were classified into 4 groups in an identical design to cohort 1. After 6 wk in the same respective conditions, body weights and intakes of feed, sugar, and total kilocalories were similar to those in cohort 1 (Supplemental Figure 1). Body composition was assessed by using a Bruker NMR Minispec LF 90II (Bruker Daltonics, Inc.). The adiposity index was calculated as [fat mass (g)/lean mass (g)] × 100.
Taxonomic classification of 16S ribosomal RNA gene sequences.
Fecal microbiome populations were identified by using next-generation high-throughput sequencing of the V3–V4 variable region of 16S ribosomal RNA (rRNA; Vaiomer SAS). Genomic DNA was isolated and collected from fecal samples, and DNA concentrations were determined by using UV spectroscopy (Nanodrop 2000; ThermoScientific). PCR amplification was performed by using 16S universal primers targeting the V3–V4 region of the bacterial 16S ribosomal gene (Vaiomer universal 16S primers), with a joint pair length encompassing 476-bp amplicons (MiSeq Reagent Kit V3, Illumina Inc.). The detection of sequencing fragments was performed by using MiSeq Illumina technology (Illumina Inc.). The 16S targeted sequences were then clustered into operational taxonomic units (OTUs) before taxonomic assignment and analyzed by using the bioinformatics pipeline as described previously (38, 39). The OTUs and taxonomy classifications were tabulated, and to account for differences in raw counts across the samples the tables were log normalized by using Equation 1:
 |
Multidimensional scaling was performed on the tables by using the “capscale” function of the R statistical software package “vegan” (40) with Bray-Curtis dissimilarity.
Statistical analysis
Two-factor ANOVA (time × group) with Holm-Sidak post hoc analyses were used to determine whether there were group differences in body weight and food, sugar, and total intakes. Data for fat mass, lean mass, and adiposity index were each statistically compared by using 1-factor ANOVA with an α level of 0.05 (GraphPad Prism, version 6.0).
The following analyses were performed by using the statistical software R (41). Bacterial taxa that were differentially abundant in pairwise analysis of dietary groups were identified by using the Kruskal-Wallis nonparametric test, followed by the Benjamini-Hochberg post hoc test with a false discovery rate of P < 0.10. The identified features were then subjected to the linear discriminant analysis (LDA) model with a threshold logarithmic LDA score set at 3.0 and ranked (42). Respective cladograms were generated with genus at the lowest level.
Differences in the abundances of bacteria classified at a given taxonomic level relative to the type of sugar consumed were determined by using the following model:
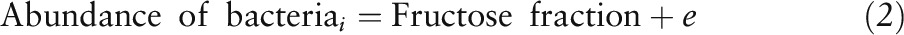 |
where “Abundance” represents log-normalized counts and the fructose fraction was a quantitative variable ranging from 35 to 65. Statistical models were only built for “nonrare taxa,” which were present in ≥25% of all samples. To determine whether the gut microbiota could explain differences in body weight (grams) or energy intake (kilocalories), a series of linear models were built, which we named intake variables, as follows:
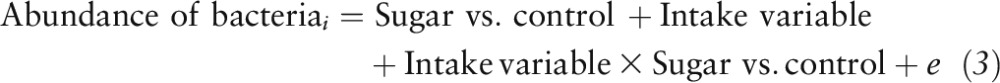 |
One model was built for each combination of the abundance of bacteria (the log-normalized counts at a phylogenetic level) and the intake variables [body weight (g), energy intake (kcal)]. Differences needed to survive the Benjamini-Hochberg post hoc test with a false discovery rate (FDR) of P < 0.10 were deemed to be significant.
Results
Effects of early-life sugar consumption on anthropometric measures.
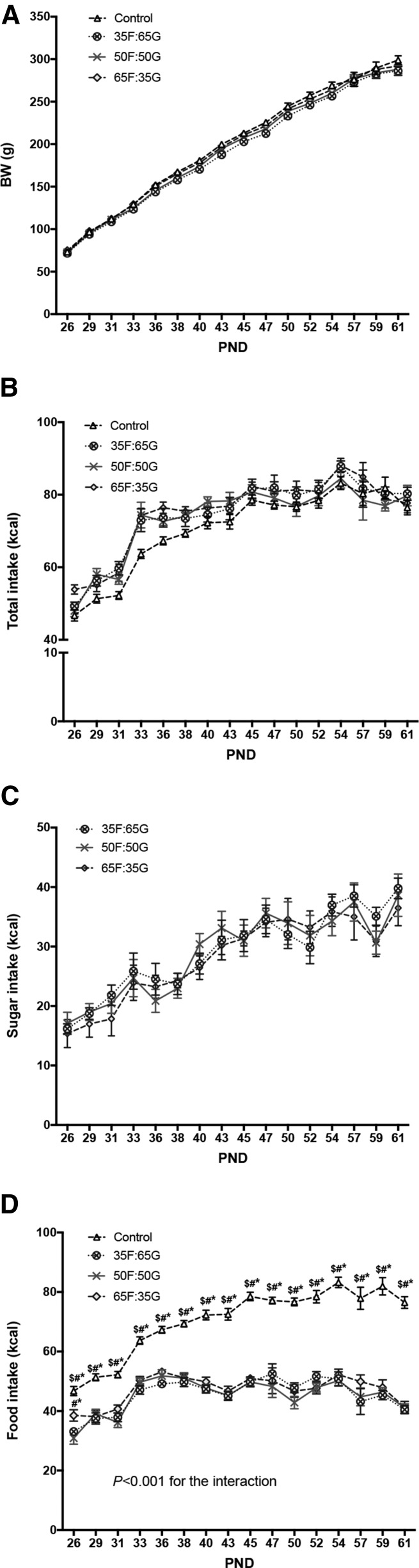
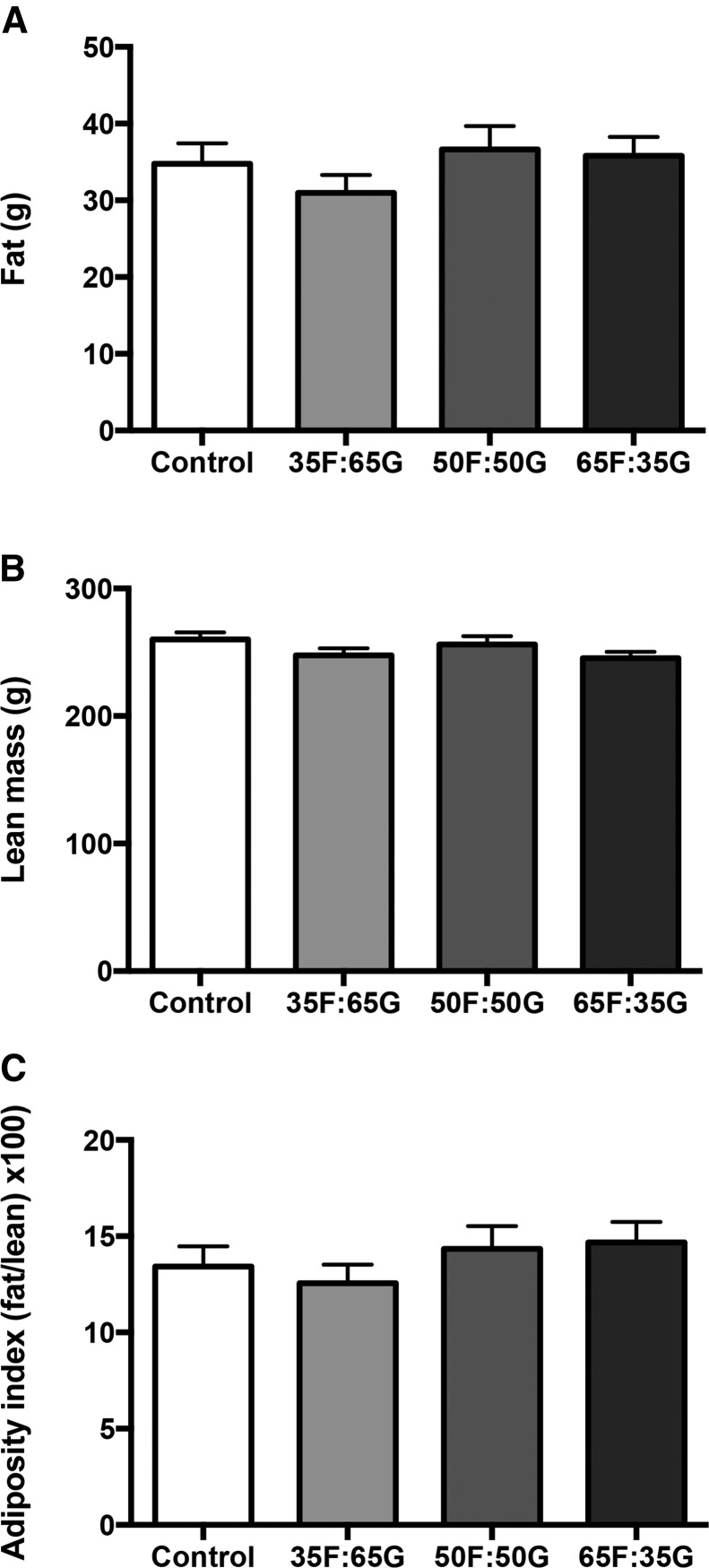
Consistent with our previous report (43), there was no effect of sugar or monosaccharide ratio on body weight gain when sugar was made available ad libitum throughout the entire juvenile and adolescent period (Figure 1A), and there were no differences in overall energy intake between the 4 groups (Figure 1B). Sugar consumption did not differ between groups fed the solutions with different monosaccharide ratios (Figure 1C). The lack of elevated weight gain in the sugar-fed groups is based, at least in part, by compensatory reductions in food intake in the sugar consumers (Figure 1D). When comparing 24-h food intake (Figure 1D), 2-factor ANOVA revealed a significant interaction (time × sugar group; P < 0.0001) with main effects of time (P < 0.0001) and sugar group (P < 0.0001). Post hoc analyses revealed that the sugar groups consumed significantly less food than did controls at each time point (P < 0.0001). Thus, the lack of elevated body weight gain in the sugar-fed groups is based, at least in part, by compensatory reductions in food intake in the sugar consumers. All 3 sugar groups consumed a significantly lower percentage of energy from fat (P < 0.0001) and protein (P < 0.0001), likely due to compensatory reductions in food intake as a result of consuming sugar. There were no differences in body fat (Figure 2A), lean mass (Figure 2B), or overall adiposity index (Figure 2C) between the 4 groups.
FIGURE 1.
Effects of consumption of rats fed 11% sugar solutions containing varying fructose-to-glucose ratios on body weight (A) and total (B), sugar solution (C), and food energy intakes (D) in male rats from PNDs 26 to 61. Values are means ± SEMs; n = 10 or 11. *Different from 35F:65G, P < 0.05; #different from 50F:50G, P < 0.05; $different from 65F:35G, P < 0.05. BW, body weight; PND, postnatal day; 35F:65G, 35% fructose:65% glucose; 50F:50G, 50% fructose:50% glucose; 65F:35G, 65% fructose:35% glucose.
FIGURE 2.
Effect of consumption of rats fed 11% sugar solutions containing varying fructose-to-glucose ratios on changes in body fat (A), lean mass (B), and adiposity index (C) in male rats from PNDs 26 to 61. Values are means ± SEMs; n = 10 or 11. PND, postnatal day; 35F:65G, 35% fructose:65% glucose; 50F:50G, 50% fructose:50% glucose; 65F:35G, 65% fructose:35% glucose.
Taxonomic classification of 16S rRNA sequence reads.
The Ribosomal Database Project classifier was used to assign taxonomy to the 16S rRNA sequence reads and QIIME was used to cluster the sequence reads into OTUs (Table 1).
TABLE 1.
Summary characteristics of 16S rRNA sequence reads from fecal samples of rats fed 11% sugar solutions containing varying fructose-to-glucose ratios1
| OTUs, n | Sequence reads, n | Reads, n/sample | Minimum reads, n/sample | Maximum reads, n/sample | |
| 16S reads generated | 1,237,456 | 29,463.24 ± 411.07 | 21,939 | 34,034 | |
| RDP classified | |||||
| Phylum | 11 | 1,208,210 | 28,766.90 ± 394.82 | 21,508 | 33,392 |
| Class | 21 | 1,191,926 | 28,379.19 ± 393.58 | 21,191 | 32,837 |
| Order | 35 | 1,187,343 | 28,270.07 ± 392.22 | 21,127 | 32,733 |
| Family | 84 | 1,086,649 | 25,872.60 ± 383.17 | 19,116 | 30,283 |
| Genus | 211 | 738,112 | 17,574.10 ± 327.48 | 12,728 | 21,924 |
| QIIME OTUs2 | 4703 | 918,964 | 21,880.10 ± 394.77 | 15,833 | 28,531 |
Values are means ± SEMs unless otherwise indicated; n = 42 samples total. The treatment groups refer to the 3 groups of rats fed sugar solutions (35F:65G, n = 11; 50F:50G, n = 10; 65F:35G, n = 11) and the control group had no access to sugar but received a second water bottle instead. OTU, operational taxonomic unit; RDP, Ribosomal Database Project; rRNA, ribosomal RNA; 35F:65G, 35% fructose:65% glucose; 50F:50G, 50% fructose:50% glucose; 65F:35G, 65% fructose:35% glucose.
More than 25% of samples.
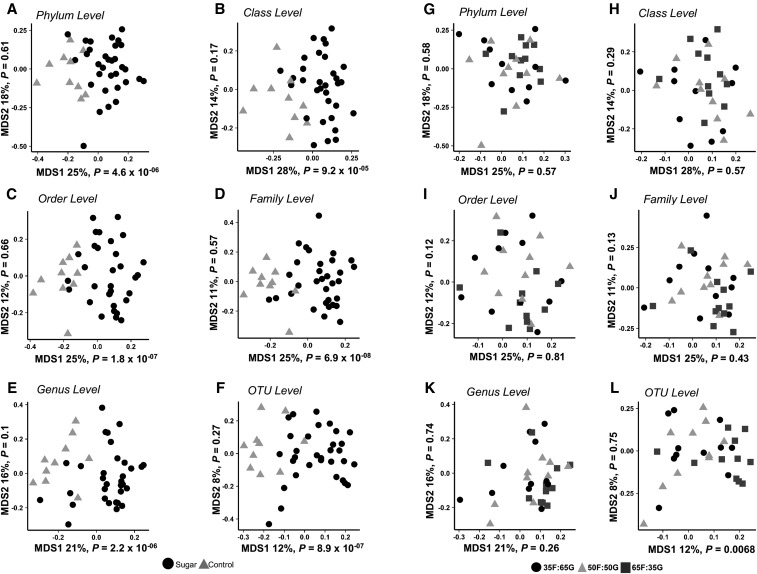
Sugar solution consumption resulted in microbial separation.
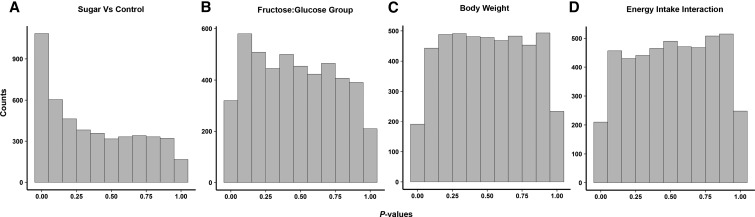
Results from our multidimensional scaling analysis on the taxonomic classification tables at all phylogenetic levels represent a summary of gut microbial composition (Figure 3). Rats fed the sugar solution (black symbols) compared with water (gray symbols) had distinct clustering patterns (Figure 3A–F). There was no clear separation based on the monosaccharide ratio of the sugar solutions administered (Figure 3G–L). The distribution of P values derived from t tests performed separately for each family showed that approximately one-quarter of nonrare bacteria at the family level were significantly different between samples from rats fed a sugar solution and control samples at a 10% FDR (Table 2). These findings are also represented in the distribution of P values derived from statistical tests at the OTU level, which, unlike the case for sugar compared with no sugar (Figure 4A), are approximately uniform (Figure 4B) when evaluating the linear models in which the fructose-to-glucose ratio is compared with the relative abundance of each OTU. Likewise, none of the family-level bacteria had a difference in abundance with respect to sugar group at a FDR threshold of 10% (Supplemental Table 1).
FIGURE 3.
Summary of clustering patterns of fecal microbiota from rats fed 11% sugar solutions containing varying fructose-to-glucose ratios or controls by using multidimensional scaling. A distinct clustering pattern of fecal microbiota was observed between rats fed a sugar solution (n = 32) and the control group that received water instead of sugar (n = 10) (A–F). No distinct clustering pattern of fecal microbiota was observed at any phylogenetic level as a result of fructose-to-glucose ratio (n = 10 or 11) (G–L; based on a significance level of P < 0.05). MDS, multidimensional scaling; OTU, operational taxonomic unit; 35F:65G, 35% fructose:65% glucose; 50F:50G, 50% fructose:50% glucose; 65F:35G, 65% fructose:35% glucose.
TABLE 2.
Comparison of fecal bacteria at the family level between rats fed sugar solution and control rats (no sugar)1
|
P |
|||||
| Bacteria family | Mean log-normalized sequence count in sugar samples ± SE | Mean log-normalized sequence count in control samples ± SE | t-Statistic2 | Sugar vs. control | BH-corrected |
| Enterobacteriaceae | 1.524 ± 0.017 | 0.477 ± 0.035 | 7.082 | <0.001 | <0.001 |
| Carnobacteriaceae | 0.900 ± 0.011 | 0.184 ± 0.025 | 7.248 | <0.001 | <0.001 |
| Corynebacteriaceae | 0.265 ± 0.010 | 0.000 ± 0.000 | 4.790 | <0.001 | <0.001 |
| Bifidobacteriaceae | 1.749 ± 0.030 | 0.609 ± 0.059 | 4.519 | <0.001 | 0.002 |
| Rikenellaceae | 2.970 ± 0.006 | 2.425 ± 0.029 | 5.511 | <0.001 | 0.002 |
| Clostridiaceae.1 | 2.611 ± 0.019 | 1.580 ± 0.057 | 4.918 | <0.001 | 0.002 |
| Cryomorphaceae | 0.318 ± 0.009 | 0.057 ± 0.012 | 4.151 | <0.001 | 0.002 |
| Lactobacillaceae | 2.684 ± 0.009 | 2.266 ± 0.025 | 4.467 | <0.001 | 0.003 |
| Moraxellaceae | 0.421 ± 0.014 | 0.086 ± 0.014 | 3.801 | <0.001 | 0.004 |
| Micrococcaceae | 0.499 ± 0.013 | 0.099 ± 0.023 | 3.866 | 0.001 | 0.004 |
| Bacteroidaceae | 3.371 ± 0.005 | 3.151 ± 0.014 | 4.102 | 0.001 | 0.005 |
| Prevotellaceae | 3.583 ± 0.007 | 3.804 ± 0.016 | −3.521 | 0.002 | 0.012 |
| Coriobacteriaceae | 1.842 ± 0.008 | 1.636 ± 0.016 | 3.075 | 0.005 | 0.029 |
| Pseudomonadaceae | 0.428 ± 0.013 | 0.191 ± 0.017 | 2.629 | 0.012 | 0.07 |
Values are presented from bacteria classified at the family level whose abundances are significantly different with a BH-corrected P < 0.10. The sugar group refers to the 3 groups of rats fed sugar solutions combined (35F:65G, n = 11; 50F:50G, n = 10; 65F:35G, n = 11; n = 32 total) and the control group had no access to sugar but received a second water bottle instead (n = 10). BH, Benjamini-Hochberg; 35F:65G, 35% fructose:65% glucose; 50F:50G, 50% fructose:50% glucose; 65F:35G, 65% fructose:35% glucose.
The t-statistic is positive when the mean abundance of the bacteria is higher in sugar samples and negative if higher in control samples.
FIGURE 4.
Differences in bacterial abundance between rats fed 11% sugar solutions and control rats (no sugar). The P distribution was obtained from t tests with the null hypothesis that there is no difference in OTU-level bacterial abundance in sugar (n = 32) and control (n = 10) samples (A) and a regression model with the null hypothesis that there is no significant difference in bacterial abundance between rats fed 1 of 3 sugar solutions differing in fructose-to-glucose ratio (B). Also presented is the P distribution obtained from dependent variables of a simple linear regression with OTU-level bacteria abundance as an independent variable and either body weight (C) or energy intake (D) as the dependent variable. OTU, operational taxonomic unit.
Relation of fecal microbiota to body weight and energy intake.
To determine how members of the microbial community are associated with body weight and calorie intake, we executed a series of linear regression models comparing these intake variables with log-normalized adjusted counts. All of these models included a categorical variable for sugar compared with nonsugar. By using Equation 3 as described in Methods at a 10% FDR threshold, there were no significant associations with body weight or food intake to any member of the microbial community and there was no association with any of the interaction terms at any phylogenetic level (Supplemental Figures 2–5). Likewise, the distribution of P values for body weight or calorie intake generated by Equation 3 produced near-uniform P values, suggesting little association (Figure 4C, D).
Effects of added sugar on abundance of fecal microbiota at different taxonomic levels.
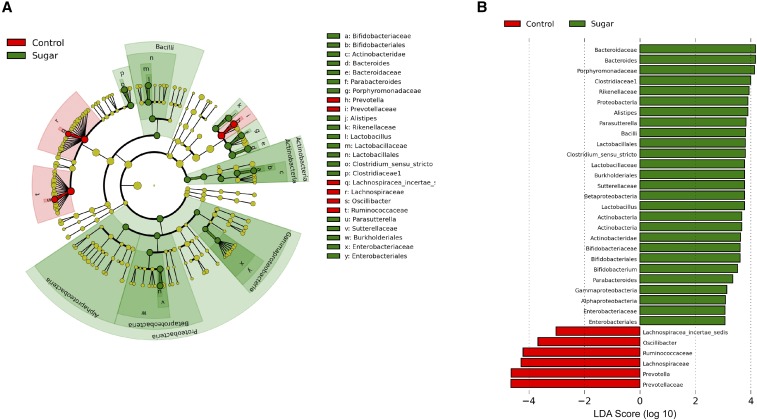
To further explore differences in the microbial community associated with sugar group, pairwise comparisons were made comparing sugar with the control at each phylogenetic level (from phylum to genus; Figure 5). At the phylum level, Proteobacteria (t-statistic = 3.44, P = 0.005) and Actinobacteria (t-statistic = 4.70, P = 0.002) were elevated in all sugar groups compared with controls. At the class level, Actinobacteria (t-statistic = 4.71, P = 0.002) and Bacilli (t-statistic = 4.48, P < 0.001) (of the phyla Actinobacteria and Firmicutes, respectively) were significantly elevated by sugar, as were Alpha- (t-statistic = 2.01, P = 0.04), Beta- (t-statistic = 2.61, P = 0.02), and Gamma- (t-statistic = 8.17, P < 0.001) Proteobacteria (of the phylum Proteobacteria). Bacteria of the orders Lactobacillales (t-statistic = 4.45, P < 0.001), Actinobacteridae (t-statistic = 3.45, P < 0.001), Burkholderiales (t-statistic = 2.59, P = 0.02), and Enterobacteriales (t-statistic = 7.07, P < 0.001) were significantly elevated by added sugar. According to our LDA effective size analysis, many taxa were significantly different between sugar and control at the family level: for example, Clostridiaceae 1 (LDA score = 3.97, P < 0.001), Lactobacillaceae (LDA score = 3.78, P < 0.001), Rikenellaceae (LDA score = 3.93, P < 0.001), Porphyromonadaceae (LDA score = 3.36, P = 0.03), Bacteroidaceae (LDA score = 4.19, P < 0.001), Bifidobacteriales (LDA score = 3.59, P = 0.001), Sutterellaceae (LDA score = 3.73, P = 0.02), and Enterobacteriaceae (LDA score = 3.09, P < 0.001) were elevated by sugar, whereas Prevotellaceae (LDA score = −4.61, P = 0.002), Ruminococcaceae (LDA score = −4.24, P = 0.02), and Lachnospiraceae (LDA score = −4.37, P = 0.04) were reduced due to sugar consumption. At the genus level, Prevotella (LDA score = −4.62, P < 0.001) and Lachnospiracea incertae sedis (LDA score = −3.01, P = 0.03) were reduced by sugar consumption, whereas Bacteroides (LDA score = 4.19, P < 0.001), Alistipes (LDA score = 3.88, P < 0.001), Lactobacillus (LDA score = 3.78, P < 0.001), Clostridium sensu stricto (LDA score = 3.77, P < 0.001), Bifidobacteriaceae (LDA score = 3.59, P = 0.001), and Parasutterella (LDA score = 3.79, P = 0.004) were all significantly elevated by sugar consumption.
FIGURE 5.
Cladogram (A) and LDA scores (B) indicating statistical differences between microbial populations in rats fed 11% sugar solution compared with control (no sugar) groups. Data are presented from the phylum to the genus level with the higher order at the outermost level (i.e., phylum, class, order, family, genus). Colors indicate the group with the highest mean of differential features in which significant differences were found. Values are means ± SEMs; n = 10 or 11. LDA, linear discriminant analysis.
Discussion
Herein we report, with the use of a rat model, that the gut microbiome is affected by added-sugar consumption during the juvenile and adolescent stage of development and that these differences are independent of obesity status and caloric intake. Moreover, the monosaccharide ratio of fructose to glucose did not significantly contribute to the overall effects of sugar consumption on microbial populations. Given that we used added sugars designed to model those commonly consumed in beverages in human populations, both in terms of caloric content and monosaccharide ratio, the present findings may have implications with regard to the relation between added sugars and the gut microbiome in humans, although the translational potential of the present results requires further epidemiologic and experimental studies in humans.
To our knowledge, this is the first investigation of how added sugars affect the gut microbiome during the juvenile and adolescent period of development, during which the brain is particularly vulnerable to the effects of sugar and other dietary factors (44). However, the effects of added dietary sugars on gut microbial populations have been investigated in adult rodents. For example, high-sucrose diets (containing 70% of kcal from carbohydrate, mainly in the form of sucrose) have previously been shown to elevate Clostridiales (a class of Firmicutes) and reduce Bacteroidales (an order of the phylum Bacteroidetes) in adult rodents (45). In the present study we did not observe changes in either one of these populations due to sugar consumption. The age of the rats (adult compared with juvenile and adolescent), the percentage of total kilocalories from sugar (70% compared with ∼40%), and the chemical composition of the sugar (disaccharide sucrose compared with free monosaccharides) may contribute to these differences. Another study found that a diet enriched in the monosaccharide fructose increases the population of the genus Coprococcus and Ruminococcus (both in the phylum Firmicutes) in adult rodents, and that either antibiotic treatment or a fecal microbiome transfer from rodents fed a healthy control diet reduces both the populations of these species as well the fructose-induced metabolic disease (46). We did not observe differences in Coprococcus or Ruminococcus in rats fed the highest dose of fructose; however, our study differed in that fructose was consumed by free choice in liquid form compared with 20.4% fructose in the feed pellets in this previous study. We did not see an effect of added sugars during the juvenile and adolescent period on body weight gain in our rodent model, nor did we observe an association between fecal microbiota and body weight. Thus, we were able to investigate the impact of added sugars on the gut microbiome during early life independently of obesity. This is an important benefit to our design, because obesity is associated with an altered composition of the gut microbiome (10, 47–49). Added sugars have been shown to contribute to obesity and metabolic disease (27–30); therefore, an understanding of how sugars affect the gut microbiome during early-life periods and independent of obesity may help identify putative causal factors for the obesogenic effects of added sugar. Related to this, we observed that early-life sugar consumption significantly elevated Proteobacteria, and more specifically within this phylum, microbes from Enterobacteriaceae were increased by added-sugar consumption. Enterobacteriaceae is an abundant family of gram-negative bacteria that are also elevated in type 2 diabetes (50) and has recently been linked with artificial sweetener consumption (9). SSB consumption is associated with type 2 diabetes (51), and thus further investigations on the impact of Enterobacteriaceae on host glucose metabolism may be a promising avenue for future research.
At the genus level, several species were elevated in all 3 sugar groups, some of which were previously associated with various health and disease processes. Parabacteroides, a genus in the phylum Bacteroidetes, were significantly elevated by sugar. Parabacteroides were previously shown to be elevated due to the consumption of soluble corn fiber, and Parabacteroides counts were negatively correlated with calcium absorption in adolescents (52). Clostridium sensu stricto was also elevated by sugar and has been correlated with the development of food allergy or food sensitization during early life (53, 54) and atopic dermatitis during childhood (55). Lactobacillus, a strain of bacteria associated with promoting regulatory T helper cells (56) and preserving tight junctions in the epithelial cells of the intestinal tract (57), was increased by sugar consumption, whereas previous studies showed that this strain is reduced in mice fed an HFD (58–61). Alistipes (genus in the family Rikenellaceae) and Bacteroides (genus in the family Bacteroidaceae) were also significantly elevated in all sugar groups compared with controls. A recent study identified both Alistipes and Bacteroides as being rapidly elevated in people who consumed an animal-based diet (consisting of meats, eggs, and cheese) compared with a plant-based diet (containing grains, legumes, fruit, and vegetables) (17). Bacteroides are highly equipped to utilize polysaccharides and contain many enzymes for hydrolyzing glycans, suggesting that they might thrive on a more polysaccharide-rich diet (62, 63). However, in our current study, rats that were fed sugar solutions consumed a greater proportion of calories from monosaccharides and a reduced contribution of calories from the polysaccharide-rich feed pellets (Lab Diet 5001). Thus, the increased amount of Bacteroides observed in the sugar groups is unexpected. Koropatkin et al. (63) postulated that elevated proportions of bacteria with glycolytic activity (e.g., Bacteroides) observed after the consumption of a high-fat and low-fiber diet could be consequent to the capacity for Bacteroides to metabolize host mucosal glycans. Thus, it is possible that the elevated consumption of simple sugars, which can be readily absorbed from the proximal intestine, promotes a competitive advantage for microbes in the distal gastrointestinal tract that are capable of finding an alternative food source (e.g., the host mucosal glycans).
Sugar consumption reduced counts of Prevotellaceae, a member of the Bacteroidetes class. This effect was primarily driven by reductions in the genus Prevotella, a gram-negative bacterium that aids in the breakdown of protein and carbohydrates (64). In humans, Prevotella amounts also correlate with regular fiber intake and are reduced with a high-protein diet (17). Thus, it is possible that the reduced Prevotella we observed after SSB consumption was due to lower intakes of complex carbohydrate and/or fiber relative to controls. Oscillibacter and Lachnospiracea incertae sedis, which are both genera of the Firmicutes phylum and the Clostridia class, were also reduced by sugar. Oscillibacter is negatively correlated with Crohn disease (65, 66) and is implicated as a potential treatment for ulcerative colitis after fecal transfer (67). Lachnospiracea incertae sedis are involved in the fermentation of starches to produce SCFAs (68). Further research is needed to determine the implications of the reduction in these intestinal health–promoting bacteria due to added sugars on colonic health and disease.
In summary, early-life sugar consumption significantly alters the gut microbiome independently of obesity and total caloric intake in a rodent model. Sugar promoted multiple differences in the microbiota at all taxonomic levels; however, there was no apparent effect of glucose-to-fructose ratio between the groups. These seemingly novel findings lay the groundwork for future studies to focus on the functional implications of these sugar-induced alternations in the gut microbiota on metabolic and cognitive disorders associated with elevated sugar consumption during the juvenile and adolescent period of development.
Acknowledgments
EEN wrote the manuscript and contributed to the research project design; TMH conducted the experiments and analyzed the data; RBJ analyzed the data and contributed to writing the manuscript; AAF analyzed the data and contributed to the project design; and MIG and SEK edited the manuscript, designed the research project, and had primary responsibility for the research content. All authors read and approved the final manuscript.
Footnotes
Abbreviations used: FDR, false discovery rate; HFD, high-fat diet; LDA, linear discriminant analysis; OTU, operational taxonomic unit; PND, postnatal day; rRNA, ribosomal RNA; SSB, sugar-sweetened beverage.
References
- 1.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science 2005;307:1915–20. [DOI] [PubMed] [Google Scholar]
- 2.Morelli L. Postnatal development of intestinal microflora as influenced by infant nutrition. J Nutr 2008;138:1791S–5S. [DOI] [PubMed] [Google Scholar]
- 3.Walker AW, Lawley TD. Therapeutic modulation of intestinal dysbiosis. Pharmacol Res 2013;69:75–86. [DOI] [PubMed] [Google Scholar]
- 4.Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu XN, Kubo C, Koga Y. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J Physiol 2004;558:263–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke G, Grenham S, Scully P, Fitzgerald P, Moloney RD, Shanahan F, Dinan TG, Cryan JF. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol Psychiatry 2013;18:666–73. [DOI] [PubMed] [Google Scholar]
- 6.Diaz Heijtz R, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A, Hibberd ML, Forssberg H, Pettersson S. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci USA 2011;108:3047–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu H, Khosravi A, Kusumawardhani IP, Kwon AH, Vasconcelos AC, Cunha LD, Mayer AE, Shen Y, Wu WL, Kambal A, et al. . Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science 2016;352:1116–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 2012;487:104–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suez J, Korem T, Zeevi D, Zilberman-Schapira G, Thaiss CA, Maza O, Israeli D, Zmora N, Gilad S, Weinberger A, et al. . Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature 2014;514:181–6. [DOI] [PubMed] [Google Scholar]
- 10.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006;444:1027–31. [DOI] [PubMed] [Google Scholar]
- 11.Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA 2004;101:15718–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, et al. . Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 2013;155:1451–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wood H. Parkinson disease. Gut reactions—can changes in the intestinal microbiome provide new insights into Parkinson disease? Nat Rev Neurol 2015;11:66. [DOI] [PubMed] [Google Scholar]
- 14.Hill JM, Bhattacharjee S, Pogue AI, Lukiw WJ. The gastrointestinal tract microbiome and potential link to Alzheimer’s disease. Front Neurol 2014;5:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dietert RR. The microbiome in early life: self-completion and microbiota protection as health priorities. Birth Defects Res B Dev Reprod Toxicol 2014;101:333–40. [DOI] [PubMed] [Google Scholar]
- 16.Murdoch TB, Detsky AS. Time to recognize our fellow travellers. J Gen Intern Med 2012;27:1704–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. . Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014;505:559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang C, Zhang M, Pang X, Zhao Y, Wang L, Zhao L. Structural resilience of the gut microbiota in adult mice under high-fat dietary perturbations. ISME J 2012;6:1848–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, Keilbaugh SA, Hamady M, Chen YY, Knight R, Ahima RS, Bushman F, Wu GD. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology 2009;137:1716–24 e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, et al. . Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 2008;455:1109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas F, Hehemann JH, Rebuffet E, Czjzek M, Michel G. Environmental and gut Bacteroidetes: the food connection. Front Microbiol 2011;2:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 2008;453:620–5. [DOI] [PubMed] [Google Scholar]
- 23.Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol 2015;33:496–503. [DOI] [PubMed] [Google Scholar]
- 24.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature 2006;444:1022–3. [DOI] [PubMed] [Google Scholar]
- 25.Bray GA, Nielsen SJ, Popkin BM. Consumption of high-fructose corn syrup in beverages may play a role in the epidemic of obesity. Am J Clin Nutr 2004;79:537–43. [DOI] [PubMed] [Google Scholar]
- 26.Ervin RB, Kit BK, Carroll MD, Ogden CL. Consumption of added sugar among U.S. children and adolescents, 2005–2008. NCHS Data Brief 2012;87:1–8. [PubMed] [Google Scholar]
- 27.Welsh JA, Sharma A, Cunningham SA, Vos MB. Consumption of added sugars and indicators of cardiovascular disease risk among US adolescents. Circulation 2011;123:249–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malik VS, Hu FB. Sweeteners and risk of obesity and type 2 diabetes: the role of sugar-sweetened beverages. Curr Diab Rep 2012;12:195–203. [DOI] [PubMed] [Google Scholar]
- 29.Malik VS, Popkin BM, Bray GA, Despres JP, Hu FB. Sugar-sweetened beverages, obesity, type 2 diabetes mellitus, and cardiovascular disease risk. Circulation 2010;121:1356–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Te Morenga L, Mallard S, Mann J. Dietary sugars and body weight: systematic review and meta-analyses of randomised controlled trials and cohort studies. BMJ 2013;346:e7492. [DOI] [PubMed] [Google Scholar]
- 31.Bray GA, Popkin BM. Dietary sugar and body weight: have we reached a crisis in the epidemic of obesity and diabetes? Health be damned! Pour on the sugar. Diabetes Care 2014;37:950–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tappy L, Le KA. Metabolic effects of fructose and the worldwide increase in obesity. Physiol Rev 2010;90:23–46. [DOI] [PubMed] [Google Scholar]
- 33.Ventura EE, Davis JN, Goran MI. Sugar content of popular sweetened beverages based on objective laboratory analysis: focus on fructose content. Obesity (Silver Spring) 2011;19:868–74. [DOI] [PubMed] [Google Scholar]
- 34.Neu J. The microbiome during pregnancy and early postnatal life. Semin Fetal Neonatal Med 2016. Jul 7 (Epub ahead of print; DOI: 10.1016/j.siny.2016.05.001). [DOI] [PubMed] [Google Scholar]
- 35.Neufeld KM, Luczynski P, Oriach CS, Dinan TG, Cryan JF. What’s bugging your teen? The microbiota and adolescent mental health. Neurosci Biobehav Rev 2016;70:300–12. [DOI] [PubMed] [Google Scholar]
- 36.Neufeld KA, Kang N, Bienenstock J, Foster JA. Effects of intestinal microbiota on anxiety-like behavior. Commun Integr Biol 2011;4:492–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diamond B, Huerta PT, Tracey K, Volpe BT. It takes guts to grow a brain: increasing evidence of the important role of the intestinal microflora in neuro- and immune-modulatory functions during development and adulthood. BioEssays 2011;33:588–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lluch J, Servant F, Paisse S, Valle C, Valiere S, Kuchly C, Vilchez G, Donnadieu C, Courtney M, Burcelin R, et al. . The characterization of novel tissue microbiota using an optimized 16S metagenomic sequencing pipeline. PLoS One 2015;10:e0142334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Païsse S, Valle C, Servant F, Courtney M, Burcelin R, Amar J, Lelouvier B. Comprehensive description of blood microbiome from healthy donors assessed by 16S targeted metagenomic sequencing. Transfusion 2016;56:1138–47. [DOI] [PubMed] [Google Scholar]
- 40.Oksanen J, Guillaume Blanchet F, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, et al. . Version R package version 2.3. 2016. [cited 2016 Oct 5]. Available from: https://cran.r-project.org, https://github.com/vegandevs/vegan.
- 41.R Development Core Team. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2015. [Google Scholar]
- 42.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol 2011;12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsu TM, Konanur VR, Taing L, Usui R, Kayser BD, Goran MI, Kanoski SE. Effects of sucrose and high fructose corn syrup consumption on spatial memory function and hippocampal neuroinflammation in adolescent rats. Hippocampus 2015;25:227–39. [DOI] [PubMed] [Google Scholar]
- 44.Noble EE, Kanoski SE. Early life exposure to obesogenic diets and learning and memory dysfunction. Curr Opin Behav Sci 2016;9:7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Magnusson KR, Hauck L, Jeffrey BM, Elias V, Humphrey A, Nath R, Perrone A, Bermudez LE. Relationships between diet-related changes in the gut microbiome and cognitive flexibility. Neuroscience 2015;300:128–40. [DOI] [PubMed] [Google Scholar]
- 46.Di Luccia B, Crescenzo R, Mazzoli A, Cigliano L, Venditti P, Walser JC, Widmer A, Baccigalupi L, Ricca E, Iossa S. Rescue of fructose-induced metabolic syndrome by antibiotics or faecal transplantation in a rat model of obesity. PLoS One 2015;10:e0134893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 2005;102:11070–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 2008;3:213–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. . A core gut microbiome in obese and lean twins. Nature 2009;457:480–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lambeth SM, Carson T, Lowe J, Ramaraj T, Leff JW, Luo L, Bell CJ, Shah VO. Composition, diversity and abundance of gut microbiome in prediabetes and type 2 diabetes. J Diabetes Obes 2015;2:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang M, Yu M, Fang L, Hu RY. Association between sugar-sweetened beverages and type 2 diabetes: a meta-analysis. J Diabetes Investig 2015;6:360–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whisner CM, Martin BR, Nakatsu CH, Story JA, MacDonald-Clarke CJ, McCabe LD, McCabe GP, Weaver CM. Soluble corn fiber increases calcium absorption associated with shifts in the gut microbiome: a randomized dose-response trial in free-living pubertal females. J Nutr 2016;146:1298–306. [DOI] [PubMed] [Google Scholar]
- 53.Ling Z, Li Z, Liu X, Cheng Y, Luo Y, Tong X, Yuan L, Wang Y, Sun J, Li L, et al. . Altered fecal microbiota composition associated with food allergy in infants. Appl Environ Microbiol 2014;80:2546–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen CC, Chen KJ, Kong MS, Chang HJ, Huang JL. Alterations in the gut microbiotas of children with food sensitization in early life. Pediatr Allergy Immunol 2016;27:254–62. [DOI] [PubMed] [Google Scholar]
- 55.Penders J, Gerhold K, Stobberingh EE, Thijs C, Zimmermann K, Lau S, Hamelmann E. Establishment of the intestinal microbiota and its role for atopic dermatitis in early childhood. J Allergy Clin Immunol 2013;132:601–7, e8. [DOI] [PubMed] [Google Scholar]
- 56.Smelt MJ, de Haan BJ, Bron PA, van Swam I, Meijerink M, Wells JM, Faas MM, de Vos P. L. plantarum, L. salivarius, and L. lactis attenuate Th2 responses and increase Treg frequencies in healthy mice in a strain dependent manner. PLoS One 2012;7:e47244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karczewski J, Troost FJ, Konings I, Dekker J, Kleerebezem M, Brummer RJ, Wells JM. Regulation of human epithelial tight junction proteins by Lactobacillus plantarum in vivo and protective effects on the epithelial barrier. Am J Physiol Gastrointest Liver Physiol 2010;298:G851–9. [DOI] [PubMed] [Google Scholar]
- 58.Lam YY, Ha CW, Campbell CR, Mitchell AJ, Dinudom A, Oscarsson J, Cook DI, Hunt NH, Caterson ID, Holmes AJ, et al. . Increased gut permeability and microbiota change associate with mesenteric fat inflammation and metabolic dysfunction in diet-induced obese mice. PLoS One 2012;7:e34233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patrone V, Ferrari S, Lizier M, Lucchini F, Minuti A, Tondelli B, Trevisi E, Rossi F, Callegari ML. Short-term modifications in the distal gut microbiota of weaning mice induced by a high-fat diet. Microbiology 2012;158:983–92. [DOI] [PubMed] [Google Scholar]
- 60.Qiao Y, Sun J, Ding Y, Le G, Shi Y. Alterations of the gut microbiota in high-fat diet mice is strongly linked to oxidative stress. Appl Microbiol Biotechnol 2013;97:1689–97. [DOI] [PubMed] [Google Scholar]
- 61.Tachon S, Lee B, Marco ML. Diet alters probiotic Lactobacillus persistence and function in the intestine. Environ Microbiol 2014;16:2915–26. [DOI] [PubMed] [Google Scholar]
- 62.Bolam DN, Koropatkin NM. Glycan recognition by the Bacteroidetes Sus-like systems. Curr Opin Struct Biol 2012;22:563–9. [DOI] [PubMed] [Google Scholar]
- 63.Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol 2012;10:323–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA 2010;107:14691–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mondot S, Kang S, Furet JP, Aguirre de Carcer D, McSweeney C, Morrison M, Marteau P, Dore J, Leclerc M. Highlighting new phylogenetic specificities of Crohn’s disease microbiota. Inflamm Bowel Dis 2011;17:185–92. [DOI] [PubMed] [Google Scholar]
- 66.Papa E, Docktor M, Smillie C, Weber S, Preheim SP, Gevers D, Giannoukos G, Ciulla D, Tabbaa D, Ingram J, et al. . Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS One 2012;7:e39242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vermeire S, Joossens M, Verbeke K, Wang J, Machiels K, Sabino J, Ferrante M, Van Assche G, Rutgeerts P, Raes J. Donor species richness determines faecal microbiota transplantation success in inflammatory Bowel disease. J Crohns Colitis 2016;10:387–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Duncan SH, Louis P, Flint HJ. Cultivable bacterial diversity from the human colon. Lett Appl Microbiol 2007;44:343–50. [DOI] [PubMed] [Google Scholar]