Abstract
Endometriosis is an estrogen-dependent, progesterone-resistant disorder largely derived from retrograde transplantation of menstrual tissue/cells into the pelvis, eliciting an inflammatory response, pelvic pain, and infertility. Eutopic endometrium (within the uterus), giving rise to pelvic disease, displays cycle-dependent transcriptomic, proteomic, and signaling abnormalities, and although its DNA methylation profiles dynamically change across the cycle in healthy women, studies in endometriosis are limited. Herein, we investigated the DNA methylome and associated gene expression in three phases of the cycle in eutopic endometrium of women with severe endometriosis versus controls, matched for ethnicity, medications, smoking, and no recent contraceptive steroid use. Genome-wide DNA methylation and gene expression were coassessed in each sample. Cycle phase was determined by histology, serum hormone levels, and unsupervised principal component and hierarchical cluster analyses of microarray data. Altered endometrial DNA methylation in endometriosis was most prominent in the midsecretory phase (peak progesterone), with disruption of the normal pattern of cycle-dependent DNA methylation changes, including a bias toward methylation of CpG islands, suggesting wide-range abnormalities of the chromatin remodeling machinery in endometriosis. DNA methylation changes were associated with altered gene expression relevant to endometrial function/dysfunction, including cell proliferation, inflammation/immune response, angiogenesis, and steroid hormone response. The data provide insight into epigenetic reprogramming and steroid hormone actions in endometrium contributing to the pathogenesis and pathophysiology of endometriosis.
Keywords: DNA methylation, endometriosis, endometrium, epigenetics
INTRODUCTION
Endometriosis is characterized by endometrial-like tissue outside the uterine cavity, derived largely from abnormal eutopic endometrium (within the uterus) refluxed during menses that implants on pelvic organs because of its enhanced survival, angiogenic, and proliferative potential, and eliciting an inflammatory response and attendant infertility and chronic pelvic pain [1]. Eutopic endometrium of women with disease has markedly different transcriptomic and proteomic profiles, abnormal steroid hormone responses (to estradiol [E2] and progesterone [P4] resistance), aberrant growth factor signaling, and a proinflammatory phenotype, compared with unaffected women [2–10]. Although the pathogenesis of these differences is not well understood, epigenetic abnormalities have been implicated [3].
Epigenetics refers to modifications of gene activity that are not accompanied by changes in gene sequence. Methylation of the 5′ carbon position of cytosines, usually in the context of CpG dinucleotides, is the main epigenetic modification of DNA with essential roles in various biological processes [11]. Up to 20% of genes display DNA methylation patterns in a tissue-specific manner, associated with tissue-specific gene expression [12]. The distribution of DNA methylation across the genome and the position of methylation in the transcriptional unit should be considered when assessing DNA methylation data. In the vertebrate genome, more than half of the genes contain CpG islands (CGIs)—short, CG-rich regions—although the rest of the genome is CpG depleted [13]. At the promoters, the CpG density has a bimodal distribution, although intermediate CpG densities also exist [13]. The position of the methylation with relation to the transcriptional unit impacts its relationship with regulation of gene expression [13]. Most CGIs are unmethylated in somatic cells, with only a minority being methylated. The CGIs near the transcription start sites of active genes are usually unmethylated, and de novo methylation at these CGIs, as shown in cancer and disease, is often associated with gene silencing. We have found in women without endometriosis that endometrial DNA methylation changes across the menstrual cycle, involving both CGIs and non-CGIs, and is associated with changes in gene expression for several loci [14]. Greatest differences were observed between the proliferative (peak E2) and the midsecretory (peak P4) phases, suggesting that epigenetic modifications play a role in normal endometrial steroid hormone responses, which can impact the tissue's functional and physiologic roles of pregnancy establishment and maintenance, tissue homeostasis after menses and regeneration, and endometrial disorders. With regard to endometriosis, aberrant promoter methylation of several genes whose products are critical for the normal endometrial P4 response (e.g., PGRB, HOXA10, ESR2, SF1) has been reported in eutopic endometrium and ectopic disease in the pelvis of women with endometriosis and in animal models of the disease, with resulting resistance to P4 action [15–17].
Herein, we investigated the global DNA methylation of proliferative, early secretory, and midsecretory endometrium of women with severe endometriosis. We also examined the association of DNA methylation changes with gene expression assessed by microarray analysis of the same samples. The results show that the DNA methylome across the cycle differs from that in controls, with most abnormalities observed in the early and midsecretory (progesterone-dominant) phases and involving mostly CGIs. Elucidating hormone-dependent epigenetic states required for normal nuclear receptor function and discovering epigenetic reprogramming events contributing to hormone resistance could have broader implications for other hormone-dependent disorders.
MATERIALS AND METHODS
Sample Collection and Processing
This study was approved by the Committee on Human Research of the University of California, San Francisco (UCSF). Only eutopic endometrial samples were studied. All patients undergoing surgery for endometriosis-related pain and/or infertility gave written informed consent through the UCSF NIH Human Endometrial Tissue and DNA Bank [18] (Supplemental Table S1; Supplemental Data are available online at www.biolreprod.org). Seventeen eutopic endometrial tissue samples were from patients with severe endometriosis (stage IV; n = 4 in proliferative [PE], n = 7 in early secretory [ESE], and n = 6 in midsecretory [MSE] phases). Disease severity was determined according to the revised American Society for Reproductive Medicine (ASRM) staging [19]. Control samples (no endometriosis; n = 6 PE, n = 5 ESE, and n = 5 MSE) were from a previous study [14]. Participants were nonsmokers (two exceptions), were not pregnant, and had had no hormonal treatments within 3 mo prior to sample acquisition. Tissue procurement, processing, and storage were conducted as described previously [14]. Menstrual cycle phase was determined by histology, serum levels of E2 and P4, and/or unsupervised principal component analysis and independent hierarchical clustering analysis of microarray gene expression data [10, 14]. Genomic DNA was extracted and stored as described previously [14].
DNA Methylation Analysis
Genomic DNA bisulfite conversion and quality controls (QCs) were conducted as described previously [14, 20]. Briefly, bisulfite conversion was done using the Zymo EZ-96 DNA methylation Kit (Zymo Research, Irvine, CA) based on the manufacturer's protocol. The QCs included a panel of MethyLight reactions [20] to assess the completeness of the bisulfite conversion and the amount of bisulfite-converted DNA. The quantity and integrity of the sample DNA after bisulfite conversion were determined by a bisulfite-dependent, methylation-independent MethyLight reaction of a multicopy ALU sequence, which is well distributed across the genome [20]. The completeness of bisulfite treatment was assessed using a panel of bisulfite-independent primers with variable bisulfite-dependent probes reflecting various degrees of bisulfite conversion in the sample, including full conversion (100% conversion), no conversion (0% conversion), and partial conversion (50% conversion) [20]. All samples, except two (one ESE and one MSE), passed all QCs and were further assayed by the quantitative Illumina Infinium HumanMethylation27K platform (Illumina, San Diego, CA) based on the manufacturer's specifications, as described previously [14]. The Illumina 27K platform interrogates DNA methylation levels at 27 578 CpG sites corresponding to 14 475 protein-coding and 110 micro-RNA coding genes.
DNA methylation values were scored as β values (ratio of methylated signal over total fluorescent signal), ranging from 0 to 1 (from no methylation to complete methylation, respectively). DNA methylation measurement quality for each probe in each sample was assessed by the detection P value, calculated based on the difference in its signal intensity compared with a set of 16 negative control probes; only probes with detection P values <0.05 (i.e., statistically significant differences from background) were retained for further analysis, and those with P > 0.05 were marked as “missing” and were excluded from further analyses. For each sample, the probe dropout rate was calculated as the percent of total number of probes with “missing” values from the total number of platform probes (27 578). Using stringent criteria, another ESE sample with highest “missing” values (3.5% dropout rate) was excluded from subsequent analyses. The dropout rates for all other samples were <1% of total platform probes (0%–0.8%). Thus, the final endometriosis samples included in the analyses were as follows: n = 4 PE, n = 5 ESE, and n = 5 MSE.
Global DNA Methylation Profile in Endometriosis
Probes with a “missing” value in more than one sample (n = 116) were removed. The remaining 27 462 probes were assessed for global DNA methylation profiles and patterns across the menstrual cycle in endometriosis. Hypermethylated CpG sites are defined as having β value >0.8; hypomethylated CpG sites, β value <0.2; and intermediately methylated CpG sites, 0.2 ≤ β ≤ 0.8. The state of methylation for each probe was determined for each cycle phase and then across other phases to determine if and which probes were hypermethylated, hypomethylated, or intermediately methylated in either all or some of the phases. The CpG sites hypomethylated or hypermethylated in all phases, intermediately methylated, or hypermethylated or hypomethylated in some but not all phases were next compared to the corresponding groups in control endometrium to determine the extent of similarities and differences in profiles, patterns, and loci in disease versus control.
Differentially Methylated CpG Sites in Cycle Phases
Median β values for each probe in each phase were calculated, and median β value differences were used to identify differentially methylated CpG sites between cycle phases. Probes with more than one missing value in each group were excluded from analysis. Cycle phase-specific median β value differences between endometriosis and controls were derived by subtracting the median β value of each probe in a specific phase in controls from the corresponding value in endometriosis, resulting in the following comparisons: PEEndo versus PEControl, ESEEndo versus ESEControl, and MSEEndo versus MSEControl. Also, comparisons across phases of the cycle were conducted for endometriosis endometrial samples (ESEEndo vs. PEEndo, MSEEndo vs. ESEEndo, and MSEEndo vs. PEEndo). Differentially methylated CpG sites in the different phase comparisons in disease were also compared to previously reported changes across the cycle in control endometrium. In addition, all endometriosis samples were compared to all control samples to investigate phase-independent differentially methylated loci between disease and controls. For all comparisons, probes were considered differentially methylated only with Δβ ≥0.136 (detectable differences of 95% confidence) [21]. As previously determined [21], smaller differences in β values may be unreliable because of background noise and platform variability.
RNA Isolation and Gene Expression Microarray
Portions of the same tissue samples were processed for total RNA isolation with DNase treatment using an RNeasy Plus Kit (Valencia, CA), according to the manufacturer's specification. RNA quality was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). RNA sample preparation, quality assessment, and hybridization to Affymetrix HU133 Plus 2.0 gene expression arrays (>38 500 genes; Affymetrix, Santa Clara, CA) was conducted as previously described [14].
Comparison of DNA Methylation and Gene Expression Data
The raw .cel expression data were GCRMA normalized using GeneSpring GX 12.0 software (Agilent Technologies). The transcription unit identifier was used to match corresponding probes from the two platforms using R (http://www.r-project.org). Every DNA methylation probe for a given locus was compared to all transcripts of the corresponding locus. DNA methylation association with gene expression was investigated using the Spearman correlation.
Gene Functional and Biological Classification Analyses
The DAVID database [22] was used to investigate functional classification of genes that were differentially methylated in the same phases of disease versus control and across the cycle in disease. Furthermore, gene lists were cross-referenced to published data of transcriptome differences in endometriosis in women [8, 23–26] and a baboon model of the disease [27]. In addition, an extensive literature review was conducted of the genes' biological characteristics and functions to evaluate genes and/or gene groups potentially important in the disease's pathophysiology.
RESULTS
DNA Methylation Profiles of Eutopic Endometrium in Endometriosis During the Menstrual Cycle
DNA methylation profiles of endometrium of patients with severe endometriosis were investigated for: 1) global profiles and patterns across the cycle; 2) phase-specific differences versus controls; and 3) differences across the cycle in disease.
Global DNA Methylation Profiles and Patterns Across the Menstrual Cycle
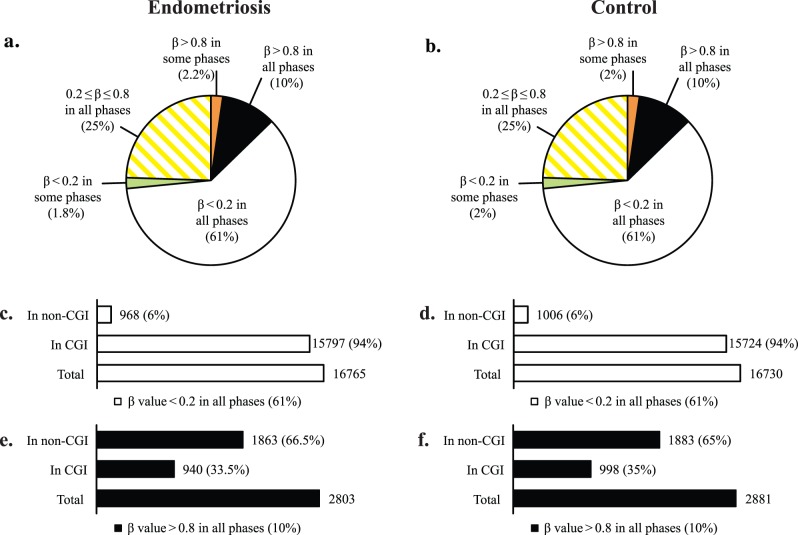
To better understand the global changes in profiles and patterns of DNA methylation across the phases of the cycle in endometriosis, we assessed the frequency of differentially methylated loci, their state of loss or gain of methylation, and their genomic and chromosomal distribution, as well as their association within or outside CGIs. In endometrium from women with endometriosis, most CpG sites (61%; 16 765 CpG sites) were hypomethylated (β value <0.2), whereas a small percentage (10%; 2803 CpG sites) were hypermethylated (β value >0.8) in all three phases (Fig. 1a). The remaining CpG sites (25%; 6795 CpG sites) had intermediate levels of methylation (0.2 ≤ β value ≤ 0.8) in all phases or were hypomethylated (β <0.2: 1.8%; 489 CpG sites) or hypermethylated (β >0.8: 2.2%; 610 CpG sites) in some but not all phases (Fig. 1a). This general pattern is very similar to that of control endometrium [14] (and compare Fig. 1, a with b); however, the genes within each group differ to various extents (see below).
FIG. 1.
Global DNA methylation pattern of eutopic endometrium in endometriosis and its comparison with normal eutopic endometrium based on both methylation levels and CpG site location within or outside CpG islands. a and b) Pie charts depict global distribution of methylation levels in eutopic endometrium of endometriosis (a), and control (b). In endometriosis (similar to control) most (61%; white section) of the platform's interrogated CpG sites are hypomethylated (β-value < 0.2) in all cycle phases, a small percentage (10%; black section) of the platform's CpG sites are hypermethylated (β-value > 0.8) in all cycle phases, about 25% (hatched yellow) have intermediate levels of methylation (0.2 ≤ β-value ≤ 0.8), and 1.8% (green section) are hypomethylated and 2.2% are hypermethylated (orange section) in some but not all phases (2% and 2% in control, respectively). c and d) Distribution of CGIs within the hypomethylated group in endometriosis and control endometrium, respectively: Most of the platform's CpG sites that are hypomethylated (61%; white) consist of nearly all (94%) CGIs, with only 6% non-CGI CpG sites. e and f) Distribution of CGIs within the hypermethylated group in endometriosis and control endometrium: the hypermethylated group in disease consists of 2803 CpG sites (10% of the platform's CpGs), but they are mostly non-CGIs (66.5%), with only 33.5% hypermethylated CGIs, comparable to control.
Association of methylation levels with location within or outside CGIs revealed that the hypomethylated group was nearly entirely (94%) comprised of CpG sites within CGIs, representing 79% of the platform's CGIs (Fig. 1c). This is in contrast to the hypermethylated group, with 33.5% of CpG sites within CGIs (Fig. 1e), representing less than 4% of the platform's CGIs. Again, this general pattern is similar to that in control endometrium [14] (and compare Fig. 1, c with d, and e with f) and similar to somatic tissue global DNA methylation patterns (i.e., hypermethylation involving non-CGI CpG sites and a small fraction of CGIs, with most CGIs remaining hypomethylated) [13, 28].
Most of the loci that are hypomethylated (β value <0.2) or hypermethylated (β value >0.8) in all cycle phases (i.e., phase independent) in disease were also hypomethylated or hypermethylated in all cycle phases in controls (disease independent). The hypomethylated group had 97% (16 538 CpG sites) and the hypermethylated group had 95% (2668 CpG sites) in common with control endometrium (Supplemental Table S2). Therefore, these loci could be regarded as both cycle and disease independent endometrial hypomethylated/hypermethylated loci. However, their endometrial uniqueness remains to be determined upon extensive comparisons to other tissues. The phase- and disease-independent hypomethylated group comprised 60% of the platform loci and therefore includes genes across the genome and with various functions. The hypermethylated group, with a smaller number of loci (9.7% of the platform), included genes in signaling and signal peptides, glycoproteins, defense response, plasma membrane, spermatogenesis, sexual reproduction, and gamete generation, among others (Supplemental Table S2).
The state of methylation differences between disease and control for the remaining loci, and with variable or intermediate methylation levels were further investigated in the context of phase-specific differences in disease versus controls as well as DNA methylation changes across the cycle in disease (see below).
Phase-Specific Differentially Methylated Loci in Endometriosis Versus Controls
Phase-specific DNA methylation profiles were compared (PEEndo vs. PEControl, ESEEndo vs. ESEControl, MSEEndo vs. MSEControl; Fig. 2) and revealed phase-specific differences in disease versus controls in the: 1) DNA methylation profiles, 2) frequencies of gain or loss of methylation, and 3) involvement of CGIs—suggestive of the interaction of the methylome with varying hormonal milieu. These are specifically addressed below.
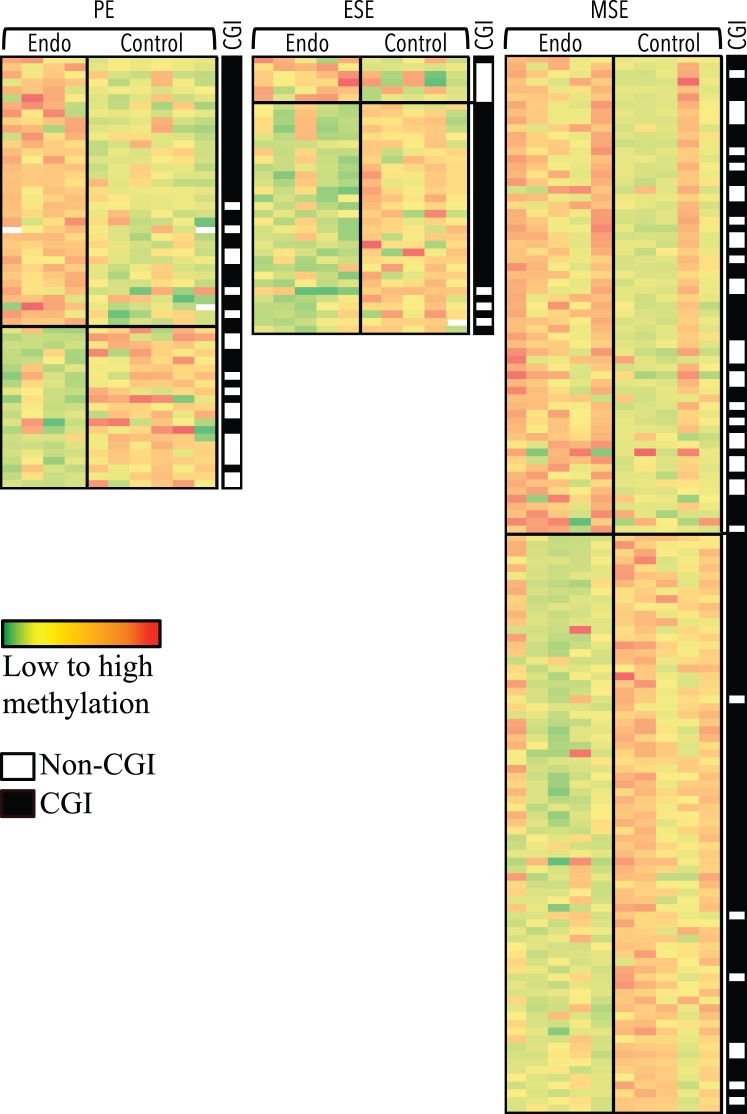
FIG. 2.
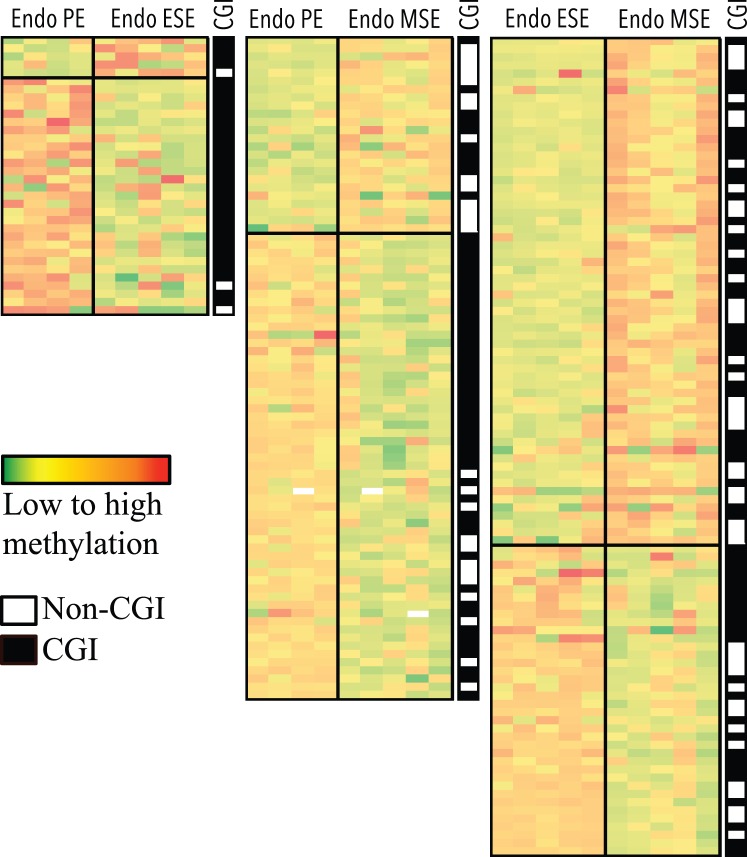
CpG sites differentially methylated by phase in disease versus control eutopic endometrium. Columns represent samples. Rows represent CpG sites. Green to red: low to high methylation. Largest differences between phases of disease (Endo) and control is observed in MSE, followed by PE and ESE. Top section of each heat map represents loci more methylated in disease versus control, and bottom section represents loci less methylated in disease versus control. Location within or outside CGIs for each locus, in each heat map is depicted in the black/white CGI column, with black representing within CGI and white representing outside CGI. Loci in each comparison heat map are unique.
DNA methylation profiles in endometriosis.
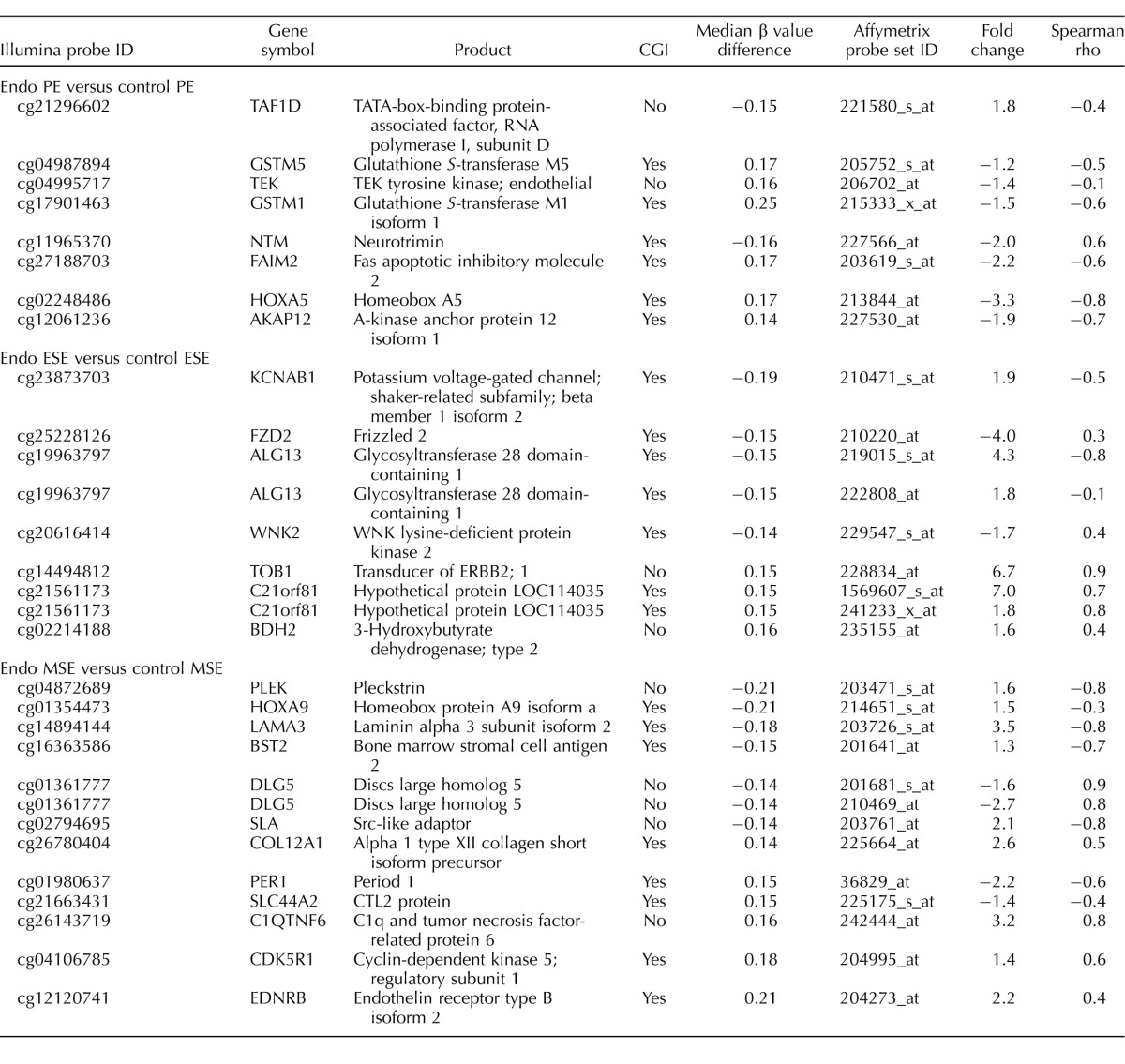
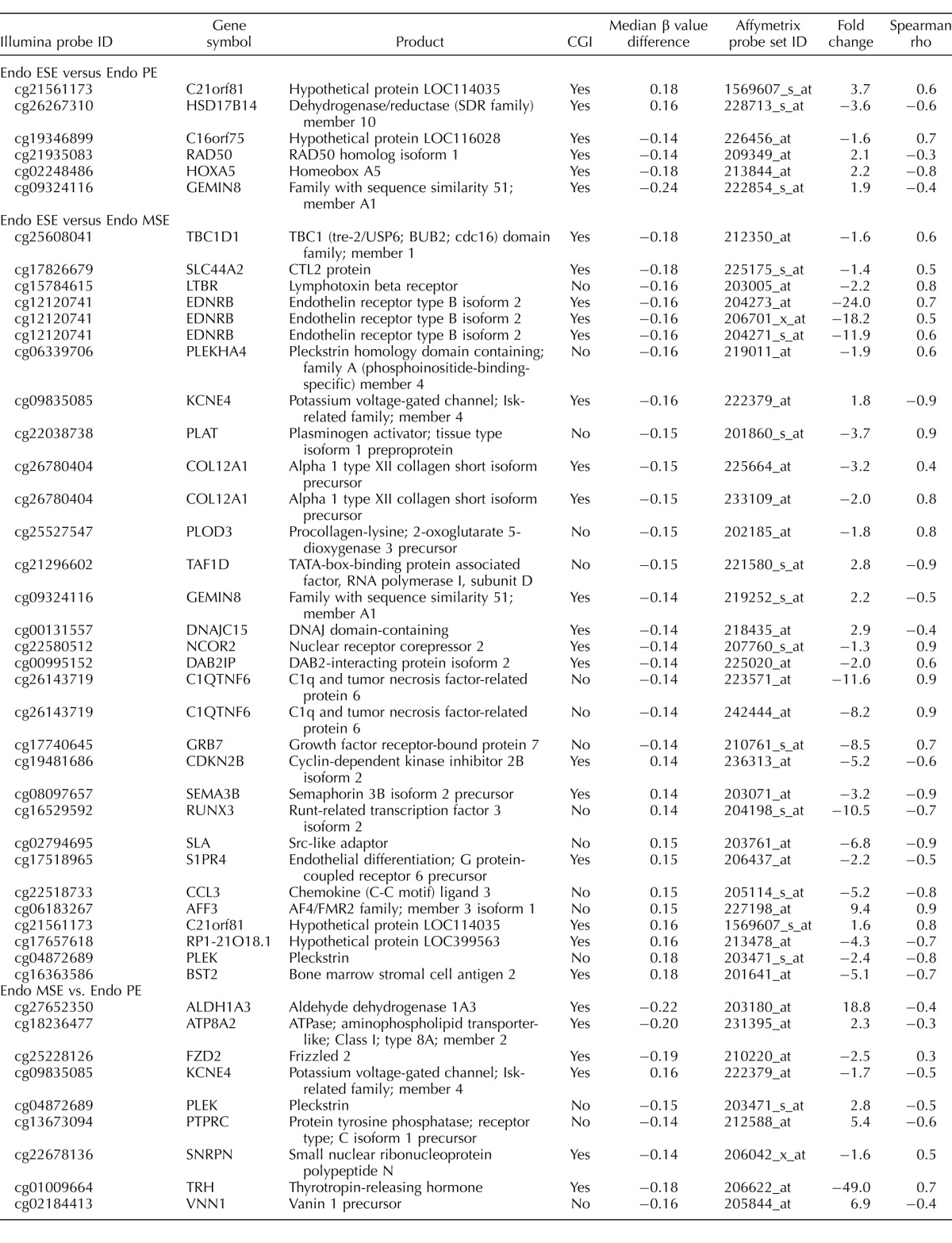
The largest differences in the number of differentially methylated CpG sites were in the P4-dominant midsecretory phase—that is, in MSEEndo versus MSEControl (137 CpG sites, corresponding to 125 loci; Fig. 2, right), followed by PEEndo versus PEControl (58 CpG sites, corresponding to 58 loci; Fig. 2, left) and ESEEndo versus ESEControl (39 CpG sites, corresponding to 36 loci; Fig. 2, middle). A representative selection of phase-specific differentially methylated CpG sites in disease versus control with biological relevance in endometrium and endometriosis is shown in Table 1 (see Supplemental Table S3 for full gene list). Differentially methylated CpG sites in disease versus control were mostly unique in each phase comparison (i.e., phase specific), with only a small number of differentially methylated loci in common between phases (Table 2).
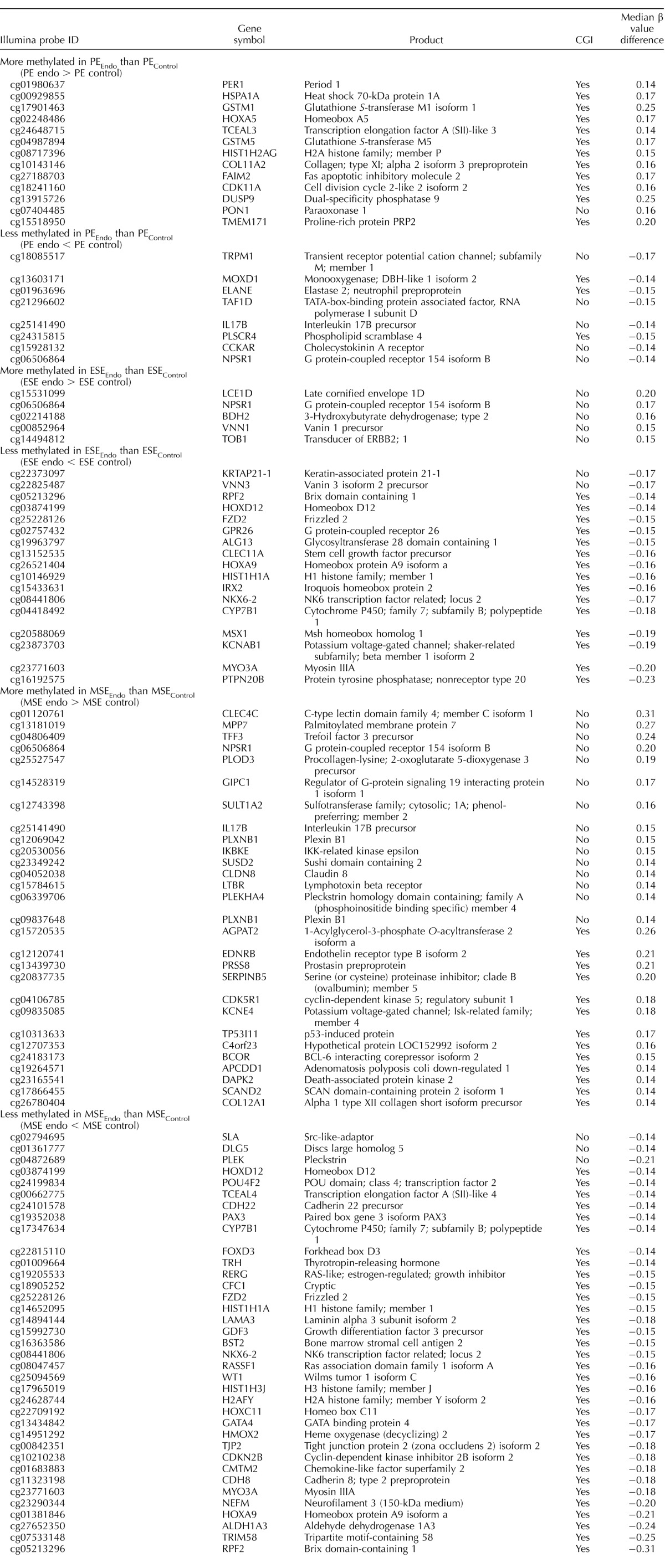
TABLE 1.
CpG sites differentially methylated in disease versus control by phase.
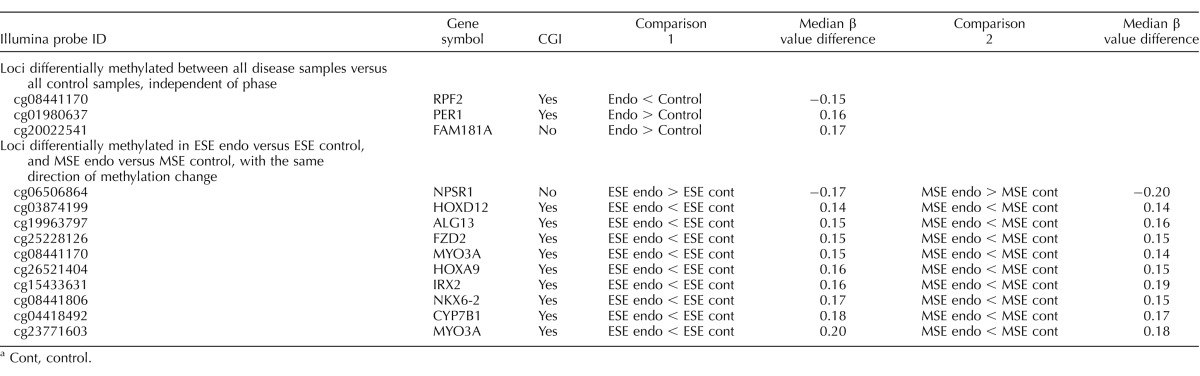
TABLE 2.
Common loci differentially methylated between phases of disease versus control.a
Cont, control.
Gain and loss of methylation.
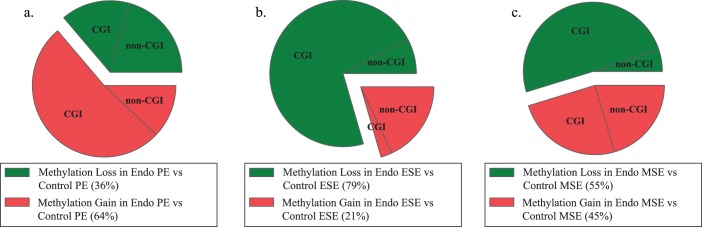
Phase-specific differential methylation in disease versus control included both gain and loss of methylation (Fig. 3), but patterns of methylation gain versus loss were different in the proliferative and secretory phases (Fig. 3). In PEEndo versus PEControl most (64%) of the differentially methylated CpG sites gained methylation in disease (Fig. 3a), whereas in ESEEndo versus ESEControl, most (79%) lost methylation in disease (Fig. 3b). In MSEEndo versus MSEControl the frequencies of gain and loss of methylation in disease were similar (Fig. 3c). Relevant to endometrium and endometriosis, some of the genes more methylated in PEEndo versus PEControl included GSTM1, GSTM5, HOXA5, and FAIM2; those less methylated in PEEndo versus PEControl included TAF1D, IL17B, and TRPM1. Gain of methylation in ESEEndo versus ESEControl included genes such as TOB1, VNN1, BDH2, and NPSR1, and loss of methylation in ESEEndo versus ESEControl included FZD2, HOXA9, HOXD12, ALG13, and CLEC11A. In MSEEndo versus MSEControl, MPP7, COL12A1, KCNE4, and EDNRB showed gain of methylation in disease, and PLEK, DLG5, HOXD12, LAMA3, and HOXA9 showed less methylation compared with controls. A more extensive list of the genes in phase comparisons is listed in Table 1, and the complete list is in Supplemental Table S3.
FIG. 3.
Phase-specific gain or loss of methylation and the association with CGIs in disease versus control. a) Differentially methylated loci divided by gain or loss of methylation in proliferative phase in disease versus control. b) Differentially methylated loci divided by gain or loss of methylation in early secretory phase in disease versus control. c) Differentially methylated loci divided by gain or loss of methylation in midsecretory phase in disease versus control. Endo, endometriosis; green, loci with loss of methylation in disease compared with control; red, loci with gain of methylation in disease compared with control; hatched, loci in CGIs. In the proliferative phase, most of the differentially methylated loci in disease gained methylation, different from ESE and MSE. Also, most of the gain of methylation in PE in disease involved CGIs, whereas most of the loss of methylation in ESE and MSE involved CGIs.
Taken together, these results indicate that: 1) the highest number of differentially methylated loci between disease and nondisease is in MSE, followed by PE and then ESE; and 2) comparing the three phases between disease and control, most differentially methylated CpGs have lower methylation in disease compared with control in the secretory phase—opposite to that of the proliferative phase.
Association with CGIs.
In general, across all phase-specific comparisons in disease versus control (Fig. 2), most differentially methylated loci were within CGIs (black and white bars in Fig. 2). However, there were marked differences in the frequency of CGI and non-CGI CpG sites when considering the loss or gain of methylation in disease versus control (Fig. 3). Loss of methylation in PE (Fig. 3a) involved CGIs and non-CGIs with similar frequency, different from loss of methylation in disease versus controls in MSE and ESE, with most CpG sites within CGIs (>90% in MSE and ESE; Fig. 3, b and c). Also, gain of methylation in disease versus controls (Fig. 3) had a different pattern in each phase. In PE most (81%) were located within CGIs (Fig. 3a), in ESE most (7 of 8; 87.5%) were outside CGIs (Fig. 3b), and in MSE were within or outside CGIs with similar frequencies (Fig. 3c). For example, of the loci mentioned above, GSTM1, GSTM5, HOXA5, and FAIM2 were located within CGIs, and TAF1D, IL17B, and TRPM1 were outside CGIs in PEEndo versus PEControl. In ESEEndo versus ESEControl, TOB1, VNN1, BDH2, and NPSR1 were outside CGIs, and FZD2, HOXA9, HOXD12, ALG13, and CLEC11A were within CGIs. In MSEEndo versus MSEControl, MPP7, PLEK, and DLG5 were located outside CGIs, and COL12A1, KCNE4, EDNRB, HOXD12, LAMA3, and HOXA9 were located inside CGIs.
Phase-independent differentially methylated loci in disease versus control.
The DNA methylome of all disease samples was compared to that of all controls across all phases, and only three CpG sites were differentially methylated, corresponding to three genes: RPF2, more methylated in controls than disease and within a CGI; and PER1 and FAM181A, both more methylated in disease than controls and within and outside a CGI, respectively (Table 2). Because of the changes of DNA methylome throughout the phases of the cycle in both disease and control, this small number was not unanticipated. Several other loci, including HOXA9, HOXD12, IRX2, NKX6-2, CYP7B1, ALG13, MYO3A, and FZD2, all within CGIs, were less methylated in disease versus controls in both ESE and MSE phases (Table 2).
Association of changes in DNA methylation with gene expression.
To investigate if phase-specific differentially methylated CpG sites in disease versus control endometrium were associated with changes in gene expression, we evaluated the gene expression profile of a portion of the same tissue samples by whole-genome microarray analysis (see Supplemental Fig. S1 for patterns of gene expression changes independent of DNA methylation). We also assessed the relationship of DNA methylation levels of each locus to the gene expression of its corresponding transcripts from the microarrays (see Materials and Methods). Because of the nonlinear nature of the relationship of DNA methylation with gene expression [29], the Spearman correlation was used.
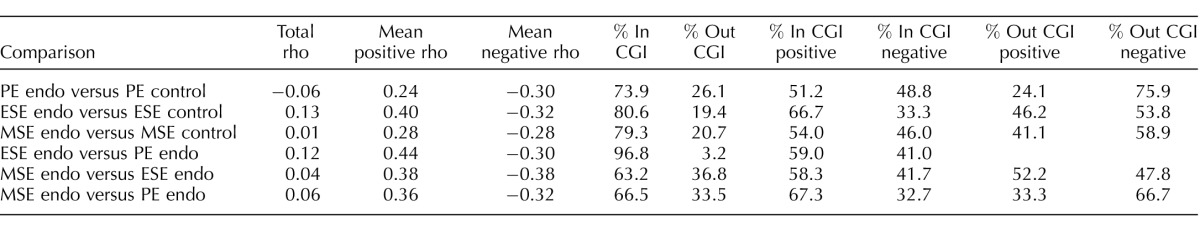
Phase-specific differentially methylated CpG sites between disease and control showed both positive and negative associations with changes in gene expression (Table 3). This is expected because DNA methylation at gene promoters, either at CGIs or non-CGIs, is usually associated with decreased gene expression, whereas gene body methylation is usually associated with increased expression [11, 30, 31]. Also, DNA methylation at CGIs usually shows both negative and positive association with gene expression, although non-CGI methylation is usually negatively associated with gene expression [13]. CpG sites differentially methylated in disease versus controls in PE and MSE showed similar strengths of positive and negative association with gene expression, with stronger negative association in PEEndo versus PEControl (+ρ = 0.24, –ρ = −0.30) and equal positive and negative association in MSEEndo versus MSEControl (+ρ = 0.28, –ρ = −0.28). Further analysis of CpG context showed that for CpG sites located within CGIs, almost equal numbers had positive or negative association with gene expression, whereas most CpG sites located outside CGIs showed negative association with gene expression. However, in ESEEndo versus ESEControl most differentially methylated CpGs were positively associated with gene expression and the positive association was stronger (+ρ = 0.40, –ρ = −0.32; Table 3). CpG island analysis showed that most differentially methylated CpG sites within CGIs were positively associated with gene expression, whereas CpG sites outside CGIs had equal numbers positively or negatively associated with gene expression. These differences between cycle phases suggest complex recruitment of sequence-specific epigenetic machinery in response to different hormonal milieu in the endometrium of endometriosis patients.
TABLE 3.
Relationship between location within or outside CGI with positive or negative association between DNA methylation and gene expression as measured by Spearman rho.
Loci with moderate/high correlation between DNA methylation and gene expression.
Differentially methylated CpG sites showing moderate/strong association with gene expression changes in phase comparisons of disease versus control involved genes in specific categories, including apoptosis regulation; cell adhesion; cell cycle regulation, growth, and differentiation; angiogenesis; transcription regulation and histones; HOX gene family members, and other genes/pathways important in endometrial biology (Table 4, select gene list). For example, TAF1D, not within a CGI, was less methylated in PEEndo versus PEControl and had a negative association with gene expression, meaning it was more expressed in PEEndo versus PEControl (FC = 1.8), whereas HOXA5, within a CGI, was more methylated in PEEndo versus PEControl, with a negative association with gene expression and therefore less expressed in PEEndo versus PEControl (FC = −3.3). ALG13, within a CGI, was less methylated in ESEEndo versus ESEControl, negatively associated with changes in gene expression, and more expressed (FC = 4.3). PLEK, not within a CGI, was less methylated in MSEEndo versus MSEControl, negatively associated with changes in gene expression, and more expressed (FC = 1.6) in MSEEndo versus MSEControl.
TABLE 4.
Select differentially methylated loci associated with gene expression in phases of disease versus control.
Differentially Methylated Loci Across the Cycle in Endometriosis
Phase comparison analyses (i.e., different hormonal milieu, ESEEndo vs. PEEndo, MSEEndo vs. PEEndo, and MSEEndo vs. ESEEndo; Fig. 4) were conducted to investigate hormone dependence/phase differences and potential abnormalities in the setting of endometriosis.
FIG. 4.
CpG sites differentially methylated in eutopic endometrium across the cycle in disease. Columns represent samples. Rows represent loci. Green to red represents low to high methylation. Largest differences between the phases of cycle in disease are observed between MSE versus ESE, with fewer changes between MSE and PE and between ESE and PE. Top portion of each heat map represents loss of methylation in the earlier phases versus the later phase depicted in each comparison heat map, and the bottom portion represents gain of methylation. The location of each locus within or outside CGIs is depicted in the black (within) and white (outside) columns near each heat map. Loci in each comparison heat map are unique. Endo, endometriosis.
Cycle-phase methylation differences.
The largest differences in DNA methylation were between MSEEndo versus ESEEndo (100 CpG sites, corresponding to 96 loci; Fig. 4, right), representing maximum P4 action (MSE) and rising P4 levels (ESE), followed by differences between MSEEndo (maximum P4) versus PEEndo (maximum E2; 81 CpG sites, corresponding to 79 loci; Fig. 4, middle). The fewest differences were observed in ESEEndo versus PEEndo (34 CpG sites, corresponding to 32 loci; Fig. 4, left; Supplemental Table S4, full gene list). For example, KRT19, CDK11A, and KCNC3 were more methylated in PEEndo versus ESEEndo; RUNX3, CDKN2B, and PLEK were more methylated in ESEEndo versus MSEEndo; and CASP8, ALDH1A3, and PTPRC were more methylated in PEEndo versus MSEEndo.
We previously found that normal endometrium displays changes between cycle phases [14]. Herein, the (above) cycle-specific differentially methylated loci were compared with those of normal endometrium. In ESE versus PE, there were no common differentially methylated loci between disease (34 CpG sites) and normal (27 CpG sites) endometrium (Supplemental Table S5). In MSE versus ESE, there was only one locus, FAM181A, in common between differentially methylated loci in disease (100 CpG sites) and normal (22 CpG sites; Supplemental Table S5) endometrium. FAM181A was more methylated in MSE than ESE in both disease and normal endometrium. In MSE versus PE, five loci are in common, but only one locus, FAM181A, showed the same direction of methylation change in both disease and normal, and the other four loci showed the opposite direction of differential methylation in MSE versus PE in disease and normal endometrium (Supplemental Table S5): FAM181A was more methylated in MSE than PE in both disease and normal endometrium; TAF1D and C21orf128 were less methylated in MSE than PE in normal, but more methylated in MSE than PE in disease; and PM20DA and ATP8A2 were more methylated in MSE than PE in normal, with the opposite pattern in disease. Aside from little overlap in cycle-phase changes with normal endometrium, the number of differentially methylated loci in cycle-phase comparisons was larger in disease than in normal. Furthermore, in normal endometrium, the least number of differences were between MSE versus ESE (22 CpG sites), and the most differences were seen in MSE versus PE (peak P4 vs. peak E2; 66 CpG sites) [14], which is different from disease, with most differences in MSE versus ESE (100 CpG sites) and least between PE and ESE (34 CpG sites). These differences, as well as methylation gain/loss pattern and CGI involvements (see below), suggest broad abnormalities in endometrial DNA methylation across the cycle in endometriosis.
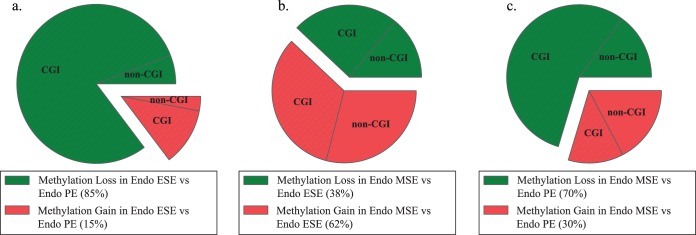
Gain and loss of methylation.
When comparing methylation changes between the proliferative with either of the secretory subphases (Fig. 5, a and c), most CpG sites showed loss of methylation (85% PE > ESE and 70% PE > MSE). However, within the secretory phase, with the highest differences in DNA methylation between ESE and MSE, loss and gain, were observed, with a higher percentage (62%) more methylated in MSE versus ESE (Fig. 5b). In control endometrium, the frequency of gain and loss of methylation is similar in all three phase comparisons [14]. This differs from disease, with more loss than gain of methylation in ESE versus PE (85% loss vs. 15% gain) and MSE versus PE (70% loss, 30% gain), and more gain than loss of methylation in MSE versus ESE (62% gain, 38% loss).
FIG. 5.
Phase comparison analysis of gain or loss of methylation across phases of disease and the association with CGIs. a) Differentially methylated loci divided by gain or loss of methylation in endometriosis (Endo) ESE versus Endo PE. b) Differentially methylated loci divided by gain or loss of methylation in Endo MSE versus Endo ESE. c) Differentially methylated loci divided by gain or loss of methylation in Endo MSE versus Endo PE. Most loci in MSE versus PE and in ESE versus PE (secretory phases vs. proliferative) show loss of methylation compared with PE, with most in CGIs. Within the secretory phase, loci with differential methylation show more gain of methylation in MSE compared with ESE involving both CGIs and non-CGIs. Green, loci with loss of methylation; red, loci with gain of methylation; hatched, loci in CGIs.
Association with CGIs.
In general, most differentially methylated CpG sites across the cycle in endometrium of women with endometriosis were within CGIs (Fig. 4). Differentially methylated CpG sites in ESEEndo versus PEEndo were mostly located in CGIs for loci, with either gain or loss of methylation in ESEEndo compared with PEEndo (Fig. 5a). In MSEEndo versus PEEndo and in MSEEndo versus ESEEndo, most CpG sites with loss of methylation in MSE were in CGIs (Fig. 5, b and c), whereas gain of methylation in MSE compared with ESEEndo or PEEndo occurred with similar frequency within or outside CGIs (Fig. 5, b and c). These observations differ from control endometrium, wherein methylation changes involved both CGIs and non-CGIs with similar frequencies in all comparisons. Also, in controls, CpG sites that had gain of methylation in MSE compared with PE were mostly located within a CGI, and CpG sites that showed loss of methylation in MSE compared with PE were mostly located outside CGIs [14]—opposite the pattern observed in disease herein.
Association of changes in DNA methylation with gene expression.
Differentially methylated CpG sites in ESEEndo versus PEEndo, MSEEndo versus ESEEndo, and MSEEndo versus PEEndo showed positive and negative associations with gene expression changes, with positive association frequency slightly higher in all three comparisons (Table 3). Also, CpG sites within CGIs showed both positive and negative associations with gene expression, with a higher frequency of positive association. For CpG sites located outside CGIs, higher frequency of negative association was observed in MSEEndo versus PEEndo, whereas equal frequency of positive and negative association was observed in MSEEndo versus ESEEndo. There were no CpG sites located outside CGIs in ESEEndo versus PEEndo associated with gene expression (Table 3).
Loci with moderate/high correlation between DNA methylation and gene expression.
Differentially methylated endometrial CpG sites showing moderate/strong association with gene expression changes across the phases in disease involved genes with specific functions, including steroid synthesis and metabolism; regulation of cell growth/differentiation; DNA repair/genomic instability; transcription regulation and histones; and genes/pathways involved in endometrial function and dysfunction (Table 5, select gene list). For example, BST2 was more methylated in ESEEndo versus MSEEndo, was negatively associated with gene expression changes, and was less expressed (FC = −5.1) in ESEEndo versus MSEEndo. ALDH1A3 was less methylated in MSEEndo versus PEEndo, was negatively associated with changes in gene expression, and therefore was more expressed in MSEEndo versus PEEndo (FC = 18.8).
TABLE 5.
Select differentially methylated loci associated with gene expression across the phases in disease.
DISCUSSION
DNA Methylome
Methylation of the carbon-5 position of cytosine, mostly in the context of CpG dinucleotides, is the main epigenetic modification of DNA [11, 13] and is essential for many biological processes [11, 13]. About 10%–20% of genes display DNA methylation patterns in a tissue-specific manner, and these are usually associated with tissue-specific patterns of gene expression [12]. Herein, in severe endometriosis, similar to other noncancerous somatic tissues including normal endometrium [14], most of the CGIs across the genome were hypomethylated, while a small group of CpG sites was hypermethylated, with most of them at non-CGIs. Of interest, most differential DNA methylation across the cycle in women with disease and in the comparison of disease versus controls involved CGIs, whereas changes in controls across the cycle involved both CGIs and non-CGIs, equally [14]. This is significant because CGI methylation is a highly controlled event in somatic tissues, and aberrant CGI methylation at key loci can alter cellular and tissue functions (e.g., tumor suppressor genes in cancer) [13, 32]. Moreover, various chromatin-remodeling components are involved in CGI methylation, and overrepresentation of abnormally methylated CGIs in endometriosis suggests wide-range abnormalities of the chromatin remodeling machinery in endometriosis.
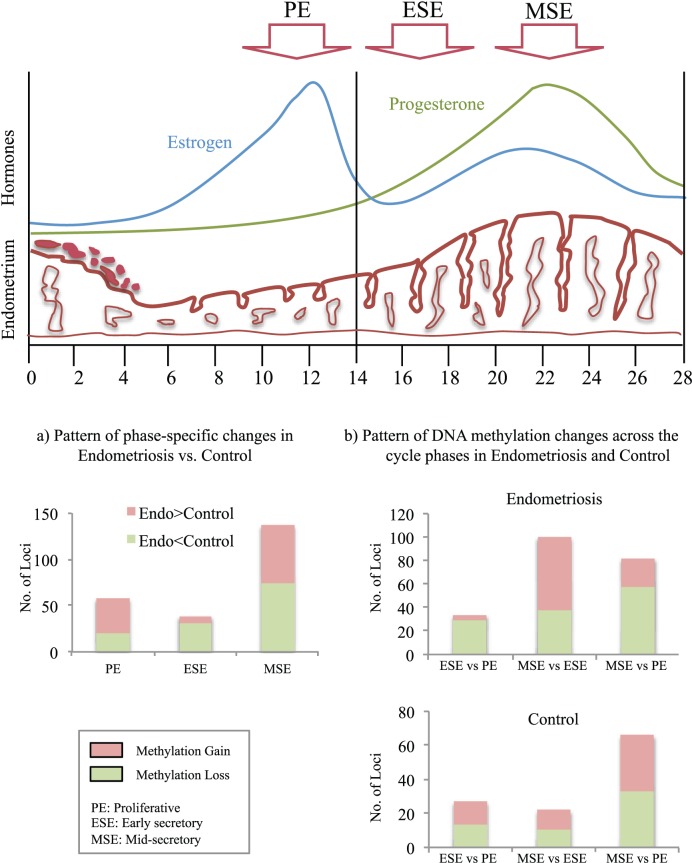
Changes in DNA methylation across the cycle could result from preexisting cis or trans epigenetic differences that, together with vast hormone-induced transcriptome changes, could further alter the methylation status of a given CpG site. Of interest with regard to different hormonal milieu were the findings of the greatest differences between MSEEndo versus MSEControl (peak P4; Fig. 6a), when resistance to P4 action is a hallmark of endometrial dysfunction in endometriosis [8]. Also, the greatest differences in disease were within the secretory phase (MSEEndo vs. ESEEndo; Fig. 6b, Endometriosis). These are different compared with controls, with greatest differences between peak P4 versus peak E2 (MSEControl vs. PEControl) and fewest differences in the secretory phase (MSEControl vs. ESEControl; Fig. 6b, Control), underscoring aberrant P4 response in endometrium of women with endometriosis (Fig. 6 summarizes these findings and in relation to hormonal and endometrial changes across the menstrual cycle).
FIG. 6.
Summary figure. Top portion is a graphic of changes in estrogen, progesterone, and endometrial characteristics across the menstrual cycle depicting the PE, ESE, and MSE phases. a) Summary of phase-specific changes in disease versus control, showing the largest differences between MSE, compared with PE and ESE, with more gain of methylation loci in disease compared with control in PE, and more loss of methylation in disease compared with control in ESE and MSE. b) Comparison of changes in DNA methylation across the cycle and the comparison with control, showing the largest difference in disease between MSE versus ESE different from control, with smallest differences between MSE versus ESE and largest differences in MSE versus PE. Also, changes involved loss and gain of methylation equally in control in all three comparisons, different from disease showing secretory phases (ESE and MSE) involving more loss of methylation compared with PE and more gain of methylation in MSE versus ESE. Endo, endometriosis.
Although we found differences between phases and across the cycle in disease versus controls, the great majority of the platform's CpG sites showed little to no difference from normal, and the differences were not very robust. Furthermore, the number of loci differentially methylated between all disease samples versus all control samples (phase independent, disease dependent) was small, similar to observations recently reported: Saare et al. [33], using endometrial tissue samples from patients with and without endometriosis and a larger DNA methylation platform, also found a small number of differentially methylated regions (DMRs) between disease and nondisease, and also with nonrobust DNA methylation changes. Yamagata et al. [34], using the same DNA methylation platform used herein, observed no separation of genome-wide profiling of cultured endometrial stromal cells from patients with and without endometriosis in cluster or principal component analyses. In our study as well as that of Saare et al. using whole-tissue samples, this could be due to the variable DNA methylation levels across the cycle, or in different cellular components in endometrium, in both disease and controls. Although a small number of disease-specific differentially methylated loci, with nonrobust DNA methylation changes, limits marker discovery potentials, abnormal methylation patterns between phases of disease and across the cycle provide molecular insights for understanding the disease pathophysiology, as discussed below. It is important to note that further functional and protein expression analyses, following this exploratory study, are required to fully assess the role of epigenetic mechanisms in regulation of gene expression in these loci, as well as their potential importance in disease pathogenesis and progression.
Insights into Endometriosis Pathophysiology
Endometriosis is an inflammatory, estrogen-dependent disorder characterized by the presence of endometrial tissue outside the uterine cavity. It is accepted that endometrial cells by retrograde menstruation establish lesions on the peritoneum and escape immune clearance, with attachment, invasion, proliferation, and neoangiogenesis necessary for continued growth and survival of the endometriotic implants. These cellular advantages may be preexisting (hereditary or acquired) in eutopic endometrial cells of endometriosis patients. This is supported by the observation that eutopic endometrium from women with endometriosis differs in transcriptome, proteome, cellular signaling, and biochemical pathways [5, 8, 35] across the menstrual cycle, and shows a persistent proliferative and proinflammatory phenotype, increased cell survival, and aberrant P4 responsiveness in humans as well as in animal models of the disease [8, 10, 36, 37]. These studies have found specific pathways, gene networks, or candidate genes to be affected in disease that are potentially advantageous in disease establishment. Herein, aberrant DNA methylation profiles in eutopic endometrium affect genes with various functions potentially important in the pathogenesis and pathophysiology of the disease—such as cell cycle regulation; inflammation and immune response; steroid hormone response; cell migration; and regulation of gene expression. These include either identical or closely related genes reported in previous transcriptomic studies [8, 24, 27].
Cell cycle, proliferation.
Our previous transcriptome studies in the eutopic endometrium of women with endometriosis versus no disease showed an enrichment of genes involved in cell cycle control and proliferation that are normally up-regulated in the proliferative phase and down-regulated in the secretory phase [8, 10]. These data suggested incomplete transition from the proliferative to the secretory phase, with enhanced cellular survival and aberrant expression of P4-regulated genes in secretory endometrium [8, 10]. Herein, we also observed abnormal DNA methylation associated with expression of genes regulating the cell cycle. For example, CDKN2B encodes a cyclin-dependent kinase inhibitor that binds specifically to CDK4 and CDK6 [38], both of which are abnormally expressed in endometrial stromal fibroblasts (eSFs) from women with endometriosis [25], and regulates cell proliferation by controlling cell cycle G1 progression [38]. The region encompassing CDKN2B and CDKN2A is affected by many signaling pathways and oncogenic agents [39], and it shows frequent copy number alterations, inactivating mutations, homozygous deletions, and aberrant DNA methylation/polycomb-associated silencing in a variety of tumors [38]. CDKN2B was more methylated in ESE compared with MSE in endometriosis and was associated with decreased expression, potentially leading to cell cycle abnormalities in ESE in disease.
Growth factors induce proliferation in steroid-dependent cells of normal endometrium. For example, the EGFR pathway is involved in eutopic and ectopic endometrial growth and differentiation, and eSF proliferation in vitro [8]. EGF is a potent mitogen for eSF in vitro [40], and MIG6, a negative regulator of EGFR signaling, is down-regulated in ESE in the endometrium of women with endometriosis [8]. Also, dysregulation of several genes in the EGFR signaling cascade was previously shown in patients [8] and in the baboon model of endometriosis [27]. Herein, we observed aberrant methylation of several EGF-like factors (TEK and LAMA3) in disease, further supporting disrupted EGF signaling in eutopic endometrium of women with endometriosis. TEK (tyrosine kinase, endothelial) plays a role in angiogenesis [41] and is more methylated in PE in disease versus controls, with decreased expression. LAMA3 (Laminin, alpha 3), an extracellular matrix component of basement membranes promoting epithelial cell attachment, migration, and organization [42], is less methylated and more expressed in MSE in disease versus control.
Inflammation and immune response.
Cytokines and growth factors effectively promote endometrial cell/tissue fragment implantation and evasion of immune-mediated clearance, playing a pivotal role in endometriosis pathophysiology [40]. Activated macrophages are more abundant, and cytokine/chemokine profiles are altered in peritoneal fluid and eutopic endometrium of endometriosis patients [43, 44] and in baboons with disease, including several aberrantly expressed genes involved in T-cell activation [27]. Transcriptomic studies demonstrate a proinflammatory phenotype in women with endometriosis versus controls without any pelvic pathology [10]. Herein, several differentially methylated genes have roles in inflammation and the immune response, including BST2, PLEK, and CCL3. BST2 (bone marrow stromal cell antigen 2, also known as CD317) is an interferon-induced protein. Inflammatory cytokines, such as interleukin 6 (IL6), induce its expression, and it is highly overexpressed in endometrial cancer [45]. It is less methylated and more expressed in MSEEndo than ESEEndo. PLEK (pleckstrin) plays an important role in proinflammatory cytokine activation (tumor necrosis factor α and IL1B) [46]. Its overexpression alters cytoskeletal organization and cell spreading [47]. It is less methylated and more expressed in MSEEndo versus ESEEndo and PEEndo, and also less methylated and more expressed in MSEEndo than MSEControl. CCL3, chemokine (C-C motif) ligand 3 (also known as macrophage inflammatory protein 1α [MIP1α]) is elevated in the peritoneal fluid of women with endometriosis [48]. In vitro studies suggest its role in inducing monocyte and T-cell migration to ectopic sites and increasing eSF invasiveness and endometriosis progression through metalloproteinase induction [48]. High E2 levels (in endometriosis) further induce its expression [48]. It is less methylated and highly upregulated in MSEEndo versus ESEEndo.
Other gene groups potentially important in disease pathophysiology and/or progression.
Differentially methylated genes affecting diverse processes at the cellular and molecular levels include those involved in cell adhesion (CDH8, CCDN8, COL12A1, and COL11A2), cell migration (G1PC1, CCKAR, and TN53), responses to hormones (PRSS8 and TFF3), protein kinase cascades (IKBKE and SLC44A2), and oxidative stress protection (ALDH1A3, CYP2A13, GSTM1, and PLOD3). Furthermore, many differentially methylated genes are involved in transcription function and regulation, and DNA binding and cation binding.
Previously Reported Genes with Abnormal Methylation in Disease
Several genes, including SF-1, ER-β, HOXA10, and PR-B, are differentially methylated in the endometrium of women with endometriosis versus controls [15–17]. CpG sites interrogated in the current platform did not show the same patterns for these genes, because CpG methylation results differ based on the interrogated location. Also, every CpG within a region may not be methylated or may show variable methylation. Aberrant methylation was found when interrogating 29 CpG sites across a 333-bp region containing exon II, intron II, exon III, and intron III (+4085 to +4337) of the SF-1 gene in endometriosis [17]. However, our platform interrogated two locations (at +17 and +127), and both probes are unmethylated in disease and control. In another study, when HOXA10 methylation was interrogated at three regions in disease (−25 to −300 and other fragments within 1.2 kb of the transcription start site), it showed no, partial, or full methylation [16]. Our platform has one probe for HOXA10 at +1461, a different location, and shows intermediate methylation in controls and disease, and no differential methylation, thus accounting for the observed differences. Naqvi et al. [49] used the Illumina platform and reported on 10 of 120 genes differentially methylated, and none of these 10 genes was in common with our data, potentially because of the use of samples from individuals with different severities of disease and/or different cycle phases, or, as they recently found, the distance of disease from the uterus [50]. Saare et al. [33], as mentioned above, have shown similar genome-wide profiles between eutopic endometrium of disease and nondisease as those reported herein. They found the largest differences in the menstrual and late secretory phases (not part of the current study) versus other cycle phases, and although they showed there are changes in methylation across the cycle, they did not find many differences between the proliferative versus the early secretory and midsecretory phases. There are several study differences that could affect the specific outcomes of our study compared with Saare et al. [33], including in their report: a smaller number of control samples in the proliferative and early secretory phases; defining and restricting the search for differential methylation to DMRs (regions smaller than 500 bp between at least three consecutive probes); a larger array platform; self-reporting of cycle date for most samples, which may affect phase determination accuracy; and the availability of samples in the menstrual and late secretory phases.
Strengths and Limitations of the Study
Strengths of our study include well-annotated participant data; strict cycle phase determination using histology, serum E2 and P4 levels, and unsupervised hierarchical clustering and principal component analyses; focus on the most severe phenotype of disease by ASRM criteria [19]; and concomitant gene expression and DNA methylation analysis on each endometrial sample from the same participants. Limitations include the small sample size, cellular heterogeneity, and the cross-sectional study design. However, despite these caveats, we observed the biggest differences in the DNA methylome in the secretory phase—the midsecretory phase showing the largest differences when comparing disease to controls and in midsecretory versus early secretory phases across the cycle in disease—in line with abnormal P4 response in endometriosis. In addition, we found differentially methylated genes associated with gene expression changes relevant to the pathophysiology of endometriosis. However, in the absence of functional analyses, our interpretation of the impact of our findings of DNA methylation differences in both gene expression regulation and disease pathogenesis is limited. Cellular heterogeneity due to the use of whole-tissue samples adds to potential variations in DNA methylation values, meaning that the data are drawn from a wide range of probability distributions (different cell types predicted to have different methylation profiles), which is particularly important with a small sample size and not normally distributed data. We removed samples from further analyses only if they failed our stringent, multistep QCs, and we used a more resilient statistical method to potential outliers that was also suitable for nonparametric data, but we did not cherry-pick the samples for a more homogenous signature. A larger sample set, collected longitudinally, with isolated cellular components and using a comprehensive genome-wide DNA methylation and gene expression platform in future studies, together with protein expression and functional analyses, will extend the understanding of epigenetic abnormalities in the eutopic endometrium of women with endometriosis.
Footnotes
Supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development, National Institutes of Health, National Centers for Translational Research in Reproduction and Infertility (NCTRI) award P50HD055764.
REFERENCES
- Giudice LC. Clinical practice: endometriosis. N Engl J Med. 2010;362:2389–2398. doi: 10.1056/NEJMcp1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulun SE. Endometriosis. N Engl J Med. 2009;360:268–279. doi: 10.1056/NEJMra0804690. [DOI] [PubMed] [Google Scholar]
- Guo SW. Epigenetics of endometriosis. Mol Hum Reprod. 2009;15:587–607. doi: 10.1093/molehr/gap064. [DOI] [PubMed] [Google Scholar]
- Arimoto T, Katagiri T, Oda K, Tsunoda T, Yasugi T, Osuga Y, Yoshikawa H, Nishii O, Yano T, Taketani Y, Nakamura Y. Genome-wide cDNA microarray analysis of gene-expression profiles involved in ovarian endometriosis. Int J Oncol. 2003;22:551–560. [PubMed] [Google Scholar]
- Kao LC, Germeyer A, Tulac S, Lobo S, Yang JP, Taylor RN, Osteen K, Lessey BA, Giudice LC. Expression profiling of endometrium from women with endometriosis reveals candidate genes for disease-based implantation failure and infertility. Endocrinology. 2003;144:2870–2881. doi: 10.1210/en.2003-0043. [DOI] [PubMed] [Google Scholar]
- Matsuzaki S, Canis M, Vaurs-Barriere C, Boespflug-Tanguy O, Dastugue B, Mage G. DNA microarray analysis of gene expression in eutopic endometrium from patients with deep endometriosis using laser capture microdissection. Fertil Steril. 2005;84(suppl 2):1180–1190. doi: 10.1016/j.fertnstert.2005.04.041. [DOI] [PubMed] [Google Scholar]
- Van Langendonckt A, Punyadeera C, Kamps R, Dunselman G, Klein-Hitpass L, Schurgers LJ, Squifflet J, Donnez J, Groothuis P. Identification of novel antigens in blood vessels in rectovaginal endometriosis. Mol Hum Reprod. 2007;13:875–886. doi: 10.1093/molehr/gam073. [DOI] [PubMed] [Google Scholar]
- Burney RO, Talbi S, Hamilton AE, Vo KC, Nyegaard M, Nezhat CR, Lessey BA, Giudice LC. Gene expression analysis of endometrium reveals progesterone resistance and candidate susceptibility genes in women with endometriosis. Endocrinology. 2007;148:3814–3826. doi: 10.1210/en.2006-1692. [DOI] [PubMed] [Google Scholar]
- Zafrakas M, Tarlatzis BC, Streichert T, Pournaropoulos F, Wolfle U, Smeets SJ, Wittek B, Grimbizis G, Brakenhoff RH, Pantel K, Bontis J, Gunes C. Genome-wide microarray gene expression, array-CGH analysis, and telomerase activity in advanced ovarian endometriosis: a high degree of differentiation rather than malignant potential. Int J Mol Med. 2008;21:335–344. [PubMed] [Google Scholar]
- Tamaresis JS, Irwin JC, Goldfien GA, Rabban JT, Burney RO, Nezhat C, DePaolo LV, Giudice LC. Molecular classification of endometriosis and disease stage using high-dimensional genomic data. Endocrinology. 2014;155:4986–4999. doi: 10.1210/en.2014-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- Bergman Y, Cedar H. DNA methylation dynamics in health and disease. Nat Struct Mol Biol. 2013;20:274–281. doi: 10.1038/nsmb.2518. [DOI] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Houshdaran S, Zelenko Z, Irwin JC, Giudice LC. Human endometrial DNA methylome is cycle-dependent and is associated with gene expression regulation. Mol Endocrinol. 2014;28:1118–1135. doi: 10.1210/me.2013-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JL, Zimbardi D, Podgaec S, Amorim RL, Abrao MS, Rainho CA. DNA methylation patterns of steroid receptor genes ESR1, ESR2 and PGR in deep endometriosis compromising the rectum. Int J Mol Med. 2014;33:897–904. doi: 10.3892/ijmm.2014.1637. [DOI] [PubMed] [Google Scholar]
- Wu Y, Halverson G, Basir Z, Strawn E, Yan P, Guo SW. Aberrant methylation at HOXA10 may be responsible for its aberrant expression in the endometrium of patients with endometriosis. Am J Obstet Gynecol. 2005;193:371–380. doi: 10.1016/j.ajog.2005.01.034. [DOI] [PubMed] [Google Scholar]
- Xue Q, Lin Z, Yin P, Milad MP, Cheng YH, Confino E, Reierstad S, Bulun SE. Transcriptional activation of steroidogenic factor-1 by hypomethylation of the 5′ CpG island in endometriosis. J Clin Endocrinol Metab. 2007;92:3261–3267. doi: 10.1210/jc.2007-0494. [DOI] [PubMed] [Google Scholar]
- Sheldon E, Vo KC, McIntire RA, Aghajanova L, Zelenko Z, Irwin JC, Giudice LC. Biobanking human endometrial tissue and blood specimens: standard operating procedure and importance to reproductive biology research and diagnostic development. Fertil Steril. 2011;95:2120–2122. doi: 10.1016/j.fertnstert.2011.01.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Society for Reproductive Medicine. Revised American Society for Reproductive Medicine classification of endometriosis: 1996 Fertil Steril 1997. 67 817 821 [DOI] [PubMed] [Google Scholar]
- Campan M, Weisenberger DJ, Trinh B, Laird PW. MethyLight. Methods Mol Biol. 2009;507:325–337. doi: 10.1007/978-1-59745-522-0_23. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Le J, Barnes B, Saedinia-Melnyk S, Zhou L, Shen R, Gunderson KL. Genome-wide DNA methylation profiling using Infinium(R) assay. Epigenomics. 2009;1:177–200. doi: 10.2217/epi.09.14. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Burney RO, Hamilton AE, Aghajanova L, Vo KC, Nezhat CN, Lessey BA, Giudice LC. MicroRNA expression profiling of eutopic secretory endometrium in women with versus without endometriosis. Mol Hum Reprod. 2009;15:625–631. doi: 10.1093/molehr/gap068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghajanova L, Hamilton A, Kwintkiewicz J, Vo KC, Giudice LC. Steroidogenic enzyme and key decidualization marker dysregulation in endometrial stromal cells from women with versus without endometriosis. Biol Reprod. 2009;80:105–114. doi: 10.1095/biolreprod.108.070300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghajanova L, Horcajadas JA, Weeks JL, Esteban FJ, Nezhat CN, Conti M, Giudice LC. The protein kinase A pathway-regulated transcriptome of endometrial stromal fibroblasts reveals compromised differentiation and persistent proliferative potential in endometriosis. Endocrinology. 2010;151:1341–1355. doi: 10.1210/en.2009-0923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghajanova L, Tatsumi K, Horcajadas JA, Zamah AM, Esteban FJ, Herndon CN, Conti M, Giudice LC. Unique transcriptome, pathways, and networks in the human endometrial fibroblast response to progesterone in endometriosis. Biol Reprod. 2011;84:801–815. doi: 10.1095/biolreprod.110.086181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afshar Y, Hastings J, Roqueiro D, Jeong JW, Giudice LC, Fazleabas AT. Changes in eutopic endometrial gene expression during the progression of experimental endometriosis in the baboon, Papio anubis. Biol Reprod. 2013;88:44. doi: 10.1095/biolreprod.112.104497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straussman R, Nejman D, Roberts D, Steinfeld I, Blum B, Benvenisty N, Simon I, Yakhini Z, Cedar H. Developmental programming of CpG island methylation profiles in the human genome. Nat Struct Mol Biol. 2009;16:564–571. doi: 10.1038/nsmb.1594. [DOI] [PubMed] [Google Scholar]
- Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10:805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Saare M, Modhukur V, Suhorutshenko M, Rajashekar B, Rekker K, Soritsa D, Karro H, Soplepmann P, Soritsa A, Lindgren CM, Rahmioglu N, Drong A, et al. The influence of menstrual cycle and endometriosis on endometrial methylome. Clin Epigenetics. 2016;8:2. doi: 10.1186/s13148-015-0168-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata Y, Nishino K, Takaki E, Sato S, Maekawa R, Nakai A, Sugino N. Genome-wide DNA methylation profiling in cultured eutopic and ectopic endometrial stromal cells. PLoS One. 2014;9:e83612. doi: 10.1371/journal.pone.0083612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May KE, Villar J, Kirtley S, Kennedy SH, Becker CM. Endometrial alterations in endometriosis: a systematic review of putative biomarkers. Hum Reprod Update. 2011;17:637–653. doi: 10.1093/humupd/dmr013. [DOI] [PubMed] [Google Scholar]
- Braundmeier AG, Fazleabas AT. The non-human primate model of endometriosis: research and implications for fecundity. Mol Hum Reprod. 2009;15:577–586. doi: 10.1093/molehr/gap057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazleabas AT. Progesterone resistance in a baboon model of endometriosis. Semin Reprod Med. 2010;28:75–80. doi: 10.1055/s-0029-1242997. [DOI] [PubMed] [Google Scholar]
- Popov N, Gil J. Epigenetic regulation of the INK4b-ARF-INK4a locus: in sickness and in health. Epigenetics. 2010;5:685–690. doi: 10.4161/epi.5.8.12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- Burney RO, Giudice LC. Pathogenesis and pathophysiology of endometriosis. Fertil Steril. 2012;98:511–519. doi: 10.1016/j.fertnstert.2012.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AA, Sandhya VK, Singh P, Parthasarathy D, Kumar A, Advani J, Gattu R, Ranjit DV, Vaidyanathan R, Mathur PP, Prasad TS, Mac Gabhann F, et al. Signaling network map of endothelial TEK tyrosine kinase. J Signal Transduct. 2014;2014:173026. doi: 10.1155/2014/173026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MC, Lee K, Miyashita Y, Carter WG. Targeted disruption of the LAMA3 gene in mice reveals abnormalities in survival and late stage differentiation of epithelial cells. J Cell Biol. 1999;145:1309–1323. doi: 10.1083/jcb.145.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margari KM, Zafiropoulos A, Hatzidaki E, Giannakopoulou C, Arici A, Matalliotakis I. Peritoneal fluid concentrations of beta-chemokines in endometriosis. Eur J Obstet Gynecol Reprod Biol. 2013;169:103–107. doi: 10.1016/j.ejogrb.2013.02.010. [DOI] [PubMed] [Google Scholar]
- Laudanski P, Szamatowicz J, Oniszczuk M. Profiling of peritoneal fluid of women with endometriosis by chemokine protein array. Adv Med Sci. 2006;51:148–152. [PubMed] [Google Scholar]
- Yokoyama T, Enomoto T, Serada S, Morimoto A, Matsuzaki S, Ueda Y, Yoshino K, Fujita M, Kyo S, Iwahori K, Fujimoto M, Kimura T, et al. Plasma membrane proteomics identifies bone marrow stromal antigen 2 as a potential therapeutic target in endometrial cancer. Int J Cancer. 2013;132:472–484. doi: 10.1002/ijc.27679. [DOI] [PubMed] [Google Scholar]
- Ding Y, Kantarci A, Badwey JA, Hasturk H, Malabanan A, Van Dyke TE. Phosphorylation of pleckstrin increases proinflammatory cytokine secretion by mononuclear phagocytes in diabetes mellitus. J Immunol. 2007;179:647–654. doi: 10.4049/jimmunol.179.1.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll RL, Bauman EM, Bennett JS, Abrams CS. Phosphorylated pleckstrin induces cell spreading via an integrin-dependent pathway. J Cell Biol. 2000;150:1461–1466. doi: 10.1083/jcb.150.6.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Wang Y, Zhou WH, Wang L, He YY, Li DJ. Combination of estrogen and dioxin is involved in the pathogenesis of endometriosis by promoting chemokine secretion and invasion of endometrial stromal cells. Hum Reprod. 2008;23:1614–1626. doi: 10.1093/humrep/den125. [DOI] [PubMed] [Google Scholar]
- Naqvi H, Ilagan Y, Krikun G, Taylor HS. Altered genome-wide methylation in endometriosis. Reprod Sci. 2014;21:1237–1243. doi: 10.1177/1933719114532841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi H, Mamillapalli R, Krikun G, Taylor HS. Endometriosis located proximal to or remote from the uterus differentially affects uterine gene expression. Reprod Sci. 2016;23:186–191. doi: 10.1177/1933719115613449. [DOI] [PMC free article] [PubMed] [Google Scholar]