Abstract
A broad biophysical analysis was performed to investigate the molecular basis of the neuroprotective action of Curcuma longa extracts in Alzheimer’s disease. By combining circular dichroism and electron paramagnetic resonance experiments with molecular modeling calculations, the minor components of Curcuma longa extracts, such as demethoxycurcumin (2, DMC), bisdemethoxycurcumin (3, BDMC) and cyclocurcumin (4, CYC), were analyzed in a membrane environment mimicking the phospholipid bilayer. Our study provides the first evidence on the relative role of single curcuminoids interacting with Aβ-peptide. When the CYC and curcumin metabolite tetrahydrocurcumin (5, THC) were inserted into an anionic lipid solution, a significant modification of the Aβ CD curves was detected. These data were implemented by EPR experiments, demonstrating that CYC reaches the inner part of the bilayer, while the other curcuminoids are localized close to the membrane interface. Computational studies provided a model for the curcuminoid-Aβ interaction, highlighting the importance of a constrained “semi-folded” conformation to interact with Aβ analogously to the pattern observed in α-helical coiled-coil peptide structures. This combined approach led to a better understanding of the intriguing in vitro and in vivo activity of curcuminoids as anti-Alzheimer agents, paving a new path for the rational design of optimized druggable analogues.
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder and the most common cause of dementia in adults. The mechanisms underlying the disease are currently in question, but the deposition of senile plaques composed by fibrillar aggregates of amyloid β peptide (Aβ) is a characteristic hallmark of the pathology and is considered fundamental in disease pathogenesis1,2,3,4. The main components of the plaques are the peptides Aβ(1–40) and Aβ(1–42), which occur as fibrillar aggregates in equilibrium with soluble oligomers and random coil or α-helical monomers5,6,7,8. Curcuma longa is a Southeast Asian plant, having an orange rhizome with a strongly aromatic smell. The rhizome is normally dried and then crushed for gastronomic use as a spice turmeric powder and also as a colorant in food (additive E100) and textile industry9. The mixture (MIX) of curcuminoids, obtained as an extract from the turmeric powder of Curcuma longa (Fig. 1A), is constituted by the most abundant curcumin (1, CUR), demethoxycurcumin (2, DMC), bisdemethoxycurcumin (3, BDMC) and cyclocurcumin (4, CYC), which is present in small amounts. They are provided with important pharmacological activities, such as antioxidant10, antiangiogenic11, antiinflammatory12, antiviral13 and anticancer14.
Figure 1.
(A) Structures of natural curcumin derivatives, curcuminoids, (1–4). (B) Structures of main curcumin metabolites (5–7).
Worldwide comparison of incidence studies promoted by the World Health Organization shows that the incidence rates of AD reported in the populations of South East Asia are lower than in Europe and North America. People living in rural compared to urban settings have lower rates of AD: United States ranks 2, Italy 21 and India 165 among 172 countries15,16, suggesting the existence of undefined protective or risk factors17.
The lower incidence rates of AD in the populations of South East Asia, the main consumers of turmeric, are associated with regular curcumin consumption, which is efficacious to counteract the neurophysiological damages occurring during the disease.
However, the low bioavailability of turmeric components greatly limits its use in therapy. The daily intake of a considerable amount of turmeric - up to 3.6 g/die - is converted into nanomolar concentrations in the plasma. Pharmacokinetic analysis of curcumin in humans showed that in the peripheral circulation, the main metabolites present were curcumin glucuronide and curcumin sulfate18,19, whereas other reduced metabolites resulting from the metabolic transformation of curcumin were detected in traces. In particular, after oral administration, the analysis of rat plasma concentrations by HPLC revealed the presence of tetrahydrocurcumin (5, THC), hexahydrocurcumin (6, HHC) and octahydrocurcumin (7, OHC) (Fig. 1B)20,21, essentially curcuminoids resulting from subsequent metabolic reactions of curcumin (1, CUR).
One of the strategies used to overcome the low bioavailability of curcumin involves the co-administration of turmeric with piperine, a component of Piper nigrum that inhibits the human enzymes CYP3A4 and P-glycoprotein, which are important for the metabolism and transport of xenobiotics and metabolites22. Moreover, as the interaction with lipid layers enhances the bioactivity of curcumin23, curcuminoids from turmeric rhizome of Curcuma longa, thus containing the four curcuminoids as in the MIX, have been commercialized as the highly standardized lecithin-formulated extract Meriva®, developed according to the Indena Phytosome® technology. Total curcuminoids absorption was about 29-fold higher for Meriva® than for its corresponding unformulated curcuminoids extract24,25,26.
Considering the implication of Aβ in oligomeric or fibrillar aggregates in the etiology and progress of Alzheimer’s disease, thousands of molecules have been screened to identify new substances that can control the formation of β amyloid fibrils, either stabilizing the soluble peptide conformation or dissolving aggregates27,28,29,30. Indeed, among the substances tested for preventing or disaggregating β-amyloid fibrils, curcumin and several synthetic curcumin derivatives have been shown to bind Aβ, affecting the process of fibril formation31,32,33,34,35,36. In particular, the analysis of curcumin derivatives highlighted the importance of keto-enol tautomerism, proving that the derivatives existing predominantly in the enol-form have higher affinity for Aβ aggregates37,38,39. Among the curcumin derivatives, minor attention has been reserved for the remaining curcuminoids present in turmeric extract and their metabolites, although increasing evidence has led to the hypothesis that the activity of each of these compounds is determinant for the ultimate biological effect of Curcuma longa extract. Indeed, i) the turmeric extract containing optimized proportions of the single curcuminoids and integrated with a given proportion of the reduced metabolite THC exhibits enhanced biological activity40; ii) the turmeric phytocomplex is therapeutic despite the unfavorable pharmacokinetic properties of curcumin alone; and iii) metabolites resulting from the metabolic transformation of CUR, such as THC, HHC and OHC (Fig. 1B), are themselves biologically active20,21.
While the structural interaction between curcumin and Aβ-peptide is known31,32,33,34,35,36, studies on the possible structural interactions with Aβ-peptide of other compounds structurally related to curcumin present in turmeric extract or deriving from curcumin metabolism are missing. To overcome this gap, using a combined approach including circular dichroism (CD) and electron paramagnetic resonance (EPR) spectroscopy, we analyzed the mixture extracted from Curcuma longa (MIX), all the single components (CUR, DMC, BDMC and CYC, Fig. 1A) and their metabolites (THC, HHC, OHC Fig. 1B) for their ability to interact with the β amyloid fragment Aβ(25–35). Using an advanced simulation protocol, we acquired data to read all the collected experimental data into molecular level.
Many β-amyloid fragments have been studied in vitro and in vivo, and are currently the focus of biophysical and computational investigations41. The choice of Aβ(25–35) was based on the evidence that this is the shortest fragment of the β-amyloid peptide that forms toxic aggregates, preserving the same activity of the full-length Aβ-peptide42,43,44,45,46,47. Moreover, our previous studies evidenced that, under several different environmental conditions, Aβ(25–35) presents the same conformational behavior as the corresponding fragment in the full length Aβ(1–42)46,48. In this respect, it is reasonable to use Aβ(25–35) for testing molecules which may act on the equilibrium monomer-oligomer-fibril of the full length peptide.
Finally, we decided to undertake our biophysical study in the membrane-mimicking environment represented by a liposomal solution of different composition. This choice was dictated by the critical role played by the membrane surface in the β-amyloid toxicity, associated with the curcuminoids’ aptitude for interaction with membranes, enhancing their biological effects23,49,50,51.
Our study provides the first evidence regarding the relative role of the individual curcuminoid interacting with Aβ-peptide in membrane-mimicking environments. Among the examined curcuminoids, only CYC and THC can modify the Aβ conformation in the liposome system. Computational studies provide a molecular model for curcuminoid-Aβ interaction, suggesting that the constrained “semi-folded” conformation of CYC is suitable for interaction with Aβ, reproducing the pattern observed in α-helical coiled-coil peptide structures.
Results
Analysis of curcuminoids
We analyzed the curcuminoids present in Meriva® using a different HPLC elution system (chromatograms obtained are shown in Supporting Information in Figure 1SI and 2SI). The chromatogram obtained under these new conditions showed the presence of CUR (retention time (rt) = 20.44 min), DMC (rt = 19.97 min), BDMC (rt = 19.50 min) and another component with the rt of 13.90 minutes. We therefore hypothesized that the new peak could correspond to cyclocurcumin. To validate our hypothesis, we embarked on the synthesis of cyclocurcumin to fully characterize the new entity via HRMS and NMR and then to evaluate its rt by HPLC analysis. The chromatogram obtained (Figure 2SI) was compared with the previous one (Figure 1SI), showing that the two had equal retention times. A quantitative analysis of curcuminoids present in the MIX, calculated as the percentage of the area under the curve, was performed, with the following results: 83% CUR, 11% DMC, 4% BDMC, and 2% CYC.
The three most abundant natural curcuminoids present in extracts 1, 2, and 3 were obtained by flash chromatography on silica gel using the eluent mixture DCM/MeOH (97:3). Each compound was subsequently characterized via HRMS and 1H-13C NMR. The purity of each isolated compound was assessed by HPLC analysis (see Supporting Information).
Synthesis of curcuminoids
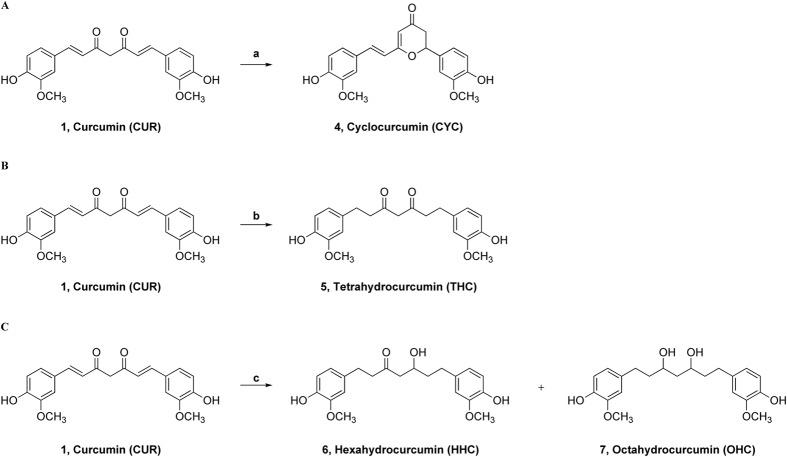
Since the quantitative analysis showed 2% cyclocurcumin in the mixture, we opted to synthesize this curcuminoid. The general reaction scheme is depicted in Fig. 2A.
Figure 2.
(A) Synthesis of cyclocurcumin, CYC (4); reagents and conditions: a) TFA, MW, 100 °C, 4 min, 10%. (B) Synthesis of THC (5); reagents and conditions: b) H2 (1 atm), Pt/C, MeOH, r.t., 1 h, 77%. (C) Synthesis of HHC (6) and OHC (7); reagents and conditions: c) H2 (6.8 atm), Pt/C, MeOH, r.t., 13 h; 6, HHC (37%); 7, OHC (55%).
Several attempts were made to obtain CYC (see Table SI1 in Supporting Information). We first reproduced the acid-catalyzed cyclization of curcumin as reported by Kiuchi and coworkers52 using benzene and TFA (trifluoroacetic acid) for 65 h in the dark. The reported reaction yield is 20%; nevertheless, in our hands, the same reaction never provided a yield higher than 5%, probably because of the difficulties shown in the purification process. We attempted to modify the acidity of the reaction medium by replacing the TFA with TfOH (trifluoromethanesulfonic acid) (Table 1SI); however, the yield did not improve. A reduction of both the reaction time and the amount of by-products was achieved using microwave-assisted synthesis in benzene. The best result was obtained through a solvent-free reaction, performing just one 4 min MW run using TFA at 100 °C. Under these conditions, the reaction yield was 10%, barely sufficient to accumulate the desired quantities despite being completed in only a few minutes.
Table 1. Calculated occurrence rates (%) of MM and PM7 conformers presenting “folded”, “semi-folded” and “extended” conformation.
| Cmp | MM |
PM7 |
||||
|---|---|---|---|---|---|---|
| Folded | Semi-folded | Extended | Folded | Semi-folded | Extended | |
| CUR (diketone) | 22.0 | 27.5 | 50.5 | 65.5 | 19.5 | 15 |
| CUR (keto-enol) | 0.0 | 0.0 | 100 | 0.0 | 5.0 | 95 |
| CYC | 0.0 | 100 | 0.0 | 0.0 | 100 | 0.0 |
| THC (diketone) | 39.5 | 47.0 | 13.5 | 56.0 | 35.0 | 9.0 |
| THC (keto-enol) | 16.0 | 59.5 | 24.5 | 37.5 | 45.0 | 17.5 |
Among the metabolites formed after the administration of curcumin, the most interesting are THC, HHC and OHC, which are obtained after the reduction in vivo of curcumin. In all these compounds, the extended conjugation is lost, and in the case of HHC and OHC, the keto-enol tautomerization is lost as well. This behavior was very interesting as a way to assess the actual importance of tautomerism in the pharmacodynamics of curcumin.
The synthesis of these three metabolites was conducted through catalytic hydrogenation, suitably varying the reaction time and H2 pressure53. THC was obtained in 77% yield using H2 (1 atm), Pt/C, and MeOH for 1 h at room temperature (Fig. 2B).
HHC and OHC were obtained in a 40:60 mixture using higher hydrogen pressures and longer reaction times in a Parr® apparatus (6.8 atm H2, Pt/C, MeOH at room temperature for 13 h (Fig. 2C). HHC and OHC were obtained with yields of 37% and 55%, respectively.
Circular dichroism analysis
Biological and biophysical experiments show that curcumin can interact with Aβ-peptide in soluble or fibrillar form31,32,33,34,35,36. The correlation between the pathological role of Aβ-peptide and its interaction with the neuronal membrane is well established23,49,50,51,54. The phytosomal formulation of curcuminoids (Meriva®) is proven to enhance the biological action of curcumin in terms of plasmatic availability24,26 and penetration in tissues characterized by amyloid plaque deposition55.
To test the hypothesis that all these actions might share a common biophysical mechanism related to the interaction of Aβ with curcumin at the biomembrane, we analyzed Aβ(25–35) in liposomal solution by CD spectroscopy (Fig. 3A,B), focusing on the variation of its conformation in the presence of the individual curcuminoids shown in Fig. 1. We chose as the liposome systems small unilamellar vesicles (SUV) composed of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and/or 1,2-dioleoyl-sn-glycero-3-phospho-rac-(1-glycerol) (DOPG). Varying the lipid constituents can suitably modulate the liposome composition. The constituents can differ in the length and number of the methylene chains, as well as the polarity and charge of the polar heads. Given the importance of the superficial charge in driving the behavior of the molecules on the membrane surface, we decided to record CD spectra in three different liposome solutions distinguished by different superficial charges. DOPC is characterized by a zwitterionic membrane surface, DOPG by a negatively charged surface, and DOPC/DOPG (90/10 molar ratio) by a zwitterionic surface doped with a small amount of negative charge.
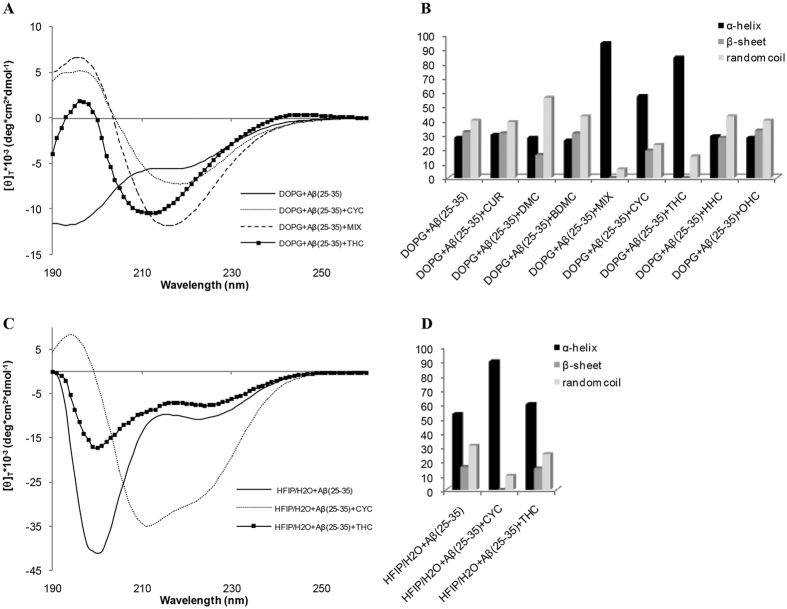
Figure 3.
(A) CD spectra of Aβ(25–35) in DOPG vesicles in the absence and the presence of ligands MIX, (B) Secondary structure quantification of Aβ(25–35) in DOPG vesicles in the presence of curcuminoid compounds. (C) CD spectra of Aβ(25–35) in HFIP/water mixture in the absence and the presence of ligands MIX, (D) Secondary structure quantification of Aβ(25–35) in HFIP/water mixture in the presence of curcuminoid compounds.
Aβ(25–35) was obtained in complete monomeric form, according to the defibrillating procedure described by Zagorski56. CD spectra of monomeric Aβ(25–35) (100 μM solution) were recorded in DOPC, DOPG and DOPC/DOPG liposomal solution (200 μM solution) (see Supporting Information). The qualitative and quantitative analysis of CD curves (DICHROWEB online server)57 indicates that Aβ(25–35), in all the examined solutions, exists in an equilibrium of random coil (slightly predominant), α-helix, and β-sheet conformations (Fig. 3B). CD spectra recorded in DOPC and DOPC/DOPG liposomal solutions after the addition of curcuminoids show no significant modification of the Aβ(25–35) conformation in DOPC and DOPC/DOPG liposome solution. Conversely, Aβ(25–35) in DOPG solution, containing CYC and THC, undergoes a marked increase in α-helix structures, as shown by the quantitative evaluation of the CD spectra reported in Fig. 3A.
To estimate the proportions in which these conformational modifications were attributable to direct Aβ(25–35)/curcuminoid interaction or were mediated by the phospholipid bilayer, we recorded CD spectra in mixture of water/organic solvents. An exhaustive conformational analysis of Aβ(25–35) in a hexafluoro-2-propanol (HFIP)/water mixture containing different proportions of organic solvent and water was previously conducted in our lab45. This study proved the ability of HFIP to stabilize monomer helical Aβ(25–35) conformations. Thus, proceeding from our previous experience, we recorded CD spectra of Aβ(25–35) in HFIP/water 80/20 v:v in the absence and in the presence of CYC and THC. Analysis of the CD spectra shows that, even in these conditions, CYC modifies the conformational behavior of Aβ(25–35) (Fig. 3C,D), whereas THC induces only modest modifications of the CD spectra. These results indicate that the ability of CYC to destabilize the conformational arrangement of Aβ(25–35) is due to a direct CYC-peptide interaction instead of a mediated mechanism involving the lipid membrane, as in the case of THC.
EPR experiments
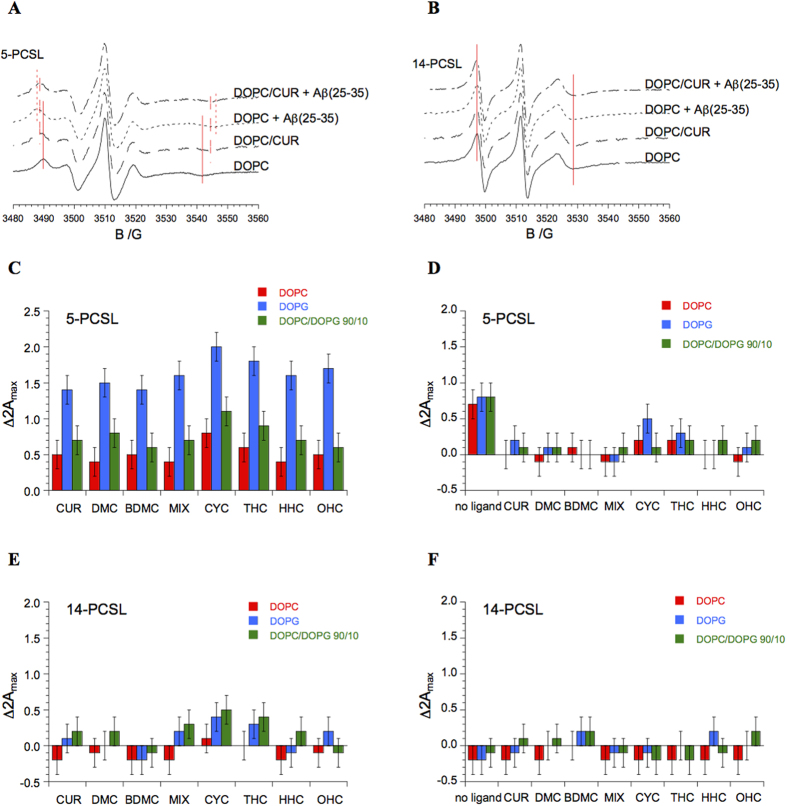
The CD experiments suggest that a significant perturbation of Aβ(25–35) conformational preferences could be induced by the peptide interaction with CYC and THC. These interactions are modulated by the presence of lipid membranes in the case of THC and MIX, whereas CYC can modify the behavior of Aβ(25–35) directly. To investigate this effect more deeply, we performed EPR analysis using experimental conditions consistent with the ones adopted for CD analysis. EPR spectroscopy requires the presence of unpaired electrons in the system to be investigated. To study microstructured systems, specific components can be selectively labeled with radical moieties, thus making it possible to obtain detailed information on the specific site where the label is positioned. In this work, we have used phospholipids specifically spin-labeled at different positions along the acyl chain. 5-PCSL is labeled very close to the hydrophilic head-group, thus allowing observation of the behavior of the bilayer region just below the interface. In contrast, 14-PCSL allows analysis of the properties of the inner part of the bilayer hydrophobic core58. Using these two spin-labels, we focused on two different aspects of the systems under consideration in this work. First, we studied the interaction of curcumin and its analogues with the lipid membranes formed by DOPC, DOPG and DOPC/DOPG (90/10 molar ratio); second, we analyzed the effects of these ligands on the interaction of the Aβ(25–35) peptide with the lipid bilayers. The 5-PCSL spectrum in DOPC bilayers (Fig. 4A, solid line) shows a clearly defined axially anisotropic behavior, as detectable by the splitting of the low- and high-field lines, indicating an ordered organization of the outer segments of the lipid acyl chains. The 14-PCSL spectrum, registered in the same system, shows an almost isotropic three-line lineshape (Fig. 4B, solid line).
Figure 4.
EPR spectra of 5-PCSL (A) and 14-PCSL (B) in lipid bilayers composed of DOPC or DOPC and CUR, in the absence and in the presence of Aβ(25–35). Red lines are guides for the eye, highlighting the shifts of the low-field maximum and high-field minimum. Increase in outer hyperfine splitting of 5-PCSL and 14-PCSL in DOPC, DOPG and DOPC/DOPG 90/10 bilayers upon inclusion of ligands (CUR, DMC, BDMC, MIX, CYC, THC, HHC and OHC) in the lipid formulation (panels C,E) and upon addition of Aβ(25/35) to the system (panels D,F).
Very similar results were obtained for the DOPG and DOPC/DOPG (90/10 molar ratio) bilayers (spectra not shown). This evidence indicates, for all the considered lipid systems, an increase flexibility in terms of the segmental chain mobility moving inward from the polar head-groups to the inner hydrophobic core, as expected for membranes in the liquid-disordered (Ld) crystalline state59. To quantitatively analyze the spectra, the values of the outer hyperfine splitting, 2Amax, were determined by measuring the difference between the low-field maximum and the high-field minimum, using a home-made MATLAB-based software routine. In general, 2Amax is dependent on both the amplitude (i.e., order) and the rate of chain rotational motion and is therefore a useful parameter for characterizing chain dynamics, as determined by local ordering, in phospholipid membranes60. It is a sensitive means for detecting and quantifying lipid bilayer interactions with guest molecules, as previously demonstrated61. The 2Amax value is much higher for 5-PCSL than for 14-PCSL (approximately 51 G vs. 32 G) and is quite insensitive to the lipid charge (for 2Amax values for 5-PCSL and 14-PCSL in DOPC, DOPG and DOPC/DOPG (90/10 molar ratio) bilayers (Table 2SI in Supporting Information). In the presence of curcumin, the anisotropy of the 5-PCSL spectrum in DOPC increases slightly but significantly, as shown by Fig. 4A, dashed line. The anisotropy increase indicates a motional restriction of the spin-labeled chain and is usually related to the presence of guest molecules61. In contrast, the 14-PCSL spectrum remains almost unaffected (Fig. 4B). These results indicate that curcumin interacts with the bilayer, positioning close to the head-group region with a limited but still detectable intercalation among the outer segments of the lipid acyl chains. This conclusion is in reasonable agreement with the “surface association” of curcumin proposed by Sun62, while confirming a certain degree of internalization as found by Barry23,54.
Qualitatively similar effects were observed for DOPG and DOPC/DOPG (90/10 molar ratio) bilayers, perturbation being slightly higher for the anionic lipid system. Consistently, the 2Amax values reported in Table 2SI show an increase for 5-PCSL, while remaining almost constant in the case of 14-PCSL. The analogues of curcumin considered in this work cause variations in the 5-PCSL and 14-PCSL 2Amax values that are qualitatively similar to the ones observed for CUR (see Table 2SI).
To easily compare the effects of the various ligands on the bilayer structure, Fig. 4 (panels C–E) reports the 2Amax variation upon ligand inclusion in all the considered systems for both spin-labels. In the case of 5-PCSL, the variation is positive in all cases and higher for the DOPG bilayers.
In the case of 14-PCSL, the 2Amax variations are almost negligible. Thus, it can be concluded that all CUR derivatives present a marked tendency to interact with the bilayers, shallowly penetrating between the lipid acyl chains. The differences between the various ligands are not dramatic, even though CYC seems more effective in perturbing the membrane inner layer. Indeed, for this ligand, even the 14-PCSL 2Amax in anionic membranes is slightly but significantly perturbed. Then, we turn our attention to the interaction of the Aβ peptide with lipid membranes, analyzing the extent to which it is affected by the presence of curcumin and its analogues.
The addition of Aβ(25–35) significantly increases the anisotropy of the 5-PCSL spectrum in DOPC (see Fig. 4A, dotted line). Accordingly, the peptide causes a clear 2Amax increase (Table 2SI), i.e., the mobility of the spin-labeled chains decreases through the interaction of the peptide with the membrane. In contrast, the addition of Aβ(25–35) to DOPC bilayers does not causes any change in the 14-PCSL spectrum (Fig. 4B). This evidence indicates that Aβ(25–35) is inserted in the bilayer positioning between the outer part of the hydrophobic core and the external hydrophilic layer. A similar solubilization site was already observed in other lipid systems58,63. When Aβ(25–35) is added to the lipid bilayers in the presence of curcumin or its analogues, the 5-PCSL spectrum is much less affected, and in some cases, no variation is observed (see Table 2SI). To better visualize the effects of the various ligands on the peptide-bilayer interaction, Fig. 4 (panels D,F) shows the 2Amax variations of the ligand-bilayer systems upon the addition of Aβ(25–35).
Inspection of the figure indicates that the presence of ligands at the bilayer interface hinders any membrane penetration of the peptide. We propose that the accumulation of a layer of bound curcumin (or its analogues) at the lipid membrane interface reduces the accessibility and number of binding sites for Aβ on the membrane. Moreover, curcumin on the membrane surface can reduce Aβ–membrane interaction by altering the lipid head-group charge and reducing the favorable electrostatic interactions between Aβ and lipid head-groups64. The inspection of Fig. 3 clearly shows that all the ligands behave similarly, with the possible exception of CYC in DOPG membrane: in this case, the further 2Amax increase due to Aβ(25–35) suggests that the peptide preserves a detectable ability to interact with the bilayer even in the presence of the ligand. This evidence supports the hypothesis that DOPG membranes create a perfect environment to enhance Aβ(25–35) interaction with CYC.
Molecular Modeling Studies
To relate the obtained CD and EPR results to the different structural features of the tested curcuminoids, a molecular modeling study was conducted. In particular, the structural features responsible for the interaction with Aβ(25–35) were investigated.
First, a comprehensive conformational analysis was performed on all possible tautomers of i) CUR, which is unable to affect the Aβ(25–35) conformational equilibrium either in a liposomal environment or in HFIP/water solution; ii) THC, which affects the Aβ(25–35) conformational equilibrium only in a liposomal environment; and iii) CYC, which perturbs the conformational behavior of Aβ(25–35), both, in DOPG and HFIP/water solution. The conformational space was sampled using a protocol including molecular dynamics (MD) simulations followed by molecular mechanic (MM) optimization, mimicking the polarity of a water medium (ε = 80*r). The identified minima were then subjected to full geometry optimization using the semi-empirical PM7 (MOPAC) method (ε = 1; for details see Experimental Section), thus obtaining a new set of conformers using a dielectric constant similar to the one for the highly hydrophobic membrane environment65. In this regard, it must be emphasized that only qualitative considerations can be deduced by comparing the results obtained from MM (ε = 80*r) and PM7 (ε = 1) calculations.
The MM and PM7 conformers obtained were ranked by their potential energy values (i.e., ΔE from the global energy minimum) and classified into different families based on the values of their torsional angles. Then, the distance between the centroids of the two aromatic rings was calculated (d1; Figures 3SI, 5SI–8SI; Tables 3SI–6SI). According to this latter parameter, the conformers were further classified as i) “folded” when d1 was ≤5 Å, ii) “semi-folded” when d1 was >5 Å and <9 Å, and iii) “extended” when d1 was ≥9 Å. Finally, the occurrence rate of each conformational family was calculated (Table 1 and Tables 3SI–6SI).
The results showed peculiar conformational features for each of CUR, CYC, and THC. The conformational behavior of CUR differed in the two tautomeric forms. Indeed, the conformers of the CUR-diketone form are distributed in two main conformational families: the “extended” and the “folded” (Tables 1 and 3SI; Figures 3SI–4SI), with the latter including the global minimum (GM) conformer. In particular, comparing PM7 with MM shows an increase in the number of “folded” conformers, i.e., presenting the two aromatic rings interacting with each other, and a concomitant decrease in the “extended” conformers can be observed. This result is due to the higher degree of accuracy of semi-empirical vs. MM methods in calculating electronic parameters, as well as to the fact that electrostatic interactions are expected to increase in non-polar (low dielectric) solvent.
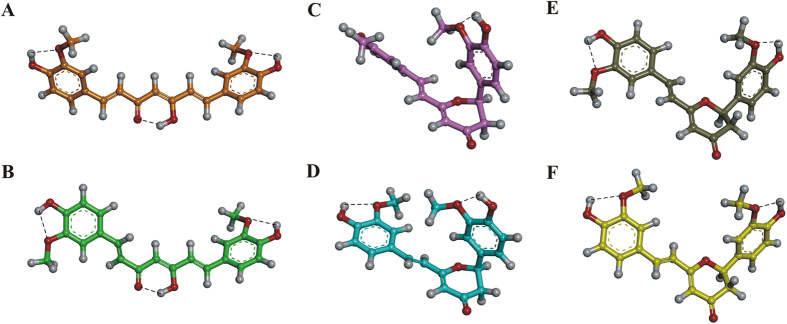
In contrast, in the keto-enol form of CUR, the conjugation of the double bonds extending from one aromatic ring to the other and the formation of a hydrogen bond between the ketone oxygen and the hydrogen of the enol function (Fig. 5A and B) strongly constrain the conformational freedom of the structure. Consequently, all CUR keto-enol conformers belong to the “extended” conformational family, resulting from both the MM and PM7 calculations (Tables 1 and 4SI; Figure 5SI). In detail, the PM7 low-energy conformers (within 5 kcal/mol from the GM) of the keto-enol form of CUR showed a specific preference for two sets of conformations, differing only for the value of the torsional angle τ1, herein named conf I (τ1 = ∼180°) and conf II (τ1 = ∼0°) (Fig. 5A and B; Table 4SI). These results are consistent with the ones obtained from the analysis of the experimentally determined structures of CUR (all in the keto-enol form) deposited in the Cambridge Structural Database (CSD) (Table 7SI). Indeed, CUR can assume either conf I (CSD codes: AXOGIE, AXOGOK, BINMEQ06, BINMEQ07, and BINMEQ08) or conf II (CSD codes: BINMEQ, BINMEQ01, BINMEQ02, BINMEQ03, BINMEQ04, and BINMEQ05), as shown by our conformational analysis.
Figure 5.
PM7 lowest energy conformers of i) keto-enol CUR conf I (A; orange) and conf II (B; green) and ii) CYC conf I (C; pink), conf II (D; cyan), conf III (E; gray), and conf IV (F; yellow). The conformers are displayed as ball & stick models and colored by atom type: O, red; H, white. Hydrogen bonds are highlighted with a black dashed line.
The presence of the dihydropyran-4-one ring in CYC determined a different conformational behavior from the observation for CUR tautomers. Indeed, all CYC conformers, both MM and PM7, fall into the “semi-folded” conformational family (Table 1 and Figure 6SI). Overall, CYC showed four possible subfamilies of conformers, differing for the values of the torsional angles τ1, τ2, and τ3 and characterized by d1 ranging from 7.00 to 9.66 Å (conf I–IV in Fig. 5C–F and Table 5SI).
THC shows yet another conformational preference compared either to CUR or CYC. According to the results of our analysis, THC conformers fall into all three conformational families regardless of the tautomeric form or the computational method used (Table 1; Figure 9SI; Table 6SI). However, either in the diketone or in the keto-enol form, the “folded” conformation is strongly stabilized by the formation of a hydrogen bond between the hydroxyl groups of the two aromatic rings (Figure 9SI). This intra-molecular H-bond may compete with the formation of H-bond interactions with solvent molecules, which are not treated explicitly in this simulation. Thus, it is likely that the actual conformational behavior of THC strongly depends on the chemical environment.
Furthermore, to investigate the experimentally observed interaction between Aβ(25–35) and CYC in HFIP/water 80/20 solution, we performed a dynamic molecular docking simulation on the CYC-Aβ(25–35) complex, mimicking the polarity of the media with a dielectric constant of 30*r. To fully explore all possible binding sites/modes during the dynamic docking procedure, the binding site was defined as the whole peptide, and all rotatable bonds of CYC and Aβ(25–35) were left fully free to move. A force constant of 10 kcal/mol/Å2 was only applied on the backbone hydrogen bonds of (Asn27-Leu34) to avoid unrealistic results during the annealing simulation (for details, see the Experimental Section). First, a Monte Carlo/minimization approach for the random generation of a maximum of 20 acceptable complexes was used. To ensure a wide variance of the input structures to be minimized afterward, an energy tolerance value of 106 kcal/mol from the previous structure was used. After the energy minimization step, the energy test, with an energy range of 50 kcal/mol, and a structure similarity check (rms tolerance = 0.3 kcal/A) was applied to select the 20 acceptable structures. To test the thermodynamic stability of the resulting docked complexes, these latter were then subjected to simulated annealing (SA) calculations. In SA the temperature is altered in time increments from an initial (500 K) to final (300 K) temperature. The temperature of 500 K was applied with the aim of surmounting torsional barriers, thus allowing a full rearrangement of the ligand and the peptide (see Experimental section for details).
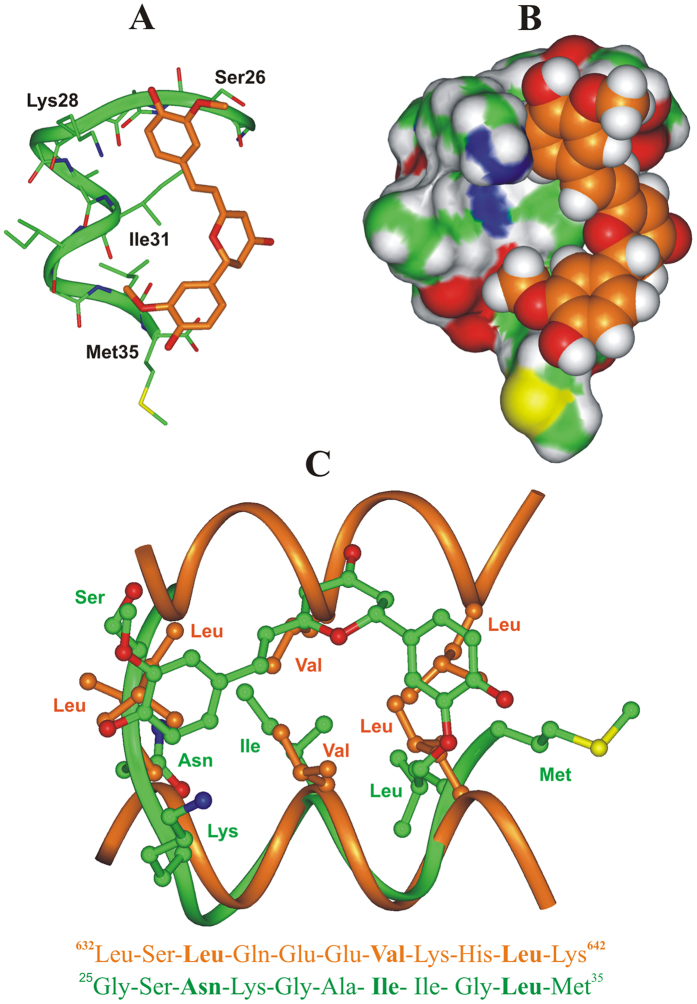
The resulting complexes were analyzed and ranked by their conformational and interaction energy values (Table 8SI). On twenty Monte Carlo generated structures, only twelve (Figure 10SI) still present CYC bound to Aβ(25–35) peptide after SA calculations. Interestingly, just one of these twelve SA complexes preserves the same binding mode resulting from Monte Carlo docking procedure, showing thermodynamic stability (Complex 1, Figure 10SI). Importantly, this complex also resulted to be the lowest energy complex and the one characterized by the most favorable interaction energy (Table 8SI). Such results strongly suggest that Complex 1 is representative for a stable and rather specific binding mode of CYC to Aβ (25–35) peptide (Fig. 6A and B).
Figure 6.
(A,B) Docked structure of CYC (orange) in complex with Aβ(25–35) peptide (green). (A) The complex is displayed as ribbon and sticks and peptide interacting residues are labeled; hydrogens are omitted for sake of clarity. (B) The Connolly surface of the peptide and the vdW volume of CYC are displayed. (C) X-ray structure of the coiled-coil dimerization motif of human ROCK I protein (carbon atoms: orange; PDB ID: 3O0Z) superimposed on the docked complex of CYC: Aβ(25–35) peptide (carbon atoms: green). Hydrogens are omitted for clarity. The corresponding 2-D sequence of human ROCK I protein and Aβ(25–35) are reported and the conserved residues of the heptad repeat motif are highlighted in bold. Molecules are colored by atom type (O: red; N: blue; S: yellow and H: white).
Interesting data provided by our docking results showed that the calculated binding mode of CYC to Aβ(25–35) resembles the canonical packing of the α-helical coiled-coil dimerization motif, where the side-chains of the “a” and “d” residues of two facing heptad repeat motifs interact with each other (Fig. 6C)66. In fact, in the consensus sequence of the heptad repeat motif, the “a” and “d” positions are occupied by hydrophobic residues (one of which can be replaced by an asparagine), favoring the formation of coiled-coil structures, where the i, i + 4, and i + 7 residues of a helix interact with the equivalent residues of another helix. As evidenced by the superimposition reported in Fig. 6C, CYC can mimic the interactions performed by such residues. In particular, the two substituted phenyl rings (d1 = 8.85 Å) can reproduce the orientation of the hydrophobic side chains of the i and i + 7 residues (distance from the side chain centroids = 8.20 Å), and at the same time, the dihydropyran-4-one ring and the ethylene bridge overlap with the central i + 4 residue (Fig. 6C).
Taken together our computational results support the hypothesis that CYC targets heptad repeat motifs like the one found in Aβ(25–35) (Fig. 6C), whose structure is preserved in full length Aβ(1–42) and which is part of the extended heptad repeat motif identified as responsible for the dimerization of APP in micellar environment (Figure 11SI)67.
Discussion
A great deal of evidence shows that curcumin and several synthetic curcumin derivatives bind beta amyloid peptides, affecting the process of fibril formation31,32,33,34,35,36. In light of the well-documented pharmacological effects, including the neuroprotective effect exerted by the less abundant curcuminoid molecules in the Curcuma Longa extract, we studied these molecules for their ability to affect the conformational arrangement of Aβ(25–35), providing a molecular basis for the observed biological effect.
In particular, our analyses were intended to understand whether the neuroprotective effect of the phytocomplex derived from Curcuma Longa can be credited to the ability of the curcuminoids to lessen lipid membrane-Aβ interactions and, by consequence, the Aβ-induced membrane-disruptive perturbations49,50,51. Accordingly, we investigated how curcuminoids, taken as single purified molecules, affect Aβ(25–35)−membrane interactions through CD and EPR experiments, using DOPC, DOPG and DOPC/DOPG liposomal solutions to mimic lipid bilayer membrane systems. The CD spectra of monomeric Aβ(25–35) in the different superficially charged liposome environments were recorded after the addition of the compounds CUR, DMC, BDMC, CYC, THC, HHC, OHC and MIX. These experiments were conducted to evaluate the effect of curcuminoids in solution on Aβ(25–35)-membrane interactions. In the presence of these curcuminoids, used pure or mixed, no significant modification of the Aβ(25–35) conformational equilibrium was observed in DOPC and DOPC/DOPG liposomal solution. In contrast, when CYC and THC were inserted into anionic DOPG lipid solution, a significant modification of the Aβ(25–35) CD curves was detected, corresponding to a perceptible increase in the soluble form of the α-helix vs. β-strand and random coil conformations. These data were obtained by EPR experiments that revealed, for both Aβ(25–35) and curcuminoids, a shallow insertion in the DOPC, DOPC/DOPG, and DOPG lipid layers, just below the outer interface (5-PCLS 2Amax shift). Only in the case of CYC in DOPG liposomal solution was a slight but significant perturbation of 14-PCSL 2Amax observed, demonstrating that the molecule reaches the inner part of the bilayer. The inclusion of the curcuminoid ligands on the bilayers prevents further interaction of the membrane with the Aβ peptide, causing only minimal variation in the ligand-bilayer systems, with the sole exception of CYC in DOPG. These results are consistent with other literature data, based on TEM (transmission electron microscopy) images and CD spectra64, which also evidenced that the presence of CUR does not inhibit Aβ40 aggregation or significantly alter the morphologies of Aβ monomers, prefibrillar or fibrillar aggregates, while it reduces the extent of cell membrane permeabilization induced by Aβ aggregates.
In the case of DOPG membrane containing CYC or THC, the presence of the guest molecules induces a significant alteration of the Aβ(25–35) conformational equilibrium, as observed in the CD spectra, and a marked perturbation of the membrane outer layer, as observed in the EPR experiments. This result may indicate a direct interaction of CYC and THC with Aβ(25–35), but it still cannot be excluded that the observed effects are mediated by the lipid membrane. To investigate this issue, CD spectra were recorded in an HFIP/water (80/20 v:v) mixture in the absence and in the presence of CYC and THC. Under these conditions, CYC was still able to modify the conformational equilibrium of Aβ(25–35) (Fig. 3C,D), whereas THC induced only minimal modification of the Aβ(25–35) secondary structure. Thus, THC does not directly interact with monomeric Aβ(25–35) in solution; nonetheless, when integrated into the DOPG system, it induces a modification in the Aβ(25–35) conformational equilibrium mediated by the lipid membrane. By contrast, CYC can modify the conformation of Aβ(25–35) either in HFIP/water solution or in anionic DOPG lipid solution, indicating that its constrained structure - naturally derived from an intramolecular cyclization - is suitable for direct interaction with Aβ(25–35) in both environments. Computational studies provided a molecular model for this interaction and a possible explanation for the peculiar ability of CYC, unlike CUR and THC, to interact directly with Aβ(25–35), either in solution or in the liposomal environment. Indeed, according to the results of our molecular modeling studies, this result is due to the constrained “semi-folded” conformation of CYC, which allows it to mimic the orientation of the hydrophobic side chains of short peptide motifs, similarly to our previous observations for other small ligands68,69. Thus, by mimicking the orientation of the i, i + 4, and i + 7 residues of an α-helix, CYC could interact with Aβ(25–35), reproducing the pattern observed in α-helical coiled-coil structures, the latter being characterized by the presence of one of the principal dimerization motifs in proteins70. Coiled-coil peptide structures are known to anchor to cellular membranes71 and are formed by heptad repeats containing hydrophobic residues at the “a” and “d” positions. Importantly, the formation of helical coiled-coil structures, through the reciprocal interaction of several heptad repeat motifs, was also observed for APP in micelles, leading to its dimerization67. In line with these results, our previous NMR studies evidenced a strong similarity between the structure of Aβ(1–42) peptide in an apolar microenvironment and the structure of the fusion domain of influenza hemagglutinin in detergent micelles (also characterized by the ability to form coiled-coil structures)72. A heptad repeat motif is still contained in the Aβ(25–35) fragment, and, similarly to previous observations, could lead to the formation of α-helical coiled-coil structures on the membrane lipid surface that are amenable to interaction with CYC. On the other hand, monomers and soluble aggregates of Aβ(25–35) in the form of α-helix are also likely to occur in HFIP/water solution, since, in this environment, the helical structure of Aβ(25–35) is strongly stabilized73. Accordingly, CYC also perturbs the conformational equilibrium of Aβ(25–35) in this environment.
The “semi-folded” conformation of CYC is poorly present in CUR (not present at all in its predominant keto-enol form) but still significantly present in the case of THC. However, due to the higher conformational flexibility of THC with respect to CYC, it represents just a fraction of the whole conformational space, with its relative weight depending on the polarity and the H-bond donor/acceptor properties of the solvent. Accordingly, CUR and other analogues characterized by a conjugated and mostly planar structure (DMC, BDMC) behave only as membrane stabilizers, forming ideal mixtures with DOPG lipids, reducing lipid flexibility, and preventing Aβ(25–35)-peptide insertion. On the contrary, CYC, which is structurally constrained in the “semi-folded” conformation, is able to bind/stabilize the monomeric helical form of Aβ(25–35). In this way, CYC may prevent not only the formation of toxic β-sheet aggregate, but also the formation of membrane disrupting aggregates formed by α-helical coiled-coil structures74,75. Finally, the more flexible THC shows a different behavior in DOPG bilayer or in the HFIP/water mixture; indeed, THC shifts the conformational equilibrium of Aβ(25–35) to the helical structure in DOPG, but this ability was not preserved in the HFIP/water solution.
Conclusions
The results of this work show that the well-documented biological activity of turmeric extract is based on a molecular mechanism in which CUR and its analogues reduce the Aβ–membrane interactions due to their marked tendency to interact with the lipid bilayer. However, this effect is differently mediated among the examined curcuminoids since i) only CYC and THC can modify the conformational behavior of Aβ(25–35) in the liposome environment, stabilizing the α-helix conformation and ii) only CYC can still modify the conformational behavior of Aβ(25–35) in the HFIP/water mixture. While the enol form of CUR (“extended” conformation) was found to be responsible for the inhibition of Aβ fibril formation, our molecular modeling studies suggest that, to interact with the helical form of Aβ and prevent the formation of membrane-disrupting aggregates (possibly consisting of α-helical coiled-coil structures), it is necessary for CUR and its derivatives to assume the partially folded conformation in which CYC is naturally constrained.
This combined approach leads to a better understanding of the intriguing in vitro and in vivo activity of curcuminoids as anti-Alzheimer agents, paving a new path for the rational design of optimized druggable analogues.
Experimental Section
All reagents and solvents were purchased from Sigma-Aldrich (St. Louis, MO, USA) and used as received. Curcumin was supplied from Indena S.p.A. (Milano, Italy) as mixture of curcuminoids extracted from Curcuma Longa. The phospholipids 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and 1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt) (DOPG) were purchased from Avanti Polar Lipids (Birmingham, AL, USA). All microwave reactions were conducted in a CEM Explorer® apparatus under monomode irradiation. Hydrogenation of curcuminoids was performed in a Parr® apparatus. All reactions were conducted under dry nitrogen atmosphere. Flash column chromatography was performed on silica gel 60, 0.040 −0.063 mm (230–400 mesh) using methanol/dichloromethane solvent mixtures. Silica gel (grade 60 PF254) was used for preparative TLC. An Agilent 1260 high-performance liquid chromatography system with a variable wavelength detector was used for analysis. NMR spectra were recorded on a 300 MHz NMR spectrometer using CDCl3 and MeOD as the solvent and TMS as the internal standard, unless otherwise specified. Conditions are specified for each spectrum (temperature 25 °C unless otherwise specified). Splitting patterns are designated as s, singlet; d, doublet; t, triplet; q, quartet; m, multiplet. Chemical shifts (δ) are given in ppm relative to the resonance of their respective residual solvent peak, MeOH (4.87 ppm, 1H; 49.00 ppm, the middle peak, 13C). High-resolution mass spectra (HRMS) were recorded on a high-resolution mass spectrometer equipped with electrospray (ESI) and nanospray sources and a quadrupole-time of flight hybrid analyzer coupled with a capillary UPLC system (Q-TOF Premier/nanoAquity, Waters) in positive mode, and protonated molecular ions [M + H]+ were used to confirm the empirical formula, unless otherwise stated.
Separation of curcuminoids
Liquid chromatography was performed on a Waters system (Milford, Massachusetts, USA) consisting of a Waters 486 Tunable Absorbance Detector and a Varian 9012 pump. Samples were prepared by dissolving curcuminoids (CUR, DMC, BDMC, CYC) and MIX in acetonitrile at 0.2 mL/min. The injection volume was 10 μL. Curcuminoids were separated on a Teknokroma C18 column (25 × 0.46 cm, 5 μm) under gradient elution at a flow rate of 0.9 mL/min. The mobile phases consisted of 2% (v/v) acetic acid in water (solvent A) and 2% (v/v) acetic acid in acetonitrile (solvent B). The following gradient was used: 0–20 min 30–70% B, 20–28 min 70–90% B, and return at initial conditions in 2 min. Detection UV at λ = 254 nm.
Curcumin (1)
Odorless orange powder, MP: 178–180 °C; 1H NMR (300 MHz, MeOD) δ 7.58 (d, J = 15.9 Hz, 2 H), 7.22 (s, 2 H), 7.11 (d, J = 7.4 Hz, 2 H), 6.82 (d, J = 7.7 Hz, 2 H), 6.63 (d, J = 15.7 Hz, 2 H), 5.97 (s, 1 H), 3.91 (s, 6 H) ppm. 13C NMR (75 MHz, MeOD) δ 185.63, 151.14, 150.07, 142.71, 129.10, 124.66, 122.76, 117.01, 112.26, 112.04, 56.50, 56.02 ppm. HRMS (ESI-Q-TOF) m/z [M + H]+ Calcd. for C21H21O6 369.1333; Found 369.1335.
Demethoxycurcumin (2)
Odorless orange powder, MP: 157–161 °C; 1H NMR (300 MHz, MeOD) δ 7.52 (dd, J = 26.6, 11.4 Hz, 4 H), 7.20 (s, 1 H), 7.09 (d, J = 7.7 Hz, 1 H), 6.81 (d, J = 7.4 Hz, 3 H), 6.73–6.49 (m, 2 H), 3.90 (s, 3 H) ppm. 13C NMR (75 MHz, MeOD) δ 185.71, 185.51, 162.05, 151.38, 150.11, 142.68, 142.45, 131.67, 128.94, 128.36, 124.61, 122.62, 122.36, 117.39, 117.05, 112.21, 111.99, 101.97, 56.47 ppm. HRMS (ESI-Q-TOF) m/z [M + H]+ Calcd. for C20H19O5 339.1227; Found 339.1230.
Bisdemethoxycurcumin (3)
Odorless orange powder, MP: 187–191 °C; 1H NMR (300 MHz, MeOD) δ 7.58 (d, J = 16.1 Hz, 3 H), 7.49 (d, J = 8.4 Hz, 4 H), 6.82 (d, J = 8.3 Hz, 5 H), 6.60 (d, J = 15.9 Hz, 2 H), 2.17 (d, J = 7.1 Hz, 3 H) ppm. 13C NMR (75 MHz, MeOD) δ 185.69, 161.94, 142.43, 131.67, 128.46, 122.44, 117.36. HRMS (ESI-Q-TOF) m/z [M + H]+ Calcd. for C19H17O4 309.1121; Found 309.1117.
Procedures for the synthesis of compounds 4, 5, 6, 7
Cyclocurcumin (4)
In a 7 mL MW vessel, curcumin 1, (20 mg, 0.054 mmol) and TFA (2.14 mmol, 164 μL) were added, and the reaction mixture was stirred in CEM Explorer®. MW Method: T = 100 °C, Power: 300 W, Hold Time: 4 min, P = 250 PSI, Power Max activated. After cooling, TFA was removed under reduced pressure, and the residue was purified by flash column chromatography (CH2Cl2/MeOH 97:3) to afford cyclocurcumin 4 as a yellowish oil (2 mg, 10%). 1H NMR (300 MHz, MeOD) δ 7.35 (d, J = 15.8 Hz, 1 H), 7.18 (s, 1 H), 7.12 (d, J = 1.7 Hz, 1 H), 7.05 (d, J = 8.3 Hz, 1 H), 6.99 (d, J = 8.2 Hz, 1 H), 6.86 (d, J = 8.1 Hz, 1 H), 6.77 (d, J = 8.2 Hz, 1 H), 6.66 (d, J = 15.8 Hz, 1 H), 5.62 (s, 1 H), 5.45 (dd, J = 13.7, 3.2 Hz, 1 H), 3.89 (d, J = 7.3 Hz, 6 H), 3.03 (dd, J = 16.9, 13.8 Hz, 1 H), 2.59 (dd, J = 16.9, 3.0 Hz, 1 H). 13C NMR (75 MHz, MeOD) δ 197.20, 150.27, 143.79, 142.37, 131.77, 124.74, 124.41, 122.41, 121.35, 119.44, 117.23, 116.70, 112.06, 111.87, 94.18, 83.43, 82.57, 56.47, 55.71 ppm. HRMS (ESI-Q-TOF) m/z [M + H]+ Calcd. for C21H21O6 369.1333; Found 369.1331.
Tetrahydrocurcumin (5)
A solution of curcumin 1 (50 mg, 0.14 mmol) in dry MeOH (5.9 mL) was introduced into an oven-dried bottle connected to a Parr apparatus. Pt/C 10% wt (2.14 mg, 0.014 mmol) was added, and the bottle was filled with H2 (1 atm) and shaken at room temperature for 1 h. The bottle was degassed, and the catalyst was filtered (ATTENTION: the Pt residue might be pyrophoric) and washed several times with MeOH. The MeOH fractions were collected, and the solvent was removed under reduced pressure to give compound 5 as a pale white oil (40 mg, 77% yield). 1H NMR (300 MHz, MeOD) δ 6.80–6.55 (m, 6 H), 3.80 (s, 6 H), 2.78 (dd, J = 15.5, 7.6 Hz, 6 H), 2.53 (t, J = 7.5 Hz, 3 H) ppm. 13C NMR (75 MHz, MeOD) δ 195.58, 149.46, 146.42, 134.21, 122.18, 116.55, 113.47, 56.38, 46.25, 41.27, 32.27, 29.93 ppm. HRMS (ESI-Q-TOF) m/z [M + H]+ Calcd. for C21H25O6 373.1646; Found 373.1648.
Hexahydrocurcumin (6) and octahydrocurcumin (7)
A solution of curcumin 1 (100 mg, 0.27 mmol) in dry MeOH (11.8 mL) was introduced into an oven-dried bottle connected to a Parr apparatus. Pt/C 10% wt (5.27 mg, 0.027 mmol) was added, and the bottle was filled with H2 (6.8 atm) and shaken at room temperature for 13 h. The bottle was degassed, and the catalyst was filtered (ATTENTION: the Pt residue might be pyrophoric) and washed several times with MeOH. The MeOH fractions were collected, and the solvent was removed under reduced pressure to give compounds 6 and 7 as a mixture (ratio 40:60). The crude mixture was purified by flash column chromatography (CH2Cl2/MeOH 97:3) to afford 6 (37 mg, 37% yield) and 7 (56 mg, 55% yield) as pale white oils. Hexahydrocurcumin (6). 1H NMR (300 MHz, MeOD) δ 6.75 (s, 7 H), 6.69 (dd, J = 8.0, 3.0 Hz, 11 H), 6.60 (d, J = 8.1 Hz, 11 H), 4.00 (dd, J = 11.2, 5.8 Hz, 5 H), 3.81 (d, J = 4.0 Hz, 32 H), 2.83–2.67 (m, 21 H), 2.57 (ddd, J = 15.7, 12.6, 6.3 Hz, 22 H), 1.68 (dd, J = 13.8, 7.6 Hz, 14 H) ppm. 13C NMR (75 MHz, MeOD) δ 212.95, 149.48, 146.33, 146.13, 135.41, 134.56, 122.24, 122.14, 116.53, 113.56, 113.50, 68.87, 68.34, 67.80, 56.89, 56.55, 56.39, 56.22, 51.29, 46.32, 40.37, 32.28, 30.13 ppm. HRMS (ESI-Q-TOF) m/z [M + H]+ Calcd. for C21H27O6 375.1802; Found 375.1805. Octahydrocurcumin (7). 1H NMR (300 MHz, MeOD) δ 6.83–6.46 (m, 3 H), 3.78 (dd, J = 20.0, 3.4 Hz, 4 H), 2.76–2.41 (m, 2 H), 1.82–1.44 (m, 3 H) ppm. 13C NMR (75 MHz, MeOD) δ 149.44, 146.06, 135.70, 122.22, 116.51, 113.54, 71.12, 68.83, 56.50, 44.81, 40.73, 32.21 ppm. HRMS (ESI-Q-TOF) m/z [M + H]+ Calcd. for C21H29O6 377.4508; Found 377.4506.
Peptide synthesis
The Aβ(25–35) peptide, (G-S-N-K-G-A-I-I-G-L-M), was manually synthesized by conventional solid-phase peptide synthesis (SPPS) using the Fmoc/tBu strategy76. The Nα-Fmoc-protected amino acids were coupled using 1-hydroxybenzotriazole and O-(benzotriazol-1-yl)-1,1,3,3-tetramethyluroniumhexa-fuorophosphate (fourfold excess) as coupling reagents, in the presence of a six-fold excess of N,N-diisopropylethylamine. Peptide-resin (Wang resin) cleavage and side-chain deprotection reactions were performed in 90% trifluoroacetic acid (TFA), 5% water and 5% triisopropylsilane (TIS). The resin was filtered and the solution added to cold diethyl ether to precipitate the peptide.
Subsequently, the peptide was purified by reversed phase chromatography (HPLC) using a Phenomenex column C18 (30 cm, 4 cm, 300 Å, 15 mm spherical particle size column). The peptide was characterized on a Finningan LCQ Deca ion trap instrument equipped with an electrospray source (LCQ Deca Finnigan, San José. CA, USA). The samples were directly infused in the ESI source using a syringe pump at a flow rate of 5.0 mL/min. The data were analyzed using the Xcalibur software. The sample purity was >98%.
Sample preparation for CD analysis
All CD experiments were recorded in small unilamellar liposome vesicles (SUVs) consisting of pure DOPC lipid, pure DOPG lipid and DOPC/DOPG mixtures at 90/10 M/M (molar ratio). Samples of DOPC, DOPG and DOPC/DOPG (90/10) small unilamellar liposome vesicles (SUVs) were prepared as follows: 200 μM of each phospholipid were dissolved in a CH2Cl2/MeOH solution, then kept in a round-bottom test tube, and a thin lipid film was produced by evaporating the solvent with dry nitrogen gas. The final traces of solvent were removed by subjecting the sample to vacuum desiccation for one night. The samples were then hydrated with an appropriate volume of 10 mM phosphate buffer at pH = 7.4 and repeatedly vortexed, obtaining a suspension. The MLV suspension thus obtained was sonicated for 5–10 minutes to obtain SUVs.
To remove aggregate states, which can be present in Aβ samples, the dry peptide was pretreated with TFA for 3 hours, followed by dilution with MilliQ water and lyophilization. The TFA pretreatment gives the Aβ peptide the properties of monomeric, random coil structures and eliminates preaggregated material77. This procedure was adopted for all CD samples, immediately before dissolution in the appropriate solvent. This procedure was also adopted before preparing Aβ(25–35) amyloid fibrils78.
To record CD experiments, Aβ(25–35) that had been previously treated according to the procedure reported above was added to SUVs (DOPC, DOPG and DOPC/DOPG (90/10)) to have a final peptide concentration of 0.1 mM (CD experiments) and a peptide-lipid molar ratio of 0.5:1 for each experiment. Similarly, ligands (CUR, DMC, BDMC, MIX, CYC, THC, HHC and OHC) were added to SUVs (DOPC, DOPG and DOPC/DOPG (90/10)) for a final ligand concentration of 0.5 mM (CD experiments).
CD measurements
Circular dichroism measurements were performed on a JASCO J-810 spectropolarimeter (Jasco, Cremella, Italy) equipped with a thermostated cell holder, using a quartz cell with a 1.0-mm path length. Spectra were collected over the wavelength range from 260–190 nm with a bandwidth of 2.0 nm and a time constant of 8 s.
All CD spectra were recorded in three different liposome vesicles (DOPC, DOPG and DOPC/DOPG (90/10)) to which Aβ(25–35) was added at a final concentration of 0.1 mM and all ligands at a final concentration of 0.5 mM. The CD spectrum of Aβ(25–35) in the absence of ligands was recorded for comparison. Each spectrum was corrected for the contribution of the liposomes containing pure ligands. To estimate the secondary structure, the CD spectra were analyzed using the CONTINN, SELCON and K2D algorithms of the DICHROWEB on-line server57.
EPR spectroscopy
For EPR experiments, multi-lamellar vesicles (MLVs) of DOPC, DOPG and DOPC/DOPG (90/10 molar ratio) were prepared by mixing appropriate amounts of lipids, dissolved in a CH2Cl2/MeOH mixture (2:1 v/v, 10 mg/mL concentration), in a round-bottom test tube. Spin-labeled phosphatidylcholines (1-palmitoyl-2-stearoyl-(n-doxyl)-sn-glycero-3-phosphocholine, n-PCSL with n = 5, 14) were added to the lipid mixture (1% by weight of the total lipid) by mixing appropriate amounts of a spin-label solution in ethanol (1 mg/mL) with the lipid organic mixture. A thin lipid film was produced by evaporating the solvents with dry nitrogen gas. The total weight of the lipids for each sample was 0.2 mg. The final traces of solvents were removed by subjecting the sample to vacuum desiccation for at least 3 h. The samples were then hydrated with 20 μL of 10 mM phosphate buffer at pH = 7.4, gently warmed and repeatedly vortexed to obtain a MLV suspension.
Samples containing curcumin or one of its analogues were prepared by the same procedure, adding appropriate amounts of the flavonoid dissolved in DMSO (10 mg/mL) to the buffer during the re-hydration step to obtain a 0.5 molar percentage with respect to the total lipids. The addition of DMSO was preventively checked to result in no perturbation of the vesicular systems.
Samples containing Aβ(25–35) were prepared in a similar manner, except that the lipid film was hydrated directly with the peptide solution in phosphate buffer, at 1:0.5 lipid/peptide weight ratio (which corresponds to approximately 20:1 mol/mol).
EPR spectra were recorded with a 9 GHz Bruker Elexys E-500 spectrometer (Bruker, Rheinstetten, Germany). Samples were placed in 25 μL glass capillaries and flame sealed. The capillaries were placed in a standard 4 mm quartz sample tube containing light silicone oil for thermal stability. All measurements were performed at 25 °C. The spectra were recorded using the following instrumental settings: sweep width, 100 G; resolution, 1024 points; time constant, 20.48 ms; modulation frequency, 100 kHz; modulation amplitude, 1.0 G; incident power, 6.37 mW. Several scans, typically 16, were accumulated to improve the signal-to-noise ratio.
Molecular modeling
Molecular modeling calculations were performed on an SGI Origin 200 8XR12000 and an E4 Server Twin 2 x Dual Xeon 5520, equipped with two nodes. Each node was 2 x Intel Xeon QuadCore E5520, 2.26 GHz, 36 GB RAM. The molecular modeling graphics were implemented on SGI Octane 2 workstations.
The apparent pKa values of CUR and its derivatives in their tautomeric forms were estimated using the ACD/Percepta software79. Accordingly, the percentages of neutral/ionized forms were computed at pH 7.2 (cytoplasm) using the Henderson-Hasselbach equation.
Conformational analysis
All compounds were built considering the prevalent ionic forms of each tautomer using the Insight 2005 Builder module (Accelrys Software, Inc., San Diego).
Atomic potentials and partial charges were assigned using the CVFF force field80. The conformational space of the compounds was sampled through 200 cycles of simulated annealing (ε = 80*r). To avoid unrealistic results, the torsional angle of the double bonds was constrained within 180° using a force constant of 100 (kcal/mol)/Å. In simulated annealing, the temperature is altered in time increments from an initial temperature to a final temperature by adjusting the kinetic energy of the structure (by rescaling the velocities of the atoms). The following protocol was applied: the system was heated to 1000 K over 2000 fs (time step of 3.0 fs); the temperature of 1000 K was applied to the system for 2000 fs (time step of 3.0 fs) to surmount torsional barriers; then, the temperature was linearly reduced to 300 K in 1000 fs (time step of 1.0 fs). The resulting conformations were then subjected to Molecular Mechanics (MM) energy minimization within the Insight 2005 Discover 3 module (CVFF force field; ε = 80*r) until the maximum RMS derivative was less than 0.001 kcal/Å, using the conjugate gradient81 as the minimization algorithm. The resulting MM conformers were ranked by (i) their potential energy values (i.e., ΔE from the global energy minimum), (ii) intra-molecular hydrogen bonds, (iii) torsional angles, and (iv) the interatomic distance between the centroids of the two aromatic rings. The centroids were determined considering the heavy atoms of the aromatic rings (Pseudo_Atom Define command, Biopolymer Module, Insight 2005). Then, the occurrence rates were calculated.
All MM conformers were subjected to a full geometric optimization by semi-empirical calculations, using the quantum mechanical method PM782 in the MOPAC2012 package82 and EF83 (eigenvector following routine) as the geometric optimization algorithm. The GNORM value was set to 0.01. To achieve full geometric optimization, the criterion for terminating all optimizations was increased by a factor of 100, using the keyword PRECISE. The resulting PM7 conformers were ranked as MM conformers.
Finally, the X-ray structures of CUR, alone (CSD codes: AXOGIE, AXOGOK, BINMEQ, BINMEQ01, BINMEQ02, BINMEQ03, BINMEQ04, BINMEQ05, BINMEQ06, BINMEQ07, BINMEQ08, BUWKUZ, GANJAG) or in complex with any metal (CSD code: RAWDOK), were selected and downloaded from the Cambridge Structural Database (CSD) using the CSDS (Cambridge Structural Database System) software Conquest 1.16. These structures were analyzed using Insight 2005 (Accelrys, San Diego, CA).
Docking procedure
To build the starting complex CYC:Aβ(25–35) for dynamic docking studies, the experimentally determined structure of Aβ(25–35) (PDB ID: 1QWP) was downloaded from the Protein Data Bank (PDB; http://www.rcsb.org/pdb/). Hydrogens were added to the PDB structure assuming a pH of 7.2. This structure was analyzed and compared with the experimentally determined structures of HDM2 protein in complex with p53 (PDB ID: 1YCR), using the Biopolymer and Homology module of Insight 2005 (Accelrys, San Diego). Accordingly, a structural superimposition was performed between the α-helix of HDM2 (A50-A65) and Aβ(25–35). The centroids of interacting residues of considered LxxLLxxL-like (i, i + 3, i + 4, i + 7) motif (B17-B29; PDB ID: 1YCR) were built, and their relative distances were calculated. The resulting values were crossed with the values of the CYC pharmacophore distances. The lowest-energy PM7 conformer of CYC was superimposed on residues i and i + 4 (i.e., Phe19 and Trp23) of the LxxLLxxL-like motif by fitting the centroids of the aromatic rings (i.e., X and Y) on the centroids of the side chains of the corresponding residues. Then, the resulting structure of the complex CYC: Aβ(25–35) was used for successive dynamic docking studies. In particular, a docking methodology was used that treats all the systems (i.e., ligand and protein) as flexible. Flexible docking was achieved using the Affinity module in the Insight 2005 suite, setting the SA_Docking procedure84 and using the Cell Multipole method for nonbonding interactions85. To mimic the experimental conditions (a mixture of HFIP and water; 80/20 v:v), throughout the calculations, a dielectric constant of 30*r was used. This value was obtained as the weighted average of the dielectric constants of water (ε = 80) and HFIP (ε = 16.7).
The docking protocol included a Monte Carlo-based conformational search of the ligand (CYC) within the Aβ(25–35) peptide. The binding domain area was defined as a subset including all residues of the Aβ(25–35) peptide. All atoms included in the binding domain area were left free to move throughout the course of the docking calculations, whereas to avoid unrealistic results, a tethering restraint was applied to the SCRs of the protein. In particular, for the α-helix Asn27-Leu34, the distance between backbone hydrogen bond donors and acceptors was restrained within 2.5 Å using a force constant of 10 kcal/mol/Å2.
A Monte Carlo/minimization approach for the random generation of a maximum of 20 acceptable complexes was used. During the first step, starting from the previously obtained roughly docked structures, the ligand was moved by a random combination of translation, rotation, and torsional changes to sample both the conformational space of the ligand and its orientation with respect to the protein (MxRChange = 3 Å; MxAngChange = 180°). During this step, the van der Waals (vdW) and Columbic terms were scaled to a factor of 0.1 to avoid very severe divergences in the vdW and Columbic energies. If the energy of a complex structure resulting from random moves of the ligand was higher by the energy tolerance parameter than the energy of the last accepted structure, it was not accepted for minimization. To ensure a wide variance in the input structures to be successively minimized, an energy tolerance value of 106 kcal/mol from the previous structure was used. After the energy minimization step (conjugate gradient; 10000 iterations; ε = 30*r), the energy test, using an energy range of 50 kcal/mol, and a structure similarity check (rms tolerance = 0.3 kcal/Å) were applied to select the 20 acceptable structures. Each subsequent structure was generated from the last accepted structure. Following this procedure, the resulting docked structures were ranked by their conformational energy and analyzed. Finally, to test the thermodynamic stability of the resulting docked complexes; these latter were subjected also to a molecular dynamics simulated annealing protocol (ε = 30*r). A tethering restraint was applied to the SCRs of the complex. The same set of structural restraints used for previous docking calculations was applied. The protocol included 5 ps of a dynamic run divided into 50 stages (100 fs each), during which the temperature of the system was linearly decreased from 500 to 300 K (Verlet velocity integrator; time step = 1.0 fs). In simulated annealing, the temperature is altered in time increments from an initial temperature to a final temperature. The temperature is changed by adjusting the kinetic energy of the structure (by rescaling the velocities of the atoms). Molecular dynamics calculations were performed using a constant temperature and constant volume (NVT) statistical ensemble, with direct velocity scaling as the temperature control method (temp window = 10 K). In the first stage, the initial velocities were randomly generated from the Boltzmann distribution, according to the desired temperature, while during the subsequent stages, the initial velocities were generated from the dynamics restart data. A temperature of 500 K was applied to surmount torsional barriers, thus allowing an unconstrained rearrangement of the “ligand” and the “protein” active site (initial vdW and Columbic scale factors = 0.1). The temperature was then linearly reduced to 300 K in 5 ps, and, concurrently, the vdW and Columbic scale factors were similarly increased from their initial values (0.1) to their final values (1.0). A final round of 105 minimization steps (ε = 30*r) followed the last dynamics steps, and the minimized structures were saved in a trajectory file.
The resulting complexes were ranked by their conformational energy and analyzed by considering the non-bond interaction energies between the ligand and the peptide (vdW and electrostatic energy contribution; Group Based method; CUT_OFF = 100; ε = 2*r; Discover_3 Module of Insight2005). The complex characterized by the lowest conformational and interaction energy was chosen as the most representative. To allow the relaxation of the whole system, this complex was further minimized with no tethering restraint (CVFF force field; Cell Multipole method for non-bond interaction) using the Conjugate Gradient algorithm (maximum rms derivative less than 0.01 kcal/Å). Finally, the selected complex was compared with the experimentally determined structures of i) HDM2 protein in complex with p53 (PDB ID: 1YCR), ii) estrogen receptor β in complex with the nuclear receptor coactivator 5 (Ncoa5; PDB ID: 2J7X), iii) the coiled-coil dimerization motif of Geminin (PDB ID: 1T6F), and iv) the coiled-coil dimerization motif of the human ROCK I protein (PDB ID: 3O0Z) using the Biopolymer and Homology module of Insight 2005 (Accelrys, San Diego).
Additional Information
How to cite this article: Randino, R. et al. Investigating the Neuroprotective Effects of Turmeric Extract: Structural Interactions of β-Amyloid Peptide with Single Curcuminoids. Sci. Rep. 6, 38846; doi: 10.1038/srep38846 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
M.R. thanks Maria Carrano for the analysis of the curcuminoids and optimization of the microwave-assisted synthesis of cyclocurcumin and curcuminoids. This work was supported by the following grants: POR Campania FESR 2007–2013 FARMABIONET (B25C1300023007), MIUR - FIRB 2012 RBFR12WB3W, and EU Project Bluegenics (Grant 311848).
Footnotes
Author Contributions M.R. and A.M.D’U. designed the work. M.P. and C.F. designed and performed the computational investigation and supervised the discussion of all results; M.G. and A.M.D’U. performed the CD experiments and peptide SPS; G.D’E. and A.D.S. performed the ESR experiments; R.R. E.C. and M.R. performed the synthesis, separation and characterization of curcuminoids. W.C., E.N. and A.R. supplied samples and provided knowhow on curcuminoids and information on pharmacokinetics. C.F., W.C., A.R., G.D’E., A.M.D’U., E.N. and M.R. wrote the manuscript. All authors reviewed the manuscript.
References
- Selkoe D. J. Alzheimer’s disease: Genes, proteins, and therapy. Physiol Rev 81, 741–766 (2001). [DOI] [PubMed] [Google Scholar]
- Walsh D. M. & Selkoe D. J. Deciphering the molecular basis of memory failure in Alzheimer’s disease. Neuron 44, 181–193 (2004). [DOI] [PubMed] [Google Scholar]
- Sipe J. D. & Cohen A. S. Review: History of the amyloid fibril. J Struct Biol 130, 88–98 (2000). [DOI] [PubMed] [Google Scholar]
- Hardy J. & Selkoe D. J. Medicine - The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science 297, 353–356 (2002). [DOI] [PubMed] [Google Scholar]
- Iversen L. L., Mortishiresmith R. J., Pollack S. J. & Shearman M. S. The Toxicity in-Vitro of Beta-Amyloid Protein. Biochem J 311, 1–16 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner D. A., Abraham C. & Selkoe D. J. X-Ray-Diffraction from Intraneuronal Paired Helical Filaments and Extraneuronal Amyloid Fibers in Alzheimer-Disease Indicates Cross-Beta Conformation. P Natl Acad Sci USA 83, 503–507 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto C., Castano E. M., Frangione B. & Inestrosa N. C. The Alpha-Helical to Beta-Strand Transition in the Amino-Terminal Fragment of the Amyloid Beta-Peptide Modulates Amyloid Formation. J Biol Chem 270, 3063–3067 (1995). [DOI] [PubMed] [Google Scholar]
- Tomaselli S. et al. The alpha-to-beta conformational transition of Alzheimer’s A beta-(1–42) peptide in aqueous media is reversible: A step by step conformational analysis suggests the location of beta conformation seeding. ChemBioChem 7, 257–267, doi: 10.1002/cbic.200500223 (2006). [DOI] [PubMed] [Google Scholar]
- Aguilar F. et al. Scientific opinion on the re-evaluation of curcumin (E 100) as a food additive. EFSA J 8, 46 (2010). [Google Scholar]
- Ruby A. J., Kuttan G., Babu K. D., Rajasekharan K. N. & Kuttan R. Antitumor and Antioxidant Activity of Natural Curcuminoids. Cancer Lett 94, 79–83 (1995). [DOI] [PubMed] [Google Scholar]
- Adams B. K. et al. Synthesis and biological evaluation of novel curcumin analogs as anti-cancer and anti-angiogenesis agents. Bioorgan Med Chem 12, 3871–3883 (2004). [DOI] [PubMed] [Google Scholar]
- Zambre A. P., Kulkarni V. M., Padhye S., Sandur S. K. & Aggarwal B. B. Novel curcumin analogs and proliferation targeting TNF-induced NF-kappa B activation in human leukemic KBM-5 cells. Bioorganic & Medicinal Chemistry 14, 7196–7204 (2006). [DOI] [PubMed] [Google Scholar]
- Vajragupta O., Boonchoong P., Morris G. M. & Olson A. J. Active site binding modes of curcumin in HIV-1 protease and integrase. Bioorganic & medicinal chemistry letters 15, 3364–3368, doi: 10.1016/j.bmcl.2005.05.032 (2005). [DOI] [PubMed] [Google Scholar]
- Surh Y. J. Cancer chemoprevention with dietary phytochemicals. Nat Rev Cancer 3, 768–780 (2003). [DOI] [PubMed] [Google Scholar]
- Rodriguez J. J. L. et al. Prevalence of dementia in Latin America, India, and China: a population-based cross-sectional survey. The Lancet 372, 464–474 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler-Graham K., Brookmeyer R., Johnson E. & Arrighi H. M. Worldwide variation in the doubling time of Alzheimer’s disease incidence rates. Alzheimer’s & Dementia 4, 316–323 (2008). [DOI] [PubMed] [Google Scholar]
- Mathuranath P. et al. Incidence of Alzheimer’s disease in India: A 10 yearsfollow-up study. Neurology India 60, 625 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcea G. et al. Detection of curcumin and its metabolites in hepatic tissue and portal blood of patients following oral administration. British journal of cancer 90, 1011–1015, doi: 10.1038/sj.bjc.6601623 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcea G. et al. Consumption of the putative chemopreventive agent curcumin by cancer patients: assessment of curcumin levels in the colorectum and their pharmacodynamic consequences. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 14, 120–125 (2005). [PubMed] [Google Scholar]
- Pan M. H., Huang T. M. & Lin J. K. Biotransformation of curcumin through reduction and glucuronidation in mice. Drug Metab Dispos 27, 486–494 (1999). [PubMed] [Google Scholar]
- Ireson C. et al. Characterization of metabolites of the chemopreventive agent curcumin in human and rat hepatocytes and in the rat in vivo, and evaluation of their ability to inhibit phorbol ester-induced prostaglandin E2 production. Cancer Research 61, 1058–1064 (2001). [PubMed] [Google Scholar]
- Shoba G. et al. Influence of piperine on the pharmacokinetics of curcumin in animals and human volunteers. Planta Med 64, 353–356 (1998). [DOI] [PubMed] [Google Scholar]
- Barry J. et al. Determining the Effects of Lipophilic Drugs on Membrane Structure by Solid-State NMR Spectroscopy: The Case of the Antioxidant Curcumin. J Am Chem Soc 131, 4490–4498 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semalty A., Semalty M., Rawat M. S. M. & Franceschi F. Supramolecular phospholipids-polyphenolics interactions: The PHYTOSOME® strategy to improve the bioavailability of phytochemicals. Fitoterapia 81, 306–314 (2010). [DOI] [PubMed] [Google Scholar]
- Giori A. & Franceschi F. Phospholipid complexes of curcumin having improved bioavailability. Indena S.P.A. United Stated patent US, 12, 296, 696, 2007 Dec 27.
- Cuomo J. et al. Comparative absorption of a standardized curcuminoid mixture and its lecithin formulation. J Nat Prod 74, 664–669, doi: 10.1021/np1007262 (2011). [DOI] [PubMed] [Google Scholar]
- De Lorenzi E. et al. Pharmaceutical strategies against amyloidosis: Old and new drugs in targeting a “protein misfolding disease”. Curr Med Chem 11, 1065–1084 (2004). [DOI] [PubMed] [Google Scholar]
- Soto C. & Estrada L. Amyloid inhibitors and beta-sheet breakers. Alzheimer’s Disease: Cellular and Molecular Aspects of Amyloid 38, 351–364 (2005). [DOI] [PubMed] [Google Scholar]
- Arkin M. R. & Wells J. A. Small-molecule inhibitors of protein-protein interactions: Progressing towards the dream. Nat Rev Drug Discov 3, 301–317 (2004). [DOI] [PubMed] [Google Scholar]
- Mason J. M., Kokkoni N., Stott K. & Doig A. J. Design strategies for anti-amyloid agents. Curr Opin Struc Biol 13, 526–532 (2003). [DOI] [PubMed] [Google Scholar]
- Porat Y., Abramowitz A. & Gazit E. Inhibition of amyloid fibril formation by polyphenols: Structural similarity and aromatic interactions as a common inhibition mechanism. Chem Biol Drug Des 67, 27–37 (2006). [DOI] [PubMed] [Google Scholar]
- Re F. et al. Beta Amyloid Aggregation Inhibitors: Small Molecules as Candidate Drugs for Therapy of Alzheimer’s Disease. Curr Med Chem 17, 2990–3006 (2010). [DOI] [PubMed] [Google Scholar]
- Ono K., Hasegawa K., Naiki H. & Yamada M. Curcumin has potent anti-amyloidogenic effects for Alzheimer’s beta-amyloid fibrils in vitro. J Neurosci Res 75, 742–750 (2004). [DOI] [PubMed] [Google Scholar]
- Narlawar R. et al. Curcumin-derived pyrazoles and isoxazoles: Swiss army knives or blunt tools’ for Alzheimer’s disease? Chemmedchem 3, 165–172, doi: 10.1002/cmdc.200700218 (2008). [DOI] [PubMed] [Google Scholar]
- Reinke A. A. & Gestwicki J. E. Structure-activity relationships of amyloid beta-aggregation inhibitors based on curcumin: Influence of linker length and flexibility. Chem Biol Drug Des 70, 206–215, doi: 10.1111/j.1747.0285.2007.00557.x (2007). [DOI] [PubMed] [Google Scholar]
- Yang F. et al. Curcumin inhibits formation of amyloid β oligomers and fibrils, binds plaques, and reduces amyloid in vivo. J Biol Chem 280, 5892–5901 (2005). [DOI] [PubMed] [Google Scholar]
- Yanagisawa D. et al. Relationship between the tautomeric structures of curcumin derivatives and their A beta-binding activities in the context of therapies for Alzheimer’s disease. Biomaterials 31, 4179–4185, doi: 10.1016/j.biomaterials.2010.01.142 (2010). [DOI] [PubMed] [Google Scholar]
- Naldi M. et al. Amyloid beta-Peptide 25–35 Self-Assembly and Its Inhibition: A Model Undecapeptide System to Gain Atomistic and Secondary Structure Details of the Alzheimer’s Disease Process and Treatment. ACS Chemical Neuroscience 3, 952–962, doi: 10.1021/cn3000982 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Airoldi C. et al. Curcumin derivatives as new ligands of A beta peptides. Journal of Biotechnology 156, 317–324, doi: 10.1016/j.jbiotec.2011.07.021 (2011). [DOI] [PubMed] [Google Scholar]
- Shytle R. D. et al. Optimized turmeric extract reduces beta-Amyloid and phosphorylated Tau protein burden in Alzheimer’s transgenic mice. Curr Alzheimer Res 9, 500–506 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamley I. The amyloid beta peptide: a chemist’s perspective. Role in Alzheimer’s and fibrillization. Chemical reviews 112, 5147–5192 (2012). [DOI] [PubMed] [Google Scholar]
- Terzi E., Holzemann G. & Seelig J. Alzheimer Beta-Amyloid Peptide-25–35 - Electrostatic Interactions with Phospholipid-Membranes. Biochemistry-Us 33, 7434–7441, doi: 10.1021/Bi00189a051 (1994). [DOI] [PubMed] [Google Scholar]
- Pike C. J., Burdick D., Walencewicz A. J., Glabe C. G. & Cotman C. W. Neurodegeneration Induced by Beta-Amyloid Peptides Invitro - the Role of Peptide Assembly State. J Neurosci 13, 1676–1687 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike C. J. et al. Structure-Activity Analyses of Beta-Amyloid Peptides - Contributions of the Beta-25–35 Region to Aggregation and Neurotoxicity. J Neurochem 64, 253–265 (1995). [DOI] [PubMed] [Google Scholar]
- D’Ursi A. M. et al. Solution structure of amyloid beta-peptide (25–35) in different media. J Med Chem 47, 4231–4238 (2004). [DOI] [PubMed] [Google Scholar]
- Esposito C. et al. Exploring interaction of β‐amyloid segment (25–35) with membrane models through paramagnetic probes. Journal of Peptide Science 12, 766–774 (2006). [DOI] [PubMed] [Google Scholar]
- Saponetti M. S. et al. Aggregation of Ass (25–35) on DOPC and DOPC/DHA bilayers: an atomic force microscopy study. PloS one 9, e115780 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementi M. E. et al. Aβ (31–35) and Aβ (25–35) fragments of amyloid beta‐protein induce cellular death through apoptotic signals: Role of the redox state of methionine‐35. FEBS letters 579, 2913–2918 (2005). [DOI] [PubMed] [Google Scholar]
- Ege C. & Lee K. Y. C. Insertion of Alzheimer’s A beta 40 peptide into lipid monolayers. Biophys J 87, 1732–1740 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi E. Y., Frey S. L. & Lee K. Y. C. Cell membrane ganglioside GM1-Mediated amyloid-beta fibrillogenesis and membrane disruption. Biophys J, 220a–220a (2007). [DOI] [PubMed] [Google Scholar]
- Chi E. Y. et al. Lipid membrane templates the ordering and induces the fibrillogenesis of Alzheimer’s disease amyloid-beta peptide. Proteins 72, 1–24 (2008). [DOI] [PubMed] [Google Scholar]
- Kiuchi F. et al. Nematocidal Activity of Turmeric - Synergistic Action of Curcuminoids. Chem Pharm Bull 41, 1640–1643 (1993). [DOI] [PubMed] [Google Scholar]
- Lee S. L., Huang W. J., Lin W. W., Lee S. S. & Chen C. H. Preparation and anti-inflammatory activities of diarylheptanoid and diarylheptylamine analogs. Bioorganic & Medicinal Chemistry 13, 6175–6181 (2005). [DOI] [PubMed] [Google Scholar]
- Karewicz A. et al. Interaction of curcumin with lipid monolayers and liposomal bilayers. Colloid Surface B 88, 231–239 (2011). [DOI] [PubMed] [Google Scholar]
- Kayabasi U., Sergott R. & Rispoli M. Retinal Examination for the Diagnosis of Alzheimer’s Disease. J Ophthalmic Pathol 3: 4. of 3, 2 (2014). [Google Scholar]
- Jao S. C., Ma K., Talafous J., Orlando R. & Zagorski M. G. Trifluoroacetic acid pretreatment reproducibly disaggregates the amyloid beta-peptide. Amyloid 4, 240–252 (1997). [Google Scholar]
- Whitmore L. & Wallace B. A. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res 32, W668–W673 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitiello G., Di Marino S., D’Ursi A. M. & D’Errico G. Omega-3 Fatty Acids Regulate the Interaction of the Alzheimer’s A beta(25–35) Peptide with Lipid Membranes. Langmuir 29, 14239–14245 (2013). [DOI] [PubMed] [Google Scholar]
- Vitiello G. et al. Fusion of raft-like lipid bilayers operated by a membranotropic domain of the HSV-type I glycoprotein gH occurs through a cholesterol-dependent mechanism. Soft matter 11, 3003–3016, doi: 10.1039/c4sm02769h (2015). [DOI] [PubMed] [Google Scholar]
- D’Errico G., Vitiello G., D’Ursi A. M. & Marsh D. Interaction of short modified peptides deriving from glycoprotein gp36 of feline immunodeficiency virus with phospholipid membranes. Eur Biophys J Biophy 38, 873–882 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedeschi A. et al. Effect of flavonoids on the A beta(25–35)-phospholipid bilayers interaction. Eur J Med Chem 45, 3998–4003 (2010). [DOI] [PubMed] [Google Scholar]
- Sun Y. et al. The bound states of amphipathic drugs in lipid bilayers: Study of curcumin. Biophys J 95, 2318–2324 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Errico G. et al. Interaction between Alzheimer’s A beta(25–35) peptide and phospholipid bilayers: The role of cholesterol. Bba-Biomembranes 1778, 2710–2716 (2008). [DOI] [PubMed] [Google Scholar]
- Thapa A. et al. Membrane-Mediated Neuroprotection by Curcumin from Amyloid-beta-Peptide-Induced Toxicity. Langmuir 29, 11713–11723 (2013). [DOI] [PubMed] [Google Scholar]
- Stern H. A. & Feller S. E. Calculation of the dielectric permittivity profile for a nonuniform system: application to a lipid bilayer simulation. The Journal of chemical physics 118, 3401–3412 (2003). [Google Scholar]
- Walther D., Eisenhaber F. & Argos P. Principles of helix-helix packing in proteins: the helical lattice superposition model. Journal of molecular biology 255, 536–553 (1996). [DOI] [PubMed] [Google Scholar]
- Nadezhdin K. D., Bocharova O. V., Bocharov E. V. & Arseniev A. S. Dimeric structure of transmembrane domain of amyloid precursor protein in micellar environment. FEBS letters 586, 1687–1692 (2012). [DOI] [PubMed] [Google Scholar]
- Ferlini C. et al. Paclitaxel directly binds to Bcl-2 and functionally mimics activity of Nur77. Cancer research 69, 6906–6914 (2009). [DOI] [PubMed] [Google Scholar]
- Persico M. et al. New anticancer agents mimicking protein recognition motifs. J Med Chem 56, 6666–6680 (2013). [DOI] [PubMed] [Google Scholar]
- Thépaut M. et al. Crystal structure of the coiled-coil dimerization motif of geminin: structural and functional insights on DNA replication regulation. Journal of molecular biology 342, 275–287 (2004). [DOI] [PubMed] [Google Scholar]
- Vagt T., Zschörnig O., Huster D. & Koksch B. Membrane Binding and Structure of De Novo Designed α‐Helical Cationic Coiled‐Coil‐Forming Peptides. Chemphyschem 7, 1361–1371 (2006). [DOI] [PubMed] [Google Scholar]
- Crescenzi O. et al. Solution structure of the Alzheimer amyloid β‐peptide (1–42) in an apolar microenvironment. European Journal of Biochemistry 269, 5642–5648 (2002). [DOI] [PubMed] [Google Scholar]
- Campiglia P. et al. A new series of 1,3-dihidro-imidazo[1,5-c]thiazole-5,7-dione derivatives: synthesis and interaction with Abeta(25–35) amyloid peptide. Chem Biol Drug Des 74, 224–233, doi: 10.1111/j.1747-0285.2009.00853.x (2009). [DOI] [PubMed] [Google Scholar]
- Lin H., Bhatia R. & Lal R. Amyloid β protein forms ion channels: implications for Alzheimer’s disease pathophysiology. The FASEB Journal 15, 2433–2444 (2001). [DOI] [PubMed] [Google Scholar]
- Rhee S. K., Quist A. P. & Lal R. Amyloid β protein-(1–42) forms calcium-permeable, Zn2+-sensitive channel. J Biol Chem 273, 13379–13382 (1998). [DOI] [PubMed] [Google Scholar]
- Merrifield R. B. Solid-Phase Peptide Syntheses. Endeavour 24, 3–7 (1965). [DOI] [PubMed] [Google Scholar]
- Zagorski M. G. et al. Methodological and chemical factors affecting amyloid beta peptide amyloidogenicity. Method Enzymol 309, 189–204 (1999). [DOI] [PubMed] [Google Scholar]
- Naiki H. & Nakakuki K. First-order kinetic model of Alzheimer’s beta-amyloid fibril extension in vitro. Laboratory Investigation 74, 374–383 (1996). [PubMed] [Google Scholar]
- Japertas P., Lanevskij K. & Sazonovas A. ACD/Percepta structure design engine: Virtual enumeration and screening of physchem properties for 10(16) compounds in real time. Abstr Pap Am Chem S 248 (2014). [Google Scholar]
- Dauber-Osguthorpe P. et al. Structure and energetics of ligand binding to proteins: Escherichia coli dihydrofolate reductase-trimethoprim, a drug-receptor system. Proteins 4, 31–47, doi: 10.1002/prot.340040106 (1988). [DOI] [PubMed] [Google Scholar]
- Fletcher R. An Overview of Unconstrained Optimization. Nato Adv Sci Inst Se 434, 109–143 (1994). [Google Scholar]
- Stewart J. J. Optimization of parameters for semiempirical methods VI: more modifications to the NDDO approximations and re-optimization of parameters. J Mol Model 19, 1–32, doi: 10.1007/s00894-012-1667-x (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker J. An Algorithm for the Location of Transition-States. J Comput Chem 7, 385–395, doi: 10.1002/jcc.540070402 (1986). [DOI] [Google Scholar]
- Senderowitz H., Guarnieri F. & Still W. C. A Smart Monte-Carlo Technique for Free-Energy Simulations of Multiconformational Molecules, Direct Calculations of the Conformational Populations of Organic-Molecules. J Am Chem Soc 117, 8211–8219, doi: 10.1021/ja00136a020 (1995). [DOI] [Google Scholar]
- Ding H. Q., Karasawa N. & Goddard W. A. Atomic Level Simulations on a Million Particles - the Cell Multipole Method for Coulomb and London Nonbond Interactions. J Chem Phys 97, 4309–4315, doi: 10.1063/1.463935 (1992). [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.