Abstract
Intermediate-filament Nestin and group B1 SOX transcription factors (SOX1/2/3) are often employed as markers for neural primordium, suggesting their regulatory link. We have identified adjacent and essential SOX and POU factor binding sites in the Nestin neural enhancer. The 30-bp sequence of the enhancer including these sites (Nes30) showed a nervous system-specific and SOX-POU-dependent enhancer activity in multimeric forms in transfection assays and was utilized in assessing the specificity of the synergism; combinations of either group B1 or group C SOX (SOX11) with class III POU proved effective. In embryonic day 13.5 mouse spinal cord, Nestin was expressed in the cells with nuclei in the ventricular and subventricular zones. SOX1/2/3 expression was confined to the nuclei of the ventricular zone; SOX11 localized to the nuclei of both subventricular (high-level expression) and intermediate (low-level expression) zones. Class III POU (Brn2) was expressed at high levels, localizing to the nucleus in the ventricular and subventricular zones; moderate expression was observed in the intermediate zone, distributed in the cytoplasm. These data support the model that synergic interactions between group B1/C SOX and class III POU within the nucleus determine Nestin expression. Evidence also suggests that such interactions are involved in the regulation of neural primordial cells.
Specific gene regulation for the determination of neural primordial cells is of high interest for its possible contribution to stem cell biology, where neural tissues are of primary importance. The majority of the neural primordial cells present in the central nervous system (CNS) from embryonic to adult stages (2, 47) have been documented to express the intermediate filament protein Nestin (11, 28, 41). Given that transcriptional regulation determining a cell type is largely reflected by the regulation of genes specific to the cell type, we focused on the regulation of the Nestin gene to better understand gene regulation that is determinative for the neural primordial state.
Previous studies of rat and human Nestin genes indicated that Nestin expression in the CNS, at least during embryonic stages, is largely regulated by a nervous system-specific enhancer located in the 3′ half of the second intron (21, 29, 58, 61). After an analysis of a 257-bp-long Nestin enhancer in the rat, Josephson et al. (21) identified putative binding sites for POU, AP2, and a hormone-responsive element; further assessment of their involvement in Nestin enhancer activity was performed using transgenic mouse embryos. Mutational alterations of the downstream POU factor sites, bound by class III POU factors in vitro, largely eliminated the activity of the enhancer; mutations of other sites altered the domain of the CNS in which the enhancer showed activity (21). These previous results indicated that the activation of the Nestin neural enhancer is fundamentally dependent on the binding of a POU factor to the downstream site and that other nuclear factors may cooperate in establishing this enhancer activity.
POU family transcription factors contain the DNA-binding POU domain, consisting of the POU-specific domain and homeodomain, and are grouped into six classes (4, 19, 27). The class III POU factors, Brn1, Brn2, Brn4, and Oct6 (also called Tst1/SCIP), are widely expressed in the developing CNS, with extensive regional overlap (1, 18), and presumably share redundant functions. Corroborating this notion, single-knockout mice lacking Brn1, Brn2, Brn4, or Oct6 do not have significant neural phenotypes, and only the simultaneous loss of Brn1 and Brn2 in double-knockout mice causes a mild CNS phenotype (4, 30, 46).
Transcription factors expressed in tight association with the neural primordia are group B1 SOX proteins, represented by SOX2 and including SOX1 and SOX3 (8, 37, 40, 45, 51, 57). SOX proteins have similar DNA-binding HMG domains (24, 36, 53) and are classified into groups A through J according to the characteristics of the HMG domain and extra-HMG domain sequences (6).
Early neurogenesis in a variety of vertebrate systems is associated with expression of group B1 Sox genes (31, 37, 45, 49). In the chicken, an enhancer for Sox2 that directly responds to neural induction signals has been identified and has been shown to be conserved among amniotes (49). In Xenopus, forced expression of Sox2, in combination with basic fibroblast growth factor, initiates the neural development of the ectoderm (31). It has also been demonstrated that group B1 SOX activity is involved in maintaining the neural primordia in the embryonic neural tube (7, 15). Taken together, these observations indicate that the expression of group B1 SOX proteins is directly involved in establishing and maintaining neural primordial cell populations.
SOX and POU transcription factors often regulate genes interdependently, forming a complex on the DNA regulatory site (12, 24). In this study, we demonstrate that group B1 and group C SOX proteins interact with POU factors and activate the Nestin neural enhancer. In the embryonic spinal cord, Nestin expression occurs in the cells expressing group B1 SOX and Brn2 in their nuclei (ventricular zone) and in those expressing group C SOX and Brn2 in the nucleus (subventricular zone). In the intermediate zone, the cells coexpress SOX11 and Brn2; however, Brn2 is largely sequestered in the cytoplasm. Thus, the participation of the POU factors in transcriptional regulation is determined by their subcellular localization. This regulated interaction between transcription factors presumably underlies the mechanisms that the determine the neural primordial cell state.
MATERIALS AND METHODS
Immunohistology.
Mouse embryos were fixed with 4% paraformaldehyde-phosphate-buffered saline and cryosectioned at a thickness of 10 to 15 μm. After treatment with 10% serum of the animal species used for secondary antibodies, sections were incubated with primary antibodies and then with Alexa Fluor 488-conjugated anti-rabbit immunoglobulin G and/or Alexa Fluor 568-conjugated anti-mouse immunoglobulin G (Molecular Probes). Observations were made using a Zeiss LSM5 Pascal laser microscope. Anti-SOX2 antibodies (demonstrating no cross-reactivity with SOX1 or SOX3) were produced in a rabbit by injecting the SOX2 N-terminal 19-amino-acid peptide linked to keyhole limpet hemocyanine; antibodies were affinity purified using peptide-conjugated beads. Anti-group B1 SOX antibodies (with demonstrated preference for SOX1 and SOX3 over SOX2 and indicated as anti-SOX1/[2]/3 in the figures) were derived from a rabbit similarly immunized with the SOX3 C-terminal 15 peptides. Anti-SOX11 and anti-Brn2 antibodies were obtained from Santa Cruz Biotechnology. Mouse anti-Nestin monoclonal antibody was obtained from BD Pharmingen.
Embryo electroporation.
Enhancer sequences were inserted into the XhoI site of the ptkEGFP vector plasmid, delivered to the area of the caudal neural fold of a stage 10 chicken embryo, and electroporated as previously described (49).
cDNAs, cell culture, and transfection.
Full-length Nes258 enhancer and octamerized Nes30 sequences were inserted into pδ51LucII (22) reporter vectors. The SOX and POU proteins were expressed by inserting cDNAs into the vector pCMV/SV in transfected cells (22). Both mouse and chicken cDNAs for SOX1, -2, -3, and -11 were used (DDBJ accession numbers for mouse sequences, AB108672, AB108673, AB108674, and AB108675), yielding essentially identical results. However, since data for SOX9, SOX14, and SOX21 were obtained by using chicken cDNAs, the data shown in the figures were generated using chicken SOX cDNAs (22, 23) for consistency. POU factor cDNAs were of mouse origin (16, 34, 35).
Liver cells from a 10-day-old chicken embryo were transfected by a calcium precipitation method (22) with 1.5 μg of DNA containing pδ51LucII reporter (1.3 μg), pCMV/SV effecter (0.1 μg), and reference pMiwZ (52) (0.1 μg). Brain cells from a 7-day-old chicken embryo and other cells, including COS7, 10T1/2, and MNS70 (32), were transfected with 3 μl of Fugene6 (Roche) and 1 μg of DNA containing 0.9 μg of reporter and 0.1 μg of reference vectors. Transfection was done at least in triplicate, and the average luciferase expression is indicated (with standard errors) in the figures.
Analysis of transcripts using reverse transcription (RT)-PCR.
Total RNAs were prepared from MNS70 or mouse embryonic spinal cords using a RiboPure kit (Ambion). cDNAs were synthesized from total RNA using ThermoScript reverse transcriptase (Invitrogen) and oligo(dT) primer and amplified by PCR using ExTaq polymerase (TaKaRa) and the following sets of forward (F) and reverse (R) primers: Nestin, F (CGCTGGAACAGAGATTGGAAGG) and R (GTCTCAAGGGTATTAGGCAAG); Sox2, F (ACCTACAGCATGTCCTACTCG) and R (GGGCAGTGTGCCGTTAATGG); Sox11, F (TCAGCTGCTGAGGCGCTACAG) and R (GAACACCAGGTCGGAGAAGTTCG); Brn2, F (GGCGCCGAGGATGTGTATGG) and R (CATTCTACTTCATTGCCTGGG); Brn4, F (ACAGCTGCCTCGAATCCCTAC) and R (ATGCAGGCTCTGTGTGGAGG); and β-actin (32), F (TGCCCATCTATGAGGGTTACG) and R (TAGAAGCATTTGCGGTGCACG). The PCR products were electrophoresed and stained with SYBR GREEN I (Molecular Probes).
EMSA and protein coprecipitation.
Recombinant proteins, tagged with glutathione S-transferase (GST) or maltose binding protein (MBP) at the N terminus and oligohistidine at the C terminus, were synthesized in Escherichia coli and affinity purified according to the method of Kamachi et al. (25). For electrophoretic mobility shift assays (EMSA), the N-terminal GST tag was removed after purification. Conditions for EMSA and protein coprecipitation assays using immunoblotting were previously described by Kamachi et al. (25). Dissociation constants (Kd) for the complexes formed between the recombinant protein and the probe DNA were estimated based on the following relationships: Kd = (α[total protein input] − [bound protein]) [free probe]/[bound probe], where α indicates the active fraction of the recombinant protein. The concentrations of total protein input and bound protein, as well as those of free and bound probes, were directly determined from experimental data. By total protein input variations, Kd and α were estimated. The Kd for SOX2(3-202) bound to Nes30 sequence was estimated to be 1.0 × 10−8 M (α = 0.75); the Kd for MBP-Brn2(215-445) bound to Nes30 was 3.6 × 10−10 M (α = 0.58).
RESULTS
Mouse Nestin gene has a 258-bp neural enhancer dependent on binding sequences for SOX and POU factors.
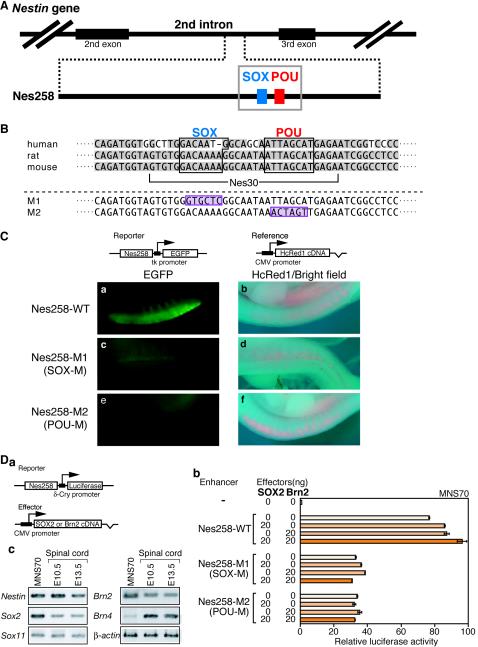
It has been demonstrated in the rat gene that Nestin expression in the neural tube is determined by an enhancer, 257 bp long, located in the second intron; the activity of the enhancer is dependent on the integrity of a small segment, including a POU factor binding site (21). We have found a 258-bp sequence in the mouse Nestin second intron that is almost identical (96% identity) to the rat neural enhancer (Fig. 1A). It is known that the second intron in the human Nestin gene also contains a conserved CNS-active enhancer (29), which is 75% identical to the mouse sequence in the corresponding 256-bp region. The essential binding site for a POU factor in the enhancer is conserved among these animal species (Fig. 1B).
FIG.1.
Involvement of SOX and POU binding sites in activation of the Nestin neural enhancer. (A) Potential SOX2 binding site found adjacent to the POU factor binding site of the Nestin neural enhancer. The 258-bp-long enhancer sequence in the second intron of the mouse Nestin gene, which is similar to rat and human enhancers, is schematically shown. The blue and red boxes represent the potential SOX binding site and the POU factor binding site, respectively. (B) Nucleotide sequence of a portion of the mouse Nestin enhancer in the gray box in panel A, which is aligned with the corresponding sequences of rat and human. Gray shading indicates conserved nucleotide sequences. The M1 mutation disrupts SOX binding motifs (24, 36, 53) and abolishes SOX2 binding (Fig. 4), while mutation M2 has been shown to eliminate Nestin neural enhancer activity (21). The mutated sequences are shaded in purple. The Nes30 core sequence used in the following analyses is also indicated. (C) Activity of the mouse Nestin neural enhancer (Nes258) and its M1-M2 mutant forms in the electroporated spinal cord of a chicken embryo. Expression vectors for enhancer-dependent EGFP (Reporter) and HcRed1 (Reference) are indicated on top. Electroporation was performed at stage 10, and observations were made after 48 h at stage 21. The HcRed1 reference vector was also included in electroporation. (a, c, and e) EGFP fluorescence in the spinal cord representing activity of Nestin enhancer. (a) Wild-type (WT) enhancer was strongly active in the spinal cord. (c and e) The mutation-containing enhancer, either M1 (c) or M2 (e), was totally inactive. (b, d, and f) HcRed1 fluorescence superimposed on bright-field images. (D) Activity of Nes258 enhancer and its dependence on the SOX and POU binding sites in the neural stem cell line MNS70 and effect of exogenous SOX2 and/or Brn2 expression on the activity. (a) Structure of Nes258 enhancer-bearing luciferase vector and expression vector for SOX2 or Brn2 effectors. CMV, cytomegalovirus. (b) Nes258 enhancer-bearing luciferase vector was transfected into MNS70 neural stem cells, displaying 75-fold activation compared to the enhancer-free vector. Exogenous expression of SOX2 or Brn2 singly augmented the level of activation to roughly 85-fold, and that of SOX2 and Brn2 together activated luciferase expression nearly 100-fold. Using mutant enhancers with defective SOX (M1) or POU (M2) binding sites, activation of the luciferase reporter fell in the range of 30- to 40-fold, and this was not augmented by exogenous SOX2 and/or Brn2 any further. The luciferase activity level generated by the reporter in the absence of enhancer was taken as 1. (b) Expression of Nestin, Sox, and Pou factor genes in the MNS70 line demonstrated by RT-PCR, in comparison with the embryonic spinal cord.
Considering the association of group B1 SOX and Nestin expression in the neural primordial cells, we searched for a potential SOX binding site in the 258-bp mouse Nestin enhancer (Fig. 1B). A candidate SOX binding sequence, GACAAAA, was found immediately upstream of the POU binding site and is perfectly conserved in the rat Nestin enhancer. In the human Nestin enhancer sequence, GACAAAA is altered to GACAATG; however, the latter sequence also provides a good binding site for group B1 SOX (24).
To confirm that the 258-bp sequence of the mouse Nestin gene has nervous system-specific enhancer activity, and to evaluate the potential contributions of SOX and POU factors to this activity, we took advantage of electroporation of chicken embryos. Wild-type or mutated 258-bp sequences were placed upstream of the herpes simplex virus thymidine kinase (tk) promoter of the tk-EGFP gene (49), and chicken embryos at stage 10 were electroporated at the caudal neural plate with this gene and an HcRed1 expression vector. The embryos were observed after 48 h at stage 21 (Fig. 1C). When the wild-type sequence was employed, the EGFP reporter gene was strongly expressed in the spinal cord (Fig. 1C, a), while coelectroporated HcRed1 was expressed in the spinal ganglia and epidermis as well (Fig. 1C). When either of the mutations, M1 or M2 (disrupting the binding sites for SOX and POU, respectively), was introduced into the 258-bp sequence, enhancer activity was attenuated (Fig. 1C, c and e). These observations indicate that the 258-bp sequence has nervous system-specific enhancer activity and that the integrity of both binding sites is essential for the enhancer.
To further confirm the dependence of the Nestin enhancer on SOX and POU binding sites, we tested the activity of the enhancer in the neural stem cell line MNS70 (32). Placing the wild-type Nestin enhancer upstream of a reporter luciferase gene (Fig. a) activated luciferase expression in transfected MNS70 cells 75-fold (Fig. 1D, b), indicating that endogenous transcription factors are sufficient for activation of this enhancer. Mutations M1 and M2, disrupting the binding sites for SOX and POU factors, respectively, significantly reduced the activity to a 30- to 40-fold activation level, thus confirming involvement of these factors in generating the activity. Exogenous expression of SOX2 or Brn2 augmented the activation level by the wild-type enhancer to 85-fold, and their simultaneous expression increased it further to 100-fold activation, indicating that these SOX and POU factors synergistically activate the Nes258 enhancer and that MNS70 cells contain nearly saturating levels of these or analogous factors. Indeed, MNS70 cells expressed Sox2 (group B1), Sox11 (group C), and Brn2/4 (class III) at a level even higher (except for Brn4) than mouse spinal cords of embryonic day 10.5 (E10.5) and E13.5 embryos, as assessed by RT-PCR (Fig. 1D, c). However, using mutant enhancers carrying either mutation M1 or M2, exogenous supply of SOX2 and/or Brn2 had no effect on the expression of luciferase reporter (Fig. 1D, b), indicating that these transcription factors act only in combination in activation of the Nes258 enhancer.
The Nestin core enhancer, composed of SOX and POU binding sites, can be activated by the synergistic action of SOX2 and Brn2.
Many cell-specific enhancers are composed of binding sites for cell-specific nuclear factors and those for factors with broader specificity (reference 14 and references therein). The core regulatory region of the enhancers provides binding sites for specific factors (25), generally essential for the entire enhancer activity, and the multimeric form demonstrates its intrinsic enhancer activity (reference 14 and references therein).
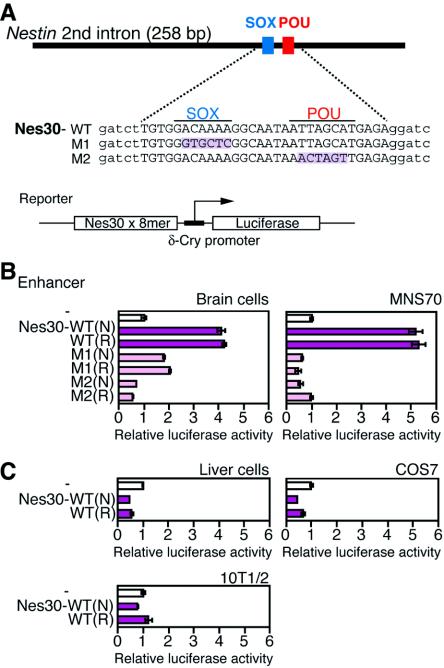
In the case of the Nestin enhancer, the SOX-POU binding region may have the property of the Nestin enhancer core sequence; the potential enhancer activity may be demonstrated by its multimerization. Thus, the 30-bp region (Nes30) of the Nestin enhancer containing SOX and POU factor binding sites (Fig. 1B and 2A) was octamerized and placed upstream of the δ51LucII reporter gene (22) (Fig. 2A). The constructs were then transfected into different cultured cells to determine whether SOX-POU-dependent enhancer activity is detectable (Fig. 2B and C).
FIG. 2.
Activities of the 30-bp core enhancer (Nes30) in several cultured cells. (A) Structures of the reporter plasmid (left) and wild-type (WT), M1 (SOX2 site mutant), and M2 (POU site mutant) Nes30 sequences. Altered nucleotides are shaded. Lowercase letters indicate linker sequences. An octamerized Nes30 sequence was inserted into the pδ51LucII reporter plasmid (22) in both normal (N) and reversed (R) orientations relative to the direction of transcription. (B) Activities of Nes30-WT, M1, and M2 in chicken primary brain cells and MNS70 neural stem cells. C. Activities of Nes30-WT in chicken primary liver cells, COS7 (kidney) cells, and 10T1/2 fibroblasts. The luciferase activity level generated by the reporter in the absence of enhancer was taken as 1.
The octamerized Nes30 sequence [Nes30(8-mer)] significantly activated luciferase expression in primary brain cells; however, this activation was absent in liver cells (Fig. 2B and C). Among the cell lines tested, the neural stem cell line MNS70 (32) supported enhancer activity of the Nes30(8-mer) sequence. In nonneural COS7 or 10T1/2 cells, the Nes30 sequence showed no activity (Fig. 2B and C). The activation of luciferase expression by Nes30(8-mer) in the brain cells and in the neural stem cells was compromised when the sequence had either the M1 or M2 mutation (Fig. 2B). These results indicate that the Nes30 sequence has nervous system-specific core enhancer activity and that this activity presumably depends on simultaneous binding of SOX and POU factors.
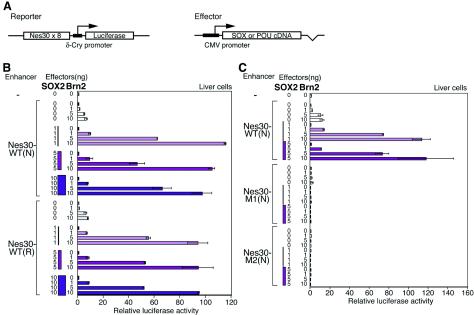
To establish the contribution of SOX and POU transcription factors in the regulation of the Nes30 core enhancer, we exogenously expressed SOX2 and Brn2 (class III POU) in liver cells, where the core enhancer has no activity (Fig. 2). When SOX2 was expressed alone, the cotransfected Nes30(8mer)-carrying luciferase gene was not activated, whereas singular Brn2 expression caused only very weak activation (5- to 10-fold). By contrast, when SOX2 and Brn2 were coexpressed, there was dramatic activation of the cotransfected luciferase gene (90- to 100-fold) (Fig. 3B). This strong activation brought about by the coexpression of SOX2 and Brn2 was extinguished when mutations M1 (SOX2 binding site) or M2 (POU binding site) were introduced into the Nes30 sequence (Fig. 3C). Thus, the Nes30 enhancer is activated by the synergistic action of SOX2 and Brn2 bound to the sequence. (Low activation of luciferase expression by exogenous Brn2 alone, in spite of its total dependence on the integrity of the SOX binding site, is now ascribed to low-level endogenous expression of SOX11 in liver cells [data not shown], which was later found to synergize with Brn2 in activation of the Nes30 enhancer [see below].)
FIG. 3.
Synergistic activation of the Nes30 enhancer by exogenous SOX2 and Brn2. (A) Schematic structure of the reporter and SOX2/Brn2-expressing effecter plasmids. The effecter plasmids had full-length cDNAs of SOX or POU factors, which are transcribed from the cytomegalovirus (CMV) promoter. (B) Activation of the Nes30 enhancer by SOX2 and Brn2. Various amounts of effecter plasmids designed to express SOX2 or Brn2, and the reporter plasmids, were transfected into liver cells. (N) and (R) indicate the orientation of the Nes30 enhancer in the reporter plasmid (normal and reversed, respectively). (C) Effect of mutations in the SOX binding site or POU factor binding site on Nes30 enhancer activity. The enhancer was in N orientation. The error bars indicate standard errors.
Binding of SOX2 and Brn2 to the Nes30 sequence in vitro.
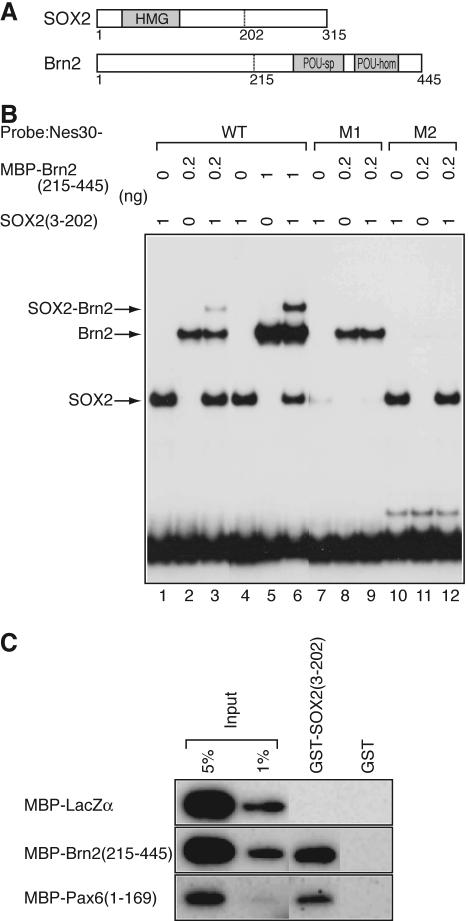
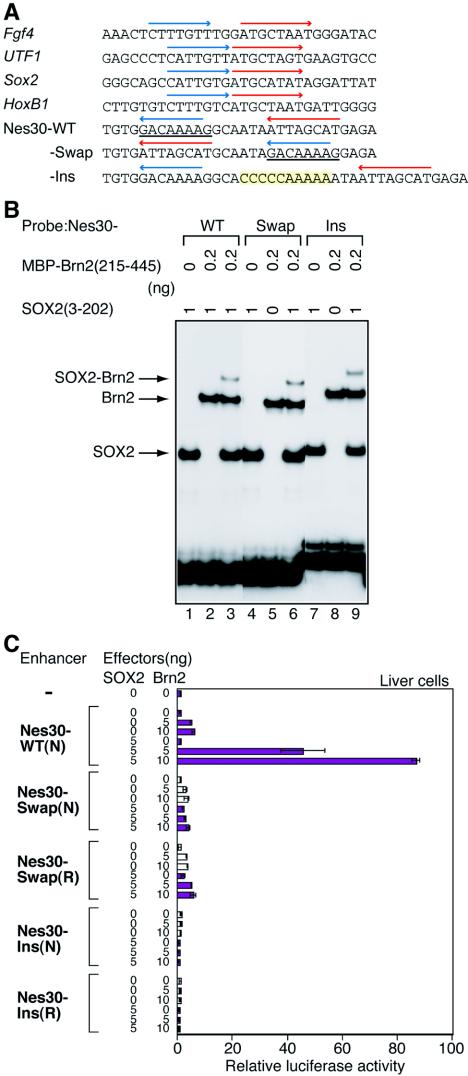
To confirm that SOX2 and Brn2 bind to the Nes30 sequence together, electrophoretic mobility shift assays (EMSA) were performed using the Nes30 sequence probe.
The region from amino acids 3 to 202 of SOX2, containing the HMG domain, and the region from amino acids 215 to 445 of Brn2, containing the POU domain, were both tagged with the MBP, synthesized (Fig. 4A), and used in EMSA (Fig. 4B). The individual protein domains bound efficiently to the probe (Fig. 4B, lanes 1, 2, 4, and 5), and the combination of these proteins yielded a new, slowly migrating band, indicating simultaneous binding of the two proteins (Fig. 4B, lane 3). With the increase of MBP-Brn2(215-445), the slowly migrating band was intensified in a predictable manner from noncooperative, stoichiometric binding of these proteins to the probe (Fig. 4B, lane 6). A dissociation constant of 1.0 × 10−8 M was estimated for binding of SOX2(3-202) to the Nes30 probe, and 3.6 × 10−10 M for binding of Brn2(215-445) to the probe, as described in Materials and Methods (data not shown). Mutation M1 eliminated binding of SOX2(3-202), while mutation M2 destroyed binding of MBP-Brn2(215-445) (Fig. 4B, lanes 7 to 12).
FIG. 4.
Binding of SOX2 and Brn2 to Nes30 DNA and their protein-protein interaction. (A) Recombinant SOX2(3-202) and Brn2(215-445) were synthesized as fusion proteins with GST and MBP, respectively. (B) Binding of SOX2(3-202) and MBP-Brn2(215-445) with GST removed to wild-type (WT) or mutant (M1 and M2) forms (Fig. 1B) of the Nes30 sequence in vitro, as assessed by EMSA. The recombinant proteins bind to the Nes30-WT sequence efficiently, either alone (lanes 1 and 2) or in combination (lane 3). In lane 3, a slowly migrating band representing a SOX2(3-202)/MBP-Brn2(215-445)/probe ternary complex is observed. A larger amount of Brn2(215-445) increased band intensity, and the formation of the ternary complex was stoichiometric (lanes 4 to 6). Mutation M1 attenuated binding of SOX2(3-202) (lanes 7 and 9), while mutation M2 disrupted the binding of Brn2(215-445) (lanes 11 and 12). (C) Protein interaction of SOX2 and Brn2 without Nes30 DNA. MBP-tagged proteins, LacZα, Brn2(215-445), and Pax6(1-169), were incubated with GST-SOX2(3-202) or GST and coprecipitated with glutathione-Sepharose, and the precipitated MBP fusion proteins were detected by immunoblotting using anti-MBP antibodies.
Given that SOX2 and Brn2 act synergistically in the activation of the Nes30 core enhancer, it is possible that these proteins interact directly without the intervention of DNA. Such direct interaction between SOX2 and Oct3/4, as well as that of SOX2 and Pax6, has been demonstrated (3, 25). To test this, GST-tagged SOX2(3-202) protein was attached to glutathione-Sepharose beads and examined to determine if MBP-tagged Brn2(215-445) coprecipitated (Fig. 4C). Attempts at synthesizing full-length proteins were hampered by extensive proteolysis during sample preparation, but previous reports indicated protein interaction sites close to HMG and POU domains (3). GST-SOX2(3-202) coprecipitated with MBP-Brn2(215-445) with an efficiency comparable to that of MBP-Pax6(1-169), which is known to bind SOX2(3-202) (25). No such binding was detected when GST alone, or MBP linked to an unrelated protein (MBP-LacZα), was employed. The observations indicate that SOX2 and Brn2 proteins interact without Nes30 DNA. More extensive protein-protein interactions might take place if full-length proteins are involved. Thus, protein-protein interaction may participate in the synergistic action of SOX2 and Brn2 in activating the enhancer.
Comparison with other cases of SOX2-POU interaction on regulatory DNA sequences, mainly involving Oct3/4, indicates that the arrangement of binding sites of SOX2 and Brn2 in the Nes30 sequence is unique, and individual binding sites are inverted relative to other cases (3, 13, 33, 48, 59) (Fig. 5A). In addition, the distance between the SOX and POU binding sites is larger in the Nestin enhancer (6 bp between the binding sites compared to 3 [Fgf4] or 0 [all other cases] bp). To see whether this arrangement of binding sites is essential for Nes30 enhancer activity, the binding site sequences were swapped (Nes30-Swap), or the space between the binding sites was increased by the insertion of 10 bp (Nes30-Ins). The alterations of the sequence did not affect binding of SOX2 and Brn2 (Fig. 5B). With these rearrangements, however, the Nes30 sequence lost the enhancer activity in SOX2/Brn2-transfected liver cells (Fig. 5C). The Nes30-Swap sequence is analogous to one of the Fgf4 enhancer mutants lacking the activity due to increased spacing between the SOX2 and POU binding sites (3). Thus, the particular orientation and spacing between the SOX2 and POU binding sites found in the Nes30 sequence represents a new functional arrangement of binding sites activated by SOX2 and a POU factor.
FIG. 5.
Effects of the arrangement of SOX and POU factor binding sites on the enhancer activity of Nes30. (A) Nucleotide sequences and arrangements of SOX2 and Oct3/4 binding sites found in the enhancers of Fgf4 (3), UTF1 (33), Sox2 (48), and HoxB1 (13) and comparison of those with SOX2 and POU binding sites in the Nes30 sequence. SOX2 and POU binding sequences are indicated by blue and red arrows, respectively. The orientations of both SOX2 and POU sites in the Nes30 sequence are in reverse, compared with the four enhancers indicated above. Two mutant sequences of Nes30 for the arrangement of SOX2 and POU binding sites were prepared. The Nes30-Swap mutant enhancer had the binding sequences in inverted orientations, where the inverted SOX binding sequence is underlined. The Nes30-Ins mutant enhancer had 10 bp inserted between SOX and POU factorbinding sites. (B) Binding of SOX2(3-202) and Brn2(215-445) to the Swap and Ins mutant sequences of Nes30. SOX2(3-202) (lanes 1, 4, and 7), MBP-Brn2(215-445) (lanes 2, 5, and 8), and the combination of SOX2(3-202) and MBP-Brn2(215-445) (lanes 3, 6, and 9) were mixed with the wild-type (WT) (lanes 1 to 3), Swap mutant (lanes 4 to 6), and Ins mutant (lanes 7 to 9). Nes30 probes and their binding were analyzed by EMSA. (C) Effects of Swap and Ins mutations on enhancer activity of Nes30 in transfected liver cells. Various amounts of plasmids to express SOX2 or Brn2 were transfected to liver cells, together with a luciferase-encoding reporter plasmid carrying an octamerized enhancer sequence. (N) and (R) indicate the orientation of the Nes30 enhancer in the reporter plasmid (normal and reversed, respectively). The Swap and Ins mutations both inactivated enhancer activity. The error bars indicate standard errors.
Differential regulation of the Nestin enhancer by various combinations of SOX and POU factors.
In addition to SOX2 and Brn2, various SOX and POU factors are expressed in the embryonic CNS, as assessed by in situ hybridization of the gene transcripts. All group B1 Sox genes are expressed in the ventricular zone, while Sox2 is additionally expressed in the roof and floor plates (37, 40, 50, 51, 57). Of the group B2 Sox genes Sox14 and Sox21, the latter is expressed primarily in the ventricular zone (50). Additionally, the group C Sox genes Sox11, Sox4, and Sox12 are strongly expressed in the subventricular and intermediate zones (9, 17, 20, 26, 51). The SOX9 (group E) protein has been shown to be expressed in the nuclei of the ventricular zone and glial precursors (44).
Among POU family transcription factor genes, those coding for class III factors, Brn1/Brn2/Brn4/Oct6, are expressed in the ventricular through intermediate zones in overlapping but slightly different anteroposterior domains of the CNS (1, 4, 18). The class V POU Oct3 (or Oct4) is expressed throughout the neural plate in very early embryos (38, 42). The class II POU protein gene Oct2 is expressed in a regionally restricted fashion (27), while the class VI POU gene Brn5 (Emb/Cns-1) is expressed throughout the CNS (10).
To learn how these SOX and POU factors may cooperate in regulating Nestin expression, analogous to SOX2 and Brn2, various combinations of SOX and POU factors were expressed in liver cells and their effects on the activity of the Nes30(8-mer) enhancer was assessed (Fig. 6).
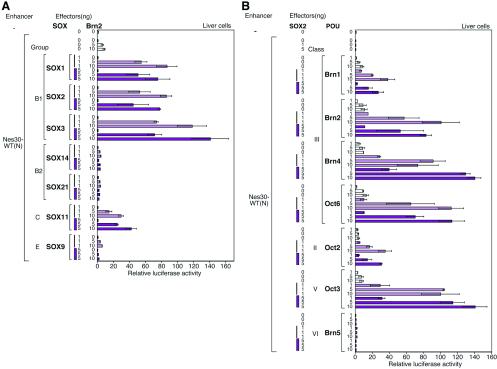
FIG. 6.
Selective combination of SOX and POU factors in activation of the Nes30 enhancer. Various amounts of plasmids to express SOX or POU factors were transfected into liver cells, together with a luciferase-encoding reporter plasmid carrying the octamerized enhancer sequence. (N) indicates the normal orientation of the Nes30 enhancer in the reporter plasmid. (A) Combinations of Brn2 with different SOX proteins. SOX1, SOX2, and SOX3 (group B1) strongly activated the enhancer, and SOX11 (group C) moderately activated the enhancer, while SOX9 (group E) and SOX14 and -21 (group B2) showed no activation. WT, wild type. The error bars indicate standard errors. (B) Combination of SOX2 with different POU factors. All class III POU factors (although Brn1 was relatively inactive) and class V Oct3/4, when combined with SOX2, activated the Nes30 enhancer by nearly 100-fold, whereas class II Oct2 did so moderately. In contrast, Brn5 did not activate the enhancer.
In combination with Brn2, all group B1 SOX proteins, SOX1, SOX2, and SOX3, activated the Nes30(8-mer) enhancer nearly 100-fold, confirming analogous activities of these factors. The group B2 SOX proteins, SOX14 and SOX21, or group E SOX9, did not activate the Nes30 enhancer (Fig. 6A).
Importantly, group C SOX11, in combination with Brn2, also activated the Nes30(8-mer) to a significant extent (30- to 40-fold activation) (Fig. 6A). This contrasts with activation of the δ-crystallin DC5 enhancer by cooperation of SOX2 with Pax6, where SOX11 could not replace SOX2 (22, 25).
Among various POU factors which were examined for the potential to activate the Nes30(8-mer) enhancer in combination with SOX2 (Fig. 6B), class III POU proteins (Brn1, Brn2, Brn4, and Oct6) strongly activated the enhancer, in which activation by Brn1 was relatively moderate. Oct3/4 (class V), in combination with SOX2, activated the enhancer comparably to class III POU factors. Class II Oct2 protein caused weak activation, whereas class VI Brn5 showed no activation with SOX2.
Expression of group B1 SOX, SOX11, and Brn2 in the spinal cord and its relationship with Nestin expression.
The observation described above indicates that group B1 and group C SOX proteins (at least SOX11) cooperate with class III POU factors and activate the Nestin enhancer through binding to the Nes30 sequence. To correlate the synergism of SOX and POU factors that occurs on the Nes30 sequence with the expression of Nestin in vivo, we examined the expression of group B1 SOX, SOX11, and Brn2 proteins in the embryonic spinal cord by immunostaining histological sections.
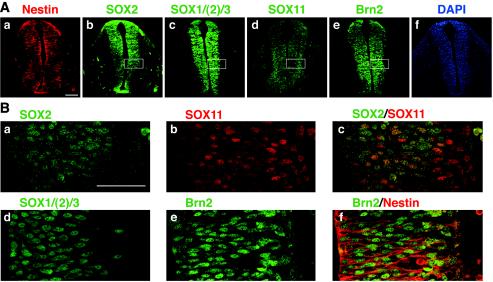
At E10.5, when virtually all cells in the neural tube expressed Nestin (Fig. 7Aa) and were largely mitotic (37), SOX2 was expressed in the major population of cells and localized to the nuclei (Fig. 7Ab and Ba), except for the most external region corresponding to the forming subventricular zone, where SOX11 was strongly expressed (Fig. 7Ad and Bb). At this stage of spinal cord development, expression of SOX2 and SOX11 overlap in a large fraction of cells (Fig. 7B and C). Immunostaining with antibodies detecting all group B1 SOX proteins (with some preference to SOX1 and SOX3) showed essentially the same distribution of the antigen, indicating that expression domains of other group B1 SOX proteins are included in that of SOX2 (Fig. 7Ac and 7Bd). On the other hand, Brn2 was distributed in all nuclei (Fig. 7Ae and Be), surrounded by Nestin-expressing cytoplasm (Fig. 7Bf).
FIG. 7.
Expression of Nestin, group B1 SOX, SOX11, and Brn2 in E10.5 spinal cord. (A) Cross sections of E10.5 spinal cord at trunk level immunostained for Nestin (a), SOX2 (b), SOX1/(2)/3 (c), SOX11 (d), and Brn2 (e) and stained with DAPI (4′,6′-diamidino-2-phenylindole) (f). The scale bar indicates 100 μm. (B) High-magnification confocal images of median regions of the spinal cord (boxed in panel A) immunostained for SOX2 (a), SOX11 (b), SOX2 (green) and SOX11 (red) (c), SOX1/(2)/3 (d), Brn2 (e), and Brn2 (green) and Nestin (red) (f). The ventricle is toward the left. The scale bar indicates 50 μm.
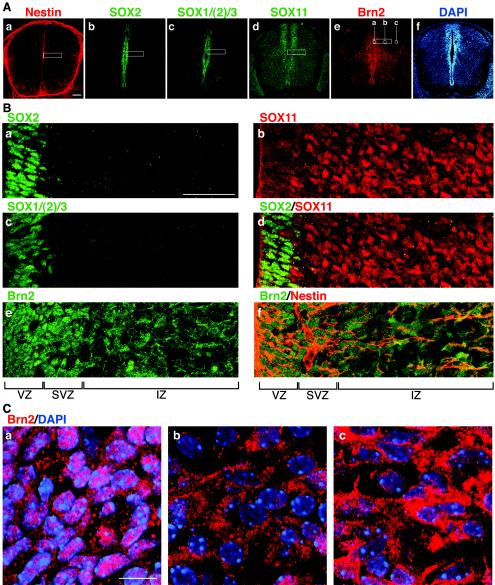
In E13.5 spinal cord, SOX2 expression was confined to the ventricular zone (Fig. 8Ab and Ba), as with other group B1 SOX proteins (Fig. 8Ac and Bc). SOX11 expression at this stage was strong in the subventricular zone, extending into the intermediate zone with a moderate expression level (Fig. 8Ad and Bb), thus making a boundary with the SOX2-expressing ventricular zone (Fig. 8Bd).
FIG. 8.
Expression of Nestin, group B1 SOX, SOX11, and Brn2 in E13.5 spinal cord. (A) Cross sections of E13.5 spinal cord at trunk level immunostained for Nestin (a), SOX2 (b), SOX1/(2)/3 (c), SOX11 (d), Brn2 (e), and DAPI (4′,6′-diamidino-2-phenylindole) (f). The scale bar indicates 100 μm. (B) High-magnification confocal images of median regions of the spinal cord roughly corresponding to the rectangles in panel A, which were immunostained for SOX2 (a), SOX11 (b), SOX1/(2)/3 (c), SOX2 (green) and SOX11 (red) (d), Brn2 (e), and Brn2 (green) and Nestin (red) (f). The scale bar indicates 50 μm. The ventricle is toward the left. The ventricular zone (VZ), subventricular zone (SVZ), and intermediate zone (IZ) are indicated. Note that subcellular localization of Brn2 is clearly different in the VZ-SVZ and IZ. (C) Further enlargement of Brn2-immunostained specimens of the areas indicated in panel A, image e, which are also stained with DAPI (blue). In the ventricular zone (a), Brn2 signals are largely identified within the nucleus, although significant signals are also observed in the cytoplasm. In the nucleus, the heterochromatic regions strongly stained with DAPI are devoid of Brn2 signal. In the intermediate-zone areas (b and c), Brn2 signals are largely within the cytoplasm. The scale bar indicates 10 μm.
Immunostaining with anti-Brn2 antibodies indicated widespread Brn2 protein expression in the neural tube. Expression was strong in the ventricular and subventricular zones and moderate in the intermediate zone (Fig. 8Ae). Examination at higher magnification revealed a remarkable transition in the subcellular localization of this protein. In the ventricular and subventricular zones, Brn2 protein was located mostly in the nuclei, whereas in the intermediate zone, its majority localization was to the cytoplasm (Fig. 8Be and C). Thus, functional cooperation of group B1 SOX and Brn2, and that of SOX11 and Brn2, can take place in the ventricular and subventricular zones, respectively, based on their nuclear colocalization. In the intermediate zone, the cooperation is likely inhibited because of the cytoplasmic localization of Brn2. This accounts for expression of Nestin in the cells of the ventricular zone (group B1 SOX and nuclear Brn2), and those of the subventricular zone (SOX11 and nuclear Brn2).
DISCUSSION
SOX- and POU-dependent Nestin enhancer and its core sequence.
To better understand gene regulation taking place in neural primordial cells, we focused on the regulation of the Nestin gene marking these cells (11, 38, 41, 43). Our study was based on the idea that transcriptional regulation determining a cell type is largely reflected by the regulation of genes specific to that cell type.
In addition to the Nestin gene, expression of group B1 Sox genes, represented by Sox2, is also regarded as an indication of the neural primordial cells (8, 43, 49, 60). Indeed, recent studies that indicated interference with group B1 SOX function in the embryonic spinal cord demonstrate the function of this class of SOX proteins in maintaining the neural primordial state (7, 15). We speculated that there would be a direct regulatory link between expression of Nestin and group B1 Sox genes and thus investigated the Nestin enhancer.
Previous studies identified the nervous system-specific enhancer of the Nestin gene in the second intron (29, 61), the activity of which is totally dependent on the binding of class III POU factors exemplified by Brn2 (21). It has also been indicated that other transcription factors, e.g., those binding to hormone-responsive elements, have subsidiary roles in augmenting activity of the enhancer in specific domains of the CNS (21, 29, 58). In this study, we demonstrated that SOX proteins binding to a site immediately upstream of the POU site are also essential for activation of the Nestin enhancer (Fig. 1 to 3), providing the functional link between the group B1 SOX and Nestin expression.
Recombinant SOX2 and Brn2 proteins bind to these sites (Fig. 4), and mutational abolition of the SOX binding site (M1) eliminated the activity of the full-length (258-bp) Nestin enhancer in electroporated chicken spinal cord and in the transfected neural stem cell line MNS70. The effect of the SOX site mutation was similar to that of POU site mutation (M2), supporting the synergistic action of SOX and POU for activation of the Nestin enhancer (Fig. 1C and D).
Many cell-type- or tissue-specific enhancers are comprised of elements with narrow specificity with essential functions (and hence called “core”) and those with broader specificity. Although individual elements do not necessarily show significant enhancer activity, their regulatory potential and specificity can be demonstrated when they are multimerized (reference 14 and references therein). The 30-bp-long region of the Nestin enhancer (Nes30), including the SOX and POU binding sites, had the features of such a core element. Indeed the octamerized Nes30 sequence showed neural specificity and absolute dependence on the SOX and POU binding sites, providing a handle on the synergistic interaction of the SOX-POU proteins (Fig. 2).
Synergistic function of SOX and POU proteins in regulation of the Nestin core enhancer.
Taking advantage of the octamerized Nes30 core sequence, we demonstrated that specific combinations of SOX and POU proteins, group B1 SOX and class III POU and group C SOX and class III POU, are involved in the activation of the Nestin enhancer. Although only SOX11 among group C SOX factors was studied here, other group C SOX factors, i.e., SOX4 and SOX12 (also called SOX22), which share many protein motifs and are known to be expressed in the subventricular and intermediate zones of the embryonic spinal cord (9, 20, 51), likely have analogous potentials for synergism. The specificity of the synergistic interaction is confirmed by the total absence of transactivation when SOX14/21 (group B2), SOX9 (group E), or Brn5 (class VI) was involved.
It is common for SOX proteins to cooperate with other factors by binding to a nearby site of target DNA sequences for their action and to employ distinct partner factors in different cells, and thus to selectively regulate a cell-specific group of genes (12, 24, 25, 56). For instance, several enhancers active in embryonic stem (ES) cells are activated by the combinatory action of SOX2-Oct3/4 (3, 13, 33, 48, 59) (Fig. 5A). Previous studies employing the Fgf4 enhancer as a model system showed that the natural pair SOX2 and Oct3/4 expressed in ES cells (3), as well as an artificial SOX11-Brn2 pair, activated the enhancer (26); the SOX2-Brn2 pair, however, failed to do so (54), supporting the model of differential and binding site-dependent SOX-POU interactions.
The arrangement of the SOX and POU factor binding site sequences in the Nes30 region was unique among known SOX-POU synergistic sites, including those of the Fgf4 enhancer (Fig. 5A). Insertion of a 10-bp sequence between the SOX and POU binding sites of Nes30, or the swapping of these sites in the 30-bp stretch, completely inactivated the core enhancer (Fig. 5C), although individual proteins bound to these sequences (Fig. 5B). Selective combinations of SOX and POU proteins in transactivation through binding to their unique configuration of the Nes30 sequence extend and confirm the diverse regulatory functions generated by the combined action of SOX and POU proteins binding to adjacent sites.
It is conceivable that there are various interactions between SOX and POU proteins to elicit gene activation synergistically, presumably employing different interfaces of the SOX and POU proteins. In support of this model, recent crystallographic analysis of SOX2-POU complexes indeed indicates recruitment of distinct interaction sites of SOX2 and Oct3/4, depending on the binding site sequences (39, 55).
Regulation of SOX and POU factors in embryonic spinal cord.
As the SOX and POU binding sites included in the Nes30 sequence are essential for the entire activity of the Nestin neural enhancer (Fig. 1B and C), the functional synergism of SOX and POU factors that takes place on the Nes30 sequence must, to a large degree, reflect the major transcriptional regulation of the Nestin neural enhancer. To validate this, the expression of transcription factors in the embryonic spinal cord was investigated.
Previous studies using in situ hybridization indicated transcript localization for these transcription factors: expression of Sox1/2/3 in the ventricular zone (40, 50, 51) and high expression of Sox11 in the subventricular zone (9, 51). Other group C Sox genes, i.e., Sox4 and Sox12, are also expressed in the CNS domains, including the subventricular zone (9, 20, 26).
It has also been shown, by using an in situ hybridization technique, that class III POU factor genes are widely expressed in the ventricular and subventricular zones of the embryonic CNS. Their spatial expression covers slightly different CNS areas along the A-P axis, but extensive overlap exists between them, presumably exerting redundant functions (1, 4, 18). The functional redundancy of the class III POU factors in the CNS is supported by the lack of major neural phenotypes in the single and double mutants lacking Brn1 and Brn2 (4, 30, 46).
Immunohistological detection of group B1 SOX and Brn2 proteins carried out in this study (Fig. 7 and 8) clearly indicated their spatial relationships. At E10.5, the majority of the cells had characteristics of the ventricular zone and expressed SOX1/2/3, and an emerging subventricular zone expressing SOX11 was not well separated (Fig. 7). Brn2 was expressed and distributed throughout the nuclei of the spinal cord (Fig. 7). Nestin was also expressed in all cells, consistent with the model of synergic interaction between group B1 SOX and class III POU, and that of group C SOX and class III POU, in activating Nestin expression.
At E13.5, Nestin expression was confined to those cells having their nuclei in the ventricular and subventricular zones (Fig. 8). At this stage, ventricular and subventricular zones were distinct as a zone of group B1 SOX expression and a zone of a high SOX11 expression, respectively (Fig. 8B), whereas expression of SOX11 at a moderate level extended to the wide intermediate zone, which is externally positioned (Fig. 8Ad and Bb and d).
On the other hand, the expression and subcellular localization of Brn2 changes dynamically between the ventricular and subventricular zones and the intermediate zone. In the ventricular and subventricular zones, Brn2 expression was very high, and the majority of Brn2 protein was localized in the nuclei of cells (Fig. 8Ca); in the intermediate zone, the expression level was lower (Fig. 8Ae), and more significantly, the Brn2 protein was localized in the cytoplasm and excluded from the nucleus (Fig. 8Cb and c).
This regulation of the subcellular localization of Brn2 results in an interaction between Brn2 and SOX11 within the nucleus that occurs only in the subventricular zone, in spite of the expression of these transcription factors in the intermediate zone as well.
Thus, the zonal and subcellular distribution of group B1 SOX, SOX11, and Brn2 supports the model that their synergism on the Nes30 core sequence is fully reflected in the activation of the Nestin gene that takes place within the ventricular and subventricular zones.
Regulation of Brn2 activity by intracellular localization of the protein adds an important item to the list of cytoplasmic sequesterings of transcription factors in differentiated cells (e.g., N-myc in mature neurons [52] and SOX2 in trophoblasts [5]). Although we investigated only Brn2 among class III POU proteins in this study, a high degree of conservation of amino acid sequences among the proteins of this class (1, 4, 18) supports the view that Brn1 and -4 and Oct6 are also under the same regulation of subcellular localization.
Two populations of Nestin-expressing cells in the embryonic CNS.
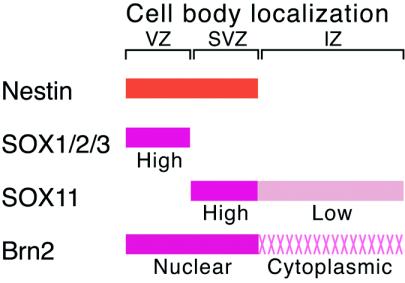
This study revealed that Nestin-expressing cells within the embryonic CNS are comprised of two major populations: (i) those expressing group B1 SOX and nuclear class III POU, located in the ventricular zone, and (ii) those expressing SOX11 (and possibly other group C SOX), together with nuclear class III POU, located in the subventricular zone (Fig. 9).
FIG. 9.
Model of Nestin gene regulation in the embryonic spinal cord. Nestin is expressed in the cells with their cell bodies in the ventricular zone (VZ) and subventricular zone (SVZ). In the ventricular zone, SOX1/2/3 (group B SOX) interact with Brn2 (and other class III POU factors) on the Nes30 sequence (Fig. 1 and 2) within the nucleus. In the subventricular zone, SOX11 (and possibly other group C SOX, i.e., SOX4 and SOX12) interacts with class III POU factors. In the cells with cell body localization in the intermediate zone (IZ), moderate expression of SOX11 and Brn2 also takes place, but sequestering the majority of Brn2 activity in the cytoplasm results in the absence of Nestin expression in these cells.
The subventricular zone is a loosely defined zone of cells that are nonproliferative and positioned immediately external to the ventricular zone. Previous in situ hybridization data of the spinal cord showed high Sox11 expression in the corresponding cell population (9). Based on the data presented in this work, it may be logical to redefine the subventricular zone (at least in the embryonic spinal cord) as the zone characterized by possession of nuclear class III POU proteins and SOX11. This argues that the shift of the cells from the ventricular zone to the subventricular zone indicates not only the change of cell state from proliferative to nonproliferative (7, 15, 37), but also the change of major transcriptional regulation from group B1 SOX dependent to group C SOX dependent. This change in the major SOX proteins may still maintain expression of a significant fraction of genes, like Nestin, but may cause altered expression of other genes; combinations of SOX2-Brn2 and SOX11-Brn2 were both effective in activation of Nestin transcription through binding to Nes30 sequence (Fig. 6) but are also distinguished in other SOX-POU cobinding sites (26, 54).
Given that Nestin expression marks neural primordial cells, they are thus comprised of two populations: group B1 SOX dependent and group C SOX dependent (Fig. 9). In addition, class III POU proteins, the synergistic partners of the SOX proteins, are under the regulation of their subcellular localization. Nestin-expressing cells in later development and in the adult CNS remain to be investigated from this viewpoint, but regulation of embryonic Nestin expression must have an important bearing on transcriptional regulation in later neural primordial cells.
Acknowledgments
We thank members of the Kondoh laboratory for stimulating discussions and Masato Nakafuku for the provision of MNS70 cells.
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (12CE2007) and from the Human Frontier Science Program (RGP0040/2001-M) to H.K. and from the National Key Basic Research and Developmental Program (G1999054000 and 2002CB713802) and the National Natural Science Foundation of China (39870283 and 39930090) to N.J.
REFERENCES
- 1.Alvarez-Bolado, G., M. G. Rosenfeld, and L. W. Swanson. 1995. Model of forebrain regionalization based on spatiotemporal patterns of POU-III homeobox gene expression, birthdates, and morphological features. J. Comp. Neurol. 355:237-295. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez-Buylla, A., B. Seri, and F. Doetsch. 2002. Identification of neural stem cells in the adult vertebrate brain. Brain Res. Bull. 57:751-758. [DOI] [PubMed] [Google Scholar]
- 3.Ambrosetti, D. C., C. Basilico, and L. Dailey. 1997. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol. Cell. Biol. 17:6321-6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersen, B., and M. G. Rosenfeld. 2001. POU domain factors in the neuroendocrine system: lessons from developmental biology provide insights into human disease. Endocr. Rev. 22:2-35. [DOI] [PubMed] [Google Scholar]
- 5.Avilion, A. A., S. K. Nicolis, L. H. Pevny, L. Perez, N. Vivian, and R. Lovell-Badge. 2003. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17:126-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowles, J., G. Schepers, and P. Koopman. 2000. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev. Biol. 227:239-255. [DOI] [PubMed] [Google Scholar]
- 7.Bylund, M., E. Andersson, B. G. Novitch, and J. Muhr. 2003. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat. Neurosci. 6:1162-1168. [DOI] [PubMed] [Google Scholar]
- 8.Cai, J., Y. Wu, T. Mirua, J. L. Pierce, M. T. Lucero, K. H. Albertine, G. J. Spangrude, and M. S. Rao. 2002. Properties of a fetal multipotent neural stem cell (NEP cell). Dev. Biol. 251:221-240. [DOI] [PubMed] [Google Scholar]
- 9.Cheung, M., M. Abu-Elmagd, H. Clevers, and P. J. Scotting. 2000. Roles of Sox4 in central nervous system development. Brain Res. Mol. Brain Res. 79:180-191. [DOI] [PubMed] [Google Scholar]
- 10.Cui, H., and R. F. Bulleit. 1998. Expression of the POU transcription factor Brn-5 is an early event in the terminal differentiation of CNS neurons. J. Neurosci. Res. 52:625-632. [DOI] [PubMed] [Google Scholar]
- 11.Dahlstrand, J., M. Lardelli, and U. Lendahl. 1995. Nestin mRNA expression correlates with the central nervous system progenitor cell state in many, but not all, regions of developing central nervous system. Brain Res. Dev. Brain Res. 84:109-129. [DOI] [PubMed] [Google Scholar]
- 12.Dailey, L., and C. Basilico. 2001. Coevolution of HMG domains and homeodomains and the generation of transcriptional regulation by Sox/POU complexes. J. Cell Physiol. 186:315-328. [DOI] [PubMed] [Google Scholar]
- 13.Di Rocco, G., A. Gavalas, H. Popperl, R. Krumlauf, F. Mavilio, and V. Zappavigna. 2001. The recruitment of SOX/OCT complexes and the differential activity of HOXA1 and HOXB1 modulate the Hoxb1 auto-regulatory enhancer function. J. Biol. Chem. 276:20506-20515. [DOI] [PubMed] [Google Scholar]
- 14.Goto, K., T. S. Okada, and H. Kondoh. 1990. Functional cooperation of lens-specific and nonspecific elements in the delta 1-crystallin enhancer. Mol. Cell. Biol. 10:958-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graham, V., J. Khudyakov, P. Ellis, and L. Pevny. 2003. SOX2 functions to maintain neural progenitor identity. Neuron 39:749-765. [DOI] [PubMed] [Google Scholar]
- 16.Hara, Y., A. C. Rovescalli, Y. Kim, and M. Nirenberg. 1992. Structure and evolution of four POU domain genes expressed in mouse brain. Proc. Natl. Acad. Sci. USA 89:3280-3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hargrave, M., E. Wright, J. Kun, J. Emery, L. Cooper, and P. Koopman. 1997. Expression of the Sox11 gene in mouse embryos suggests roles in neuronal maturation and epithelio-mesenchymal induction. Dev. Dyn. 210:79-86. [DOI] [PubMed] [Google Scholar]
- 18.He, X., M. N. Treacy, D. M. Simmons, H. A. Ingraham, L. W. Swanson, and M. G. Rosenfeld. 1989. Expression of a large family of POU-domain regulatory genes in mammalian brain development. Nature 340:35-41. [DOI] [PubMed] [Google Scholar]
- 19.Herr, W., and M. A. Cleary. 1995. The POU domain: versatility in transcriptional regulation by a flexible two-in-one DNA-binding domain. Genes Dev. 9:1679-1693. [DOI] [PubMed] [Google Scholar]
- 20.Jay, P., I. Sahly, C. Goze, S. Taviaux, F. Poulat, G. Couly, M. Abitbol, and P. Berta. 1997. SOX22 is a new member of the SOX gene family, mainly expressed in human nervous tissue. Hum. Mol. Genet. 6:1069-1077. [DOI] [PubMed] [Google Scholar]
- 21.Josephson, R., T. Muller, J. Pickel, S. Okabe, K. Reynolds, P. A. Turner, A. Zimmer, and R. D. McKay. 1998. POU transcription factors control expression of CNS stem cell-specific genes. Development 125:3087-3100. [DOI] [PubMed] [Google Scholar]
- 22.Kamachi, Y., K. S. Cheah, and H. Kondoh. 1999. Mechanism of regulatory target selection by the SOX high-mobility-group domain proteins as revealed by comparison of SOX1/2/3 and SOX9. Mol. Cell. Biol. 19:107-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamachi, Y., M. Uchikawa, J. Collignon, R. Lovell-Badge, and H. Kondoh. 1998. Involvement of Sox1, 2 and 3 in the early and subsequent molecular events of lens induction. Development 125:2521-2532. [DOI] [PubMed] [Google Scholar]
- 24.Kamachi, Y., M. Uchikawa, and H. Kondoh. 2000. Pairing SOX off with partners in the regulation of embryonic development. Trends Genet. 16:182-187. [DOI] [PubMed] [Google Scholar]
- 25.Kamachi, Y., M. Uchikawa, A. Tanouchi, R. Sekido, and H. Kondoh. 2001. Pax6 and SOX2 form a co-DNA-binding partner complex that regulates initiation of lens development. Genes Dev. 15:1272-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhlbrodt, K., B. Herbarth, E. Sock, J. Enderich, I. Hermans-Borgmeyer, and M. Wegner. 1998. Cooperative function of POU proteins and SOX proteins in glial cells. J. Biol. Chem. 273:16050-16057. [DOI] [PubMed] [Google Scholar]
- 27.Latchman, D. S. 1999. POU family transcription factors in the nervous system. J. Cell Physiol. 179:126-133. [DOI] [PubMed] [Google Scholar]
- 28.Lendahl, U., L. B. Zimmerman, and R. D. McKay. 1990. CNS stem cells express a new class of intermediate filament protein. Cell 60:585-595. [DOI] [PubMed] [Google Scholar]
- 29.Lothian, C., and U. Lendahl. 1997. An evolutionarily conserved region in the second intron of the human nestin gene directs gene expression to CNS progenitor cells and to early neural crest cells. Eur. J. Neurosci. 9:452-462. [DOI] [PubMed] [Google Scholar]
- 30.McEvilly, R. J., M. O. de Diaz, M. D. Schonemann, F. Hooshmand, and M. G. Rosenfeld. 2002. Transcriptional regulation of cortical neuron migration by POU domain factors. Science 295:1528-1532. [DOI] [PubMed] [Google Scholar]
- 31.Mizuseki, K., M. Kishi, M. Matsui, S. Nakanishi, and Y. Sasai. 1998. Xenopus Zic-related-1 and Sox-2, two factors induced by chordin, have distinct activities in the initiation of neural induction. Development 125:579-587. [DOI] [PubMed] [Google Scholar]
- 32.Nakagawa, Y., T. Kaneko, T. Ogura, T. Suzuki, M. Torii, K. Kaibuchi, K. Arai, S. Nakamura, and M. Nakafuku. 1996. Roles of cell-autonomous mechanisms for differential expression of region-specific transcription factors in neuroepithelial cells. Development 122:2449-2464. [DOI] [PubMed] [Google Scholar]
- 33.Nishimoto, M., A. Fukushima, A. Okuda, and M. Muramatsu. 1999. The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol. Cell. Biol. 19:5453-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okamoto, K., H. Okazawa, A. Okuda, M. Sakai, M. Muramatsu, and H. Hamada. 1990. A novel octamer binding transcription factor is differentially expressed in mouse embryonic cells. Cell 60:461-472. [DOI] [PubMed] [Google Scholar]
- 35.Okamoto, K., M. Wakamiya, S. Noji, E. Koyama, S. Taniguchi, R. Takemura, N. G. Copeland, D. J. Gilbert, N. A. Jenkins, M. Muramatsu, et al. 1993. A novel class of murine POU gene predominantly expressed in central nervous system. J. Biol. Chem. 268:7449-7457. [PubMed] [Google Scholar]
- 36.Pevny, L. H., and R. Lovell-Badge. 1997. Sox genes find their feet. Curr. Opin. Genet. Dev. 7:338-344. [DOI] [PubMed] [Google Scholar]
- 37.Pevny, L. H., S. Sockanathan, M. Placzek, and R. Lovell-Badge. 1998. A role for SOX1 in neural determination. Development 125:1967-1978. [DOI] [PubMed] [Google Scholar]
- 38.Reim, G., and M. Brand. 2002. Spiel-ohne-grenzen/pou2 mediates regional competence to respond to Fgf8 during zebrafish early neural development. Development 129:917-933. [DOI] [PubMed] [Google Scholar]
- 39.Remenyi, A., K. Lins, L. J. Nissen, R. Reinbold, H. R. Scholer, and M. Wilmanns. 2003. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 17:2048-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rex, M., A. Orme, D. Uwanogho, K. Tointon, P. M. Wigmore, P. T. Sharpe, and P. J. Scotting. 1997. Dynamic expression of chicken Sox2 and Sox3 genes in ectoderm induced to form neural tissue. Dev. Dyn. 209:323-332. [DOI] [PubMed] [Google Scholar]
- 41.Rietze, R. L., H. Valcanis, G. F. Brooker, T. Thomas, A. K. Voss, and P. F. Bartlett. 2001. Purification of a pluripotent neural stem cell from the adult mouse brain. Nature 412:736-739. [DOI] [PubMed] [Google Scholar]
- 42.Scholer, H. R., A. K. Hatzopoulos, R. Balling, N. Suzuki, and P. Gruss. 1989. A family of octamer-specific proteins present during mouse embryogenesis: evidence for germline-specific expression of an Oct factor. EMBO J. 8:2543-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwartz, P. H., P. J. Bryant, T. J. Fuja, H. Su, D. K. O'Dowd, and H. Klassen. 2003. Isolation and characterization of neural progenitor cells from post-mortem human cortex. J. Neurosci. Res. 74:838-851. [DOI] [PubMed] [Google Scholar]
- 44.Stolt, C. C., P. Lommes, E. Sock, M. C. Chaboissier, A. Schedl, and M. Wegner. 2003. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 17:1677-1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Streit, A., S. Sockanathan, L. Perez, M. Rex, P. J. Scotting, P. T. Sharpe, R. Lovell-Badge, and C. D. Stern. 1997. Preventing the loss of competence for neural induction: HGF/SF, L5 and Sox-2. Development 124:1191-1202. [DOI] [PubMed] [Google Scholar]
- 46.Sugitani, Y., S. Nakai, O. Minowa, M. Nishi, K. Jishage, H. Kawano, K. Mori, M. Ogawa, and T. Noda. 2002. Brn-1 and Brn-2 share crucial roles in the production and positioning of mouse neocortical neurons. Genes Dev. 16:1760-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Temple, S. 2001. The development of neural stem cells. Nature 414:112-117. [DOI] [PubMed] [Google Scholar]
- 48.Tomioka, M., M. Nishimoto, S. Miyagi, T. Katayanagi, N. Fukui, H. Niwa, M. Muramatsu, and A. Okuda. 2002. Identification of Sox-2 regulatory region which is under the control of Oct-3/4-Sox-2 complex. Nucleic Acids Res. 30:3202-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uchikawa, M., Y. Ishida, T. Takemoto, Y. Kamachi, and H. Kondoh. 2003. Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev. Cell. 4:509-519. [DOI] [PubMed] [Google Scholar]
- 50.Uchikawa, M., Y. Kamachi, and H. Kondoh. 1999. Two distinct subgroups of Group B Sox genes for transcriptional activators and repressors: their expression during embryonic organogenesis of the chicken. Mech. Dev. 84:103-120. [DOI] [PubMed] [Google Scholar]
- 51.Uwanogho, D., M. Rex, E. J. Cartwright, G. Pearl, C. Healy, P. J. Scotting, and P. T. Sharpe. 1995. Embryonic expression of the chicken Sox2, Sox3 and Sox11 genes suggests an interactive role in neuronal development. Mech. Dev. 49:23-36. [DOI] [PubMed] [Google Scholar]
- 52.Wakamatsu, Y., Y. Watanabe, H. Nakamura, and H. Kondoh. 1997. Regulation of the neural crest cell fate by N-myc: promotion of ventral migration and neuronal differentiation. Development 124:1953-1962. [DOI] [PubMed] [Google Scholar]
- 53.Wegner, M. 1999. From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res. 27:1409-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiebe, M. S., T. K. Nowling, and A. Rizzino. 2003. Identification of novel domains within Sox-2 and Sox-11 involved in autoinhibition of DNA binding and partnership specificity. J. Biol. Chem. 278:17901-17911. [DOI] [PubMed] [Google Scholar]
- 55.Williams, D. C., Jr., M. Cai, and G. M. Clore. 2004. Molecular basis for synergistic transcriptional activation by Oct1 and Sox2 revealed from the solution structure of the 42-kDa Oct1.Sox2.Hoxb1-DNA ternary transcription factor complex. J. Biol. Chem. 279:1449-1457. [DOI] [PubMed] [Google Scholar]
- 56.Wilson, M., and P. Koopman. 2002. Matching SOX: partner proteins and co-factors of the SOX family of transcriptional regulators. Curr. Opin. Genet. Dev. 12:441-446. [DOI] [PubMed] [Google Scholar]
- 57.Wood, H. B., and V. Episkopou. 1999. Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech. Dev. 86:197-201. [DOI] [PubMed] [Google Scholar]
- 58.Yaworsky, P. J., and C. Kappen. 1999. Heterogeneity of neural progenitor cells revealed by enhancers in the nestin gene. Dev. Biol. 205:309-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yuan, H., N. Corbi, C. Basilico, and L. Dailey. 1995. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 9:2635-2645. [DOI] [PubMed] [Google Scholar]
- 60.Zappone, M. V., R. Galli, R. Catena, N. Meani, S. De Biasi, E. Mattei, C. Tiveron, A. L. Vescovi, R. Lovell-Badge, S. Ottolenghi, and S. K. Nicolis. 2000. Sox2 regulatory sequences direct expression of a (beta)-geo transgene to telencephalic neural stem cells and precursors of the mouse embryo, revealing regionalization of gene expression in CNS stem cells. Development 127:2367-2382. [DOI] [PubMed] [Google Scholar]
- 61.Zimmerman, L., B. Parr, U. Lendahl, M. Cunningham, R. McKay, B. Gavin, J. Mann, G. Vassileva, and A. McMahon. 1994. Independent regulatory elements in the nestin gene direct transgene expression to neural stem cells or muscle precursors. Neuron 12:11-24. [DOI] [PubMed] [Google Scholar]