Abstract
The genetic model plant Arabidopsis thaliana (arabidopsis) has been instrumental to recent advances in our understanding of the molecular function of the plant immune system. However, this work has not yet included plant associated and phytopathogenic yeasts largely due to a lack of yeast species known to interact with arabidopsis. The plant phylloplane is a significant habitat for neutral-residents, plant-growth and health-promoting species, and latent-pathogenic species. However, yeast phylloplane residents of arabidopsis remain underexplored. To address this, resident yeasts from the phyllosphere of wild arabidopsis collected in field conditions have been isolated and characterized. A total of 95 yeast strains representing 23 species in 9 genera were discovered, including potentially psychrophilic and pathogenic strains. Physiological characterization revealed thermotolerance profiles, sensitivity to the arabidopsis phytoalexin camalexin, the production of indolic compounds, and the ability to activate auxin responses in planta. These results indicate a rich diversity of yeasts present in the arabidopsis phylloplane and have created culture resources and information useful in the development of model systems for arabidopsis-yeast interactions.
Plants constantly interact with a large number of microorganisms, including bacteria, oomycetes, filamentous fungi, and yeasts. Bacteria are the best studied plant-associated microbes and yeasts have received the least attention. These microbes colonize all plant surfaces, where each plant compartment or structure has its own distinct microbiome. The rhizosphere and endophytic microbiota are the most thoroughly explored and offer numerous examples of microbes modulating plant growth, mineral absorption, and immunity1,2,3,4. The phyllosphere is termed as the total surface of aerial parts of plants that is colonized by microorganisms. A variety of bacteria, yeasts and filamentous fungi have been isolated from the phyllosphere of several plant species5,6,7,8,9,10. Physical barriers and multiple chemical factors limit the growth and survival of microbes in the phyllosphere. However, the energy content of leaf surface components and simple sugars leached from the interior tissues11,12 make the phyllosphere a considerable microbial habitat. Microbes residing in the phyllosphere can have various lifestyles and modes of interaction with the host, being neutral residents, latent pathogens, or plant-health and -growth promoters.
Many plant associated microorganisms, including both growth-promoting and pathogenic microbes, have the ability to synthesize the plant developmental hormone auxin, including the most common auxin, indole-3-acetic acid (IAA) which is responsible for division, elongation and differentiation of plant cells and tissues. Increased attention has been paid to the role of microbial IAA in plant-microbe interactions. It was suggested that auxin production capability might be an important colonization strategy for the phyllosphere environment13. IAA-producing bacteria and fungi are able to promote plant growth14,15. Therefore, they are suggested as potential of bio-fertilizers.
The genetic model plant, Arabidopsis thaliana (arabidopsis) is a well characterized model for plant microbe interactions. This encompasses host and non-host pathogen-interaction systems, root-associated microbiome studies, and endophytic fungi. Indeed, arabidopsis has been instrumental in defining the basis of plant immunity, including mechanisms engaged in the perception microbe associated molecular patterns (MAMPs) in MAMP-triggered immunity and Resistance (R)-gene function in signalling of effector-triggered immunity16,17. These studies have focused on fungi, bacteria and oomycetes, but not yeasts. In mammals, a distinct set of immune receptors responsible for perception of yeast-specific MAMPs have been found18. Thus, it can be expected that in plants yeasts engage a distinct, and yet unknown, set of immune receptors. The ability of yeast cell wall components to induce plant immunity has been demonstrated19. Specifically, baker’s yeast cell-wall and glucopeptide components exhibited elicitor activity in arabidopsis. Knowledge of phyllosphere yeasts and, in general, of plant-yeast interactions is required for a comprehensive view of plant health. However, the power of arabidopsis as a genetic model organism has not been utilized in the study of plant-yeast interactions due to a lack of yeasts known to interact with arabidopsis. In this study arabidopsis phyllosphere resident yeasts were isolated, identified, and physiologically characterized to clarify the nature of their interaction with arabidopsis and create resources for genetic studies of plant-yeast interactions.
Results
Isolation of yeasts from the arabidopsis phyllosphere
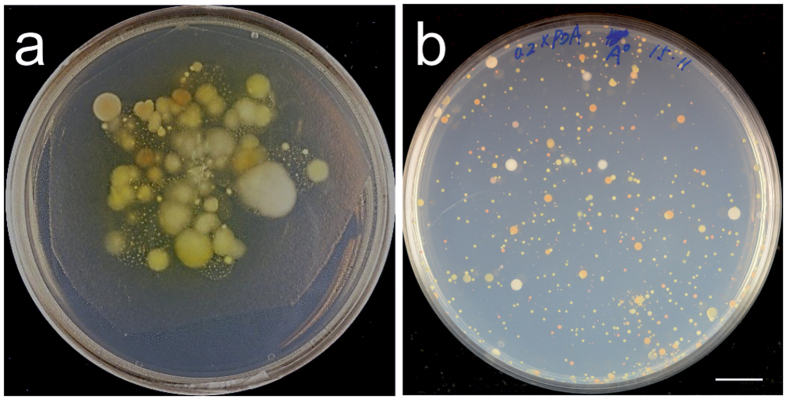
Few naturally occurring yeasts that are associated with, or pathogenic on, arabidopsis are currently described. To rapidly assess the presence of yeasts, whole rosette imprints of wild arabidopsis collected in the field were cultured using a medium that favours the growth of yeasts. These cultures (Fig. 1a) indicate a high number and apparent wide variety of microbes were present, the majority of which had a colony morphology consistent with yeasts. This result prompted further investigation of arabidopsis associated yeasts.
Figure 1.
(a), The colonies of leaf resident filamentous fungi (5–15 mm in diameter), yeasts and bacteria (0.1–2 mm in diameter) visualized by leaf printing onto a 0.2 × PDA medium plate from a wild Arabidopsis thaliana. (b), Typical yeast colonies on 0.2 × PDA medium, seven days after plating 20 μL of leaf-wash solution from sample A (Table 1). Similar plates with lower colony density from plating 1:10 or 1:100 dilutions of leaf-wash were used for colony picking. Scale bar: 1 cm.
Wild arabidopsis were collected from three sites in Helsinki; site name, location and samples collected are listed in Table 1. Two sites (Kulosaari and Mustikkamaa) are on islands of the Baltic coast and separated by circa 300 metres, while the third site (Kivikko) was circa 5 km inland. Multiple samples from the same site were always collected from plants at least two metres apart. Serial dilutions of leaf wash solutions were plated and grown at room temperature. A typical plate is depicted in Fig. 1b. Dilute (0.2 x) potato dextrose agar (PDA) was selected as a culture medium with the reasoning that a nutrient-poor, plant-based medium lacking high levels of exogenous amino acids would mimic the conditions in the phyllosphere11,20. Plates were monitored daily over two weeks and colonies marked and picked as they appeared. Typically 3–5 colonies with similar colour and morphology were picked and streaked on 0.2 x PDA plates to isolate pure single colonies. Isolated colonies were regrown in replicated microtitre plates for storage and DNA isolation. DNA samples were used to PCR amplify rDNA internal transcribed spacer (ITS) sequences for identification. Isolates producing no ITS PCR product were observed by light microscopy to eliminate prokaryotes and PCR was retested with new cultures and DNA samples. Samples failing these tests (60.9% of colonies picked) were eliminated from further analysis. ITS PCR products were subjected to restriction fragment polymorphism analysis to classify the yeasts into operational taxonomic units (OTUs).
Table 1. Leaf samples collected from three different sites in Helsinki.
| Date | Collection sitea | GPS coordinatesb | Sample | Colonies picked | Yeast strains |
|---|---|---|---|---|---|
| May 2013 | Kivikko | Lat: 60.23270 Lon: 25.06191 | A | 47 | 13 |
| B | 51 | 41 | |||
| C | 57 | 16 | |||
| Dec 2013 | Kulosaari | Lat: 60.18376 Lon: 24.99241 | F | 18 | 1 |
| F’ | 10 | 0 | |||
| Mustikkamaa | Lat: 60.18070 Lon: 24.99463 | G | 28 | 1 | |
| H | ndc | nd | |||
| Apr 2015 | Kulosaari | Lat: 60.18376 Lon: 24.99241 | M | 32 | 23 |
| N | nd | nd |
aCollection sites are all districts in Helsinki, Finland.
bLat: latitude; Lon: longitude.
cnd: no data.
For isolate identification and definition of OTUs, restriction patterns and ITS sequencing were used (Supplementary Table S1). For Samples A, B and C collected from Kivikko in May of 2013, there were totally 70 novel yeast strains isolated and identified out of 155 picked colonies. From a total of 56 colonies picked from the November 2013 samples, only one new OTU from Kulosaari and one from Mustikkamaa were found. The low number of new yeasts found in the November samples was partly due to the reappearance of previously isolated OTUs (Table 1 and Supplementary Table S1). However, it was apparent that the diversity of yeasts was much higher in the samples collected in spring than those from autumn. In the M sample from Kulosaari in April 2015, 23 new OTUs were discovered from 32 colonies.
Identification of yeasts
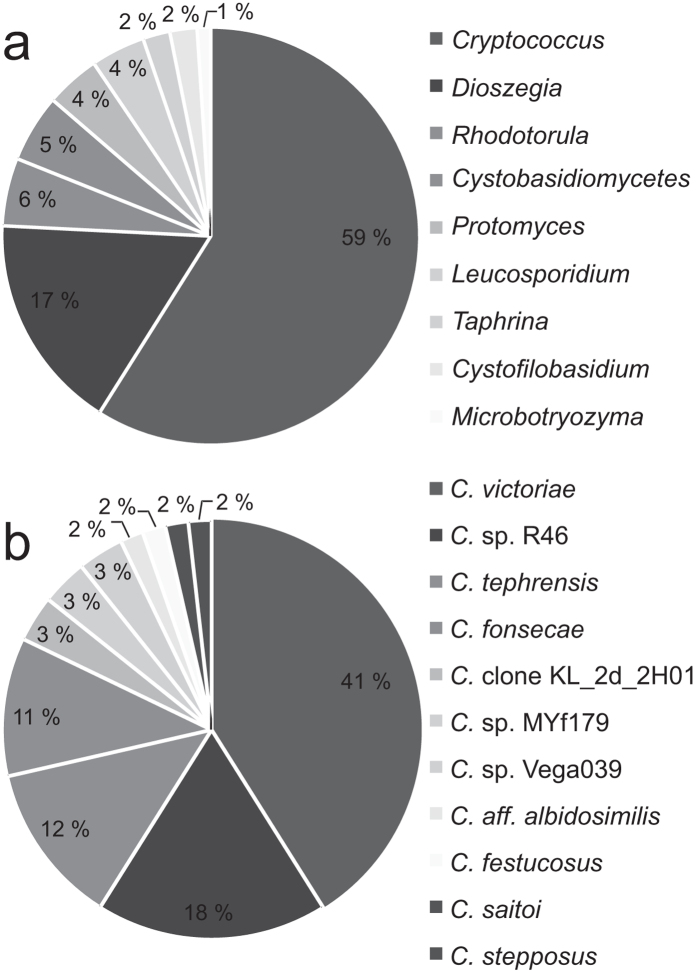
The ITS region from all unique OTUs was sequenced and compared to the NCBI database identifying 95 yeast strains representing 23 species in nine genera (Table 2). There were two species in two genera of the phylum Ascomycota and 21 species in seven genera of the phylum Basidiomycota. Ascomycete yeasts represented 6.3% of total and 93.7% were Basidiomycete. Cryptococcus was by far the dominant genus in the arabidopsis phyllosphere, accounting over half of all isolates. OTUs similar to C. victoriae strains accounted for 41% of total isolates within the genus Cryptococcus (Fig. 2). However, there was a rich diversity of Cryptococcus present with a total of 20 OTUs represented.
Table 2. Yeast OTUs isolated from the wild arabidopsis phyllosphere.
| OTUsa | Closest speciesb (strain/isolate) | No.c | ITS lend | Sube | ITS simf | Gaps | Acc no.g |
|---|---|---|---|---|---|---|---|
| Ascomycota | |||||||
| Taphrinomycetes | |||||||
| 1 | Protomyces inouyei JCM 22201 | 3(3) | 462 | 19/469 | 96% | 10/469 | LT602858 |
| 2 | Protomyces inouyei JCM 22201 | 1(1) | 463 | 4/470 | 95% | 11/470 | LT602859 |
| 3 | Taphrina carnea strain CBS 332.55 | 2(2) | 578 | 4/579 | 99% | 1/579 | LT602860 |
| Basidiomycota | |||||||
| Tremellomycetes | |||||||
| 4 | Cryptococcus sp. R46 | 4(3) | 458 | 1/458 | 99% | 1/458 | LT602861 |
| 5 | Cryptococcus festucosus CBS 11757 | 1(1) | 474 | 2/474 | 99% | 2/474 | LT602862 |
| 6 | Cryptococcus fonsecae DSM 26992 | 2(2) | 457 | 1/457 | 99% | 0/457 | LT602863 |
| 7 | Cryptococcus sp. MYf180 | 1(1) | 439 | 1/439 | 99% | 1/439 | LT602864 |
| 8 | Cryptococcus fonsecae KF921 | 4(4) | 453 | 1/453 | 99% | 0/453 | LT602865 |
| 9 | Cryptococcus aff. albidosimilis GW_OTU40 | 1(1) | 538 | 1/539 | 99% | 1/539 | LT602866 |
| 10 | Cryptococcus saitoi P01D003 | 1(1) | 561 | 2/563 | 99% | 2/563 | LT602867 |
| 11 | Cryptococcus tephrensis | 6(3) | 435 | 0/435 | 100% | 0/435 | LT602868 |
| 12 | Cryptococcus stepposus P25B002 | 1(1) | 568 | 2/570 | 99% | 2/570 | LT602869 |
| 13 | Cryptococcus tephrensis | 1(1) | 450 | 58/455 | 87% | 7/455 | LT602870 |
| 14 | Cryptococcus clone KL_2d_2H01 | 2(2) | 409 | 0/409 | 100% | 0/409 | LT602871 |
| 15 | Cryptococcus victoriae HB 1221 | 11(1) | 447 | 1/448 | 99% | 1/448 | LT602872 |
| 16 | Cryptococcus sp. MYf179 | 1(1) | 421 | 8/421 | 98% | 0/405 | LT602873 |
| 17 | Cryptococcus victoriae M5-10C-5 | 9(5) | 432 | 0/432 | 100% | 0/432 | LT602874 |
| 18 | Cryptococcus victoriae M5-10C-5 | 1(1) | 431 | 1/431 | 99% | 0/431 | LT602875 |
| 19 | Cryptococcus sp. Vega039 | 2(1) | 432 | 1/432 | 99% | 0/432 | LT602876 |
| 20 | Cryptococcus victoriae M5-10C-5 | 1(1) | 432 | 2/432 | 99% | 0/432 | LT602877 |
| 21 | Cryptococcus victoriae HB 1221 | 1(1) | 431 | 0/431 | 100% | 0/431 | LT602878 |
| 22 | Cystofilobasidium capitatum ATCC 24507 | 2(1) | 518 | 0/518 | 100% | 0/518 | LT602879 |
| 23 | Dioszegia crocea DBVPG 5030 | 16(7) | 366 | 0/366 | 100% | 0/366 | LT602880 |
| 24 | Cryptococcus sp. R46 | 3(3) | 512 | 0/513 | 99% | 1/513 | LT602881 |
| 25 | Cryptococcus sp. R46 | 3(1) | 475 | 4/476 | 99% | 1/476 | LT602882 |
| Microbotryomycetes | |||||||
| 26 | Leucosporidium creatinivorum JCM10699 | 3(2) | 511 | 4/511 | 99% | 4/511 | LT602883 |
| 27 | Leucosporidium golubevii PYCC 5759T | 1(1) | 523 | 1/524 | 99% | 1/524 | LT602884 |
| Urediniomycetes | |||||||
| 28 | Microbotryozyma collariae ATCC MYA-4666 | 1(1) | 528 | 3/528 | 86% | 28/528 | LT602885 |
| 29 | Rhodotorula glacialis AB23-2 | 2(2) | 523 | 2/524 | 99% | 1/524 | LT602886 |
| 30 | Rhodotorula sp. YSAR13 | 1(1) | 514 | 9/514 | 98% | 0/514 | LT602887 |
| 31 | Rhodotorula sonckii | 2(2) | 472 | 60/446 | 87% | 16/446 | LT602888 |
| Cystobasidiomycetes | |||||||
| 32 | Cystobasidiomycetes sp. KSS-2008 5-19 | 2(2) | 563 | 34/568 | 94% | 10/568 | LT602889 |
| 33 | Cystobasidiomycetes sp. DSM 28479 | 3(3) | 538 | 1/537 | 99% | 0/537 | LT602890 |
aOperational taxonomic units (OTUs) identified in this study.
bClosest known related species as determined by best BLAST hit with ITS sequences; results represent searches from NCBI databases in June 2016.
cNo.: Number of isolates (numbers of isolates sequenced).
dITS len: ITS length (bp).
eSub: Substitution.
fITS sim: ITS similarity.
gITS sequences have been deposited in European Nucleotide Archive under the listed accession numbers (Acc no.).
Figure 2.
Composition of arabidopsis surface yeasts isolated from the view of genus (a) and the species or strain composition of the dominant genus Cryptococcus (b).
The behaviour of a selected subset of isolated yeasts was monitored in several physiological assays in order to understand aspects of yeast lifestyle, support the assertion that these species are specifically associated with arabidopsis, and characterize the nature of their host-yeast interaction. The following isolates were selected, as a representative set of genera in this study: Cryptococcus sp. OTU 9, Cryptococcus sp. OTU 4, Leucosporidium sp. OTU 26, Taphrina sp. OTU 3, Protomyces sp. OTU 1, Dioszegia sp. OTU 23 strain 1, Dioszegia sp. OTU 23 strain 2, Cryptococcus sp. OTU 17, Cystobasidiomycetes sp. OTU 32, Microbotryozyma sp. OTU 28, Leucosporidium sp. OTU 27. Additionally, Baker’s yeast (Saccharomyces cerevisiae L40) and fission yeast (Schizosaccharomyces pombe FY7519) were used as non-pathogenic control species and the mustard pathogen, Eremothecium sinecaudum BSL-1, was used as a known pathogen of Brassicaceae family plants21.
Yeast production of indolic compounds
Both pathogenic and growth-promoting microbes are known to produce the plant hormone auxin, prompting examination of this trait in our selected yeasts. Indolic compounds, including indole acetic acid (IAA) and other auxins, were detected using the Salkowski reagent colorimetric assay with supernatants from yeast cultures in media with varied amounts of the auxin precursor tryptophan. Among the 11 selected strains, eight produced indolic compounds when cultivated in glucose yeast peptone (GYP) medium, which contains moderate levels of tryptophan derived from yeast extract and peptone (Fig. 3). Indole compound production, measured in IAA equivalents (IAA eq.), ranged from 7.25 ± 0.60 to 24.98 ± 0.75 μg/ml. When cultivated in GYP supplemented with 0.1% tryptophan, production increased, ranging from 11.48 ± 1.80 to 41.45 ± 6.45. For conditions lacking exogenous tryptophan, a minimal medium of nitrogen base with glucose was used for cultivation. Interestingly, three isolates displayed indole production in minimal medium (Fig. 3), suggesting the ability to produce indoles in the absence of exogenous tryptophan, which better mimics the conditions in the arabidopsis phyllosphere.
Figure 3. Production of indolic compounds by yeasts.
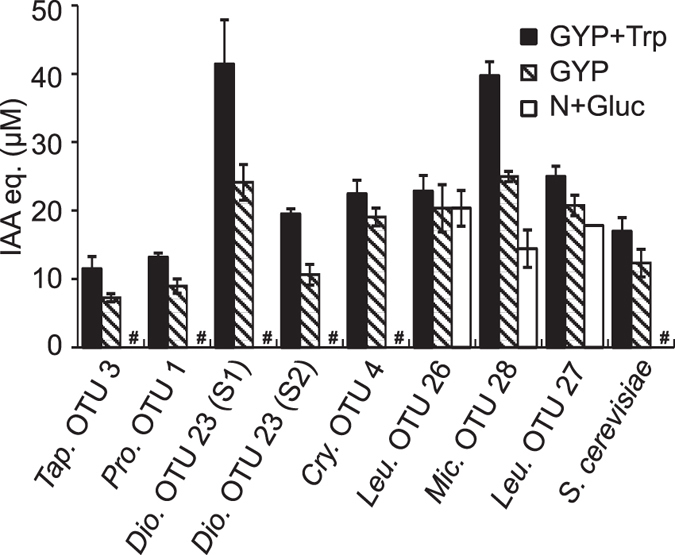
Selected isolates were cultivated in nitrogen base with 1% glucose (N + gluc), glucose-yeast-peptone (GYP) media with and without 0.1% L-tryptophan (Trp). Standard curve was calibrated by using indole acetic acid (IAA) and results are presented as equivalents of IAA (IAA eq.) concentration. #Not detected. Three independent biological replicates were conducted with similar results.
Activation of auxin responses in planta
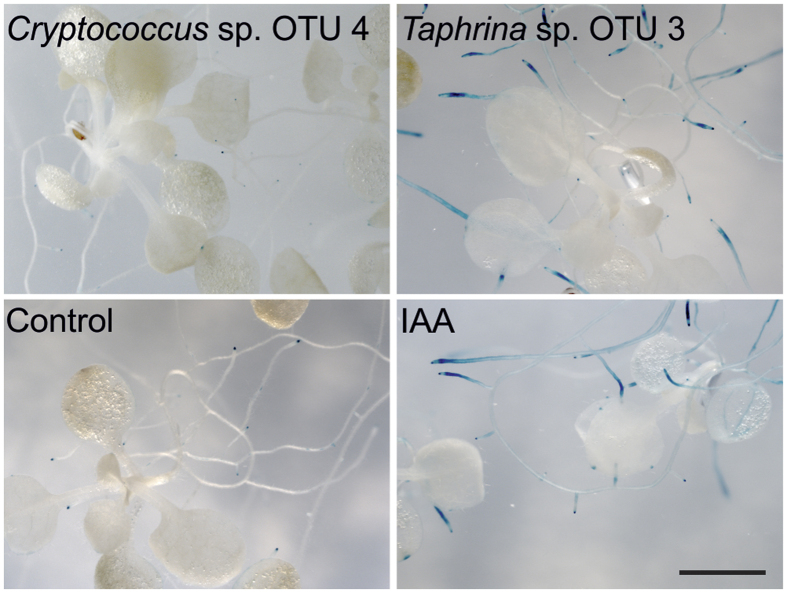
The artificial auxin-responsive promoter DR5 is commonly used to monitor the activation of auxin transcriptional response in arabidopsis. To investigate if yeast-derived indolic compounds had auxin activity in planta, arabidopsis roots bearing a construct with the DR5 promoter fused to the β-glucuronidase reporter gene (DR5::GUS) in the Col-0 accession were incubated with culture filtrates from five-day-old yeast cultures and monitored by GUS histochemical staining. A 5 μM IAA control treatment and the culture supernatant of Taphrina sp. OTU 3, as a representative examples of a positive responses, both exhibit strong blue GUS stains in the roots, indicating induction of DR5-promoter activity (Fig. 4). Similar GUS activity was detected in seedlings treated with culture supernatant of Taphrina sp. OTU 3, Protomyces sp. OTU 1, Dioszegia sp. OTU 23 strain 1, Saccharomyces cerevisiae, Eremothecium sinecaudum, as well as light GUS expressions in seedlings treated with Leucosporidium sp. OTU 26 and Leucosporidium sp. OTU 27 (Supplementary Fig. S1), suggesting the auxin transcriptional response was activated by exogenous auxin from the yeast culture.
Figure 4. Effect of indolic compounds produced by yeasts on gene expression of auxin regulated genes, monitored as expression of the artificial auxin responsive DR5 promoter.
Two-week-old in vitro grown DR5::GUS arabidopsis seedlings were treated with the filtered supernatants of five day old yeast cultures for 15 hours. As an example of a positive response, strong GUS expression were detected in seedling roots treated with Taphrina sp. OTU 3 supernatant and in the positive control 5 μM IAA treatment. As representative negative results, only light root tip staining was seen with Cryptococcus sp. OTU 4 supernatant and the negative control GYP medium treatments. Similar results were observed in three independent biological replicate experiments. Scale bar = 2 mm. See also Supplementary Fig. S1 for results of all treatments.
Plants treated with an uncultured GYP medium (negative control) or supernatant of Cryptococcus sp. OTU 4, are presented as representative examples of negative staining results, which exhibited only light staining of root tips (Fig. 4). Similar negative GUS staining was seen in plants treated with supernatants from Cryptococcus sp. OTU 9, Cryptococcus sp. OTU 17, Cystobasidiomycetes sp. OTU 32, Dioszegia sp. OTU 23 strain 2, Cryptococcus sp. OTU 4, Microbotryozyma sp. OTU 28 and Schizosaccharomyces pombe (Supplementary Fig. 1). The indolic compounds produced by strains Dioszegia sp. OTU 23 strain 2, Cryptococcus sp. OTU 4, and Microbotryozyma sp. OTU 28 (Fig. 3) did not trigger auxin transcriptional responses (Supplementary Fig. 1), indicating the presence of indolic compounds with no IAA activity in these species.
Camalexin sensitivity
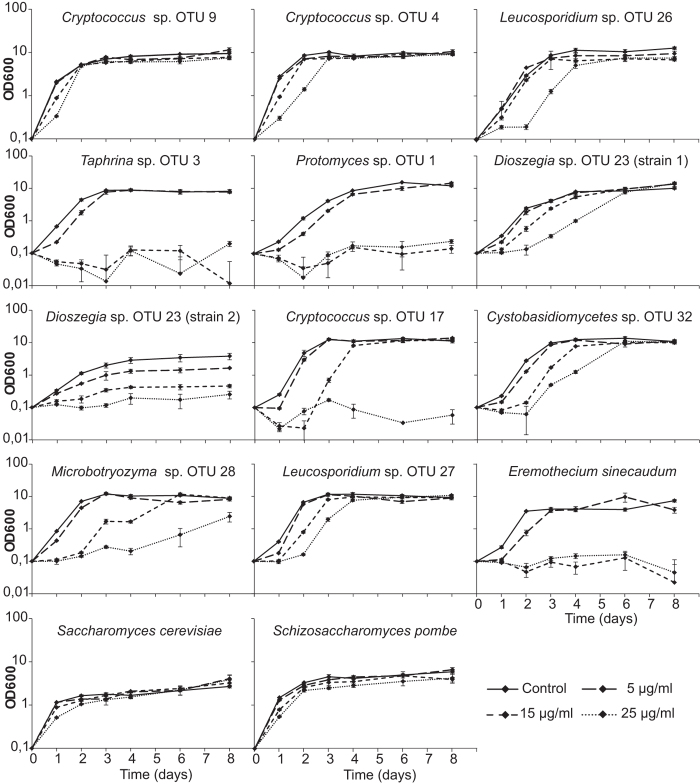
We reasoned that pathogenic and non-pathogenic yeasts may differ in their sensitivity to growth inhibition by the arabidopsis antimicrobial phytoalexin, camalexin. Yeasts were cultured in liquid media supplemented with 5, 15 and 25 μg/ml camalexin, revealing high variability in the level of camalexin sensitivity (Fig. 5). Many isolates, including control isolates, exhibited a delay at the initial period of their growth, which suggests these species have inducible camalexin-tolerance mechanisms. Two isolates from genera known to contain plant pathogens (Taphrina sp. OTU 3 and Protomyces sp. OTU 1) and the control plant pathogen (Eremothecium sinecaudum) were sensitive to camalexin at concentrations higher than 15 μg/ml. Isolates from genera not previously known to be phytopathogens were highly resistant to camalexin of the concentration up to 25 μg/ml (Fig. 5).
Figure 5. Effect of arabidopsis phytoalexin camalexin on yeast growth.
Yeasts were cultured in liquid GYP medium containing 0, 5, 15, 25 μg/ml camalexin, from starting cell density OD = 0.1. Similar results were observed from three independent biological replicate experiments.
Temperature tolerance
Most yeasts are mesophilic and grow optimally at temperatures between 20 and 25 °C, but still many species can grow between 2 and 10 °C. All the strains in this study were isolated at 21 °C from plants growing at low temperatures. In order to characterize the thermotolerance profiles and identify cold-adapted yeasts, growth under four temperature conditions (8, 21, 30, 37 °C) was tested. Seven out of 11 selected strains grew at 8 °C, indicating a high number of cold-adapted yeasts among our isolates. OTU 4 and OTU 26 grew better at low temperature than at room temperature. Only one strain (OTU 9) grew at 30 °C and at 37 °C (Fig. 6).
Figure 6. Effect of temperature on yeast growth.
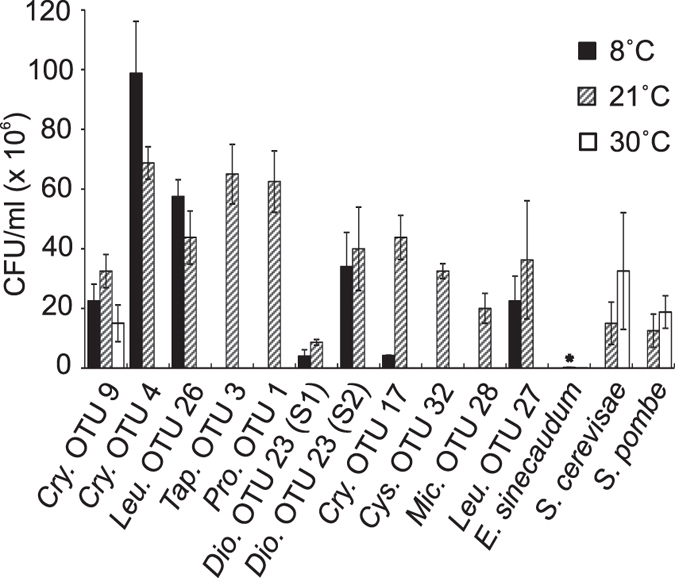
Yeasts were first grown on liquid GYP medium for three days and then diluted to OD = 1. Serial of dilutions (OD = 10−2, 10−3, 10−4, 10−5, 10−6, 10−7) were plated on GYP agar medium for grown at different temperature conditions (8 °C, 21 °C, 30 °C, 37 °C). Colony forming units (CFUs) were counted to quantify growth after seven days. No isolate grew at 37 °C. Similar results were observed from three independent biological replicate experiments.
Discussion
In this study we have isolated yeasts from the arabidopsis phylloplane. Among the yeast isolates presented here, 22 OTUs belong to the phylum Basidiomycota, which was much more abundant than the phylum Ascomycota with only two OTUs. The Ascomycete and Basidiomycete ratio is less than those of yeasts isolated in tropical regions5,7,22, subtropical areas8 and temperate areas23,24. However, Ascomycete yeasts may also dominate in the phyllosphere of plants in tropical regions, when the isolation was performed using a higher temperature7. Interestingly, a study of seasonal dynamics demonstrated the abundance of Ascomycetes increased gradually from spring to autumn23.
In Finland and other northern counties, arabidopsis seeds germinate in the autumn and grow until usually November, when temperatures drop, overwintering as a small rosette, often under snow cover, until growth resumes in the spring, typically in April. The lifecycle is then completed with seed set in late May or early June. Accordingly, a high percentage of low-temperature adapted yeasts were isolated from plants sampled in May, whose rosettes had overwintered under snow. Some plant pathogens are specialized for infection during winter and there is currently no genetically tractable model pathosystem for such interactions. Further studies are required, however, some of the isolates presented here may have potential for the development of a low temperature adapted pathogen model system.
Auxin production by a plant-associated microorganism was first reported by Kaper and Veldstra25. Auxin of microbial origin has been shown to play a wide variety of roles26. Within and between microbes, IAA can function as a signalling molecule regulating microbial gene expression, promoting growth-form switching, and potentially as a quorum sensing molecule26,27,28.
In pathogenic microbes, IAA functions in tumour or gall formation, suppression of plant immune-signalling and aiding pathogen ingress via induction of plant cell wall expansion and weakening29. Remarkably, two potentially pathogenic OTUs isolated here (OTU 1 and 3), both of which exhibited the ability to produce indolic compounds and induce an auxin response in planta, both come from genera known to induce plant tumours as disease symptoms (Protomyces and Taphrina). In plant-growth promoting microorganisms, IAA results in enhanced root-proliferation, increased mineral adsorption, and increased basal defence9,14. Given the variety of these responses, diverse model systems are needed for the study IAA in plant-microbe interactions in different contexts. Although IAA-producing yeasts have been characterized from many other plants, and IAA-producing bacteria and fungi that interact with arabidopsis are known, this study offers the first opportunity for a yeast-arabidopsis interaction involving this plant hormone. In this study, we isolated several potentially arabidopsis-associated yeasts with the ability to produce indolic compounds. Among the tested eleven yeast isolates, eight isolates exhibited significant production of indolic compounds when cultivated in various media with different levels of exogenous tryptophan. Tryptophan is the main precursor for IAA biosynthesis pathway in most organisms and there are five different biosynthesis pathways known leading from tryptophan to IAA26. Additionally, a tryptophan-independent pathway exists in bacteria and fungi27,30. Three isolates (Leucosporidium sp. OTU 26, Microbotryozyma sp. OTU 28 and Leucosporidium sp. OTU 27) exhibited the production of indolic compounds in the absence of tryptophan in the growth medium. Culture filtrates from two of these isolates (OTU 26 and OTU 27) had in planta auxin activity (Supplementary Fig. S1). Most studies reporting microbial auxin production utilize growth media containing exogenous tryptophan or containing yeast extract or peptone, which contain high levels of tryptophan. In contrast, tryptophan was reported as a low abundance amino acid in oat and barley leaf exudates20. Here we demonstrate yeasts with the ability to produce auxins or other indolic compounds under conditions more similar to the low tryptophan levels expected in the phylloplane. These yeasts may use either endogenous de novo tryptophan biosynthesis or a non-tryptophan-dependent IAA/indole biosynthesis pathway.
In general, most of the assayed yeast-culture filtrates that contained indolic compounds were also shown to have auxin activity in the induction of a transcriptional marker in arabidopsis suggesting that they contain auxins. It has been proposed that indolic compound production by yeasts is a general adaptation strategy for plant phyllosphere environment9. Since our yeast strains were isolated from wild arabidopsis plants, without any disease symptoms, indolic compound production may be an important indicator for plant associated beneficial or neutral microbes.
The indole alkaloid, camalexin (3-thiazol-2′-yl-indole), is the major anti-microbial phytoalexin in arabidopsis. Like other low molecular weight antimicrobial compounds, camalexin synthesis is rapidly induced by plant pathogens31,32,33,34. Several arabidopsis pathogens are known to be sensitive to camalexin35, which contributes to arabidopsis pathogen immunity36,37. Cell membrane disruption is considered as the primary mode of camalexin toxicity against bacterial, fungal and plant cells38. Sensitivity to camalexin is not restricted to strictly to pathogenic species; both resistant and sensitive isolates were found within Botrytis cinerea36. However, fungal pathogens generally have a much lower toxicity threshold than bacterial pathogens and host plant cells38. In this study, the high sensitivity of three potentially pathogenic yeast isolates (Fig. 5) confirmed the low toxicity threshold of pathogenic fungi. It has been shown that culture media supplemented with 20 μg/ml camalexin caused a significant decrease in spore germination and hyphal growth of Botrytis cinerea isolates36, which is in accordance to the sensitivity of three potentially pathogenic yeast isolates to 15 μg/ml camalexin.
Several fungi are reported to have tolerance against camalexin by its active degradation into less toxic compounds39,40. Similarly, the non-pathogenic yeast strains exhibited high tolerance in cultures with camalexin concentrations up to 25 μg/ml. Although they might be sensitive higher camalexin concentrations38, these are not realistic to the situation in the phyllosphere of arabidopsis, where it has been estimated that camalexin accumulation reached a maximum of 8 μg/g leaf fresh weight in arabidopsis after inoculation with a bacterial pathogen41.
Remarkably, in the control experiments, higher camalexin tolerance was a characteristic of the non-pathogenic species (S. pombe and S. cerevisiae), while the control pathogen (Eremothecium) was more sensitive. This trend also held among the strains isolated here, genera previously known to be plant pathogens were sensitive, while genera with no previous report of plant pathogenicity tended to be tolerant (see Fig. 5 and discussion of the individual genera below). This is consistent with the pathogen lifestyle and virulence strategy; pathogens do not require camalexin tolerance as they can avoid camalexin, via subverting immune signalling pathways with effector proteins and other virulence mechanisms. Thus we contend that, although not a perfect indicator all of the time, camalexin sensitivity can be used as a guide in the identification of potential latent pathogens among phylloplane resident microbes.
All the yeast genera isolated from the arabidopsis phyllosphere have been previously demonstrated to be associated with plants. Several have also been previously found to be associated with arabidopsis. Recently, Alger et al.42 characterized the arabidopsis phyllosphere using metagenomics. This study independently verified the presence of OTUs in the genera Protomyces, Dioszegia, Leucosporidium, and Rhodotorula, in the phyllosphere of arabidopsis collected from two distant sites in Germany. This suggests that these yeasts may indeed belong to the recurring core phyllosphere of arabidopsis.
The characteristics of each genus found in our isolations are individually discussed below.
Cryptococcus is the dominant genus in phyllosphere of arabidopsis in this study. Members of the genus Cryptococcus are widely distributed and were described as a dominant species in the phyllosphere of other plants, such as carnivorous plant9, spruce and birch23. Another group of yeast commonly found in the phyllosphere of many plants, Dioszegia spp. have been previously detected on plant leaves and roots, and even polar desert soil10,43,44,45. Some novel psychrophilic species in this genus isolated from Antarctica have been previously described45. The world-wide distribution of the Cryptococcus and Dioszegia species may due to the extreme temperature tolerance and the ability to utilize the nutrients from harsh environments. Among 11 tested isolates, two Dioszegia isolates and two Cryptococcus isolates performed active growth at low temperature, suggesting the possible cold adaptation in northern climates. A Dioszegia sp. was previously found in the arabidopsis phyllosphere42 and found to act as a so called “hub” microbe, which are species that have a role in organizing the microbiome community. Specifically, presumably via direct microbe-microbe interactions, Dioszegia spp. shaped the prokaryotic microbiome of the plants on which they resided42. Due to the importance of these species, we have sequenced the genome of the Dioszegia sp. OTU 23 isolated in this study. Analysis of this genome will be presented elsewhere.
The genus Rhodotorula is a group of pigmented yeasts, widely variant in colour, being cream to orange, red, pink or yellow. As a common environmental microbe, Rhodotorula species have been cultured from water, milk, fruit juice, soil and even air samples46. Interestingly, R. glacialis has been studied as an oleaginous yeast, which might be used for the production of single cell oils47. The studies also demonstrated that under appropriate conditions, this yeast accumulates high amount of lipids. Additionally, siderophores produced by Rhodotorula strains exhibited antifungal activity against plant pathogens including Botrytis cinerea48. Holtermannia is a group of yeast with wide-spread distribution. Holtermannia strains have been isolated from many natural and artificial substrates from different parts of the world49.
Two of the potentially pathogenic yeasts isolated are from the genera, Taphrina and Protomyces, which both belong to the Taphrinomycotina. Members of this early diverging Ascomycete subphylum are considered to be ancient lineages and as such are important for understanding fungal evolution and the evolution of pathogenicity. Established by Unger in 1832, the genus Protomyces was described as pathogens that cause galls on stems, leaves or fruits on host plants in the families Compositae and Umbelliferae. Protomyces has been strictly defined based morphologically on cell sizes and on their host range, where only yeasts pathogenic on Compositae and Umbelliferae belong to Protomyces50. This is among the first reports to isolate or detect a Protomyces sp., as defined by ITS sequences, from a plant outside of these two host families. ITS sequences of the Protomyces spp. described here (OTU 1 and OTU 2), place them firmly within the Protomyces. However, these ITS sequences are only 97% and 95% similar to that of their closest BLAST hit Protomyces inouyei. This suggests that OTU 1 and 2 are novel Protomyces species with hosts outside of the defined range. Many yeasts morphologically similar to Protomyces have been characterized but excluded from the genus based on their host plants50; however, descriptions of these species did not include molecular data (ITS sequences). Taken together, these data suggest that the definition of the genus Protomyces should be revised. It has been previously noted that genus and species demarcation in Protomyces and the related genera, Burenia, Protomycopsis, Taphridium, and Volkartia, are unclear and likely incorrect and that extensive molecular comparisons are needed51. Relationships within Protomyces are currently being addressed by sequencing the genome of OTU 1 and several reference Protomyces species (Wang, Sipilä and Overmyer, unpublished data). This work will be presented elsewhere.
As a genus closely related to Protomyces, Taphrina contains nearly 100 described species that are parasitic on different families of primarily woody plants52. Some Taphrina species cause diseases on fruit trees, of which the symptoms are diverse malformations of leaves and other infected tissue53.
Species of genus Leucosporidium were originally transferred from Rhodotorula54. Most of the species of this genus were isolated from cold climates51, some of which were described for the ability to degrade phenol and phenol-related compounds55. One Leucosporidium isolated here is suggested to be cold-adapted because of its active growth at low temperature. The capacity for biodegradation of phenol and phenol-related compounds by Leucosporidium isolates found in this study may be of biotechnological interest and warrants further study. The genus Leucosporidium is phylogenetically closely related to Leucosporidiella. One of the isolates discovered in this genus (OTU 27), which was most closely related to Leucosporidium golubevii isolated from river water in Portugal54, grew actively at low temperature in accordance with the psychrophilic nature of this genus. In addition, an unknown isolate was collected with 86% ITS identity to Microbotryozyma collariae, which was described recently as a novel species56.
The yeast species in Cystofilobasidium are widely distributed mainly in cold climates. Interestingly, a C. capitatum strain (PPY-1) was isolated from soil and was suggested may produce novel enzymes that can degrade pectin at low temperature57. Subsequently, a cold-active extracellular pectin lyase from the strain PPY-1 has been purified58. The ability to produce plant cell wall degrading enzymes may be a survival strategy for several microbe groups. Production of pectin lyase and related enzymes may be relevant to our strains, future testing for enzymes with these characteristics may have practical application.
Metagenomics is currently the state of the art for the study of plant microbiota and application of this technique to explore the fungi and yeasts in the arabidopsis phyllosphere microbiome gives a more comprehensive picture of the species present42 and is complementary to this study. The purpose of the present study was to make an initial survey of yeast in the arabidopsis phyllosphere, isolate strains for use in future studies and physiologically characterize these strains to aid in selecting appropriate strains for developing new model systems with arabidopsis. Currently, the majority of work on molecular plant-microbe interactions with arabidopsis utilize only a small number of pathogenic species, including mostly bacteria and fungi59,60, but no yeasts. There is a need to diversify the study of plant pathology and include molecular work on plant yeast interactions. The strain resources and information generated in this study will contribute significantly to this effort.
Methods
Sample collection and yeast isolation
Healthy rosettes of arabidopsis (Arabidopsis thaliana) were collected from three distinct sites (in the Kivikko, Kulosaari and Mustikkamaa districts) in Helsinki, Finland (Table 1). The month average temperature of Helsinki in April, May and November are 4 °C, 10 °C and 0 °C (Finnish Meteorological Institute; http://en.ilmatieteenlaitos.fi). Plants were collected by sterile scissor and forceps and were placed in sterile 50 ml centrifuge tubes. Samples were kept at cold-room (4 °C) until yeast isolation procedures. As a pre-wash to remove surface dust, the leaves were vortexed two times in a 2 ml tube containing 1 ml sterile water for 3 seconds. Leaves were transferred into another 2 ml tube containing 1 ml MQ water with 0.025% Silwet-L77 and were shaken gently for 4 hours on a rocking platform. Serial dilutions of leaf wash solution were plated onto 0.2 × PDA (potato dextrose agar, Sigma-Aldrich) medium. Single colonies were picked and were streaked twice on 0.2 × PDA medium with 100 μg/ml Ampicillin for purification. Long-term storage of wash solutions as well as purified strains was performed in 50% sterile glycerol at −80 °C.
DNA extraction
Yeast were cultivated in 2 ml 0.2 × PDB (Potato dextrose broth, Sigma-Aldrich) for 5 days. Cells were pelleted by centrifugation for 5 min at 12,000 g and the pellet was suspended in 200 μl lysis buffer (100 mM Tris-HCl pH8.0, 50 mM EDTA, 500 mM NaCl). Glass beads (0.3 g) and 200 μl phenol/chloroform/isoamyl alcohol were added and vortexed at high speed for 3 min. Cells were briefly vortexed after adding 200 μl TE buffer. The samples were centrifuged 5 min at maximum speed. The aqueous layer was transferred to a clean 2 ml centrifuge tube, ethanol precipitated, pelleted by centrifugation and resuspended in 0.4 ml TE buffer. Following DNase-free RNase A treatment (30 μl of 1 mg/ml, 5 min at 37 °C) samples were ethanol precipitated as above and resuspended in 100 μl TE buffer.
Yeast identification
Sequences of the ITS (internal transcribed spacer) region were determined by polymerase chain reaction (PCR) products from yeast genomic DNA. PCR products were obtained using forward primer ITS3 (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and reverse primer ITS4 (5′-TCCTCCGCTTATTGATATGC-3′). Each reaction mixture contained 2 μl 10 × fire polymerase buffer B (Solis BioDyne), 1.6 μl MgCl2 (25 mM), 0.4 μl dNTP, 0.4 μl of each primer (10 mM). After denaturation of DNA at 94 °C for 5 minutes, 35 cycles of amplification with the following thermocycling program: denaturation at 94 °C for 20 s, followed by 10 s at 50 °C for primer annealing and 30 s at 72 °C for extension. Final extension 5 minutes at 72 °C was used. The PCR products were observed by separating the fragments with 1.5% agarose gel electrophoresis. ExoSAP (Thermo Scientific™ Exonuclease I, Shrimp Alkaline Phosphatase) treatment of individual PCR products was performed to remove residual primers.
Restriction enzymes HaeIII and TaqI (Thermo scientific) were chosen for ITS fragment digestion, following the manufacturer’s instructions and fragments were observed by separation by electrophoresis using a 3% agarose gel. Isolates were classified as OTUs (operational taxonomic units) on the basis of a unique ITS restriction pattern with the enzymes HaeIII and TaqI and corresponding ITS amplicon length. Representatives of each OTU were selected for sequencing with ITS4 and ITS1 (GGAAGTAAAAGTCGTAACAAGG) primers. The complete ITS region was assembled from the sequencing results (Chromas Lite; Ape). ITS comparison to described fungi species was performed using BLAST (Basic Local Alignment Search Tool) at the NCBI (National Centre for Biotechnology Information) database.
Indolic compound production and GUS staining
The production of indolic compounds of selected strains was monitored using Salkowski reagent61. Three different media (GYP, GYP with 0.1% L-tryptophan, and nitrogen base with 1% glucose) were used. Yeasts were cultured in liquid medium in 96 deep-well plate (P8116, Sigma-Aldrich) for three days from starting cell density OD = 0.1. Breathable film (A9224, Sigma-Aldrich) was used for sealing the plates. One ml yeast cells were centrifuged at 12,000 g for 5 min, and 0.5 ml supernatant was mixed well with 0.5 ml Salkowski regent (12 g of FeCl3 per litre in 7.9 M H2SO4). The mixture was kept at darkness for 30 min. The red colour development was quantified spectrophotometrically (Agilent 8453) as absorbance at wavelength 530 nm. A calibration curve was established using indole-3-acetic acid (IAA) with concentrations of 1, 2, 4, 5, 6, 8, 10 μg/ml. Experiments were repeated independently three times. An arabidopsis DR5::GUS transgenic line in the Col-0 accession (obtained from the Nottingham Arabidopsis Stock Centre; NASC; http://arabidopsis.info) was cultivated in 0.5 × Murashige and Skoog15 (M5519, Sigma, USA) with addition of 1% (w/v) agar and 1% (w/v) sucrose (pH = 5.7). Seeds were surface sterilized by washing with 70% ethanol with 2% Triton X-100 for 5 min, and washed two times with absolute ethanol. Seedlings were grown in 6 well cell culture plates (CLS3516, Corning Costar), with 10 seeds in each well. Plants were placed in growth chamber (16/8, 24 °C). Ten-day-old DR5::GUS seedlings were treated with 1 ml filtered supernatants of 5 day yeast cultures (GYP with 0.1% L-tryptophan) overnight. Positive control group was treated with 1 ml 5 μM IAA, and 1 ml medium for negative control group. For histochemical staining, seedlings were fixed with ice-cold 90% acetone for 1 h, washed three times with ice-cold wash solution (36 mM Na2HPO4, 14 mM NaH2PO4, pH 7.2), 30 min for each wash. Seedlings were vacuum infiltrated and kept at room temperature in GUS staining solution (1 mM 5-bromo-4-chloro-3-indolyl b-D-glucuronide dissolved in methanol, 5 mM potassium ferricyanide and 5 mM potassium ferrocyanide in 50 mM sodium phosphate buffer, pH 7.2). Stained seedlings were washed three times with absolute ethanol and were kept in 70% ethanol. Experiments were repeated independently three times.
Camalexin and temperature tolerance assays
Eleven selected strains and three control strains were cultivated in liquid GYP medium in 96 deep-well plate, containing 0, 5, 15, 25 μg/ml camalexin (SML1016, Sigma-Aldrich). The mustard pathogen, Eremothecium sinecaudum BSL-1 (CBS 8199) used as a control was obtained from CBS-KNAW (http://www.cbs.knaw.nl). Plates were shaken at 850 rpm at 21 °C in a growth chamber. Absorbance density at 600 nm of cultures were measured at several time points (day 0, 1, 2, 3, 4, 6, 8) as indication of yeast density. Yeast growth at different camalexin concentrations were measured from starting cell density OD = 0.1. Experiments were repeated independently three times. For temperature growth experiments, yeast strains were first cultivated in liquid GYP medium for three days, then diluted to OD = 1 with medium. Serial dilutions (10−1 to 10−7) were prepared. Drop plating with 2 μl of each dilution was conducted in square GYP agar plates. The strains were cultivated at 8 °C, 21 °C, 30 °C and 37 °C conditions. Colony forming units (CFUs) were counted to determine growth after seven days. Experiments were repeated independently three times.
Additional Information
How to cite this article: Wang, K. et al. The isolation and characterization of resident yeasts from the phylloplane of Arabidopsis thaliana. Sci. Rep. 6, 39403; doi: 10.1038/srep39403 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
K.W. is a member of the University of Helsinki Doctoral Program in Plant Science (DPPS). Leena Grönholm is acknowledged for excellent technical support and thank Fuqiang Cui and Xin Wang for their kind assistance with GUS staining. Members of Plant Stress Meta-Group are acknowledged for many fruitful discussions. This work was supported by the following grants: Academy of Finland Fellowship (decisions no. 251397, 256073 and 283254) to K.O. Academy of Finland Centre of Excellence in Primary Producers 2014–2019 (decision #271832). The Nottingham Arabidopsis Stock Centre (NASC) is acknowledged for arabidopsis seeds and the CBS-KNAW Fungal Biodiversity Centre for the Eremothecium sinecaudum strain.
Footnotes
Author Contributions K.O. conceived the research, K.W. performed all experiments, K.W. and K.O., wrote the manuscript, all authors designed experiments, analysed data, edited and approved the manuscript.
References
- Conn V. M., Walker A. R. & Franco C. M. M. Endophytic Actinobacteria Induce Defense Pathways in Arabidopsis thaliana. Molecular Plant-Microbe Interactions 21, 208–218, doi: 10.1094/MPMI-21-2-0208 (2008). [DOI] [PubMed] [Google Scholar]
- van der Lelie D. et al. Poplar and its Bacterial Endophytes: Coexistence and Harmony. Critical Reviews in Plant Sciences 28, 346–358, doi: 10.1080/07352680903241204 (2009). [DOI] [Google Scholar]
- Rodriguez R. J. et al. Stress tolerance in plants via habitat-adapted symbiosis. ISME J 2, 404–416 (2008). [DOI] [PubMed] [Google Scholar]
- Marschner H., Römheld V., Horst W. J. & Martin P. Root-induced changes in the rhizosphere: Importance for the mineral nutrition of plants. Zeitschrift für Pflanzenernährung und Bodenkunde 149, 441–456, doi: 10.1002/jpln.19861490408 (1986). [DOI] [Google Scholar]
- Nakasel’s T. et al. Ballistoconidium—Forming Yeasts Found in the Phyllosphere of Thailand. Group 500 (1987). [Google Scholar]
- Limtong S., Kaewwichian R., Yongmanitchai W. & Kawasaki H. Diversity of culturable yeasts in phylloplane of sugarcane in Thailand and their capability to produce indole-3-acetic acid. World Journal of Microbiology and Biotechnology 30, 1785–1796 (2014). [DOI] [PubMed] [Google Scholar]
- Limtong S. & Kaewwichian R. The diversity of culturable yeasts in the phylloplane of rice in Thailand. Annals of Microbiology 65, 667–675 (2015). [DOI] [PubMed] [Google Scholar]
- Inácio J. et al. Estimation and diversity of phylloplane mycobiota on selected plants in a mediterranean–type ecosystem in Portugal. Microbial Ecology 44, 344–353 (2002). [DOI] [PubMed] [Google Scholar]
- Sun P. F. et al. Indole-3-acetic acid-producing yeasts in the phyllosphere of the carnivorous plant Drosera indica L. PloS one 9, e114196, doi: 10.1371/journal.pone.0114196 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inácio J., Portugal L., Spencer-Martins I. & Fonseca Á. Phylloplane yeasts from Portugal: seven novel anamorphic species in the Tremellales lineage of the Hymenomycetes (Basidiomycota) producing orange-coloured colonies. FEMS yeast research 5, 1167–1183 (2005). [DOI] [PubMed] [Google Scholar]
- Ryffel F. et al. Metabolic footprint of epiphytic bacteria on Arabidopsis thaliana leaves. The ISME journal (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier J. & Lindow S. Role of leaf surface sugars in colonization of plants by bacterial epiphytes. Applied and Environmental Microbiology 66, 369–374 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl M. & Lindow S. Contribution of indole-3-acetic acid production to the epiphytic fitness of Erwinia herbicola. Applied and environmental microbiology 64, 3256–3263 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras-Cornejo H. A., Macías-Rodríguez L., Cortés-Penagos C. & López-Bucio J. Trichoderma virens, a plant beneficial fungus, enhances biomass production and promotes lateral root growth through an auxin-dependent mechanism in Arabidopsis. Plant Physiology 149, 1579–1592 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad F., Ahmad I. & Khan M. Screening of free-living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiological research 163, 173–181 (2008). [DOI] [PubMed] [Google Scholar]
- Jones J. D. & Dangl J. L. The plant immune system. Nature 444, 323–329 (2006). [DOI] [PubMed] [Google Scholar]
- Dangl J. L. & Jones J. D. G. Plant pathogens and integrated defence responses to infection. Nature 411, 826–833 (2001). [DOI] [PubMed] [Google Scholar]
- Cheng S. C., Joosten L. A., Kullberg B. J. & Netea M. G. Interplay between Candida albicans and the mammalian innate host defense. Infection and immunity 80, 1304–1313, doi: 10.1128/iai.06146-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raacke I. C., von Rad U., Mueller M. J. & Berger S. Yeast Increases Resistance in Arabidopsis Against Pseudomonas syringae and Botrytis cinerea by Salicylic Acid-Dependent as Well as -Independent Mechanisms. Molecular Plant-Microbe Interactions 19, 1138–1146, doi: 10.1094/MPMI-19-1138 (2006). [DOI] [PubMed] [Google Scholar]
- Weibull J., Ronquist F. & Brishammar S. Free Amino Acid Composition of Leaf Exudates and Phloem Sap: A Comparative Study in Oats and Barley. Plant Physiology 92, 222–226 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman C. P. Relationships among the genera Ashbya, Eremothecium, Holleya and Nematospora determined from rDNA sequence divergence. J Ind Microbiol 14, 523–530 (1995). [DOI] [PubMed] [Google Scholar]
- de Azeredo L. A. I., Gomes E. A. T., Mendonça-Hagler L. C. & Hagler A. N. Yeast communities associated with sugarcane in Campos, Rio de Janeiro, Brazil. International Microbiology 1, 205–208 (2010). [PubMed] [Google Scholar]
- Maksimova I. A. & Chernov I. [Community structure of yeast fungi in forest biogeocenoses]. Mikrobiologiia 73, 558–566 (2004). [PubMed] [Google Scholar]
- SlÁviková E., Vadkertiová R. & Vránová D. Yeasts colonizing the leaves of fruit trees. Annals of microbiology 59, 419–424 (2009). [Google Scholar]
- Kaper J. & Veldstra H. On the metabolism of tryptophan by Agrobacterium tumefaciens. Biochimica et biophysica acta 30, 401–420 (1958). [DOI] [PubMed] [Google Scholar]
- Spaepen S., Vanderleyden J. & Remans R. Indole‐3‐acetic acid in microbial and microorganism‐plant signaling. FEMS microbiology reviews 31, 425–448 (2007). [DOI] [PubMed] [Google Scholar]
- Rao R. P., Hunter A., Kashpur O. & Normanly J. Aberrant Synthesis of Indole-3-Acetic Acid in Saccharomyces cerevisiae Triggers Morphogenic Transition, a Virulence Trait of Pathogenic Fungi. Genetics 185, 211–220, doi: 10.1534/genetics.109.112854 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague G. F. & Winans S. C. Eukaryotes learn how to count: quorum sensing by yeast. Genes & development 20, 1045–1049 (2006). [DOI] [PubMed] [Google Scholar]
- Spaepen S. & Vanderleyden J. Auxin and plant-microbe interactions. Cold Spring Harbor perspectives in biology 3, a001438 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinsen E., Costacurta A., Michiels K., Vanderleyden J. & Van Onckelen H. Azospirillum brasilense indole-3-acetic acid biosynthesis: evidence for a non-tryptophan dependent pathway. Molecular Plant Microbe Interactions 6, 609–609 (1993). [Google Scholar]
- Glazebrook J. et al. Phytoalexin-deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics 146, 381–392 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. & Ausubel F. M. Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proceedings of the National Academy of Sciences 91, 8955–8959 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway A. et al. Characterization of cauliflower mosaic virus (CaMV) resistance in virus-resistant ecotypes of Arabidopsis. MPMI-Molecular Plant Microbe Interactions 9, 810–818 (1996). [DOI] [PubMed] [Google Scholar]
- Roetschi A., Si‐Ammour A., Belbahri L., Mauch F. & Mauch‐Mani B. Characterization of an Arabidopsis–Phytophthora pathosystem: resistance requires a functional PAD2 gene and is independent of salicylic acid, ethylene and jasmonic acid signalling. The Plant Journal 28, 293–305 (2001). [DOI] [PubMed] [Google Scholar]
- Thomma B. P., Nelissen I., Eggermont K. & Broekaert W. F. Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. The Plant Journal 19, 163–171 (1999). [DOI] [PubMed] [Google Scholar]
- Kliebenstein D. J., Rowe H. C. & Denby K. J. Secondary metabolites influence Arabidopsis/Botrytis interactions: variation in host production and pathogen sensitivity. The Plant Journal 44, 25–36 (2005). [DOI] [PubMed] [Google Scholar]
- Bohman S., Staal J., Thomma B. P., Wang M. & Dixelius C. Characterisation of an Arabidopsis–Leptosphaeria maculans pathosystem: resistance partially requires camalexin biosynthesis and is independent of salicylic acid, ethylene and jasmonic acid signalling. The Plant Journal 37, 9–20 (2004). [DOI] [PubMed] [Google Scholar]
- Rogers E. E., Glazebrook J. & Ausubel F. M. Mode of action of the Arabidopsis thaliana phytoalexin camalexin and its role in Arabidopsis-pathogen interactions. Molecular Plant Microbe Interactions 9, 748–757 (1996). [DOI] [PubMed] [Google Scholar]
- Soledade M., Pedras C. & Khan A. Q. Unprecedented detoxification of the cruciferous phytoalexin camalexin by a root phytopathogen. Bioorganic & Medicinal Chemistry Letters 7, 2255–2260 (1997). [Google Scholar]
- Pedras M. S. C. & Khan A. Q. Biotransformation of the phytoalexin camalexin by the phytopathogen Rhizoctonia solani. Phytochemistry 53, 59–69 (2000). [DOI] [PubMed] [Google Scholar]
- Tsuji J., Jackson E. P., Gage D. A., Hammerschmidt R. & Somerville S. C. Phytoalexin accumulation in Arabidopsis thaliana during the hypersensitive reaction to Pseudomonas syringae pv syringae. Plant Physiology 98, 1304–1309 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agler M. T. et al. Microbial Hub Taxa Link Host and Abiotic Factors to Plant Microbiome Variation. PLoS Biol 14, e1002352, doi: 10.1371/journal.pbio.1002352 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.-M., Jia J.-H. & Bai F.-Y. Diversity of basidiomycetous phylloplane yeasts belonging to the genus Dioszegia (Tremellales) and description of Dioszegia athyri sp. nov., Dioszegia butyracea sp. nov. and Dioszegia xingshanensis sp. nov. Antonie van Leeuwenhoek 93, 391–399 (2008). [DOI] [PubMed] [Google Scholar]
- Renker C., Blanke V., Börstler B., Heinrichs J. & Buscot F. Diversity of Cryptococcus and Dioszegia yeasts (Basidiomycota) inhabiting arbuscular mycorrhizal roots or spores. FEMS yeast research 4, 597–603 (2004). [DOI] [PubMed] [Google Scholar]
- Connell L. B. et al. Dioszegia antarctica sp. nov. and Dioszegia cryoxerica sp. nov., psychrophilic basidiomycetous yeasts from polar desert soils in Antarctica. International journal of systematic and evolutionary microbiology 60, 1466–1472 (2010). [DOI] [PubMed] [Google Scholar]
- Wirth F. & Goldani L. Z. Epidemiology of Rhodotorula: an emerging pathogen. Interdisciplinary perspectives on infectious diseases 2012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaretti A. et al. Single cell oils of the cold-adapted oleaginous yeast Rhodotorula glacialis DBVPG 4785. Microbial cell factories 9, 1 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvente V., De Orellano M., Sansone G., Benuzzi D. & De Tosetti M. S. Effect of nitrogen source and pH on siderophore production by Rhodotorula strains and their application to biocontrol of phytopathogenic moulds. Journal of Industrial Microbiology and Biotechnology 26, 226–229 (2001). [DOI] [PubMed] [Google Scholar]
- Wuczkowski M. et al. Description of Holtermanniella gen. nov., including Holtermanniella takashimae sp. nov. and four new combinations, and proposal of the order Holtermanniales to accommodate tremellomycetous yeasts of the Holtermannia clade. International journal of systematic and evolutionary microbiology 61, 680–689 (2011). [DOI] [PubMed] [Google Scholar]
- Reddy M. S. & Kramer C. A taxonomic revision of the Protomycetales (Burenia, Fungi). Mycotaxon (1975). [Google Scholar]
- Kurtzman C., Fell J. W. & Boekhout T. The yeasts: a taxonomic study (Elsevier, 2011). [Google Scholar]
- Rodrigues M. G. & Fonseca Á. Molecular systematics of the dimorphic ascomycete genus Taphrina. International journal of systematic and evolutionary microbiology 53, 607–616 (2003). [DOI] [PubMed] [Google Scholar]
- Mix A. J. A monograph of the genus Taphrina. University of Kansas Science Bulletin 33 (1949). [Google Scholar]
- Sampaio J. P., Gadanho M., Bauer R. & Weiß M. Taxonomic studies in the Microbotryomycetidae: Leucosporidium golubevii sp. nov., Leucosporidiella gen. nov. and the new orders Leucosporidiales and Sporidiobolales. Mycological Progress 2, 53–68 (2003). [Google Scholar]
- Bergauer P., Fonteyne P.-A., Nolard N., Schinner F. & Margesin R. Biodegradation of phenol and phenol-related compounds by psychrophilic and cold-tolerant alpine yeasts. Chemosphere 59, 909–918 (2005). [DOI] [PubMed] [Google Scholar]
- Suh S.-O., Maslov D. A., Molestina R. E. & Zhou J. J. Microbotryozyma collariae gen. nov., sp. nov., a basidiomycetous yeast isolated from a plant bug Collaria oleosa (Miridae). Antonie van Leeuwenhoek 102, 99–104 (2012). [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Nagaoka T., Taniguchi S., Miyaji T. & Tomizuka N. Isolation and characterization of psychrophilic yeasts producing cold‐adapted pectinolytic enzymes. Letters in Applied Microbiology 38, 383–387 (2004). [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Nagaoka T., Miyaji T. & Tomizuka N. A cold‐active pectin lyase from the psychrophilic and basidiomycetous yeast Cystofilobasidium capitatum strain PPY‐1. Biotechnology and applied biochemistry 42, 193–196 (2005). [DOI] [PubMed] [Google Scholar]
- Mansfield J. et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol 13, 614–629, doi: 10.1111/j.1364-3703.2012.00804.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean R. et al. The Top 10 fungal pathogens in molecular plant pathology. Molecular Plant Pathology 13, 414–430, doi: 10.1111/j.1364-3703.2011.00783.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickmann E. & Dessaux Y. A critical examination of the specificity of the salkowski reagent for indolic compounds produced by phytopathogenic bacteria. Applied and environmental microbiology 61, 793–796 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.