Abstract
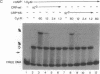
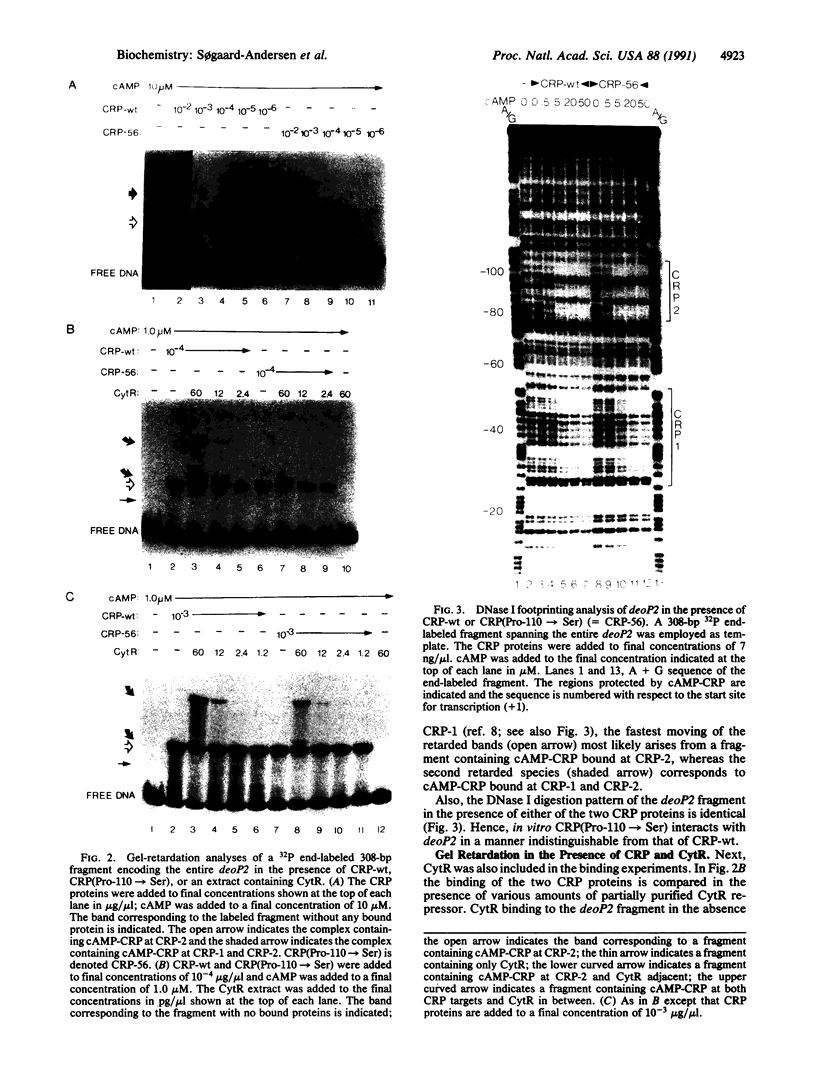
Promoters in Escherichia coli that are negatively regulated by the CytR repressor are also activated by the cAMP receptor protein (CRP) complexed to cAMP; as a characteristic, these promoters encode tandem binding sites for cAMP-CRP. In one such promoter, deoP2, CytR binds to the region between the tandem CRP binding sites with a relatively low affinity; in the presence of cAMP-CRP, however, the repressor and activator bind cooperatively to the DNA. Here we have investigated this cooperativity by isolating mutants of the CRP protein that abolish CytR regulation without exhibiting a concomitant loss in their ability to activate transcription. Four different, single amino acid substitutions in CRP give rise to this phenotype. These amino acids lie in close proximity on the surface of the CRP tertiary structure in a portion of the protein that is not in contact with the DNA. In vitro analyses of one of the CRP mutants show that it interacts with the DNA in a manner indistinguishable from wild-type CRP, whereas its interaction with CytR is perturbed. These results strongly indicate that cooperative DNA binding of CytR and cAMP-CRP is achieved through protein-protein interactions.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiba H., Fujimoto S., Ozaki N. Molecular cloning and nucleotide sequencing of the gene for E. coli cAMP receptor protein. Nucleic Acids Res. 1982 Feb 25;10(4):1345–1361. doi: 10.1093/nar/10.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiba H., Nakamura T., Mitani H., Mori H. Mutations that alter the allosteric nature of cAMP receptor protein of Escherichia coli. EMBO J. 1985 Dec 1;4(12):3329–3332. doi: 10.1002/j.1460-2075.1985.tb04084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A., Sprague G. F., Jr MAT alpha 1 protein, a yeast transcription activator, binds synergistically with a second protein to a set of cell-type-specific genes. Cell. 1987 Aug 28;50(5):681–691. doi: 10.1016/0092-8674(87)90326-6. [DOI] [PubMed] [Google Scholar]
- Benezra R., Davis R. L., Lockshon D., Turner D. L., Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990 Apr 6;61(1):49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- Bremer E., Gerlach P., Middendorf A. Double negative and positive control of tsx expression in Escherichia coli. J Bacteriol. 1988 Jan;170(1):108–116. doi: 10.1128/jb.170.1.108-116.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandanell G., Valentin-Hansen P., Larsen J. E., Hammer K. Long-range cooperativity between gene regulatory sequences in a prokaryote. 1987 Feb 26-Mar 4Nature. 325(6107):823–826. doi: 10.1038/325823a0. [DOI] [PubMed] [Google Scholar]
- Fried M. G., Crothers D. M. Equilibrium studies of the cyclic AMP receptor protein-DNA interaction. J Mol Biol. 1984 Jan 25;172(3):241–262. doi: 10.1016/s0022-2836(84)80025-x. [DOI] [PubMed] [Google Scholar]
- Friedman D. I. Integration host factor: a protein for all reasons. Cell. 1988 Nov 18;55(4):545–554. doi: 10.1016/0092-8674(88)90213-9. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garges S., Adhya S. Sites of allosteric shift in the structure of the cyclic AMP receptor protein. Cell. 1985 Jul;41(3):745–751. doi: 10.1016/s0092-8674(85)80055-6. [DOI] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach P., Valentin-Hansen P., Bremer E. Transcriptional regulation of the cytR repressor gene of Escherichia coli: autoregulation and positive control by the cAMP/CAP complex. Mol Microbiol. 1990 Mar;4(3):479–488. doi: 10.1111/j.1365-2958.1990.tb00614.x. [DOI] [PubMed] [Google Scholar]
- Ghosaini L. R., Brown A. M., Sturtevant J. M. Scanning calorimetric study of the thermal unfolding of catabolite activator protein from Escherichia coli in the absence and presence of cyclic mononucleotides. Biochemistry. 1988 Jul 12;27(14):5257–5261. doi: 10.1021/bi00414a046. [DOI] [PubMed] [Google Scholar]
- Goodman S. D., Nash H. A. Functional replacement of a protein-induced bend in a DNA recombination site. Nature. 1989 Sep 21;341(6239):251–254. doi: 10.1038/341251a0. [DOI] [PubMed] [Google Scholar]
- Hahn S., Dunn T., Schleif R. Upstream repression and CRP stimulation of the Escherichia coli L-arabinose operon. J Mol Biol. 1984 Nov 25;180(1):61–72. doi: 10.1016/0022-2836(84)90430-3. [DOI] [PubMed] [Google Scholar]
- Irwin N., Ptashne M. Mutants of the catabolite activator protein of Escherichia coli that are specifically deficient in the gene-activation function. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8315–8319. doi: 10.1073/pnas.84.23.8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Bruist M. F., Simon M. I. Host protein requirements for in vitro site-specific DNA inversion. Cell. 1986 Aug 15;46(4):531–539. doi: 10.1016/0092-8674(86)90878-0. [DOI] [PubMed] [Google Scholar]
- Josephsen J., Hammer-Jespersen K. Fusion of the lac genes to the promotor for the cytidine deaminase gene of Escherichia coli K-12. Mol Gen Genet. 1981;182(1):154–158. doi: 10.1007/BF00422783. [DOI] [PubMed] [Google Scholar]
- Keleher C. A., Goutte C., Johnson A. D. The yeast cell-type-specific repressor alpha 2 acts cooperatively with a non-cell-type-specific protein. Cell. 1988 Jun 17;53(6):927–936. doi: 10.1016/s0092-8674(88)90449-7. [DOI] [PubMed] [Google Scholar]
- Koch C., Kahmann R. Purification and properties of the Escherichia coli host factor required for inversion of the G segment in bacteriophage Mu. J Biol Chem. 1986 Nov 25;261(33):15673–15678. [PubMed] [Google Scholar]
- Lovett M. A., Helinski D. R. Method for the isolation of the replication region of a bacterial replicon: construction of a mini-F'kn plasmid. J Bacteriol. 1976 Aug;127(2):982–987. doi: 10.1128/jb.127.2.982-987.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Ptashne M. The carboxy-terminal 30 amino acids of GAL4 are recognized by GAL80. Cell. 1987 Jul 3;50(1):137–142. doi: 10.1016/0092-8674(87)90670-2. [DOI] [PubMed] [Google Scholar]
- McKay D. B., Weber I. T., Steitz T. A. Structure of catabolite gene activator protein at 2.9-A resolution. Incorporation of amino acid sequence and interactions with cyclic AMP. J Biol Chem. 1982 Aug 25;257(16):9518–9524. [PubMed] [Google Scholar]
- Musso R. E., Di Lauro R., Adhya S., de Crombrugghe B. Dual control for transcription of the galactose operon by cyclic AMP and its receptor protein at two interspersed promoters. Cell. 1977 Nov;12(3):847–854. doi: 10.1016/0092-8674(77)90283-5. [DOI] [PubMed] [Google Scholar]
- Raibaud O., Vidal-Ingigliardi D., Richet E. A complex nucleoprotein structure involved in activation of transcription of two divergent Escherichia coli promoters. J Mol Biol. 1989 Feb 5;205(3):471–485. doi: 10.1016/0022-2836(89)90218-0. [DOI] [PubMed] [Google Scholar]
- Shapira S. K., Chou J., Richaud F. V., Casadaban M. J. New versatile plasmid vectors for expression of hybrid proteins coded by a cloned gene fused to lacZ gene sequences encoding an enzymatically active carboxy-terminal portion of beta-galactosidase. Gene. 1983 Nov;25(1):71–82. doi: 10.1016/0378-1119(83)90169-5. [DOI] [PubMed] [Google Scholar]
- Søgaard-Andersen L., Martinussen J., Møllegaard N. E., Douthwaite S. R., Valentin-Hansen P. The CytR repressor antagonizes cyclic AMP-cyclic AMP receptor protein activation of the deoCp2 promoter of Escherichia coli K-12. J Bacteriol. 1990 Oct;172(10):5706–5713. doi: 10.1128/jb.172.10.5706-5713.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Søgaard-Andersen L., Møllegaard N. E., Douthwaite S. R., Valentin-Hansen P. Tandem DNA-bound cAMP-CRP complexes are required for transcriptional repression of the deoP2 promoter by the CytR repressor in Escherichia coli. Mol Microbiol. 1990 Sep;4(9):1595–1601. [PubMed] [Google Scholar]
- Valentin-Hansen P., Holst B., Josephsen J., Hammer K., Albrechtsen B. CRP/cAMP- and CytR-regulated promoters in Escherichia coli K12: the cdd promoter. Mol Microbiol. 1989 Oct;3(10):1385–1390. doi: 10.1111/j.1365-2958.1989.tb00120.x. [DOI] [PubMed] [Google Scholar]
- Valentin-Hansen P., Svenningsen B. A., Munch-Petersen A., Hammer-Jespersen K. Regulation of the deo operon in Escherichia coli: the double negative control of the deo operon by the cytR and deoR repressors in a DNA directed in vitro system. Mol Gen Genet. 1978 Feb 16;159(2):191–202. doi: 10.1007/BF00270893. [DOI] [PubMed] [Google Scholar]
- Valentin-Hansen P. Tandem CRP binding sites in the deo operon of Escherichia coli K-12. EMBO J. 1982;1(9):1049–1054. doi: 10.1002/j.1460-2075.1982.tb01295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Crombrugghe B., Busby S., Buc H. Cyclic AMP receptor protein: role in transcription activation. Science. 1984 May 25;224(4651):831–838. doi: 10.1126/science.6372090. [DOI] [PubMed] [Google Scholar]