Abstract
Xist is an X-linked gene responsible for cis induction of X chromosome inactivation. Studies have indicated that Xist is abnormally activated in the active X chromosome in cloned mouse embryos due to loss of the maternal Xist-repressing imprint following enucleation during somatic cell nuclear transfer (SCNT). Inhibition of Xist expression by injecting small interfering RNA (siRNA) has been shown to enhance the in vivo developmental efficiency of cloned male mouse embryos by more than 10-fold. The purpose of this study was to investigate whether a similar procedure can be applied to improve the cloning efficiency in pigs. We first found that Xist mRNA levels at the morula stage were aberrantly higher in pig SCNT embryos than in in vivo fertilization-derived pig embryos. Injection of a preselected effective anti-Xist siRNA into 1-cell-stage male pig SCNT embryos resulted in significant inhibition of Xist expression through the 16-cell stage. This siRNA-mediated inhibition of Xist significantly increased the total cell number per cloned blastocyst and significantly improved the birth rate of cloned healthy piglets. The present study contributes useful information on the action of Xist in the development of pig SCNT embryos and proposes a new method for enhancing the efficiency of pig cloning.
Keywords: Cloning, Pig, RNAi, Somatic cell nuclear transfer (SCNT), Xist
Xist is a non-protein coding RNA gene located on the mammalian X chromosomes. The major function of Xist is to trigger X chromosome inactivation (XCI). During normal development of mammalian embryos, two forms of XCI occur, specifically imprinted XCI and random XCI [1, 2]. Xist is responsible for cis induction of both the forms of XCI [3]. Therefore, Xist plays an important role in mammalian development.
Many studies have shown that Xist is aberrantly expressed in cloned mammalian embryos or in animals generated by somatic cell nuclear transfer (SCNT) [4,5,6,7,8]. Inoue et al. [9] found that Xist was ectopically expressed from the active X chromosome at the pre-implantation stage in both male and female mouse SCNT embryos. Xist expression results in significantly lower expression of global X-linked genes in cloned embryos of both the sexes than that observed in in vitro fertilization-derived embryos. Early mouse embryos cloned from donor somatic cells ectopically express Xist because of the lack of the maternal Xist-repressing imprint, which is established at the last stage of oogenesis, but is removed by enucleation during SCNT [10]. Suppression of aberrant Xist expression in mouse SCNT embryos via knockout of the Xist allele on the active X chromosome not only abolished the dysregulation of X-linked genes, but also improved the cloning efficiency by 8–9-fold [9]. Furthermore, inhibition of ectopic Xist expression in early cloned male mouse embryos by RNA interference (RNAi) also increased the SCNT efficiency by more than 10-fold [11]. These studies show that the maternal Xist allele in cloned mouse embryos is abnormally activated at the pre-implantation stage, and inhibition of this erroneous Xist expression can significantly enhance SCNT efficiency in mice.
Recently, it has been demonstrated that Xist mRNA expression at the blastocyst [18], fetal, and postnatal stages in cloned pigs is higher than that in in vivo fertilization (IVV)-derived pigs [19, 20]. The purpose of this study was to investigate whether RNAi can repress the abnormally elevated Xist expression in early stages of cloned pig embryos, and whether RNAi-mediated suppression of Xist expression can improve the developmental rate of cloned pig embryos. We showed that the Xist expression at the morula stage was significantly higher in pig SCNT embryos than in IVV-derived embryos. Microinjection of a preselected effective anti-Xist small interfering RNA (siRNA) into cloned male pig embryos significantly inhibited Xist expression and improved the developmental efficiency of the male pig SCNT embryos.
Materials and Methods
Ethics statements
This study was conducted in Guangdong province of China in strict accordance with the “Guidelines with Respect to Caring for Laboratory Animals”, issued by the Ministry of Science and Technology of China. The protocol for animal experiments was approved by the Institutional Animal Care and Use Committee of South China Agricultural University. All precautions were taken to minimize animal suffering.
Preparation of embryos
Male and female SCNT embryos were produced as previously described [12]. Briefly, the mature oocyte was aspirated firmly into a holding pipette (outer diameter = 100–120 µm, inner diameter = 20–30 µm) to ensure immobility. The enucleation pipette (inner diameter = 15 µm) was inserted through the zona pellucida. The first polar body and adjacent cytoplasm, presumably containing all the chromosomes, were aspirated into the enucleation pipette. Then a single fibroblast cell, which was separated by pipetting after digestion with 0.256% trypsin, was microinjected into the perivitelline space of the oocytes. The oocyte-donor cell complexes were cultured in porcine zygote medium 3 (PZM3) at 39°C, 5% CO2, 5% O2, 90% N2, and 100% humidity for 1.5 h. The cell complexes were activated to fuse in a medium containing 250 mM mannitol, 0.1 mM CaCl2.2H2O, 0.1 mM MgCl2.6H2O, 0.5 mM HEPES, and 0.01% polyvinyl alcohol, by two successive DC pulses at 1.2 kv/cm for 30 µsec using an electro-fusion instrument (model: CF-150/B, Biological Laboratory Equipment Maintenance and Service, Budapest, Hungary). The activated cloned embryos were then cultured in PZM3 containing Cytochalasin B (5 µg/ml) for 4 h. After the post-activation treatment, the reconstructed embryos were cultured in PZM3 at 39°C, 5% CO2, 7% O2, 88% N2, and 100% humidity. In vivo fertilized embryos of the 1-cell stage were collected from artificially inseminated sows in accordance with our previous description [13] and cultured in vitro in PZM3 (at 39°C, 5% CO2, 7% O2, 88% N2, and 100% humidity).
Identification of porcine Xist genomic DNA sequence
The genomic DNA sequence of porcine Xist was identified by electronic cloning, based on the already known partial genomic DNA sequence of porcine Xist, the porcine expressed sequence tag (EST) and UniGene libraries, and the conserved genomic DNA sequences among human, bovine, and mouse Xist. The identified porcine Xist genomic DNA sequence was verified by RT-PCR, 3′ rapid amplification of cDNA ends (RACE), followed by sequencing, and was submitted to GenBank (Accession Number: KC149530).
Preparation of siRNAs
Three siRNA duplexes were designed according to the cDNA sequence of porcine Xist and synthesized by GenePharma (Shanghai, China). Their sequences were as follows: siRNA1 sense 5′-GCCUGGUUAAGAUGAAUAATT-3′, antisense: 5′-UUAUUCAUCUUAACCAGGCTT-3′; siRNA2 sense 5′-GUGCCUGCUAAUUGAAAGATT-3′, antisense: 5′-UCUUUCAAUUAGCAGGCACTT-3′; siRNA3 sense 5′-CCCUUCAUAUUUGCCAAAUTT-3′, antisense: 5′-AUUUGGCAAAUAUGAAGGGTT-3′; Mock siRNA sense: 5′-UUCUCCGAACGUGUCACGUTT-3′, antisense: 5′-ACGUGACACGUUCGGAGAATT-3′.
Real-time PCR
Total RNA was extracted from the embryos or cells using an E.Z.N.A. Total RNA Kit I (Omega, GA, USA). PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa, Tokyo, Japan) was used to synthesize cDNA. Real-time PCR was performed using SYBR Premix Ex Taq (TaKaRa) and an Eco Real-time PCR system (Illumina, CA, USA). All the reactions were performed at an annealing temperature of 60°C for 50 cycles. The primer sequences were—Xist forward 5′-GCAGCTCTAAGAAGT-TCCGCATTGA-3′, reverse 5′-TGCCCCATCTCCACCTAAGG-3′; and β-actin forward 5′-GCCGACAGGATGCAGAAGGA-3′, reverse 5′-GGGGCCGGACTCGTCGTACT-3′.
Screening for the most effective anti-Xist siRNA
Human embryonic kidney-derived HEK 293T cells were grown at 37°C in Dulbecco’s modified Eagle medium (DMEM; Sigma, USA) containing 10% heat-inactivated fetal bovine serum (FBS; Gibco-BRT, NY, USA) supplemented with L-glutamine (1 mmol/l), streptomycin (100 µg/ml), and penicillin (100 U/ml). The cells were seeded into 24-well plates at a density of 0.5–2.0 × 105 cells/well with fresh medium (500 μl/well) without antibiotics, 24 h prior to transfection.
HEK 293 cells were transfected with 0.5 μl of 20 μM siRNA and 0.8 μg of target-EGFP fusion gene expression plasmid with Lipofectamine 2000 (Invitrogen, USA) in accordance with the manufacturer’s instructions. The cells were observed under a fluorescence microscope 48 h after co-transfection. For flow cytometry analysis, cells were harvested 48 h after co-transfection and digested with 0.25% trypsin, washed twice with phosphate buffered saline (PBS), and resuspended in PBS to measure the fluorescence intensity using a FACScan flow cytometer with emission at 507 nm and excitation at 488 nm (BD, NJ, USA). About 105 cells per sample were counted and analyzed with CellQuest software, using non-transfected HEK 293T cells as control.
Female pig embryonic fibroblasts were grown at 39°C in DMEM (Sigma, USA) containing 10% heat-inactivated FBS (Gibco-BRT, NY, USA) supplemented with L-glutamine (1 mmol/l), streptomycin (100 µg/ml), and penicillin (100 U/ml). The cells were seeded into 24-well plate at a density of 0.5–2.0 × 105 cells/well with fresh medium (500 μl/well) without antibiotics 24 h prior to transfection. Cells were transfected with 0.5 µl of 20 µM siRNA mixed with Lipofectamine 2000 (Invitrogen, USA) in accordance with the manufacturer’s instructions. The cells were collected for real-time PCR analysis 48 h after transfection.
siRNA injection
Microinjection of siRNA1 was performed with a micropipette driven by a Piezo (PiezoXpert, Eppendorf, Germany). SCNT embryos at 6–7 h post-activation were injected with 10 pl of 5 µM [11] or 50 µM siRNA. Injected embryos were cultured in vitro in PZM3 at 39°C, 5% CO2, 7% O2, 88% N2, and 100% humidity.
Embryo culture and analysis of in vitro developmental indexes
Embryos were cultured under similar conditions and in accordance with the protocol described below. Injected cloned embryos were transferred to PZM3 and cultured at 39°C, 5% CO2, 7% O2, 88% N2, and 100% humidity. The time of embryo activation was defined as 0 h. The cleavage rate and blastocyst rate of cultured embryos was assessed at 24 h and 168 h. The total cell number of blastocysts was counted at 168 h by staining the embryos with 1 μg/ml Hoechst 33342 stain and viewing the cell nuclei under a fluorescence microscope.
Embryo transfer
After culturing under similar conditions for 4–6 h (at 14–16 h post-activation), siRNA1-injected and non-injected cloned embryos at the 1–2 cell stage were loaded into a transparent transfer tube and kept in a portable incubator (Minitube, WI, USA.) during transportation to the farm where the recipient sows were housed. Landrace sows in parity 2–5, from a same farm, with a similar genetic background, and which showed a natural standing estrus within 40–42 h prior to embryo transfer were used as embryo recipients. The sows were anesthetized with an anesthetic (Quanmianbao) consisting of ketamine (25 mg/kg body weight) and xylazine (1.1 mg/kg body weight) for induction and 3% isoflurane for maintenance. The ovaries and oviducts were exposed by making ~7-cm incision along the midline of the sow’s abdomen between the last two pairs of teats. Follicles in the ovaries were examined to determine the recipient’s ovulation status by criteria similar to that previously reported [14]. The cloned embryos with 0.1 ml culture medium were delivered directly into the recipient’s oviduct (at 2/3 length of the oviduct) using a 1-ml syringe attached to a transparent transfer tube. The transfer tube was examined subsequently under a microscope to ensure the transfer of all the embryos.
Diagnosis of recipient pregnancy and delivery of cloned piglets
The pregnancy status of the recipient sows was monitored using an ultrasound machine equipped with a convex transducer at approximately one month after embryo transfer. If spontaneous farrowing did not occur until gestation day 116, the recipient sows were injected with a prostaglandin analogue (cloprostenol, 200 µg/recipient) to induce delivery. After 24 h of injection, if the recipients still did not start to farrow, caesarean section was performed to deliver the cloned piglets. The number of total born/born alive/healthy cloned piglets in each litter was recorded.
Statistical analysis
For analysis of cleavage rate, blastocyst rate, pregnancy rate, and farrowing rate of recipients; and rate of transferred cloned embryos developing into full term/live/healthy cloned piglets, the GENMOD procedure using a Poisson distribution and logit link function in the SAS software (SAS Insititute, Cary, NC, USA) was used to determine differences between two groups. The number of cells per blastocyst was reported as the mean ± standard error of the mean (SEM). Differences between the two groups in the total number of cells per blastocyst were analyzed by t-test.
Results
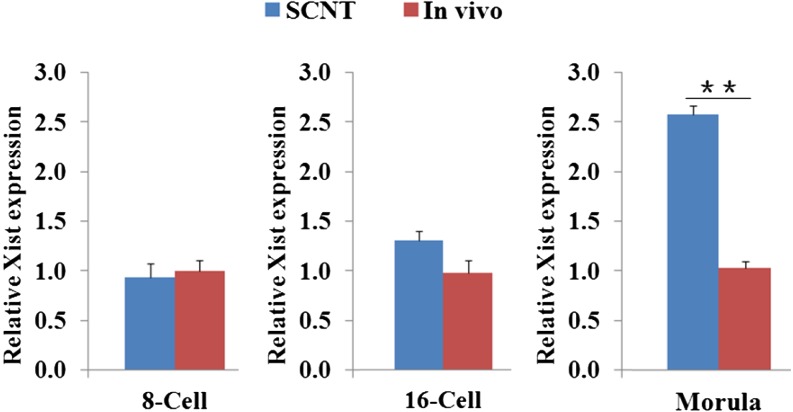
To investigate whether the Xist expression level is higher in cloned pig embryos than in IVV-derived embryos, the relative Xist mRNA abundance at the 8-cell, 16-cell, and morula stages was compared between pig SCNT embryos and IVV embryos (Fig. 1). The results indicated that at the morula stage, the Xist mRNA level in SCNT embryos was significantly higher (P < 0.01) than that in the IVV embryos. This suggests that Xist expression is abnormally elevated in early pig SCNT embryos, similar to that in mouse SCNT embryos [9].
Fig. 1.
Comparison of Xist mRNA levels at different pre-implantation stages in pig SCNT and IVV-derived embryos. Xist mRNA levels in the IVV group were measured from 10–15 embryos (in theory, containing a mixture of male and female embryos at a ratio of 1:1), which were collected at the 8-cell (72 h post fertilization), 16-cell (96 h post fertilization), and morula (120 h post fertilization) stages. Xist mRNA levels in the SCNT group were measured from a mixture of 10–18 embryos (male:female = 1:1), which were collected at the 8-cell (72 h post activation), 16-cell (96 h post activation), and morula (120 h post activation) stages. At each stage, the Xist mRNA level of the SCNT group was normalized to that of the IVV group, which was defined as 1. ** The mean values, calculated from 3 replicates, are significantly different between the 2 groups (P < 0.01).
To design siRNAs for knockdown of porcine Xist expression, we identified the genomic DNA sequence of porcine Xist by electronic cloning, and submitted the identified sequence to GenBank (Accession Number: KC149530). Three siRNAs were designed that matched with the sequences on exon 1, 5, and 6 of porcine Xist, respectively (Fig. 2A).
Fig. 2.
Screening for the most effective anti-Xist siRNA. A: Structural illustration of the porcine Xist gene showing the target site of the 3 designed anti-Xist siRNAs. B: Structural illustration of the plasmid expressing anti-Xist siRNA target sequences fused with the EGFP gene. The anti-Xist siRNA target site is about 400 bp, composed of partial sequences from exons 1, 5, and 6 of the pig Xist, and covers the target sequences of 3 designed anti-Xist siRNAs. C: Fluorescence microscopy analysis of EGFP expression, 48 h after co-transfection with anti-Xist siRNA target-EGFP fusion gene expression plasmid and siRNAs into HEK 293 cells. D: Flow cytometry of relative EGFP+ cell percentage, 48 h after co-transfection with the anti-Xist siRNA target-EGFP fusion gene expression plasmid and siRNAs into HEK 293 cells. E: Inhibition of Xist expression in female pig fibroblasts by 3 different siRNAs. ** The mean values, calculated from 3 replicates, are significantly different between the 2 groups (P < 0.01).
To determine the most effective anti-Xist siRNA, a target plasmid that expresses the siRNA target sequences fused with the EGFP gene was constructed (Fig. 2B), and was co-transfected with anti-Xist siRNA into HEK 293 cells to compare the interfering effect of 3 different siRNAs. The results showed that siRNA1 and siRNA2 blocked the expression of their target by about 55–65%, while siRNA3 only inhibited the expression of its target by about 30% (Fig. 2C and 2D).
The effects of the 3 siRNAs on suppression of porcine Xist expression were observed after transfecting them into female porcine fibroblasts (Fig. 2E). We found that siRNA1 was the most effective siRNA among the 3 tested anti-Xist siRNAs, because it decreased porcine Xist expression by ~80% (Fig. 2E). According to the results shown in Figs. 2C, 2D, and 2E, siRNA1 was chosen for the subsequent experiments.
RNAi-mediated knockdown of Xist could remarkably enhance the developmental competence of only the cloned male mouse embryos, whereas it did not affect the cloned female mouse embryos due to an inability to mimic normal monoallelic Xist expression [11, 15]. Therefore, we used only male pig SCNT embryos to test the effects of siRNA1 on reduction of Xist expression level, in accordance with the previously described procedure [11]. The results indicated that the siRNA1 (5 µM) injected into SCNT embryos effectively blocked Xist expression by about 73% and 48% at the 4-cell and 16-cell stages, respectively, but not at the morula stage (Fig. 3). When the concentration of injected siRNA1 was increased to 50 µM, a similar result was noted (Fig. 3). These results suggest that the injected anti-Xist siRNA1 has an interfering effect that lasts for approximately 4 d (from 1-cell stage to 16-cell stage), but it is not able to significantly inhibit Xist expression in injected embryos at 5 d post-injection (i.e., at the morula stage).
Fig. 3.
Inhibition of Xist expression at different pre-implantation stages of male pig SCNT embryos injected with anti-Xist siRNA1. Xist mRNA levels in each group were measured from a mixture of 10–20 embryos collected at the 4-cell (48 h post activation), 16-cell (96 h post activation), and morula (120 h post activation) stages. At each stage, Xist mRNA levels in the 5 µM and 50 µM siRNA1-injected groups were normalized to that in the non-injected (control) group, which was defined as 1. ** The mean values, calculated from 3 replicates, are significantly different between the 2 groups (P < 0.01).
Chemical modification (CM) has been shown to enhance the stability of siRNA, and hence, prolong its silencing effect in mammalian cells [16, 17]. Therefore, we synthesized CM anti-Xist siRNA1 and injected it into cloned male porcine embryos to investigate whether this would decrease Xist mRNA level. The results showed that the CM siRNA1 significantly downregulated Xist expression at the 4-cell and 16-cell stages, but lost the blocking activity at the morula stage (Fig. 4). This result suggests that CM of siRNA1 is unable to prolong its effect on Xist expression in cloned embryos.
Fig. 4.
Inhibition of Xist expression at different pre-implantation stages of male pig SCNT embryos injected with CM siRNA1. Xist mRNA levels in each group were measured from a mixture of 10–20 embryos collected at the 4-cell (48 h post activation), 16-cell (96 h post activation), and morula (120 h post activation) stages. At each stage, Xist mRNA levels in the CM siRNA1-injected group were normalized to that in the non-injected (control) group, which was defined as 1. * The mean values, calculated from 3 replicates, are significantly different between the 2 groups (P < 0.05); ** The mean values, calculated from 3 replicates, are significantly different between the 2 groups (P < 0.01).
Although the above results show that injected anti-siRNA1 was unable to decrease Xist mRNA levels at the morula stage of cloned porcine embryos, it might still affect the developmental potential of injected SCNT embryos, because it has the ability to significantly inhibit Xist expression at 4-cell and 16-cell stages (Fig. 3). To investigate the influence of knockdown of Xist expression on the in vitro developmental ability of cloned pig embryos, anti-Xist siRNA1 (5 µM) was microinjected into 1-cell-stage male SCNT embryos, which were then cultured in vitro for 7 days to evaluate their developmental indexes (Table 1). The results indicated that non-injected and siRNA1-injected cloned embryos were not different in cleavage rate or blastocyst rate, yet SCNT embryos injected with siRNA1 had a significantly higher total cell number at the blastocyst stage (47.93 ± 4.52) than did the control SCNT embryos (31.29 ± 3.92; Table 1). This suggests that inhibition of Xist expression can enhance the in vitro developmental potential of cloned male pig embryos.
Table 1. Comparison of in vitro developmental rate between non-injected and siRNA1-injected male pig SCNT embryos.
| Groups | No. of cloned embryos | No. of cleaved embryos (%) | No. of blastocysts (%) | Average total cell No. per blastocyst |
| Non-injected | 264 | 184 (69.69) | 42 (15.91) | 31.29 ± 3.92 A |
| siRNA1-injected | 243 | 189 (77.77) | 38 (15.64) | 47.93 ± 4.52 B |
Values in the same column with different superscripts are statistically different (P < 0.05) from each other.
To examine the effect of blockage of Xist expression on the in vivo full term developmental rate of cloned pig embryos, 5 µM siRNA1-injected male SCNT embryos and non-injected control male SCNT embryos were transferred into recipient sows to compare their developmental efficiency. The results showed that surrogate sows who received siRNA1-injected SCNT embryos tended to have higher rates of pregnancy (69.23%) and farrowing (38.46%) than the sows who received the control SCNT embryos did (61.54% and 30.77%, respectively). The siRNA1-injected cloned embryos also tended to have a higher rate of developing into full term (0.53%), live (0.43%), or healthy (0.17%) cloned piglets than the control SCNT embryos did (0.48%, 0.32%, and 0.08%, respectively). However, the differences in all these measured developmental indexes between siRNA1-injected and non-injected cloned embryos did not reach statistical significance (Table 2).
Table 2. Comparison of in vivo developmental rate between non-injected and siRNA1-injected male pig SCNT embryos.
| Groups | Total transferred cloned embryo No.a |
Total/pregnant/ farrowed recipient No. | Pregnancy/ farrowing rate (%) | Total born/born aliveb/ born healthy cloned piglet No.c |
Rate of transferred cloned embryos developing into full term/ live/healthy cloned piglets (%) |
| Non-injected | 6223 | 26/16/8 | 61.54/30.77 | 30/20/5 | 0.48/0.32/0.08 |
| siRNA1-injected | 6058 | 26/18/10 | 69.23/38.46 | 32/26/10 | 0.53/0.43/0.17 |
a Each recipient was transferred with 210–250 cloned embryos at 1-2 cell stage. b Total born = born alive + still born; c Born healthy = new born piglets show no malformations and with a birth weight over 0.8 kg. Values in the same column without superscripts are not statistically different (P > 0.05) from each other.
The present study used non-injected cloned embryos as control (Table 2), unlike the mock siRNA-injected SCNT embryos reported elsewhere [11]. This might result in underestimation of the effect of siRNA1-mediated Xist knockdown on the efficiency of pig cloning, since microinjection usually has an injurious effect on the development of manipulated embryos. To investigate the true effects of siRNA-based knockdown of Xist on the success rate of pig cloning, full term developmental competence between mock siRNA-injected and anti-Xist siRNA1-injected male pig SCNT embryos was compared in vivo. The results indicated that the birth rate of healthy cloned piglets in the siRNA1 injection group (0.26%) was significantly higher than that of the mock siRNA injection group (P = 0.0396), although other measured developmental indexes were not statistically significant between the two groups (Table 3). These results suggest that knockdown of Xist expression by injection of siRNA has a minor, yet statistically significant, effect on increasing the in vivo developmental rate of male pig SCNT embryos.
Table 3. Comparison of in vivo developmental rate between mock siRNA-injected and siRNA1-injected male pig SCNT embryos.
| Groups | Total transferred cloned embryo No.a |
Total/pregnant/ farrowed recipient No. |
Pregnancy/ farrowing rate (%) |
Total born/born aliveb/ born healthy cloned piglet No.c |
Rate of transferred cloned embryos developing into full term/ live/healthy cloned piglets (%) |
| Mock siRNA-injected | 2337 | 10/6/2 | 60/20 | 7/5/0 | 0.30/0.21/0 A |
| siRNA1-injected | 2309 | 10/7/4 | 70/40 | 9/7/6 | 0.39/0.30/0.26 B |
a Each recipient was transferred with 210–250 cloned embryos at 1-2 cell stage. b Total born = born alive + still born; c Born healthy = new born piglets show no malformations and with a birth weight over 0.8 kg. Values in the same column with different superscripts are statistically different (P < 0.05) from each other.
Discussion
In this study, we found that Xist transcript levels at the pre-implantation stage of pig SCNT embryos were significantly higher than that at the pre-implantation stage of IVV-derived embryos. A similar finding in cloned pig embryos was reported by another study [18]. These data indicate abnormal upregulation of Xist in the early-stage cloned pig embryos, similar to that in cloned mouse embryos. The aberrant elevation of Xist mRNA levels in pre-implanted cloned pig embryos may be caused by the loss of maternal Xist-repressing imprint, which is generated during oogenesis but is absent in SCNT embryos due to enucleation of oocytes [10].
Although both mouse and pig SCNT embryos have abnormal Xist expression profile at the pre-implantation stage as compared to IVV-derived embryos, mouse SCNT embryos naturally normalize the Xist expression after implantation [11, 15], while in cloned pigs, the abnormal Xist expression pattern persists at the post-implantation stage of the embryos and even at the postnatal stage [19, 20]. This suggests that the system for regulation of Xist expression during the post-implantation stage is different between mouse and pig SCNT embryos.
Both mouse and pig SCNT embryos exhibited significantly higher Xist transcript levels at the morula stage than did the IVV-derived embryos. However, the silencing effect of anti-Xist siRNA injected at the 1-cell stage needed to be maintained for at least 3 days in mouse and 5 days in pig SCNT embryos for effective Xist knockdown at the morula stage. This is because mouse and pig embryos required 3 and 5 days, respectively, to develop from the 1-cell stage to the morula stage. Studies have shown that the effect of CM or non-CM siRNA on repression of target gene expression can last only for 2–5 days [21,22,23]. In the present study, the interfering effect of anti-Xist siRNA lasted only for a maximum of 4 days in the cloned pig embryos. Therefore, it is not surprising that anti-Xist siRNA injected at the 1-cell stage can effectively inhibit abnormal Xist expression at the morula stage in mouse SCNT embryos [11], but not in pig SCNT embryos, as observed in the present study. This may be the major reason why siRNA-mediated Xist repression remarkably increased the mouse cloning efficiency by more than 10-fold [11], but only slightly improved the survival rate of cloned pig embryos in the present study.
To prolong the silencing effect of injected anti-Xist siRNA, we increased the concentration of injected anti-Xist siRNA by 10-fold, or injected CM anti-Xist siRNA. However, these methods still could not effectively suppress Xist expression at the morula stage in cloned pig embryos. Since short-hairpin RNA (shRNA) expressed from plasmids can provide more persistent and stable gene silencing than siRNA [24, 25], the use of anti-Xist shRNA expression plasmids to replace anti-Xist siRNA for injection may result in significant blockage of Xist expression at the morula stage of pig SCNT embryos. In addition, postponing the injection of anti-Xist agents from the 1-cell to the 2- or 4-cell stage may also effectively decrease Xist transcript levels at the morula stage of cloned pig embryos. Disruption of Xist expression at the morula stage can also be achieved by knockout of the Xist gene in pig SCNT embryos, similar to that in mouse SCNT embryos [9], although mutation of the Xist gene is undesired when wild-type cloned animals are needed for subsequent use. These approaches to effectively inhibit Xist expression at the morula stage of pig SCNT embryos need further testing in future studies for more effective enhancement of pig cloning efficiency.
In summary, we have demonstrated that pig SCNT embryos, similar to mouse SCNT embryos, express higher Xist mRNA levels at the pre-implantation stage than the IVV-derived embryos do. Injection of anti-Xist siRNA into pig SCNT embryos at the 1-cell stage effectively inhibits Xist expression through the 16-cell stage, which significantly increases the number of cells per blastocyst and improves the birth rate of healthy cloned piglets. This study provides valuable data for understanding the role of Xist in SCNT embryo development, and presents a method for increasing the success rate of pig SCNT.
Acknowledgments
This work was supported by 4 grants received from Department of Science and Technology of Guangdong province, China (Grant numbers: 2015TX01N081, 2014A030313464, 2011B060400029, 2016B020233006).
References
- 1.Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 1961; 190: 372–373. [DOI] [PubMed] [Google Scholar]
- 2.Takagi N, Sasaki M. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature 1975; 256: 640–642. [DOI] [PubMed] [Google Scholar]
- 3.Augui S, Nora EP, Heard E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat Rev Genet 2011; 12: 429–442. [DOI] [PubMed] [Google Scholar]
- 4.Xue F, Tian XC, Du F, Kubota C, Taneja M, Dinnyes A, Dai Y, Levine H, Pereira LV, Yang X. Aberrant patterns of X chromosome inactivation in bovine clones. Nat Genet 2002; 31: 216–220. [DOI] [PubMed] [Google Scholar]
- 5.Bao S, Miyoshi N, Okamoto I, Jenuwein T, Heard E, Azim Surani M. Initiation of epigenetic reprogramming of the X chromosome in somatic nuclei transplanted to a mouse oocyte. EMBO Rep 2005; 6: 748–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li S, Li Y, Du W, Zhang L, Yu S, Dai Y, Zhao C, Li N. Aberrant gene expression in organs of bovine clones that die within two days after birth. Biol Reprod 2005; 72: 258–265. [DOI] [PubMed] [Google Scholar]
- 7.Nolen LD, Gao S, Han Z, Mann MR, Gie Chung Y, Otte AP, Bartolomei MS, Latham KE. X chromosome reactivation and regulation in cloned embryos. Dev Biol 2005; 279: 525–540. [DOI] [PubMed] [Google Scholar]
- 8.Shao GB, Ding HM, Gong AH, Xiao DS. Inheritance of histone H3 methylation in reprogramming of somatic nuclei following nuclear transfer. J Reprod Dev 2008; 54: 233–238. [DOI] [PubMed] [Google Scholar]
- 9.Inoue K, Kohda T, Sugimoto M, Sado T, Ogonuki N, Matoba S, Shiura H, Ikeda R, Mochida K, Fujii T, Sawai K, Otte AP, Tian XC, Yang X, Ishino F, Abe K, Ogura A. Impeding Xist expression from the active X chromosome improves mouse somatic cell nuclear transfer. Science 2010; 330: 496–499. [DOI] [PubMed] [Google Scholar]
- 10.Oikawa M, Inoue K, Shiura H, Matoba S, Kamimura S, Hirose M, Mekada K, Yoshiki A, Tanaka S, Abe K, Ishino F, Ogura A. Understanding the X chromosome inactivation cycle in mice: a comprehensive view provided by nuclear transfer. Epigenetics 2014; 9: 204–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matoba S, Inoue K, Kohda T, Sugimoto M, Mizutani E, Ogonuki N, Nakamura T, Abe K, Nakano T, Ishino F, Ogura A. RNAi-mediated knockdown of Xist can rescue the impaired postimplantation development of cloned mouse embryos. Proc Natl Acad Sci USA 2011; 108: 20621–20626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi J, Zhou R, Luo L, Mai R, Zeng H, He X, Liu D, Zeng F, Cai G, Ji H, Tang F, Wang Q, Wu Z, Li Z. Influence of embryo handling and transfer method on pig cloning efficiency. Anim Reprod Sci 2015; 154: 121–127. [DOI] [PubMed] [Google Scholar]
- 13.Li Z, Zeng F, Meng F, Xu Z, Zhang X, Huang X, Tang F, Gao W, Shi J, He X, Liu D, Wang C, Urschitz J, Moisyadi S, Wu Z. Generation of transgenic pigs by cytoplasmic injection of piggyBac transposase-based pmGENIE-3 plasmids. Biol Reprod 2014; 90: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koo OJ, Park HJ, Kwon DK, Kang JT, Jang G, Lee BC. Effect of recipient breed on delivery rate of cloned miniature pig. Zygote 2009; 17: 203–207. [DOI] [PubMed] [Google Scholar]
- 15.Oikawa M, Matoba S, Inoue K, Kamimura S, Hirose M, Ogonuki N, Shiura H, Sugimoto M, Abe K, Ishino F, Ogura A. RNAi-mediated knockdown of Xist does not rescue the impaired development of female cloned mouse embryos. J Reprod Dev 2013; 59: 231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braasch DA, Jensen S, Liu Y, Kaur K, Arar K, White MA, Corey DR. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry 2003; 42: 7967–7975. [DOI] [PubMed] [Google Scholar]
- 17.Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA 2003; 9: 1034–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park CH, Jeong YH, Jeong YI, Lee SY, Jeong YW, Shin T, Kim NH, Jeung EB, Hyun SH, Lee CK, Lee E, Hwang WS. X-linked gene transcription patterns in female and male in vivo, in vitro and cloned porcine individual blastocysts. PLoS ONE 2012; 7: e51398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan L, Wang A, Yao C, Huang Y, Duan F, Lv Q, Wang D, Ouyang H, Li Z, Lai L. Aberrant expression of Xist in aborted porcine fetuses derived from somatic cell nuclear transfer embryos. Int J Mol Sci 2014; 15: 21631–21643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang L, Lai L, Samuel M, Prather RS, Yang X, Tian XC. Expression of X-linked genes in deceased neonates and surviving cloned female piglets. Mol Reprod Dev 2008; 75: 265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tuschl T. Expanding small RNA interference. Nat Biotechnol 2002; 20: 446–448. [DOI] [PubMed] [Google Scholar]
- 22.Holen T, Amarzguioui M, Wiiger MT, Babaie E, Prydz H. Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res 2002; 30: 1757–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amarzguioui M, Holen T, Babaie E, Prydz H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res 2003; 31: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu S, Jin L, Zhang F, Huang Y, Grimm D, Rossi JJ, Kay MA. Thermodynamic stability of small hairpin RNAs highly influences the loading process of different mammalian Argonautes. Proc Natl Acad Sci USA 2011; 108: 9208–9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh S, Narang AS, Mahato RI. Subcellular fate and off-target effects of siRNA, shRNA, and miRNA. Pharm Res 2011; 28: 2996–3015. [DOI] [PubMed] [Google Scholar]