Abstract
Background
Little is known about the role of the microbiome in primary sclerosing cholangitis.
Aim
Our goal was to explore the mucosa-associated microbiota in PSC patients across different locations in the gut, and to compare it with IBD-only patients and healthy controls.
Methods
Biopsies from the terminal ileum, right colon, and left colon were collected from patients and healthy controls undergoing colonoscopy. Microbiota profiling using bacterial 16S rRNA sequencing was performed on all biopsies.
Results
Forty-four patients were recruited: 20 with PSC (19 with PSC-IBD and one with PSC-only), 15 with IBD-only, and 9 healthy controls. The overall microbiome profile was similar throughout different locations in the gut. No differences in the global microbiome profile were found. However, we observed significant PSC-associated enrichment in Barnesiellaceae at the family level, and in Blautia and an unidentified Barnesiellaceae at the genus level. At the operational taxa unit (OTU) level, most shifts in PSC were observed in Clostridiales and Bacteroidales orders, with approximately 86% of shifts occurring within the former order.
Conclusion
The overall microbiota profile was similar across multiple locations in the gut from the same individual regardless of disease status. In this study, the mucosa associated-microbiota of PSC patients was characterized by enrichment of Blautia and Barnesiellaceae and by major shifts in OTUs within Clostridiales order.
Keywords: Gut microbiota, Primary sclerosing cholangitis, inflammatory bowel disease cholestasis
INTRODUCTION
Primary sclerosing cholangitis (PSC) is a chronic idiopathic cholestatic liver disease, characterized by progressive inflammation and fibrosis of bile ducts, strongly associated with inflammatory bowel disease (IBD), especially with ulcerative colitis (UC)1–3. PSC-IBD patients have a distinctive phenotype as compared to IBD alone. They typically present asymptomatic or mildly symptomatic extensive colitis, a quiescent clinical course, more right-sided inflammation, rectal sparing, backwash ileitis, increased risk of pouchitis, and an increased risk for developing colorectal neoplasia4–10. Neither the cause of PSC11, nor the mechanisms by which IBD patients develop this unique phenotype when they have concomitant PSC are known.
Some data from basic and clinical studies support the hypothesis that the intestinal microbiota may have a role in disease pathogenesis12, 13. For example, animal models of surgically-induced small intestinal bacterial overgrowth develop abnormalities in the intra- and extra-hepatic bile ducts that resemble PSC by cholangiography and histology12. The improvement of liver enzymes following a trial of antibiotic therapy, and a recent case reporting laboratorial and histological abnormalities reversal following vancomycin treatment in a liver transplant PSC recurrence, also suggest a role for the gut flora in disease pathogenesis14–16.
With the advent of culture-independent techniques, a better understanding of how the gut microbiome can affect and modulate the development of liver diseases has emerged. Indeed microbiota alterations in cirrhosis are now well-documented17. A prior study has observed a lower abundance of uncultured Clostridiales in PSC patients as compared to IBD and healthy controls, not providing however more in-depth analysis18. Being a cholestatic liver disease with profound alterations in bile acid pool19, and acknowledging the reciprocal relationship between gut flora and bile acid metabolism20–23, it is very likely that patients with PSC-IBD exhibit alterations in the gut microbiota. The main objective of this study was to further explore the mucosa-associated microbiota in PSC patients, and to compare it with IBD-only and healthy controls.
MATHERIAL AND METHODS
Ethical considerations
This study was approved by the Institutional Review Boards of Icahn School of Medicine at Mount Sinai and the University of Chicago. All patients and healthy controls signed an informed consent form.
Subjects and sampling
Between November 2011 and November 2014, patients with PSC, patients with IBD and healthy controls undergoing colonoscopy at the Mount Sinai Medical Center and at the University of Chicago, were prospectively recruited, within a collaborative multicentre program for integrated studies in IBD (Sinai-Helmsley Alliance for Research Excellence (SHARE) Network). Sample collection protocol was standardized across two institutions to avoid possible bias during sample collection and processing. The inclusion criteria were age greater than 18 years, confirmed diagnosis of PSC based on histology and/or abnormal cholangiogram (ERCP or MRCP), and confirmed diagnosis of IBD by conventional endoscopic and histological criteria24. Patients with a personal history of colectomy, a diagnosis of secondary sclerosing cholangitis, or concomitant infectious colitis at the time of colonoscopy were excluded. Patients with newly diagnosed PSC, who were scheduled for their initial colonoscopy to screen for IBD, and healthy controls undergoing screening colonoscopy, were also recruited.
Demographical and clinical information were recorded for every patient. During colonoscopy, disease severity was recorded, and biopsies for colorectal neoplasia screening were obtained, according to current guidelines. On all subjects, biopsies were collected from the left colon (LC) for microbiota analysis, and in a subset of patients, biopsies were also collected from the terminal ileum (TI) and right (RC) for comparison of the microbiota features across different colonic locations. Biopsies were either snap frozen or stored in RNAlater® (Qiagen, Valencia CA) for subsequent analysis. All samples were analysed at Icahn School of Medicine at Mount Sinai, New York.
Tissue DNA extraction and 16S ribosomal RNA (rRNA) amplification
Tissue samples were transferred into bead tubes (MO-BIO, Carlsbad, CA) and homogenized using bead beating method. Homogenized tissue samples were further processed using the Qiagen DNeasy Blood & Tissue Kit following the manufacturer’s protocol (Qiagen, Valencia, CA). Total DNA concentration was determined with Qubit 2.0 Fluorometer (Life Technologies, Norwalk, CT). The phylogenetically informative V3-V4 region of 16S ribosomal RNA (rRNA) gene was amplified using universal primer set 347F/803R25. The primers were synthesized by IDT (Integrated DNA Technology, Coralville, IA). We used a dual-barcoding approach to label the 16S rRNA amplicons from each sample as described previously. Briefly, the 6-mer barcodes were attached on the 5’ ends of both forward and reverse PCR primers so that 16S rRNA PCR amplicons from each sample contained a unique dual barcode combination. The 16S rRNA amplicons were further pooled with equal molarity and submitted for MiSeq 2×300 pair-end sequencing at high depth. The paired sequence readings were merged and filtered by size (>400bp) and quality score (>Q30) using CLC genomics workbench version 7. The processed readings were further split by dual barcode for each sample and assigned taxonomic classification using QIIME pipeline 1.8.026. Repeated measurements of the same sample were made to assess sequencing reproducibility. After processing, QIIME provided detailed operational taxa unit (OTU) tables containing the microbiota composition and abundance for each individual sample.
Metagenomic 16S rRNA data analysis
To characterise the gut microbiota, firstly the overall microbiota dissimilarities among all samples, also known as beta-diversity, were accessed using the Bray-Curtis distance matrices and visualized by non-metric multiple dimensional scaling (nMDS) plots. The PerMANOVA test27, with the maximum number of permutations = 999, was performed using the [Adonis] function of the R package vegan 2.0–528, 29 to test the significance of the overall microbiota differences between groups by PSC and IBD status. Secondly, the diversity of the microbial community within each sample, so-called alpha-diversity, was measured using the Shannon Index as a metric to represent the species diversity30. Next, significant differential taxa features at the family and genus levels were selected using random forest algorithm, a supervised machine learning approach, using R package rfPermute and confirmed by Boruta feature selection (R package Boruta)31, 32. Only features that were consistent in both analyses were selected. The significance of the selected taxa was further tested by t-test. In addition, at the OTU level, we performed the log likelihood ratio test (QIIME command group_significance.py using g-test statistics) to further identify significant differential OTUs between PSC and the healthy controls using LC samples only. The resulted p-values were adjusted by the FDR methods. We also compared the PSC vs. IBD-only at LC, RC and TI locations using the differential OTUs selected from LC samples.
Blautia-specific long 16S rRNA sequencing
Long 16S rRNA reads can further improve the taxa OTU inference33, 34. Therefore, we designed 16-base-barcoded 404F/1263R primer pairs (Supplementary Table 1) specifically for the Blautia genus based on 16S rRNA reference sequence of the Blautia genus. The ~860 bp-sized PCR amplicons were pooled for sequencing on the PacBio RS II. Sequencing data from PacBio was processed using the manufacturer provided program smrtanalysis v.2.1.1 (https://github.com/PacificBiosciences/SMRT-Analysis/). Circular consensus sequencing (CCS) reads were then filtered by size (>800bp) and the quality score (>Q30) using CLC genomics workbench version 7. After split by barcode for each sample, all filtered reads were processed using QIIME pipeline 1.8.035. The generated OTUs were filtered to only keep OTUs assigned to the Blautia genus and with more than 100 counts of reads. We performed the G-test of independence35 to determine whether Blautia OTU presence/absence is associated with PSC status at LC samples only, in which we combined healthy control and non-PSC IBD together as non-PSC group to compare with PSC. Representative sequences from significantly differential Blautia OTUs were further aligned with the Blautia reference sequences to construct the maximum likelihood phylogenetic tree using UPGMA method and performed a pair-wise sequence alignment comparison
RESULTS
Study population
Between November 2011 and November 2014, 46 patients were enrolled at both centres: 20 with PSC (19 of which had concomitant IBD), 16 with IBD and 10 healthy controls. There were 27 males (61%) and the median age for the whole population was 47 years (IQR 33.5, 58). The mean age of each group was as following: 43.8 years for healthy controls, 50.3 for the IBD group, and 45.3 for the PSC group. No patient was on antibiotics at the time of colonoscopy.
Samples from two subjects (one IBD patient and one healthy control) were eliminated from further analysis due to over-contamination (>90% relative abundance) with Escherichia coli). Therefore, 44 patients were included in the final analysis: 20 patients with PSC (19 with PSC-IBD and one with PSC-only), 15 patients with IBD and 9 healthy controls (Table 1). Biopsies from different colonic locations (TI, RC and LC) were available in 18 subjects (11 with PSC, 6 with IBD-only and 1 with PSC only), except for two cases, where the TI could not be intubated. The clinical characteristics of PSC and IBD patients are described in Table 1. The patient with PSC-only was analysed in the PSC group. One of the PSC-IBD patients had a history of liver transplant and another had a history of choledochojejunostomy for a dominant stricture. There was no history of colon surgery among any of the participants.
Table 1.
Clinical characteristics of PSC and IBD (the patient presenting PSC-only was included in the PSC group together with PSC-IBD).
| PSC (n=20) | IBD (n=15) | |
|---|---|---|
| Male (n, %) | 16 (80%) | 9 (60%) |
| Age (y) Median, (IQR) |
47 (33.5, 59.3) | 48 (34.5, 59.5) |
| Smoking status | ||
| Never | 13 (65%) | 10 (67%) |
| Ever | 6 (30%) | 5 (33%) |
| Unknown | 1 (5%) | |
| Type of IBD | ||
| UC/IBDU | 13 (65%) | 13 (87%) |
| CD | 6 (30%) | 2 (13%) |
| No IBD | 1 (5%) | |
| Extent of IBD |
UC/IBDU Extensive colitis – 100% |
UC Extensive colitis- 12 (92%) Left colitis – 1 (8%) |
|
CD Colonic disease: 3 (50%) Ileocolonic disease:3(50%) |
CD Colonic disease: 2(100%) |
|
|
PSC duration; median years (IQR) |
4 (2, 12.3) | - |
|
IBD duration, median years, (IQR) |
9 (4.8, 18.9) | 9 (4.75, 22) |
|
PSC Mayo score*, median, (IQR) |
0.03(−0.63, 0.42) | - |
|
Endoscopic score of inflammation¶ |
||
| Normal/quiescent | 11 | 11 |
| Mild | 5 | 3 |
| Moderate | 4 | 1 |
|
Medications at the time of colonoscopy |
||
| Unknown/No medications | 5 | - |
| 5-ASA | 8 | 10 |
| Thiopurines | 5 | 8 |
| Anti-TNF | 5 | 3 |
| Ursodeoxycholic acid | 10 | - |
| Tacrolimus | 1 | - |
| Cholestyramine | 1 | - |
The PSC Mayo score could not be calculated in two patients due to lack of information.
For purposes of simplicity the endoscopic score CD-SES was replaced in this table by the subjective impression of the endoscopist performing the colonoscopy into mild or moderate inflammation (the median CD-SES for the patients with CD in this study was 11.5, IQR 10–13).
IQR: interquartile range, IBDU: IBD unclassified
Samples
16S rRNA amplicons from 81 samples (44 LC, 18 RC, 16 TI and 3 technical repeats) were sequenced and a total of 9.3 million reads were generated after filtering by size and quality, as described in the methods section. On average, each sample contained ~110,000 reads. The repeated measurements showed Pearson correlation to be 99% at the genus level. We used the mean of the repeated measurements for further analysis.
The mucosa-associated microbiota is stable across different locations within each individual
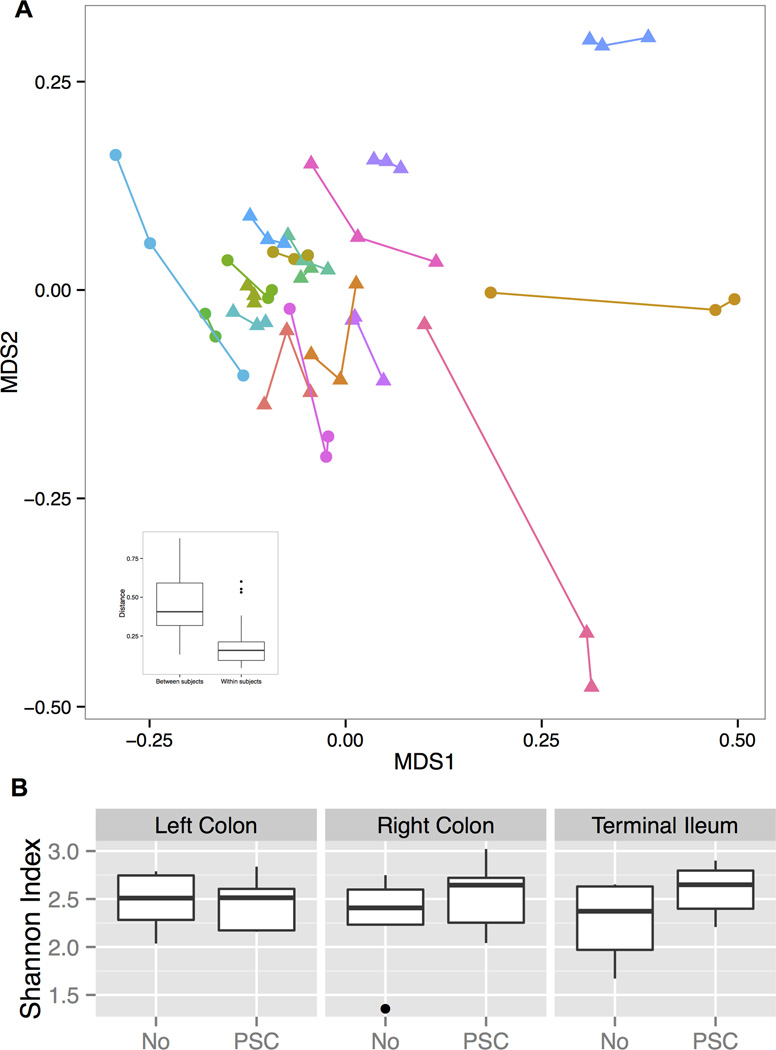
Taking into account the distinct phenotype of more inflammation and a higher prevalence of neoplasia in the right colon observed in PSC-IBD patients, we specifically assessed if there were any differences in the spatial distribution of the mucosa-associated microbiota between the RC, LC and TI. Therefore, we compared the microbiota composition at three biopsy locations in the 18 subjects, from whom multiple locations were available. Our results (Fig. 1A.) showed that although a few samples from the same subjects showed substantial variations, in general, the overall microbiota was consistent across all three sampling locations. The distance across locations within the same subject was significantly smaller than the distance between samples from the same location but from different subjects (mean=0.18 and 0.45, SD=0.13 and 0.17, respectively, p-value<0.05). No significant differences in the species richness (measured by the Shannon index) were observed between PSC and IBD-only patients in all three locations (Fig. 1B.).
Figure 1. Microbiota profiles across multiple biopsy locations.
(A) Overall microbiota dissimilarities between samples. Dissimilarities were measured by distance and visualized using non-parametric multidimensional scaling plot. Lines link the multiple sampling locations from the same subject. Insert at lower left corner shows the mean and variance of the distance between the samples from the same or different subjects. Triangles represent PSC patients; Circles represent non-PSC, IBD patients. (B) The boxplots show the mean and variance of the richness of the microbial community within each sample, showing no significant difference across multiple locations.
PSC associated left colon mucosa microbiota features
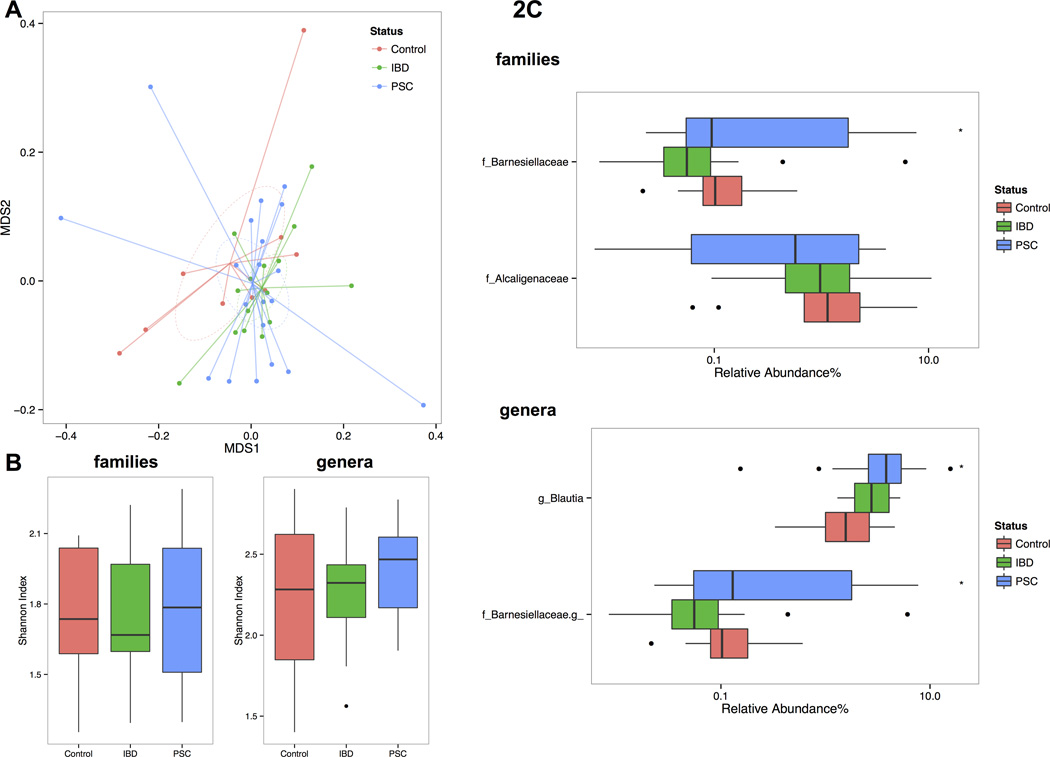
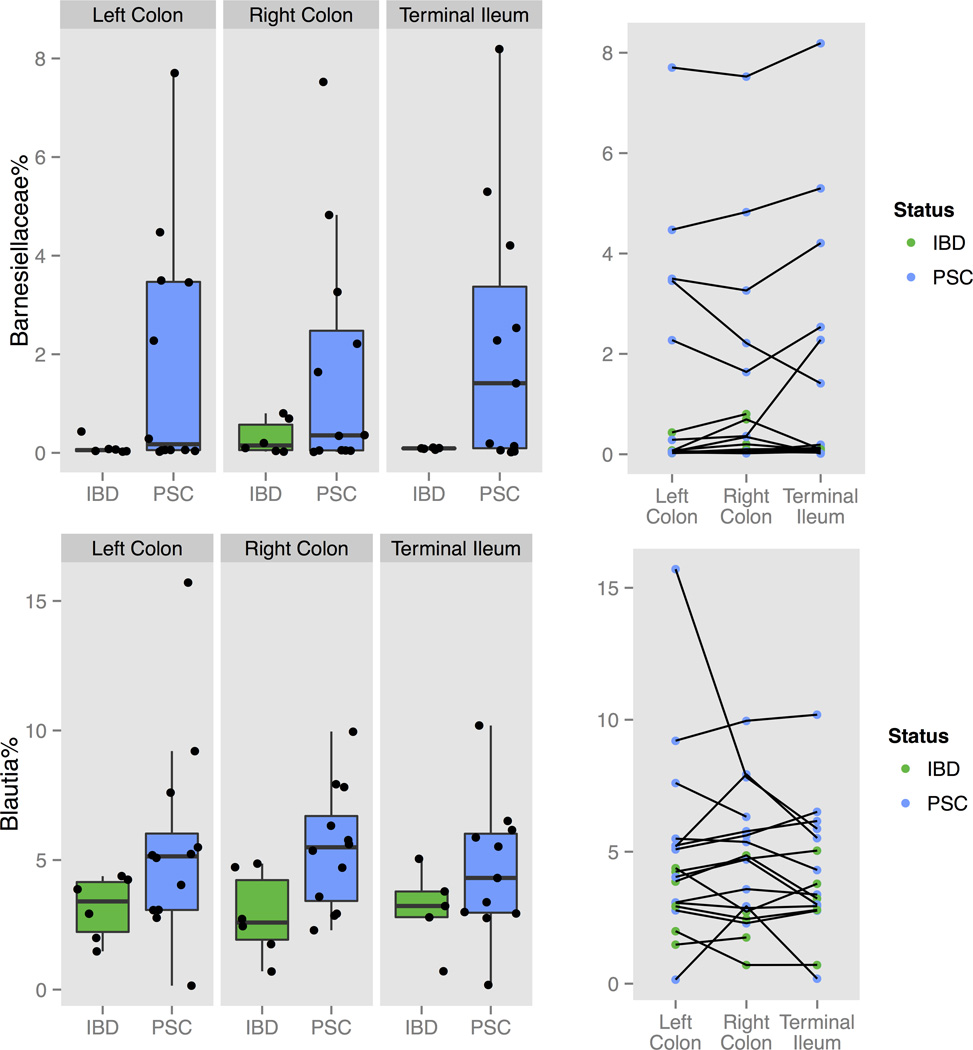
Observing that there were no overall differences in the microbiota diversity between TI, RC and LC, we next analysed the microbiota composition only in the LC of all subjects. The overall microbiota dissimilarities among all 44 LC samples (20 PSC, 15 IBD patients and 9 healthy controls) grouped by PSC and IBD status were accessed using the Bray-Curtis distance matrices (Fig. 2A.). Although we noticed a separation between the healthy control and the IBD or PSC-IBD, the PerMANOVA test did not find a significant difference in the global LC microbiota profile by disease status. We also did not observe a significant difference among controls, IBD and PSC samples in species richness using Shannon Index (Fig. 2B.). At the taxa level, two families including Barnesiellaceae and Alcaligenaceae, as well as two genera including Blautia and an unidentified genus from Barnesiellaceae family were selected by random forest algorithm using R package rfPermute and confirmed by Boruta feature selection (R package Boruta) (Fig. 2C.). Among those selections, we observed significant enrichment of Barnesiellaceae family and its further assigned unidentified genus (mean abundance=1.3% in PSC samples, 0.48% in IBD and 0.16% in healthy controls; p-values=0.44, 0.025, respectively by t-test) and Blautia (mean=4.5% in PSC samples, 2.9% in IBD and 2.1% in healthy controls; p-values=0.22 and 0.02, respectively by t-test) in PSC samples compared to healthy controls. To test whether or not the PSC-associated taxa features found in LC were consistent in the RC and TI, we further compared the abundance of Barnesiellaceae family and Blautia genus at three locations in the available samples from these locations. We found that not only did the enrichment of both taxa in PSC patients occur in all locations, but also that the abundance of those taxa was consistent across multiple locations within the same individual (Fig. 3.). Further analysis excluding the patients with PSC with an history of OLT and an history of choledochojejunostomy did not change results (data not shown).
Figure 2. Microbiota dissimilarities by PSC and IBD status in left colon samples.
(A) Overall microbiota dissimilarities between samples grouped by PSC and IBD status. Dissimilarities were measured by distance and visualized using non-parametric multidimensional scaling plot. (B) The boxplots showed the mean and variance of the richness of the microbial community between different disease status. (C) The log-scaled boxplots showed the differential taxa features selected at family and genus level by health status. The asteroid indicates the p-value <0.05 between PSC and healthy controls.
Figure 3. The relative abundance of Barnesiellaceae and Blautia in PSC and non-PSC-IBD at multiple locations.
Boxplots (left panel) showed the mean and variance of the relative abundance; Dot plots (right panel) showed the relative abundance for each individual samples and the lines link the samples from the same subject. Green: non-PSC IBD; blue: PSC
We then assessed whether there were any differences in microbiota composition according to PSC severity score. Based on the ranked PSC Mayo score, the PSC patients were assigned to low risk (Mayo score <0), intermediate risk (Mayo score from 0–2) and high-risk score (Mayo score>2). There were no patients with severe disease; however, among the low and intermediate risk, groups had a similar global microbiota composition (Supplementary Fig. 1A.) and taxa richness (Supplementary Fig. 1B.). At the taxa level, the level of Blautia was not different between two risk groups. The low risk group showed higher median level of Barnesiellaceae family compared to the intermediate risk group (Supplementary Fig. 1C.), but this did not reach statistical significance.
Differential OTUs by PSC status
We performed de novo OTU picking using QIIME pipeline. Based on the 97% similarity of the 16S rRNA sequencing reads, all sequencing reads were clustered into individual OTUs. After removing rare OTUs (relative abundance < 0.1% in all samples), we compared 2439 OTUs and selected 80 and 15 OTUs significantly (p<0.05 by parametric t-test, not adjusted) different between PSC, healthy control (Supplementary Table S2) or IBD (Supplementary Fig. 2.). We found that when compared with healthy controls, most of the PSC-associated shifts in the bacterial composition were observed in the Clostridiales and Bacteroidetes orders, with 86% in the former order. In agreement with our above findings, several PSC-enriched OTUs belonged to the Blautia genus and the Barnesiellaceae family. When compared to IBD, many OTUs were from the Blautia genus. Similar enrichments and reductions at the PSC-associated OTUs selected from LC samples (Supplementary Fig. 2.) were observed in RC and TI locations.
Differential OTUs at Blautia genus between PSC and non-PSC by long-read 16S rRNA sequencing
Our results showed that both Barnesiellaceae and Blautia genus were enriched in PSC patients. But unlike Barnesiellaceae, Blautia comprised >2% of the entire microbiota regardless of the disease status, so it is plausible to specifically enrich this genus and further use the long 16S rRNA reads to perform additional taxa OTU inference at Blautia genus in both patient and control samples. After processing, 2967 OTUs were assigned to Blautia genus. Among those, 135 OTUs had reads of more than 100 counts. The G-test of independence further selected 7 out of 135 OTUs significantly different between PSC and non-PSC (unadjusted p-values<0.05). We aligned those Blautia OTUs with Blautia reference sequences and constructed the maximum likelihood phylogenetic tree using UPGMA method (Supplementary Fig.3A.). We also performed the pair-wise sequence alignment comparison and showed the number of difference (upper) and the percent identity (lower) between pairs. (Supplementary Fig.3C.). Our results showed that 4 OTUs (denovo17640, denovo28452, denovo13317, denovo6236) were enriched and 3 OTUs (denovo25871, denovo781, denovo18792) were reduced in PSC samples (Supplementary Fig. 3B.). We also found that the OTUs denovo17640, denovo13317 and denovo28452 showed >99% percent identity to reference strains from species of Blautia wexlerae, Ruminococcus obeum and Blautia faecis, respectively (Supplementary Fig. 3C.).
DISCUSSION
Herein we compared the mucosa-associated bacterial flora of patients with PSC to IBD alone and to normal subjects. Because faecal microbiota does not necessarily reflects the mucosa-associated microbiota36, 37, we chose to examine the mucosa-associated microbiota, rather than stool, in order to gain more insight into the unique phenotype of PSC-IBD patients who tend to develop more right-sided inflammation, and proximal neoplasia. We thought there might be some differences in the microbiota composition throughout different locations of the ileo-colon in this specific clinical context, given the phenotypic associations with disease location for PSC5. However, and in line with previous observations in healthy subjects38, no significant site-specific differences in the microbial composition were found throughout the colon. Although this analysis may have been limited by sample size, it is possible that other mechanisms may be operating and interacting differently with the microbiota in the right as opposed to the left colon39 in patients with PSC and IBD. Rossen et al18 in a prior study, also described similar hierarchical clustering between samples from the TI and RC from the same subject from between PSC, IBD and healthy controls. However, no comparisons with the mucosa-associated from the left colon were available. The same authors described a lower diversity and abundance of uncultured Clostridiales II at the genus level compared to UC and healthy controls. However, this study, was limited by a smaller sample size and, by the use of a probe-based approach, the HITChip18, 40 that only allows to detect phylotypes present in the chip at the genus-like level. Our study, using a longer read 16S sequencing, allowed us to provide deeper taxonomic analysis that could inform on specific microbiome shifts associated with PSC. Another recently published study41, assessing differences in microbial composition PSC-UC and UC patients, did not report differences in overall microbial diversity between PSC-IBD and UC, however different geographical provenience of samples was the main driver for microbial composition in this study. We observed a trend in the compositional dissimilarity of the overall microbiota between healthy controls, and PSC-IBD, but this did not reach statistical significance. This could be due to the modest sample size, underrepresentation of healthy controls, and disease remission for most of the IBD patients, since shifts in the microbiota can vary according to disease activity42. Using bacterial 16S rRNA next-generation sequencing, we found, across all colonic locations, a consistent PSC-enrichment in Blautia and Barnesiellaceae genera and shifts under the Clostridiales, and less frequently under the Bacteroidales order. Deeper taxa analysis at the OTU level was consistent with these findings, also showing several enriched OTUs particularly from Blautia and Barnesiellaceae genera. Specifically, around 86% of the relative changes in the microbiota occurred within the Clostridiales order, with reduction in 3 and enrichment in 66 OTUs. This is interesting, as shifts in these taxa have been observed both in IBD and also in cirrhosis20, 43. Bacteroidetes species play an important role in protein metabolism44 as well in bile acid deconjugation45. The Clostridiales order encompasses bacteria from Lachnospiraceae family, Ruminococcaceae family and Blautia genus, which are able to perform 7α-dehydroxylation23, an important step in converting primary to secondary bile acids in the intestine. Furthermore, it has been demonstrated that certain Clostridum spp. can affect number, function and differentiation of colonic Treg cells, therefore playing a crucial role in colonic homeostasis46. In advanced cirrhosis, a shift in the gut flora towards the enrichment of Enterobacteriaceae and the reduction of Clostridiales and Bacteroidetes in parallel with a reduced level of faecal secondary bile acids has been described20. Conversely, Islam and colleagues showed that feeding animal models with bile acids leads to an enrichment of colonic bile acids, which in turns results in the expansion of the Firmicutes phylum, specifically within the Clostridia class, with Blautia spp expanding significantly47. Being a cholestatic liver disease, characterized by scarring of the bile ducts, PSC is expected to lead to a reduction of the flow of bile acids from the liver to the intestine, at least in the more advanced stages of the disease. However, there also are data suggesting that during obstructive cholestasis, the apical sodium dependent bile acid transporter ASBT is down-regulated48, 49, as a feedback anti-cholestatic mechanism. This could hypothetically lead to a relative increase of BA in the proximal colon, which could in turn lead to an enrichment of bacterial species involved in bile acid handling23, 48. Since in our study we did not have patient´s bile acid profiles we couldn´t test for this hypothesis.
A limitation of our study is its relatively small sample size, so we were unable and unpowered to make any associations between disease severity and microbial composition, or to take into account any impact of medication, diet, or disease course. Despite this limitation, and compared with previous studies, we were able to compare microbiota composition from multiple sites41, had representation of PSC-IBD patients, IBD and normal controls41, and a reasonable sample size18, that allowed to perform deeper taxonomic analyses18. We observed that the mucosa-associated microbiota was consistent among all locations, and that the Blautia and Barnesiellaceae enrichment was consistently found not only in the left colon samples but also in other locations. Consistent with findings at the family and genus level, deeper OTU level analyses also found enrichment of Blautia and Barnesiellaceae OTUs in PSC. Therefore, in that sense we validated our findings. The cross-sectional nature of the study does not allow us to conclusively determine a causal link between the abundance of these species and its role in PSC. It is possible that the shifts in the microbiota features we observed in PSC-IBD are a consequence, rather than the cause, of the interaction between cholestasis and colonic inflammation. Furthermore, in the absence of a non-PSC liver disease control group, it is difficult to appreciate if the microbiota shifts we observed are specific to PSC or belong to a broader dysbiosis observed in chronic liver diseases17, 20. Whether these changes contribute to the special phenotype observed in PSC-IBD patients can only be speculated upon at this stage, and merits further investigation. Future studies investigating the role of microbiota in PSC should aim in collecting larger samples sizes, that could allow adjustment for clinical and analytical variables that could influence microbial composition in PSC such disease duration and stage, medication intake, diet, geographical location of patients, impact of liver transplant and or biliary surgery, as well as serum and faecal bile acid pool.
Supplementary Material
Acknowledgments
We thank the OCS genome technology centre of New York University Langone Medical Center for the Illumina Miseq library preparation and sequencing service. We thank the genomic core facility at Icahn School of Medicine at Mount Sinai for the long-read 16S rRNA sequencing using the Pacific Biosciences RS II for single molecule sequencing.
Financial support: This study was supported by funds from National Institute of Health, NIDDK 1K01DK094986-01(JH); The Chemotherapy Foundation (SI), and The Helmsley Foundation (SI, BJ, IP).
Authors' declaration of personal interests:
Joana Torres has received speaking fees from Falk and consulting fees from Abbvie. Joseph Odin is an advisory board member for Intercept Pharmaceuticals and has received research funding from Gilead, NIDDK. J.-F. Colombel has served as consultant, advisory board member or speaker for Abbvie, AB Science, Amgen, Bristol Meyers Squibb, Celltrion, Danone, Ferring, Genentech, Giuliani SPA, Given Imaging, Janssen, Immune Pharmaceuticals, Medimmune, Merck & Co., Millenium Pharmaceuticals Inc., Neovacs, Nutrition Science Partners Ltd., Pfizer Inc. Prometheus Laboratories, Protagonist, Receptos, Sanofi, Schering Plough Corporation, Second Genome, Shire, Takeda, Teva Pharmaceuticals, Tigenix, UCB Pharma, Vertex, and Dr. August Wolff GmbH & Co.Steve Itzkowitz received grant support and fees for serving on a scientific advisory board from Exact Sciences.
List of abbreviations
- PSC
primary sclerosing cholangitis
- IBD
inflammatory bowel disease
- UC
ulcerative colitis
- ERCP
endoscopic retrograde cholangiopancreatography
- MRCP
magnetic resonance cholangiopancreatography
- CD
Crohn´s disease
- TI
terminal ileum
- RC
right colon
- LC
left colon
- SHARE
Sinai-Helmsley Alliance for Research Excellence Network
- DNA
deoxyribonucleic acid
- RNA
Ribonucleic acid
- PCR
polymerase chain reaction
- OTU
operational taxonomic unit
Footnotes
Authors´ Contributions: JT, JFC, JH and SI have contributed to the study design and concept. All authors have contributed to the acquisition of data. JT, JH and SI analysed, interpreted the data and drafted the manuscript. All authors have reviewed versions of the manuscript and provided critical comments. SI, BJ, JH obtained funding support.
REFERENCES
- 1.Fausa O, Schrumpf E, Elgjo K. Relationship of Inflammatory Bowel Disease and Primary Sclerosing Cholangitis. Semin Liver Dis. 1991;11(01):31, 39. doi: 10.1055/s-2008-1040420. [DOI] [PubMed] [Google Scholar]
- 2.Rasmussen HH, Fallingborg JF, Mortensen PB, Vyberg M, Tage-Jensen U, Rasmussen SN. Hepatobiliary dysfunction and primary sclerosing cholangitis in patients with Crohn's disease. Scand J Gastroenterol. 1997;32(6):604–610. doi: 10.3109/00365529709025107. [DOI] [PubMed] [Google Scholar]
- 3.Olsson R, Danielsson A, Jarnerot G, et al. Prevalence of primary sclerosing cholangitis in patients with ulcerative colitis. Gastroenterology. 1991;100(5 Pt 1):1319–1323. [PubMed] [Google Scholar]
- 4.Rahman M, Desmond P, Mortensen N, Chapman R. The clinical impact of primary sclerosing cholangitis in patients with an ileal pouch–anal anastomosis for ulcerative colitis. Int J Colorectal Dis. 2011:1–7. doi: 10.1007/s00384-011-1140-9. [DOI] [PubMed] [Google Scholar]
- 5.Loftus EV, Jr, Harewood GC, Loftus CG, et al. PSC-IBD: a unique form of inflammatory bowel disease associated with primary sclerosing cholangitis. Gut. 2005;54(1):91–96. doi: 10.1136/gut.2004.046615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lundqvist K, Broome U. Differences in colonic disease activity in patients with ulcerative colitis with and without primary sclerosing cholangitis: a case control study. Dis Colon Rectum. 1997;40(4):451–456. doi: 10.1007/BF02258391. [DOI] [PubMed] [Google Scholar]
- 7.Sokol H, Cosnes J, Chazouilleres O, et al. Disease activity and cancer risk in inflammatory bowel disease associated with primary sclerosing cholangitis. World J Gastroenterol. 2008;14(22):3497–3503. doi: 10.3748/wjg.14.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Navaneethan U, Choudhary M, Venkatesh PG, et al. The effects of liver transplantation on the clinical course of colitis in ulcerative colitis patients with primary sclerosing cholangitis. Aliment Pharmacology Ther. 2012;35(9):1054–1063. doi: 10.1111/j.1365-2036.2012.05067.x. [DOI] [PubMed] [Google Scholar]
- 9.Torres J, Pineton de Chambrun G, Itzkowitz S, Sachar DB, Colombel JF. Review article: colorectal neoplasia in patients with primary sclerosing cholangitis and inflammatory bowel disease. Aliment Pharmacol Ther. 2011;34(5):497–508. doi: 10.1111/j.1365-2036.2011.04753.x. [DOI] [PubMed] [Google Scholar]
- 10.Eaton JE, Smyrk TC, Imam M, et al. The fate of indefinite and low-grade dysplasia in ulcerative colitis and primary sclerosing cholangitis colitis before and after liver transplantation. Aliment Pharmacology Ther. 2013;38(8):977–987. doi: 10.1111/apt.12469. [DOI] [PubMed] [Google Scholar]
- 11.Eaton JE, Juran BD, Atkinson EJ, et al. A comprehensive assessment of environmental exposures among 1000 North American patients with primary sclerosing cholangitis, with and without inflammatory bowel disease. Aliment Pharmacology Ther. 2015;41(10):980–990. doi: 10.1111/apt.13154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tabibian JH, O'Hara SP, Lindor KD. Primary sclerosing cholangitis and the microbiota: current knowledge and perspectives on etiopathogenesis and emerging therapies. Scan J Gastroenterol. 2014;49(8):901–908. doi: 10.3109/00365521.2014.913189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tabibian JH, Talwalkar JA, Lindor KD. Role of the microbiota and antibiotics in primary sclerosing cholangitis. Biomed Res Int. 2013;2013:389537. doi: 10.1155/2013/389537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tabibian JH, Weeding E, Jorgensen RA, et al. Randomised clinical trial: vancomycin or metronidazole in patients with primary sclerosing cholangitis - a pilot study. Aliment Pharmacology Ther. 2013;37(6):604–612. doi: 10.1111/apt.12232. [DOI] [PubMed] [Google Scholar]
- 15.Paterson JC, Dillon JF. Commentary: vancomycin or metronidazole in patients with primary sclerosing cholangitis? Aliment Pharmacology Ther. 2013;37(9):915. doi: 10.1111/apt.12282. [DOI] [PubMed] [Google Scholar]
- 16.Davies YK, Tsay CJ, Caccamo DV, Cox KM, Castillo RO, Cox KL. Successful treatment of recurrent primary sclerosing cholangitis after orthotopic liver transplantation with oral vancomycin. Case Rep Transplant. 2013;2013:314292. doi: 10.1155/2013/314292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schnabl B, Brenner DA. Interactions between the intestinal microbiome and liver diseases. Gastroenterology. 2014;146(6):1513–1524. doi: 10.1053/j.gastro.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rossen NG, Fuentes S, Boonstra K, et al. The mucosa-associated microbiota of PSC patients is characterized by low diversity and low abundance of uncultured Clostridiales II. J Crohns Colitis. 2015;9(4):342–348. doi: 10.1093/ecco-jcc/jju023. [DOI] [PubMed] [Google Scholar]
- 19.Sinakos E, Marschall H-U, Kowdley KV, Befeler A, Keach J, Lindor K. Bile acid changes after high-dose ursodeoxycholic acid treatment in primary sclerosing cholangitis: Relation to disease progression. Hepatology. 2010;52(1):197–203. doi: 10.1002/hep.23631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kakiyama G, Pandak WM, Gillevet PM, et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol. 2013;58(5):949–955. doi: 10.1016/j.jhep.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bajaj JS, Ridlon JM, Hylemon PB, et al. Linkage of gut microbiome with cognition in hepatic encephalopathy. Am J Physiol Gastrointest Liver Physiol. 2012;302(1):G168–G175. doi: 10.1152/ajpgi.00190.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. Curr Opin Gastroenterol. 2014;30(3):332–338. doi: 10.1097/MOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47(2):241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 24.Lennard-Jones JE. Classification of inflammatory bowel disease. Scan J Gastroenterol. Supplement. 1989;170:2–6. doi: 10.3109/00365528909091339. discussion 16-9. [DOI] [PubMed] [Google Scholar]
- 25.Ahn J, Yang L, Paster BJ, et al. Oral microbiome profiles: 16S rRNA pyrosequencing and microarray assay comparison. PloS One. 2011;6(7):e22788. doi: 10.1371/journal.pone.0022788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu J, Franzen O, Pei Z, Itzkowitz S, Peter I. P-237 Multiple Double-barcoding 16S Sequencing on the MiSeq Platform to Study the Gut Microbiome in Ashkenazi Jews with Crohn's Disease. Inflamm Bowel Dis. 2013;19:S119–S121. [Google Scholar]
- 27.Tong M, McHardy I, Ruegger P, et al. Reprograming of gut microbiome energy metabolism by the FUT2 Crohn's disease risk polymorphism. The ISME journal. 2014 doi: 10.1038/ismej.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Price LB, Liu CM, Johnson KE, et al. The effects of circumcision on the penis microbiome. PLoS One. 2010;5(1):e8422. doi: 10.1371/journal.pone.0008422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, Bittinger K, Charlson ES, et al. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics. 2012;28(16):2106–2113. doi: 10.1093/bioinformatics/bts342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oksanen J. Multivariate Analysis of Ecological Communities in R: vegan tutorial. 2013 [Google Scholar]
- 31.Aagaard K, Riehle K, Ma J, et al. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PloS one. 2012;7(6):e36466. doi: 10.1371/journal.pone.0036466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sze MA, Tsuruta M, Yang SW, et al. Changes in the bacterial microbiota in gut, blood, and lungs following acute LPS instillation into mice lungs. PloS one. 2014;9(10):e111228. doi: 10.1371/journal.pone.0111228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morgan XC, Huttenhower C. Chapter 12: Human Microbiome Analysis. Plos Comput Biol. 2012;8(12):e1002808. doi: 10.1371/journal.pcbi.1002808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shannon CE. A Mathematical Theory of Communication. At&T Tech J. 1948;27(4):623–656. [Google Scholar]
- 35.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morgan XC, Tickle TL, Sokol H, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13(9):R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stearns JC, Lynch MD, Senadheera DB, et al. Bacterial biogeography of the human digestive tract. Sci Rep. 2011;1:170. doi: 10.1038/srep00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zoetendal EG, von Wright A, Vilpponen-Salmela T, Ben-Amor K, Akkermans AD, de Vos WM. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Applied and environmental microbiology. 2002;68(7):3401–3407. doi: 10.1128/AEM.68.7.3401-3407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torres J, Bao X, Iuga AC, et al. Farnesoid X receptor expression is decreased in colonic mucosa of patients with primary sclerosing cholangitis and colitis-associated neoplasia. Inflamm Bowel Dis. 2013;19(2):275–282. doi: 10.1097/MIB.0b013e318286ff2e. [DOI] [PubMed] [Google Scholar]
- 40.Karlsson F, Tremaroli V, Nielsen J, Backhed F. Assessing the human gut microbiota in metabolic diseases. Diabetes. 2013;62(10):3341–3349. doi: 10.2337/db13-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kevans D, Tyler AD, Holm K, et al. Characterization of Intestinal Microbiota in Ulcerative Colitis Patients with and without Primary Sclerosing Cholangitis. J Crohns Colitis. 2015 doi: 10.1093/ecco-jcc/jjv204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bornigen D, Morgan XC, Franzosa EA, et al. Functional profiling of the gut microbiome in disease-associated inflammation. Genome Med. 2013;5(7):65. doi: 10.1186/gm469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gevers D, Kugathasan S, Denson Lee A, et al. The Treatment-Naive Microbiome in New-Onset Crohn’s Disease. Cell Host Microbe. 15(3):382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rajilic-Stojanovic M, de Vos WM. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev. 2014;38(5):996–1047. doi: 10.1111/1574-6976.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Narushima S, Itoha K, Miyamoto Y, et al. Deoxycholic acid formation in gnotobiotic mice associated with human intestinal bacteria. Lipids. 2006;41(9):835–843. doi: 10.1007/s11745-006-5038-1. [DOI] [PubMed] [Google Scholar]
- 46.Atarashi K, Tanoue T, Oshima K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500(7461):232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 47.Islam KB, Fukiya S, Hagio M, et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011;141(5):1773–1781. doi: 10.1053/j.gastro.2011.07.046. [DOI] [PubMed] [Google Scholar]
- 48.Hruz P, Zimmermann C, Gutmann H, et al. Adaptive regulation of the ileal apical sodium dependent bile acid transporter (ASBT) in patients with obstructive cholestasis. Gut. 2006;55(3):395–402. doi: 10.1136/gut.2005.067389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sauer P, Stiehl A, Fitscher BA. Downregulation of ileal bile acid absorption in bile-duct-ligated rats. J Hepatol. 2000;33(1):2–8. doi: 10.1016/s0168-8278(00)80152-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.