Abstract
Cohesins containing a meiosis-specific α-kleisin subunit, RAD21L or REC8, play roles in diverse aspects of meiotic chromosome dynamics including formation of axial elements (AEs), assembly of the synaptonemal complex (SC), recombination of homologous chromosomes (homologs), and cohesion of sister chromatids. However, the exact functions of individual α-kleisins remain to be elucidated. Here, we examined the localization of RAD21L and REC8 within the SC by super-resolution microscopy, 3D-SIM. We found that both RAD21L and REC8 were localized at the connection sites between lateral elements (LEs) and transverse filaments (TFs) of pachynema with RAD21L locating interior to REC8 sites. RAD21L and REC8 were not symmetrical in terms of synaptic homologs, suggesting that the arrangement of different cohesins is not strictly fixed along all chromosome axes. Intriguingly, some RAD21L signals, but not REC8 signals, were observed between unsynapsed regions of AEs of zygonema as if they formed a bridge between homologs. Furthermore, the signals of recombination intermediates overlapped with those of RAD21L to a greater degree than with those of REC8. These results highlight the different properties of two meiotic α-kleisins, and strongly support the previous proposition that RAD21L is an atypical cohesin that establishes the association between homologs rather than sister chromatids.
Keywords: Cohesin, Meiosis, Recombination, Synapsis, The synaptonemal complex
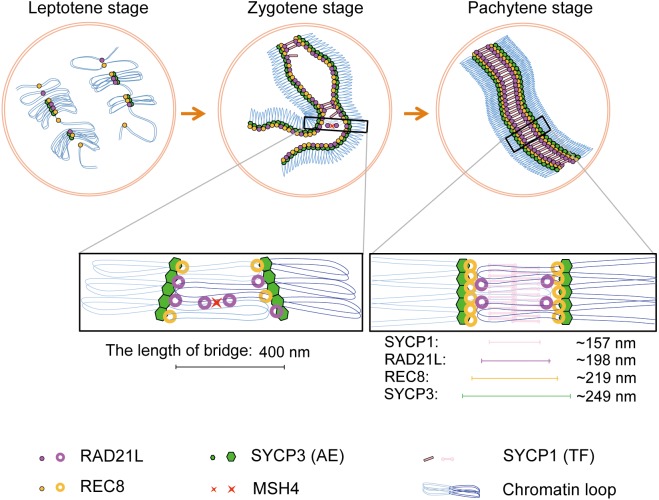
Meiosis is a special type of cell division that is required for sexual reproduction, in which the chromosome number is halved by two successive meiotic divisions following a single round of DNA replication. Meiosis I differs from mitosis or meiosis II; during meiosis I, homologous chromosomes (homologs) segregate from each other, whereas in meiosis II, sister chromatids segregate. To ensure successful homolog segregation in meiosis I, it is a prerequisite for homologs to establish a connection with their partners. This is achieved by three meiosis-specific events that occur during prophase I, namely pairing, synapsis, and recombination of homologs [1, 2]. These events occur in parallel with the assembly of a tripartite structure called the synaptonemal complex (SC). SC assembly is initiated by the formation of axial elements (AEs) along each chromosome during the leptotene stage. Two AEs of homologous chromosomes begin to synapse through a connection between transverse filaments (TFs) during the zygotene stage, and complete synapsis along the entire length of chromosomes by the pachytene stage. The two AEs connected by TFs are called lateral elements (LEs). Finally, during the diplotene stage, the SC starts to disassemble [2, 3].
In eukaryotes, meiotic recombination is initiated by the generation of DNA double-strand breaks (DSB), which is catalyzed by SPO11 [4,5,6,7]. The generation of DSBs causes a DNA damage response, which is accompanied by the phosphorylation of histone variant H2AX (γH2AX) [8]. DSBs are resected to generate 3´ single-stranded overhangs, to which DNA repair proteins such as RAD51 and DMC1 are recruited [9]. Single-stranded DNA then invades the double-stranded DNA of the homolog. As meiotic recombination proceeds, the early recombination intermediates, containing RAD51 and DMC1, are replaced by middle intermediates containing MSH4, and subsequently by late intermediates containing MLH1 in a step-wise manner [10, 11]. During this process, double Holliday junctions are formed and ultimately resolved as either crossover or non-crossover recombinations [12]. After SC disassembly, crossover recombination plays an important role in maintaining the connection between homologs until anaphase I, with the help of sister chromatid cohesion distal to the chiasmata.
Cohesin, a multi-subunit protein complex that is well conserved from yeasts to mammals plays a pivotal role in sister chromatid cohesion [13, 14]. The cohesin complex consists of four different subunits: two structural maintenance of chromosome subunits (SMC1 and SMC3), an α-kleisin subunit (RAD21), and either SA1/STAG1 or SA2/STAG2 [15−17], forming a ring-like structure in which sister chromatids could be held [18, 19]. Mammals have several meiosis-specific paralogs of cohesin subunits, including SMC1β [20], REC8 [21, 22] or RAD21L [23, 24], and STAG3 [25]. Meiotic cohesins function not only in sister chromatid cohesion but also for formation of AEs, assembly of the SC, and crossover recombination during prophase I [26,27,28,29,30,31,32,33]. Considering that two meiotic α-kleisins, RAD21L and REC8, are conserved among many vertebrate species, it is reasonable to speculate that each of these subunits has its own function in meiotic chromosome dynamics. In fact, there are some phenotypic differences between Rad21L KO and Rec8 KO mice. Rad21L KO mice show sexual dimorphism in fertility [31], whereas both sexes of Rec8 KO mice are infertile [28]. In addition, synapsis occurs between non-homologous chromosomes in Rad21L KO mice [31], whereas synapsis occurs mainly between sister chromatids in Rec8 KO mice [29]. Therefore, it seems that RAD21L or REC8 has specific and distinct important roles in different aspects of chromosomal events. However, their precise roles remain to be elucidated. To investigate their individual functions, further characterization of these two meiotic α-kleisins is needed.
In the present study, we examined the localization of two meiotic α-kleisins in the SC at high resolution by three dimensional structured illumination microscopy (3D-SIM). 3D-SIM has a resolution of 120 nm along the x-y directions, and is advantageous for the localization of molecules of interest with a great robustness in sample preparation [34]. Moreover, immunoreactivity is generally better preserved in the samples fixed with paraformaldehyde and used for 3D-SIM than in those fixed with glutaraldehyde commonly used for electron microscopy [35]. Thus, 3D-SIM is suitable for analyzing the precise localization of proteins in the SC. Here, we showed for the first time that both RAD21L and REC8 are located at the connection sites between AEs and TFs. We also identified the different properties of RAD21L and REC8, and finally proposed potential roles for these individual cohesin subunits.
Materials and Methods
Mice
C56BL/6J mice were used for the analysis of chromosomal localization of meiotic cohesins. This study was approved by the Institutional Animal Care and Committee (Permission number: 25-03-02) and performed according to the Kobe University Animal Experimentation Regulation.
Antibodies
The primary antibodies used in this study were as follows: rabbit and rat polyclonal anti-RAD21L antibodies [24], rabbit polyclonal anti-REC8 [24], mouse polyclonal anti-REC8 antibody [22], mouse polyclonal anti-SYCP3 antiserum [22], rabbit polyclonal anti-MSH4 antibody (ab58666, Abcam, Cambridge, MA, USA), rabbit polyclonal anti-SYCP1 antibody (ab15087, Abcam), and rabbit polyclonal anti-RAD51 antibody (sc-8349, Santa Cruz Biotechnology, Dallas, TX, USA). The antigen-antibody complexes were detected by appropriate secondary antibodies conjugated with Alexa Fluor 488 or 568 (Molecular Probes, Eugene, OR, USA).
Immunofluorescence analysis
A testicular cell suspension from adult mice was prepared according to the method described by Heyting and Dietrich [36]. The cells were placed on poly-L-lysine-coated coverslips and fixed with 2% paraformaldehyde in buffer H [120 mM HEPES-KOH (pH 7.6), 30 mM HEPES-NaOH, 2 mM EDTA-3Na, 0.5 mM EGTA, 0.5 mM spermidine 3HCl, 0.2 mM spermine 4HCl, 15 mM 2-mercaptoethanol] for 10 min [37]. Testicular cells on coverslips were permeabilized with 0.2% triton X-100 in PBS for 5 min, and washed in PBS three times for 10 min each. The cells were incubated with primary antibodies at appropriate dilutions in blocking buffer at 4°C overnight. Cells were then washed in PBS for 10 min, in detergent solution for 10 min, and in PBS for 10 min. Cells were then incubated with secondary antibodies at appropriate dilutions in blocking buffer. DNA was counterstained with DAPI (Wako, Osaka, Japan). Samples were mounted with VECTASHIELD Mounting Medium (Vector Laboratories, Burlingame, CA, USA). Images were acquired with a DeltaVision OMX microscope (GE Healthcare, Buckinghamshire, UK) with a 100 × UPlanSApo NA1.40 oil immersion objective lens (Olympus, Tokyo, Japan) for 3D-SIM or with a confocal laser scanning microscope (LSM; Olympus, Lake Success, NY, USA) with a Plan Apochromat 10 × and 100 × /1.46 oil DIC objective lens. All Images were processed with DeltaVision SoftWorx software (GE Healthcare). Some of the images shown were projected views of z-stacks.
Image analysis
For every measurement, images were acquired as z-sections encompassing the entire nuclei. To measure the distance between two signals on the SC, 3D images of synapsed chromosomes were linearized (Fig. 1H). For linearization, binary images were first made by automatic iterative thresholding [37]. Next, individual SCs in binary 3D images were identified by our program using functions from the “ndimage” module of the Scipy package (http://www.scipy.org). Subsequently, the lines of the SC in binary, single-color images were traced with the “Trace” function in Priism suite (http://msg.ucsf.edu/IVE/). Finally, using the “Trace” output function as the model file, the original multicolor 3D images were linearized using the “Straight” function of Priism suite. The distance between two fluorescence peaks on the synaptic LEs was measured in at least five areas. The statistical significance between different samples was tested using a Student’s t-test.
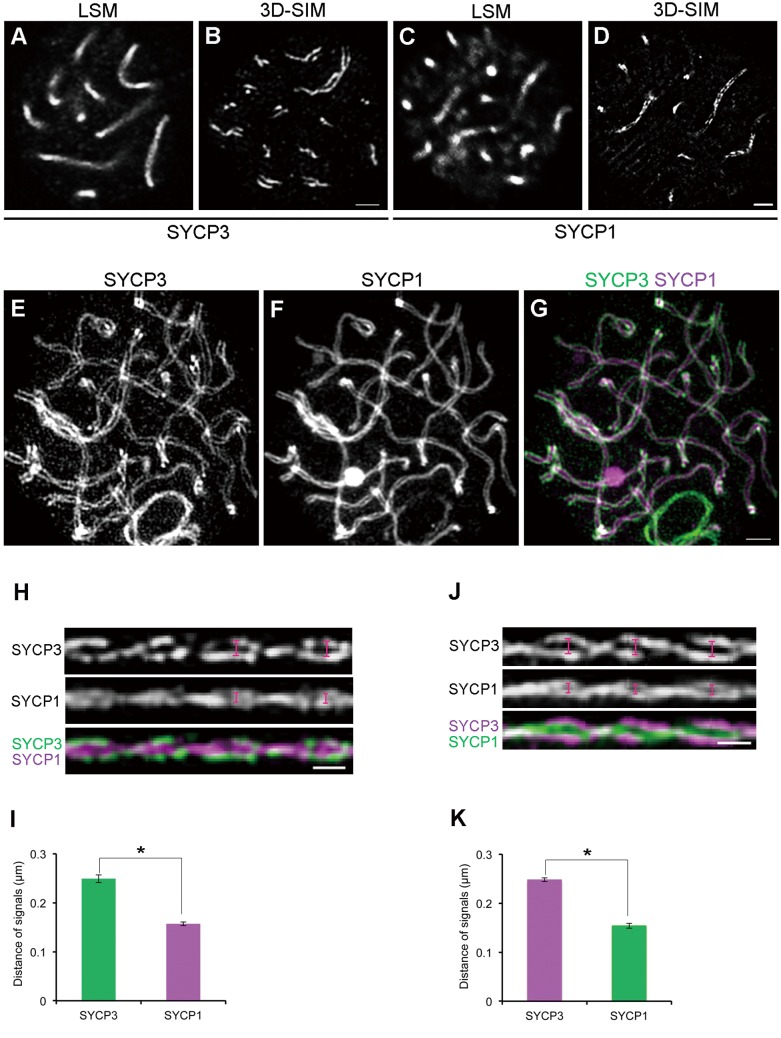
Fig. 1.
Comparison of the resolution power between conventional confocal microscopy and 3D-SIM. (A–D) Mouse spermatocytes at the pachytene stage were immunofluorescently labeled with anti-SYCP3 (A, B) or anti-SYCP1 antibodies (C, D). Subsequently, the signals were observed either by conventional confocal microscopy (LSM) (A, C) or by 3D-SIM (B, D). The nuclei in A and B were from different samples. A single optical section is shown. Scale bars: 1 µm. (E–G). The 3D image of the SC in an entire nucleus of a pachytene spermatocyte was visualized using the “Volume Viewer” function of Priism suite. In the merged image, SYCP3 and SYCP1 signals appear in green and purple, respectively. Scale bar: 1 µm. (H) Linearized SC images from three-dimensional z-stacks obtained by 3D-SIM. The top, middle, and bottom panels show SYCP3, SYCP1, and the merged image (SYCP3 in green and SYCP1 in purple), respectively. Scale bar: 0.5 µm. (I) The average spacing of SYCP3 or SYCP1 (n = 98 homologs, * P < 0.01 by t-test). (J, K) In a similar manner as shown in H and I, the experiments were repeated, exchanging the fluorescent dyes that were conjugated to secondary antibodies. In the merged image, SYCP3 and SYCP1 signals appear in purple and green, respectively. (n = 72 homologs, * P < 0.01 by t-test).
Co-localization (overlap) was analyzed in individual nuclei of 3D-SIM images. We used automatic thresholding (mentioned previously) with the same value for the “delta” coefficient for automatic thresholding to keep the threshold visually constant for all images [37]. Then, using the threshold, Mandars co-localization coefficients [38] were measured. The ratios of the α-kleisin-recombination intermediates-double-positive areas (overlapping areas) to the recombination intermediates-positive areas were calculated and are shown in Fig. 4. We corrected chromatic shifts using our adaptive image registration algorithm [37]. The statistical significance between different samples was tested using a Student’s t-test.
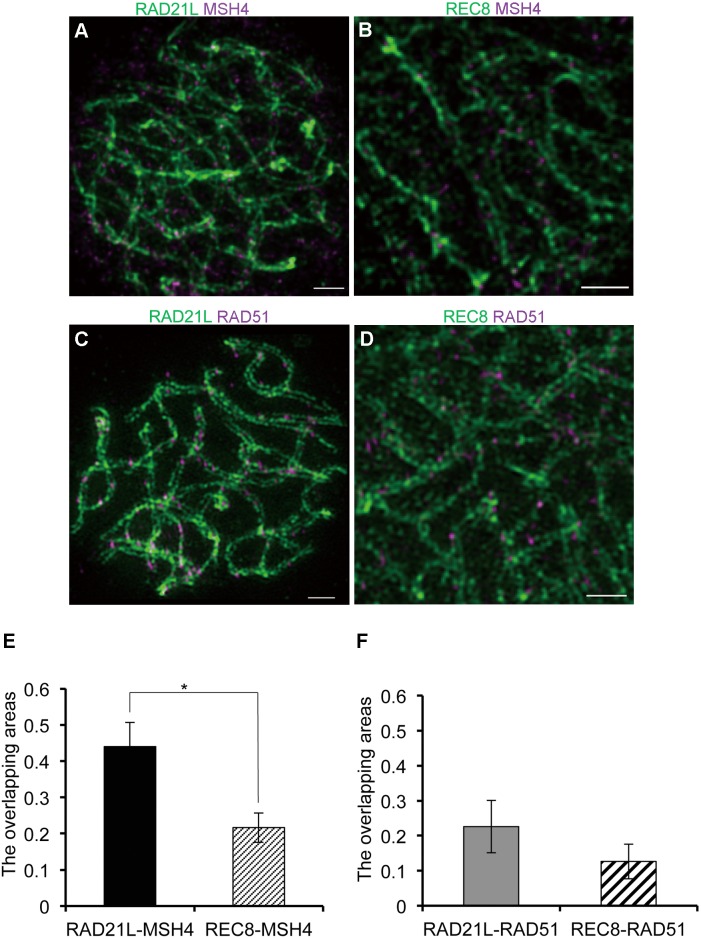
Fig. 4.
RAD21L signals overlap with recombination intermediates to a greater extent than REC8 signals. (A–D) The volume viewers of mouse spermatocytes at the zygotene stage were immunofluorescently labeled with both anti-RAD21L and anti-MSH4 antibodies (A), with both anti-REC8 and anti-MSH4 antibodies (B), with both anti-RAD21L and anti-RAD51 antibodies (C), or with both anti-REC8 and anti-RAD51 antibodies (D). (A–D) White arrows indicate the signals of MSH4 or RAD51 at the synaptic regions of the SC. Scale bars: 1 μm. (E) The ratios of overlapping areas of RAD21L-MSH4 signals and REC8-MSH4 signals relative to MSH4 signals were calculated and compared (n = 60 homologs, * P < 0.01 by t-test). (F) The ratios of overlapping areas of RAD21L-RAD51 signals and REC8-RAD51 signals relative to RAD51 signals were calculated and compared (n = 80 homologs).
Results
The arrangement of SYCP3 and SYCP1 in the pachytene stage of mouse spermatocytes visualized by 3D-SIM
To identify the framework structure of SCs at the resolution of our imaging system using 3D-SIM, and to validate the experimental workflow, we first examined the localization of SYCP3 and SYCP1, which are well-defined components of AEs/LEs and TFs, respectively, of the SC [39,40,41]. At the pachytene stage, when homologs were completely synapsed, SYCP3 and SYCP1 signals were observed as a single line on each bivalent using a confocal laser-scanning-microscope (LSM) (Fig. 1A and C). In contrast, SYCP3 signals were observed as two distinct lines representing LEs by 3D-SIM (Fig. 1B), as observed in previous 3D-SIM analyses in animal and plant meiocytes [42, 43]. SYCP1 signals were also observed as two lines on each bivalent (Fig. 1D), which was unexpected as SYCP1 is localized along the TFs formed between LEs. Because SYCP1 forms a homodimer through its N-terminus and since the anti-SYCP1 antibody used was raised against a C-terminal peptide, the two separate lines indicated that we were able to resolve the C-terminal ends of the SYCP1 homodimer. Thus, the spatial resolution in our study was high enough to distinguish two ends of a single SYCP1 homodimer. From 3D images, we could observe that such fine structures of SYCP1 and SYCP3 were oriented in various directions with curving and twisting in the nucleus of pachytene spermatocytes (Fig. 1E–G).
To analyze the structure of SCs in more detail, 3D images were computationally linearized (Fig. 1H and I). This facilitated the measurement of spacing between two linear signals of a protein of interest and could be applicable to a quantitative analysis (Fig. 1H and J). In this way, we measured the spacing between synapsed LEs (two linear SYCP3 signals) and between both ends of the TFs (two linear SYCP1 signals) and compared these values. Since the meiotic chromosome axes were twisting, the spacing between signals on paired homologs was measured at the widest regions of the helices (indicated by scale marks in Fig. 1H and J). As expected, the spacing of SYCP1 signals (mean value ± SD = 0.157 ± 0.004 µm) was significantly shorter than that of SYCP3 signals (0.249 ± 0.008 µm) (n = 98 homologs, P < 0.01) (Fig. 1I). Because differences in fluorescent emission wavelengths might influence the measurement, we repeated the experiments after swapping fluorophores on secondary antibodies. Regardless of the wavelengths tested, similar results were obtained: the spacing of SYCP3 was 0.245 ± 0.004 µm and the spacing of SYCP1 was 0.154 ± 0.005 µm (n = 72 homologs, P < 0.01) (Fig. 1J and K). This proved that our measurements were not affected by the specific fluorophores and that the results were reproducible within approximately 5 nm.
RAD21L is interior to REC8 in the SC
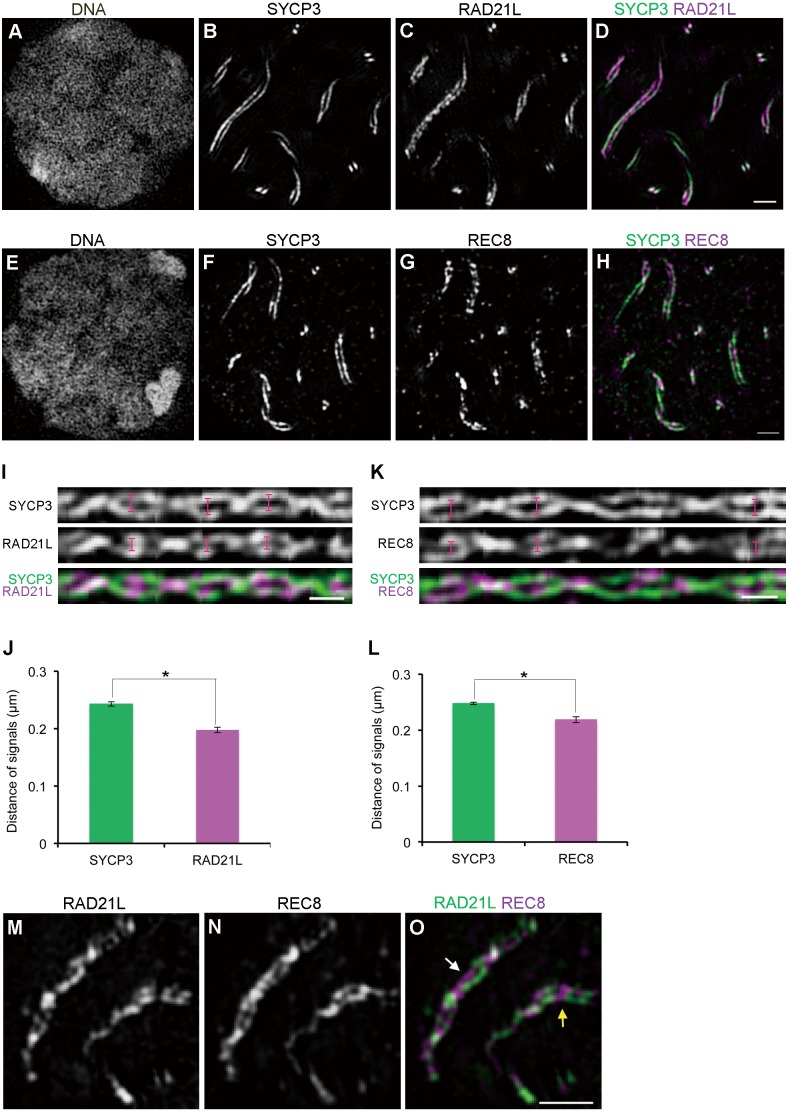
It has been shown that all cohesin subunits examined so far are localized along the AEs/LEs during mammalian meiosis [20,21,22,23,24,25, 44, 45]. However, their precise localization in the SC remains elusive. Therefore, we examined the precise positioning of RAD21L and REC8 in the SC. Mouse spermatocytes were immunofluorescently labeled with antibodies against either RAD21L or REC8 and SYCP3 and analyzed by 3D-SIM. At the pachytene stage, both RAD21L and REC8 were discontinuously localized along the LEs (Fig. 2C and G). To determine the location of cohesin in the SC, we measured the spacing between the RAD21L/REC8 signals on the synaptic LEs in the same way as in Fig. 1H (Fig. 2I and K). Since this method requires a continuous linear signal along each chromosome axis for automatic isolation of individual SCs, we used SYCP3 signals as a reference for the measurement of two cohesin subunits detected as discontinuous signals along chromosome axes. The spacing of RAD21L signals (0.198 ± 0.005 µm) was significantly shorter than that of SYCP3 signals (0.243 ± 0.004 µm) (n = 87 homologs, P < 0.01) (Fig. 2J). The spacing of REC8 signals (0.219 ± 0.005 µm) was also significantly shorter than that of SYCP3 signals (0.248 ± 0.002 µm) (n = 239 homologs, P < 0.01) (Fig. 2L). These results revealed that cohesin cores are located at the connection sites between AEs and the end lines of TFs, with RAD21L localized to a more interior region in the SC compared to the location of REC8. It has been reported that RAD21L and REC8 are distributed symmetrically along AEs/LEs between homologs [23]. In contrast, we observed that most of these meiotic cohesin signals were localized asymmetrically on the two synapsed homologs (white arrow in Fig. 2O), although some signals were distributed symmetrically (yellow arrow in Fig. 2O).
Fig. 2.
RAD21L is localized to a more interior position than REC8 in the synaptonemal complex. (A–H) Mouse spermatocytes at the pachytene stage were immunofluorescently labeled with anti-SYCP3 (B and F) and either anti-RAD21L (C) or anti-REC8 (G) antibodies. (A and E) DNA was counterstained with DAPI. (D) SYCP3 (green) and RAD21L (purple) staining in a merged image is shown. Scale bars: 1 μm. (I) Linearized SC images from three-dimensional z-stacks as in Fig. 1H. The top, middle, and bottom panels show signals of SYCP3, RAD21L, and the merged image (SYCP3 in green and RAD21L in purple), respectively. Scale bar: 0.5 μm. (J) The average spacing of SYCP3 or RAD21L (n = 87 homologs, * P < 0.01 by t-test). (K) Linearized SC images from three-dimensional z-stacks as described above. The top, middle, and bottom panels show signals of SYCP3, REC8, and the merged image (SYCP3 in green and REC8 in purple), respectively. Scale bar: 0.5 μm. (L) The average spacing of SYCP3 or REC8 (n = 239 homologs, * P < 0.01). (M–O) Mouse spermatocytes at the pachytene stage were immunofluorescently labeled with anti-RAD21L (M) and anti-REC8 (N) antibodies. (O) The merged image is shown (RAD21L in green and REC8 in purple). RAD21L and REC8 show both asymmetric (white arrow in O) and symmetric localization (yellow arrow in O) between homologous axes. Scale bars: 1 μm.
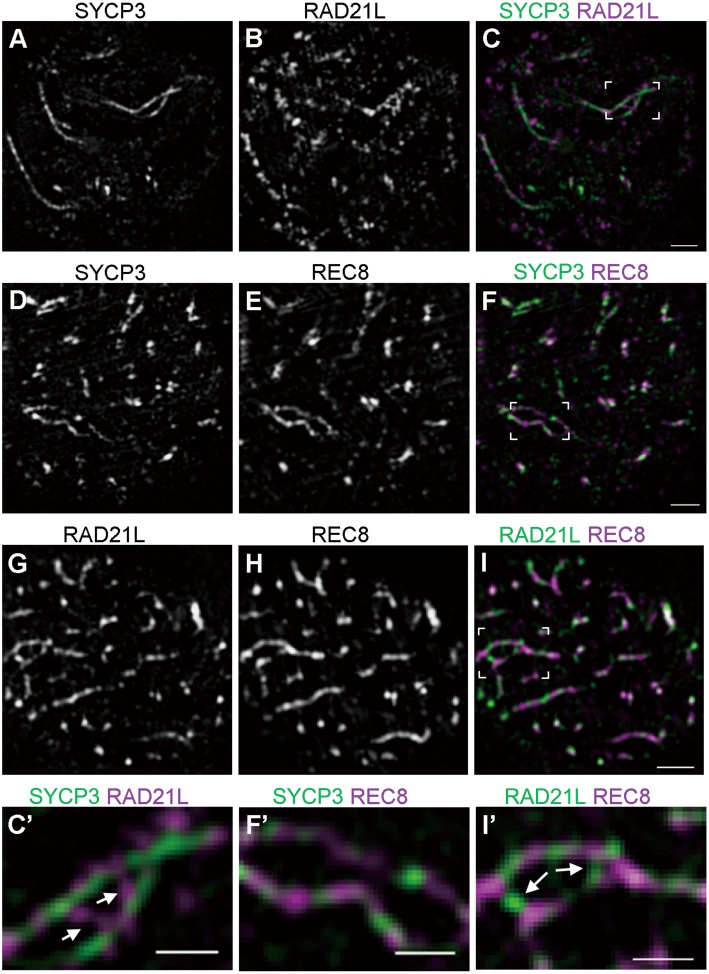
We also examined the localization of the two α-kleisins at the zygotene stage (Fig. 3). Interestingly, RAD21L foci were occasionally observed between unsynapsed regions of AEs (mean occurrence per cell ± SD = 2.286 ± 2.430, n = 7 cells) (Fig. 3C, I, C’ and I’; also see Supplementary movies S1 and S3). In contrast, the detection of such REC8 foci between unsynapsed regions of AEs was scarce (mean occurrence per cell ± SD = 0.500 ± 0.548, n = 6 cells) (Fig. 3F, I, F’ and I’; also see Supplementary movies S2 and S3).
Fig. 3.
The chromosomal localization of RAD21L and REC8 in zygotene spermatocytes. (A–I) Mouse spermatocytes at the zygotene stage were immunofluorescently labeled with either anti-SYCP3 and anti-RAD21L antibodies (A–C), with anti-SYCP3 and anti-REC8 antibodies (D–F), or with anti-RAD21L and anti-REC8 antibodies (G–I). The merged images are shown (C, F, and I). Also shown at the bottom are magnified images of unsynapsed chromosomes co-labeled with the above antibodies. (C’, F’, I’) RAD21L forms bridges between two linear axes (white arrows). Scale bars: 1 μm for main panels and 0.5 μm for magnified images.
RAD21L is associated with recombination foci
Because RAD21L and REC8 are essential for the recombination of homologs during meiosis [28, 29, 31], we examined the relationship between α-kleisins and recombination intermediates. Mouse spermatocytes were immunofluorescently labeled with antibodies against α-kleisins (RAD21L or REC8) and recombination intermediate molecules (RAD51 or MSH4), and were analyzed by 3D-SIM. During the zygotene stage, some MSH4 and RAD51 signals were detected between synaptic AEs, and thus appeared to overlap with RAD21L or REC8 signals (Fig. 4A–D). To further investigate this localization, we calculated the ratio of overlapping areas (α-kleisins and recombination intermediates-double-positive areas) to recombination intermediates-positive areas. We discovered that the ratio of overlapping areas of MSH4-RAD21L (mean value ± SD = 0.440 ± 0.067) was significantly greater than that of MSH4-REC8 (mean value ± SD = 0.217 ± 0.041, n = 80 homologs, P < 0.01) (Fig. 4E). The ratio of overlapping areas of RAD51-RAD21L (mean value ± SD = 0.226 ± 0.075) was greater than that of RAD51-REC8 although this was not significant (mean value ± SD = 0.127 ± 0.051, n = 80 homologs) (Fig. 4F). These results suggest that RAD21L locates more closely to recombination intermediates than REC8.
Discussion
The assembly of the SC with its constituent components and cohesins in mammals
It is well known that the SC is assembled in two steps. First, AEs/LEs composed of SYCP2/SYCP3 heterodimers are formed for each homolog [39, 41, 46]. Then, TFs composed of SYCP1 molecules form a bridge between two AEs/LEs of paired homologs [47,48,49]. However, it remains elusive as to how AEs/LEs are connected by TFs because a direct interaction between LE and TF components has never been reported. Although previous studies using KO mice have suggested that RAD21L and REC8 function redundantly in the formation of cohesin cores that are essential for AE formation [32], little is known about how cohesins might function in SC assembly after AE formation. In the present study using 3D-SIM, we have shown for the first time the geometry of the meiotic α-kleisin subunits of cohesin, RAD21L and REC8, relative to AEs/LEs (SYCP3) and TFs (SYCP1) in the SC in mouse spermatocytes. Our study provides new evidence that both meiotic α-kleisins are localized to connection sites (interspace) between LEs and TFs with RAD21L localizing more centrally in the SC. The spacing between respective molecular axes in the SC is summarized in Fig. 5. RAD21L and REC8 are located at the innermost sides of two LEs of the pachynema, in agreement with a previous electron microscopy-based analysis [33]. Meiotic α-kleisins have been co-immunoprecipitated with SYCP3 and/or SYCP1 [22, 24, 33]. Therefore, when considering previous findings, our results suggest that cohesin cores are positioned at the connection sites and mediate associations between AEs and TFs during SC assembly. A recent study utilizing 3D-SIM in barley has revealed two different SC configurations that contain one or two axes of the TF component ZYP1 for each homolog [42]. However, in the present study, such different configurations of TFs were not observed. This discrepancy might be attributed to differences in TF components or in TF arrangements between these two species.
Fig. 5.
The precise positioning of meiotic cohesins in the synaptonemal complex and the mechanism of synapsis and recombination during prophase I in mammals.
The distinct roles of RAD21L and REC8 in meiotic chromosome dynamics
The discovery of the second meiotic α-kleisin RAD21L, in addition to REC8, suggested that the two types of meiotic cohesins might play distinctive roles in specific aspects of meiotic chromosome dynamics. In this regard, at least after the diplotene stage, REC8 but not RAD21L should be responsible for sister chromatid cohesion because RAD21L is expressed only in meiotic prophase I until mid pachytene, whereas REC8 is present throughout meiosis up to metaphase II [22, 24]. In fact, using mice carrying TEV protease-cleavable REC8 or RAD21, it was demonstrated that REC8 is essential for maintaining sister chromatid cohesion at both centromeres and arm regions during meiosis [50]. In contrast, from leptotene to mid-pachytene, both subunits are expressed and localized to the SC; hence, it is not easy to distinguish their respective functions. In the present study, we found several differences between RAD21L and REC8, providing evidence to address this issue. RAD21L and REC8 were observed at different positions in the longitudinal axes of chromosomes, and these homologs did not form a mirror image, suggesting that the chromosomal positions were flexible rather than fixed. If this is true, it argues against a symmetric distribution of two types of α-kleisin along chromosomal axes that contributes to the recognition of homologs for pairing and synapsis [23]. However, the sparseness of antibody labeling using fixed samples is often a major limitation for 3D-SIM in visualizing the entire fraction of molecules present in cells. Although it is difficult to label all molecules without affecting the physiology of the cell, these results need to be clarified. We also observed bridge-like signals for RAD21L, but rarely for REC8, between unsynapsed AEs spaced ~400 nm apart (Fig. 3; Supplementary movies S1, S2, and S3); these structures are reminiscent of inter-axis bridges corresponding to sites of DSB-mediated interhomolog interactions that are formed when homologs are co-aligned in various organisms [2]. Thus, it is probable that RAD21L is localized to the DSB-mediated interhomolog bridges of zygonema. In support of this notion, RAD21L was observed closer to recombination intermediates than REC8 (Fig. 4). It seems that RAD21L-containing cohesin might localize close to the recombination intermediates either through direct or indirect association with the recombination intermediate molecules or by recognizing the configuration of the DNA (chromatin) during the process of recombination from zygotene to mid pachytene. Alternatively, but not mutually exclusively, RAD21L might have an intrinsic feature allowing it to make and/or maintain the connection between non-sister chromatids rather than between sister chromatids. In support of the latter case, recent studies have suggested that homolog recognition and pairing occur in a DSB-independent manner before AE formation [33, 51]. In addition, this early recognition and pairing are mainly dependent on the function of RAD21L but only partly on REC8 since homolog co-alignment was observed in Rec8/Spo11 double KO but not Rad21L/Spo11 double KO spermatocytes [33]. Our observation that RAD21L locates to a position interior to the site of REC8 in the SC (Fig. 2) might reflect the fact that RAD21L but not REC8 can couple the DNA of non-sister chromatids from homologs irrespective of recombination position (Fig. 5). A recent work using 3D-SIM in fission yeast meiosis also suggests that Rec8, the only meiotic α-kleisin in this species, plays an essential role in building a platform to support the chromosome architecture necessary for the spatial alignment of homologs [52]. Therefore, it seems that the role of meiosis-specific cohesin subunits in establishing the linkage between homologs is conserved among eukaryotes.
In summary, the present study highlights the different properties of two meiotic α-kleisins, strongly supporting the view that RAD21L is an atypical cohesin that establishes an association between homologs rather than between sister chromatids [24, 33].
Supplementary
A serial z-sectioning image of SYCP3 and RAD21L at the zygotene stage. Mouse spermatocytes were immunofluorescently labeled with anti-SYCP3 (green) and anti-RAD21L (purple) antibodies. Some RAD21L foci were detected between unsynapsed regions of AEs as if they formed a bridge. A scale bar: 1 μm.
A serial z-sectioning image of SYCP3 and REC8 at the zygotene stage.Mouse spermatocytes were immunofluorescently labeled with anti-SYCP3 (green) and anti-REC8 antibodies (purple). No inter-axis bridge-like signals for REC8 were observed in the unsynapsed regions of AEs although such signals were detected in the synapsed regions. Scale bar: 1 μm.
A serial z-sectioning image of RAD21L and REC8 at the zygotene stage. Mouse spermatocytes at the zygotene stage were immunofluorescently labeled with anti-RAD21L (green) and anti-REC8 antibodies (purple). Some RAD21L foci, but not REC8 foci, were detected between unsynapsed regions of AEs. Scale bar: 1 μm.
Acknowledgments
We are grateful to the members of our laboratory for their valuable support and discussions. This work was supported in part by JSPS KAKENHI (Grant Numbers 26292169 to JL, 15K14500 and 16H01440 to AM, and 26251037 to YH).
References
- 1.Jordan P. Initiation of homologous chromosome pairing during meiosis. Biochem Soc Trans 2006; 34: 545–549. [DOI] [PubMed] [Google Scholar]
- 2.Zickler D, Kleckner N. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb Perspect Biol 2015; 7: a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Page SL, Hawley RS. The genetics and molecular biology of the synaptonemal complex. Annu Rev Cell Dev Biol 2004; 20: 525–558. [DOI] [PubMed] [Google Scholar]
- 4.Keeney S. Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol 2001; 52: 1–53. [DOI] [PubMed] [Google Scholar]
- 5.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 1997; 88: 375–384. [DOI] [PubMed] [Google Scholar]
- 6.Baudat F, Manova K, Yuen JP, Jasin M, Keeney S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell 2000; 6: 989–998. [DOI] [PubMed] [Google Scholar]
- 7.Romanienko PJ, Camerini-Otero RD. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell 2000; 6: 975–987. [DOI] [PubMed] [Google Scholar]
- 8.Mahadevaiah SK, Turner JM, Baudat F, Rogakou EP, de Boer P, Blanco-Rodríguez J, Jasin M, Keeney S, Bonner WM, Burgoyne PS. Recombinational DNA double-strand breaks in mice precede synapsis. Nat Genet 2001; 27: 271–276. [DOI] [PubMed] [Google Scholar]
- 9.Tarsounas M, Morita T, Pearlman RE, Moens PB. RAD51 and DMC1 form mixed complexes associated with mouse meiotic chromosome cores and synaptonemal complexes. J Cell Biol 1999; 147: 207–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plug AW, Peters AH, Keegan KS, Hoekstra MF, de Boer P, Ashley T. Changes in protein composition of meiotic nodules during mammalian meiosis. J Cell Sci 1998; 111: 413–423. [DOI] [PubMed] [Google Scholar]
- 11.Santucci-Darmanin S, Walpita D, Lespinasse F, Desnuelle C, Ashley T, Paquis-Flucklinger V. MSH4 acts in conjunction with MLH1 during mammalian meiosis. FASEB J 2000; 14: 1539–1547. [DOI] [PubMed] [Google Scholar]
- 12.Hunter N, Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell 2001; 106: 59–70. [DOI] [PubMed] [Google Scholar]
- 13.Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol 2006; 7: 311–322. [DOI] [PubMed] [Google Scholar]
- 14.Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet 2009; 43: 525–558. [DOI] [PubMed] [Google Scholar]
- 15.Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev 1998; 12: 1986–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tóth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1p (Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev 1999; 13: 320–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM. Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol 2000; 151: 749–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haering CH, Löwe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell 2002; 9: 773–788. [DOI] [PubMed] [Google Scholar]
- 19.Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell 2003; 112: 765–777. [DOI] [PubMed] [Google Scholar]
- 20.Revenkova E, Eijpe M, Heyting C, Gross B, Jessberger R. Novel meiosis-specific isoform of mammalian SMC1. Mol Cell Biol 2001; 21: 6984–6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eijpe M, Offenberg H, Jessberger R, Revenkova E, Heyting C. Meiotic cohesin REC8 marks the axial elements of rat synaptonemal complexes before cohesins SMC1beta and SMC3. J Cell Biol 2003; 160: 657–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J, Iwai T, Yokota T, Yamashita M. Temporally and spatially selective loss of Rec8 protein from meiotic chromosomes during mammalian meiosis. J Cell Sci 2003; 116: 2781–2790. [DOI] [PubMed] [Google Scholar]
- 23.Ishiguro K, Kim J, Fujiyama-Nakamura S, Kato S, Watanabe Y. A new meiosis-specific cohesin complex implicated in the cohesin code for homologous pairing. EMBO Rep 2011; 12: 267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee J, Hirano T. RAD21L, a novel cohesin subunit implicated in linking homologous chromosomes in mammalian meiosis. J Cell Biol 2011; 192: 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prieto I, Suja JA, Pezzi N, Kremer L, Martínez-A C, Rufas JS, Barbero JL. Mammalian STAG3 is a cohesin specific to sister chromatid arms in meiosis I. Nat Cell Biol 2001; 3: 761–766. [DOI] [PubMed] [Google Scholar]
- 26.Klein F, Mahr P, Galova M, Buonomo SB, Michaelis C, Nairz K, Nasmyth K. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 1999; 98: 91–103. [DOI] [PubMed] [Google Scholar]
- 27.Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature 1999; 400: 461–464. [DOI] [PubMed] [Google Scholar]
- 28.Bannister LA, Reinholdt LG, Munroe RJ, Schimenti JC. Positional cloning and characterization of mouse mei8, a disrupted allelle of the meiotic cohesin Rec8. Genesis 2004; 40: 184–194. [DOI] [PubMed] [Google Scholar]
- 29.Xu H, Beasley MD, Warren WD, van der Horst GT, McKay MJ. Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev Cell 2005; 8: 949–961. [DOI] [PubMed] [Google Scholar]
- 30.Novak I, Wang H, Revenkova E, Jessberger R, Scherthan H, Höög C. Cohesin Smc1beta determines meiotic chromatin axis loop organization. J Cell Biol 2008; 180: 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herrán Y, Gutiérrez-Caballero C, Sánchez-Martín M, Hernández T, Viera A, Barbero JL, de Álava E, de Rooij DG, Suja JÁ, Llano E, Pendás AM. The cohesin subunit RAD21L functions in meiotic synapsis and exhibits sexual dimorphism in fertility. EMBO J 2011; 30: 3091–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Llano E, Herrán Y, García-Tuñón I, Gutiérrez-Caballero C, de Álava E, Barbero JL, Schimenti J, de Rooij DG, Sánchez-Martín M, Pendás AM. Meiotic cohesin complexes are essential for the formation of the axial element in mice. J Cell Biol 2012; 197: 877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishiguro K, Kim J, Shibuya H, Hernández-Hernández A, Suzuki A, Fukagawa T, Shioi G, Kiyonari H, Li XC, Schimenti J, Höög C, Watanabe Y. Meiosis-specific cohesin mediates homolog recognition in mouse spermatocytes. Genes Dev 2014; 28: 594–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carlton PM. Application of advanced fluorescence microscopy to the structure of meiotic chromosomes. Biophys Rev 2013; 5: 313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riederer BM. Antigen preservation tests for immunocytochemical detection of cytoskeletal proteins: influence of aldehyde fixatives. J Histochem Cytochem 1989; 37: 675–681. [DOI] [PubMed] [Google Scholar]
- 36.Heyting C, Dietrich AJ. Meiotic chromosome preparation and protein labeling. Methods Cell Biol 1991; 35: 177–202. [DOI] [PubMed] [Google Scholar]
- 37.Matsuda A, Chikashige Y, Ding DQ, Ohtsuki C, Mori C, Asakawa H, Kimura H, Haraguchi T, Hiraoka Y. Highly condensed chromatins are formed adjacent to subtelomeric and decondensed silent chromatin in fission yeast. Nat Commun 2015; 6: 7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manders EMM, Verbeek FJ, Aten JA. Measurement of co‐localization of objects in dual‐colour confocal images. J Microsc 1993; 169: 375–382. [DOI] [PubMed] [Google Scholar]
- 39.Dobson MJ, Pearlman RE, Karaiskakis A, Spyropoulos B, Moens PB. Synaptonemal complex proteins: occurrence, epitope mapping and chromosome disjunction. J Cell Sci 1994; 107: 2749–2760. [DOI] [PubMed] [Google Scholar]
- 40.Lammers JH, Offenberg HH, van Aalderen M, Vink AC, Dietrich AJ, Heyting C. The gene encoding a major component of the lateral elements of synaptonemal complexes of the rat is related to X-linked lymphocyte-regulated genes. Mol Cell Biol 1994; 14: 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Offenberg HH, Schalk JAC, Meuwissen RLJ, van Aalderen M, Kester HA, Dietrich AJJ, Heyting C. SCP2: a major protein component of the axial elements of synaptonemal complexes of the rat. Nucleic Acids Res 1998; 26: 2572–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phillips D, Nibau C, Wnetrzak J, Jenkins G. High resolution analysis of meiotic chromosome structure and behaviour in barley (Hordeum vulgare L.). PLoS ONE 2012; 7: e39539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiao H, Chen JK, Reynolds A, Höög C, Paddy M, Hunter N. Interplay between synaptonemal complex, homologous recombination, and centromeres during mammalian meiosis. PLoS Genet 2012; 8: e1002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eijpe M, Heyting C, Gross B, Jessberger R. Association of mammalian SMC1 and SMC3 proteins with meiotic chromosomes and synaptonemal complexes. J Cell Sci 2000; 113: 673–682. [DOI] [PubMed] [Google Scholar]
- 45.Prieto I, Pezzi N, Buesa JM, Kremer L, Barthelemy I, Carreiro C, Roncal F, Martinez A, Gomez L, Fernandez R, Martinez-A C, Barbero JL. STAG2 and Rad21 mammalian mitotic cohesins are implicated in meiosis. EMBO Rep 2002; 3: 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang F, De La Fuente R, Leu NA, Baumann C, McLaughlin KJ, Wang PJ. Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. J Cell Biol 2006; 173: 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meuwissen RL, Offenberg HH, Dietrich AJ, Riesewijk A, van Iersel M, Heyting C. A coiled-coil related protein specific for synapsed regions of meiotic prophase chromosomes. EMBO J 1992; 11: 5091–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmekel K, Meuwissen RL, Dietrich AJ, Vink AC, van Marle J, van Veen H, Heyting C. Organization of SCP1 protein molecules within synaptonemal complexes of the rat. Exp Cell Res 1996; 226: 20–30. [DOI] [PubMed] [Google Scholar]
- 49.de Vries FA, de Boer E, van den Bosch M, Baarends WM, Ooms M, Yuan L, Liu JG, van Zeeland AA, Heyting C, Pastink A. Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev 2005; 19: 1376–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tachibana-Konwalski K, Godwin J, van der Weyden L, Champion L, Kudo NR, Adams DJ, Nasmyth K. Rec8-containing cohesin maintains bivalents without turnover during the growing phase of mouse oocytes. Genes Dev 2010; 24: 2505–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boateng KA, Bellani MA, Gregoretti IV, Pratto F, Camerini-Otero RD. Homologous pairing preceding SPO11-mediated double-strand breaks in mice. Dev Cell 2013; 24: 196–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ding DQ, Matsuda A, Okamasa K, Nagahama Y, Haraguchi T, Hiraoka Y. Meiotic cohesin-based chromosome structure is essential for homologous chromosome pairing in Schizosaccharomyces pombe. Chromosoma 2016; 125: 205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A serial z-sectioning image of SYCP3 and RAD21L at the zygotene stage. Mouse spermatocytes were immunofluorescently labeled with anti-SYCP3 (green) and anti-RAD21L (purple) antibodies. Some RAD21L foci were detected between unsynapsed regions of AEs as if they formed a bridge. A scale bar: 1 μm.
A serial z-sectioning image of SYCP3 and REC8 at the zygotene stage.Mouse spermatocytes were immunofluorescently labeled with anti-SYCP3 (green) and anti-REC8 antibodies (purple). No inter-axis bridge-like signals for REC8 were observed in the unsynapsed regions of AEs although such signals were detected in the synapsed regions. Scale bar: 1 μm.
A serial z-sectioning image of RAD21L and REC8 at the zygotene stage. Mouse spermatocytes at the zygotene stage were immunofluorescently labeled with anti-RAD21L (green) and anti-REC8 antibodies (purple). Some RAD21L foci, but not REC8 foci, were detected between unsynapsed regions of AEs. Scale bar: 1 μm.