Abstract
Late embryogenesis abundant (LEA) proteins, a diverse family, accumulate during seed desiccation in the later stages of embryogenesis. LEA proteins are associated with tolerance to abiotic stresses, such as drought, salinity and high or cold temperature. Here, we report the first comprehensive survey of the LEA gene family in Dendrobium officinale, an important and widely grown medicinal orchid in China. Based on phylogenetic relationships with the complete set of Arabidopsis and Oryza LEA proteins, 17 genes encoding D. officinale LEAs (DofLEAs) were identified and their deduced proteins were classified into seven groups. The motif composition of these deduced proteins was correlated with the gene structure found in each LEA group. Our results reveal the DofLEA genes are widely distributed and expressed in tissues. Additionally, 11 genes from different groups were introduced into Escherichia coli to assess the functions of DofLEAs. Expression of 6 and 7 DofLEAs in E. coli improved growth performance compared with the control under salt and heat stress, respectively. Based on qPCR data, all of these genes were up-regulated in various tissues following exposure to salt and heat stresses. Our results suggest that DofLEAs play an important role in responses to abiotic stress.
In nature, abiotic stresses, such as drought, high temperature, salinity, cold, and heavy metal pollution, have a serious effect on plant growth and development1, with drought, high temperature, and salinity being associated with water limitation. High temperature damages cell membrane integrity, inhibits photosynthesis, and leads to cellular aging and death. Salt stress decreases total biomass, limits photosynthesis and affects the metabolic system1,2,3. Higher plants have evolved a wide variety of defense mechanisms in response to adverse conditions, often manifesting in the synthesis of a series of functional proteins to reduce damage and protect cells. For example, accumulation of late embryogenesis abundant (LEA) proteins is an important response to abiotic stress4,5.
LEA proteins were first shown to accumulate in seeds during the later stages of embryogenesis. Since then, these proteins have been detected in seedlings, stems, and roots and also in anhydrobiotic bacteria and invertebrates6. LEA proteins comprise a group of small and hydrophilic polypeptides (MW = 10–30 kDa). They are predominantly composed of a repeating arrangement of hydrophilic amino acids that form a highly hydrophilic structure with high heat stability. In addition, LEA proteins have been shown to protect plant metabolism against abiotic stresses, with properties that include antioxidant activity, metal ion binding, membrane and protein stabilization, hydration buffering, and DNA and RNA interactions4,6.
Based on repeated motifs, amino acid composition and phylogenetic relationships, plant LEA proteins can be classified into 8 subgroups (LEA1, LEA2, LEA3, LEA4, LEA5, LEA6, dehydrin and SMP), as opposed to 6 or 7 subgroups suggested by prior studies7,8. However, LEA family proteins have thus far only been investigated in a few plant species, such as Arabidopsis, rice, poplar, Chinese plum, legume, tomato, potato, barley and Pinus9,10,11,12,13,14,15,16. In contrast, very few LEA genes have been identified in Orchidaceae, and studies on LEA family evolutionary relationships and functional characteristics are scarce.
Dendrobium officinale, a herbaceous perennial plant, has great ornamental and economic value as one of the most important plants used in traditional Chinese medicine. D. officinale is a typical epiphytic orchid. It grows compatibly on tree trunks in primeval forests or on damp rock of mountain climates at 500–1600 meters in warm and humid environments17. However, this orchid is easily influenced by abiotic stress, such as drought, high temperature, and salinity, causing an extremely low natural reproduction rate and slow growth in the wild18,19,20. Therefore, it is important to understand the mechanisms underlying the response of D. officinale to abiotic stress. The recent publication of the D. officinale genome has enabled the identification and characterization of the complete repertoire of LEAs in this plant. In the present study, we performed genome-wide analysis of LEA genes in D. officinale. We also investigated the characteristics of the LEA gene family in D. officinale (DofLEA), including evolutionary relationships, putative functions, expression patterns in various tissues and responses to different abiotic stresses.
Results
LEA genes in the D. officinale genome
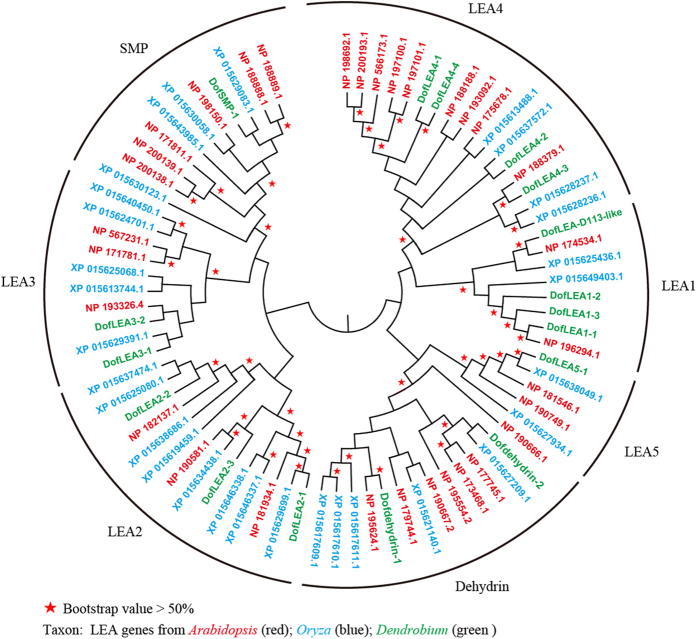
Based on the D. officinale genome and seed transcriptome, TBLASTN searches using Arabidopsis and Oryza LEA proteins as query sequences identified 30 different LEA genes (DofLEAs). Subsequently, we designed primers and successfully cloned 15 and 2 LEA genes from D. officinale seeds and roots, respectively (listed in Supplementary Table S1). Phylogenetic analysis of the predicted proteins revealed that the 17 DofLEA genes could be classified into seven groups together with the Arabidopsis and Oryza LEAs, including 4 LEA1, 3 LEA2, 2 LEA3, 4 LEA4, 1 LEA5, 2 dehydrin and 1 SMP protein-encoding genes (Fig. 1).
Figure 1. Phylogenetic relationships of D. officinale, Oryza and Arabidopsis LEA proteins.
Analysis of physicochemical properties (Table 1) revealed that the DofLEA proteins have molecular weights ranging from 8.8 to 54.8 kDa, with the smallest proteins belonging to the LEA5 group (~8.8 kDa) and the largest proteins belonging to the LEA2 group (~54.8 kDa). The calculated GRAVY values suggested that all DofLEA proteins are quite hydrophilic. Subcellular localization prediction indicated that the LEA1 proteins are located exclusively in the nucleus and mitochondrion; the majority of the LEA2 and SMP proteins are in the cytoplasm, most of the LEA3 proteins in the chloroplast, and the LEA5 and dehydrin proteins in the nucleus.
Table 1. Characteristics of the putative LEA proteins from Dendrobium officinale.
| Group | Protein | Protein length(aa) | Molecular weight(kDa) | GRAVY value | Predicted Subcellular localization |
|---|---|---|---|---|---|
| LEA1 | DofLEA1-1 | 143 | 15.0 | −0.964 | Mitochondrial |
| LEA1 | DofLEA1-2 | 128 | 13.9 | −0.995 | Mitochondrial |
| LEA1 | DofLEA1-3 | 148 | 15.7 | −0.874 | Nuclear |
| LEA1 | DofLEA-D113 | 168 | 17.3 | −0.682 | Nuclear |
| LEA2 | DofLEA2-1 | 307 | 34.3 | −0.392 | Cytoplasmic |
| LEA2 | DofLEA2-2 | 159 | 17.6 | −0.113 | Cytoplasmic |
| LEA2 | DofLEA2-3 | 515 | 54.8 | −0.182 | Cytoplasmic |
| LEA3 | DofLEA3-1 | 102 | 11.1 | −0.064 | Chloroplast |
| LEA3 | DofLEA3-2 | 93 | 10.2 | −0.431 | Chloroplast |
| LEA4 | DofLEA4-1 | 102 | 10.4 | −0.947 | Mitochondrial |
| LEA4 | DofLEA4-2 | 207 | 22.3 | −0.988 | Chloroplast |
| LEA4 | DofLEA4-3 | 238 | 26.9 | −1.616 | Nuclear |
| LEA4 | DofLEA4-4 | 93 | 9.4 | −0.954 | Mitochondrial |
| LEA5 | DofLEA5-1 | 83 | 8.8 | −1.229 | Nuclear |
| SMP | DofSMP-1 | 257 | 27.1 | −0.429 | Cytoplasmic |
| dehydrin | Dofdehydrin-1 | 181 | 18.7 | −1.127 | Nuclear |
| dehydrin | Dofdehydrin-2 | 250 | 27.7 | −1.179 | Nuclear |
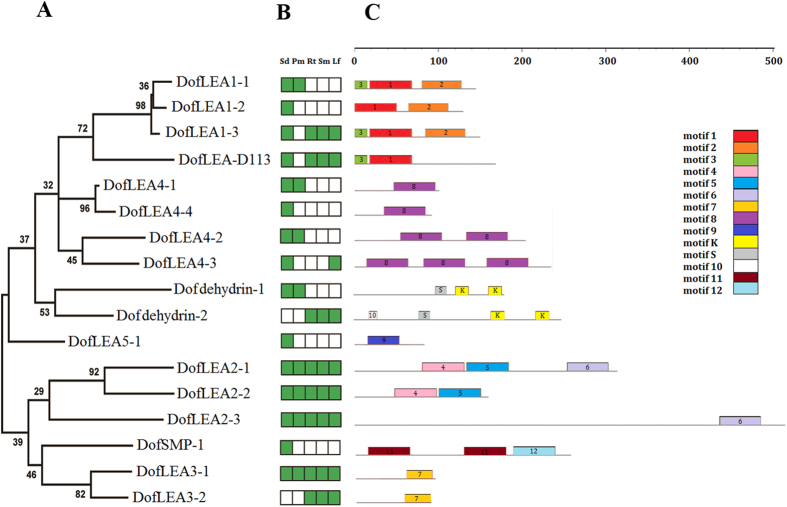
Motif analysis of the DofLEA proteins showed that members of each LEA group possess several group-specific conserved motifs (Figs 2-C and S1). Similar characteristics have also been reported for LEA proteins in Arabidopsis, Prunus, poplar and tomato12,13,16,21. For example, an important conserved motif in the dehydrin group is the repeated motif, EKKGIMDKIKEKLPG (motif K, richness in lysine residues). In addition, the LEA4 group harbors several repeats of conserved motif 8. The DofSMP-1 protein contains repeated group-specific motif 11, with two repetitions.
Figure 2.
Phylogenetic relationships (A), expression patterns (B) and motif structure (C) of D. officinale LEAs. In B, the green box indicates positive detection of gene expression in the corresponding tissue: seed (Sd), protocorm (Pm), root (Rt), stem (Sm) and leaf (Lf). In C, the motif sequences are provided in Supplementary Figure S1.
Expression patterns of DofLEA genes in different tissues
We collected five different tissues (seed, protocorm, root, stem and leaf, Fig. 2-B) to analyze the expression pattern of D. officinale LEA genes. Based on qPCR results (Figs 2-B and S2), four DofLEA genes (DofLEA2-1, DofLEA2-2, DofLEA2-3 and DofLEA3-1) were found to be expressed in all tissues. Four DofLEA genes (DofLEA1-2, DofLEA4-4, DofLEA5-1 and DofSMP-1) were only expressed in the seed. In contrast, no detectable seed expression was found for two DofLEA genes (DofLEA3-2 and Dofdehydrin-2); thus, we cloned these genes from the D. officinale roots. The majority of DofLEA genes (15 genes) were expressed in the seed, suggesting that LEA genes play an important role in seed maturation in D. officinale. Nine DofLEA genes were expressed in vegetative organs (root, stem or leaf), indicating that LEA genes are also involved in normal plant growth and development. Overall, our results showed that the LEA gene family is widely distributed and expressed in this orchid.
Enhancement of the heat and salt tolerance of transformed E. coli (pET28a-DofLEA)
To determine the function of LEA proteins in D. officinale under stress conditions, we selected 1–2 genes from each group (11 genes in total, shown in Supplementary Figure S3), transformed constructs into E. coli (pET28a-DofLEA), and applied heat and salt treatments. The cell viability ratio of E. coli transformed with the pET28a-DofLEA vector and the control containing the empty vector (pET28a) were measured under salt and heat stress.
In the group exposed to heat treatment, strains carrying 6 DofLEA genes (DofLEA2-2, DofLEA2-3, DofLEA4-3, DofLEA1-3, DofLEA5-1 and Dofdehydrin-1) had mean viability ratios 2-10-fold higher than those of the control strain under heat stress (Figs 3 and S4). Interestingly, these DofLEA genes belong to different groups. Among these genes, DofLEA4-3 showed the highest viability ratio after 50 °C induction for 180 and 240 min; after 50 °C induction for 120 min, DofLEA2-3 had a mean viability ratio ~3-fold higher than that of the control. In addition, the viability ratios for DofLEA2-2, DofLEA1-3, DofLEA5-1 and Dofdehydrin-1 were higher than that of the control strain after 50 °C induction for 180 min. There were no differences in mean viability ratios between other LEA genes and the control strain.
Figure 3.
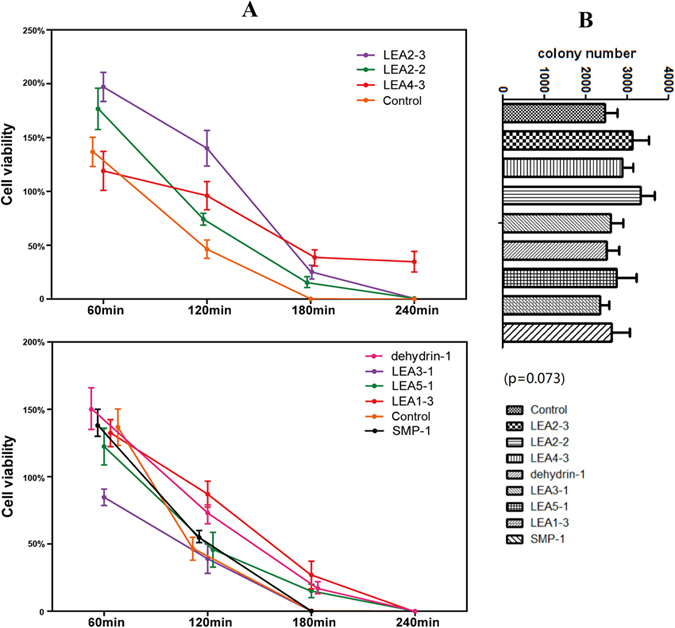
Cell viability ratio of E. coli with pET28a-LEA and pET28a (as control gourp) under heat treatment (A) and the initial colony number without heat treatment (B). (A) Cell viability ratio = (colony number on heated plate/colony number on unheated plate) × 100%. The procedure of heat treatment is shown in Figure 6S. The values are the mean ± SE of three samples. (B) The initial colony number of un-treated plates (without heat treatment), n = 3, means ± SE (one-way ANOVA, p > 0.05).
After salt treatment (Figs 4 and S5), our results demonstrated that strains carrying 7 of the 11 genes showed signs of increased salt tolerance, with viability ratios higher than that of the control strain. In this case, 6 genes (DofLEA2-2, DofLEA3-1, DofLEA4-2, DofLEA5-1, DofSMP-1 and Dofdehydrin-1) from different groups, but not DofLEA1-1, resulted in notably increased salt tolerance. Among these genes, DofSMP-1 (2-5-fold), DofLEA5-1 (~3.5-fold), and Dofdehydrin-1 (2-7-fold) had significantly higher survival ratios compared to the other genes under different concentrations of salt.
Figure 4.
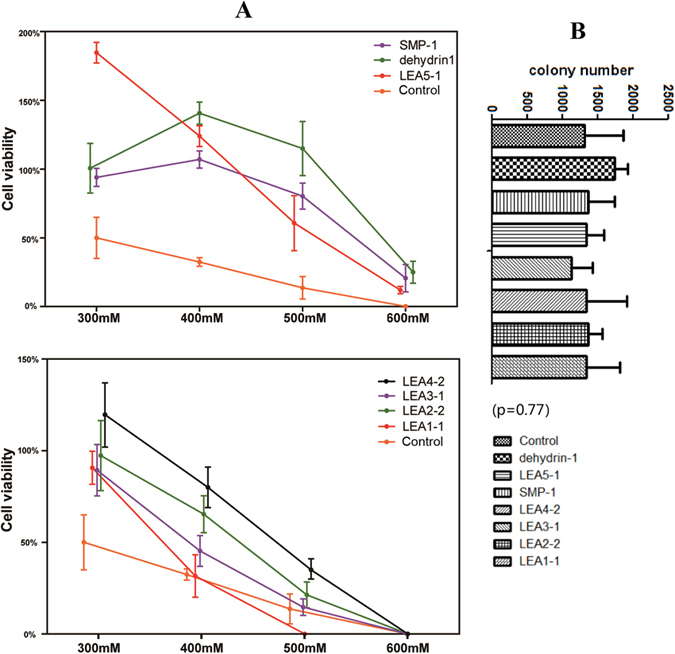
Cell viability ratio of E. coli with pET28a-LEA and pET28a (as control group) under salt treatment (A) and the initial colony number without salt treatment (B). (A) Cell viability ratio = (colony number on salted plate/colony number on unsalted plate) × 100%. The procedure of salt treatment is shown in Figure 6S. The values are the mean ± SE of three samples. (B) The initial colony number of un-treated plates (without salt treatment), n = 3, means ± SE (one-way ANOVA, p > 0.05).
Expression patterns of DofLEA genes under stress treatments
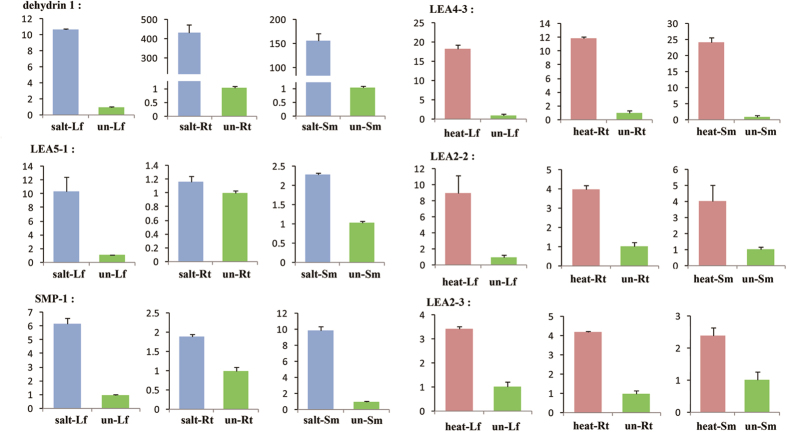
To identify DofLEA genes with a potential role in abiotic stress response, we investigated the expression patterns of 11 candidate genes using qPCR analysis in D. officinale plantlets exposed to heat and salinity. The results of changes in expression of 6 genes are shown in Fig. 5.
Figure 5. Quantitative PCR analysis of the selected genes.
Abbreviations: salt- (salt-treated), heat- (heat-treated), un- (un-treated), Rt (root), Sm (stem) and Lf (leaf), e.g., salt-Lf is the leaf from salt-treated plantlets. Y-axis: relative expression (2−ΔΔCT).
Analysis of DofLEA gene expression following heat treatment showed induction of DofLEA4-3, DofLEA2-2 and DofLEA2-3 mRNA expression in the leaf, stem, and root. Our results also indicated that salt treatment induced the expression of Dofdehydrin-1, DofLEA5-1 and DofSMP-1 in the leaf and stem. Overall, the results of expression patterns of most DofLEAs under heat or salt stress were consistent with the transformed E. coli data, indicating the validity of our experimental results.
Discussion
Orchidaceae is one of the largest and most widespread families of flowering plants (more than 250,000 species), and the genus Dendrobium is one of the largest genera (more than 1000 species). D. officinale, an important medicinal herb in China, is critically endangered in the wild17. In nature, it grows as an epiphyte on trees or on rocks. Abiotic stress, such as drought, high temperature, and salinity, strongly influences the growth and development of D. officinale18,19. LEA genes have been shown to play important roles in abiotic stress responses of Arabidopsis, Oryza, Populus and other plants11,12,16. In contrast, to date, few LEA genes have been described in Orchidaceae. In this study, we identified 30 DofLEA genes in the D. officinale genome, a number similar to that reported in the genomes of rice (34), Arabidopsis (51), Prunus (30), poplar (53), tomato (27) and potato (29)11,12,13,14,16,21. In total, we successfully cloned 17 LEA genes from D. officinale. Based on phylogenetic analysis, these 17 DofLEA genes belong to seven groups of the LEA gene family.
Motif analysis of the proteins encoded by DofLEA genes showed that members of each LEA group contain conserved motifs (Fig. 2) that have been previously identified in other plant species, including Prunus, potato, tomato and Arabidopsis13,14,16,21. These results suggest that DofLEAs encode functional LEA proteins that have group-specific functions. Additionally, the conserved motifs observed within each LEA group indicate that their members likely originated from gene expansion within the groups.
Our results also indicated that DofLEA proteins are rather hydrophilic. Similar characteristics have also been reported for LEA proteins in Arabidopsis, Prunus, poplar and tomato, indicating that these proteins are evolutionarily conserved in higher plants13,14,16,21.
Subcellular localization analyses showed the present of DofLEA proteins in all subcellular compartments, including nucleus, mitochondria, chloroplast, and cytoplasm, as also reported for Arabidopsis and tomato LEAs16,21. There is a strong inference that LEA proteins from the principal groups are ubiquitous within cells and their respective tissues, suggesting that their function is required in all cellular compartments during stress4. While our results showed that all the DofLEA2 were in the cytoplasm. In the case of the maize LEA2 protein, distribution between nucleus and cytoplasm is controlled by phosphorylation of its serine stutter: removal of this sequence results in lack of phosphorylation and retention in the cytoplasm4. In addition, our result also showed that two DofLEA1 proteins were in the nucleus. Prior research classified all of the group 1 LEA proteins as nuclear, and one of these as DNA-binding. Moreover, most of the DofLEA3 proteins were in the chloroplast. Similarly, CD and FT-IR spectroscopic analyses have revealed that group 3 LEA proteins localize in the cytoplasm, mitochondria, or chloroplasts, and are pre-dominantly unstructured in solution and become mainly α-helical upon desiccation4,22,23.
The development of orthodox seeds concludes by a desiccation phase. The dry seeds then enter a phase of dormancy, also called the afterripening phase, and become competent for germination. At this stage in the development process, seeds acquire the ability to withstand extreme dehydration, LEA proteins have been associated with desiccation tolerance. From our result, 15 DofLEA genes were expressed in the seed, suggesting that LEA genes play an important role in seed maturation. Early and middle phases of seed maturation are dominated by the action of abscisic acid (ABA). Subsequently, ABA levels decline and late maturation follows characterised by the synthesis of LEA proteins, associated to the dehydration process and acquisition of desiccation tolerance. Previous studies showed that when ABA was used to activate LEA protein gene expression prematurely in immature soybean seeds, an corresponding improvement in cell integrity was noted after desiccation stress. The plant hormone ABA plays essential roles in several aspects of plant growth and development, including seed maturation, seed dormancy, and adaptation to environmental stresses such as drought and high salinity4,24,25.
Nine DofLEA genes were expressed in vegetative organs (root, stem or leaf), indicating that LEA genes are also involved in normal plant growth and development. Among them, three DofLEA2 and one DofLEA3 genes were found to be expressed in all tissues. The expression profile of these genes is different from that of the original prototypes (seed-specific or stress-induced expression). Similarly, one dehydrin Xero1 from the group 2 LEA proteins appears to be constitutively expressed. It is not inducible by desiccation or by application of ABA. Prior research has also shown that Q064 31_BETVE, a homologue of the group 3 LEA protein, is itself only constitutively expressed and at low levels4.
To date, functional expression screening of LEA proteins from various plant species has been successfully performed in E. coli exposed to abiotic stress. Previous study showed that a Brassica napus LEA4 gene enhanced the tolerance of E. coli to high temperature and salt stresses26. Another study showed that an SMP protein from tea could enhance E. coli tolerance to various stresses27. Liu et al.28 investigated a maize LEA3 gene expressed in E. coli and reported enhanced tolerance to low temperature. Previous studies have primarily focused on the major groups LEA3, LEA4, SMP and dehydrin, but no Orchidaceae LEA genes have been examined. In this study, 6 (~55%) and 7 (~64%) of the DofLEAs analyzed increased the tolerance of E. coli to heat and salt stresses, respectively. These genes are members of almost all groups of the LEA family, which indicates that these genes have a function in stress response. Thus, we showed for the first time that D. officinale LEA proteins from different LEA subgroups enhance tolerance to salt and heat exposure. Our results indicate that LEA proteins play an important role in plant acclimation to stress conditions.
The LEA subgroups represent diverse adaptation to heat and salt stresses. Based on our results, LEA1 increased bacterial resistance to heat and salt, but not significantly; the viability ratio was 1-2-fold higher than that of the control strain. Dehydrin, SMP and LEA5 induced higher tolerance to salt stress (more than 3-fold higher than the control strain), and LEA2 and LEA4 induced higher tolerance to heat (more than 2-fold higher than the control strain), similar with previous studies26,27. A recent study investigated the functions of six subgroups of LEA proteins from Arabidopsis expressed in Saccharomyces cerevisiae exposed to desiccation stress, with the results showing that LEA2, LEA4 and dehydrin enhanced tolerance to desiccation stress29. While, another functional analysis of three genes from the LEA1, LEA3 and dehydrin groups performed using an E. coli expression system supported the hypothesis that LEA1 and LEA3 increased salt tolerance in soybean30.
We also used qPCR to investigate the expression patterns of 11 candidate genes in D. officinale plantlets under heat and salt stresses. The results indicated that most of the LEA groups tested are expressed in different plant tissues in response to abiotic stresses (heat and salt). Similarly, several LEA1, LEA3, LEA4 and dehydrin genes were also induced by drought stress in Arabidopsis, whereas certain LEA3, LEA4, LEA5 and dehydrin genes were induced by salt16,29,31. Additionally, 5 genes from the LEA1, LEA2, LEA4 and dehydrin groups were up-regulated by drought and salt stresses in tomato21. In rice, the LEA2, LEA3 and dehydrin groups showed strong responses to osmotic stress, salt and abscisic acid11,32,33.
In conclusion, we for the fisrt time identified and characterized LEA genes in the D. officinale genome. The results indicate that LEA proteins are a large family in D. officinale and exhibit diverse sequences, motif composition, chromosomal locations and expression patterns. We preliminarily elucidated their functional role and explored their effects on tolerance to abiotic stress (heat and salt).
Methods
Plant material and stress treatments
Seeds and tissue-cultured plantlets of D. officinale were purchased from Changzhou of Jiangsu. The seed germination procedure for D. officinale was described previously17. Then, samples of seeds, protocorms, roots, stems and leaves were prepared. The roots, stems and leaves from unstressed plants were used as the control group. For temperature stress, plantlets were heated at 37 °C for 48 h and the heat-treated tissues (roots, stems and leaves) were collected. For salinity stress, plantlets were treated with 0.5 M NaCl for 48 h and the salt-treated tissues (roots, stems and leaves) were then collected.
Identification of LEA genes in the D. officinale genome
The D. officinale genome (downloaded from ftp://202.203.187.112/genome/dendrobe/) and the D. officinale seed transcriptome (unpublished) were queried using 51 Arabidopsis and 36 Oryza LEA protein sequences (downloaded from http://www.ncbi.nlm.nih.gov/) with the TBLASTN tool. All the identified candidates were analyzed using the Pfam database (http://pfam.sanger.ac.uk) to confirm LEA conserved domains. Finally, we retrieved 30 DofLEA genes, allowing us to design 30 primers for cloning LEA genes from D. officinale.
Molecular cloning of D. officinale LEA genes
Total RNA was extracted from D. officinale seeds and roots using an RNeasy® Plant Mini Kit (QIAGEN, Germany) according to the manufacturer’s instructions. First strand cDNA was synthesized using a PrimeScript RT reagent PCR Kit (TaKaRa, Japan), and the synthesized cDNAs were used as templates for PCR with specific primers designed using Primer Premier 6.0 (shown in Supplementary Table S2). The parameters of PCR were as follows: 95 °C for 3 min, 30 cycles of 95 °C for 15 s, and 57 °C for 30 s, followed by 72 °C for 1 min. The PCR products were purified using an Agarose Gel DNA Purification Kit (AidLab, China), inserted into the pTOPO vector (AidLab, China), and sequenced in both directions to verify the LEA sequences. Finally, we successfully cloned 15 and 2 DofLEA genes from seeds and roots, respectively. Pfam was used to confirm that these 17 sequences possess LEA conserved domains.
Bioinformatics analysis
The molecular weight (MW) and grand average of hydropathy (GRAVY) of putative proteins were predicted using PROTPARAM (http://web.expasy.org/protparam/). Multiple sequence alignment was performed using Clustalx with default parameters34. Subcellular localization of the proteins was predicted by WoLF_PSORT (http://www.genscript.com/psort/wolf_psort.html). Protein conserved motif analysis was conducted using the program MEME/MAST (http://meme-suite.org). Phylogenetic analysis was performed using the maximum likelihood and neighborjoining methods (bootstrap value = 1000) with MEGA 6.0 software35.
Expression analysis of DofLEAs in different tissues
Total RNA was isolated from seeds, protocorms, stems, roots and leaves as previously described36. Primers designed using Primer Premier 6.0 are shown in Supplementary Table S3. PrimeScript RT reagent Kit (TaKaRa, Japan) was used for reverse transcription. Subsequently, qPCR was performed in a 15 μL reaction mixture containing 7.5 μL 2 × SYBR® Premix Ex TaqTM II (TaKaRa, Japan), 1 μL cDNA template, 0.3 μL each gene-specific primer and 5.9 μL ddH2O. We performed three biological replicates and three technical replicates using the LightCycler® 480 II RT-PCR System (Roche, Switzerland) with its relative quantification software ver. 1.5. The parameters of reactions were as follows: 95 °C for 30 s, 40 cycles of 95 °C for 5 s, and 60 °C for 30 s. The 18S rRNA reference gene was used as an internal control.
Expression of LEA proteins in E. coli
We selected 11 DofLEA genes representing each group and transformed them into E. coli. The transformed E. coli (pET28a-DofLEA) cell were grown in our lab as described previously10,30,37. Briefly, DofLEA genes were inserted into the pET28a vector and then transformed into the E. coli host strain BL21 (DE3). The primers are shown in Supplementary Table S4. Recombinant proteins were induced by the addition of isopropyl β-D-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM, and incubation continued at 37 °C for 4 h. The bacterial cells were harvested by centrifugation, resuspended in PBS buffer, lysed by sonication, and centrifuged at 12,000 rpm for 10 min. The clarified cell extract was analyzed by 12% SDS-PAGE.
Heat and salt treatments of transformed E. coli cells
Heat and salt tolerance assays were performed as described previously10,30,37 (shown in Figure 6S). The transformed E. coli (pET28a-DofLEA) colonies grown on Luria-Bertani (LB)+ kanamycin plates were used to inoculate starting cultures, which were incubated at 37 °C for 12–16 h. The cultures were diluted 50-fold using fresh LB medium, and incubation continued at 37 °C until the mid-log phase (2–3 h, OD600 = 0.6). After IPTG induction, the cultures were diluted and transferred to 50 °C, sampled at 60, 120, 180 and 240 min, and plated onto LB+ kanamycin plates. For the salt treatment, after IPTG induction, LB+ kanamycin plates (containing 171 mM NaCl) were supplemented with 300 mM, 400 mM, 500 mM, and 600 mM NaCl. Each 20 μl sample was plated onto LB+ kanamycin plates and then incubated overnight at 37 °C, followed by counting the number of colonies formed to determine cell viability. The colony number on each plate was recorded using CellProfiler software38.
The viability ratio of the transformants under heat and salt conditions was calculated according to the following formula10: Cell viability ratio = (colony number on stressed plate/colony number on unstressed plate) × 100%. For all experiments (heat and salt), the means of three experiments were determined from three independent transformants. Here, E. coli with the empty vector (pET28a) is the control group.
Expression analysis of DofLEAs under stress treatments
Quantitative real-time RT-PCR (qPCR) was used to measure changes in expression in DofLEAs between unstressed plantlets and heat-treated or salt-treated plantlets. Total RNA and RT-PCR products were treated as previously described.
Additional Information
How to cite this article: Ling, H. et al. Functional insights into the late embryogenesis abundant (LEA) protein family from Dendrobium officinale (Orchidaceae) using an Escherichia coli system. Sci. Rep. 6, 39693; doi: 10.1038/srep39693 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This study was financially supported by the National Natural Science Foundation of China (No. of 81573526) and CAMS basic research fund for central public research institutes (2016ZX350062). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author Contributions Conceived and designed the experiments: H.L., X.Z., and S.X.G. Performed the experiments: H.L. and X.Z. Analyzed the data: X.Z. Wrote the paper: X.Z. All authors read and approved the final manuscript.
References
- Wang W., Vinocur B. & Altman A. Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218, 1–14 (2003). [DOI] [PubMed] [Google Scholar]
- Mittler R. Abiotic stress, the field environment and stress combination. Trends in Plant Science 11, 15–19 (2006). [DOI] [PubMed] [Google Scholar]
- Howarth C. J. Genetic improvements of tolerance to high temperature. (Howarth Press, 2005). [Google Scholar]
- Tunnacliffe A. & Wise M. J. The continuing conundrum of the LEA proteins. Naturwissenschaften 94, 791–812 (2007). [DOI] [PubMed] [Google Scholar]
- Umezawa T., Fujita M., Fujita Y., Yamaguchi-Shinozaki K. & Shinozaki K. Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Current Opinion in Biotechnology 17, 113–122 (2006). [DOI] [PubMed] [Google Scholar]
- Shao H. B., Liang Z. S. & Shao M. A. LEA proteins in higher plants: structure, function, gene expression and regulation. Colloids and Surfaces B: Biointerfaces 45, 131–135 (2005). [DOI] [PubMed] [Google Scholar]
- Hunault G. & Jaspard E. LEAPdb: a database for the late embryogenesis abundant proteins. BMC Genomics 11, 1 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wu Y., Yang X., Guo X. & Cao X. SmLEA2, a gene for late embryogenesis abundant protein isolated from Salvia miltiorrhiza, confers tolerance to drought and salt stress in Escherichia coli and S. miltiorrhiza. Protoplasma 1–12 (2016). [DOI] [PubMed] [Google Scholar]
- Choi D. W., Zhu B. & Close T. The barley (Hordeum vulgare L.) dehydrin multigene family: sequences, allele types, chromosome assignments, and expression characteristics of 11 Dhn genes of cv Dicktoo. Theoretical and Applied Genetics 98, 1234–1247 (1999). [Google Scholar]
- Gao J. & Lan T. Functional characterization of the late embryogenesis abundant (LEA) protein gene family from Pinus tabuliformis (Pinaceae) in Escherichia coli. Scientific Reports 6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. S. et al. Genome-scale identification and analysis of LEA genes in rice (Oryza sativa L.). Plant Science 172, 414–420 (2007). [Google Scholar]
- Lan T., Gao J. & Zeng Q. Y. Genome-wide analysis of the LEA (late embryogenesis abundant) protein gene family in Populus trichocarpa. Tree Genetics and Genomes 9, 253–264 (2013). [Google Scholar]
- Du D. et al. Genome-wide identification and analysis of late embryogenesis abundant (LEA) genes in Prunus mume. Molecular Biology Reports 40, 1937–1946 (2013). [DOI] [PubMed] [Google Scholar]
- Charfeddine S., Saïdi M. N., Charfeddine M. & Gargouri-Bouzid R. Genome-wide identification and expression profiling of the late embryogenesis abundant genes in potato with emphasis on dehydrins. Molecular Biology Reports 42, 1163–1174 (2015). [DOI] [PubMed] [Google Scholar]
- Battaglia M. & Covarrubias A. A. Late Embryogenesis Abundant (LEA) proteins in legumes. Frontiers in Plant Science 4, 190 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundertmark M. & Hincha D. K. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9, 1 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M. M., Zhang G., Zhang D. W., Hsiao Y. Y. & Guo S. X. ESTs analysis reveals putative genes involved in symbiotic seed germination in Dendrobium officinale. PLoS One 8, e72705 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang D., Sun L. & Wei L. Effects of exogenous salicylic acid on the physiological characteristics of Dendrobium officinale under chilling stress. Plant Growth Regulation 79, 199–208 (2016). [Google Scholar]
- He C. et al. Identification of genes involved in biosynthesis of mannan polysaccharides in Dendrobium officinale by RNA-seq analysis. Plant Molecular Biology 88, 219–231 (2015). [DOI] [PubMed] [Google Scholar]
- Akula R. & Ravishankar G. A. Influence of abiotic stress signals on secondary metabolites in plants. Plant Signaling and Behavior 6, 1720–1731 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J. & Li X. Identification and phylogenetic analysis of late embryogenesis abundant proteins family in tomato (Solanum lycopersicum). Planta 241, 757–772 (2015). [DOI] [PubMed] [Google Scholar]
- Boswell L. C. & Hand S. C. Intracellular localization of group 3 LEA proteins in embryos of Artemia franciscana. Tissue and Cell 46, 514–519 (2014). [DOI] [PubMed] [Google Scholar]
- Hatanaka R. et al. Biochemical and structural characterization of an endoplasmic reticulum-localized late embryogenesis abundant (lea) protein from the liverwort marchantia polymorpha. Biochemical and Biophysical Research Communications 454, 588–593 (2014). [DOI] [PubMed] [Google Scholar]
- Angelovici R., Galili G., Fernie A. R. & Fait A. Seed desiccation: a bridge between maturation and germination. Trends in Plant Science 15, 211–218 (2010). [DOI] [PubMed] [Google Scholar]
- Vicente-Carbajosa J. & Carbonero P. Seed maturation: developing an intrusive phase to accomplish a quiescent state. International Journal of Developmental Biology 49, 645–651 (2005). [DOI] [PubMed] [Google Scholar]
- Dalal M., Tayal D., Chinnusamy V. & Bansal K. C. Abiotic stress and ABA-inducible Group 4 LEA from Brassica napus plays a key role in salt and drought tolerance. Journal of Biotechnology 139, 137–145 (2009). [DOI] [PubMed] [Google Scholar]
- Paul A., Singh S., Sharma S. & Kumar S. A stress-responsive late embryogenesis abundant protein 7 (CsLEA7) of tea [Camellia sinensis (L.) O. Kuntze] encodes for a chaperone that imparts tolerance to Escherichia coli against stresses. Molecular Biology Reports 41, 7191–7200 (2014). [DOI] [PubMed] [Google Scholar]
- Liu Y., Liang J., Sun L., Yang X. & Li D. Group 3 LEA protein, ZmLEA3, is involved in protection from low temperature stress. Frontiers in Plant Science 7, 190 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang N. X., Popova A. V., Hundertmark M. & Hincha D. K. Functional characterization of selected LEA proteins from Arabidopsis thaliana in yeast and in vitro. Planta 240, 325–336 (2014). [DOI] [PubMed] [Google Scholar]
- Lan Y., Cai D. & Zheng Y. Z. Expression in Escherichia coli of three different soybean late embryogenesis abundant (LEA) genes to investigate enhanced stress tolerance. Journal of Integrative Plant Biology 47, 613–621 (2005). [Google Scholar]
- Hundertmark M., Buitink J., Leprince O. & Hincha D. K. The reduction of seed-specific dehydrins reduces seed longevity in Arabidopsis thaliana. Seed Science Research 21, 165–173 (2011). [Google Scholar]
- Sengupta S. & Majumder A. L. Insight into the salt tolerance factors of a wild halophytic rice, Porteresia coarctata: a physiological and proteomic approach. Planta 229, 911–929 (2009). [DOI] [PubMed] [Google Scholar]
- Yu J., Lai Y., Wu X., Wu G. & Guo C. Overexpression of OsEm1 encoding a group I LEA protein confers enhanced drought tolerance in rice. Biochemical and Biophysical Research Communications 478, 703–709 (2016). [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F. & Higgins D. G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25, 4876–4882 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. S. et al. Comparative transcriptome analysis of genes involved in GA-GID1-DELLA regulatory module in symbiotic and asymbiotic seed germination of Anoectochilus roxburghii (Wall.) Lindl. (Orchidaceae). International Journal of Molecular Sciences 16, 30190–30203 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T., Zhou N., Fu M., Qin J. & Huang X. Characterization of OsLEA1a and its inhibitory effect on the resistance of E. coli to diverse abiotic stresses. International Journal of Biological Macromolecules 91, 1010–1017 (2016). [DOI] [PubMed] [Google Scholar]
- Carpenter A. E. et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biology 7, R100 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.