Abstract
Antibodies that bind RNA represent an unrealized source of reagents for synthetic biology and for characterizing cellular transcriptomes. However, facile access to RNA binding antibodies requires engineering of effective Fab libraries guided by knowledge of the principles that govern RNA recognition. Here we describe a Fab identified from a YSGR minimalist synthetic library during phage display against a branched RNA target. The Fab (BRG) binds with 20 nM dissociation constant to a single-stranded RNA sequence adjacent to the branch site and can block the action of debranchase enzyme. We report the crystal structure in complex with RNA target at 2.38Å. The Fab traps the RNA in a hairpin conformation that contains a two base-pair duplex capped by a tetraloop. The paratope surface consists of residues located in four CDRs including a major contribution from H3, which adopts a helical structure that projects into a deep, wide groove formed by the RNA. The amino acid composition of the paratope reflects the library diversity, consisting mostly of tyrosine and serine residues and a small, but significant contribution from a single arginine residue. This structure, involving recognition of single-stranded RNA via a stem-loop conformation, together with our two previous structures involving recognition of an RNA hairpin loop and an RNA tertiary structure, reveals the capacity of YSG(R)-biased, minimalist libraries to form binding surfaces for specific RNA conformations and distinct levels of RNA structural hierarchy.
Keywords: Fab, tetraloop, RNA-protein interactions, phage display, crystallography chaperone, RNA binding Fab, Crystal Structure, ssRNA, YSGR library
Graphical abstract
Introduction
As key protein components of the mammalian immune systems, antibodies serve critical roles in identifying, neutralizing, and eliminating foreign substances. Within the molecular context of the immunoglobulin architecture, variation of the CDR amino acid composition allows the immune system to generate antibodies against virtually any target with high affinity and exquisite specificity. This vast, functional capacity has led to a rapidly growing number of antibodies for medical applications as diagnostic tools and therapeutic agents and has empowered a variety of methodologies for research applications ranging from imaging techniques to chaperone assisted crystallization1; 2; 3. Their versatility and widespread utility has fueled the development of methods to obtain antibodies, including hybridoma approaches and display technologies using natural repertoires derived from immune systems or man-made repertoires derived from synthetic oligonucleotides of designed diversity4; 5.
Most antibodies used for medical or research applications, target proteins of the terrestrial proteome and associated posttranslational modifications6; 7. Nevertheless, biological systems also possess a highly complex and dynamic cellular RNA population, collectively known as the transcriptome, and antibodies that target RNA could provide equally valuable reagents for medicine and research. Transcriptomes play significant roles in gene regulation, cell growth, differentiation, and disease8; 9; 10; 11. A small fraction of these RNAs serve as mRNAs for protein synthesis, but the biological role for the majority of transcripts is still unclear12. Despite their biological and medical importance, our understanding of transcriptomes and associated posttranscriptional modifications remains in its infancy, due in part to limited methods for elucidating the transcriptome’s structure and function. Currently, large-scale identification and analysis of these non-coding RNAs (ncRNAs) rely predominantly on deep-sequencing data13, sometimes following immunoprecipitation of specific proteins14; 15. Following ncRNA identification, other conventional techniques such as hybridization and antisense can be deployed to locate, image or isolate specific RNAs for further research16; 17. Nevertheless many ncRNAs mediate their functions through diverse secondary and tertiary structures, which cannot be characterized readily using available techniques10. Conformation-specific RNA-binding antibodies, as highly engineerable proteins with diversity in their complementarity determining regions (CDRs), can potentially serve this purpose well.
The blood serum from human patients or animal models with autoimmune disease may contain antibodies that bind to nucleic acids18; 19; 20; 21; 22; 23; 24. However, healthy immune systems deploy components of the innate immune system, such as Toll-like receptors and protein kinases, in response to the presence of non-self RNA25. Thus, injection of structured RNA into the blood of a host animal likely would not elicit production of antibodies that target the injected RNA. Moreover, the presence of nucleases in blood serum likely would limit the lifetime of the injected RNA, precluding the use of immunization approaches to obtain antibodies.
In recent years, display technologies have enabled access to recombinant antibodies without the need for host immunization, providing an opportunity to circumvent exposure to nucleases and thereby offering a platform to engineer and isolate antibodies that bind to specific RNAs6. These display technologies have used natural immune repertoires26, but minimalist repertoires built from designed diversity have also proven effective against protein antigens1; 27. These synthetic libraries have undergone successive rounds of optimization based on information from antibody:protein interactions derived from analysis of functional antibody sequences and structures28; 29. For RNA targets, our recent work has demonstrated that minimalist libraries within the Fab 4D5 framework can yield Fabs that bind RNAs with high affinity and specificity30. We have also established proof-of-principle that these Fabs can serve as chaperones for RNA crystallization30; 31; 32. Nevertheless, compared with selections against protein antigens using optimized libraries, selections against RNA targets have yielded relatively few high affinity Fabs, reflecting limited knowledge of library design principles with respect to CDR sequence diversity, length, and structure.
The limitations of currently available libraries underscore the need to investigate structural and energetic principles of Fab:RNA complexes for a range of RNA types in order to elucidate the principles and minimum requirements underlying protein-RNA recognition and to inform future library design. Previously we described the structures of two Fab-RNA complexes, one revealing recognition of an RNA tertiary structure and the other revealing recognition of an RNA hairpin loop 30; 32. In this study, we describe a Fab capable of recognizing a stem-loop RNA conformation of a single-stranded RNA (ssRNA). Using a minimalist library we obtained a Fab that binds to a 10-nt ssRNA sequence with high affinity and specificity, and we solved the crystal structure of the Fab:RNA complex at 2.38Å resolution. The new structure together with the two previous structures reveals principles of RNA recognition that govern binding surfaces generated from minimalist libraries enriched in Y, S, G, and R and suggest strategies to improve the design of future libraries.
Results
Selection and characterization of Fabs directed to ssRNA
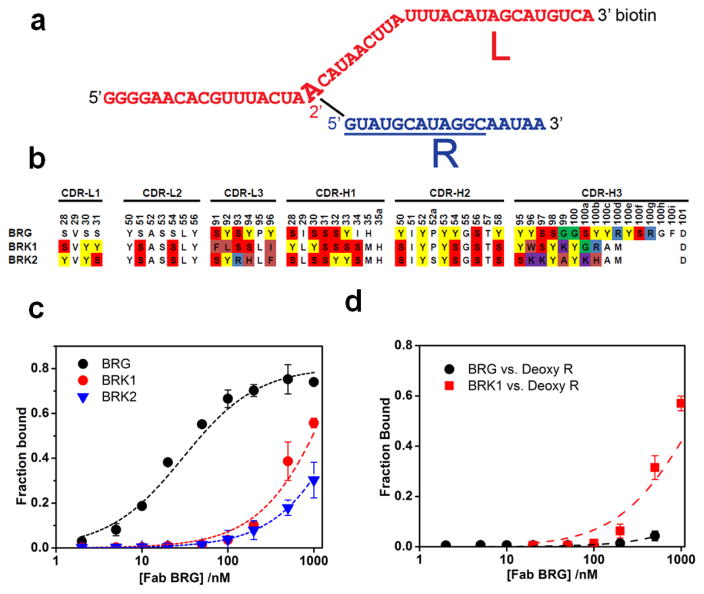
We initially set out to isolate Fabs that bind to branched RNAs, which arise from the chemistry of nuclear premessenger RNA splicing catalyzed by the spliceosome. We used a deoxyribozyme to construct a branched RNA corresponding in sequence to the branch region of the pre-mRNA intron derived from the YBL059W gene from S. cerevisiae33; 34. For target immobilization, strand L of the branched RNA was biotinylated on the 3’-end during solid-phase oligonucleotide synthesis (Fig. 1a). For in vitro selection using phage display, we deployed two synthetic antibody libraries that carry a “reduced genetic code”29; 32. One library, termed YSGR, encodes equal proportions of Y and S at variable positions in CDR-L3, CDR-H1, and CDR-H2. CDR-H3 encodes 38% Y, 25% S, 25% G, and 12% R28; 30. The second library, termed YSGRKX, encodes diversity in all six CDRs. CDR-L1 and –L2 contain equal proportions of Y and S at variable positions; CDR-H1 and H2 contain equal proportions of Y, F, and S; and CDR-L3 and H3 encode 25% Y, 15% S, 10% G, 12.5% R, 7.5% K and allows for 30% of all other amino acids except C, I, and M. In addition, four of the CDRs in the YSGRKX library have variable loop sizes: L1 (5–6 residues), L3 (2–8 residues), H1 (3–8 residues) and H3 (4 to 17 residues)35. We conducted selections against the branched RNA target and identified three unique Fab sequences from selected pools after three rounds of panning: BRG from YSGR library, BRK1 and BRK2 from YSGRKX library (Fig. 1b).
Figure 1.
The YBL059W branched RNA target and binding by Fabs selected from synthetic phage-displayed Fab libraries. (a). Sequence and structure of the target. The RNA is composed of two strands, L (red) and R (blue), connected by a chemical bond between 5’-end of R and A18 2’-OH of L. The RNA contains a biotin moiety on the 3’-end of L for antigen immobilization during selection. (b). CDR sequences of three selected Fab clones (Fab-BRG, -BRK1, and –BRK2. Only positions with designed diversity are colored according to amino acid type. (c). Fab binding to branched RNA revealed by filter binding assays. Fraction bound reflects the fraction of RNA retained on a nitrocellulose filter as a result of incubation with the indicated Fab. For Fab-BRG, a fit of the data to a binding equation gave KD = 21 ± 3 nM. All binding assays contained PBS at pH 7.4, 0.2 mM EDTA (d). BRG and BRK1 binding to Deoxy R.
We subsequently expressed these Fab sequences as soluble proteins and measured their affinity to the target RNA using a nitrocellulose filter binding assay. BRK1 and BRK2 expressed poorly in E. coli (< 1 mg/L) with BRK2 giving only trace yield. These poor expression yields could reflect in part the significant number of positively charged residues in the CDR loops (2 in BRK1 and 4 in BRK2), which can negatively impact Fab expression36. Fab BRG bound the YBL059W branched RNA with high affinity (KD = 21 ± 3 nM); BRK1 and BRK2 bound with significantly weaker affinity (KD > 500 nM; Fig. 1c). We also tested the ability of BRG and BRK1 Fabs to bind a deoxynucleotide version of the R strand (Deoxy R). BRG exhibited no detectable binding, but BRK1 appeared to retain Deoxy R on nitrocellulose with a concentration dependence similar to that observed for the parent branched RNA (Fig. 1d). Because Fab BRG appeared to have RNA-specific binding, we characterized it further.
RNA binding epitope and specificity of Fab BRG
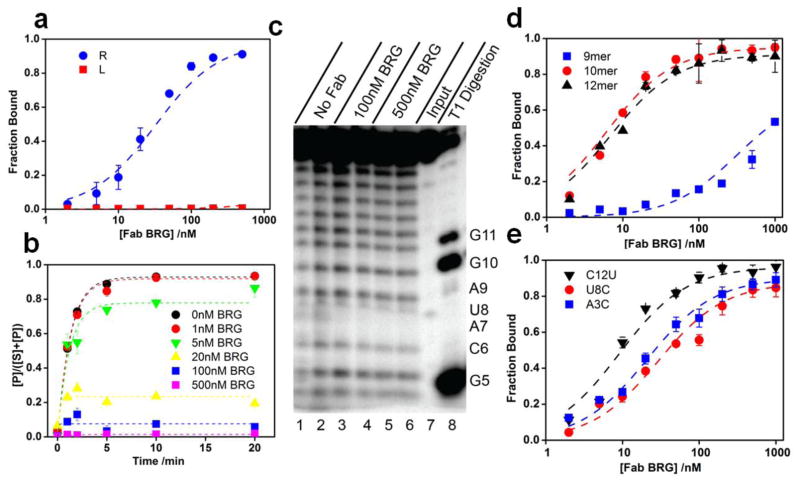
To identify the region of the branched RNA involved in binding Fab BRG, we tested the ability of Fab BRG to bind non-branched oligonucleotide strands, L and R, corresponding to sequences found in YBL059W branched RNA. Fab BRG exhibited no detectable binding to the L strand, but its affinity to the R strand was similar to that for the branched RNA, suggesting that the entire binding epitope may reside solely within the R strand (Fig. 2a). We also anticipated that Fab binding to the branched RNA might inhibit the debranching reaction catalyzed by debranchase Dbr1 from S. cerevisiae37; 38, as Dbr1 recognizes the branch point and immediately adjacent nucleotides on strand R39. Our data show that Fab BRG inhibits Dbr1-catalyzed debranching in a concentration-dependent manner with Ki close to the Kd (Fig. 2b; Supplementary Fig. 1), consistent with competitive inhibition. Concentrations of Fab BRG above 100 nM abolished the reaction almost completely.
Figure 2.
Location of the Fab-BRG binding site within the branched RNA. (a). Fab BRG binds strand R with full affinity (KD = 26 ± 1 nM). (b). Fab BRG inhibits Dbr1 debranching of the RNA target ([Dbr1] = 1.4 nM). See Supplementary Fig. 1 for original gel figures. (c). Hydroxyl radical footprinting on strand R in the presence and absence of Fab BRG. Input – control samples without Fe-EDTA treatment; T1 Digestion – ladder generated by treating samples with ribonuclease T1. (d). Fab BRG binds a 10-mer oligonucleotide (A3 to C12) with full affinity (KD = 10 ± 2 nM). (e). Fab BRG binds 3 of the 10 scanning mutagenesis 10-mer constructs with nearly full affinity (See Table 1 for more details).
We next sought to identify the minimal RNA sequence able to bind to Fab. First we performed hydroxyl radical footprinting of the R strand in the presence of Fab BRG (Fig. 2c). The Fab gave strong protection from hydroxyl radical cleavage at nucleotide A7, suggesting that Fab binding renders the ribose at this position solvent inaccessible. Next, we analyzed binding of a series of strand R mutants starting with a truncation of the five 3’-terminal nucleotides to give a 12-mer RNA oligonucleotide (R-12: 5’-GUAUGCAUAGGC-3’). Fab BRG bound to R-12 with the same affinity as it bound to R (Fig. 2d). Truncation of R-12 from 5’-end showed that the two 5’-terminal nucleotides (G1 and U2) can be removed with no loss of affinity (R-10: 5’-AUGCAUAGGC-3’); however, deletion of A3 as well (R-9: 5’-UGCAUAGGC-3’) decreased the affinity by more than 10-fold (Fig. 2d; Table 1 – Truncation). The proximity of the binding site to the branch site accounts for the inhibitory effect of Fab BRG on debranching. However, Dbr1 substrate recognition involves only two nucleotides downstream of the branch site40, suggesting that Fab BRG sterically occludes the enzyme rather than competing directly for binding to the same nucleotides. In addition, in the context of R-10, mutation at positions other than A3, U8 and C12 abolished detectable binding (Fig. 2e; Table 1 - Mutation). These data collectively demonstrate a minimal Fab-binding sequence of 10 nucleotides (N3U4G5C6A7N8A9G10G11N12), with apparent stringent nucleobase specificity at seven of the positions.
Table 1.
Summary of oligonucleotide sequences tested and their KD values#
| Name | Sequence | KD to BRG/nM* | ||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Targets | L | 5’ | G | G | G | G | A | A | C | A | C | G | U | U | U | A | C | U | A | A | C | A | U | A | A | C | U | N.B. |
| U | U | A | U | U | U | A | C | A | U | A | G | C | A | U | G | U | C | A | 3’ | |||||||||
| R | 5’ | G | U | A | U | G | C | A | U | A | G | G | C | A | A | U | A | A | 3’ | 32 ± 6 | ||||||||
| Deoxy R | 5’ | d | ( | G | U | A | U | G | C | A | U | A | G | G | C | A | A | U | A | A | ) | 3’ | N.B. | |||||
| Truncations | R-12 (Crystallized) | 5’ | G | U | A | U | G | C | A | U | A | G | G | C | 3’ | 12 ± 1 | ||||||||||||
| R-10 | 5’ | A | U | G | C | A | U | A | G | G | C | 3’ | 10 ± 2 | |||||||||||||||
| R-9 | 5’ | U | G | C | A | U | A | G | G | C | 3’ | > 500 nM | ||||||||||||||||
| Mutations | A3C | 5’ | C | U | G | C | A | U | A | G | G | C | 3’ | 15 ± 2 | ||||||||||||||
| U4C | 5’ | A | C | G | C | A | U | A | G | G | C | 3’ | N.B. | |||||||||||||||
| G5A | 5’ | A | U | A | C | A | U | A | G | G | C | 3’ | N.B. | |||||||||||||||
| C6U | 5’ | A | U | G | U | A | U | A | G | G | C | 3’ | N.B. | |||||||||||||||
| A7G | 5’ | A | U | G | C | G | U | A | G | G | C | 3’ | N.B. | |||||||||||||||
| U8C | 5’ | A | U | G | C | A | C | A | G | G | C | 3’ | 21 ± 4 | |||||||||||||||
| A9G | 5’ | A | U | G | C | A | U | G | G | G | C | 3’ | N.B. | |||||||||||||||
| G10A | 5’ | A | U | G | C | A | U | A | A | G | C | 3’ | N.B. | |||||||||||||||
| G11A | 5’ | A | U | G | C | A | U | A | G | A | C | 3’ | N.B. | |||||||||||||||
| C12U | 5’ | A | U | G | C | A | U | A | G | G | U | 3’ | 29 ± 3 | |||||||||||||||
| Deletions | ΔU8 | 5’ | A | U | G | C | A | - | A | G | G | C | 3’ | N.B. | ||||||||||||||
| ΔC12 | 5’ | A | U | G | C | A | U | A | G | G | - | 3’ | N.B. | |||||||||||||||
| Hairpins | GC-swap | 5’ | A | U | G | G | A | U | A | G | C | C | 3’ | N.B. | ||||||||||||||
| plusGC | 5’ | A | U | G | C | C | A | U | A | G | G | G | C | 3’ | N.B. | |||||||||||||
| 5bp | 5’ | G | U | A | U | G | C | A | U | A | G | G | C | A | U | A | 3’ | 145 ± 10 | ||||||||||
(N.B. – No binding, less than 50% bound even at highest Fab concentration)
Names in bold letters indicate constructs with KD < 100 nM. Italic letters in sequences
indicate mutated or additional nucleotides.
All binding assays were performed using 5’-32P-radiolabeled RNA
Crystal structure of Fab BRG in complex with the R-12 RNA
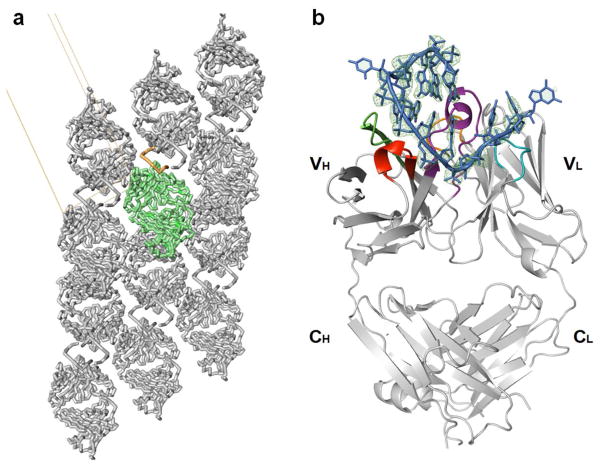
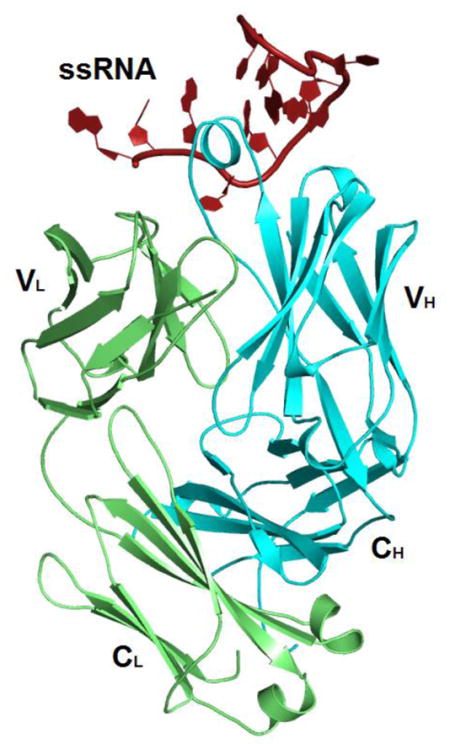
We purified Fab BRG in complex with the R-12 RNA (Supplementary Fig. 2) using size-exclusion chromatography (SEC) and subsequently crystallized it for X-ray diffraction. Initial phases were obtained by molecular replacement using Fab BL3-6 (Protein Data Bank accession code: 3IVK) minus four of the variable CDR loops (L3, H1, H2 and H3) as a search model. After model building and refinement at 2.38Å resolution, the final values of Rfree and Rwork were 0.2338 and 0.2094, respectively (Table 2). The complex crystallized with a single Fab and a single RNA in each asymmetric unit in a C2 space group lattice. Within the lattice, the complex interacts via RNA with its adjacent symmetric mate (Fig. 3a). We observed similar interaction in the crystal lattices of our two previous RNA-Fab complexes30; 32. Within the epitope-paratope interface, Fab-RNA interactions buried 928Å2 of solvent-accessible Fab surface area, mostly involving the heavy chain (723Å2). For comparison, the interface between Fab BL3-6 and its hairpin epitope buries slightly more surface area (821Å2)31; 41 and that between Fab2 and the P4-P6 RNA domain buries substantially more (1,316Å2). For comparison Fab-protein complexes bury on average 777 ± 135Å2 of surface area32. All other intermolecular interactions in the Fab BRG-R12 crystal lattice, including Fab-Fab and Fab-RNA contacts buried an additional 1601 Å2 of surface area. The dimerization interface buried 768Å2, including 522Å2 involving RNA-RNA interactions. Overall, Fab BRG contributes 83% to the total buried surface area in the epitope and lattice, consistent with its hypothesized role in facilitating crystallization2; 31.
Table 2.
Crystallographic statistics
| Data Collection | |
| Wavelength (Å) | 0.9795 |
| Space Group | C 1 2 1 |
| Cell dimensions | |
| a, b, c (Å), | 199.5, 63.4, 57.1 |
| α, β, γ (°) | 90.0, 97.5, 90.0 |
| Resolution (Å) | 60.38–2.38(2.46–2.38) |
| No. Reflections | 28135 |
| Rmerge (%) | 8.4(51.8) |
| I/σI | 11.5 (1.3) |
| Completeness (%) | 98.5 (97.1) |
| Redundancy | 3.3 (3.1) |
| Refinement | |
| Resolution (Å) | 60.38–2.38 |
| Rwork | 0.2094 |
| Rfree | 0.2338 |
| R. M. S deviations | |
| Bond Length (Å) | 0.006 |
| Bond Angles (degree) | 1.427 |
| Number of atoms | 3715 |
| Protein | 3384 |
| RNA | 255 |
| Water | 76 |
| B-factor | |
| Overall | 57.26 |
| Protein | 58.02 |
| RNA | 48.59 |
| Water | 52.20 |
| Maximum-Likelihood based | |
| Coordinate Error (Å) | 0.33 |
Values in parentheses are for highest-resolution shell.
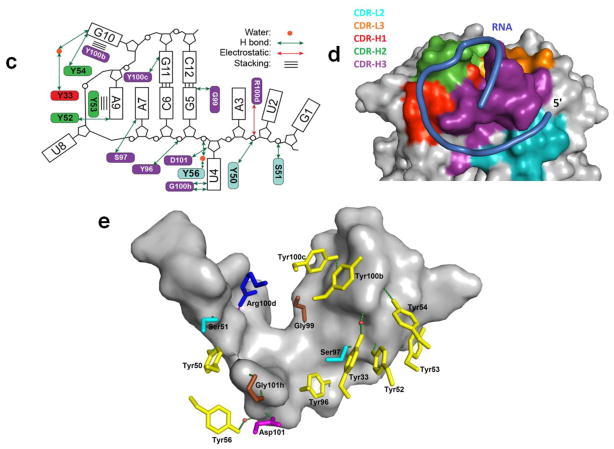
Figure 3.
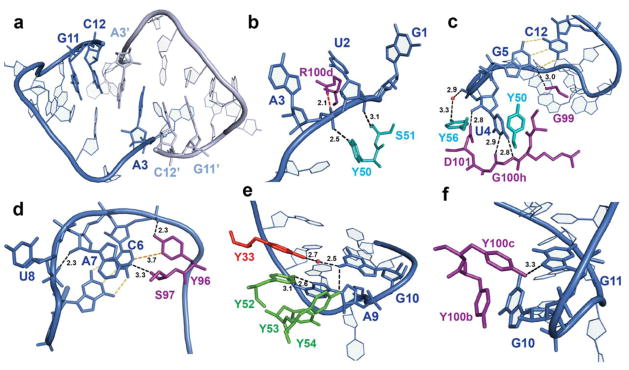
Crystal structure of the Fab-BRG:R-12 RNA complex. (a). Packing of the Fab-RNA complex (Fab: green; RNA: orange) in the crystal lattice with a unit cell displayed. Symmetry-related molecules are shown in grey. (b). Overall structure of the Fab-RNA complex (Color code: blue – RNA, cyan –CDR-L2, orange – CDR-L3, red – CDR-H1, green – CDR-H2, purple – CDR-H3). RNA is contoured with the |Fobs|-|Fcal| difference map (green mesh, 3.5σ) from a simulated annealing refinement omitting the R-12 RNA. (c). Schematic summary of Fab-RNA interactions on R-12 hairpin. Rectangles represent nucleobases and ovals represent CDR residues colored as in (d). (d). Molecular surface of the Fab-BRG variable domain in complex with the RNA. (e). Interaction between the 12mer RNA and the CDR residues. Orange spheres represent water molecules that mediate hydrogen bonds. Green dashes represent hydrogen bonds, and the red dash represents a salt bridge. Colors correspond to amino acid types (Tyr: yellow, Gly: green, Ser: red, Arg: blue, and others: magenta).
As expected by design, the structure shows that Fab BRG has an identical variable domain scaffold (excluding the six CDR loops) as its parent, Fab 4D5 (PDB code: 1FVD) with an RMSD value of 0.484 Å32; 42 and R-12 forms a hairpin secondary structure with a short stem of only two GC pairs (G5·C12 and C6·G11; Fig. 3b). Fab BRG recognizes R-12 using mostly tyrosine, serine, glycine, and arginine residues in its CDR loops to achieve shape complementarity and mediate stacking, hydrogen bonding and electrostatic interactions (Fig. 3c). These residues predominantly come from diversified positions within the library, with the only exceptions being G100h and D101, located at the end of CDR-H3, and Y50 and Y56, located at each end of CDR-L2. In general, VH residues recognize the stem-loop, and VL residues mostly interact with the single-stranded region (Fig. 3b). Within the heavy chain CDR loops, 11 Tyr residues emerged from randomized positions (Fig. 1b); seven of these along with two Tyr residues from the constant CDR-L2, provide direct contacts to the RNA (Fig. 3e). The longest CDR loop, H3, forms a helical knob that inserts into a pocket created by the major groove of the RNA hairpin and the single-stranded region (Fig. 3b and Supplementary Fig. 3). CDR H3 and the other CDRs together form clefts through which the RNA backbone traverses (Fig. 3d).
In the crystal lattice, nucleobase stacking involving G11, C12, A3’, A3, C12’ and G11’ predominantly mediates RNA-RNA interactions between the complexes (Fig. 4a). According to dynamic light scattering data, the fitted Stokes radius of the Fab-RNA complex (3.44nm; MW=52 kDa) in solution exceeds that of Fab BRG alone (3.29nm; MW=48 kD) by less than 5%, consistent with a monomeric RNA:Fab complex (Supplementary Fig. 4). We conclude that the complex exists in solution predominately as a monomer as expected.
Figure 4.
Interactions within the Fab-RNA binding interface. (a). Dimerization of the oligonucleotide observed in crystal lattice. The symmetry-related molecule is shown in pale color, with primes after its nucleotide numbers. (b–f) Close-up views of Fab-RNA interactions involving (b) CDR-L2, U2 and A3; (c) CDR-H3, U4, G5 and C12 (d) CDR-H3, C6, A7 and U8; (e) CDR-H2, A9 and G10; (f) CDR-H3, G10 and G11. Dashed lines with numbers indicating distances in Å: black – Fab-RNA hydrogen bonds, yellow – GC-pair hydrogen bonds, red – electrostatic, orange – hydrophobic. Residue color code matches Figure 3b.
RNA-protein interactions within the R-12:Fab BRG interface
Here we provide a detailed description of the RNA contacts, progressing 5’ to 3’ along the chain. CD-L2 mediates interactions with the backbone of the single-stranded region of the RNA near its 5’-terminus (Fig. 4b). G1, which is dispensable for binding, has little electron density and may be disordered. The phosphodiesters at U2 and A3 form hydrogen bonds to the side chains of CDR-L2 residues S51 and Y50, respectively, although the tight binding of R-10 (KD = 10 ± 2 nM), which lacks G1 and U2, suggests that the U2 phosphodiester interaction may make no net energetic contribution to binding (Fig. 2d). The A3 phosphodiester also interacts electrostatically with CDR-H3 residue R100d, which may explain the more than 10-fold decrease in affinity of the Fab for binding R-9 compared to R-12 (Fig. 2d).
Beginning with U4, the RNA backbone traverses CDR-H3 and makes contacts with it (Fig. 4c, d). U4, located 5’ of the RNA stem, projects its nucleobase away from the helical stack and into a pocket created by CDR-H3 and partially by CDR-L2 (Figs. 3d and 4c, Supplementary Fig. 5). This allows A3 to stack with A3’ of the symmetry-related molecule (Fig. 4a). Within the pocket, two tyrosine residues Try50 and Tyr 56 at the termini of CDR-L2 form edge-on π–interactions with the U4 nucleobase, and two residues from CDR-H3 form hydrogen-bonding interactions with U4 (Fig. 4c, Supplementary Fig. 5). The G100h backbone atoms form hydrogen binds to the nucleobase and D101 forms a hydrogen bond to the U4 2’-OH. The former interaction accounts for the observed absence of binding by the U4C mutant (Table 1). G5 and C6 form base pairs with G11 and C12, respectively, to form the stem. Aligning G11C12 to one side of a standard, two base-pair duplex shows the base planes of the complementary nucleotides G5 and C6 rotated about 30 relative to the base planes of the complementary nucleotides in the standard duplex, respectively (Supplementary Fig. 6).
The Fab makes four hydrogen bond interactions with the duplex region of the RNA, three via CDR-H3 and one via CDR-L2. The latter involves a water-mediated interaction between the phosphate of G5 and the hydroxyl group of Y56 (Fig. 4c). From CDR-H3 the backbone NH at G99 contacts the O6 keto group of G5, (Fig. 4c); at Y96 the hydroxyl group contacts the C6 phosphodiester and the aromatic ring locates in Van der Waals contact distance to the C6 nucleobase. On the opposite strand of the duplex, the Y100c hydroxyl group interacts with N7 of G11. The structure accounts for the observed effects of mutations in this region (Table 1). Mutation of either G5 or G11 in the duplex to adenosine abolishes binding, likely by disrupting base pairing. The C12U mutation affects binding minimally, suggesting that the complex may accommodate a G5:U12 wobble pair. C12 participates in the stacking that mediates dimerization (Fig. 4a) but makes no contact with the Fab. In contrast, the C6U mutation disrupts binding. Possibly U6:G11 wobble pairing alters the position of G11 and disrupts the interaction with Y108.
Progressing along the RNA sequence into the loop, the A7 nucleobase points toward and resides within hydrogen bonding distance of S97. U8 adopts the syn conformation about the glycosidic bond, which directs the nucleobase away from the RNA and Fab towards solvent and allows the phosphodiester backbone to make a reverse turn (Fig. 4d). The turn at U8 positions the 2’-OH of A7 within hydrogen bonding distance (2.4 Å) of the A9 phosphate, consistent with the observed protection from hydroxyl radical cleavage at A7 in the presence of Fab (Fig. 2c). Mutation of U8 had no effect on binding but its deletion abolished binding, likely by altering the backbone conformation (Fig. 2e; Table 1 – Deletion).
CDR-H1 and CDR-H2 play significant roles in contacting RNA loop nucleotides after the turn at U8 (Fig. 4e). The hydroxyl groups of Y52 and Y54 of CDR-H2 form hydrogen bonds with the A9 and G10 nucleobases, respectively, and CDR-H1 Y33 stacks on Y52 and forms another water-mediated hydrogen bond with G10. Y53 stacks on A9, a stack that continues through A7, C6 and G5 nucleobases (Fig. 4e). Following A9, the RNA backbone loops back towards CDR-H3, forming a stacking interaction between Y100b and G10 (Fig. 4f). Notably, all contacts in this region from A9 to G11 involve exclusively six tyrosine residues from VH.
Fab BRG stabilizes the hairpin conformation within the R-12 RNA
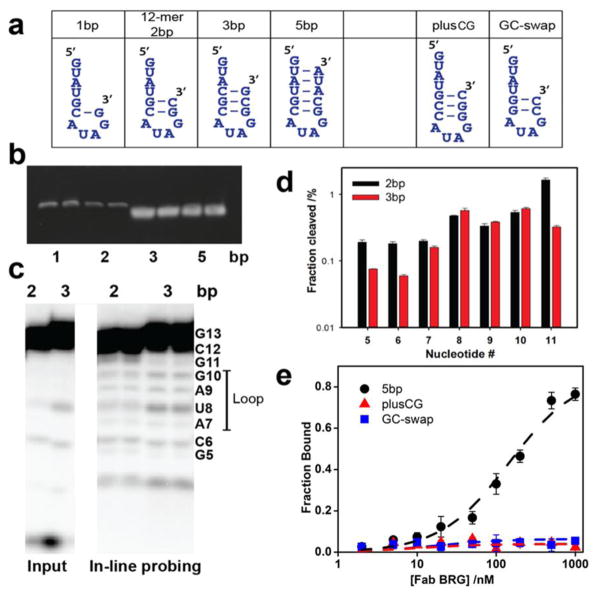
The finding that R-12 interacts with the antibody paratope as a hairpin led us to assess whether the free RNA adopts this conformation on its own. Secondary structure predictions using mFold with different parameters gave ambiguous outcomes (Tm = 44 °C and 27 °C, respectively)43; 44; 45. We therefore conducted experiments on variants designed to have stems of from 1 to 5 putative base pairs (Fig. 5a). In a non-denaturing polyacrylamide electrophoresis gel, the variants with one and two potential base pairs migrated similarly, whereas the 3-bp and 5-bp variants (mFold Tm > 55 °C) migrated more rapidly. These results suggest that the 3-bp and 5-bp variants possess a greater degree of compaction than the other variants, consistent with formation of the hairpin conformation in variants with at least 3 base pairs (Fig. 5b)46. We also conducted in-line probing assays, which assess base pairing of RNA nucleotides by relative resistance to spontaneous RNA cleavage47. The putative loop nucleotides (7, 8, 9, and 10) showed a similar cleavage fraction in the 2-bp and 3-bp variants. However, putative stem nucleotides 5, 6, and 11 showed more cleavage in the 2-bp than in the 3-bp variant (Fig. 5c, d), suggesting that these nucleotides in R-12 experience base pairing less frequently. Overall, these data show that R-12 RNA possesses little secondary structure in the unbound state. We infer that stable population of the hairpin conformation occurs as a result of Fab binding.
Figure 5.
R-12 RNA undergoes a shift in conformation upon Fab-BRG binding. (a). RNA oligonucleotides drawn in their putative hairpin conformations. All variants contain the AUAG tetraloop nucleotides were designed to differ in the number helical base pairs (b). EMSA assays (20% acrylamide) for RNA oligonucleotides containing 1, 2, 3 and 5 putative base pairs. The 2-bp hairpin migrates in a manner similar to 1-bp oligonucleotides but slower than the 3-bp and 5-bp oligonucleotide. (c). In-line probing analysis of 2-bp and 3-bp oligonucleotides. Input – control samples without probing buffer treatment (d). Quantitative analysis of the in-line probing results from (c). Positions 5, 6, and 11 react faster in the 2-bp oligonucleotide compared to 3-bp, whereas position 7, 8, 9, and 10 react to similar extent. Degradation counts from input are subtracted from each band in quantification. Data represent mean values ± s.d. from two experiments. (e). Effect of the number and identity of stem base-pairs on Fab-BRG binding (See Table 1 for more details).
We further investigated the sequence specificity of Fab BRG through binding analysis of three additional variants designed to retain the capacity to adopt a hairpin conformation. (Fig. 5a, e). First, an RNA able to form a 2-bp stem but with C6 and G11 swapped (“GC-swap”) resulted in complete loss of binding, possibly by disrupting the Y100c-G11 interaction (Fig. 4f). Additionally, an RNA designed to form a hairpin with an additional CG base pair inserted (“plusCG”) had no binding. These results indicate that the sequence of the R-12 stem has significance for binding beyond the capacity to form duplex. Second, we designed an RNA to have a 5-bp stem (“R-5bp”) that contains the original R-12 sequence but with three nucleotides added to the 3’-end to involve U2, A3, and U4 in base-pairing. R-5bp retained the ability to bind Fab-BRG but with about 10-fold lower affinity. We conclude that Fab BRG has specificity for the RNA primary sequence in the stem-loop conformation and based on the R-5bp results, favors the absence of secondary structure in the upstream nucleotides.
Discussion
Fab BRG recognizes a specific conformation of an ssRNA
Our previous work has established that Fabs derived from synthetic libraries can specifically recognize RNA secondary and tertiary structure motifs. For example, Fab2 recognizes the tertiary structure of the P4–P6 domain derived from the Tetrahymena group I intron32. Fab BL3-6 recognizes an AAACA hairpin loop closed by a GC base pair30. RNA recognition in these two cases includes nucleotide-specific interactions in the context of the folded RNA. The current work demonstrates another mode of RNA association in which Fab BRG binds to a specific 10-nt ssRNA sequence with nanomolar affinity through stabilization of a 2-bp hairpin conformation closed by an A7U8A9G10 loop. The binding interactions have stringent specificity for the nucleobases with little tolerance for mutation. The A7U8A9G10 tetraloop does not correspond to common tetraloops found in biological systems neither in sequence nor structure, and the first and fourth nucleotides do not form a non-canonical base pairing interaction as observed in the common tetraloops (Fig. 3c)48. However, the A7U8A9G10 tetraloop does share some features in common with other tatraloops48. Specifically, the first and the third nucleobases (A7 and A9) stack over the closing base of the stem at the 5’-end. In addition, as observed in the common tetraloops, the A7U8A9G10 tetraloop contains a reverse turn, in this case at the second nucleotide U8, which adopts the syn conformation about the glycosidic bond, allowing the ribose-phosphate backbone to reverse direction by 180° on the 3’-side of U8. Fab BRG recognition of the stem loop involves predominantly hydrogen bonding and extensive π-π interactions with tyrosine residues. The use of extensive π-π interactions occurs infrequently in natural tetraloop-protein interactions48, possibly reflecting evolutionary adaptation for specialized biological functions, whereas Fab BRG reflects a binding solution from restricted amino acid diversity without biological constraints49. In the absence of Fab, the RNA likely samples a dynamic ensemble of conformations, including the 2-bp hairpin conformation observed in our structure. In this respect, Fab BRG provides proof-of-concept for the use of Fabs to isolate and crystallize specific conformers among conformationally dynamic populations of RNA.
Tyrosine Dominates the Fab BRG paratope
The paratope involves CDRs L2, H1, H2, and H3. CDR-H3 plays the dominant role (Supplementary Fig. 7), just as it does for Fabs that bind protein antigens50. Protein-binding Fabs also frequently make use of the adjacent CDR-L350; 51. In contrast, CDR-L3 in Fab BRG makes no direct contact to the RNA but may support CDR-H3 structure indirectly through a hydrogen bond between the backbone carbonyl of Ser91 and the side chain of Arg 100g. Instead CDR-L2, which contributes to protein binding interfaces only occasionally, makes a substantial contribution to the RNA binding interface (19% of the buried surface area) despite having had no encoded diversity in the phage library (Supplementary Fig. 7). Previously, we reported a substantial CDR-L2 contribution from Fab2 in the P4-P6 interface. (CDR-L2 contributes 28% to the buried surface area)32. In addition, the U4 pocket between CDR-L2 and H3 in Fab BRG resembles the U130 pocket at the same location of Fab2, both accommodating the uridine nucleobase using CDR-H3 residues G100h and D101 and CDR-H2 residues Tyr50 and Tyr56 (Fig. 4c and Supplementary Fig. 5). This nucleobase pocket also forms in Fab BL3-6 but resides outside the RNA binding paratope.
With the exception of Asp 101, which resides within hydrogen bonding distance of the U4 2’OH, Fab BRG binding to the RNA involves exclusively interactions with Tyr, Ser, Gly, and Arg as dictated by the prescribed amino acid diversity (Fig. 3e, Supplementary Fig. 7). CDR-H3 contains two Arg residues; only Arg100d makes direct contact with the RNA phosphodiester backbone, whereas Arg100g engages in the aforementioned hydrogen bond with a serine residue in CDR-L3. Two Ser and two Gly residues also contribute to the interface, but Tyr dominates the binding surface, making both hydrogen bonding and stacking interactions (Fig. 3c).
Comparison to previous Fab:RNA complex structures
We have thus far conducted structural studies on three unique Fabs (Fab2, BL3-6 and BRG) and used them to solve four different RNA structures, including this work (Supplementary Fig. 7)30; 31; 32. In our structural studies, these Fab molecules have served as effective crystallization chaperones to mediate at least 70% of intermolecular contacts in the lattice (including the binding interface). Moreover, they show the capacity to adopt a range of elbow angles (between the variable and constant domain) to accommodate lattice packing (Supplementary Fig. 7, 8a)52. In all three cases, the a interfaces buried at least as much surface area as for Fab complexes with proteins (777Å2)41 but substantially less surface area than natural protein-RNA interactions (2,545Å2)53. Despite the smaller average surface burial compared to RNA-binding proteins, the Fabs still achieved mid-to-low-nanomolar affinity, underscoring both the efficiency of antibodies in target binding and their utility as engineerable designer RNA-binding proteins54.
The paratopes of these Fab-RNA complexes involve multiple CDRs (four or more; Supplementary Fig. 7, Supplementary Table 1). Common to all are contributions from CDR H2 and a dominant contribution from H3. CDRs H1, L2 and L3 each make contributions in two of the three complexes. As expected, in these structures all Fabs including the parent Fab 4D5 adopt an identical variable domain conformation excluding the CDR loops (Supplementary Fig. 9a,b; RMSD < 1 Å between each other), indicating its structural robustness to extensive mutations in the CDRs. As the Fab libraries used lack diversity in the L1 and L2 CDRs, CDR-L1 and CDR-L2 have identical sequences (Supplementary Fig. 8c) and adopt highly similar peptide backbone conformations in Fab2, BL3-6 and BRG (Supplementary Fig. 9). CDR-L1 makes the least, if any, contribution to the paratope surfaces of the three Fabs. In contrast, L2 contributes significantly in both Fab BRG and Fab2, although the extensive RNA interactions mediated by CDR L2 in Fab2 (2RBS) and BRG (5E08) have little influence on the L2 CDR conformation (Supplementary Fig. 9). CDR-L3 and -H1, diversified with Y and S at selected positions, have slightly different sequences among the Fabs and adopt modestly different conformations (Supplementary Fig. 9). Again, RNA interactions appear to have little influence on the CDR-L3 conformation (Supplementary Fig. 9). On the other hand, CDR-H2, also diversified with Y and S at selected positions, contributes significantly to all three paratopes and adopts a distinct conformation in the case of Fab2 (Supplementary Fig 9). The conformational difference in Fab2 likely arises from mutation of a non-paratope residue, P53aS that generally contributes to CDR conformation. CDR-H3, the loop designed with the highest diversity and length variability, plays the most significant role in all three Fab-RNA interfaces and shows the greatest sequence and structural diversity.
In both Fab2 and Fab BL3-6, Tyr plays a significant role in RNA binding though not as dominant as in Fab BRG (Supplementary Fig. 7). In Fab 2, the contribution of Ser in terms of buried surface area nearly equals that of Tyr (31% and 29% for Tyr and Ser, respectively)32 and in Fab BL3-630; 31, the contribution of Arg to surface burial exceeds that of Tyr (38% and 31%, respectively). Among the four paratope Arg residues in Fab BL3-6, three contact the RNA and one, Arg100, contacts a residue (Asp101) at the carboxy terminus of CDR-H3 in the Fab-BL3-6 RNA ligase structure30 but points away from the Fab surface toward solvent in the BL3-6:Spinach RNA aptamer structure31. In comparison to Fabs selected for protein binding from minimalist libraries, our three structures together suggest that serine plays a more prominent role in RNA binding Fabs, making as many direct contacts to the RNA as tyrosine does, in addition to enabling conformational plasticity. Focusing on CDR-H3 only, the 22 direct interactions to RNA within the three Fab:RNA complexes follow the order Try (7) > Gly (6) > Arg (5) > Ser (4).
Conclusion and Outlook
In this work, we obtained a synthetic antibody Fab BRG that specifically binds a 10 nt ssRNA sequence and solved the crystal structure of the Fab:RNA complex to 2.38Å resolution. The structure and biochemical analysis unveiled an RNA hairpin conformation in the complex that does not form stably in the absence of Fab, analogous to other RNA-binding proteins and enzymes55; 56; 57. In this respect, Fabs might serve as powerful reagents for structural analysis to capture dynamic RNAs in distinct conformational states. Comparison to the structures of two other Fab:RNA complexes confirms the capacity of Tyr, Ser, and Gly biased CDRs, aided by the presence of Arg, to mediate RNA interactions and shows that the versatility of Tyr seen in molecular recognition of proteins extends to recognition of RNA32; 49. Nevertheless, analysis of the CDR-H3 paratope composition suggests that our current libraries may underrepresent Arg. However, adding more Arg could lead to less specific Fabs28, suggesting the need for systematic investigation to determine the interplay between library output and specificity. Our new structure also provides a second example in which CDR-L2 contributes significantly to the paratope surface despite the lack of diversity and conformational plasticity for this CDR in the starting library. These findings implicate both Arg content and CDR-L2 diversity as important parameters to evaluate in tailoring Fab libraries for RNA binding.
To achieve recognition of ten consecutive nucleotides, natural RNA binding proteins combine multiple RNA binding modules that individually recognize a shorter stretch of nucleotides58. For example, Pumilio family proteins consist of linked three-helix bundle domains that individually recognize a single nucleotide and require tandem linkage of these domains to achieve recognition of longer sequence elements59; 60; 61; 62. The transcription factor NusA brings together two K-homology domains to create an extended RNA-binding surface that allows the recognition of an 11 nt RNA63. The capacity of the Fab scaffold to support a sequence specific binding surface to a ten-nucleotide stretch of RNA illustrates the potential to create synthetic RNA binding proteins. These synthetic affinity reagents may serve as reagents for investigations of the biological roles of RNA binding proteins and for applications in biotechnology and synthetic biology54; 64.
Materials and Methods
RNA oligonucleotides preparation
For crystallization trials, the R-12 oligonucleotide (5’-GUAUGCAUAGGC-3’) was purchased from Dharmacon Inc. and was deprotected and desalted according to the manufacturer’s instructions. The RNA oligo was dissolved and denatured in water by heating at 90 C for 1 min. The mixture was diluted with 1x PBS buffer (pH 7.4, containing 137 mM NaCl) and used directly for forming complex with Fab.
All other oligonucleotides for biochemical assays were prepared in house by solid phase synthesis on a 1 μmol scale using an Expedite Nucleic Acid Synthesis System (8900) by following standard RNA synthesis protocols. Oligonucleotides were released from solid support with 3:1 NH4OH/EtOH, 55 C 17 hrs, desilylated with 300μL 6:3:4 N-methylpyrrolidinone/triethylamine (TEA)/TEA·3HF, 65 C 1.5h, and precipitated by n-BuOH. All oligonucleotides were 5’-radiolabeled using γ-32P-adenosine triphosphate (Perkin Elmer) and dPAGE purified.
Fab selection using phage display
YSGR Fab library was constructed at Genentech according to published procedures, and YSGRKX library was constructed in an analogous manner29. Phage selection against the branch RNA followed previous established protocols32. In order to immobilize RNA targets onto streptavidin beads, the branch RNA was chemically 3’-biotinylated on its L-chain. In the first round, 0.5 nmol of biotinylated branch RNA was immobilized on magnetic beads (Promega) and incubated with approximately 1012–13 cfu of phages separately from each library for 15 min in 0.5 mL of PTE buffer [1x PBS pH 7.4, 0.05% Tween 20, 2.5 mM EDTA, 5% glycerol], supplemented with 0.1 mg/ml BSA, 0.1 mg/ml streptavidin, and 1 U/μL murine RNase inhibitor (NEB). The solution was then removed, and the beads were washed twice with PTE buffer. In the subsequent rounds, amplified and purified phage pools were first incubated with streptavidin beads in PTE buffer for 30 min, and the supernatant was used in the selection on a KingFisher magnetic particle processor (Thermo Electron Corporation). Approximately 1010–11 cfu phages were incubated for 15 min with 50 nM biotinylated branch RNA target in 100 μL of PTE buffer, supplemented with 0.1 mg/mL BSA, 1 unit/μL murine RNase inhibitor, and 1.5 debranched RNA mix as specific competitors. The beads were then added to the solution for 15 min to capture the RNA together with the bound phages, then blocked with 50 μM biotin, washed five times with PTE buffer, and eluted in 50 μL of elution buffer (1x PBS, 5% glycerol, and 1 μg/mL biotinylated RNase A). After each round of selection, recovered phages were amplified as described65. After selection, individual clones from enriched output were grown in a 96-well format in 500 μL of 2YT broth supplemented with ampicillin and M13-KO7 helper phage. The culture supernatants were then used in phage ELISA65.
Fab expression and purification
Output clones were sequenced, and reformatted with introduction of stop codon on phagemids as described29. The desired Fab proteins were then expressed as soluble proteins according to published protocols66. Collected cell pellets were lysed, and Fab proteins were purified using the AKTAxpress FPLC purification system (Amersham) as described30. Purified protein was dialyzed into 1x PBS pH 7.4, concentrated and analyzed by 12% SDS-PAGE using Coomassie Blue R-250 staining for visualization. Aliquots of Fab samples were tested for RNase activity using the RNase-alert kit (Ambion).
Nitrocellulose filter binding assay
Approximately 10,000 cpm of 5’-radiolabeled oligonucleotide were diluted to a total of 900 μL with filter binding buffer (PBS pH 7.4, plus 0.2 mM EDTA), supplemented with 0.5 mg/mL heparin and 0.1 U/uL RNase inhibitor (Amersham). The final concentration of oligonucleotide was less than 1 nM. Filter binding assay was performed as reported previously31.
Hydroxyl radical Footprinting
Footprinting reactions contained 1 μL approximately 100,000 cpm/μL of 5'-labeled oligonucleotides, diluted into aliquots containing 9 μL 1x PBS pH 7.4 and 0.5 mg/mL heparin. 1 μL of 10x Fab (20 μM) was added, and the RNA-Fab mixture to incubated at R.T. for 30 min to allow for Fab binding. Footprinting assay was then performed as described31.
Debranching assay
Dbr1 debranching assay was performed as previously described37; 38. All experiments were performed in PBS at pH 7.4 with 1.4 nM yeast Dbr1 debranchase. Reactions in 10 μL aliquots contained approximately 20,000 cpm 5’-radiolabeled branch RNA per each lane of loading, and were allowed to incubate with Dbr1 for 0, 1, 2, 5, 10 and 20 minutes. All reactions were subsequently quenched by the addition of 5 μL of a thiourea stop solution, and then fractionated by 10% dPAGE (10% acrylamide, 0.5x TBE, 7 M urea). Gels were exposed to Phosphorimager screens, which were scanned on a Typhoon Trio imager (GE Healthcare), and images were processed by ImageQuant software. For debranching assay in presence of Fab BRG, each concentration of Fab was added into lariat RNA and allowed to incubate for 30 min at R.T. prior to debranching by Dbr1.
Secondary structure in-line probing
In-line probing assay was performed as previously reported47. Approximately 50,000 cpm per aliquot of each 5’-radiolabeled oligonucleotide was folded in 1x PBS at pH = 7.4, 37 °C for 30 min, and then treated with in-line probing buffer (50 mM Tris – HCl at pH 8.3, 20 mM MgCl2, and 100 mM KCl) for 48 hours at R.T., before fractionating with 20% denaturing PAGE for footprinting. Gels were exposed to Phosphorimager screens, which were scanned on a Typhoon Trio imager (GE Healthcare), and images were processed by ImageQuant software.
Crystallization of the Fab-oligonucleotide complex
BRG Fab-RNA complex was formed by incubating 20 nmol of the 12-mer oligonucleotide (Dharmacon) in 1x PBS buffer at pH 7.4 with 1.1 eq of BRG Fab, at R.T. for 30 min. The mixture was then loaded onto HiLoad 16/60 Superdex 200pg gel filtration column (GE) equilibrated with 1 x PBS running buffer and elution was monitored by the AKTA FPLC purification system (Amersham). All gel filtration runs were carried out at 4 °C. The Fab-RNA complex was fractionated at about 90 ml of elution, collected and concentrated to 10 mg/mL using an Amicon Ultra-15 column (10 kDa molecular weight cut-off). To decrease the number of nucleation events during crystallization trials67, sample was passed over Millipore centrifugal filter units (0.2 μm cutoff) immediately before tray setup. Mosquito liquid handling robot (TTP Labtech) was used to set up high-throughput hanging drop vapor diffusion crystallization screens using 284 conditions provided by Hampton, plus RNA crystallization screen (Sigma). High diffracting crystals were observed in one condition straightly from the screens: 1.8 M Ammonium citrate. Crystals grew to full size within 4–5 days on hanging-drop 24-well plates at room temperature. For cryoprotection, drops bearing suitable crystals were brought to 1.8M Ammonium citrate plus 20% glycerol, then flash-frozen into liquid nitrogen for diffraction.
Data Collection and Processing
X-ray diffraction and all data collection were performed at the Advanced Photon Source (APS) NE-CAT beamline 24-ID-C. All datasets were integrated and scaled using HKL2000/HKL3000 software suite68. Initial phases were obtained by molecular replacement (MR) with Fab BL3-6 minus CDR-L3, H1, H2 and H3 (PDB code: 3IVK) using Phaser on Phenix69, which located one Fab-RNA complex per asymmetric unit. The initial phases obtained from molecular replacement were sufficient to trace the RNA backbone (12 nt). Model building was completed with COOT70, and then refinement was carried out with the Phenix package71. Solvent accessible surface area and area of interaction were calculated using PDBePISA72. All figures were made in Pymol (Schrodinger).
Supplementary Material
HIGHLIGHTS.
Fabs that bind RNA have great potential as biomedical research tools.
Fab BRG binds to a 12 nt single-stranded RNA with high affinity and specificity.
The Fab BRG stabilizes a hairpin conformation of the RNA.
CDR-H3 and tyrosine play dominant roles in RNA recognition.
Fab provides a versatile scaffold for RNA binding.
Acknowledgments
The work is supported by US National Institutes of Health (NIH) grant R01 GM102489 to J.P. and R21 HG005262 to J.S., and the Defense Threat Reduction Agency grant HDTRA1-13-1-0004 to J.P. This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P41 GM103403). The Pilatus 6M detector on 24-ID-C beam line is funded by a NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Footnotes
Accession code
Coordinates for the Fab BRG-12mer RNA complex has been deposited in the Protein Data Bank with accession code: 5E08.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sidhu SS, Fellouse FA. Synthetic therapeutic antibodies. Nat Chem Biol. 2006;2:682–8. doi: 10.1038/nchembio843. [DOI] [PubMed] [Google Scholar]
- 2.Koide S. Engineering of recombinant crystallization chaperones. Curr Opin Struct Biol. 2009;19:449–57. doi: 10.1016/j.sbi.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–24. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 4.Bradbury AR, Marks JD. Antibodies from phage antibody libraries. J Immunol Methods. 2004;290:29–49. doi: 10.1016/j.jim.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Ohara R, Knappik A, Shimada K, Frisch C, Ylera F, Koga H. Antibodies for proteomic research: comparison of traditional immunization with recombinant antibody technology. Proteomics. 2006;6:2638–46. doi: 10.1002/pmic.200500579. [DOI] [PubMed] [Google Scholar]
- 6.Colwill K, Graslund S Renewable Protein Binder Working G. A roadmap to generate renewable protein binders to the human proteome. Nat Methods. 2011;8:551–8. doi: 10.1038/nmeth.1607. [DOI] [PubMed] [Google Scholar]
- 7.Taussig MJ, Stoevesandt O, Borrebaeck CA, Bradbury AR, Cahill D, Cambillau C, de Daruvar A, Dubel S, Eichler J, Frank R, Gibson TJ, Gloriam D, Gold L, Herberg FW, Hermjakob H, Hoheisel JD, Joos TO, Kallioniemi O, Koegl M, Konthur Z, Korn B, Kremmer E, Krobitsch S, Landegren U, van der Maarel S, McCafferty J, Muyldermans S, Nygren PA, Palcy S, Pluckthun A, Polic B, Przybylski M, Saviranta P, Sawyer A, Sherman DJ, Skerra A, Templin M, Ueffing M, Uhlen M. ProteomeBinders: planning a European resource of affinity reagents for analysis of the human proteome. Nat Methods. 2007;4:13–7. doi: 10.1038/nmeth0107-13. [DOI] [PubMed] [Google Scholar]
- 8.Ostankovitch M, Pyle AM. Noncoding RNAs: A story of networks and long-distance relationships. J Mol Biol. 2013;425:3577–81. doi: 10.1016/j.jmb.2013.07.032. [DOI] [PubMed] [Google Scholar]
- 9.Novikova IV, Hennelly SP, Tung CS, Sanbonmatsu KY. Rise of the RNA machines: exploring the structure of long non-coding RNAs. J Mol Biol. 2013;425:3731–46. doi: 10.1016/j.jmb.2013.02.030. [DOI] [PubMed] [Google Scholar]
- 10.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–7. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 11.Mercer TR, Mattick JS. Understanding the regulatory and transcriptional complexity of the genome through structure. Genome Res. 2013;23:1081–8. doi: 10.1101/gr.156612.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moran VA, Niland CN, Khalil AM. Co-Immunoprecipitation of long noncoding RNAs. Methods Mol Biol. 2012;925:219–28. doi: 10.1007/978-1-62703-011-3_15. [DOI] [PubMed] [Google Scholar]
- 15.Quinn JJ, Ilik IA, Qu K, Georgiev P, Chu C, Akhtar A, Chang HY. Revealing long noncoding RNA architecture and functions using domain-specific chromatin isolation by RNA purification. Nat Biotechnol. 2014;32:933–40. doi: 10.1038/nbt.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amann R, Fuchs BM. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat Rev Microbiol. 2008;6:339–48. doi: 10.1038/nrmicro1888. [DOI] [PubMed] [Google Scholar]
- 17.Engreitz J, Lander ES, Guttman M. RNA antisense purification (RAP) for mapping RNA interactions with chromatin. Methods Mol Biol. 2015;1262:183–97. doi: 10.1007/978-1-4939-2253-6_11. [DOI] [PubMed] [Google Scholar]
- 18.Eilat D, Asofsky R, Laskov R. A hybridoma from an autoimmune NZB/NZW mouse producing monoclonal antibody to ribosomal-RNA. J Immunol. 1980;124:766–8. [PubMed] [Google Scholar]
- 19.Epstein P, Lidsky M, Reddy R, Tan E, Busch H. Identification of three different anti-4S RNA sera associated with autoimmune disease. Biochem Biophys Res Commun. 1982;109:548–55. doi: 10.1016/0006-291x(82)91756-9. [DOI] [PubMed] [Google Scholar]
- 20.Pokkuluri PR, Bouthillier F, Li Y, Kuderova A, Lee J, Cygler M. Preparation, characterization and crystallization of an antibody Fab fragment that recognizes RNA. Crystal structures of native Fab and three Fab-mononucleotide complexes. J Mol Biol. 1994;243:283–97. doi: 10.1006/jmbi.1994.1654. [DOI] [PubMed] [Google Scholar]
- 21.van Venrooij WJ, Hoet R, Castrop J, Hageman B, Mattaj IW, van de Putte LB. Anti-(U1) small nuclear RNA antibodies in anti-small nuclear ribonucleoprotein sera from patients with connective tissue diseases. J Clin Invest. 1990;86:2154–60. doi: 10.1172/JCI114954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalmakoff J, Maskill WJ, Thongkrajai P, Palmer DG. Antibodies against double-stranded RNA in patients with rheumatoid arthritis, osteoarthrosis and Paget's disease of bone. Aust N Z J Med. 1981;11:173–8. doi: 10.1111/j.1445-5994.1981.tb04227.x. [DOI] [PubMed] [Google Scholar]
- 23.Ohosone Y, Matsumura M, Chiba J, Nagaoka S, Matsuoka Y, Irimajiri S, Mimori T. Anti-transfer RNA antibodies in two patients with pulmonary fibrosis, Raynaud's phenomenon and polyarthritis. Clin Rheumatol. 1998;17:144–7. doi: 10.1007/BF01452262. [DOI] [PubMed] [Google Scholar]
- 24.Gilliam AC, Steitz JA. Rare scleroderma autoantibodies to the U11 small nuclear ribonucleoprotein and to the trimethylguanosine cap of U small nuclear RNAs. Proc Natl Acad Sci U S A. 1993;90:6781–5. doi: 10.1073/pnas.90.14.6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karpala AJ, Doran TJ, Bean AG. Immune responses to dsRNA: implications for gene silencing technologies. Immunol Cell Biol. 2005;83:211–6. doi: 10.1111/j.1440-1711.2005.01331.x. [DOI] [PubMed] [Google Scholar]
- 26.Rothe C, Urlinger S, Lohning C, Prassler J, Stark Y, Jager U, Hubner B, Bardroff M, Pradel I, Boss M, Bittlingmaier R, Bataa T, Frisch C, Brocks B, Honegger A, Urban M. The human combinatorial antibody library HuCAL GOLD combines diversification of all six CDRs according to the natural immune system with a novel display method for efficient selection of high-affinity antibodies. J Mol Biol. 2008;376:1182–200. doi: 10.1016/j.jmb.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 27.Adams JJ, Sidhu SS. Synthetic antibody technologies. Curr Opin Struct Biol. 2014;24:1–9. doi: 10.1016/j.sbi.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 28.Birtalan S, Zhang Y, Fellouse FA, Shao L, Schaefer G, Sidhu SS. The intrinsic contributions of tyrosine, serine, glycine and arginine to the affinity and specificity of antibodies. J Mol Biol. 2008;377:1518–28. doi: 10.1016/j.jmb.2008.01.093. [DOI] [PubMed] [Google Scholar]
- 29.Fellouse FA, Esaki K, Birtalan S, Raptis D, Cancasci VJ, Koide A, Jhurani P, Vasser M, Wiesmann C, Kossiakoff AA, Koide S, Sidhu SS. High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J Mol Biol. 2007;373:924–40. doi: 10.1016/j.jmb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 30.Koldobskaya Y, Duguid EM, Shechner DM, Suslov NB, Ye J, Sidhu SS, Bartel DP, Koide S, Kossiakoff AA, Piccirilli JA. A portable RNA sequence whose recognition by a synthetic antibody facilitates structural determination. Nat Struct Mol Biol. 2011;18:100–6. doi: 10.1038/nsmb.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang H, Suslov NB, Li NS, Shelke SA, Evans ME, Koldobskaya Y, Rice PA, Piccirilli JA. A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore. Nat Chem Biol. 2014;10:686–91. doi: 10.1038/nchembio.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye JD, Tereshko V, Frederiksen JK, Koide A, Fellouse FA, Sidhu SS, Koide S, Kossiakoff AA, Piccirilli JA. Synthetic antibodies for specific recognition and crystallization of structured RNA. Proc Natl Acad Sci U S A. 2008;105:82–7. doi: 10.1073/pnas.0709082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coppins RL, Silverman SK. A deoxyribozyme that forms a three-helix-junction complex with its RNA substrates and has general RNA branch-forming activity. J Am Chem Soc. 2005;127:2900–7. doi: 10.1021/ja044881b. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Silverman SK. Efficient one-step synthesis of biologically related lariat RNAs by a deoxyribozyme. Angew Chem Int Ed Engl. 2005;44:5863–6. doi: 10.1002/anie.200501643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suslov NB. Dissertation. The University of Chicago; 2012. The crystal structure of the Varkud Satellite ribozyme: Implications for RNA catalysis, biology and evolution. [Google Scholar]
- 36.Ippolito GC, Schelonka RL, Zemlin M, Ivanov II, Kobayashi R, Zemlin C, Gartland GL, Nitschke L, Pelkonen J, Fujihashi K, Rajewsky K, Schroeder HW., Jr Forced usage of positively charged amino acids in immunoglobulin CDR-H3 impairs B cell development and antibody production. J Exp Med. 2006;203:1567–78. doi: 10.1084/jem.20052217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chapman KB, Boeke JD. Isolation and characterization of the gene encoding yeast debranching enzyme. Cell. 1991;65:483–92. doi: 10.1016/0092-8674(91)90466-c. [DOI] [PubMed] [Google Scholar]
- 38.Khalid MF, Damha MJ, Shuman S, Schwer B. Structure-function analysis of yeast RNA debranching enzyme (Dbr1), a manganese-dependent phosphodiesterase. Nucleic Acids Res. 2005;33:6349–60. doi: 10.1093/nar/gki934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Montemayor EJ, Katolik A, Clark NE, Taylor AB, Schuermann JP, Combs DJ, Johnsson R, Holloway SP, Stevens SW, Damha MJ, Hart PJ. Structural basis of lariat RNA recognition by the intron debranching enzyme Dbr1. Nucleic Acids Res. 2014;42:10845–55. doi: 10.1093/nar/gku725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nam K, Hudson RH, Chapman KB, Ganeshan K, Damha MJ, Boeke JD. Yeast lariat debranching enzyme. Substrate and sequence specificity. J Biol Chem. 1994;269:20613–21. [PubMed] [Google Scholar]
- 41.Jones S, Thornton JM. Principles of protein-protein interactions. Proc Natl Acad Sci U S A. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eigenbrot C, Randal M, Presta L, Carter P, Kossiakoff AA. X-ray structures of the antigen-binding domains from three variants of humanized anti-p185HER2 antibody 4D5 and comparison with molecular modeling. J Mol Biol. 1993;229:969–95. doi: 10.1006/jmbi.1993.1099. [DOI] [PubMed] [Google Scholar]
- 43.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–40. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 44.Walter AE, Turner DH, Kim J, Lyttle MH, Muller P, Mathews DH, Zuker M. Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc Natl Acad Sci U S A. 1994;91:9218–22. doi: 10.1073/pnas.91.20.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jacques JP, Susskind MM. Use of electrophoretic mobility to determine the secondary structure of a small antisense RNA. Nucleic Acids Res. 1991;19:2971–7. doi: 10.1093/nar/19.11.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Regulski EE, Breaker RR. In-line probing analysis of riboswitches. Methods Mol Biol. 2008;419:53–67. doi: 10.1007/978-1-59745-033-1_4. [DOI] [PubMed] [Google Scholar]
- 48.Thapar R, Denmon AP, Nikonowicz EP. Recognition modes of RNA tetraloops and tetraloop-like motifs by RNA-binding proteins. Wiley Interdiscip Rev RNA. 2014;5:49–67. doi: 10.1002/wrna.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koide S, Sidhu SS. The importance of being tyrosine: lessons in molecular recognition from minimalist synthetic binding proteins. ACS Chem Biol. 2009;4:325–34. doi: 10.1021/cb800314v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Persson H, Ye W, Wernimont A, Adams JJ, Koide A, Koide S, Lam R, Sidhu SS. CDR-H3 diversity is not required for antigen recognition by synthetic antibodies. J Mol Biol. 2013;425:803–11. doi: 10.1016/j.jmb.2012.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson IA, Stanfield RL. Antibody-antigen interactions: new structures and new conformational changes. Curr Opin Struct Biol. 1994;4:857–67. doi: 10.1016/0959-440x(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 52.Stanfield RL, Zemla A, Wilson IA, Rupp B. Antibody elbow angles are influenced by their light chain class. J Mol Biol. 2006;357:1566–74. doi: 10.1016/j.jmb.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 53.Barik ACN, Pilla SP, Bahadur RP. Molecular architecture of protein-RNA recognition sites. J Biomol Struct Dyn. 2015:1–14. doi: 10.1080/07391102.2015.1004652. [DOI] [PubMed] [Google Scholar]
- 54.Filipovska A, Rackham O. Designer RNA-binding proteins: New tools for manipulating the transcriptome. RNA Biol. 2011;8:978–83. doi: 10.4161/rna.8.6.17907. [DOI] [PubMed] [Google Scholar]
- 55.Hoang C, Chen J, Vizthum CA, Kandel JM, Hamilton CS, Mueller EG, Ferre-D'Amare AR. Crystal structure of pseudouridine synthase RluA: indirect sequence readout through protein-induced RNA structure. Mol Cell. 2006;24:535–45. doi: 10.1016/j.molcel.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 56.Shao Y, Richter H, Sun S, Sharma K, Urlaub H, Randau L, Li H. A Non-Stem-Loop CRISPR RNA Is Processed by Dual Binding Cas6. Structure. 2016;24:547–54. doi: 10.1016/j.str.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williamson JR. Induced fit in RNA-protein recognition. Nat Struct Biol. 2000;7:834–7. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
- 58.Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8:479–90. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zamore PD, Williamson JR, Lehmann R. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA. 1997;3:1421–33. [PMC free article] [PubMed] [Google Scholar]
- 60.Wang X, Zamore PD, Hall TM. Crystal structure of a Pumilio homology domain. Mol Cell. 2001;7:855–65. doi: 10.1016/s1097-2765(01)00229-5. [DOI] [PubMed] [Google Scholar]
- 61.Wang X, McLachlan J, Zamore PD, Hall TM. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110:501–12. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- 62.Edwards TA, Pyle SE, Wharton RP, Aggarwal AK. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell. 2001;105:281–9. doi: 10.1016/s0092-8674(01)00318-x. [DOI] [PubMed] [Google Scholar]
- 63.Beuth B, Pennell S, Arnvig KB, Martin SR, Taylor IA. Structure of a Mycobacterium tuberculosis NusA-RNA complex. EMBO J. 2005;24:3576–87. doi: 10.1038/sj.emboj.7600829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen Y, Varani G. Engineering RNA-binding proteins for biology. FEBS J. 2013;280:3734–54. doi: 10.1111/febs.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sidhu SS, Lowman HB, Cunningham BC, Wells JA. Phage display for selection of novel binding peptides. Methods Enzymol. 2000;328:333–63. doi: 10.1016/s0076-6879(00)28406-1. [DOI] [PubMed] [Google Scholar]
- 66.Paduch M, Koide A, Uysal S, Rizk SS, Koide S, Kossiakoff AA. Generating conformation-specific synthetic antibodies to trap proteins in selected functional states. Methods. 2013;60:3–14. doi: 10.1016/j.ymeth.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chayen R. Rigorous filtration for protein crystallography. Journal of Applied Crystallography. 2009;42:2. [Google Scholar]
- 68.Otwinowski ZMW. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:20. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 69.McCoy AJGK, RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:17. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 71.Afonine PVGK, RW, Adams PD. The Phenix refinement framework. CCP4 Newsletter. 2005;42:1. [Google Scholar]
- 72.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–97. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.