Summary
Over 90% of drugs with preclinical activity fail in human trials, largely due to insufficient efficacy. We hypothesized that adequately powered trials of patient-derived xenografts (PDX) in mice could efficiently define therapeutic activity across heterogeneous tumors. To address this hypothesis, we established a large, publically available repository of well-characterized leukemia and lymphoma PDXs that undergo orthotopic engraftment called the Public Repository of Xenografts (PRoXe; www.proxe.org). PRoXe includes all de-identified information relevant to the primary specimens and the PDXs derived from them. Using this repository, we demonstrate that large studies of acute leukemia PDXs that mimic human randomized clinical trials can characterize drug efficacy and generate transcriptional, functional and proteomic biomarkers in both treatment-naïve and relapsed/refractory disease.
Introduction
Essentially all oncology therapeutic candidates tested in humans have preclinical activity yet over 90% of these agents fail during clinical development (Kola and Landis, 2004; Ledford, 2011; Mak et al., 2014). This lamentable status quo results largely from a lack of efficacy in clinical trials. These trials, which generally evaluate agents in unselected patient populations with relapsed and refractory disease, are large, expensive and empiric. Thus, there is a desperate need for efficient and broadly applicable methods for preclinical assessment that have improved predictive value for human testing (Mak et al., 2014).
Cancer cell lines have significant limitations in their ability to model the biology and therapeutic responsiveness of cancers in their native microenvironment (Abaan et al., 2013; Gillet et al., 2011; Hausser and Brenner, 2005). The diversity of cancer, based on extensive genomic and transcriptional studies, is remarkably underrepresented by the number of available cell lines. This is even more problematic for transgenic murine models, which exist for a very small number of genetically-defined cancer subtypes. For example, there are over 100 different diagnostic subtypes of hematologic malignancies alone and almost all of these encompass multiple distinct genetic entities based on the presence of well-defined chromosomal rearrangements, aneuploidies and/or single/oligonucleotide sequence alterations (Jaffe et al., 2008). Among the cell lines that do exist, adaptation to in vitro culture and passaging for hundreds or even thousands of generations exerts substantial selective pressure that is not reverted by subcutaneous or even orthotopic xenografting (Daniel et al., 2009; Gillet et al., 2011; Hausser and Brenner, 2005). Nearly all cell lines are derived from patients who were previously untreated and/or from sites (e.g. pleural effusions) that are very uncommonly involved by their tumor types. For these reasons, the available cell lines are not representative of either the genetic abnormalities or treatment status of most patient tumors that will receive treatment in early phase trials. Finally, trials of in vivo therapeutics performed by subcutaneous xenografting of cell lines into the mouse flank fail to capture microenvironmental interactions that can modulate therapeutic efficacy (Aparicio et al., 2015).
Patient-derived xenografts (PDXs) established within highly immunocompromised mice overcome many of these shortcomings (Bertotti et al., 2011; Fichtner et al., 2008; Hidalgo et al., 2014; Julien et al., 2012; Reyal et al., 2012; Zhang et al., 2013). PDXs are passaged only in vivo and thereby avoid the selective pressures from ex vivo culture. They can be collected from patients with typical presentations of disease, either upfront or in the relapsed/refractory setting. Because engraftment rates are high for a variety of tumor types, very large repositories can be established to more broadly capture the range of human cancer. For example, a bank of over 1,000 solid tumor PDXs (mostly treatment-naïve) was recently reported (Gao et al., 2015). Large therapeutic studies of small molecule inhibitors in these PDXs recapitulated population-based response frequencies that were observed in clinical trials. In addition, the synergy identified between IGF1R inhibitors and multiple agents in cell lines was not observed in PDXs (Gao et al., 2015), a proof-of-principle that in vivo studies with PDXs may challenge results from cell lines.
In some settings, primary cancers can be orthotopically xenografted to recapitulate microenvironmental interactions within patients. The study by Gao et al. utilized subcutaneous flank xenografts of solid tumors, and as such, therapeutic efficacy was based on reduced growth or regression relative to vehicle-treated animals (Gao et al., 2015). In contrast, acute leukemias and other bone marrow-resident disorders readily undergo orthotopic engraftment after tail-vein or intra-osseous injection (Liem et al., 2004). As a result, therapeutic trials in mice engrafted with these diseases can utilize endpoints like overall survival or time to disease progression, just as in human trials. Mice can be treated until they progress on therapy, which allows for the development of acquired resistance. Samples can be taken from the peripheral blood or by sacrificing sentinel animals at multiple timepoints to establish biomarkers of response and resistance. Passaging in adequate numbers of animals also generates essentially unlimited numbers of primary cells for agnostic and targeted discovery efforts.
The central concern over PDXs is that they may fail to capture phenotypic, transcriptional, genetic and other characteristics of the tumors from which they were derived (Aparicio et al., 2015; Klco et al., 2014). Despite this concern, multiple entities now offer xenografting of patient tumors followed by in vivo drug testing, which they market as predictive of clinical response. We do not consider that each PDX is directly representative of the clinical sample from which it was derived. Instead, we hypothesize that using PDXs to conduct statistically powered, randomized trials in mice can efficiently define therapeutic activity across a broad range of genetically distinct tumor xenografts and inform the design and execution of human trials. At the same time, these preclinical trials can be used to establish biomarkers predictive of response, to generate models of drug resistance after in vivo exposure, and to interrogate aspects of in vivo biology, including tropism. In order to address this hypothesis, we aimed to establish a large repository of well-characterized PDXs of hematologic malignancies and implement these PDXs in comprehensive preclinical studies of candidate therapeutics in mice.
Results
Establishment of Patient Derived Xenografts
Primary bone marrow and blood specimens from patients with leukemia and lymphoma were xenografted into Nod.Cg-PrkdcscidIL2rg<tm1Wjl>/SzJ (NSG) mice. Success rates for engraftment in first passage (i.e., P0) after tail-vein injection varied based on disease subtype: B-cell acute lymphoblastic leukemia (B-ALL; 67.5%), T-cell ALL (T-ALL; 46.7%); acute myelogenous leukemia (AML; 23.2%), and all lymphomas (20.3%). Engraftment into P1 recipients also varied by disease, with very high rates of engraftment for B-ALL (95.3%) and lower rates for AML (65.7%), T-ALL (75%), and all lymphomas (76.9%) by 8 months after injection. Thirteen (30.2%) of 43 additional lymphomas engrafted in P0 within 6 months after tumor seeds obtained from tissue biopsies were implanted under the renal capsule.
Table 1 outlines the current repository based on the 2008 World Health Organization classification (Jaffe et al., 2008). In addition to a range of acute leukemia subtypes, we established multiple PDXs representative of lymphomas with no or very few available cell lines or in vivo models, including angioimmunoblastic T-cell lymphoma (AITL; Figure S1A,B), extranodal NK/T-cell lymphoma (Figure S1C), breast implant-associated anaplastic large cell lymphoma (Figure S1D), peripheral T-cell lymphoma, not otherwise specified (PTCL-NOS; Figure S1E), blastic plasmacytoid dendritic cell neoplasm (BPDCN; Figure S1F), and adult T-cell leukemia/lymphoma (ATLL; Figure S1G).
Table 1.
PDXs established in first (P0) or later passages classified according to the 2008 World Health Organization Classification1. See also Figure S1 and Movie S1.
| ≥P0 | ≥P1 | |
|---|---|---|
| Acute myeloid leukemia | 71 | 40 |
|
| ||
| AML with MDS-related changes | 11 | 6 |
| AML with inv(16) CBFB-MYH11 | 1 | 0 |
| AML with t(9;11) MLLT3-MLL | 1 | 1 |
| AML with MLL rearrangement (details N/A) | 2 | 2 |
| Therapy-related AML | 1 | 1 |
| AML with recurrent gene mutations | 16 | 8 |
|
| ||
| FLT3 ITD+/−TKD and NPM1 mutated | 9 | 6 |
| FLT3 ITD only, NPM1 unknown | 2 | 1 |
| FLT3 ITD only, NPM1 negative | 1 | 1 |
| NPM1 mutated only | 3 | 0 |
| CEBPA mutated only | 1 | 0 |
| AML NOS | 16 | 10 |
|
| ||
| Acute myelomonocytic leukemia | 7 | 3 |
| Acute monocytic leukemia | 1 | 1 |
| No specific subcategory | 8 | 6 |
| Blastic plasmacytoid dendritic cell neoplasm | 9 | 8 |
| Subclassification pending annotation | 14 | 4 |
|
| ||
| Myelodysplastic syndrome | 1 | 1 |
|
| ||
| Refractory cytopenia with multilineage dysplasia | 1 | 1 |
|
| ||
| Acute lymphoid leukemia | 144 | 74 |
|
| ||
| B-ALL NOS | 53 | 28 |
| B-ALL with t(12;21) TEL-AML1 (ETV6-RUNX1) | 17 | 3 |
| B-ALL with t(9;22) BCR-ABL1 | 16 | 6 |
| B-ALL with t(v;11q23) MLL rearranged | 12 | 7 |
| B-ALL with t(1;19) E2A-PBX1 (TCF3-PBX1) | 3 | 1 |
| B-ALL with hypodiploidy | 6 | 1 |
| B-ALL with hyperdiploidy | 4 | 0 |
| T-ALL | 32 | 28 |
| Subclassification pending annotation | 1 | 0 |
|
| ||
| Acute leukemia of ambiguous lineage | 4 | 3 |
|
| ||
| Mixed phenotype acute leukemia with t(v;11q23) MLL rearranged | 2 | 2 |
| Mixed phenotype acute leukemia with t(9;22) BCR-ABL1 | 1 | 0 |
| B/myeloid acute leukemia | 1 | 1 |
|
| ||
| Mature B-cell neoplasms | 17 | 13 |
|
| ||
| Mantle cell lymphoma | 6 | 4 |
| DLBCL NOS | 4 | 4 |
| Follicular lymphoma | 3 | 2 |
| B-cell lymphoma, unclassifiable, with features intermediate between DLBCL and BL | 2 | 2 |
| Extranodal marginal zone lymphoma | 1 | 1 |
| Subclassification pending annotation | 1 | 0 |
|
| ||
| Mature T- and NK-cell neoplasms | 9 | 7 |
|
| ||
| Angioimmunoblastic T-cell lymphoma | 3 | 2 |
| Anaplastic large cell lymphoma, ALK positive | 2 | 2 |
| Anaplastic large cell lymphoma, ALK negative | 1 | 1 |
| Adult T-cell leukemia/lymphoma | 1 | 1 |
| Extranodal NK/T-cell lymphoma | 1 | 1 |
| Primary cutaneous CD30 positive T-cell lymphoproliferative disorder | 1 | 0 |
|
| ||
| Hodgkin Lymphoma | 1 | 0 |
|
| ||
| Subclassification pending annotation | 1 | 0 |
|
| ||
| All Diagnoses | 248 | 138 |
Multiple models demonstrated tropism within recipient mice that mimicked the clinical disease. For example, Sézary Syndrome is a form of cutaneous T-cell lymphoma with peripheral blood involvement. We injected peripheral blood from a patient with Sézary Syndrome into the tail-vein of NSG mice and the cells trafficked to and involved the mouse skin (Figure S1H), establishing a unique model of human lymphoma epidermotropism. Similarly, a diffuse large B-cell lymphoma (DLBCL) from a patient who did not have central nervous system (CNS) involvement engrafted under the renal capsule in P0. That tumor was dissociated into a single-cell suspension and injected by tail-vein into P1 mice. All 5 (100%) injected mice succumbed with CNS involvement by DLBCL (Figure S1I–K). Despite the difficulty of developing low-grade lymphoma models in vivo, a marginal zone lymphoma engrafted as a low-grade, CD20+ lymphoma after implantation under the renal capsule (Figure S1L) and a follicular lymphoma engrafted with small BCL2-positive, CD20-positive lymphocytes. Similarly, two different mantle cell lymphomas (MCLs) that involved blood and bone marrow in patients also involved the blood, bone marrow and spleen of xenografted mice (Figure S1M). Even more remarkably, MCL obtained from the blood of a patient with blood, nodal and gastrointestinal involvement trafficked to each of these sites after tail vein injection (Movie S1).
In early passage, some PDXs maintained human, non-malignant cell populations that were present in the original tumor. For example, the PTCL-NOS engrafted in P0 with populations of non-malignant CD4+ and CD8+ T-cells as well as a rare population of EBV-positive B-cells (Figure S1E). Some PTCLs harbor a small subset of EBV-positive B-cells (Jaffe et al., 2008) but the role of these cells in disease development and persistence has not been defined.
Transcriptional, genetic and phenotype characterization of PDXs confirms subtype fidelity
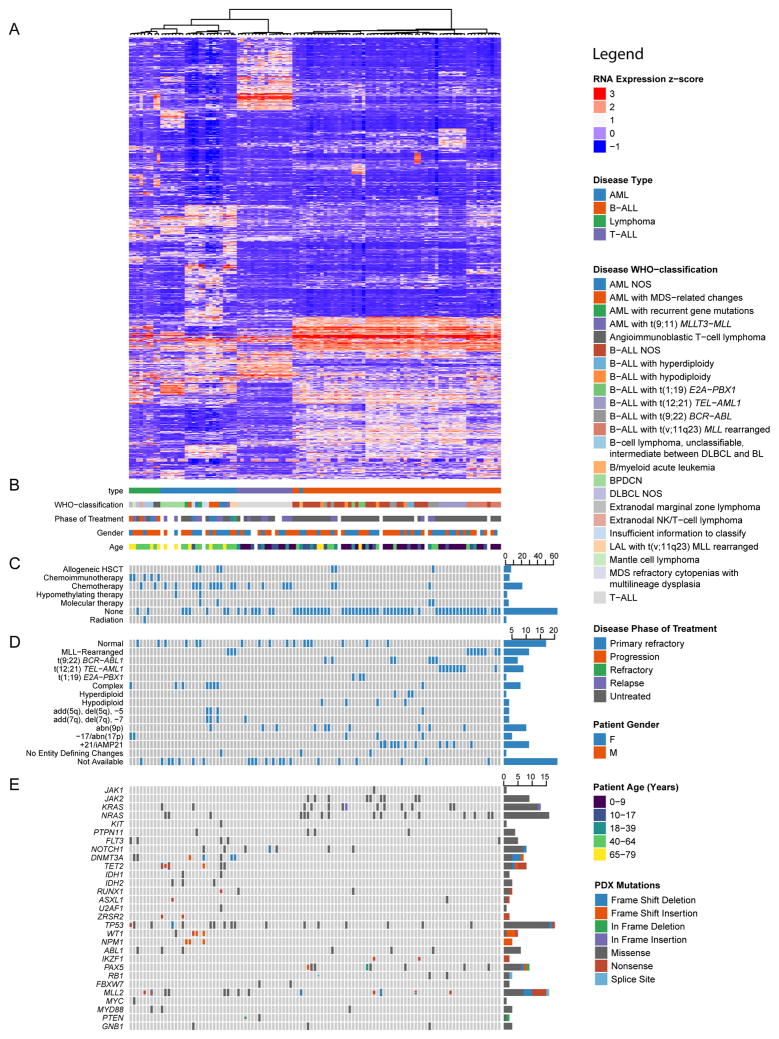
We confirmed lineage fidelity in 157 PDXs by flow cytometry for markers including human CD3 (T cell), CD19 (B cell), CD33 (early myeloid), CD34 (hematopoietic progenitor) and CD45 (pan-hematopoietic). Whole transcriptome sequencing (RNAseq) was performed on 107 PDXs in addition to targeted DNA sequencing of all exons of 205 hematologic malignancy-associated genes (Odejide et al., 2014) (Table S1, S2). Analysis of RNAseq data was implemented as a Snakemake (Koster and Rahmann, 2012) workflow. To permit similar analyses by other users, the complete workflow documenting all utilized parameters and tools is available at: https://bitbucket.org/cfce/weinstock-leukemia. Unsupervised hierarchical clustering based on disease type (AML, T-ALL, B-ALL, BPDCN, or lymphoma (Figure 1, Table S3) correctly clustered 105 (98%) of the 107 PDXs. A broad diversity of genetic alterations is captured within the PDXs, including targetable lesions (e.g. mutations of IDH1, IDH2 or JAK2, rearrangements involving MLL or ABL).
Figure 1. Integrative analysis of leukemia and lymphoma PDXs.
(A) Unsupervised hierarchical clustering over expression of 1000 genes with the greatest variance-to-mean ratios among 107 PDXs representing all WHO diagnostic categories encompassed by our repository. (B) Key clinical characteristics of patients and their tumors from which PDXs were derived. Patient age in years reflects the time when the xenografted tumor specimen was obtained. Phases of treatment are defined as: Untreated, prior to therapy directed at the xenografted tumor (n.b., does not include therapy directed at prior malignancies); Primary refractory, failed to respond to all tumor-directed therapy to date; Relapse, recurred by standard disease-specific criteria after achievement of a complete remission; Refractory, disease that is progressing during or shortly after the administration of salvage therapy for relapsed disease; Progression, specific to lymphomas progressing by clinical or radiographic criteria after a period of stable disease or partial remission. (C) Binary matrix of prior therapies to which patient was exposed prior to sampling of the referenced tumor, if known. (D) Selected cytogenetic features of patient tumors from which PDXs were derived. (E) OncoPrint of selected mutations detected in PDXs by targeted exon sequencing of a panel of 205 genes (Odejide et al., 2014). See also Figure S2 and Tables S1–S3.
Among B-ALL PDXs, interrogation of RNAseq data for gene fusions rediscovered 28 (96.6%) of 29 fusions identified by clinical cytogenetics analysis of the patient samples, including BCR-ABL1, MLL, ETV6-AML1, and TCF3-PBX1 rearrangements (Figure S2A). In addition, previously unrecognized fusions involving PAX5, JAK2, KDM6A, RUNX1, CRLF2 and SPI1 were recovered from the PDXs, with fusion transcripts from both derivative chromosomes identified in multiple PDXs (Figure S2A). Interrogation of the transcriptomes from AML PDXs similarly identified both known and unrecognized fusions (Figure S2B).
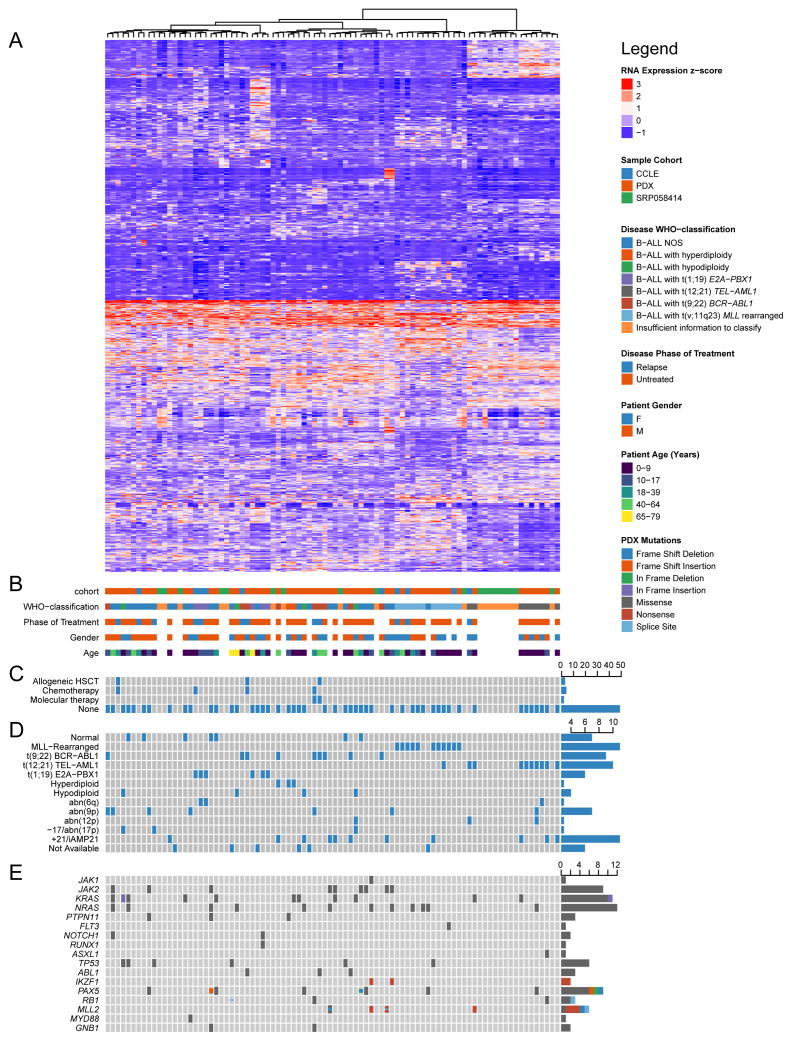
Next, we performed unsupervised clustering of transcriptomes from B-ALL PDXs in combination with transcriptomes of primary B-ALLs and B-ALL cell lines from the Cancer Cell Line Encyclopedia (Figure 2, Table S4) (Barretina et al., 2012). To permit analysis of the aggregated data, quantile normalization (Limma 3.26.1 (Ritchie et al., 2015)) was first performed to adjust for library depth and platform-specific differences. Principle components analysis revealed that the primary source of post-normalization variation derived from batch effects. These batch effects within each general diagnostic category (AML, B-ALL, T-ALL, lymphoma) were successfully removed using the ComBat approach from SVA 3.18.00 (Leek and Storey, 2007) agnostic of additional clinical and molecular covariates. Hierarchical clustering with Ward’s linkage and Euclidean distance over the 1000 genes with the greatest variance-to-mean ratio was performed over all diagnostic categories (PDX dataset only) and for each diagnostic category separately (all three datasets) (see Supplemental Experimental Procedures for details).
Figure 2. Integrative analysis of B-ALL PDXs, primary samples and cell lines.
(A) Unsupervised hierarchical clustering of RNA expression profiles among 60 B-ALL PDXs (PDX cohort), 19 primary pre-B ALL samples (SRP058414), and 10 B-cell leukemia cell lines (CCLE), using the same methods as for Figure 1. (B) Key clinical characteristics of cell lines and primary samples, including those from which PDXs were derived. (C) Binary matrix of prior therapies to which patient was exposed prior to sampling of the referenced tumor, if known. (D) Selected cytogenetic features of cell lines and primary samples, including those from which PDXs were derived. (E) OncoPrint of selected mutations detected in PDXs by targeted exon sequencing, or reported in cell lines by ATCC or DSMZ. Mutation data for the primary samples (SRP058414) were not available. See also Table S4.
Compared with cell lines, PDXs spanned a broader diversity of WHO diagnostic categories, patient demographic characteristics, phases of treatment, prior therapies, cytogenetic profiles, and genotypes (Figure 2). At present, there are 28 B-cell leukemia cell lines available through the German cell line repository DSMZ, of which only 10 have RNA expression data available through the CCLE. In contrast, we have generated 115 B-ALL PDX lines, of which 82 have RNA expression data already available. Unsupervised clustering underscored the biologic influence of canonical fusion genes such as those involving MLL, BCR-ABL1, TEL-AML1, and E2A-PBX1. Although fewer cases were analyzed, separate clustering of AML, T-ALL and lymphomas identified distinct clades in each disease (Figures S2C–E). Lymphoma samples segregated by cell lineage (B vs. T), with canonical fusion genes correlating with a number of derivative clades, including CCND1-IGH+ MCL, NPM1-ALK+ ALCL, and DLBCLs with concurrent rearrangements of MYC and BCL2 (Figure S2E).
PRoXe: A resource to facilitate studies of leukemia and lymphoma PDXs
To massively expand the widespread use of these models and the data derived from them, we established an open source web portal called the Public Repository of Xenografts (PRoXe; www.proxe.org). We created PRoXe using Shiny, a web application library for R that permits the fluid integration and analysis of heterogeneous data types in a graphical user interface. All underlying code for PRoXe is freely available on GitHub (https://bitbucket.org/scottkall/proxe).
PRoXe provides extensive patient, tumor, PDX, and germline administrative information for each line. Any of the 56 characteristics that are categorical or quantitative can be visualized interactively via histograms, barplots, scatterplots, and boxplots, allowing the user to dynamically interrogate individual variables as well as interactions between these variables. Flow cytometry plots, full-color immunohistochemistry images, detailed pathology reports, and class I HLA alleles inferred from RNAseq (Shukla et al., 2015) are available for a subset of lines. RNA expression in the form of log-transformed RPKM derived from RNAseq can be visualized as bar plots or heat maps for individual genes or panels of genes for selected PDX lines. Curated mutations among panels of genes identified by targeted exon sequencing can be visualized in matrix form using a publically available OncoPrint script (https://gist.github.com/armish/564a65ab874a770e2c26). RPKMs and mutation calls are also being uploaded to the cBio portal (www.cbioportal.org).
Randomized phase II-like trial in PDXs
We tested the utility of our large PDX repository by designing and performing a phase II-like preclinical trial following the same approaches utilized in human trials. Small molecule inhibitors that disrupt the MDM2-p53 interaction have proven effective in a subset of tumor models and patient tumors with wild-type p53 (Andreeff et al., 2015; Jeay et al., 2015; Lv et al., 2015; Weisberg et al., 2015). In contrast, tumors with pathogenic TP53 mutations or biallelic deletions are typically resistant to MDM2 inhibition. The MDM2 inhibitor CGM097 is currently in phase I clinical trials for patients with solid tumors that have wild-type TP53 (Jeay et al., 2015; Weisberg et al., 2015). Approximately 85% of B-ALLs lack TP53 alterations (Pui et al., 2004), suggesting that this agent may have broad activity across a heterogeneous panel of PDXs.
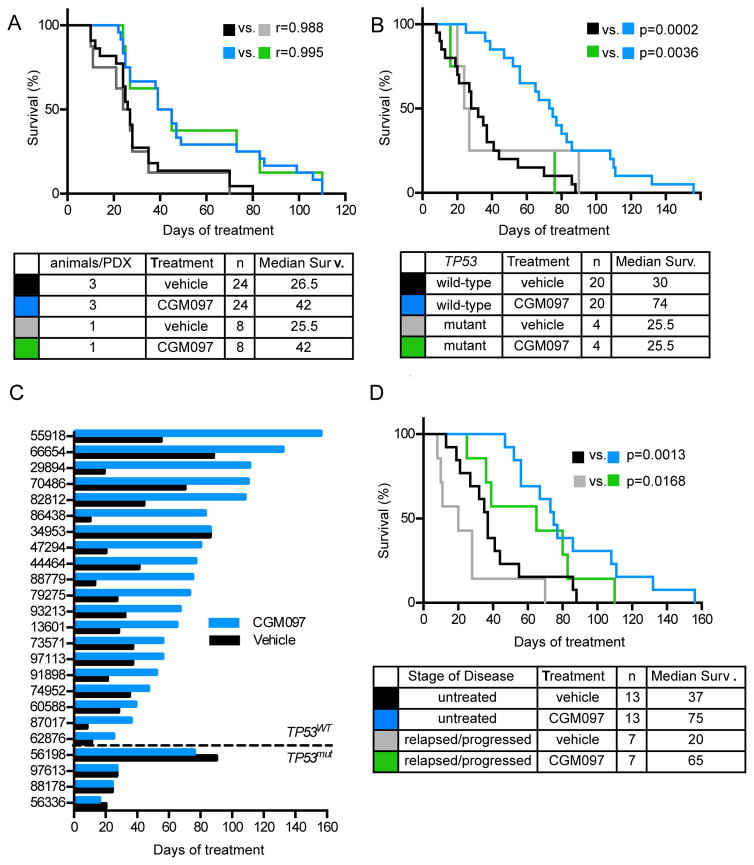
One significant benefit to performing trials in PDXs (compared with human trials) is that each xenograft can be injected into multiple animals, which allows for direct comparison between drug and control in the same cancer. To test the reproducibility of using only one vehicle-treated and one CMG097-treated mouse per PDX, we performed a pilot study on eight PDXs, in which six mice were injected with each PDX. Of the six, three mice were randomized to receive vehicle and three to receive CGM097 100 mg/kg by oral gavage beginning upon engraftment (defined as ≥2% peripheral blood hCD45+ cells). One of the three from each cohort was randomly selected prior to beginning therapy and considered a distinct cohort. Survival from the 3 vs. 3 study and the 1 vs. 1 study showed outstanding correlation for both drug-treated and vehicle-treated arms (Figure 3A). This strongly supports a 1 mouse/cohort study design in B-ALL PDXs.
Figure 3. The MDM2 inhibitor CGM097 extends survival of mice engrafted with TP53-wild-type B-ALL PDXs in a randomized phase II-like trial.
(A) Kaplan-Meier survival analysis for overall survival of mice engrafted with 8 B-ALL PDXs, with 3 animals per treatment arm or 1 randomly selected animal from each arm. (B) Kaplan-Meier survival analysis for overall survival of vehicle- and CGM097 treated cohorts of mice engrafted with TP53 wild-type (n=20) and TP53 mutant (n=4) B-ALL PDXs. (C) Time to sacrifice after start of treatment for vehicle and CGM097 treated animals for each PDX. (D) Kaplan-Meier analysis of PDXs derived from patients who had relapsed or progressed on any form of therapy (relapsed/progressed) versus patients who had not received treatment (untreated). See also Figure S3.
The primary pre-defined endpoint of the full phase II-like study was overall survival in animals treated with CGM097 daily compared to animals treated with vehicle. The trial was designed to detect a 40% difference in survival at 50 days of treatment assuming 60% alive in the treated group versus 20% alive in the untreated group using a one-sided Fisher exact test at the 0.05 level with 84% power. These estimates were based on the 8 PDX pilot study (Figure 3A). Pre-defined subset analyses included comparisons of median survival between vehicle and CGM097 treatment of leukemias with wild-type TP53 from patients who are: age <18 years old, age ≥18 years old, male sex, female sex, untreated for B-ALL, or have relapsed/refractory disease after treatment.
We injected 29 PDXs into four mice each and monitored animals weekly for engraftment (Figure S3A). Two PDXs were not included in the final analysis due to failure to engraft, one PDX was excluded because of death of one animal prior to engraftment, and two additional PDXs were censored from the survival analysis due to failure of the vehicle-treated mouse to meet criteria for sacrifice by day +120 of treatment (Figure S3A). Thus, the final cohort included 24 different PDXs, including 4 that harbored TP53 mutations (Table 2). Engraftment across the four mice for each PDX was very similar (Figure S3B). Upon engraftment, two mice from each PDX were randomly selected to receive vehicle and two mice received CGM097. After 26 hours (two hours after the second dose), one animal from each cohort was sacrificed to assess pharmacodynamic (PD) markers and the remaining two animals were followed until they met criteria for sacrifice.
Table 2.
Patient characteristics and survival data of PDXs in CGM097 survival study.
| Patient characteristics | Survival Data | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| PDX | TP53 genotype | Sex (m/f) | Age (years) | WHO | Phase of Treatment | Vehicle (days) | CGM097 (days) |
| 86438 | WT | f | 48 | MLLr | Persistent, post-induction | 10 | 83 |
| 29894 | WT | f | 5 | NOS | Untreated | 19 | 111 |
| 88779 | WT | f | 5 | NOS | Untreated | 13 | 75 |
| 87017 | WT | f | 8 | NOS | Relapsed | 8 | 36 |
| 47294 | WT | m | 50 | NOS | Relapsed, post-allogeneic transplant | 20 | 80 |
| 55918 | WT | 1 | MLLr | Untreated | 55 | 156 | |
| 79275 | WT | f | 12 | NOS | Untreated | 24 | 73 |
| 91898 | WT | f | 83 | NOS | Untreated | 21 | 53 |
| 82812 | WT | f | 5 | NOS | Untreated | 44 | 108 |
| 13601 | WT | f | 59 | Ph+ | Relapsed, post-allogeneic transplant | 28 | 65 |
| 62876 | WT | m | 36 | Ph+ | Relapsed | 11 | 25 |
| 93213 | WT | 0.3 | MLLr | Untreated | 32 | 67 | |
| 44464 | WT | m | 3 | hypodiploid | Untreated | 41 | 77 |
| 70486 | WT | m | 15 | NOS | Relapsed | 70 | 110 |
| 97113 | WT | f | 54 | MLLr | Untreated | 37 | 56 |
| 73571 | WT | m | 26 | NOS | Untreated | 37 | 56 |
| 66654 | WT | m | 30 | MLLr | Untreated | 88 | 132 |
| 60588 | WT | f | 12 | NOS | Relapsed | 28 | 39 |
| 74952 | WT | f | 44 | Ph+ | Untreated | 35 | 47 |
| 34953 | WT | f | 20 | B/myeloid | Untreated | 86 | 86 |
| 88178 | N239D | m | 15 | MLLr | Untreated | 24 | 24 |
| 56198 | Y220C | m | 71 | hypodiploid | Untreated | 90 | 76 |
| 56336 | D61fs | m | 13 | hypodiploid | Untreated | 20 | 16 |
| 97613 | C275G | f | 2 | hypodiploid | Untreated | 27 | 27 |
| 97626 | WT | m | 6 | NOS | Untreated | >120 | >120 |
| 44038 | WT | m | 59 | NOS | Untreated | >120 | >120 |
WHO = World Health Organization classification. MLLr = MLL-rearranged, NOS= Not Otherwise Specified, Ph+= Philadelphia-positive, hypodiploid= <44 chromosomes, B/myeloid= mixed phenotype
CGM097 extends survival across a diverse panel of B-ALL PDXs with wild-type TP53
CGM097 conferred no survival benefit in the 4 PDXs with TP53 mutations (Figure 3B–C, Table 2). In contrast, CGM097 conferred a survival benefit of ≥10 days in 19 of 20 PDXs with wild-type TP53 (Figure 3C). For the one exception (PDX 34953), both animals required sacrifice at day 86 of treatment due to dermatitis and lethargy but neither had splenomegaly or significant numbers of B-ALL cells upon necropsy, suggesting that mortality was unrelated to leukemia progression. The pre-defined primary endpoint (overall survival at day 50 of treatment) was statistically in favor of CGM097 treatment (80% versus 20% among the 20 TP53 wild-type PDXs; p=0.0004 by two-sided Fisher’s exact test). The difference remained statistically significant (p=0.0012) if all 24 PDXs were included.
Among the 20 PDXs with wild-type TP53, median survival was improved by a median value of 44 days in the CGM097 cohort (Figure 3B). The gradual downward slope of the survival curve for the 20 mice treated with vehicle demonstrates the heterogeneity in disease progression across the B-ALL PDXs. Similarly, the gradual separation between the vehicle-treated and CGM097-treated curves for the 20 PDXs demonstrates heterogeneity in the extent of disease control by CGM097. This data is also captured in a “randomized Swimmer’s plot” in Figure 3C, which further shows the broad variability in both absolute and relative survival benefit conferred by CGM097 treatment across PDXs.
Pre-defined subset analyses demonstrated statistical superiority for the CGM097 treatment arm in 5 of 6 cohorts with wild-type TP53: age <18 years old, male sex, female sex, (Figure S3C) previously untreated, and relapsed/refractory (Figure 3D). In fact, treatment with CGM097 improved survival of mice engrafted with PDXs from relapsed/refractory B-ALL by 45 days (65 vs. 20 days for vehicle) (Figure 3D). This provides strong preclinical evidence for testing CGM097 in patients who have received extensive treatment for B-ALL but maintain wild-type TP53, including children.
PDX studies allow for the development of pharmacodynamic endpoints and biomarkers predictive of response
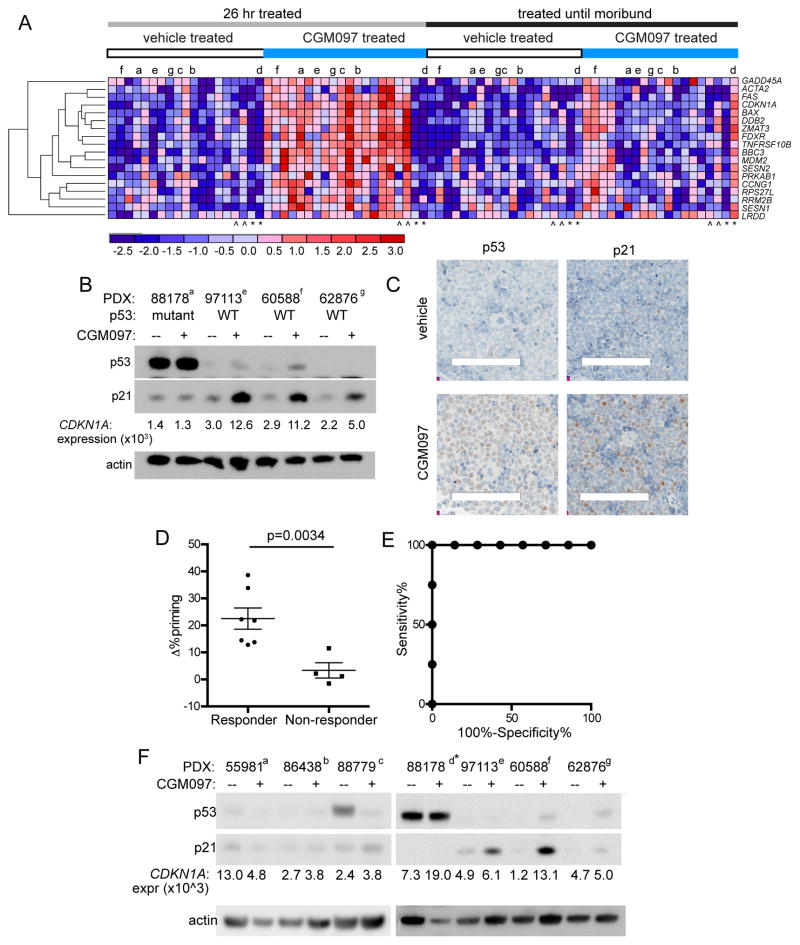
To assess transcriptional correlates of CGM097 activity, we designed a custom NanoString panel that included p53 targets and modulators of p53 activity (Table S5). We applied this assay to purified PDX cells obtained from mice treated with either vehicle or CGM097 at both the PD and survival (SUR) timepoints. Nine genes were differentially expressed in CGM097-treated PDXs with wild-type TP53 at the PD timepoint (≥1.2-fold over vehicle-treated, false discovery rate (FDR) adjusted p value <0.05) (Table S6). In addition, all vehicle-treated and CGM097-treated samples at both timepoints were subjected to agglomerative clustering on expression of a curated set of p53 target genes (Figure S4A) (GenePattern) (Riley et al., 2008). Visual inspection of the resulting heatmap revealed a cluster of genes strongly expressed in 10 CGM097-treated PD samples, 3 CGM097-treated SUR samples and 1 vehicle-treated SUR sample. The genes in this cluster included GADD45A, BBC3, MDM2, ACTA2, FAS, BAX, DDB2, CDKN1A, FDXR, TNFRSF10B, PRKAB1, CCNG1, SESN1 RPS27L and RRM2B. This p53 target cluster included 7 of 9 genes identified solely by differential expression analysis in PD samples, as well as 10 of 13 genes identified in a signature that predicted response to CGM097 in cell lines and solid tumor xenografts (Jeay et al., 2015). Combining the two analyses generated an 18 gene set of CGM097-dependent p53 target genes (Figure S4B, Figure 4A). The 18 genes included all 7 genes (BAX, BBC3, CDKN1A, FDXR, MDM2, TNFRSF10B and ZMAT3) that were previously reported to increase expression in patient tumors after treatment with the MDM2 inhibitor RG7112 (Andreeff et al., 2015). This overlap is remarkable, as the previous signatures (Andreeff et al., 2015; Jeay et al., 2015) were established with untreated specimens (i.e. as predictive biomarkers) while our gene set was established on treated leukemias (i.e. as a pharmacodynamic biomarker). As expected, expression of the 18 gene set did not differ between vehicle- and CGM097-treated samples from PDXs with TP53 mutations (Figure 4A).
Figure 4. Transcriptional, protein-based and functional biomarkers for response and resistance to CGM097 in B-ALL PDXs.
(A) Agglomerative clustering by gene expression of CGM097-induced genes in each treatment group. Samples from censored animals indicated by ^. PDX samples with TP53 mutations are indicated by *. Samples used for immunoblotting in Figure 4F are indicated with lower-case letters (a–g). (B) Immunoblotting performed on purified splenic B-ALL cells at the 26 hour timepoint. Transcript levels of CDKN1A (encodes p21) were determined on the same samples. (C) Immunohistochemistry for p53 and p21 in vehicle and CGM097-treated spleens collected on day 6 of treatment. Scale bars depict 0.1mm. (D) Δ% priming in 26 hour vehicle- and CGM097-treated groups was determined by dynamic BH3 profiling using a PUMA BH3 peptide in 11 PDXs. The 4 “non-responder” models harbor TP53 mutations and had no improvement in survival between CGM097- and vehicle-treated mice. Error bars represent standard error of the mean (S.E.M.) (E) R.O.C. curve analysis for dynamic BH3 profiling using the PUMA BH3 peptide. (F) Immunoblotting for p53 and p21 in CGM097- and vehicle-treated splenic B-ALL cells collected at the SUR timepoint. See also Figure S4, Tables S5–S7.
Induction of both p53 and p21 protein expression by CGM097 in TP53 wild-type PDXs was confirmed by immunoblotting and IHC in spleen (Figure 4B–C). Expression levels of p21 protein closely paralleled the normalized levels of CDKN1A (which encodes p21) transcript (Figure 4B).
The Letai laboratory previously demonstrated that dynamic BH3 profiling can predict the response of tumor cells to both chemotherapy and targeted agents in vitro and in vivo (Montero et al., 2015). To determine the accuracy of dynamic BH3 profiling as a predictive assay for in vivo response to CGM097, we harvested cells from spleens of vehicle-treated animals at the SUR timepoint and treated ex vivo for 18 hours with 2 μM CGM097 or DMSO. Cells were then analyzed by FACS-based dynamic BH3 profiling using BH3 peptides derived from PUMA or NOXA, and the increase in priming from CGM097 treatment (expressed by Δ% priming values) was established for each PDX. Of note, dynamic BH3 profiling was performed blinded as to in vivo response. The Δ% priming by PUMA, which is capable of binding to multiple antiapoptotic proteins from the BCL-2 family, perfectly discriminated B-ALL PDXs with ≥12 days improvement in overall survival from CGM097 treatment (p=0.0034; area under curve for receiver operating characteristic = 1.00) (Figure 4D–E). In contrast, Δ% priming with a NOXA peptide that specifically binds to the antiapoptotic protein MCL-1 did not predict survival benefit (p=0.384). Thus, dynamic BH3 profiling using a PUMA peptide is an excellent binary predictor to accurately determine CGM097 sensitivity in B-ALL PDXs. Of note, BBC3, which encodes PUMA, was one of the signature genes induced by CGM097 treatment at the PD timepoint (Table S6).
Of the nine genes upregulated in CGM097-treated PDXs at the PD timepoint, six were significantly (nominal p value <0.05) reduced in the CGM097-treated SUR samples relative to the CGM097-treated PD samples (Table S7). When clustering was performed on the 18 gene set by timepoint and treatment arm, there was a clear reduction in expression of these genes in the majority of CGM097-treated SUR samples. However, in 4 samples that progressed on CGM097, induction of p53 targets was sustained at the SUR timepoint, suggesting at least two classes of CGM097 resistant samples. In class I, expression of CGM097-dependent p53 targets was lost upon disease progression on drug. In class II, expression of these genes was sustained and remained elevated despite progression of disease in vivo. This dichotomy was supported by immunoblotting for p21, which remained at high levels in a subset of PDXs at the SUR timepoint (Figure 4F).
Discussion
Countless previous studies, reviews, and perspectives have discussed the high rate of failure, the long time from discovery to clinical benefit, and the massive cost of developing effective cancer therapeutics. In order to facilitate preclinical drug studies that are more informative, we have established a resource to support randomized PDX trials in mice. We demonstrate that adequately powered studies in PDXs can be used to develop transcriptional, proteomic and functional biomarkers of response and resistance while defining the efficacy of therapeutics across genetically diverse subtypes of leukemia. Unlike most clinical trials, we had no difficulties in capturing large numbers of specimens at each timepoint. We also generated a large number of models with acquired in vivo resistance, a unique facet of phase II-like trials in mice.
As we have shown, leukemias and lymphomas offer several advantages that make them amenable to this approach. First, many subtypes of leukemia and lymphoma readily engraft into NSG mice or similarly immunocompromised strains by tail-vein injection. Second, bone marrow and blood specimens from patients with these diseases are easily obtained in comparison with most solid tumors. Third, tail-vein injection results in orthotopic disease growth, both in the bone marrow and in extramedullary sites that are commonly involved by these diseases (e.g. skin, CNS, lymph nodes, gastrointestinal tract). Finally, studies of orthotopic leukemia PDXs allow for survival or disease progression, rather than tumor shrinkage, to be used as the primary endpoint. Previous discordances between tumor shrinkage in preclinical models and survival in human studies suggest that tumor shrinkage alone can be misleading (Couzin-Frankel, 2014).
It is important to note that direct and adequately powered comparisons for predictive ability between cell lines and PDXs are lacking. This has largely resulted from the inadequate number of available cell lines for an individual disease subtype. It remains possible that cell lines would be equally predictive as PDXs for many therapeutics if adequate numbers of cell lines were available. Similarly, other types of in vivo models have clear advantages over PDXs. Insertional mutagenesis and chemical carcinogenesis models are highly diverse within an individual mouse and across a population of tumors (Westcott et al., 2015). Both these models and transgenic models have significant advantages over PDXs for modeling microenvironmental and tumor-immune interactions (Aparicio et al., 2015). Not all human tumors are easily engrafted orthotopically, which further limits the ability to study the effects of tumor microenvironment on therapeutic response (Tabe and Konopleva, 2015). Another clear limitation of PDXs is the bottleneck that tumor cells must navigate to engraft in immunocompromised mice. Although leukemias and lymphomas have relatively high ‘take rates’, certain subsets have proven completely intractable to xenografting, while others only xenograft in a limited fraction of cases (Klco et al., 2014). This may introduce an important bias that limits applicability of PDX studies to unselected patient populations.
As a result of these limitations, we make no claims that each individual model in PRoXe is representative of the leukemia or lymphoma from which it was derived. Careful studies of AML PDXs have demonstrated quite clearly that different leukemias can engraft in myriad different ways, as assessed by the relative representation of subclones, even when injected as biologic replicates into congenic recipients (Klco et al., 2014). Further complicating the diversity of models are differences in recipient strain, approach to transplantation (e.g. tail-vein injection, intraosseous injection, radiation of recipient mice), timing of injection (e.g. from fresh specimens versus after freezing) and patient demographics between centers that generate PDXs. Our intention was never to analyze the fidelity of individual models to their starting material. Instead, we developed a resource to facilitate the pre-clinical evaluation of therapeutics across a diversity of different malignancies. The observation that 3 different mice engrafted with the same PDX and treated with the same agent have almost exactly the same survival comes as little surprise, as previous studies from our groups (Ott et al., 2012; Weigert et al., 2012; Zelenetz et al., 2015) and many others have shown that treatment of large cohorts of mice injected with the same PDX typically results in survival curves that “fall off a cliff”. Although these curves, in which all mice treated in the same way die within a day or two of each other, provide robust statistical p values, they fail to inform the field about the true activity of a therapeutic across diverse tumors.
Randomized phase II-like studies of PDXs in mice are applicable to a range of therapeutic agents, especially those that act through cancer cell-intrinsic mechanisms. These studies may markedly reduce the time and financial expense of drug development while mitigating the very real human cost of administering toxic and ineffective therapies to patients during the drug development process. In our study of CGM097 across 24 different B-ALL PDXs, several findings are noteworthy. First and foremost, the improvement in median survival far exceeded that noted from a variety of different agents, including our studies of BRD4, HSP90, and JAK2 inhibitors (Ott et al., 2012; Weigert et al., 2012; Zelenetz et al., 2015). Second, this improvement was highly similar in PDXs developed from treatment-naive samples and samples acquired from patients after treatment. Typical regimens for B-ALL include multiple cytotoxic chemotherapies administered over the course of 2 years. Yet, if a PDX engrafted with wild-type TP53, this prior therapy appeared to have no significant effect on response to CGM097.
In order to foster the maximum impact from the PDX repository, we developed PRoXe as an open source website. We are currently in negotiations with multiple different academic centers to incorporate their PDXs into PRoXe or to develop Shiny-based web portals for their own PRoXe sites. Incorporation of clinical-, pathologic- and PDX-level data from the >1,000 solid tumor PDXs recently published by Gao et al. (2015) is underway. We encourage others to consider adding their models. Limitations clearly exist in the willingness of centers to share models, based largely on concerns over liability, hopes for financial compensation and confidentiality-related issues germane to unfettered access. Addressing these is an essential next step if the true value of PDXs is ever going to be realized. In addition, academic centers are ill-suited to bear the burden of housing, expanding, archiving, characterizing and disseminating PDXs to investigators (academic and industrial) across the world. As a result, we are licensing a large portion of the models to the Jackson Laboratories, which will allow for industry-scale expansion, quality control, characterization and distribution, much as Addgene has provided this service for DNA plasmids.
Experimental Procedures
Generation and Use of PDXs
Primary bone marrow and blood specimens were collected from patients with leukemia and lymphoma at Dana-Farber Cancer Institute, Brigham and Women’s Hospital, and Boston Children’s Hospital. We also imported and transplanted primary leukemia and lymphoma specimens from 8 additional centers as well as 72 total PDXs that were already established by participating investigators. De-identified patient samples were obtained with informed consent and xenografted under Dana-Farber/Harvard Cancer Center Institutional Review Board (IRB)-approved protocols #13-351, Nod.Cg-PrkdcSCIDIL2Rγ<tm1Wjl>/SzJ (NSG) mice were purchased from Jackson Laboratories (Bar Harbor, ME) and handled according to Dana-Farber Cancer Institute’s Institutional Animal Care and Use Committee-approved protocol #13-034.
Hemoseq
Targeted exon capture and next-generation sequencing of all coding exons of 205 genes (Table S1) were performed as previously described (Odejide et al., 2014). A total of 115 PDX models, 45 of which had matched patient germline tissue available, were genotyped. Single nucleotide variants (SNVs) and small insertions/deletions (InDels) identified using the Genome Analysis Toolkit (GATK, Broad Institute) and curated as likely pathologic mutations were individually visualized with the Integrative Genome Viewer (IGV, Broad Institute) to ensure biphasic and heterogeneous read distribution. The final set of curated mutations is shown in Table S2 (provided as a separate excel file).
RNASeq
Messenger RNA was extracted from PDX leukemia or lymphoma cells using magnetic microbeads (μMACS mRNA Isolation Kit, Mitenyi Biotec). Non-stranded RNASeq libraries were generated using the True-SeqRNA Sample Prep kit (Illumina) on a Sciclone platform (Perkin Elmer). mRNA underwent fragmentation, cDNA synthesis, and next generation library synthesis via end repair, adenylation, adapter ligation and PCR amplification. Libraries were sequenced on a Next-Seq instrument (Illumina) using a paired-end protocol.
CGM097 Study
Viably frozen B-ALL xenograft cells were thawed and changed into 1X PBS before tail-vein injection at 0.5–2.0×106 per mouse. Engraftment was monitored every 7 days, starting 3 weeks post-injection, by flow cytometry of 50 μl of peripheral blood. Blood was processed with Red Blood Cell Lysis Buffer (Qiagen) before staining with antibodies against human CD45 (APC-conjugated, eBioscience 17-0459-42) and human CD19 (PE-conjugated eBioscience 12-0193-82) in 1XPBS with 2mM EDTA to assess engraftment. Flow cytometry data was analyzed with FlowJo. When engraftment was determined to be ≥ 2% of the live cell population, the remaining two animals with the same PDX were bled for analysis before starting treatment. Animals were dosed daily with 100 mg/kg CGM097 or methylcellulose vehicle by oral gavage. Kaplan Meier survival analysis was conducted using GraphPad Prism 6.0 (GraphPad Software Inc.).
Pharmacodynamic Analyses
26 hour treated splenocytes were harvested by mechanical disruption followed by Red Blood Cell lysis (Qiagen) and depletion of mouse cells by magnetic bead enrichment (Millitenyi). Cells were divided into 1–5×106/cell aliquots and snap frozen for further analysis. Frozen cell pellets were thawed on ice and resuspended in 1ml Trizol® (Life Technologies) and extracted using a modified RNeasy Kit® (Qiagen) extraction procedure, before aliquoting and snap freezing. Cell lysate was prepared for western blotting by lysing with RIPA lysis buffer (Westnet), followed by separation by SDS-PAGE and transfer to PVDF membrane. Blots were probed with antibodies against p53 (DO-1, Santa Cruz Biotechnology), p21 (DCS60, Cell Signaling Technologies) and β-actin (13E5, Cell Signaling Technologies). Dynamic BH3 profiling is a technique used to quantify the ability of a therapeutic agent to promote apoptosis. Dynamic BH3 profiling was performed as previously described (Montero et al., 2015). Unsupervised clustering analysis was performed using Pearson correlation with average link clustering. Differential expression between experimental criteria was determined using raw counts and normalization procedures within the DEseq package in R based on a negative binomial distribution. The FDR (Benjamini and Hochberg) method was used to adjust for multiple comparisons after filtering for those with ≥1.2 fold change. Both unsupervised and supervised clustering were performed using GenePattern. Additional details are available in the Supplemental Experimental Procedures.
Supplementary Material
Significance.
Preclinical drug development of cancer therapeutics largely depends on in vitro and transgenic models that have significant limitations. We have generated a large, publically available repository of patient-derived xenografts of leukemias and lymphomas that we call the Public Repository of Xenografts (ProXe). This repository will enable investigators in both academic and industrial laboratories to conduct therapeutic testing in orthotopically engrafted patient-derived samples of diverse hematologic malignancies, including diseases for which there are no currently available models. The size of this repository also enables drug testing on a scale comparable to human clinical trials. Using this repository, we demonstrate that the MDM2 inhibitor CGM097 has potent activity against patient-derived B-ALL xenografts established from patients with both untreated and relapsed/refractory disease.
Acknowledgments
These studies were made possible through the generosity of a large number of anonymous donors, as well as the patients who provided their samples for research. We thank Kevin Shannon for thoughtful comments. A.G. is supported by the Damon Runyon Cancer Research Foundation and Gabrielle’s Angel Foundation for Cancer Research. D.M.W. is a Leukemia and Lymphoma Society Scholar.
Footnotes
Accession Numbers
Primary sequencing data are being made publicly available through the NCBI Sequence Read Archive (Submission ID SUB1369293, BioProject ID PRJNA314605).
Author Contributions
E.C.T and M.A.M. conceived and performed experiments and wrote the manuscript. A.C., A.L.C., J.K., T.A.D., E.A.M., P.R., R.V., S.P.K., H.L., S-C.W., M.H.H., O.P., M.I., M.Mur., J.M., Z.T.H., R.K., N.K., L.L., S.Y.N., R.R.P., J.A.R., A.R.T., P.vH., A.A.L. conceived and performed experiments. A.S.L, E.H., S.J., H.L., J.C.A., A.L., M.B., M.A.S., and J.D.G. conceived experiments and provided essential reagents. K.E.S. and D.N. analyzed data. P.A., K.K.B., S.C., R.A.C., V.G.C., M.S.D., D.J.D., D.M.D., B.L.E., J.E., B.F., D.C.F., A.S.F., I.A.G., J.S.G., H.G., F. G-O., T.A.G., A.G., S.M.H., G.I., A.M.I., S.I., E.D.J., C.A.J., I.J., M.A.K., M.K., A.L.K., T.S.K., N.L., E.L, A.T.L., M.Mus, M.A., P.B-R., S.M.K., O.O., S.H.O., A.E.P., J.E.R., S.E.S., B.S., L.B.S., R.J.S., D.P.S., K.S., R.M.S., J.T., M.Wa., A.P.W., M.We., J.U.W., D.A.W., B.M.W., J.R. and M.M.B. provided essential reagents. H.E. provided project administration. D.M.W. secured funding, conceived experiments and wrote the manuscript
References
- Abaan OD, Polley EC, Davis SR, Zhu YJ, Bilke S, Walker RL, Pineda M, Gindin Y, Jiang Y, Reinhold WC, et al. The exomes of the NCI-60 panel: a genomic resource for cancer biology and systems pharmacology. Cancer Res. 2013;73:4372–4382. doi: 10.1158/0008-5472.CAN-12-3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeff M, Kelly KR, Yee KW, Assouline SE, Strair R, Popplewell L, Bowen D, Martinelli G, Drummond MW, Vyas P, et al. Results of the Phase 1 Trial of RG7112, a Small-molecule MDM2 Antagonist in Leukemia. Clin Cancer Res. 2015 doi: 10.1158/1078-0432.CCR-15-0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio S, Hidalgo M, Kung AL. Examining the utility of patient-derived xenograft mouse models. Nat Rev Cancer. 2015;15:311–316. doi: 10.1038/nrc3944. [DOI] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertotti A, Migliardi G, Galimi F, Sassi F, Torti D, Isella C, Cora D, Di Nicolantonio F, Buscarino M, Petti C, et al. A molecularly annotated platform of patient-derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov. 2011;1:508–523. doi: 10.1158/2159-8290.CD-11-0109. [DOI] [PubMed] [Google Scholar]
- Couzin-Frankel J. The littlest patient. Science. 2014;346:24–27. doi: 10.1126/science.346.6205.24. [DOI] [PubMed] [Google Scholar]
- Daniel VC, Marchionni L, Hierman JS, Rhodes JT, Devereux WL, Rudin CM, Yung R, Parmigiani G, Dorsch M, Peacock CD, et al. A primary xenograft model of small-cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro. Cancer Res. 2009;69:3364–3373. doi: 10.1158/0008-5472.CAN-08-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichtner I, Rolff J, Soong R, Hoffmann J, Hammer S, Sommer A, Becker M, Merk J. Establishment of patient-derived non-small cell lung cancer xenografts as models for the identification of predictive biomarkers. Clin Cancer Res. 2008;14:6456–6468. doi: 10.1158/1078-0432.CCR-08-0138. [DOI] [PubMed] [Google Scholar]
- Gao H, Korn JM, Ferretti S, Monahan JE, Wang Y, Singh M, Zhang C, Schnell C, Yang G, Zhang Y, et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med. 2015;21:1318–1325. doi: 10.1038/nm.3954. [DOI] [PubMed] [Google Scholar]
- Gillet JP, Calcagno AM, Varma S, Marino M, Green LJ, Vora MI, Patel C, Orina JN, Eliseeva TA, Singal V, et al. Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti-cancer drug resistance. Proc Natl Acad Sci U S A. 2011;108:18708–18713. doi: 10.1073/pnas.1111840108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausser HJ, Brenner RE. Phenotypic instability of Saos-2 cells in long-term culture. Biochemical and biophysical research communications. 2005;333:216–222. doi: 10.1016/j.bbrc.2005.05.097. [DOI] [PubMed] [Google Scholar]
- Hidalgo M, Amant F, Biankin AV, Budinska E, Byrne AT, Caldas C, Clarke RB, de Jong S, Jonkers J, Maelandsmo GM, et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov. 2014;4:998–1013. doi: 10.1158/2159-8290.CD-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe ES, Harris NL, Stein H, Isaacson PG. Classification of lymphoid neoplasms: the microscope as a tool for disease discovery. Blood. 2008;112:4384–4399. doi: 10.1182/blood-2008-07-077982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeay S, Gaulis S, Ferretti S, Bitter H, Ito M, Valat T, Murakami M, Ruetz S, Guthy DA, Rynn C, et al. A distinct p53 target gene set predicts for response to the selective p53-HDM2 inhibitor NVP-CGM097. eLife. 2015;4 doi: 10.7554/eLife.06498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien S, Merino-Trigo A, Lacroix L, Pocard M, Goere D, Mariani P, Landron S, Bigot L, Nemati F, Dartigues P, et al. Characterization of a large panel of patient-derived tumor xenografts representing the clinical heterogeneity of human colorectal cancer. Clin Cancer Res. 2012;18:5314–5328. doi: 10.1158/1078-0432.CCR-12-0372. [DOI] [PubMed] [Google Scholar]
- Klco JM, Spencer DH, Miller CA, Griffith M, Lamprecht TL, O’Laughlin M, Fronick C, Magrini V, Demeter RT, Fulton RS, et al. Functional heterogeneity of genetically defined subclones in acute myeloid leukemia. Cancer Cell. 2014;25:379–392. doi: 10.1016/j.ccr.2014.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nature reviews Drug discovery. 2004;3:711–715. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- Koster J, Rahmann S. Snakemake--a scalable bioinformatics workflow engine. Bioinformatics. 2012;28:2520–2522. doi: 10.1093/bioinformatics/bts480. [DOI] [PubMed] [Google Scholar]
- Ledford H. Translational research: 4 ways to fix the clinical trial. Nature. 2011;477:526–528. doi: 10.1038/477526a. [DOI] [PubMed] [Google Scholar]
- Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:1724–1735. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liem NL, Papa RA, Milross CG, Schmid MA, Tajbakhsh M, Choi S, Ramirez CD, Rice AM, Haber M, Norris MD, et al. Characterization of childhood acute lymphoblastic leukemia xenograft models for the preclinical evaluation of new therapies. Blood. 2004;103:3905–3914. doi: 10.1182/blood-2003-08-2911. [DOI] [PubMed] [Google Scholar]
- Lv PC, Sun J, Zhu HL. Recent advances of p53-MDM2 small molecule inhibitors (2011-present) Current medicinal chemistry. 2015;22:618–626. doi: 10.2174/0929867322666141128162557. [DOI] [PubMed] [Google Scholar]
- Mak IW, Evaniew N, Ghert M. Lost in translation: animal models and clinical trials in cancer treatment. American journal of translational research. 2014;6:114–118. [PMC free article] [PubMed] [Google Scholar]
- Montero J, Sarosiek KA, DeAngelo JD, Maertens O, Ryan J, Ercan D, Piao H, Horowitz NS, Berkowitz RS, Matulonis U, et al. Drug-induced death signaling strategy rapidly predicts cancer response to chemotherapy. Cell. 2015;160:977–989. doi: 10.1016/j.cell.2015.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odejide O, Weigert O, Lane AA, Toscano D, Lunning MA, Kopp N, Kim S, van Bodegom D, Bolla S, Schatz JH, et al. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood. 2014;123:1293–1296. doi: 10.1182/blood-2013-10-531509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott CJ, Kopp N, Bird L, Paranal RM, Qi J, Bowman T, Rodig SJ, Kung AL, Bradner JE, Weinstock DM. BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia. Blood. 2012;120:2843–2852. doi: 10.1182/blood-2012-02-413021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350:1535–1548. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- Reyal F, Guyader C, Decraene C, Lucchesi C, Auger N, Assayag F, De Plater L, Gentien D, Poupon MF, Cottu P, et al. Molecular profiling of patient-derived breast cancer xenografts. Breast cancer research: BCR. 2012;14:R11. doi: 10.1186/bcr3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nature reviews Molecular cell biology. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla SA, Rooney MS, Rajasagi M, Tiao G, Dixon PM, Lawrence MS, Stevens J, Lane WJ, Dellagatta JL, Steelman S, et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat Biotechnol. 2015;33:1152–1158. doi: 10.1038/nbt.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabe Y, Konopleva M. Role of Microenvironment in Resistance to Therapy in AML. Current hematologic malignancy reports. 2015;10:96–103. doi: 10.1007/s11899-015-0253-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigert O, Lane AA, Bird L, Kopp N, Chapuy B, van Bodegom D, Toms AV, Marubayashi S, Christie AL, McKeown M, et al. Genetic resistance to JAK2 enzymatic inhibitors is overcome by HSP90 inhibition. J Exp Med. 2012;209:259–273. doi: 10.1084/jem.20111694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg E, Halilovic E, Cooke VG, Nonami A, Ren T, Sanda T, Simkin I, Yuan J, Antonakos B, Barys L, et al. Inhibition of Wild-Type p53-Expressing AML by the Novel Small Molecule HDM2 Inhibitor CGM097. Mol Cancer Ther. 2015;14:2249–2259. doi: 10.1158/1535-7163.MCT-15-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westcott PM, Halliwill KD, To MD, Rashid M, Rust AG, Keane TM, Delrosario R, Jen KY, Gurley KE, Kemp CJ, et al. The mutational landscapes of genetic and chemical models of Kras-driven lung cancer. Nature. 2015;517:489–492. doi: 10.1038/nature13898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelenetz AD, Gordon LI, Wierda WG, Abramson JS, Advani RH, Andreadis CB, Bartlett N, Byrd JC, Czuczman MS, Fayad LE, et al. Chronic lymphocytic leukemia/small lymphocytic lymphoma, version 1.2015. J Natl Compr Canc Netw. 2015;13:326–362. doi: 10.6004/jnccn.2015.0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Claerhout S, Prat A, Dobrolecki LE, Petrovic I, Lai Q, Landis MD, Wiechmann L, Schiff R, Giuliano M, et al. A renewable tissue resource of phenotypically stable, biologically and ethnically diverse, patient-derived human breast cancer xenograft models. Cancer Res. 2013;73:4885–4897. doi: 10.1158/0008-5472.CAN-12-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.