Abstract
The testis-specific cytoplasmic poly(A) polymerase PAPOLB/TPAP is essential for spermatogenesis. Although this enzyme is responsible for poly(A) tail extension of a subset of mRNAs in round spermatids, the stability and translational efficiency of these mRNAs are unaffected by the absence of PAPOLB. To clarify the functional importance of this enzyme’s adenylation activity, we produced PAPOLB-null mice expressing a polyadenylation-defective PAPOLB mutant (PAPOLBD114A), in which the catalytic Asp at residue 114 was mutated to Ala. Introducing PAPOLBD114A failed to rescue PAPOLB-null phenotypes, such as reduced expression of haploid-specific mRNAs, spermiogenesis arrest, and male infertility. These results suggest that PAPOLB regulates spermatogenesis through its adenylation activity.
Keywords: Cytoplasmic polyadenylation, PAPOLB, Poly(A), Spermatogenesis
Poly(A) tails present at the 3'-end of eukaryotic mRNAs are central in the life of mRNAs by regulating stability, translation initiation and termination, and degradation [1,2,3,4]. Thus, controlling poly(A) tail length is one of the post-transcriptional regulations of gene expression. In mammalian cells, primary transcripts are generally cleaved within 30 nucleotides (nt) downstream of the polyadenylation signal, AAUAAA, followed by the addition of ~250 nt poly(A) tails by poly(A) polymerase α (PAPOLA) in a co-transcriptional manner [5, 6]. Besides nuclear polyadenylation, cytoplasmic polyadenylation occurs during oocyte maturation and early embryogenesis, where transcription is entirely quiescent. Maternal mRNAs transcribed in earlier stages are deadenylated upon their exit from the nucleus (~A10) and are stored in a translationally inactive state. During specific developmental stages, subsets of mRNAs undergo cytoplasmic poly(A) tail extension (~A200) as a prerequisite for translational activation [7,8,9,10,11].
Spermatogenesis is a specialized cellular differentiation process that produces spermatozoa, which comprises spermatogonial proliferation, two meiotic divisions, and drastic morphological remodeling during spermiogenesis, the haploid phase of spermatogenesis. This process is accomplished by a controlled, stage-specific gene expression program regulated at the transcriptional, post-transcriptional, and translational levels [12,13,14,15,16,17]. At least two distinct patterns of poly(A) tail length changes have been described during spermatogenesis. Haploid-specific mRNAs encoding structural proteins required for the formation of flagella and highly compacted nuclei are transcribed in round spermatids and are stored as translationally inert messenger ribonucleoprotein particles (mRNPs) with long poly(A) tracts (~A180) for several days. Subsequent translational activation of the mRNAs during later spermiogenesis accompanies the generation of partially deadenylated species ranging from A150 to A30 [18, 19]. Conversely, poly(A) tails of some mRNAs are 50–150 nt longer in round spermatids than in meiotic pachytene spermatocytes [13, 20,21,22,23].
We previously reported that ablating the testis-specific cytoplasmic poly(A) polymerase PAPOLB/TPAP results in impaired spermiogenesis and male infertility, which can be recovered by transgenic re-expression [23, 24]. This enzyme is responsible for cytoplasmic poly(A) tail extension of certain transcription factor mRNAs in round spermatids [23]. However, additional poly(A) tail elongation appeared not to be responsible for either stability or translational efficiency of the substrate mRNAs (Fig. 1) [23]. These findings prompted the hypothesis that PAPOLB polyadenylation activity is dispensable for spermatogenesis. To address this, we produced and analyzed PAPOLB-null mice expressing a polyadenylation-defective PAPOLB mutant.
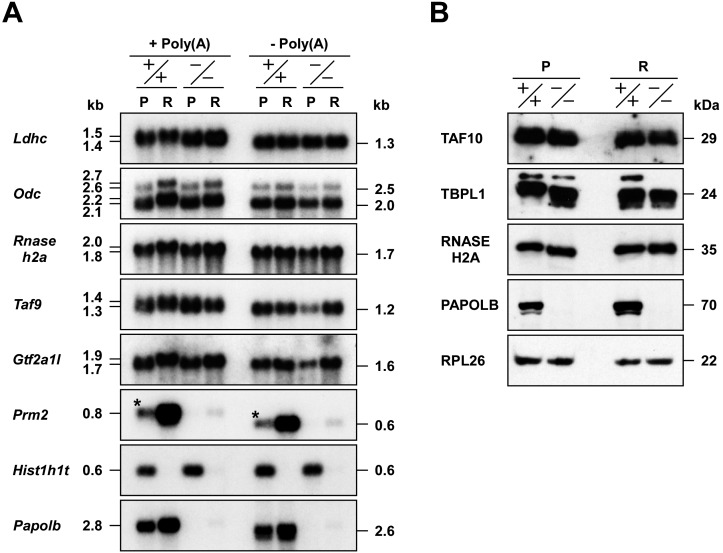
Fig. 1.
Additional poly(A) tail extension by PAPOLB and its effect on mRNA metabolism. Pachytene spermatocytes (P) and round spermatids (R) of wild-type (+/+) and PAPOLB-null (–/–) mice were purified by unit gravity sedimentation on 2–4% BSA gradients. (A) Northern blot analysis. Total RNAs were incubated with RNase H in the presence [–Poly(A)] or absence [+Poly(A)] of oligo(dT)15 and subjected to Northern blot analysis with the cDNA probes indicated. Hist1h1t and Prm2 were used as meiosis- and haploid-specific markers, respectively. Asterisks represent Prm2 mRNA derived from round spermatids contaminated during germ cell fractionation. (B) Immunoblot analysis. Protein extracts were analyzed by immunoblotting using the antibodies indicated. RPL26 was used as a control.
Materials and Methods
Plasmid construction
Expression plasmids encoding N-terminally FLAG- and HA-tagged PAPOLB and polyadenylation-defective PAPOLBD114A mutant, in which Asp at position 114 (one of the three putative catalytic Asp residues) was replaced with Ala, were constructed as follows. The cDNA fragment encoding the entire open reading frame of PAPOLB was amplified by polymerase chain reaction (PCR) using the primers 5'-GGAATTCATGATGCCATTTGCGTGACC-3' and 5'-TCCTCGAGCTAGACTCCTAGTATAGGATTGG-3', and the cloned mouse cDNA as a template [22]. After digestion with EcoRI and XhoI, the fragment was inserted into pcDNA3-FLAGHAN, which is designed to express the protein with N-terminal FLAG and HA tags (PAPOLB/pcDNA3-FLAGHAN). The plasmid encoding FLAG- and HA-tagged PAPOLBD114A (PAPOLBD114A/pcDNA3-FLAGHAN) was generated by PCR-based mutagenesis using the primer pair (mutated nucleotides are underlined): 5'-CGAAAGGTGCAGCTATCGATGCTTTG-3' and 5'-CAAAGCATCGATAGCTGCACCTTTCG-3' and PAPOLB/pcDNA3-FLAGHAN as a template.
Polyadenylation assay
Poly(A) polymerase activity was measured as previously described [22]. Briefly, recombinant enzymes were produced in rabbit reticulocyte lysates using a TNT T7 Quick Coupled Transcription/Translation system (Promega, Madison, WI, USA). PAPOLB/pcDNA3-FLAGHAN or PAPOLBD114A/pcDNA3-FLAGHAN (0.5 µg) was used as a template in a 50-μl reaction mixture. The polyadenylation assay was carried out in 100 μl of 50 mM Tris/HCl, pH 8.3, 7% glycerol, 0.3% polyvinyl alcohol, 40 mM KCl, 0.5 mM MnCl2, 25 mM (NH4)2SO4, 0.25 mM dithiothreitol (DTT), 50 units of RNase OUT (Thermo Fischer Scientific, Waltham, MA, USA), 0.1 mM ATP, 4 μl of [α-32P]ATP (29.6 TBq/mmol; PerkinElmer, Waltham, MA, USA), 4 μg of oligo(A)12, and 12 μl of in vitro translation product. The mixtures were incubated at 30°C and aliquots (5 μl) of reaction mixtures were spotted onto Whatman DE-81 papers, dried, and washed five times with 0.1 M sodium phosphate buffer, pH 7.0, and once with ethanol. The incorporation of radiolabeled AMP was quantified by a liquid scintillation counter (Beckman Coulter, Indianapolis, IN, USA).
Transgenic construct
The 7.1-kbp transgenic construct was prepared as follows. The 2.0-kbp SalI-BamHI and the 3.0-kbp HindIII-EcoRI fragments at the 5'- and 3'-ends, respectively, were cut from the wild-type Papolb transgenic construct [24]. The 0.9-kbp BamHI-ApaI fragment was prepared by restriction digestion of a 1.0-kbp fragment amplified by PCR using the primers 5'-TTGTCAGCAACAGGAGTGAAACTGG-3' and 5'-AAGGGCCCCTCTAGCTCCCGCGCCG-3'. The 1.2-kbp ApaI-HindIII fragment encoding the N-terminal region of FLAG- and HA-tagged PAPOLBD114A was prepared by digesting a 2.0-kbp PCR product amplified by using the primers 5'-AAGGGCCCGCCACCATGGACTACAAGGACGATGATG-3' and 5'-TCCTCGAGCTAGACTCCTAGTATAGGATTGG-3' and PAPOLBD114A/pcDNA3-FLAGHAN as a template. The 2.0-kbp SalI-BamHI and the 0.9-kbp BamHI-ApaI fragments were cloned into pBluescriptII SK (+) digested with SalI and ApaI, whereas the 1.2-kbp ApaI-HindIII and the 3.0-kbp HindIII-EcoRI fragments were inserted into the ApaI and EcoRI sites of pBluescriptII SK (+) followed by digestion with ApaI and NotI. The resulting 2.9-kbp SalI-ApaI and the 4.2-kbp ApaI-NotI fragments were cloned into SalI and NotI sites of pBluescriptII SK (+). The 7.1-kbp SalI-NotI fragment was purified by agarose gel electrophoresis after restriction digestion.
Generation of transgenic mice
The 7.1-kbp SalI-NotI fragment was injected into male pronuclei of C57BL/6 fertilized eggs at the Laboratory Animal Resource Center, University of Tsukuba. Transgenesis was verified by PCR of tail DNA using the primers 5'-ATGGACTACAAGGACGATGATG-3' and 5'-CGAAGCTTGCACCTTTCGTGTGTACTCC-3'. Screening of 72 male pups identified five mice positive for the transgene. The founder mice were bred with C57BL/6 females (Japan SLC, Shizuoka, Japan) to produce F1 heterozygotes. Two lines of transgenic mice, designated as Tg16 and Tg66, were maintained by mating with C57BL/6 mice. To produce PAPOLB-deficient mice (Papolb–/–) expressing PAPOLBD114A, Tg16 males were initially crossed with Papolb–/– females. The resulting Tg16+/–/Papolb+/– F1 males were further mated with Papolb–/– females to obtain Tg16+/–/Papolb–/– F2 mice.
Antibodies
Antibodies against mouse RNASEH2A and mouse TAF10 were produced in rabbits by immunizing with recombinant proteins followed by affinity-purification. Briefly, 6 × His- and thioredoxin (TRX)-tagged mouse RNASEH2A at residues Met45-Leu301 or the entire region of mouse TAF10 was produced in Escherichia coli BL21 (DE3). The recombinant proteins were purified on a Ni-NTA His column (Merck Millipore, Billerica, MA, USA), emulsified with Freund’s complete or incomplete adjuvant (Becton Dickinson, Franklin Lakes, NJ, USA), and injected intradermally into female New Zealand White rabbits (Japan SLC) [25]. Each antiserum was fractionated with ammonium sulfate (0–40% saturation), followed by immunoaffinity chromatography on a Sepharose 4B (GE Healthcare, Piscataway, NJ, USA) column conjugated with the same protein region fused to glutathione S-transferase (GST) [26]. The anti-PAPOLB antibody was prepared as described previously [22]. A mouse monoclonal antibody against mouse A-kinase anchor protein 4 (AKAP4; sc-135827) and goat polyclonal antibodies against human phosphoglycerate kinase 2 (PGK2; sc-133905), human protamine 2 (PRM2; sc-23104), and mouse transition protein 2 (TNP2; sc-21106) were purchased from Santa Cruz Biotechnology (Dallas, TX, USA). A mouse monoclonal antibody against β-actin (ACTB; A5441), rabbit polyclonal antibody against human ribosomal protein L26 (RPL26; IHC-00093), rabbit polyclonal antibody against human TATA-binding protein-like 1 (TBPL1; GTX104694), and rat monoclonal antibody against hemagglutinin peptide (HA; 11867423001) were obtained from Sigma-Aldrich (St. Louis, MO, USA), Bethyl Laboratories (Montgomery, TX, USA), GeneTex (Hsinchu, Taiwan), and Roche Life Science (Indianapolis, IN, USA), respectively.
Northern blot analysis
Total RNAs were prepared from mouse testicular tissues or purified populations of spermatogenic cells [27] using ISOGEN kit (Nippon Gene, Toyama, Japan). RNase H digestion of poly(A) RNA in the presence of oligo(dT)15, which generates deadenylated mRNA, was carried out as described previously [22]. Briefly, total RNAs (10 μg) were incubated in a 50-μl mixture containing 40 mM Tris/HCl, pH 7.8, 4 mM MgCl2, 100 mM KCl, 1 mM DTT, 4% glycerol, 1 unit of RNase H (TAKARA Bio., Otsu, Japan), and 0.5 μg of oligo(dT)15. After incubation at 37°C for 1 h, the RNAs were extracted with phenol/chloroform, and precipitated with ethanol. The RNA samples (2–5 µg) were denatured with glyoxal, separated on agarose gels, and transferred onto Hybond-N+ nylon membranes (GE Healthcare) [22]. The blots were probed with 32P-labeled cDNA fragments.
Immunoblot analysis
Testicular tissues or purified germ cells were homogenized at 4°C in 20 mM Tris/HCl, pH 7.5, 150 mM NaCl, 0.5% Nonidet P-40, supplemented with 1 µg/ml leupeptin, 1 µg/ml pepstatin A, 1 mM DTT, and 0.5 mM phenylmethanesulfonyl fluoride (PMSF), using a Teflon-glass homogenizer (750 rpm, 10 strokes). After incubation at 4°C for 4 h, the homogenates were centrifuged at 13,400 × g for 10 min at 4°C. The supernatant solutions were used as protein extracts. Protein concentration was determined by using a Coomassie protein assay reagent kit (Thermo Fisher Scientific). Protein samples (5 µg) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes (Merck Millipore). After blocking with 2% skim milk or gelatin, the blots were probed with primary antibodies followed by incubation with horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA). The immunoreactive bands were visualized by an ECL or an ECL Prime Western blot detection kit (GE Healthcare).
Histological analysis
Testicular and epididymal tissues were fixed with Bouin’s fixative and embedded in paraffin. Paraffin sections (4 µm thick) were prepared by a MICROM HM340E (Microedge Instruments, White Rock, BC, Canada), mounted on slides, deparaffinized in xylene, and hydrated in a graded ethanol series. After staining with hematoxylin and eosin (Wako, Osaka, Japan), the slides were observed under a DM IRBE microscope (Leica Microsystems, Wetzlar, Germany).
Statistical analysis
Data are presented as mean ± SEM (n ≥ 3), unless stated otherwise. The Student’s t-test was used for statistical analysis; significance was assumed at P < 0.05.
Ethics statement
All animal experiments were approved and performed in accordance with the Guide for the Care and Use of Laboratory Animals at the University of Tsukuba (approval numbers 14-022 and 15-015).
Results
We previously found that PAPOLB catalyzes additional poly(A) tail extension of specific transcription factor mRNAs in round spermatids [23]. To examine whether cytoplasmic polyadenylation by PAPOLB is specific for transcription factor mRNAs, we analyzed the lactate dehydrogenase C subunit (Ldhc) and two forms of ornithine decarboxylase (Odc) mRNAs. Northern blot analysis showed that these mRNAs are ~100 nt longer in round spermatids than in pachytene spermatocytes of wild-type mice (Papolb+/+, +/+), as described previously [21, 28]. In contrast, no (for Ldhc) or slight (for Odc) increase in mRNA length was observed in round spermatids of PAPOLB-null mice (Papolb–/–, –/–). After RNase H digestion in the presence of oligo(dT)15, each mRNA decreased in length and their size was uniform among four cell types, indicating that the size differences are due to poly(A) tail elongation (Fig. 1A). Extensive analysis revealed that the ribonuclease H2 A subunit (Rnaseh2a), a male germ cell-specific paralog of general transcription factor IIA α/β subunit (Gtf2a1l/Alf/TfIIaτ), and TATA-binding protein-associated factor subunit 9 (Taf9) mRNAs are also regulated by PAPOLB. These results indicate that PAPOLB is capable of extending poly(A) tails of different mRNA classes. The levels of substrate mRNAs in round spermatids of Papolb–/– mice were comparable to those in Papolb+/+ mice, despite the incomplete poly(A) tail extension (Fig. 1A). We next examined protein levels encoded by two previously identified target mRNAs (Taf10 and Tbpl1) and Rnaseh2a mRNA. Though it has long been believed that there is a positive correlation between poly(A) tail length and translational efficiency [9], the amount of TAF10, TBPL1, and RNASEH2A protein in round spermatids was similar between Papolb+/+ and Papolb–/– mice (Fig. 1B). Sucrose gradient analyses also showed that additional polyadenylation of Tbpl1 and Rnaseh2a mRNAs in round spermatids did not greatly increase the ratio of mRNAs associated with translationally active polyribosomes (Supplementary Fig. 1: online only), as was reported for Ldhc mRNA [21]. Collectively, these results suggest that additional poly(A) tail extension by PAPOLB is not responsible for enhancing either stability or translation of the mRNAs examined.
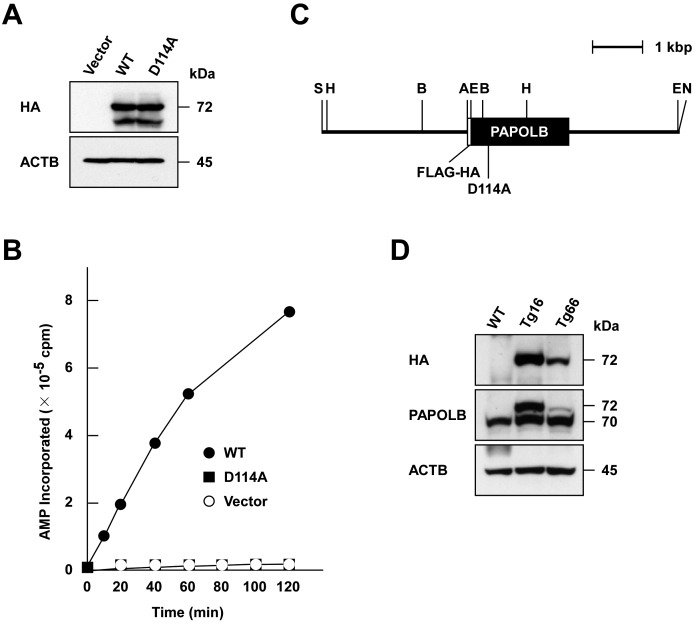
The results described above led us to speculate that PAPOLB regulates spermiogenesis independently of its polyadenylation activity. To address this possibility, we sought to examine whether PAPOLB-null phenotypes can be recovered by transgenically introducing polyadenylation-defective PAPOLB. First, we tested whether Asp at residue 114 (Asp114), one of three putative catalytic Asp residues conserved in the DNA polymerase β-like nucleotidyltransferase superfamily [22], is essential for polyadenylation activity. The N-terminally FLAG- and HA-tagged PAPOLBD114A mutant (hereafter, abbreviated as PAPOLBD114A), where Asp114 was mutated to Ala, was assayed for non-specific polyadenylation activity [22]. When the same amount of in vitro translation product was analyzed, wild-type PAPOLB was capable of adding AMP residues on oligo(A)12, whereas PAPOLBD114A mutant only showed activity comparable to background levels (Fig. 2A and B). We thus generated transgenic mice expressing PAPOLBD114A under control of the Papolb promoter (Fig. 2C). Immunoblot analysis of testicular extracts from two transgenic mouse lines, designated as Tg16 and Tg66, detected a 72-kDa protein band reactive to anti-HA antibody, which corresponds to FLAG- and HA-tagged PAPOLBD114A (Fig. 2D). Because Tg16 male mice produced mutant protein at levels equivalent to endogenous 70-kDa PAPOLB, we mainly used Tg16 for further experiments.
Fig. 2.
Generation of transgenic mice expressing polyadenylation-defective PAPOLBD114A. (A) Immunoblot analysis of proteins synthesized in vitro. N-terminally FLAG- and HA-tagged PAPOLB (WT) and PAPOLBD114A (D114A) were synthesized in rabbit reticulocyte lysates and were analyzed by immunoblotting using anti-HA antibody. ACTB was used as a control. (B) Polyadenylation assay. The polyadenylation activity of wild-type PAPOLB (closed circle) and PAPOLBD114A (closed square) was monitored by measuring the incorporation of AMP from [α-32P]ATP into an oligo(A)12 primer at the incubation times indicated. Empty vector (open circle) was used as a control. (C) Physical map of the transgenic construct. The protein-coding regions of FLAG and HA tags and PAPOLBD114A are indicated by open and filled boxes, respectively. The restriction enzymes are denoted as follows: A, ApaI; B, BamHI; E, EcoRI; H, HindIII; S, SalI; N, NotI. (D) Immunoblot analysis. Testicular protein extracts of wild-type (WT) and two transgenic mouse lines (Tg16 and Tg66) were probed with anti-HA and anti-PAPOLB antibodies. ACTB served as a control.
Both Tg16 male and female mice grew normally and were apparently normal in behavior, body weight, and health condition. When male fertility was tested by mating with wild-type females, the average litter size from Tg16 males (8.2 ± 0.4 pups/litter, n = 5) was comparable to that of wild-type matings (8.4 ± 0.4 pups/litter, n = 5, Fig. 3A). The testicular weights of Tg16 and wild-type littermates were also similar (90.3 ± 5.3 and 91.7 ± 4.0 mg in Tg16 and wild-type mice, respectively, n = 6, Fig. 3B). Histological analysis indicated that spermatogenesis and sperm migration, respectively, in the testis and epididymis of Tg16 mice were unaffected (Supplementary Fig. 2: online only). Normal progression of spermatogenesis to elongating/elongated spermatids in transgenic mice was also evidenced by the unaltered expression of postmeiotic mRNAs and the presence of translation-coupled deadenylated mRNA species (0.4 kb for Prm1 and Tnp1 and 0.6 kb for Prm2 and Tnp2, Fig. 3C) [18, 19]. Furthermore, the abundance of four testis-specific proteins, PRM2, TNP2, PGK2, and AKAP4, was similar among wild-type and two transgenic mouse lines (Fig. 3D). These results indicate that the transgenic expression of PAPOLBD114A in wild-type mice has no dominant-negative effect on spermatogenesis.
Fig. 3.
Characterization of transgenic mice expressing PAPOLBD114A. Three- to four-month-old mice were analyzed. (A) Fertility of male mice. Wild-type (WT) and Tg16 males were mated with 8-week-old C57BL/6 females and the pups were counted (5 litters). (B) Testicular weight. Testicular tissues of WT and Tg16 mice were weighed (n = 6). (C) Northern blot analysis. Total testicular RNAs of wild-type (WT) and two transgenic mouse lines (Tg16 and Tg66) were subjected to Northern blot analysis with the haploid-specific cDNA probes indicated. Actb served as a control. (D) Immunoblot analysis. Testicular protein extracts were examined by immunoblotting with the antibodies indicated. ACTB served as a control.
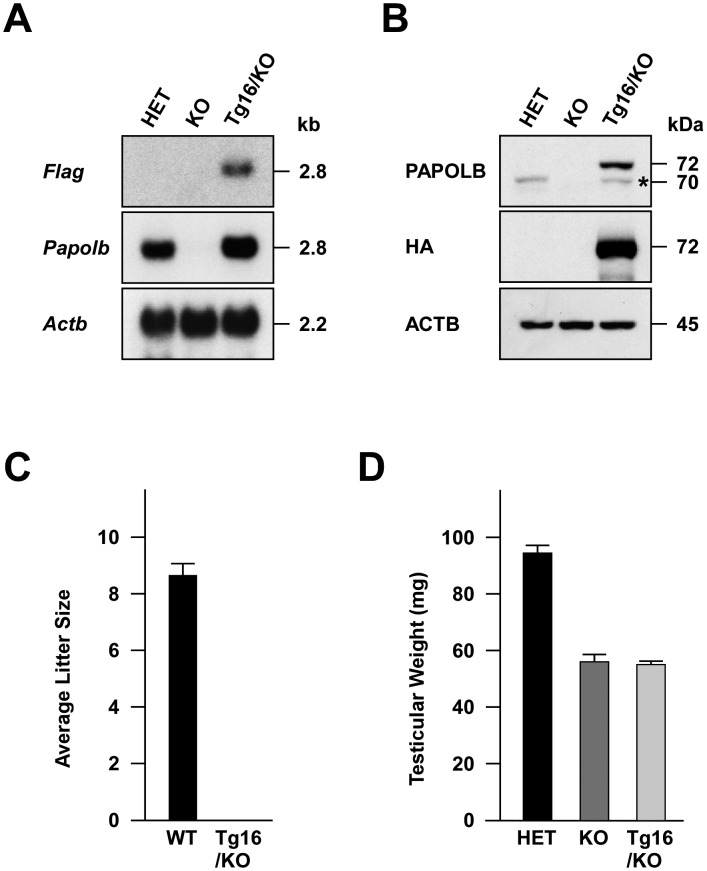
We then crossed Tg16 male mice with Papolb–/– females. The resulting Tg16+/–/Papolb+/– males were successively mated with Papolb–/– females to produce Tg16+/–/Papolb–/– mice. Northern blot analysis of testicular tissues using a Flag probe revealed transgene expression in Tg16+/–/Papolb–/– (Tg16/KO) but not in Papolb+/– (HET) and Papolb–/– (KO) mice (Fig. 4A). When hybridized with a Papolb-specific probe, the signal intensity of PapolbD114A mRNA was approximately two-fold stronger than that of the endogenous Papolb mRNA in Papolb+/– testes. Correspondingly, the 72-kDa FLAG- and HA-tagged PAPOLBD114A in Tg16+/–/Papolb–/– testis was more abundant than 70-kDa wild-type PAPOLB in Papolb+/– testis (Fig. 4B). To examine whether introducing polyadenylation-defective PAPOLBD114A is able to restore male infertility of PAPOLB-null mice, Tg16+/–/Papolb–/– males were mated to wild-type females. In contrast to wild-type male mice (8.7 ± 0.4 pups/litter, n = 6), Tg16+/–/Papolb–/– and Papolb–/– males failed to produce any pregnancies, despite the formation of copulation plugs in females mated (n = 6, Fig. 4C). Testes of Tg16+/–/Papolb–/– and Papolb–/– animals were similar in weight (55.1 ± 1.2 and 56.1 ± 2.8 mg in Tg16+/–/Papolb–/– and Papolb–/– mice, respectively, n = 5), but were 42% smaller than those of heterozygous mice (94.5 ± 2.8 mg, n = 5, Fig. 4D). Histological analysis showed disordered spermatogenesis in Tg16+/–/Papolb–/– mice, including the complete absence of late haploid cells (Supplementary Fig. 3: online only). As a consequence, cauda epididymis of Tg16+/–/Papolb–/– mice contained no detectable sperm but a small number of round-shaped cells possibly corresponding to degenerated spermatogenic cells. These data indicate that transgenic introduction of PAPOLBD114A into PAPOLB-null mice fails to rescue abnormal spermatogenesis and infertility, suggesting that the polyadenylation activity of PAPOLB is indispensable for spermiogenesis.
Fig. 4.
Generation and characterization of PAPOLB-null mice expressing PAPOLBD114A. Three- to four-month-old mice were used for analysis. (A) Northern blot analysis. Total testicular RNAs from Papolb+/– (HET), Papolb–/– (KO), and Tg16+/–/Papolb–/– (Tg16/KO) mice were subjected to Northern blot analysis using Flag and Papolb cDNA probes. Actb served as a control. (B) Immunoblot analysis. Testicular protein extracts were probed with anti-PAPOLB and anti-HA antibodies. ACTB was used as a control. Band marked with an asterisk likely represents PAPOLBD114A lacking FLAG and HA tags, which was translated from the initiation codon of Papolb. (C) Fertility of male mice. Wild-type (WT) and Tg16+/–/Papolb–/– (Tg16/KO) males were crossed with 8-week-old C57BL/6 females and the pups were counted (6 litters). (D) Testicular weight. Testicular tissues of Papolb+/– (HET), Papolb–/– (KO), and Tg16+/–/Papolb–/– (Tg16/KO) mice were weighed (n = 5).
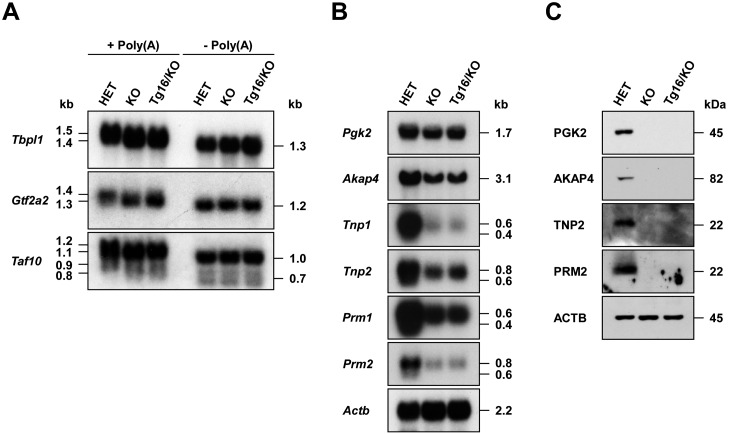
To ascertain whether PAPOLBD114A is inactive in vivo, we compared the poly(A) tail length of target mRNAs. Northern blot analysis of total testicular RNAs revealed that the sizes of three substrate mRNAs were identical between Tg16+/–/Papolb–/– and Papolb–/– testes, but were approximately ~100 nt smaller than those in Papolb+/– mice (Fig. 5A). Poly(A) tail removal by RNase H digestion in the presence of oligo(dT)15 diminished respective mRNAs to the same length among Papolb+/–, Papolb–/–, and Tg16+/–/Papolb–/– testes, indicating that the size decrease was attributed to defective polyadenylation (Fig. 5A). We next investigated if spermiogenesis arrest in Tg16+/–/Papolb–/– mice accompanies impaired testicular mRNA expression. The amount of Pgk2 and five postmeiotic mRNAs was significantly decreased in Tg16+/–/Papolb–/– and Papolb–/– mice compared with levels in Papolb+/– mice (Fig. 5B). In addition, deadenylated forms of Tnp1, Tnp2, Prm1, and Prm2 mRNAs (0.4 kb for Tnp1 and Prm1 and 0.6 kb for Tnp2 and Prm2), which are characteristic of late haploid cells, were undetectable in mutant mice lacking wild-type PAPOLB. Consistent with the absence of poly(A)-shortened mRNA forms [18, 19], four proteins PGK2, AKAP4, TNP2, and PRM2 were totally absent in Tg16+/–/Papolb–/– and Papolb–/– mice (Fig. 5C). These results further support that spermatogenesis is arrested prior to the spermatid elongation stage in Tg16+/–/Papolb–/– mice.
Fig. 5.
Poly(A) tail length of substrate mRNAs and expression and translation of haploid-specific mRNAs in Tg16+/–/Papolb–/– mice. (A) Poly(A) tail length of target mRNAs. Total testicular RNAs were obtained from Papolb+/– (HET), Papolb–/– (KO), and Tg16+/–/Papolb–/– (Tg16/KO) mice, incubated with RNase H in the presence [–Poly(A)] or absence [+Poly(A)] of oligo(dT)15, and subjected to Northern blot analysis with the cDNA probes indicated. (B) Expression of testis-specific mRNAs. Testicular total RNAs were analyzed by Northern blotting using the cDNA probes indicated. Actb was used as a control. (C) Translation of testis-specific mRNAs. Testicular protein extracts were subjected to immunoblot analysis with the antibodies indicated. ACTB served as a control.
Discussion
This study describes the essential role of PAPOLB polyadenylation activity in spermatogenesis. The enzyme is capable of extending poly(A) tails of not only transcription factor mRNAs but also mRNAs in other classes (Fig. 1). In addition, poly(A) tail extension is unlikely to enhance stability and translational efficiency of the substrate mRNAs examined because the levels of mRNAs and their translation products were not compromised by the absence of PAPOLB (Fig. 1). These observations raised the possibility that PAPOLB possesses a critical function besides poly(A) polymerase activity. Nevertheless, transgenic introduction of polyadenylation-defective PAPOLBD114A in PAPOLB-null mice failed to compensate for loss of endogenous PAPOLB (Figs. 4 and 5), implying that spermatogenesis indeed requires cytoplasmic polyadenylation catalyzed by PAPOLB, or at least its terminal nucleotidyltransferase activity.
Efficient translation of eukaryotic mRNAs is accomplished by cap and poly(A) tail synergy mediated by three proteins: cytoplasmic poly(A)-binding protein (PABPC) bound to poly(A) tails associates with the scaffold protein eukaryotic initiation factor (EIF) 4G, which is also capable of interacting with EIF4E, a cap-binding protein. This “closed-loop” structure is thought to be critical for efficient translation by promoting ribosome recycling and/or enhancing the interaction between initiation factors and mRNAs, as well as for protecting mRNAs from deadenylation [29,30,31,32]. Given that transcription is entirely silent during oocyte maturation and early embryogenesis, mRNAs encoding proteins required for progression of these processes are transcribed in earlier stages and are stored as deadenylated forms after transport to the cytoplasm [7,8,9,10,11]. These maternal mRNAs with short poly(A) tails (~A10) are translationally incompetent, where PABPC is unable to bind mRNAs, and are later activated by cytoplasmic polyadenylation. However, poly(A) tail extension by PAPOLB differs from the case during oocyte maturation and early embryogenesis in that the substrate mRNAs identified to date already possess poly(A) tract lengths (~100 nt) sufficient for PABPC-binding. Additional poly(A) tail extension by PAPOLB had no impact on translation and steady-state level of the mRNAs and is thus considered to be non-specific or a side effect in view of function (Fig. 1) [23]. This is consistent with the recent findings that the median length of poly(A) tails is 50–100 nt in mammalian cells, which is shorter than previously thought (150–250 nt), and that additional poly(A) tail elongation does not further contribute to translational enhancement except on maternal mRNAs [33,34,35]. Thus, if cytoplasmic polyadenylation by PAPOLB governs spermiogenesis, it is expected that the “true” target(s) possesses maternal mRNA-like short poly(A) tail(s). It is also conceivable that PAPOLB, like noncanonical poly(A) polymerase PAPD4/GLD-2, is involved in 3'-end monoadenylation of certain microRNAs, thereby regulating their stability [36, 37].
It is interesting to note that the poly(A) tails of some mRNAs, including odc (Fig. 1A) and Taf10 [23], are slightly longer in round spermatids than in pachytene spermatocytes of PAPOLB-deficient mice, but are shorter in round spermatids of wild-type mice. These observations imply that poly(A) tail extension of the mRNAs is carried out not only by PAPOLB but also by another poly(A) polymerase, such as PAPD4 [38]. Indeed, poly(A) tails of Acr, Acrbp-W, Acrbp-V5, and Zpbp1 mRNAs were still elongated in round spermatids even in the absence of PAPOLB (Supplementary Fig. 4: online only) [23]. Identifying the polyadenylation pathway responsible for these processes will be helpful to understand poly(A) tail dynamics during mammalian spermatogenesis.
Although PAPOLB polyadenylation activity is most likely essential for spermatogenesis, we cannot rule out the possibility that Asp114 of PAPOLB is involved in another yet unknown activity, as is the case for protein arginine methyltransferase 8 (PRMT8), in which Ser120 is required for both arginine methylation and phosphatidylcholine hydrolysis activities [39]. At any rate, further studies, including the identification of the “true” target(s), are necessary to elucidate PAPOLB function.
Supplementary
Acknowledgments
We thank Dr Satoru Takahashi at the Laboratory Animal Resource Center, University of Tsukuba, for producing transgenic mice. This work was partly supported by the Japan Society for the Promotion of Science (grant # 22580384 to SK) and the Ministry of Education, Culture, Sports, Science and Technology (grant # 23013005 to SK).
References
- 1.Sachs A, Wahle E. Poly(A) tail metabolism and function in eucaryotes. J Biol Chem 1993; 268: 22955–22958. [PubMed] [Google Scholar]
- 2.Wickens M, Anderson P, Jackson RJ. Life and death in the cytoplasm: messages from the 3′ end. Curr Opin Genet Dev 1997; 7: 220–232. [DOI] [PubMed] [Google Scholar]
- 3.Brook M, Gray NK. The role of mammalian poly(A)-binding proteins in co-ordinating mRNA turnover. Biochem Soc Trans 2012; 40: 856–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith RW, Blee TK, Gray NK. Poly(A)-binding proteins are required for diverse biological processes in metazoans. Biochem Soc Trans 2014; 42: 1229–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev 1997; 11: 2755–2766. [DOI] [PubMed] [Google Scholar]
- 6.Chan S, Choi E-A, Shi Y. Pre-mRNA 3′-end processing complex assembly and function. Wiley Interdiscip Rev RNA 2011; 2: 321–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richter JD. Cytoplasmic polyadenylation in development and beyond. Microbiol Mol Biol Rev 1999; 63: 446–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Méndez R, Richter JD. Translational control by CPEB: a means to the end. Nat Rev Mol Cell Biol 2001; 2: 521–529. [DOI] [PubMed] [Google Scholar]
- 9.Weill L, Belloc E, Bava F-A, Méndez R. Translational control by changes in poly(A) tail length: recycling mRNAs. Nat Struct Mol Biol 2012; 19: 577–585. [DOI] [PubMed] [Google Scholar]
- 10.Charlesworth A, Meijer HA, de Moor CH. Specificity factors in cytoplasmic polyadenylation. Wiley Interdiscip Rev RNA 2013; 4: 437–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reyes JM, Ross PJ. Cytoplasmic polyadenylation in mammalian oocyte maturation. Wiley Interdiscip Rev RNA 2016; 7: 71–89. [DOI] [PubMed] [Google Scholar]
- 12.Steger K. Transcriptional and translational regulation of gene expression in haploid spermatids. Anat Embryol (Berl) 1999; 199: 471–487. [DOI] [PubMed] [Google Scholar]
- 13.Kleene KC. A possible meiotic function of the peculiar patterns of gene expression in mammalian spermatogenic cells. Mech Dev 2001; 106: 3–23. [DOI] [PubMed] [Google Scholar]
- 14.Steger K. Haploid spermatids exhibit translationally repressed mRNAs. Anat Embryol (Berl) 2001; 203: 323–334. [DOI] [PubMed] [Google Scholar]
- 15.Kimmins S, Kotaja N, Davidson I, Sassone-Corsi P. Testis-specific transcription mechanisms promoting male germ-cell differentiation. Reproduction 2004; 128: 5–12. [DOI] [PubMed] [Google Scholar]
- 16.Idler RK, Yan W. Control of messenger RNA fate by RNA-binding proteins: an emphasis on mammalian spermatogenesis. J Androl 2012; 33: 309–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleene KC. Connecting cis-elements and trans-factors with mechanisms of developmental regulation of mRNA translation in meiotic and haploid mammalian spermatogenic cells. Reproduction 2013; 146: R1–R19. [DOI] [PubMed] [Google Scholar]
- 18.Kleene KC, Distel RJ, Hecht NB. Translational regulation and deadenylation of a protamine mRNA during spermiogenesis in the mouse. Dev Biol 1984; 105: 71–79. [DOI] [PubMed] [Google Scholar]
- 19.Kleene KC. Poly(A) shortening accompanies the activation of translation of five mRNAs during spermiogenesis in the mouse. Development 1989; 106: 367–373. [DOI] [PubMed] [Google Scholar]
- 20.Kilpatrick DL, Borland K, Jin DF. Differential expression of opioid peptide genes by testicular germ cells and somatic cells. Proc Natl Acad Sci USA 1987; 84: 5695–5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujimoto H, Erickson RP, Toné S. Changes in polyadenylation of lactate dehydrogenase-X mRNA during spermatogenesis in mice. Mol Reprod Dev 1988; 1: 27–34. [DOI] [PubMed] [Google Scholar]
- 22.Kashiwabara S, Zhuang T, Yamagata K, Noguchi J, Fukamizu A, Baba T. Identification of a novel isoform of poly(A) polymerase, TPAP, specifically present in the cytoplasm of spermatogenic cells. Dev Biol 2000; 228: 106–115. [DOI] [PubMed] [Google Scholar]
- 23.Kashiwabara S, Noguchi J, Zhuang T, Ohmura K, Honda A, Sugiura S, Miyamoto K, Takahashi S, Inoue K, Ogura A, Baba T. Regulation of spermatogenesis by testis-specific, cytoplasmic poly(A) polymerase TPAP. Science 2002; 298: 1999–2002. [DOI] [PubMed] [Google Scholar]
- 24.Zhuang T, Kashiwabara S, Noguchi J, Baba T. Transgenic expression of testis-specific poly(A) polymerase TPAP in wild-type and TPAP-deficient mice. J Reprod Dev 2004; 50: 207–213. [DOI] [PubMed] [Google Scholar]
- 25.Kimura M, Ishida K, Kashiwabara S, Baba T. Characterization of two cytoplasmic poly(A)-binding proteins, PABPC1 and PABPC2, in mouse spermatogenic cells. Biol Reprod 2009; 80: 545–554. [DOI] [PubMed] [Google Scholar]
- 26.Kashiwabara S, Tsuruta S, Okada K, Saegusa A, Miyagaki Y, Baba T. Functional compensation for the loss of testis-specific poly(A)-binding protein, PABPC2, during mouse spermatogenesis. J Reprod Dev 2016; 62: 305–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kashiwabara S, Arai Y, Kodaira K, Baba T. Acrosin biosynthesis in meiotic and postmeiotic spermatogenic cells. Biochem Biophys Res Commun 1990; 173: 240–245. [DOI] [PubMed] [Google Scholar]
- 28.Alcivar AA, Hake LE, Mali P, Kaipia A, Parvinen M, Hecht NB. Developmental and differential expression of the ornithine decarboxylase gene in rodent testis. Biol Reprod 1989; 41: 1133–1142. [DOI] [PubMed] [Google Scholar]
- 29.Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 γ and the translational repressors 4E-binding proteins. Mol Cell Biol 1995; 15: 4990–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J 1998; 17: 7480–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell 1998; 2: 135–140. [DOI] [PubMed] [Google Scholar]
- 32.Kahvejian A, Svitkin YV, Sukarieh R, M’Boutchou M-N, Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev 2005; 19: 104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang H, Lim J, Ha M, Kim VN. TAIL-seq: genome-wide determination of poly(A) tail length and 3′ end modifications. Mol Cell 2014; 53: 1044–1052. [DOI] [PubMed] [Google Scholar]
- 34.Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 2014; 508: 66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park J-E, Yi H, Kim Y, Chang H, Kim VN. Regulation of poly(A) tail and translation during the somatic cell cycle. Mol Cell 2016; 62: 462–471. [DOI] [PubMed] [Google Scholar]
- 36.Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T, Suzuki T. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev 2009; 23: 433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D’Ambrogio A, Gu W, Udagawa T, Mello CC, Richter JD. Specific miRNA stabilization by Gld2-catalyzed monoadenylation. Cell Rep 2012; 2: 1537–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwak JE, Wang L, Ballantyne S, Kimble J, Wickens M. Mammalian GLD-2 homologs are poly(A) polymerases. Proc Natl Acad Sci USA 2004; 101: 4407–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J-D, Park K-E, Ishida J, Kako K, Hamada J, Kani S, Takeuchi M, Namiki K, Fukui H, Fukuhara S, Hibi M, Kobayashi M, Kanaho Y, Kasuya Y, Mochizuki N, Fukamizu A. PRMT8 as a phospholipase regulates Purkinje cell dendritic arborization and motor coordination. Sci Adv 2015; 1: e1500615. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.