Abstract
Phenotypes frequently vary across and within species. The connection between specific phenotypic effects and function, however, is less understood despite being essential to our understanding of the adaptive process. Snake venoms are ideal for identifying functionally important phenotypic variation because venom variation is common, and venoms can be functionally characterized through simple assays and toxicity measurements. Previous work with the eastern diamondback rattlesnake (Crotalus adamanteus) used multivariate statistical approaches to identify six unique venom phenotypes. We functionally characterized hemolytic, gelatinase, fibrinogenolytic, and coagulant activity for all six phenotypes, as well as one additional venom, to determine if the statistically significant differences in toxin expression levels previously documented corresponded to differences in venom activity. In general, statistical differences in toxin expression predicted the identified functional differences, or lack thereof, in toxic activity, demonstrating that the statistical approach used to characterize C. adamanteus venoms was a fair representation of biologically meaningful differences. Minor differences in activity not accounted for by the statistical model may be the result of amino-acid differences and/or post-translational modifications, but overall we were able to link variation in protein expression levels to variation in function as predicted by multivariate statistical approaches.
Keywords: Venom, Toxic activities, Expression variation
1. Introduction
Natural selection can vary spatially due to genotype-by-environment interactions (Novembre and Rienzo, 2009; Whitlock and Guillaume, 2009), leading to extensive phenotypic variation (Grant and Grant, 2002; Linnen et al., 2009; Barrett et al., 2011; Margres et al., 2015a). Identifying phenotypic variation is usually straightforward, but determining the functional significance of this variation relative to the ecology and evolution of a particular lineage can often be difficult (Gibbs and Mackessy, 2009). Snake venoms provide an ideal system for identifying functionally important phenotypic variation because venom variation is common (Chippaux et al., 1991; Daltry et al., 1996; Mackessy et al., 2003; Pung et al., 2005; Mackessy, 2008; Casewell et al., 2014), although not ubiquitous (Margres et al., 2015a), and venoms can be functionally characterized through simple assays and experimental tests (Huang and Perez, 1980; Bernheimer et al., 1987; da Silva and Aird, 2001; Sánchez et al., 2003; Kini, 2005; Sánchez et al., 2005; Bello et al., 2006; Calvete et al., 2007; Núñez et al., 2009; Salazar et al., 2009; Calvete et al., 2010; Suntravat et al., 2010; Biardi and Coss, 2011; Bohlen et al., 2011; Ciscotto et al., 2011; Salazar et al., 2011).
Snake venoms are modular, polygenic traits that are critical for feeding (Biardi et al., 2000) and defense (Jansa and Voss, 2011), resulting in strong selective pressures on the genes encoding venom components at the sequence (Kordiš and Gubenšek, 2000; Lynch, 2007; Gibbs and Rossiter, 2008; Juárez et al., 2008; Rokyta et al., 2011; Margres et al., 2013; Vonk et al., 2013) and expression levels (Daltry et al., 1996; Barlow et al., 2009; Rokyta et al., 2015; Margres et al., 2016). These strong selective pressures are believed to be responsible for the extensive interspecific (Mackessy, 2008; Sanz et al., 2008; Rokyta et al., 2013; McGivern et al., 2014), intraspecific (Daltry et al., 1996; Creer et al., 2003; Gibbs et al., 2009; Boldrini-França et al., 2010; Margres et al., 2015a, 2016; Rokyta et al., 2015), and ontogenetic venom variation (Mackessy, 1988; Mackessy et al., 2003; Guércio et al., 2006; Madrigal et al., 2012; Zelanis et al., 2012; Durban et al., 2013; Margres et al., 2015b; Wray et al., 2015) because venom variation has been shown to be associated with dietary shifts (Daltry et al., 1996; Barlow et al., 2009). These spatial and temporal shifts in resource use presumably lead to shifts in phenotypic optima and should be accompanied by functional divergence (Jensen et al., 2012; Margres et al., 2015b).
Crotalus adamanteus is the largest rattlesnake species and exclusively consumes endothermic prey, including rats, squirrels, and rabbits (Klauber, 1997). Although historically native to seven states in the southeastern United States, this species has been extirpated from Louisiana, is listed as endangered in North Carolina (Palmer and Braswell, 1995), and is currently under consideration for listing as threatened under the Endangered Species Act (United States Fish and Wildlife Service, 2012). Margres et al. (2014; 2015a; 2016) used transcriptomics and proteomics to construct a genotype-phenotype map for the venom system of C. adamanteus. This map was used to identify extensive protein expression variation across seven adult populations, identifying five unique phenotypes, as well as the specific loci being differentially expressed across populations (Margres et al., 2015a). Margres et al. (2015b) identified significant expression variation across age classes and found that ontogenetic effects explained more of the toxin expression variation in C. adamanteus than geographic effects. Although these studies identified candidate functional variants, it has yet to be shown if these statistical differences in toxin expression equate to functional differences and, therefore, are biologically meaningful.
We functionally characterized four toxic activities for six of the seven adult venoms (i.e., populations) from Margres et al. (2015a) as well as a juvenile venom (Margres et al., 2015b) to determine if the statistically significant differences in toxin expression levels previously documented corresponded to differences in venom activity. These six adult populations represent all five unique phenotypes identified as well as one redundant phenotype from an island population (SAP/Ca7); this population did not possess a significantly different venom from the two mainland populations found in panhandle Florida (i.e., ANF/Ca2 in this study; Margres et al., 2015a). The inclusion of this population allowed us to determine whether venoms that were not statistically different possess similar or different functional activities, and characterizing all seven venoms allowed us to evaluate the statistical model (i.e., whether the statistical approach was too conservative, too sensitive, or a fair representation of biologically meaningful differences).
2. Materials and methods
2.1. Venoms
All assays were performed using pooled venom samples; population designations were consistent with those of Margres et al. (2015a; 2015b): Ca2 adult venom (n= 10; ANF, Apalachicola National Forest and the surrounding region east of the Apalachicola River and west of the Suwannee River, Florida), Ca2 juvenile venom (n = 13), Ca3 adult venom (n = 11; SH, Spring Hill, Florida), Ca4 adult venom (n = 3; ENP, Everglades National Park, Florida), Ca5 adult venom (n =9; LSG, Little St. George Island, Florida), Ca6 adult venom (n= 5; CAL, Caladesi Island, Florida), and Ca7 adult venom (n = 8; SAP, Sapelo Island, Georgia; Fig. 1). Because of the ontogenetic shift in venom composition (Margres et al., 2015b), specimens ≥102 cm were classified as adults and individuals <102 cm were classified as juveniles (Waldron et al., 2013), consistent with previous analyses (Margres et al., 2015a, b; 2016). All venom samples included in this study had been analyzed previously (i.e., reversed-phase high-performance liquid chromatography (RP-HPLC) and statistical analyses; Margres et al., 2015a, b, 2016). Samples were collected under the following permits: Florida Fish and Wildlife Conservation Commission (FWC) LSSC-13-00004 and LSSC-09-0399, Eglin Air Force Base 96 SFS/SFOIRP, Everglades National Park - EVER-2012-SCI-0053, Florida Department of Environmental Protection - Permit # 04101310, and St. Vincent National Wildlife Refuge - Permit # 41650-2012-08. The above procedures were approved by the Florida State University Institutional Animal Care and Use Committee (IACUC) under protocols #0924 and #1333.
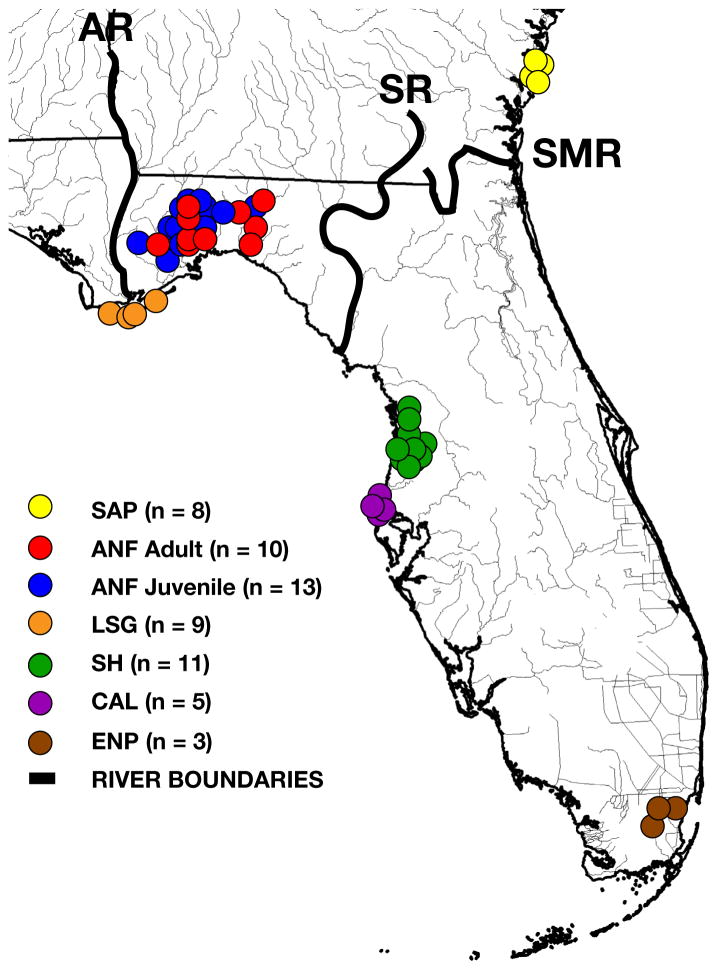
Fig. 1.
Sampling of Crotalus adamanteus for venom pools. We collected venom samples from 46 adult C. adamanteus across six populations as well as from 13 juveniles from a single population. Population designations are consistent with those of Margres et al. (2015a; 2015b): SAP (Ca7, Sapelo Island, Georgia), ANF adult and juvenile (Ca2, Apalachicola National Forest and the surrounding region east of the Apalachicola River and west of the Suwannee River, Florida), LSG (Ca5, Little St. George Island, Florida), SH (Ca3, Spring Hill, Florida), CAL (Ca6, Caladesi Island, Florida), and ENP (Ca4, Everglades National Park, Florida). AR, Apalachicola River; SMR, Saint Mary’s River; SR, Suwannee River.
2.2. Reversed-phase high-performance liquid chromatography
Reversed-phase high-performance liquid chromatography was performed on a Beckman System Gold HPLC (Beckman Coulter, Fullerton, CA) as described previously for each pooled venom as well as a reference venom sample from a single individual (Margres et al., 2015a, 2016, b; Wray et al., 2015). Briefly, 100 μg of total protein were injected onto a Jupiter C18 column, 250 × 4.6 mm (Phenomenex, Torrence, CA) using the solvent system of A =0.1% trifluoroacetic acid (TFA) in water and B = 0.075% TFA in acetonitrile. After five minutes at 5% B, a 1% per minute linear gradient of A and B was run to 25% B, followed by a 0.25% per minute gradient from 25 to 65% B at a flow rate of 1 mL per minute. Column effluent was monitored at 220 and 280 nm.
2.3. SDS-PAGE analysis
A NuPAGE 4–12% Bis-Tris SDS-PAGE gel (1.0 mm × 10 well, Invitrogen) was loaded with 25 μg of total protein for each pooled, crude venom. Briefly, 2.5 μL of sample (10 mg/mL) were combined with 11 μL of SDS buffer solution and 5 μL NuPAGE reducing agent (Invitrogen) and then heated to 70 °C for 10 min. Samples were run at 100 V for 60 min; SeeBlue Plus2 Protein Standard 1X (Invitrogen) was used as the ladder. Gels were stained and destained with Simply Blue SafeStain (Invitrogen) according to the manufacturer’s protocol. Protein identification was based on approximate molecular weights as described in Mackessy (2008).
2.4. Indirect hemolytic activity
We modified the approach of Habermann and Hardt (1972) as described by Salazar et al. (2009) to test each pooled venom for indirect hemolytic activity, a proxy for phospholipase A2 activity. Briefly, 0.3 mL of packed human erythrocytes was washed three times with 0.85% saline solution. Next, 25 mL of 0.8% agarose dissolved in phosphate-buffered saline solution (pH 8.1) were heated; 0.3 mL of fresh egg yolk diluted 1:4 with 0.85% saline solution and 0.25 mL of a 0.01 M CaCl2 solution were added to the agarose solution. Once the solution cooled to ~38 °C, the packed human erythrocytes were added. The mixture was then poured into plastic Petri dishes and allowed to solidify. After the mixture solidified, wells were prepared and filled with 25 μL of crude venom solution at two concentrations: 1 mg/mL (25 μg venom) and 0.4 mg/mL (10 μg venom). Three replicates for the high venom concentration and two replicates for the low venom concentration were run with Crotalus atrox venom serving as the positive control. All plates were incubated at 37 °C for 5 h and the diameters of the hemolytic haloes were measured. To test for interpopulation variation in hemolytic activity, we ran individual ANOVAs on the halo diameters for each venom concentration set.
2.5. Gelatinase activity
We modified the approach of Huang and Perez (1980) as described by Sánchez et al. (2003) to test each pooled venom for gelatinase activity. Because gelatin is a denatured form of collagen, this assay measures proteolytic (i.e., proteinase) activity; metalloproteinases are the main enzymes responsible for extracellular matrix protein degradation. We tested 20 μL of crude venom solution at two concentrations in duplicate: 1 mg/mL (20 μg venom) and 2 mg/mL (40 μg venom). Twenty μL of crude venom solution were placed on a Kodak X-OMAT scientific imaging film with a gelatin coating. The film was incubated at 37 °C for 4 h and then washed with distilled water. Hydrolysis of the gelatin coat (i.e., a clearing of the gelatin on the film) was recorded as a positive result and indicated proteolytic activity; lack of hydrolysis was recorded as a negative result.
2.6. Fibrinogenolytic activity
We tested each pooled venom for fibrinogenolytic activity, a proxy for proteinase activity. Briefly, 0.005 g of fibrinogen (without plasminogen; HYPHEN BioMed) was dissolved in 1mL of water and incubated at 37 °C for 30 min. Following incubation, 20 μL of the fibrinogen solution were separately added to 10 μL of each C. adamanteus venom (1 mg/mL, 10 μg protein), 10 μL of 0.85% saline solution (negative control), and 10 μL of C. atrox venom (1 mg/mL, 10 μg protein; positive control). All samples were incubated at 37 °C for 24 h. Next, 11 μL of SDS buffer solution and 5 μL NuPAGE reducing agent (Invitrogen) were added and each sample was heated to 70 °C for 10 min. Fifteen μL of the sample mixture were run on a NuPAGE 4–12% Bis-Tris SDS-PAGE gel (1.0 mm × 10 well, Invitrogen) at 200 V for 35 min; SeeBlue Plus2 Protein Standard 1X (Invitrogen) was used as the ladder. Gels were stained and destained with Simply Blue SafeStain (Invitrogen) according to the manufacturer’s protocol. Absence of the α, β, and/or γ chain(s) indicated hydrolysis of that chain and was recorded as a positive result (i.e., fibrinogenolytic activity).
2.7. Coagulation assay
We followed the approach of Suntravat et al. (2010) and Sánchez et al. (2010) to test the effects of each pooled venom on the coagulation cascade. Pooled normal citrated plasma was prepared by drawing a blood sample from M.J.M; 40 mL of blood was collected by gravity into a 50 mL plastic test tube containing 4 mL of 3.8% sodium citrate to prevent coagulation. The citrated blood was incubated at 37 °C during the assays. The procoagulant and anticoagulant activities of all venoms were measured using the Sonoclot analyzer, which measures the activated clotting time (ACT), the clot rate (CR), and platelet function (PF) of whole blood (von Kaulla et al., 1975). The ACT is the time that the plasma remains a liquid (i.e., time in which fibrin formation begins), the CR is the rate of fibrin polymerization and is defined as the change in clot signal over time (e.g., kinetic measure of fibrin formation and clot development), and the PF is the timing and quality of clot retraction. Briefly, a red cuvette from the activated Sonoclot kit (i.e., with glass beads) was placed into the cuvette holder to maintain a temperature of 37 °C. A pre-warmed 10 μL sample (37 °C) of 0.3 M CaCl2 was added to one side of the cuvette, a 10 μL sample of each venom at a concentration of 100 ng/μL (1 μg protein) was added to the other side of the cuvette, and a constant volume of 330 μL of whole blood was also added. The ACT, CR, and PF were measured; 10 μL of 0.85% saline solution replaced the venom sample in the positive control. Each venom was run in triplicate with a control run between different venoms (e.g., three adult ANF replicates were run, followed by a control, and then three juvenile ANF replicates, etc.). To determine if any of the venoms differed from the control and significantly affected coagulation, we ran individual ANOVAs on each of the three variables measured (i.e., ACT, CR, and PF). If these results were significant for a particular variable, we conducted pairwise t-tests versus the control for each sample/variable combination and used a Bonferonni correction to account for multiple tests. To test for interpopulation variation in coagulation activity, we ran individual ANOVAs on each of the three variables but only included the venom samples (i.e., excluded the control).
3. Results
3.1. SDS-PAGE analysis
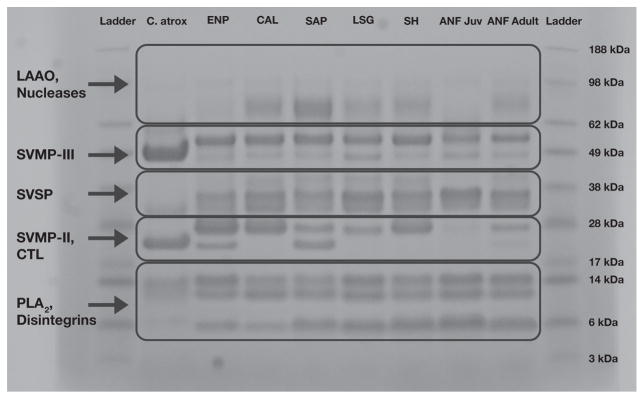
SDS-PAGE analysis demonstrated variability across venoms in the abundance of particular protein classes, consistent with previous work (Fig. 2; Margres et al., 2015a, b; 2016). Snake venom type III metalloproteinases were prominent in all venoms. Snake venom type II metalloproteinases, however, exhibited variation across venoms; these toxins were down-regulated in juveniles and an additional band was present in the ENP and SAP populations. Phospholipases A2 and snake venom serine proteinases were prominent in all venoms with relatively conserved abundances.
Fig. 2.
Comparison of the seven pooled Crotalus adamanteus venoms used in the toxicity assays. Protein abundances were largely similar, but snake venom type II metalloproteinases (SVMP-II) were down-regulated in juveniles and an additional band was present in the ENP and SAP populations. Protein identification was based on approximate molecular weights and venom labels are consistent with Fig. 1. Crotalus atrox venom was used as a positive control. Abbreviations are as follows: LAAO, L-amino acid oxidase; SVMP-III, Snake venom type III metalloproteinase; SVSP, Snake venom serine proteinase; SVMP-II, Snake venom type II metalloproteinase; CTL, C-type lectin; PLA2, Phospholipase A2.
3.2. Hemolytic activity
To test for interpopulation variation in hemolytic (i.e., phospholipase A2) activity, we ran individual ANOVAs on each venom concentration set. All venoms demonstrated hemolytic activity, and no significant differences were detected among venoms at either concentration (P25μg =0.079; P10μg =0.681; Table 1).
Table 1.
Hemolytic and proteolytic activities of Crotalus adamanteus venoms. All venoms possessed hemolytic activity, but these activities were not significantly different. Venoms differed in their gelatinase activity; the SAP and ENP populations were the only venoms to test positive for gelatinase activity at the lower dose, and all venoms except for the juvenile venom exhibited gelatinase activity at the higher dose. Hemolytic activity was measured in millimeters; the mean value and the standard deviation are listed. Gelatinase activity is indicated as present (+) or absent (−); each concentration was tested in duplicate.
| Population | Hemolytic (25 μg) | Hemolytic (10 μg) | Gelatinase (20 μg) | Gelatinase (40 μg) |
|---|---|---|---|---|
| ANF adult | 16.67 ± 0.58 | 14.50 ± 0.71 | −/− | +/+ |
| ANF juvenile | 16.67 ± 0.58 | 13.50 ± 0.71 | −/− | −/− |
| LSG adult | 17.33 ± 0.58 | 14.50 ± 0.71 | −/− | +/+ |
| SH adult | 15.33 ± 1.53 | 14.00 ± 1.41 | −/− | +/+ |
| SAP adult | 15.00 ± 1.00 | 13.00 ± 1.41 | +/+ | +/+ |
| ENP adult | 16.33 ± 0.58 | 15.00 ± 1.41 | +/+ | +/+ |
| CAL adult | 15.67 ± 1.15 | 14.00 ± 1.41 | −/− | +/+ |
| C. atrox (+control) | 17.33 ± 0.58 | 16.00 ± 0.00 | +/+ | +/+ |
| Negative control | NA | NA | −/− | −/− |
3.3. Gelatinase activity
All venoms, except the ANF juvenile population, demonstrated gelatinase/proteolytic activity at the highest venom concentration tested. Only the SAP and ENP populations, however, tested positive for gelatinase activity at the lower venom concentration. The level of gelatinase activity in descending order was as follows: SAP = ENP > ANF Adult = LSG = SH= CAL > ANF juvenile (Table 1).
3.4. Fibrinogenolytic activity
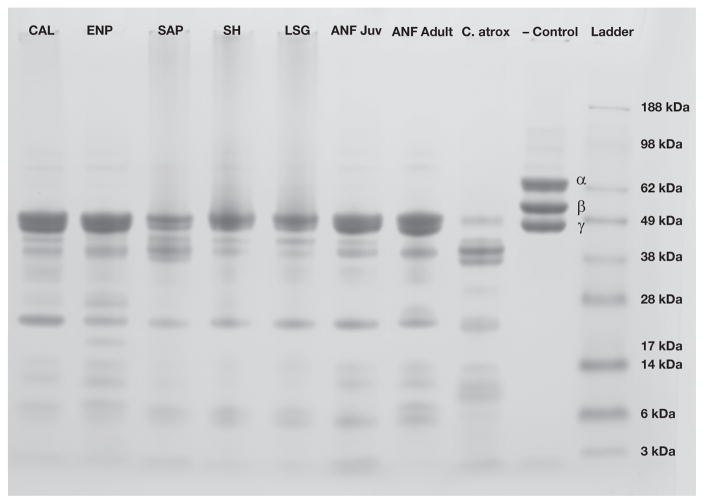
All C. adamanteus venoms hydrolyzed the α and β chain fibrinogen degradation products; the SAP, SH and LSG populations also had minor effects on the γ chain (Fig. 3).
Fig. 3.
The fibrinogenolytic assay identified significant proteinase activity in the venoms of Crotalus adamanteus. All C. adamanteus venoms hydrolyzed both α and β chain fibrinogen degradation products; the SAP, SH and LSG populations had minor effects on the γ chain. α, β, and γ chain fibrinogen degradation products are present and labeled in the negative control; absence of a particular chain indicates hydrolysis. Crotalus atrox venom was used as a positive control.
3.5. Coagulation assay
To determine if any of the venoms differed from the control and significantly affected coagulation, we ran individual ANOVAs on each of the three variables measured and all were significant (PACT = 0.029; PCR<0.001; PPF =0.034), indicating that at least one venom for all three variables significantly differed from the control. To determine which particular venoms significantly differed from the control, we conducted pairwise t-tests versus the control for each sample/variable combination and used a Bonferonni correction to account for multiple tests within each variable (i.e., α= 0.007). For the ACT, only the ANF juvenile venom was significantly different from the control (P =0.002) and demonstrated anticoagulant activity. For CR, five of the seven venoms were significantly different from the control (PANFAdult < 0.001; PANFJuvenile < 0.001; PLSG < 0.001; PSH= 0.004; PCAL =0.005) and demonstrated anticoagulant activity; only the SAP and ENP populations were not significantly different (PSAP = 0.027; PENP =0.780). For PF, only the ANF adult venom was significantly different from the control (P = 0.001) and affected the quality of clot retraction (Table 2). To test for interpopulation variation in coagulation activity, we ran individual ANOVAs on each of the three variables, excluding the control, and only the CR analysis was significant (PACT =0.097; PCR= 0.007; PPF= 0.139). An additional ANOVA limited to the five venoms that significantly differed from the CR control, however, was not significant (P = 0.071), indicating an absence of interpopulation variation among the five venoms that significantly affected the CR (i.e., the only significant variation in CR was the presence/absence of activity).
Table 2.
Coagulant properties of Crotalus adamanteus venoms. The procoagulant and anticoagulant activities of all venoms were measured using the Sonoclot analyzer; the activated clotting time is the time that plasma remains a liquid (i.e., time in which fibrin formation begins), the clot rate is defined as the change in clot signal over time (e.g., kinetic measure of fibrin formation and clot development), and platelet function is the timing and quality of clot retraction. At least one venom significantly differed from the control for each variable; significant values are indicated by an *.
| Population | Activated clotting time (s) | Clot rate | Platelet function |
|---|---|---|---|
| ANF adult | 139.67 ± 12.71 | 21.33 ± 2.87* | 2.37 ± 0.53* |
| ANF juvenile | 144.67 ± 6.13* | 21.00 ± 1.41* | 3.50 ± 0.45 |
| LSG adult | 134.67 ± 4.19 | 22.67 ± 0.47* | 2.83 ± 0.70 |
| SH adult | 123.00 ± 13.88 | 24.00 ± 4.97* | 3.00 ± 0.93 |
| SAP adult | 137.33 ± 7.41 | 29.33 ± 2.36 | 3.23 ± 0.82 |
| ENP adult | 128.67 ± 1.25 | 32.33 ± 6.18 | 3.63 ± 0.38 |
| CAL adult | 134.00 ± 2.16 | 28.00 ± 2.16* | 3.87 ± 0.41 |
| Control | 130.71 ± 2.37 | 33.14 ± 1.55 | 3.59 ± 0.08 |
4. Discussion
4.1. Conserved phospholipase A2 expression leads to conserved indirect hemolytic activity
Phospholipase A2 enzymes hydrolyze glycerophospholipids, releasing lysophospholipids and free fatty acids. These enzymes are known to cause a variety of pharmacological effects, including presynaptic and postsynaptic neurotoxicity, myotoxicity, and cardiotoxicity among others (Kini, 2003; Doley et al., 2010). This protein family was identified as the most abundant toxin family in the venom proteome of a sub-adult ANF female C. adamanteus (Margres et al., 2014). Only a single phospholipase A2 toxin was identified in the venom, and this toxin was the only protein present in RP-HPLC peak 10 (Fig. 4; Margres et al., 2016). This protein peak was the second most conserved peak across adult populations (Fig. 5; Margres et al., 2015a) and the fourth most conserved peak across age classes (Margres et al., 2015b), consistent with our SDS-PAGE results (Fig. 2). We, therefore, expected to see significant indirect hemolytic activity across all venoms, but did not expect significant variation in hemolytic activities across populations or age classes, consistent with our results. All venoms possessed hemolytic activity, but none demonstrated significant differences in this activity. Our findings suggest that phospholipase A2 activity in C. adamanteus venoms was dependent on protein expression, and nonsynonymous substitutions or post-translational modifications were either absent or did not significantly alter the function of the enzyme, at least as measured by this assay (e.g., neurotoxicity and non-toxic, chaperone-like activity, as seen in the subunits of crotoxin, would not be detected in this assay; Calvete, 2011).
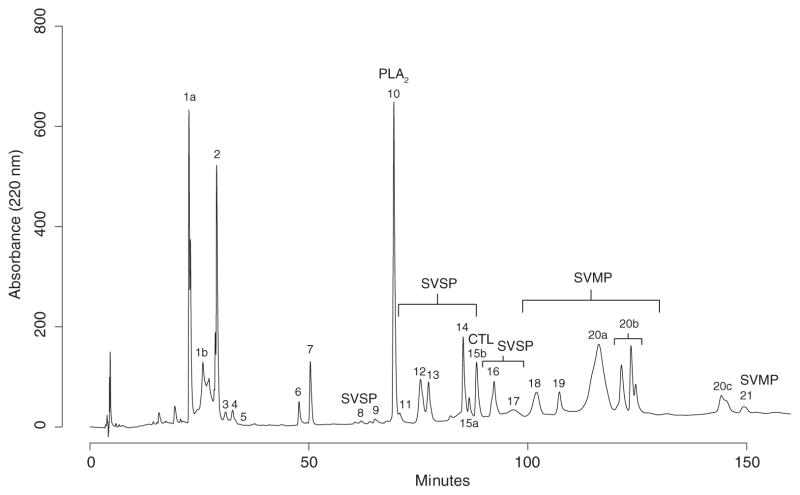
Fig. 4.
Reversed-phase high-performance liquid chromatography profile of a representative Crotalus adamanteus venom sample. The 25 major peaks identified by Margres et al. (2015a) are labeled. The toxin families of interest to the activities characterized in this study are also labeled. Abbreviations are as follows: CTL, C-type lectin; PLA2, Phospholipase A2; SVMP, Snake venom metalloproteinase; SVSP, Snake venom serine proteinase.
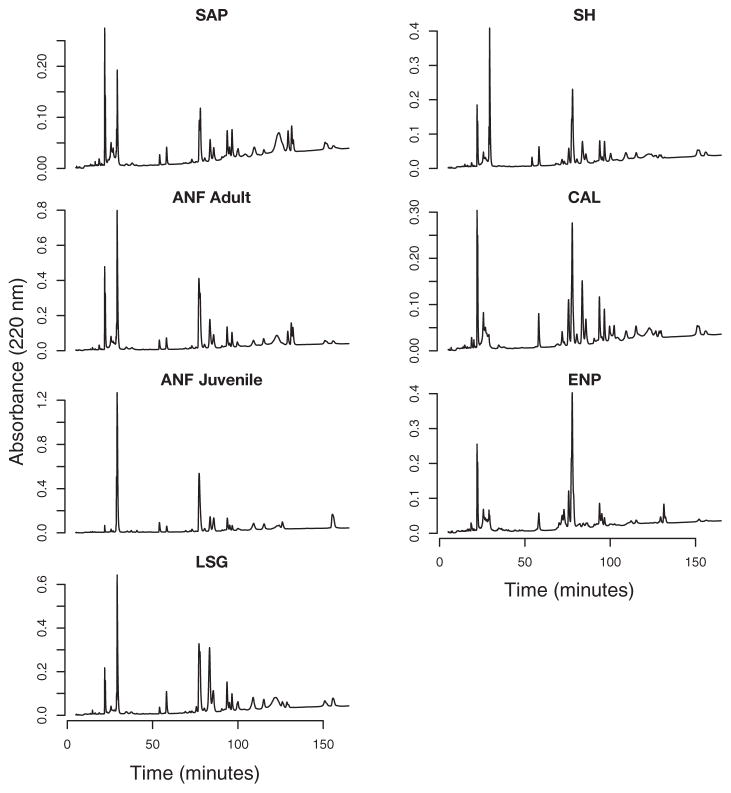
Fig. 5.
Reversed-phase high-performance liquid chromatography profiles of all seven pooled Crotalus adamanteus venom samples. Venom labels are consistent with Fig. 1.
4.2. Metalloproteinase expression differentiation predicts variability in gelatinase activity
Snake-venom metalloproteinases are often associated with extensive hemorrhaging but can also be myonecrotic, fibrinolytic, and can result in the degradation of the extracellular matrix (Fox and Serrano, 2010). This protein family was identified as the fourth most abundant toxin family in the venom proteome of a subadult ANF female C. adamanteus (Margres et al., 2014) and was one of the more diverse toxin groups identified (Rokyta et al., 2012; Margres et al., 2014). Five snake-venom metalloproteinases were identified in five RP-HPLC peaks: 18, 19, 20a, 20b, and 21 (Fig. 4; Margres et al., 2016). Peaks 20b and 21 explained the second and third most variation of any RP-HPLC peak across age classes, with peak 20b (snake venom type II metalloproteinase-2) being expressed at much higher levels in adults and peak 21 (snake venom type II metalloproteinase-1) being much more highly expressed in juveniles (Fig. 5; Margres et al., 2015b). Overall, juvenile venoms expressed the majority of metalloproteinases at much lower levels than adults (Margres et al., 2015b). We, therefore, expected that adult venoms would display significantly more activity than the juvenile venom tested, consistent with our results; the juvenile venom was the only venom to lack activity at the higher venom concentration tested (Table 1), and our SDS-PAGE analysis revealed that snake venom type II metalloproteinases were expressed at much lower levels in juveniles relative to adults (Fig. 2).
Among adult venoms, all populations possessed activity at the higher concentration tested, but only the SAP and ENP venoms possessed activity at the lower venom concentration (Table 1). These two venoms were the only venoms to possess an additional electrophoretic band with a molecular weight consistent with snake venom type II metalloproteinases (Fig. 2), suggesting that these toxins were responsible for the heightened gelatinase activity of these venoms.
Snake venom type II metalloproteinase-2 (peak 20b), the metalloproteinase present in adults and mostly absent in juveniles, was most highly expressed among adults by the ENP population (Margres et al., 2015a), suggesting that the expression level of this particular toxin may be responsible for the differences in metalloproteinase activity and represent the additional band identified in our SDS-PAGE analysis. The SAP population, which also possessed this band in the SDS-PAGE gel, did not possess a venom significantly different than that of the ANF population; these two populations, along with samples from the western Florida panhandle, were combined into a single population based on a shared venom phenotype (Margres et al., 2015a). Snake venom type II metalloproteinase-2 was also highly expressed in this combined group (Margres et al., 2015a), but SAP venom demonstrated activity at the lower venom concentration and possessed the additional electrophoretic band, and ANF venom did not. Because venom is a polygenic, complex phenotype, multivariate statistical approaches (i.e., MANOVA) are the most appropriate tests for identifying significant variation. Multivariate statistical approaches, however, compare vectors of means, and extensive variation at a single peak may not necessarily result in a significant outcome if the majority of other proteins are expressed at similar levels across populations.
To determine if the statistical model used by Margres et al. (2015a; 2015b; 2016) failed to detect significant metalloproteinase expression variation because of conserved expression patterns at the majority of other loci, we compared the mean centered logratio expression levels for the five RP-HPLC peaks containing metalloproteinases across the SAP and ANF venoms to identify the most variable (difference >2) and conserved (difference <1) loci as previously described (Margres et al., 2015a, b). Expression of peaks 18, 19, 20a, and 20b (i.e., snake venom type II metalloproteinase-2) was conserved, particularly for peak 20b (difference = 0.36), and no loci had a difference >2. We know from the SDS-PAGE analysis, however, that the SAP population possessed more snake venom type II metalloproteinases than the ANF adult population (Fig. 2). This lack of significant variation based on the RP-HPLC results suggested that other proteins may have co-eluted with metalloproteinases, and expression variation in these others proteins may have obscured metalloproteinase expression variation in the RP-HPLC analyses.
4.3. Minor differences in fibrinogenolytic activity not predicted by expression levels
Snake-venom serine proteinases are known to affect a diversity of substrates that are involved in the coagulation, fibrinolytic, fibrinogenolytic, and kallikrein-kinin systems, as well as affecting platelets, resulting in an imbalance of the hemostatic system of prey (Serrano and Maroun, 2005). This protein family was identified as the second most abundant toxin family in the venom proteome of a sub-adult ANF female C. adamanteus (Margres et al., 2014) and was one of the more diverse toxin groups identified (Rokyta et al., 2012; Margres et al., 2014). Four serine proteinases were identified in eight RP-HPLC peaks: 8, 11, 12, 13, 14, 15a, 16 and 17 (Fig. 4; Margres et al., 2016). Of these eight peaks, only peaks 12 (snake venom serine proteinase 4) and 17 (snake venom serine proteinases 4 and 7) exhibited significant variation across adult venoms, with peak 12 being down-regulated in ENP venoms and peak 17 being down-regulated in LSG venoms (Fig. 5; Margres et al., 2015a); no significant variation in serine proteinase expression was found across age classes (Margres et al., 2015b), and our SDS-PAGE analysis demonstrated that these toxins were abundant in each venom but did not identify any obvious differences in serine proteinase abundance (Fig. 2). We, therefore, expected to see significant fibrinogenolytic activity across all venoms and, if variation in activity was identified, it would be present as reduced activity in LSG and/or ENP if the expression of snake venom serine proteinases 4 and 7 affected fibrinogenolytic activity. We found that all C. adamanteus venoms hydrolyzed the α and β chain fibrinogen degradation products, consistent with our expectations, but the SAP, SH and LSG populations also had minor effects on the γ chain (Fig. 3). Snake venom serine proteinase expression was mostly conserved, with the only significant variation identified being the down-regulation of particular loci in the LSG and ENP populations. The down-regulation of snake venom serine proteinases 4 and 7 could not explain the additional, albeit minor, activity detected in the LSG population, and the SAP and SH populations had serine proteinase expression profiles very similar to all other populations. These small differences in activity may have been the result of amino-acid differences and/or post-translational modifications (Garcia et al., 2004; Casewell et al., 2014), but could also be the result of population-specific toxins that were not detected in the original transcriptome and, therefore, excluded from the genotype-phenotype map. As stated above, the original transcriptome for this species was that of a sub-adult ANF female (Margres et al., 2014). The SAP, SH, and LSG populations may possess toxins not found in the ANF population.
Fibrinogenolytic toxins are often common in snake venoms and directly affect fibrinogen, but can do so in various ways. These toxins may split fibrinogen to fibrin and the resulting degradation products, or may result in incomplete splitting, producing a form of fibrinogen that is no longer effective (White, 2005). A fibrinogenolytic thrombin-like toxin, Crotalase, had previously been identified in the venom of C. adamanteus; this toxin belongs to the venombin A group, preferentially releasing the α chain with β chain release being much slower (Markland, 1976, 1988; Henschen-Edman et al., 1999). The results of this assay most likely demonstrate Crotalase activity, or that of a closely-related homolog, in all seven venoms analyzed, but it should be noted that snake venom metalloproteinases can also possess fibrinogenolytic activity (Matsui et al., 2000).
4.4. Significant anticoagulant activity in Crotalus adamanteus venoms
Metalloproteinases (Fox and Serrano, 2005), serine proteinases (Markland,1988), C-type lectins (Jansa and Voss, 2011), disintegrins (Calvete et al., 2005), and other toxic molecules can affect hemostasis (Mackessy, 2008), the process of clot formation through the coagulation cascade. Viperid venoms have been shown to possess pro-and anticoagulant properties, and these properties can vary across individuals within a species; Suntravat et al. (2010) used 10 ng of crude venom for multiple C. adamanteus from Florida and did not detect any coagulant activity. We used 1 μg of crude venom and identified significant anticoagulant activity (i.e., extended ACT and reduced CR; Suntravat et al., 2010) in some, but not all, of the venoms tested, indicating significant intraspecific variation in anticoagulant activity within C. adamanteus.
The ANF juvenile venom was the only venom that significantly extended the ACT and demonstrated anticoagulant activity for this measure. Ontogenetic expression variation is well-documented in snake venoms (Mackessy, 1988; Mackessy et al., 2003; Guércio et al., 2006; Madrigal et al., 2012; Zelanis et al., 2012; Durban et al., 2013; Margres et al., 2015b; Wray et al., 2015). Juvenile venoms, for the most part, are often less complex versions of the adult phenotype (Mackessy, 1988; Margres et al., 2015b); most toxins found in juveniles are found in adults, but not all toxins found in adults are found in juveniles (Fig. 2; but see Durban et al., 2013). This appears to be the case in C. adamanteus (Margres et al., 2015b), and this juvenile-specific toxic activity may reflect the compositional nature of the venom phenotype. Because the assay was standardized by protein amounts, and juvenile venoms are simpler than adult venoms (Margres et al., 2015b), 1 μg of juvenile venom may contain more anticoagulant toxins than 1 μg of adult venom because several adult toxins are down-regulated or absent from juvenile venom, changing the composition of that 1 μg. For example, if juvenile venoms contained 20 toxins and five of these toxins possessed anticoagulant activity, and adult venoms contained 30 toxins and six of these toxins possessed anticoagulant activity, a 1 μg juvenile sample would contain a larger percentage of anticoagulant toxins than a 1 μg adult sample (25% and 20%, respectively). This age-specific difference in toxic activity could also indicate that ontogenetic variation in toxin expression may not be unidirectional, and that juvenile-specific toxins may affect the coagulation cascade in ways that adult-specific toxins do not. The specific toxic molecules responsible for this age-specific effect, however, was unclear because a number of toxin families can affect the coagulation cascade (e.g., C-type lectins and phospholipases A2). C-type lectins can possess significant anticoagulant activity, but these toxins were much more abundant in adults (Margres et al., 2015b), although variation in C-type lectin expression may not be apparent when using RP-HPLC to quantify protein abundances (Eichberg et al., 2015). C-type lectins and snake venom type II metalloproteinases have a similar molecular weight, making it difficult to differentiate these toxins in our SDS-PAGE analysis (Fig. 2). The differences identified in this region, however, were consistent with metalloproteinase activity/the results of the gelatinase assay rather than the coagulation assay.
Five of the seven venoms significantly reduced the CR and demonstrated anticoagulant activity for this measure; only the SAP and ENP populations did not reduce the CR. Without knowing the specific toxins responsible for this anticoagulant activity, interpreting this result was difficult. The SAP and ENP venoms were the only venoms to possess gelatinase activity at the lower venom concentration tested, suggesting that there may be an inverse relationship between gelatinase and anticoagulant activities. C-type lectins can possess significant anticoagulant activity; peak 15b contained two C-type lectin toxins (Margres et al., 2016), and this peak was least abundant in the ENP population (Margres et al., 2015a). As stated earlier, these differences in activity could also be the result of amino-acid differences and/or post-translational modifications.
The ANF adult venom was the only venom to significantly affect PF relative to the control (although it should be noted that the PF value for this venom (2.37 ± 0.53) is considered within “normal” range); no obvious expression differences appeared related to this result. The ANF population possessed different anticoagulant activities unique to each age class (i.e., ANF juvenile venom was the only venom that significantly extended the ACT, and ANF adult venom was the only venom to significantly affect PF), demonstrating that ontogenetic variation in protein expression can substantially affect venom activity. These two venoms were the only venoms to demonstrate significant anticoagulant activity in more than one measure, suggesting that anticoagulant activity may be a more important aspect of prey incapacitation in this population relative to the other six.
5. Conclusion
We investigated the functional consequences of protein expression variation and found that, overall, statistical differences in toxin expression equated to functional differences in toxic activity. Demonstrating that significantly different phenotypes at a statistical level reflect meaningful ecological/evolutionary differences can often be difficult (Gibbs and Mackessy, 2009), but connecting phenotypic variation to corresponding functional differences is necessary for understanding the dynamics of the adaptive process. This study demonstrates that the unique venom phenotypes identified by Margres et al. (2015a; 2015b; 2016) do possess unique activities, and that the statistical approach used in these studies and others (Wray et al., 2015) was a fair representation of biologically meaningful differences. Minor differences in activity not accounted for by the statistical model may be the result of amino-acid differences and/or post-translational modifications (Garcia et al., 2004; Casewell et al., 2014) unless very minor, nonsignificant changes in expression level result in detectable functional differences. Given the general consistency of activity and protein expression levels identified in this study, we suggest that the minor variations in activity are the result of amino-acid differences and/or post-translational modifications; work is currently underway to identify population-specific sequence variants. Overall, we linked variation in phenotype (i.e., protein expression levels) to variation in function and provided proof-of-concept for the statistical approach described by Margres et al. (2015a; 2015b; 2016).
Acknowledgments
This work was supported by the National Science Foundation (DEB 1145987 to D.R.R.), the Robert B. Short Scholarship in Zoology from the Department of Biological Science at Florida State University (to M.J.M.), the National Institutes of Health/ORIP (5P40OD010960 to E.E.S), and the Robert A. Welch Foundation (AC-0006 to TAMUK Department of Chemistry). The authors thank Karalyn Aronow, Richard Bartlett, Jeffrey Fobb, Nathanael Herrera, Pierson Hill, Chip Leavine, Charles Looney, Jacob Loyacano, John Malone, Mark S. Margres, Jim Mendenhall, Moses Michelson, Flavio Morrissiey, Joe Pfaller, Michael Rochford, Nathan Schuessler, Jordan Sirosky, and Ken Wray for help in acquiring venom samples. The authors thank Megan Lamb and Rebecca Bernard with the Florida DEP and Apalachicola River NERR, Dorset Hurley and Doug Samson with the Sapelo Island NERR, Brett Williams and the Department of the Air Force, and Carl Calhoun and Rick Coosey at Caladesi Island State Park for access to field sites. The authors thank James McGivern and Dragana Sanader for assistance in the laboratory.
Footnotes
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Ethical statement
All techniques involving vertebrate animals were approved by the Florida State University Institutional Animal Care and Use Committee (IACUC) under protocols #0924 and #1333.
Transparency document
Transparency document related to this article can be found online at http://dx.doi.org/10.1016/j.toxicon.2016.05.005.
References
- United States Fish & Wildlife Service. Endangered and threatened wildlife and plants: 90-day finding on a petition to list the eastern diamondback rattlesnake as threatened. Fed Reg. 2012;77:27403–27411. [Google Scholar]
- Barlow A, Pook CE, Harrison RA, Wüster W. Coevolution of diet and prey-specific venom activity supports the role of selection in snake venom evolution. Proc R Soc Lond B Biol Sci. 2009;276(1666):2443–2449. doi: 10.1098/rspb.2009.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RDH, Paccard A, Healy TM, Bergek S, Schulte PM, Schluter D, Rogers SM. Rapid evolution of cold tolerance in stickleback. Proc R Soc Lond B Biol Sci. 2011;278:233–238. doi: 10.1098/rspb.2010.0923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello CA, Hermogenes ALN, Magalhaes A, Veiga SS, Gremski LH, Richardson M, Sanchez EF. Isolation and biochemical characterization of a fibrinolytic proteinase from Bothrops leucurus (white-tailed jararaca) snake venom. Biochimie. 2006;88:189–200. doi: 10.1016/j.biochi.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Bernheimer A, Linder R, Weinstein S, Kim KS. Isolation and characterization of a phospholipase B from venom of collett’s snake, pseudechis colletti. Toxicon. 1987;25(5):547–554. doi: 10.1016/0041-0101(87)90290-x. [DOI] [PubMed] [Google Scholar]
- Biardi J, Coss R. Rock squirrel (spermophilus variegatus) blood sera affects proteolytic and ehomlytic activities of rattlesnake venoms. Toxicon. 2011;57:323–331. doi: 10.1016/j.toxicon.2010.12.011. [DOI] [PubMed] [Google Scholar]
- Biardi JE, Coss RG, Smith DG. California ground squirrel (Spermophilus beecheyi) blood sera inhibits Crotalid venom proteolytic activity. Toxicon. 2000;38:713–721. doi: 10.1016/s0041-0101(99)00179-8. [DOI] [PubMed] [Google Scholar]
- Bohlen C, Chesler A, Sharif-Naeini R, Medzihradszky K, Zhou S, King D, Sanchez E, Burlingame A, Basbaum A, Julius D. A heteromeric texas coral snake toxin targets acid-sensing ion channels to produce pain. Nature. 2011;479(7373):410–414. doi: 10.1038/nature10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldrini-França J, Corrêa-Netto C, Silva MMS, Rodrigues RS, Torre PDL, Pérez A, Soares AM, Zingali RB, Nogueira RA, Rodrigues VM, Sanz L, Calvete JJ. Snake venomics and antivenomics of Crotalus durissus subspecies from Brazil: assessment of geographic variation and its implication on snakebite management. J Proteom. 2010;73:1758–1776. doi: 10.1016/j.jprot.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Calvete JJ. Proteomics in venom research: a focus on PLA2 molecules. Acta Chim Slov. 2011;58:629–637. [PubMed] [Google Scholar]
- Calvete JJ, Marcinkiewicz C, Monleón D, Esteve V, Celda B, Juárez P, Sanz L. Snake venom disintegrins: evolution of structure and function. Toxicon. 2005;45:1063–1074. doi: 10.1016/j.toxicon.2005.02.024. [DOI] [PubMed] [Google Scholar]
- Calvete JJ, Marcinkiewicz C, Sanz L. Snake venomics of Bitis gabonica gabonica. Protein family composition, subunit organization of venom toxins, and characterization of dimeric disintegrins bitisgabonin-1 and bitisgabonin-2. J Proteome Res. 2007;6:326–336. doi: 10.1021/pr060494k. [DOI] [PubMed] [Google Scholar]
- Calvete JJ, Sanz L, Cid P, de la Torre P, Flores-Díaz M, Santos MCD, Borges A, Bremo A, Angulo Y, Lomonte B, Alape-Girón A, Gutiérrez JM. Snake venomics of the Central American Rattlesnake Crotalus simus and the south american Crotalus durissus complex points to neurotoxicity as an adaptive paedomorphic trend along Crotalus dispersal in South America. J Proteome Res. 2010;9:528–544. doi: 10.1021/pr9008749. [DOI] [PubMed] [Google Scholar]
- Casewell N, Wagstaff S, Wuster W, Cook D, Bolton F, King S, Pia D, Sanz L, Calvete J, Harrison R. Medically important differences in snake venom composition are dictated by distinct postgenomic mechanisms. P Natl Acad Sci U S A. 2014;111(25):9205–9210. doi: 10.1073/pnas.1405484111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chippaux J, Williams V, White J. Snake venom variability: methods of study, results and interpretation. Toxicon. 1991;29:1279–1303. doi: 10.1016/0041-0101(91)90116-9. [DOI] [PubMed] [Google Scholar]
- Ciscotto P, Rates B, Silva D, Richardson M, Silva L, Andrade H, Donato M, Cotta G, Maria W, Rodrigues R, Sanchez E, De Lima M, Pimenta A. Venomic analysis and evaluation of antivenom cross-reactivity of south american Micrurus species. J Proteom. 2011;74:1810–1825. doi: 10.1016/j.jprot.2011.07.011. [DOI] [PubMed] [Google Scholar]
- Creer S, Malhotra A, Thorpe RS, Stöcklin R, Favreau P, Chou WH. Genetic and ecological correlates of intraspecific variation in pitviper venom composition detected using matrix-assisted laser desorption time-of-flight mass spectrometry (MALDI-TOF-MS) and isoelectric focusing. J Mol Evol. 2003;56:317–329. doi: 10.1007/s00239-002-2403-4. [DOI] [PubMed] [Google Scholar]
- da Silva N, Jr, Aird S. Prey specificity, comparative lethality and compositional differences in coral snake venoms. Comp Biochem Physiol. 2001;128:425–456. doi: 10.1016/s1532-0456(00)00215-5. [DOI] [PubMed] [Google Scholar]
- Daltry JC, Wüster W, Thorpe RS. Diet and snake venom evolution. Nature. 1996;379:537–540. doi: 10.1038/379537a0. [DOI] [PubMed] [Google Scholar]
- Doley R, Zhou X, Manjunatha Kini R. Handbook of Venoms and Toxins of Reptiles. CRC Press; Boca Raton, Florida: 2010. pp. 173–205. Ch. Snake Venom Phospholipase A2 Enzymes. [Google Scholar]
- Durban J, Perez A, Sanz L, Gomez A, Bonilla F, Rodriguez S, Chacon D, Sasa M, Angulo Y, Gutierrez J, Calvete J. Integrated “omics” profiling indicates that mirnas are modulators of the ontogenetic venom composition shift in the central american rattlesnake, Crotalus simus simus. BMC Genomics. 2013;14(1):234. doi: 10.1186/1471-2164-14-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichberg S, Sanz L, Calvete J, Pla D. Constructing comprehensive venom proteome reference maps for integrative venomics. Expert Rev Proteom. 2015;12(5):557–573. doi: 10.1586/14789450.2015.1073590. [DOI] [PubMed] [Google Scholar]
- Fox JW, Serrano SMT. Structural considerations of the snake venom metalloproteinases, key members of the M12 reprolysin family of metalloproteinases. Toxicon. 2005;45:969–985. doi: 10.1016/j.toxicon.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Fox J, Serrano S. Handbook of Venoms and Toxins of Reptiles. CRC Press; Boca Raton, Florida: 2010. pp. 95–113. Ch. Snake Venom Metalloproteinases. [Google Scholar]
- Garcia L, Parreiras e Silva L, Ramos O, Carmona A, Bersanetti P, Selistre-de Araujo H. The effect of post-translational modifications on the hemorrhagic activity of snake venom metalloproteinases. Comp Biochem Physiol Part C Toxicol Pharmacol. 2004;138(1):23–32. doi: 10.1016/j.cca.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Gibbs HL, Mackessy SP. Functional basis of a molecular adaptation: prey-specific toxic effects of venom from Sistrurus rattlesnakes. Toxicon. 2009;53:672–679. doi: 10.1016/j.toxicon.2009.01.034. [DOI] [PubMed] [Google Scholar]
- Gibbs HL, Rossiter W. Rapid evolution by positive selection and gene gain and loss: PLA2 venom genes in closely related Sistrurus rattlesnakes with divergent diets. J Mol Evol. 2008;66:151–166. doi: 10.1007/s00239-008-9067-7. [DOI] [PubMed] [Google Scholar]
- Gibbs HL, Sanz L, Calvete JJ. Snake population venomics: proteomics-based analyses of individual variation reveals significant gene regulation effects on venom protein expression in Sistrurus rattlesnakes. J Mol Evol. 2009;68:113–125. doi: 10.1007/s00239-008-9186-1. [DOI] [PubMed] [Google Scholar]
- Grant P, Grant B. Unpredictable evolution in a 30-year study of darwin’s finches. Science. 2002;296:707–711. doi: 10.1126/science.1070315. [DOI] [PubMed] [Google Scholar]
- Guércio RAP, Shevchenko A, Schevchenko A, López-Lozano JL, Paba J, Sousa MV, Ricart CAO. Ontogenetic variations in the venom proteome of the Amazonian snake Bothrops atrox. Proteome Sci. 2006;4:11. doi: 10.1186/1477-5956-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermann E, Hardt K. A sensitive and specific plate test for the quantitation of phospholipases. Anal Biochem. 1972;50(1):163–173. doi: 10.1016/0003-2697(72)90495-2. [DOI] [PubMed] [Google Scholar]
- Henschen-Edman A, Theodor I, Edwards B, Pirkle H. Crotalase, a fibrinogen-clotting snake venom enzyme: primary structure and evidence for a fibrinogen recognition exocite different from thrombin. Thromb Haemost. 1999;81:81–86. [PubMed] [Google Scholar]
- Huang S, Perez J. Comparative study on hemorrhagic and proteolytic activities of snake venoms. Toxicon. 1980;18(4):421–426. doi: 10.1016/0041-0101(80)90049-5. [DOI] [PubMed] [Google Scholar]
- Jansa SA, Voss RS. Adaptive evolution of the venom-targeted vWF protein in opossums that eat pitvipers. PLoS One. 2011;6(6):e20997. doi: 10.1371/journal.pone.0020997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen H, Kiljunen M, Amundsen P. Dietary ontogeny and niche shift to piscivory in lacustrine brown trout Salmo trutta revealed by stomach content and stable isotope analyses. J Fish Biol. 2012;80:2448–2462. doi: 10.1111/j.1095-8649.2012.03294.x. [DOI] [PubMed] [Google Scholar]
- Juárez P, Comas I, González-Candelas F, Calvete JJ. Evolution of snake venom disintegrins by positive Darwinian selection. Mol Biol Evol. 2008;25(11):2391–2407. doi: 10.1093/molbev/msn179. [DOI] [PubMed] [Google Scholar]
- Kini RM. Excitement ahead: structure, function and mechanism of snake venom phospholipase A2 enzymes. Toxicon. 2003;42:827–840. doi: 10.1016/j.toxicon.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Kini RM. Structure-function relationships and mechanism of anticoagulant phospholipase A2 enzymes from snake venoms. Toxicon. 2005;45:1147–1161. doi: 10.1016/j.toxicon.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Klauber LM. Rattlesnakes: Their Habits, Life Histories, and Influence on Mankind. 2. University of California Press; Berkeley, California: 1997. [Google Scholar]
- Kordiš D, Gubenšek F. Adaptive evolution of animal toxin multigene families. Gene. 2000;261:43–52. doi: 10.1016/s0378-1119(00)00490-x. [DOI] [PubMed] [Google Scholar]
- Linnen CR, Kingsley EP, Jensen JD, Hoekstra HE. On the origin and spread of an adaptive allele in deer mice. Science. 2009;325:1095–1098. doi: 10.1126/science.1175826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch VJ. Inventing an arsenal: adaptive evolution and neofunctionalization of snake venom phospholipase A2 genes. BMC Evol Biol. 2007;7:2. doi: 10.1186/1471-2148-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackessy SP. Venom ontogeny in the Pacific rattlesnakes Crotalus viridis helleri and C. v. oreganus. Copeia. 1988;1988(1):92–101. [Google Scholar]
- Mackessy SP. Venom composition in rattlesnakes: trends and biological significance. In: Hayes WK, Beaman KR, Cardwell MD, Bush SP, editors. The Biology of Rattlesnakes. Loma Linda University Press; Loma Linda, CA: 2008. pp. 495–510. [Google Scholar]
- Mackessy SP, Williams K, Ashton KG. Ontogenetic variation in venom composition and diet of Crotalus oreganus concolor: a case of venom paedomorphosis? Copeia. 2003;2003(4):769–782. [Google Scholar]
- Madrigal M, Sanz L, Flores-Diaz M, Sasa M, Nunez V, Alape-Giron A. Snake venomics across genus lachesis. ontogenetic changes in the venom composition of lachesis stenophrys and comparative proteomics of the venoms of adult Lachesis melanocephala and Lachesis acrochorda. J Proteom. 2012;77:280–297. doi: 10.1016/j.jprot.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Margres M, Aronow K, Loyacano J, Rokyta D. The venom-gland transcriptome of the eastern coral snake (Micrurus fulvius) reveals high venom complexity in the intragenomic evolution of venoms. BMC Genomics. 2013;14:531. doi: 10.1186/1471-2164-14-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margres M, McGivern J, Wray K, Seavy M, Calvin K, Rokyta D. Linking the transcriptome and proteome to characterize the venom of the eastern diamondback rattlesnake (Crotalus adamanteus) J Proteom. 2014;96:145–158. doi: 10.1016/j.jprot.2013.11.001. [DOI] [PubMed] [Google Scholar]
- Margres M, McGivern J, Seavy M, Wray K, Facente J, Rokyta D. Contrasting modes and tempos of venom expression evolution in two snake species. Genetics. 2015a;199(1):165–176. doi: 10.1534/genetics.114.172437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margres M, Wray K, Seavy M, McGivern J, Sanader D, Rokyta D. Phenotypic integration in the feeding system of the eastern diamondback rattlesnake (Crotalus adamanteus) Mol Ecol. 2015b;24(13):3405–3420. doi: 10.1111/mec.13240. [DOI] [PubMed] [Google Scholar]
- Margres M, Wray K, Seavy M, McGivern J, Herrera N, Rokyta D. Expression differentiation is constrained to low-expression proteins over ecological timescales. Genetics. 2016;202:273–283. doi: 10.1534/genetics.115.180547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markland F. Crotalase. Methods Enzymol. 1976;45:223–236. doi: 10.1016/s0076-6879(76)45022-x. [DOI] [PubMed] [Google Scholar]
- Markland F. Hemostasis and Animal Venoms. Marcel Dekker; New York: 1988. pp. 149–172. Ch. Fibrinogenolytic enzymes from snake venoms. [Google Scholar]
- Matsui T, Fujimura Y, Titani K. Snake venom proteases affecting hemostasis and thrombosis. Biochim Biophys Acta. 2000;1477:146–156. doi: 10.1016/s0167-4838(99)00268-x. [DOI] [PubMed] [Google Scholar]
- McGivern J, Wray K, Margres M, Couch M, Mackessy S, Rokyta D. RNA-seq and high-definition mass spectrometry reveal the complex and divergent venoms of two rear-fanged colubrid snakes. BMC Genomics. 2014;15(1):1061. doi: 10.1186/1471-2164-15-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J, Rienzo AD. Spatial patterns of variation due to natural selection in humans. Nat Rev Genet. 2009;10:745–755. doi: 10.1038/nrg2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Núñez V, Cid P, Sanz L, Torre PDL, Angulo Y, Lomonte B, Gutiérrez JM, Calvete JJ. Snake venomics and antivenomics of Bothrops atrox venoms from Colombia and the Amazon regions of Brazil, Perú and Ecuador suggest the occurrence of geographic variation of venom phenotype by a trend towards paedomorphism. J Proteom. 2009;73:57–78. doi: 10.1016/j.jprot.2009.07.013. [DOI] [PubMed] [Google Scholar]
- Palmer WM, Braswell AL. Reptiles of North Carolina. University of North Carolina Press; Chapel Hill, North Carolina: 1995. [Google Scholar]
- Pung YF, Wong PTH, Kumar PP, Hodgson WC, Kini RM. Ohanin, a novel protein from king cobra venom, induces hypolocomotion and hyperalgesia in mice. J Biol Chem. 2005;280(13):13137–13147. doi: 10.1074/jbc.M414137200. [DOI] [PubMed] [Google Scholar]
- Rokyta DR, Wray KP, Lemmon AR, Lemmon EM, Caudle SB. A high-throughput venom-gland transcriptome for the eastern diamondback rattlesnake (Crotalus adamanteus) and evidence for pervasive positive selection across toxin classes. Toxicon. 2011;57:657–671. doi: 10.1016/j.toxicon.2011.01.008. [DOI] [PubMed] [Google Scholar]
- Rokyta DR, Lemmon AR, Margres MJ, Aronow K. The venom-gland transcriptome of the eastern diamondback rattlesnake (Crotalus adamanteus) BMC Genomics. 2012;13:312. doi: 10.1186/1471-2164-13-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokyta D, Wray K, Margres M. The genesis of an exceptionally deadly venom in the timber rattlesnake (Crotalus horridus) revealed through comparative venom–gland transcriptomics. BMC Genomics. 2013;14(1):394. doi: 10.1186/1471-2164-14-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokyta D, Wray K, McGivern J, Margres M. The transcriptomic and proteomic basis for the evolution of a novel venom phenotype within the Timber Rattlesnake (Crotalus horridus) Toxicon. 2015;98:34–48. doi: 10.1016/j.toxicon.2015.02.015. [DOI] [PubMed] [Google Scholar]
- Salazar A, Guerrero B, Cantu B, Cantu E, Rodriguez-Acosta A, Perez J, Galan J, Tao A, Sanchez E. Venom variation in hemostasis of the southern Pacific rattlesnake (Crotalus oreganus helleri): isolation of hellerase. Comp Biochem Physiol Part C Toxicol Pharmacol. 2009;149(3):307–316. doi: 10.1016/j.cbpc.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar A, Vivas J, Sánchez E, Rodríguez-Acosta A, Ibarra C, Gil A, Carvajal Z, Girón M, Estrella A, Navarrete L, Guerrero B. Hemostatic and toxinological diversities in venom of micrurus tener tener, micrurus fulvius fulvius and micrurus isozonus coral snakes. Toxicon. 2011;58:35–45. doi: 10.1016/j.toxicon.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez E, Ramirez M, Galan J, Lopez G, Rodriguez-Acosta A, Perez J. Cross reactivity of three antivenoms against North American snake venoms. Toxicon. 2003;41(3):315–320. doi: 10.1016/s0041-0101(02)00293-3. [DOI] [PubMed] [Google Scholar]
- Sánchez EE, Galán JA, Powell RL, Reyes SR, Soto JG, Russell WK, Russell DH, Pérez JC. Disintegrin, hemorrhagic, and proteolytic activities of Mojave rattlesnake, crotalus scutulatus scutulatus venoms lacking Mojave toxin. Comp Biochem Physiol Part C. 2005;141:124–132. doi: 10.1016/j.cca.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Sánchez E, Lucena S, Reyes S, Soto J, Cantu E, Lopez-Johnston J, Guerrero B, Salazar A, Rodriguez-Acosta A, Galan J, Tao W, Perez J. Cloning, expression, and hemostatic activities of a disintegrin, r-mojastin 1, from the mohave rattlesnake (Crotalus scutulatus scutulatus) Thromb Res. 2010;126:e211–e219. doi: 10.1016/j.thromres.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L, Escolano J, Ferritti M, Biscoglio MJ, Rivera E, Crescenti EJ, Angulo Y, Lomonte B, Gutiérrez JM, Calvete JJ. Snake venomics of the South and Central American bushmasters. Comparison of the toxin composition of Lachesis muta gathered from proteomic versus transcriptomic analysis. J Proteom. 2008;71:46–60. doi: 10.1016/j.jprot.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Serrano SMT, Maroun RC. Snake venom serine proteinases: sequence homology vs. substrate specificity, a paradox to be solved. Toxicon. 2005;45:1115–1132. doi: 10.1016/j.toxicon.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Suntravat M, Nuchprayoon I, Perez J. Comparative study of anticoagulant and procoagulant properties of 28 snake venoms from families Elapidae, Viperidae, and purified Russell’s viper venom-factor X activator(RVV-X) Toxicon. 2010;56(4):544–553. doi: 10.1016/j.toxicon.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kaulla K, Ostendorf P, von Kaulla E. The impedance machine: a new bedside coagulation recording device. J Med. 1975;6:73–88. [PubMed] [Google Scholar]
- Vonk F, Casewell N, Henkel C, Heimberg A, Jansen H, McCleary R, Kerkkamp H, Vos R, Guerreiro I, Calvete J, Wuster W, Woods A, Logan J, Harrison R, Castoe T, Jason de Koning A, Pollock D, Yandell M, Calderon D, Renjifo C, Currier R, Salgado D, Pla D, Sanz L, Hyder A, Ribeiro J, Arntzen J, van den Thillart G, Boetzer M, Pirovano W, Dirks R, Spaink H, Duboule D, McGlinn E, Kini R, Richardson M. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. P Natl Acad Sci U S A. 2013;110(51):20651–20656. doi: 10.1073/pnas.1314702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron JL, Welch SM, Bennett SH, Kalinowsky WG, Mousseau TA. Life history constraints contribute to the vulnerability of a declining North American rattlesnake. Biol Conserv. 2013;159:530–538. [Google Scholar]
- White J. Snake venoms and coagulopathy. Toxicon. 2005;45(8):951–967. doi: 10.1016/j.toxicon.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Whitlock MC, Guillaume F. Testing for spatially divergent selection: comparing Qst to Fst. Genetics. 2009;183:1055–1063. doi: 10.1534/genetics.108.099812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray K, Margres M, Seavy M, Rokyta D. Early significant ontogenetic changes in snake venoms. Toxicon. 2015;96:74–81. doi: 10.1016/j.toxicon.2015.01.010. [DOI] [PubMed] [Google Scholar]
- Zelanis A, Andrade-Silva D, Rocha MM, Furtado MF, Serrano SMT, de Azevedo ILMJ, Ho PL. A transcriptomic view of the proteome variability of newborn and adult Bothrops jararaca snake venoms. PLoS Negl Trop Dis. 2012;6(3):e1554. doi: 10.1371/journal.pntd.0001554. [DOI] [PMC free article] [PubMed] [Google Scholar]