Abstract
Pregnancies resulting from fresh in vitro fertilization (IVF) cycles exposed to supraphysiologic estrogen levels have been associated with higher rates of low birth weight and small for gestational age babies. We identified GATA3, a transcription factor selectively expressed in the trophectoderm during the blastocyst stage of embryo development, in an upstream analysis of genes that were differentially methylated in chorionic villus samples between IVF and non-IVF infertility treatment pregnancies. In this study, we investigate the hypothesis that GATA3 is hormonally regulated and plays an important functional role in trophoblast migration, invasion, and placentation. We found that GATA3 expression was hormonally regulated by estradiol in HTR8/SVneo first trimester trophoblast cells; however, no change in expression was seen with progesterone treatment. Furthermore, GATA3 knockdown resulted in decreased HTR8/SVneo cell migration and invasion compared with controls. RNA sequencing of GATA3 knockdown cells demonstrated 96 differentially regulated genes compared with controls. Genes known to play an important role in cell-cell and cell-extracellular matrix interactions, cell invasion, and placentation were identified, including CTGF, CYR61, ADAMTS12, and TIMP3. Our results demonstrate estradiol down-regulates GATA3, and decreased GATA3 expression leads to impaired trophoblast cell migration and invasion, likely through regulation of downstream genes important in placentation. These results are consistent with clinical data suggesting that supraphysiologic estrogen levels seen in IVF pregnancies may play an important role in attenuated trophoblast migration, invasion, and impaired placentation. GATA3 appears to be an important regulator of placentation and may play a role in impaired outcomes associated with fresh IVF cycles.
Keywords: ART (assisted reproductive technology), differential methylation, estrogen regulation, GATA3, HTR8/SVneo cells, placentation, trophoblast migration
INTRODUCTION
Pregnancies conceived using assisted reproductive technologies (ART) have higher rates of adverse outcomes including low birth weight and small for gestational age babies, placental abnormalities, and preterm labor than pregnancies conceived spontaneously [1–5]. Abnormal placentation caused by failure of first trimester trophoblast invasion into the placental bed and inadequate vascular remodeling are at the core of the pathogenesis of many of these adverse outcomes, including placental perfusion-related intrauterine growth restriction, which significantly contributes to preterm delivery [6, 7]. Many factors can affect abnormal placentation and lead to adverse fetal outcomes, including epigenetic changes and differential gene expression (DEG) in the placenta, both of which have been linked to ART [8–11]. Further understanding of the mechanisms behind ART-associated risks becomes increasingly important as the number of babies conceived using in vitro fertilization (IVF) continues to grow, with ART currently contributing to 1.5% of live births in the United States [12].
It is important to distinguish whether the risks associated ART result from the procedures themselves or from the underlying infertility and its causes. Pregnancies conceived using fertility treatments, either IVF or non-IVF infertility treatment (NIFT), do not have an increased incidence of cytogenetic abnormalities compared with those conceived spontaneously [13]. Studies looking at infertile couples with matched sibling IVF and spontaneous gestations provide evidence that perhaps it is the underlying infertility that contributes to adverse outcomes [14, 15]. However, aspects of the IVF procedures themselves may contribute to adverse outcomes by affecting epigenetic changes through mechanisms such as embryo culture conditions or embryo transfer into a hyperstimulated endometrium exposed to supraphysiologic estrogen levels [9, 16, 17]. Notably, epigenetic changes have been found to play an important role in the regulation of placentation [18]. During pre-implantation embryo development, global DNA methylation reprogramming occurs, and both rodent and choriocarcinoma cell line studies have shown that specific patterns of methylation are important for normal placental function and that aberrations in methylation negatively affect placentation [19–21].
In our prior study, in an attempt to control for underlying infertility, we compared genomewide DNA methylation from chorionic villus samples (CVS) from matched singleton pregnancies conceived using either fresh IVF or NIFT to determine if IVF induces epigenetic changes in the placenta during early fetal development [22]. We found similar global methylation among the groups. However, significant differential methylation was found between fresh IVF and NIFT pregnancies at 34 CpG sites, including multiple loci at three genes, ANAPC2, CXCL14, and RIMS1. Analysis of these genes was performed, examining how these genes tie into relevant pathways and networks that have implications for placentation. Further upstream analysis was performed for the current study, and GATA3 was identified as a potentially important regulatory gene.
GATA3 is a transcription factor that is selectively expressed in the trophectoderm during the blastocyst stage of embryo development. It has also been shown to play a role in tumor cell metastasis, migration, and invasion. However, there is very little data examining the role and regulation of GATA3 in trophoblast cell invasion and placentation. Based on the identification of GATA3 as a potentially important regulatory gene of differentially methylated genes in fresh IVF and NIFT conceptions and the known placenta-related adverse outcomes associated with fresh IVF cycles, we hypothesize that the hormonal milieu at the time of fresh embryo transfer may play a role in GATA3 regulation leading to alterations in placentation.
MATERIALS AND METHODS
Sampling of CVS and Patient Selection
As previously described, collection of CVS was performed between 11 and 13 wk gestation at the Cedars-Sinai Prenatal Diagnostic Center by the same physician [22]. Leftover tissue not used for clinical genetic testing was collected under an institution review board-approved protocol at our institution. All patients provided informed written consent for the procurement and use of this leftover material prior to CVS collection. Briefly, 5–15 mg of villous tissue was collected and placed in RNAlater RNA Stabilization Reagent (Qiagen) and then stored at −80°C in our Prenatal Biorepository. Ten CVS were analyzed, which included five singleton pregnancies from pregnancies conceived by IVF and by NIFT. Subjects were matched based on gestational age, race, and sex of the fetus. All subjects in the IVF group underwent conventional IVF without intracytoplasmic sperm injection and fresh embryo transfer. All subjects in the NIFT group underwent superovulation with clomiphene citrate and/or gonadotropins and intrauterine insemination. The indication for CVS in all subjects was advanced maternal age. All investigations using experimental human tissue were conducted in accordance with the Society for the Study of Reproduction's specific guidelines and standards.
Ingenuity Pathway Analysis Upstream Regulator Analysis
The upstream regulator analysis in Ingenuity Pathway Analysis (IPA) (Ingenuity) is used to predict upstream transcriptional regulators that can be used to explain observed gene expression or methylation changes identified in a dataset. First, methylation data from CVS-derived DNA was obtained using the Infinium HumanMethylation450 BeadChip Array (Illumina). Differential methylation patterns were seen in several genes, including at multiple CpG sites in three genes (ANAPC2, CXCL14, and RIMS1) when comparing samples of CVS from IVF and NIFT patients [22]. IPA was then used to visualize the network of transcriptional regulators of this gene network to explain how regulators interact with one another. The identified target genes were used to provide testable hypotheses for gene regulatory networks. Overlapping P-values were used to identify transcriptional regulators that can explain observed methylation differences between study groups. The overlapping P-value measures where there is a statistically significant overlap between the genes in the dataset and the genes that are regulated by particular transcription regulators. This calculation was performed using Fisher exact test, and the significance level was set at P < 0.05.
Hormone Treatment of HTR8/SVneo Cells
The immortalized human first trimester trophoblast cell line HTR8/SVneo (a gift from Dr. C. Graham; Queen's University, Kingston, Ontario, Canada), was maintained in RPMI 1640 with Hepes and L-glutamine (Life Technologies) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin in a humidified environment (5% CO2 and 95% air) at 37°C. HTR8/SVneo cells were plated on 6-well plates at a concentration of 2.5 × 105 cells/ml and cultured until they reached ∼70% confluency. Hormone stock solutions of 17-β estradiol (E2257; Sigma-Aldrich) and progesterone (P0130; Sigma-Aldrich) were freshly prepared for each experiment in absolute ethanol and dimethyl sulfoxide (Sigma), respectively. The appropriate vehicle controls were included in all experiments. HTR8/SVneo cells were subsequently treated with 7.5 nM of 17-β estradiol or 10 μM progesterone, and vehicles diluted in a modified medium composed of phenol red-free RPMI 1640 with Hepes and L-glutamine (Life Tech) supplemented with 10% charcoal-stripped FBS (Life Tech) and 1% penicillin/streptomycin to avoid any uncontrolled hormone contamination [23]. The cells were treated for 48 h at 37°C and 5% CO2, at which point they were harvested for RNA isolation. This protocol for estrogen and progesterone treatment of HTR8/SVneo cells was based on several prior studies [23–25]. The duration of estrogen and progesterone treatment was based on previous studies that treated HTR8/SVneo cells with hormone for between 18 and 48 h, depending on the group and experiment performed [23–25]. However, the only direct comparison between 24 versus 48 h of incubation by Chen et al. [25] showed the most significant difference in wound healing assay with 48 h of treatment. Thus, we chose 48 h based on this study showing maximal results at this time point.
Small Interfering RNA Transfection
HTR8/SVneo cells were plated in 6-well plates at a concentration of 2.5 × 105 cells/ml and cultured until ∼70% confluent. They were transfected with 50nM of Silencer Select GATA3 small interfering RNA (siRNA) (4392420; Thermo Fisher Scientific) or nonsense scrambled siRNA (4390843; Thermo Fisher), using Lipofectamine 2000 Transfection Reagent (Life Tech) and Opti-MEM I Reduced Serum medium (Life Tech), according to the manufacturer's instructions. Transfection and gene knockdown efficiency was assessed by quantitative real-time PCR (qRT-PCR), as described below.
Reverse Transcription and Quantitative Real-Time PCR
Total RNA was extracted from HTR8/SVneo cells using RNeasy Mini kit according to the manufacturer's protocol (Qiagen), and 1 μg of RNA was reverse transcribed using iScript cDNA Synthesis kit (Bio-Rad Laboratories) to synthesize cDNA. qRT-PCR was performed using iQ SYBR Green Supermix (Bio-Rad) and MyiQ Thermal Cycler (Bio-Rad). The PCR was performed in two-step reactions: 95°C for 10 sec and 60°C for 45 sec for 40 cycles. Gene expression levels were compared to that of an internal control, GAPDH. While we did not use multiple housekeeping genes, there was little variation in the mean cycle threshold (Ct) value for GAPDH with a mean of 14.6 and a range of 14.2 and 15.3 over five replicates. Gene specific primers were designed using National Center for Biotechnology Information/Primer-BLAST (www.ncbi.nlm.nih.gov/tools/primer-blast/) and are as follows: GATA3 forward: 5′-GCTCTTCGCTACCCAGGTGA-3′, reverse 3′-AAAAAGGGGCGACGACTCTG-5′; GADPH forward 5′-GAAGGTGAAGGTCGGACTC-3′, reverse 3′-GAAGATGGTGATGGGATTTC-5′.
Transwell Migration Assay and Matrigel Invasion Assay
Migration assays were performed as previously described [26, 27] using 6.5mm Transwell plates with 8.0 μm pore polycarbonate membrane inserts (Corning). Invasion assays were performed using BioCoat invasion chambers with 8.0 μm pore and precoated with Matrigel (Corning). For both assays, HTR8/SVneo cells transfected with either GATA3 siRNA or scrambled siRNA were serum-starved overnight, trypsinized, and seeded at a density of 1 × 105 cells in 100 μl of serum-free RPMI 1640 medium in the upper chamber of the Transwell plate. The lower chambers of the Transwell plate were filled with 600 μl of RPMI medium containing 10% FBS. The cells were incubated at 37°C and 5%CO2 for 48 h to allow cell migration/invasion. The inserts were then washed in PBS, and the nonmigrating cells in the upper chamber were removed with a cotton swab. The inserts containing the migrated/invaded cells were then fixed with methanol and Wright-Giemsa stained using the QUICK III Stainkit (Astral Diagnostics). The filters were excised and mounted on slides. The migrated cells on the lower surface of the membrane were counted in six random fields at a magnification of 200×. The assay was repeated at least three times, and the results are represented as fold change in cell migration/invasion of GATA3 siRNA knockdown compared to the scrambled siRNA control.
RNA Sequencing of GATA3 Knockdown Cells
RNA was extracted after siRNA transfection as previously described using the RNeasy Minikits (Qiagen), and RNA concentration was quantified using spectroscopy (Eppendorf BioPhotometer). Total RNA was quantified using both the NanoDrop to assess sample contamination by proteins or carryover reagents from RNA isolation and the Qubit fluorometer (Invitrogen). Samples were then qualified using the Fragment Analyzer (Advanced Analytical Technologies) that analyzes the integrity of the total RNA by measuring the ratio between the 18S and 28S ribosomal peaks. RNAseq libraries maintaining strand-specificity were made using 1 μg of material per sample with the Illumina TruSeq Stranded mRNA kit (Illumina) according to the manufacturer's instructions. Final libraries were again quantitated with the Qubit fluorometer and checked for size via the Fragment Analyzer. Libraries were diluted to 4 nM and pooled in equal volumes for denaturation, hybridization, and sequencing on the Illumina NextSeq 500 (Illumina) with single end 75 bp sequencing chemistry. On average, about 20 million reads were generated from each sample.
RNA Sequencing Data Analysis
Raw reads obtained from RNAseq were aligned to the transcriptome using STAR (version 2.2.1) / RSEM (version 1.2.20) [28] with default parameters, using a custom human GRCh38 transcriptome reference downloaded from http://www.gencodegenes.org, containing all protein coding and long noncoding RNA genes based on GENCODE version 23 annotation. Expression counts for each gene in all samples were normalized by a modified trimmed mean of the M-values normalization method and the unsupervised principal component analysis was performed with DESeq2 Bioconductor package version 1.10.1 in R version 3.2.2. Each gene was fitted into a negative binomial generalized linear model, and the Wald test was applied to assess the differential expressions between two sample groups by DESeq2. Benjamini and Hochberg procedure was applied to adjust for multiple hypothesis testing, and DEG candidates were selected with a false discovery rate less than 0.01. For visualization of coordinated gene expression in samples, a two-way hierarchical clustering with Pearson correlation distance matrix was performed with samples and DEG candidates using the Bioconductor g-plots package (version 2.14.2) in R. The pathway enrichment analysis was performed on these candidates with DAVID (https://david.ncifcrf.gov/summary.jsp) and IPA (Qiagen).
Networks and Bio Functions Analysis
Comparative analyses of networks and cellular functions of the identified DEGs from RNA sequencing of GATA3 knockdown versus control HTR8/SVneo cells were performed using IPA software. DEG results were first filtered as significant at Padj value < 0.05 and uploaded to IPA as a dataset. Core analysis, which interprets data in the context of pathways, networks, and biological functions, was performed. In these analyses, the Fischer exact test was used to calculate the P values. Based on connections found in the Ingenuity knowledge base, IPA queries the dataset to identify direct and indirect interactions and functional categories.
Statistics
Statistical analysis for the qRT-PCR, siRNA GATA3 knockdown, and migration/invasion assays was performed using the Wilcoxon signed-rank (nonparametric) test or one-way ANOVA followed by Dunnet multiple comparison tests for post hoc analysis when appropriate. Analyses were performed using JMP12 (SAS Institute). For RNA sequencing, statistical analysis was performed as described above.
RESULTS
Clinical Characteristics of CVS
All CVS were from Caucasian, singleton male fetuses with a normal karyotype. There were no significant differences between the two groups (IVF and NIFT) in terms of maternal age, prepregnancy body mass index, or gestational age at the time of CVS (Table 1). All pregnancies resulted in term deliveries with no difference in gestational age at the time of delivery or in birth weight. The indication for CVS was advanced maternal age in all cases. All samples in the IVF group were pregnancies resulting from fresh embryo transfers.
TABLE 1.
Clinical characteristics of chorionic villus samples (CVS).
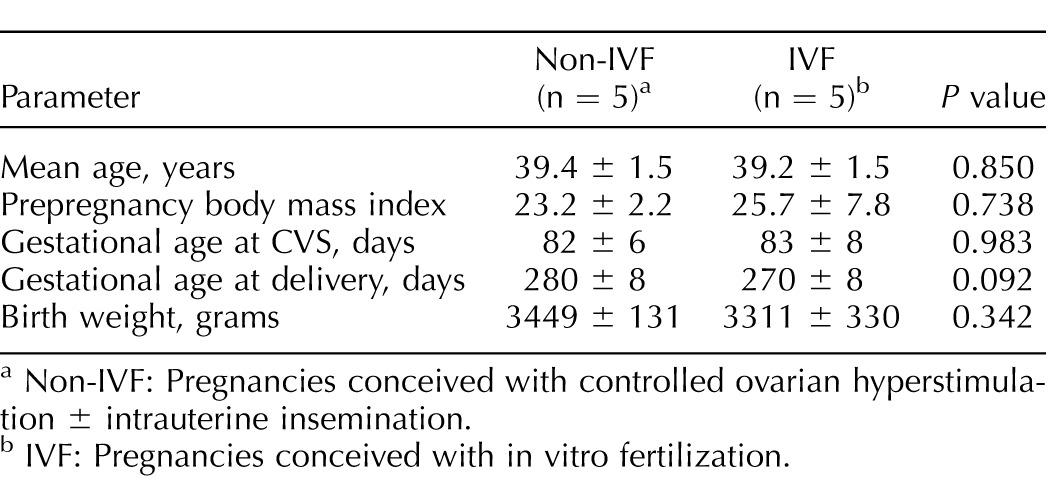
Non-IVF: Pregnancies conceived with controlled ovarian hyperstimulation ± intrauterine insemination.
IVF: Pregnancies conceived with in vitro fertilization.
Upstream IPA Analysis
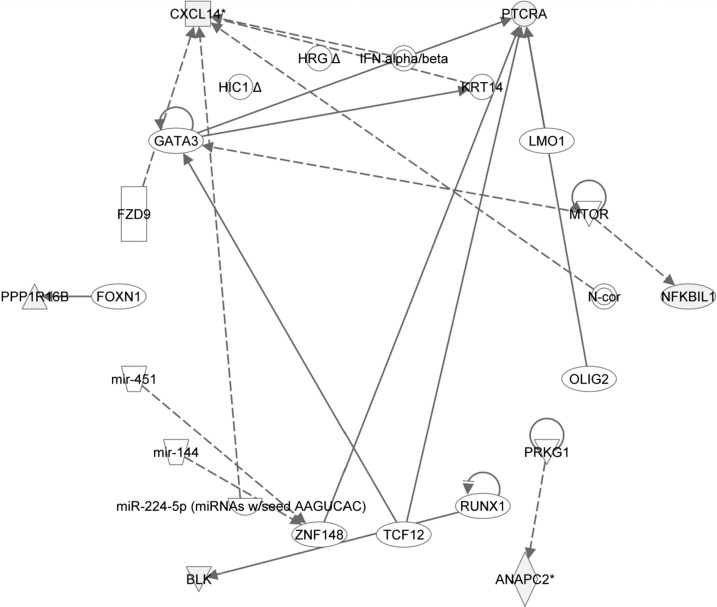
The IPA upstream analysis predicted GATA3 to be a transcriptional regulator based on differential methylation of samples of CVS from IVF and NIFT pregnancies (overlapping P values < 0.05). This upstream analysis of differentially methylated CpG sites is displayed as a network, demonstrating how GATA3 may directly, via PTCRA, or indirectly, via CXCL14 (which regulates another transcription factor KRT14), affect the methylation changes of the targets identified by our dataset (Fig. 1). GATA3 may also be self-regulated or may function as a downstream target for two other predicted transcriptional factors MTOR and TCF12. Taken together, GATA3 appears to be a critical factor involved in several biological activities occurring in samples of CVS, suggesting its potential important role in normal fetal development.
FIG. 1.
Differentially methylated genes between IVF and non-IVF infertility pregnancies (shaded in gray) were input for an upstream analysis using IPA software to visualize a network of upstream transcriptional regulators.
Impact of Estrogen and Progesterone Treatment on GATA3 Expression in HTR8 Cells
HTR8/SVneo cells were treated with 7.5 nM 17-β-estradiol and compared to a vehicle control. After 48 h of estradiol treatment, RNA extraction was performed and GATA3 expression as determined by qRT-PCR was significantly decreased compared with the ethanol control (P < 0.05; Fig. 2A). In a similar fashion, HTR8/SVneo cells were treated with 10 μM progesterone and compared to a vehicle control. After 48 h of treatment, no significant difference was seen GATA3 expression (Fig. 2B).
FIG. 2.
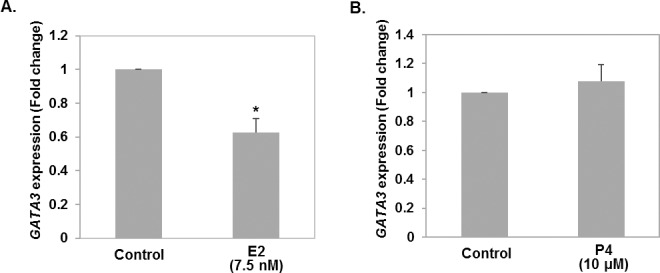
A) Effect of 17-β estradiol (E2) on the expression of GATA3 in HTR8/SVneo cells. B) Effect of progesterone (P4) treatment on the expression of GATA3 in HTR8/SVneo cells. HTR8/SVneo cells were treated with 7.5 nM 17-β-estradiol or 10 μM P4 for 48 h, after which point GATA3 expression was determined by qRT-PCR and compared to vehicle control groups. All results are expressed in fold-change and are the means ± SEM of at least three representative independent experiments, each analyzed in triplicate. P < 0.05, one-way ANOVA followed by Dunnet multiple comparison post hoc test. Asterisk (*) denotes statistically significant difference.
Migration and Invasion of GATA3 Knockdown in HTR8 Cells
To assess the functional significance of GATA3 in first trimester trophoblasts, we generated a knockdown model of GATA3 in HTR8/SVneo cells. Transient transfection of HTR8/SVneo cells with GATA3 siRNA resulted in ∼60% knockdown of GATA3 expression (P = 0.03) compared to cells transfected with scrambled siRNA (Fig. 3). As the primary function of the first trimester extravillous trophoblasts is to migrate and burrow into the uterine decidua in order to establish blood supply to the fetus, we examined the impact of the decrease in GATA3 expression on the migratory and invasive ability of HTR8/SVneo cells in a Transwell assay. GATA3 knockdown resulted in a 51% reduction of migration (P = 0.01) and 81% reduction of invasion (P = 0.01) when normalized to that of scrambled siRNA-transfected control cells (Fig. 4).
FIG. 3.
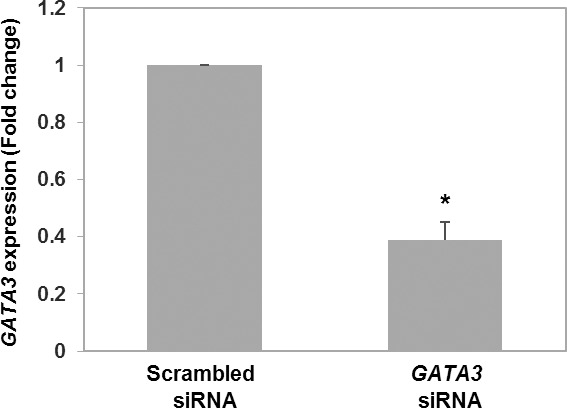
Expression of GATA3 in cells transfected with GATA3 siRNA or scrambled siRNA was assessed with qRT-PCR. Results are means ± SEM. P = 0.03, Wilcoxin signed-rank test. Asterisk (*) denotes statistically significant difference.
FIG. 4.
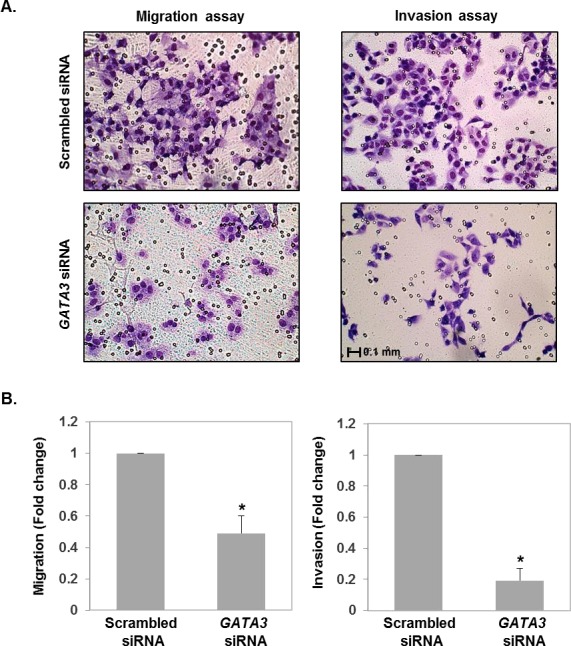
A) Representative images of Transwell migration (left panels) and Matrigel invasion (right panels) assays are shown. Top panels show migration/invasion of cells transfected with scrambled siRNA. Bottom panels show migration/invasion of GATA3 knockdown cells. B) Cell migration and invasion was quantified by the migration/invasion index indicated as the ratio to that of control cells. Results are means ± SEM. P < 0.01, Wilcoxin signed-rank test. Asterisk (*) denotes statistically significant difference.
Differential Gene Expression in GATA3 Knockdown HTR8 Cells
To identify downstream genes involved in GATA3 regulation of trophoblast function, we performed RNA sequencing of GATA3 knockdown HTR8/SVneo cells. Following GATA3 knockdown, 200 genes were significantly differentially expressed with a 1.5-fold or greater difference; 97 genes were up-regulated and 103 genes were down-regulated (Supplemental Table S1; all Supplemental Data are available online at www.biolreprod.org). To elucidate the diseases and biofunctions regulated by these genes, we performed IPA core analysis with RNA sequencing results. The top diseases and cellular functions identified were those involved in cellular movement, cell death and survival, cellular development, the cell cycle, tissue morphology, connective tissue development and function, and cancer, all processes actively occurring in first trimester trophoblasts during early placentation. To highlight the most influential subset of genes, those DEGs with fold change of 2 or greater and select genes with fold change of 1.5–2 that play known roles in extracellular matrix (ECM) remodeling and cell cycle regulation are presented in Table 2.
TABLE 2.
Differentially expressed genes (DEGs) in GATA3 knockdown HTR8/SVneo cells with fold change (FC) of 2 or greater and select DEGs with FC of 1.5–2 that are extracellular matrix and cell cycle regulatory molecules.
DISCUSSION
In this study, we examine the functional significance and hormonal regulation of GATA3, an upstream transcriptional regulator of genes that we found to be differentially methylated between IVF and non-IVF infertility pregnancies. Gene expression of GATA3 was significantly down-regulated by estradiol in HTR8/SVneo cells, providing a possible mechanism for GATA3 regulation secondary to supraphysiologic estrogen levels. Conversely, GATA3 gene expression was not significantly impacted by progesterone exposure. Functional studies of migration and invasion of HTR8/SVneo cells show decreased cell migration and invasion in GATA3 siRNA knockdowns, demonstrating the significant role for GATA3 in this model of trophoblast cell migration and invasion. Lastly, we used RNA sequencing in GATA3 knockdown HTR8/SVneo cells to more fully elucidate the network of downstream genes regulated by GATA3 and found genes within connective tissue development and function and cell cycle functional networks, among others, which play an important role in trophoblast invasion and establishment of placentation.
GATA is a family of key transcription factors that play a critical role in determination of developmental fates during early mammalian embryo development. In mammalian blastocysts, GATA is expressed in a spatially restricted manner to orchestrate the differentiation of three distinct germ layers via regulation of its target genes. More recently, GATA3 has also been shown to be an essential regulator of trophectoderm differentiation in mammalian blastocysts. GATA3 expression is high in the trophectoderm of blastocysts, and GATA3 expression is specifically up-regulated within the trophectoderm but excluded from the inner cell mass [29, 30]. Additionally, up-regulation of GATA3 induces embryonic stem cells to differentiate into trophoblast cells in vitro and expression is high in trophoblast stem cells [31, 32]. Underscoring its role in the regulation of trophoblast establishment and maintenance, the reduction of GATA3 expression in blastocysts resulted in decreased embryo hatching and implantation rates in mice [33].
In addition to its role in the determination of the trophectoderm lineage, GATA3 has also been implicated in other stages of trophoblast differentiation and subsequent placental development. GATA2 and GATA3 are required for trophoblast giant cell differentiation, secretion of proliferin and placental lactogen I, and regulation of expression of syncytin [34–36]. GATA2-null mouse placentas exhibit low proliferin expression, a regulatory factor known to play a role in the establishment of fetal vascular connections [37]. Low proliferin expression subsequently leads to decreased angiogenic activity in GATA2-null mice [35]. GATA2 and GATA3 also regulate syncytin, a fusogenic glycoprotein that is required for fusion of cytotrophoblasts to form the syncytial layer of the placenta [36, 38, 39]. Interestingly, dysregulation of syncytin expression is found in pre-eclamptic placentas [40], another state of altered placental function. Thus, GATA3 appears to be a significant transcription factor regulating trophoblast differentiation and function, and any alterations in its expression and function may lead to abnormal placental development.
Multiple factors besides underlying infertility are unique to IVF pregnancies and may contribute to adverse outcomes. These include embryo culture, embryo manipulation, epigenetic changes, and the endometrial environment into which the embryo is transferred [16, 17]. Samples of CVS from IVF and NIFT pregnancies were compared in an attempt to better control for underlying infertility. While we acknowledge that the comparison between IVF and NIFT could also present a model for cultured versus uncultured conditions, it also can be a model for the effects of estrogen on the endometrium and placentation. A single follicle produces a serum level of estradiol between 200 and 400 pg/ml whereas an IVF cycle in a moderate to good responder typically produces a serum level of 1500–3000 pg/ml [41]. While some NIFT cycles may produce a slightly higher estradiol level than a natural, monofollicular cycle, the goal is to generate between one and three ovulatory follicles, and this would on average result in an estradiol level significantly lower the typical IVF cycle [42]. Thus, the comparison between IVF and NIFT represents an appropriate model to examine the molecular mechanisms behind the role of supraphysiologic estrogen on the endometrium and placentation.
In fresh IVF cycles, embryos are transferred into a hyperstimulated endometrium that has been exposed to supraphysiologic levels of estrogen, with levels typically in the 2000–3000 pg/ml range and sometimes even higher. Fresh embryo transfers in singleton IVF gestations, exposed to high levels of estrogen, have been associated with adverse obstetric and perinatal outcomes, including low birth weight, fetal growth restriction, pre-eclampsia, and abnormal placentation, when compared with frozen embryo transfers and cycles with lower peak estradiol levels [17, 43–45]. The more physiologic endometrium resulting from lower estradiol exposure has led to a shift in clinical practice toward more routine use of frozen embryo transfers [46].
The exact mechanisms underlying these differences in fetal birth weight and other adverse outcomes are still under investigation and involve impaired trophoblast invasion and impaired angiogenesis, possibly resulting from underlying epigenetic and genetic modifications. In normal placentation, trophoblast cell invasion is required for spiral artery remodeling, allowing for vascular dilation and increased nutrient and oxygen delivery to the intravillous space [6, 47]. Animal studies in the baboon have shown attenuation of extravillous trophoblast invasion after exposure to elevated levels of estradiol, mediated by impaired angiogenesis [48]. Additionally, microarray studies have shown a large discrepancy in gene expression between natural and hyperstimulated endometrium sampled at time of embryo implantation [49, 50]. In our study, treatment of HTR8/SVneo cells with estradiol provides a model for the high estrogen exposure of trophoblasts cells during implantation at the time of a fresh IVF transfer. We have shown GATA3 is down-regulated by estradiol and that decreased expression of GATA3 leads to inhibition of HTR8/SVneo cell migration and invasion. While prior studies have evaluated the impact of estradiol on trophoblast cell lines, some demonstrating impaired invasion, our study takes this a step further, assessing the impact on our target gene of interest, GATA3 [23, 24, 51].
The DEG analysis results from GATA3 knockdown trophoblasts shed light on the biological functions regulated by GATA3 downstream targets. Among the down-regulated genes were various ECM molecules (CYR61, CTGF, THBS1, THBS2, ADAMTS12, and TIMP3), which are involved in cell-to-cell and cell-to-ECM interactions via modulations of ECM components, such as cell surface integrins, laminins, collagens, and focal adhesion kinases. They mediate endothelial cell growth, adhesion, migration, and survival as well as tumor growth and angiogenesis. Notably, CTGF and CYR61 knockout mice exhibit vascular defects during embryogenesis and fetal development [52], and reduced levels of CYR61 were found in early-onset pre-eclampsia [53]. Interestingly, in a mouse retinal neovascularization model, knocking down CYR61 resulted in a reduction in neovascularization [54]. ADAMTS12 and TIMP3, which belong to a family of matrix metalloproteinases and their inhibitors, have been identified as modulators of trophoblast invasiveness in first trimester trophoblast cell lines [55–57]. Our analysis of DEGs showed up-regulation of cell cycle regulatory genes (E2F2, BTG2, and CDKN1A) and cytoskeleton-associated genes (ANK1 and SYNE2) in GATA3 knockdown trophoblasts. Cellular functions analysis of networks that include these DEGs identified cellular movement, cell death and survival, cell cycle, cellular development, and cellular growth and proliferation as the most represented functions in GATA3 knockdown trophoblasts, all of which are hallmark processes occurring in first trimester trophoblasts. Together, these findings lend support to the hypothesis that GATA3 not only regulates differentiation of trophectoderm lineage, but also underlies regulation of trophoblast function by affecting expression of downstream genes involved in major functions of first trimester trophoblasts, including migration and invasion toward the decidua and establishment of a blood supply to the fetus.
One limitation of the current study is that we were not able to completely knockdown GATA3 expression with about 40% of GATA3 expression remaining in the knockdown cells, possibly limiting identification of additional downstream genes that would be identified by a more complete knockdown of GATA3. However, even with a knockdown of 60%, we were able to identify a significant number of genes that are differentially regulated and likely play an important role in trophoblast cell migration and invasion.
In this study, we investigated the functional role and regulation of GATA3, identified through preliminary studies as a potential upstream transcriptional regulator of differentially methylated genes between IVF and non-IVF infertility pregnancies. We demonstrated that GATA3 is altered by the hormonal milieu and GATA3 down-regulation inhibits normal trophoblast migration and invasion, which impacts normal placentation, likely through other downstream genes found to be important in placentation. Because supraphysiologic levels of estradiol are present in IVF, and pregnancies conceived through IVF are at increased risk of placental defects associated with abnormal placentation, GATA3, through estradiol, is likely a significant factor contributing to these alterations and subsequent abnormal placentation.
Footnotes
The research was supported by the NICHD of the National Institutes of Health under award number R01HD074368. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Data have been deposited in GEO (accession number: GSE85995; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=wdklyogghxydvmd&acc=GSE85995). Presented at the 49th Annual Meeting of the Society for the Study of Reproduction, 16–20 July 2016, San Diego, California.
REFERENCES
- Jackson RA, Gibson KA, Wu YW, Croughan MS. Perinatal outcomes in singletons following in vitro fertilization: a meta-analysis. Obstet Gynecol. 2004;103:551–563. doi: 10.1097/01.AOG.0000114989.84822.51. [DOI] [PubMed] [Google Scholar]
- Schieve LA, Meikle SF, Ferre C, Peterson HB, Jeng G, Wilcox LS. Low and very low birth weight in infants conceived with use of assisted reproductive technology. N Engl J Med. 2002;346:731–737. doi: 10.1056/NEJMoa010806. [DOI] [PubMed] [Google Scholar]
- Pinborg A, Wennerholm UB, Romundstad LB, Loft A, Aittomaki K, Soderstrom-Anttila V, Nygren KG, Hazekamp J, Bergh C. Why do singletons conceived after assisted reproduction technology have adverse perinatal outcome? Systematic review and meta-analysis. Hum Reprod Update. 2013;19:87–104. doi: 10.1093/humupd/dms044. [DOI] [PubMed] [Google Scholar]
- Romundstad LB, Romundstad PR, Sunde A, von During V, Skjaerven R, Vatten LJ. Increased risk of placenta previa in pregnancies following IVF/ICSI; a comparison of ART and non-ART pregnancies in the same mother. Hum Reprod. 2006;21:2353–2358. doi: 10.1093/humrep/del153. [DOI] [PubMed] [Google Scholar]
- Kaser DJ, Melamed A, Bormann CL, Myers DE, Missmer SA, Walsh BW, Racowsky C, Carusi DA. Cryopreserved embryo transfer is an independent risk factor for placenta accreta. Fertil Steril. 2015;103:1176–1184. doi: 10.1016/j.fertnstert.2015.01.021. [DOI] [PubMed] [Google Scholar]
- Pijnenborg R, Vercruysse L, Hanssens M. The uterine spiral arteries in human pregnancy: facts and controversies. Placenta. 2006;27:939–958. doi: 10.1016/j.placenta.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Khong TY, De Wolf F, Robertson WB, Brosens I. Inadequate maternal vascular response to placentation in pregnancies complicated by pre-eclampsia and by small-for-gestational age infants. Br J Obstet Gynaecol. 1986;93:1049–1059. doi: 10.1111/j.1471-0528.1986.tb07830.x. [DOI] [PubMed] [Google Scholar]
- Turan N, Ghalwash MF, Katari S, Coutifaris C, Obradovic Z, Sapienza C. DNA methylation differences at growth related genes correlate with birth weight: a molecular signature linked to developmental origins of adult disease? BMC Med Genomics. 2012;5:10. doi: 10.1186/1755-8794-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katari S, Turan N, Bibikova M, Erinle O, Chalian R, Foster M, Gaughan JP, Coutifaris C, Sapienza C. DNA methylation and gene expression differences in children conceived in vitro or in vivo. Hum Mol Genet. 2009;18:3769–3778. doi: 10.1093/hmg/ddp319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed N, Choufani S, Wilkins-Haug LE, Koren G, Weksberg R. Comparison of genome-wide and gene-specific DNA methylation between ART and naturally conceived pregnancies. Epigenetics. 2015;10:474–483. doi: 10.4161/15592294.2014.988041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen EC, van Montfoort AP, Dumoulin JC, Evers JL. Epigenetics and the placenta. Hum Reprod Update. 2011;17:397–417. doi: 10.1093/humupd/dmq052. [DOI] [PubMed] [Google Scholar]
- Sunderam S, Kissin DM, Crawford SB, Folger SG, Jamieson DJ, Barfield WD. Centers for Disease Control and Prevention. Assisted reproductive technology surveillance–United States, 2011. MMWR Surveill Summ. 2014;63:1–28. [PubMed] [Google Scholar]
- Conway DA, Liem J, Patel S, Fan KJ, Williams J, III, Pisarska MD. The effect of infertility and assisted reproduction on first-trimester placental and fetal development. Fertil Steril. 2011;95:1801–1804. doi: 10.1016/j.fertnstert.2010.12.010. [DOI] [PubMed] [Google Scholar]
- Basso O, Baird DD. Infertility and preterm delivery, birthweight, and Caesarean section: a study within the Danish National Birth Cohort. Hum Reprod. 2003;18:2478–2484. doi: 10.1093/humrep/deg444. [DOI] [PubMed] [Google Scholar]
- Romundstad LB, Romundstad PR, Sunde A, von During V, Skjaerven R, Gunnell D, Vatten LJ. Effects of technology or maternal factors on perinatal outcome after assisted fertilisation: a population-based cohort study. Lancet. 2008;372:737–743. doi: 10.1016/S0140-6736(08)61041-7. [DOI] [PubMed] [Google Scholar]
- Song S, Ghosh J, Mainigi M, Turan N, Weinerman R, Truongcao M, Coutifaris C, Sapienza C. DNA methylation differences between in vitro- and in vivo-conceived children are associated with ART procedures rather than infertility. Clin Epigenetics. 2015;7:41. doi: 10.1186/s13148-015-0071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra SK, Ratcliffe SJ, Coutifaris C, Molinaro T, Barnhart KT. Ovarian stimulation and low birth weight in newborns conceived through in vitro fertilization. Obstet Gynecol. 2011;118:863–871. doi: 10.1097/AOG.0b013e31822be65f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakovic B, Rakyan V, Ng HK, Manuelpillai U, Dewi C, Wong NC, Morley R, Down T, Beck S, Craig JM, Saffery R. Specific tumour-associated methylation in normal human term placenta and first-trimester cytotrophoblasts. Mol Hum Reprod. 2008;14:547–554. doi: 10.1093/molehr/gan046. [DOI] [PubMed] [Google Scholar]
- Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- Rahnama F, Shafiei F, Gluckman PD, Mitchell MD, Lobie PE. Epigenetic regulation of human trophoblastic cell migration and invasion. Endocrinology. 2006;147:5275–5283. doi: 10.1210/en.2006-0288. [DOI] [PubMed] [Google Scholar]
- Choux C, Carmignac V, Bruno C, Sagot P, Vaiman D, Fauque P. The placenta: phenotypic and epigenetic modifications induced by assisted reproductive technologies throughout pregnancy. Clin Epigenetics. 2015;7:87. doi: 10.1186/s13148-015-0120-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Barlow GM, Cui J, Wang ET, Lee B, Akhlaghpour M, Kroener L, Williams J III, Rotter JI, Chen YI, Goodarzi MO, Pisarska MD. Comparison of genome-wide and gene-specific DNA methylation profiling in first-trimester chrionic villi from pregnancies conceived with infertility treatments Reprod Sci (in press). DOI: 10.1177/1933719116675056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S, Kilburn B, Imudia A, Armant DR, Skafar DF. Estradiol elicits proapoptotic and antiproliferative effects in human trophoblast cells. Biol Reprod. 2015;93:74. doi: 10.1095/biolreprod.115.129114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZK, Liu HY, Fang WN, Yang Y, Wang HM, Peng JP. Insulin-like growth factor binding protein 7 modulates estrogen-induced trophoblast proliferation and invasion in HTR-8 and JEG-3 cells. Cell Biochem Biophys. 2012;63:73–84. doi: 10.1007/s12013-012-9342-5. [DOI] [PubMed] [Google Scholar]
- Chen JZ, Wong MH, Brennecke SP, Keogh RJ. The effects of human chorionic gonadotrophin, progesterone and oestradiol on trophoblast function. Mol Cell Endocrinol. 2011;342:73–80. doi: 10.1016/j.mce.2011.05.034. [DOI] [PubMed] [Google Scholar]
- Chang WL, Yang Q, Zhang H, Lin HY, Zhou Z, Lu X, Zhu C, Xue LQ, Wang H. Role of placenta-specific protein 1 in trophoblast invasion and migration. Reproduction. 2014;148:343–352. doi: 10.1530/REP-14-0052. [DOI] [PubMed] [Google Scholar]
- Shan N, Zhang X, Xiao X, Zhang H, Tong C, Luo X, Chen Y, Liu X, Yin N, Deng Q, Qi H. Laminin alpha4 (LAMA4) expression promotes trophoblast cell invasion, migration, and angiogenesis, and is lowered in preeclamptic placentas. Placenta. 2015;36:809–820. doi: 10.1016/j.placenta.2015.04.008. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George KM, Leonard MW, Roth ME, Lieuw KH, Kioussis D, Grosveld F, Engel JD. Embryonic expression and cloning of the murine GATA-3 gene. Development. 1994;120:2673–2686. doi: 10.1242/dev.120.9.2673. [DOI] [PubMed] [Google Scholar]
- Home P, Ray S, Dutta D, Bronshteyn I, Larson M, Paul S. GATA3 is selectively expressed in the trophectoderm of peri-implantation embryo and directly regulates Cdx2 gene expression. J Biol Chem. 2009;284:28729–28737. doi: 10.1074/jbc.M109.016840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston A, Cox BJ, Nishioka N, Sasaki H, Chea E, Rugg-Gunn P, Guo G, Robson P, Draper JS, Rossant J. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development. 2010;137:395–403. doi: 10.1242/dev.038828. [DOI] [PubMed] [Google Scholar]
- Xu RH, Chen X, Li DS, Li R, Addicks GC, Glennon C, Zwaka TP, Thomson JA. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol. 2002;20:1261–1264. doi: 10.1038/nbt761. [DOI] [PubMed] [Google Scholar]
- Saha B, Home P, Ray S, Larson M, Paul A, Rajendran G, Behr B, Paul S. EED. and KDM6B coordinate the first mammalian cell lineage commitment to ensure embryo implantation. Mol Cell Biol. 2013;33:2691–2705. doi: 10.1128/MCB.00069-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng YK, George KM, Engel JD, Linzer DI. GATA factor activity is required for the trophoblast-specific transcriptional regulation of the mouse placental lactogen I gene. Development. 1994;120:3257–3266. doi: 10.1242/dev.120.11.3257. [DOI] [PubMed] [Google Scholar]
- Ma GT, Roth ME, Groskopf JC, Tsai FY, Orkin SH, Grosveld F, Engel JD, Linzer DI. GATA-2 and GATA-3 regulate trophoblast-specific gene expression in vivo. Development. 1997;124:907–914. doi: 10.1242/dev.124.4.907. [DOI] [PubMed] [Google Scholar]
- Cheng YH, Handwerger S. A placenta-specific enhancer of the human syncytin gene. Biol Reprod. 2005;73:500–509. doi: 10.1095/biolreprod.105.039941. [DOI] [PubMed] [Google Scholar]
- Jackson D, Volpert OV, Bouck N, Linzer DI. Stimulation and inhibition of angiogenesis by placental proliferin and proliferin-related protein. Science. 1994;266:1581–1584. doi: 10.1126/science.7527157. [DOI] [PubMed] [Google Scholar]
- Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, Keith JC, Jr, McCoy JM. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- Yu C, Shen K, Lin M, Chen P, Lin C, Chang GD, Chen H. GCMa regulates the syncytin-mediated trophoblastic fusion. J Biol Chem. 2002;277:50062–50068. doi: 10.1074/jbc.M209316200. [DOI] [PubMed] [Google Scholar]
- Lee X, Keith JC, Jr, Stumm N, Moutsatsos I, McCoy JM, Crum CP, Genest D, Chin D, Ehrenfels C, Pijnenborg R, van Assche FA, Mi S. Downregulation of placental syncytin expression and abnormal protein localization in pre-eclampsia. Placenta. 2001;22:808–812. doi: 10.1053/plac.2001.0722. [DOI] [PubMed] [Google Scholar]
- Pereira N, Reichman DE, Goldschlag DE, Lekovich JP, Rosenwaks Z. Impact of elevated peak estradiol levels during controlled ovarian hyperstimulation on birth weight of term singletons from fresh IVF-ET cycles. J Assist Reprod Genet. 2015;35:527–532. doi: 10.1007/s10815-015-0434-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey RP, Taylor SN, Lu PY, Sartor BM, Rye PH, Pryrak R. Relationship of follicle numbers and estradiol levels to multiple implantation in 3,608 intrauterine insemination cycles. Fertil Steril. 2001;75:69–77. doi: 10.1016/s0015-0282(00)01631-9. [DOI] [PubMed] [Google Scholar]
- Imudia AN, Awonuga AO, Doyle JO, Kaimal AJ, Wright DL, Toth TL, Styer AK. Peak serum estradiol level during controlled ovarian hyperstimulation is associated with increased risk of small for gestational age and preeclampsia in singleton pregnancies after in vitro fertilization. Fertil Steril. 2012;97:1374–1379. doi: 10.1016/j.fertnstert.2012.03.028. [DOI] [PubMed] [Google Scholar]
- Farhi J, Ben-Haroush A, Andrawus N, Pinkas H, Sapir O, Fisch B, Ashkenazi J. High serum oestradiol concentrations in IVF cycles increase the risk of pregnancy complications related to abnormal placentation. Reprod Biomed Online. 2010;21:331–337. doi: 10.1016/j.rbmo.2010.04.022. [DOI] [PubMed] [Google Scholar]
- Royster GD, Krishnamoorthy K, Csokmay JM, Yauger BJ, Chason RJ, DeCherney AH, Wolff EF, Hill MJ. Are intracytoplasmic sperm injection and high serum estradiol compounding risk factors for adverse obstetric outcomes in assisted reproductive technology? Fertil Steril. 2016;106:363–370. doi: 10.1016/j.fertnstert.2016.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans J, Hannan NJ, Edgell TA, Vollenhoven BJ, Lutjen PJ, Osianlis T, Salamonsen LA, Rombauts LJ. Fresh versus frozen embryo transfer: backing clinical decisions with scientific and clinical evidence. Hum Reprod Update. 2014;20:808–821. doi: 10.1093/humupd/dmu027. [DOI] [PubMed] [Google Scholar]
- Burton GJ, Woods AW, Jauniaux E, Kingdom JC. Rheological and physiological consequences of conversion of the maternal spiral arteries for uteroplacental blood flow during human pregnancy. Placenta. 2009;30:473–482. doi: 10.1016/j.placenta.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonagura TW, Pepe GJ, Enders AC, Albrecht ED. Suppression of extravillous trophoblast vascular endothelial growth factor expression and uterine spiral artery invasion by estrogen during early baboon pregnancy. Endocrinology. 2008;149:5078–5087. doi: 10.1210/en.2008-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horcajadas JA, Riesewijk A, Polman J, van Os R, Pellicer A, Mosselman S, Simon C. Effect of controlled ovarian hyperstimulation in IVF on endometrial gene expression profiles. Mol Hum Reprod. 2005;11:195–205. doi: 10.1093/molehr/gah150. [DOI] [PubMed] [Google Scholar]
- Haouzi D, Assou S, Mahmoud K, Hedon B, De Vos J, Dewailly D, Hamamah S. LH/hCGR gene expression in human cumulus cells is linked to the expression of the extracellular matrix modifying gene TNFAIP6 and to serum estradiol levels on day of hCG administration. Hum Reprod. 2009;24:2868–2878. doi: 10.1093/humrep/dep263. [DOI] [PubMed] [Google Scholar]
- Chen JZ, Sheehan PM, Brennecke SP, Keogh RJ. Vessel remodelling, pregnancy hormones and extravillous trophoblast function. Mol Cell Endocrinol. 2012;349:138–144. doi: 10.1016/j.mce.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Brigstock DR. Regulation of angiogenesis and endothelial cell function by connective tissue growth factor (CTGF) and cysteine-rich 61 (CYR61) Angiogenesis. 2002;5:153–165. doi: 10.1023/a:1023823803510. [DOI] [PubMed] [Google Scholar]
- Kipkeew F, Kirsch M, Klein D, Wuelling M, Winterhager E, Gellhaus A. CCN1 (CYR61) and CCN3 (NOV) signaling drives human trophoblast cells into senescence and stimulates migration properties. Cell Adh Migr. 2016;10:163–178. doi: 10.1080/19336918.2016.1139265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Y, Zhang Y, Nie Q, Chen X. CCN1/Cyr61-PI3K/AKT signaling promotes retinal neovascularization in oxygen-induced retinopathy. Int J Mol Med. 2015;36:1507–1518. doi: 10.3892/ijmm.2015.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beristain AG, Zhu H, Leung PC. Regulated expression of ADAMTS-12 in human trophoblastic cells: a role for ADAMTS-12 in epithelial cell invasion? PLoS One. 2011;6:e18473. doi: 10.1371/journal.pone.0018473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Wang YL, Zhu SJ, Luo SY, Piao YS, Zhuang LZ. Expression of matrix metalloproteinase-2, -9, and -14, tissue inhibitors of metalloproteinase-1, and matrix proteins in human placenta during the first trimester. Biol Reprod. 2000;62:988–994. doi: 10.1095/biolreprod62.4.988. [DOI] [PubMed] [Google Scholar]
- Staun-Ram E, Goldman S, Gabarin D, Shalev E. Expression and importance of matrix metalloproteinase 2 and 9 (MMP-2 and -9) in human trophoblast invasion. Reprod Biol Endocrinol. 2004;2:59. doi: 10.1186/1477-7827-2-59. [DOI] [PMC free article] [PubMed] [Google Scholar]