Abstract
Sgt1p is a well-conserved protein proposed to be involved in a number of cellular processes. Genetic studies of budding yeast suggest a role for SGT1 in signal transduction, cell cycle advance, and chromosome segregation. Recent evidence has linked Sgt1p to HSP90 chaperones, although the precise relationship between these proteins is unclear. To further explore the role of Sgt1p in these processes, we have characterized the interactions among Sgt1p, the inner kinetochore complex CBF3, and HSP90 chaperones. We show that the amino terminus of Sgt1p interacts with CBF3 subunits Skp1p and Ctf13p. HSP90 interacts with Sgt1p and, in combination with the carboxy terminus of Sgt1p, regulates the interaction between Sgt1p and Skp1p in a nucleotide-dependent manner. While the Sgt1p-Skp1p interaction is required for CBF3 assembly, mutations that stabilize this interaction prevent the turnover of protein complexes important for CBF3 assembly. We propose that HSP90 and Sgt1p act together as a molecular switch, maintaining transient interactions required to balance protein complex assembly with turnover.
Protein complexes consisting of multiple subunits must assemble and remain stable to carry out their cellular functions. However, many biological processes, including but not limited to signal transduction and cell cycle advance, require that protein complexes are maintained only transiently (20). Multiple solutions for maintaining the proper balance of protein complexes have been described, including linking protein phosphorylation to protein turnover as in the case of cell cycle advance. In other cases, such as nuclear hormone receptors, the assembly of protein complexes appears to be directly linked to their turnover (10, 11). This direct coupling may provide quality control, ensuring that misfolded protein complexes do not interfere with normal cellular functions. Alternatively, such a direct coupling of protein assembly and turnover has been proposed to contribute to the plasticity of signaling pathways, allowing the levels of assembled complexes to be tightly coupled to regulatory cues.
The HSP90 class of chaperones has been implicated in both the assembly of multiprotein complexes and their turnover (for a review, see reference 16). While believed to aid in client protein folding, HSP90 chaperone activity appears to be more restricted than that of other classes of chaperones. A special class of interacting proteins termed cochaperones regulates the activity of HSP90 chaperones (4, 5, 12). Cochaperones can regulate the ATP hydrolysis of chaperones, thereby influencing their affinity for client proteins, or can target HSP90 chaperones to their clients or to a particular subcellular compartment. In some cases cochaperones appear to have a chaperone activity independent of HSP90. Two classes of cochaperones have been identified on the basis of their domain structure: (i) those that include tetratricopeptide repeats (TPR) (e.g., HOP1) and (ii) those that contain a non-TPR fold, such as the HSP20/α-crystallin fold found in the p23 cochaperone (13, 26, 32). Interestingly, Sgt1p is a protein that has been proposed to have both a TPR domain and homology to an HSP20/α-crystallin fold (7, 21). The recent demonstration that Sgt1p can interact with HSP90 further implicates Sgt1p in HSP90 function (17, 35). However, little is known concerning the biochemical function of Sgt1p and the significance of its interaction with HSP90 remains unclear.
Sgt1p is a highly conserved protein that functions in an array of biological processes, apparently through direct interaction with different multiprotein complexes. In yeast, phenotypic analyses of mutations in SGT1 suggest that it is required for the activity of the ubiquitin ligase SCF, for the assembly of the inner kinetochore complex CBF3, and for the activity of adenylyl cyclase (7, 21). Sgt1p is physically linked to SCF and CBF3 through its interaction with Skp1p, a subunit of both complexes. This interaction appears conserved, as Sgt1p in plants has also been reported to interact with SCF. In addition, Sgt1p in plants has been proposed to associate with proteins in other complexes, including proteins important for R gene-mediated disease resistance (1, 2, 17, 36) and proteins associated with the COP9 signalosome, a complex that removes the ubiquitin-like molecule Nedd8p (2, 6). Consistent with the ability of Sgt1p to interact with multiple complexes, two-hybrid analysis has identified a number of potential interactors, characterized by the presence of a leucine-rich repeat domain (7). Given the number of pathways proposed to require SGT1, it is tempting to speculate that Sgt1p serves a general role in each of these pathways perhaps by linking a specific protein complex subunit to HSP90 function.
In the case of CBF3, Sgt1p is required for the function of the core CBF3 subunit, Ctf13p. The finding that HSP82 (an HSP90 chaperone in yeast) is also important for Ctf13p function (34) supports a possible link between Sgt1p and HSP90. However, the target of HSP90 is not clear; HSP90 may act directly on Ctf13p to facilitate its interaction with other CBF3 subunits or may act indirectly to control the interaction between Sgt1p and Skp1p, an interaction that is essential for Ctf13p function and CBF3 assembly. Interestingly, the ability of Sgt1p and Skp1p to dissociate may be equally important, as mutations that stabilize their interaction prevent turnover of CBF3 (29). Previous work has argued that controlling the levels of CBF3 is important to maintain the selectivity of kinetochore formation on centromeric DNA (18). Together, these results suggest that Sgt1p and Skp1p interactions must be carefully regulated to maintain selective assembly of kinetochores and contribute to the orderly segregation of chromosomes.
To explore how Sgt1p, Skp1p, and HSP90 contribute to CBF3 assembly, we have characterized the interactions between Sgt1p and other subunits of the CBF3 complex. Our data show that Skp1p and Ctf13p interact with domains in the amino terminus of Sgt1p. These interactions are inhibited by a negative regulatory domain in the carboxy terminus of Sgt1p. Mutations that stabilize the interaction between Sgt1p and Skp1p compromise CBF3 assembly and generally interfere with normal cell growth. We show that HSP90 interacts with Sgt1p and that the nucleotide-bound state of HSP90 determines the affinity of Sgt1p for Skp1p. Together these results argue that CBF3 homeostasis requires HSP90 to maintain the transient interaction between Sgt1p and Skp1p. We speculate that Sgt1p generally functions in the assembly of multiprotein complexes by transiently linking HSP90 chaperones to client proteins and uses HSP90 to recycle from these complexes.
MATERIALS AND METHODS
Plasmids, yeast strain construction, and antibodies.
All of the yeast strains used in this study were constructed in the S288C background (Table 1). SGT1 mutations were introduced by PCR-mediated site-directed mutagenesis (sequences of mutagenic oligonucleotides are available on request), and the integrity of the coding region was verified by DNA sequencing. Plasmids were introduced into yeast strains by the TRAFO transformation technique (14), and a plasmid shuffle strategy was used to introduce novel SGT1 alleles over a chromosomal deletion (3). Sgt1p variants containing the indicated carboxy-terminal fusions were created by homologous recombination with the chromosomal copy of SGT1 by standard PCR-based targeting techniques (23) and confirmed by PCR analyses of genomic DNA. Antisera against Skp1p and Sgt1p were made as previously described (18, 30). The monoclonal antibody (12CA5) to the hemagglutinin (HA) tag was purchased from Boehringer Mannheim. HSP90 antibody (which recognizes both HSP90 gene products) was a generous gift from Y. Kimura (Tokyo Metropolitan Institute of Medical Science). The immunoblots shown do not resolve the two closely migrating HSP90 chaperones.
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| YPH500 | matα ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 | 32a |
| YKK39 | matα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 sgt1Δ1::HIS31 pRS316-SGT1 | 21 |
| YKK45 | mata ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 sgt1-5::LEU2 CFIII (CEN3.L.YPH983) TRP1 SUP11 | 21 |
| YKK54 | mataura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 sgt1-3LEU2 CFIII (CEN3.L.YPH983) TRP1 SUP11 | 21 |
| KSC451 | matα ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 SGT1-TAP::TRP | This study |
| KSC669 | matα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 sgt1 Δ1::HIS31 pRS315-SGT1 | This study |
| KSC670 | matα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 sgt1 Δ1::HIS31 pRS315-SGT1(L31P) | This study |
| KSC671 | matα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 sgt1 Δ1::HIS31 pRS315-SGT1(F99L) | This study |
| KSC672 | matα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 sgt1 Δ1::HIS31 pRS315-SGT1(N213I) | This study |
| KSC673 | matα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 sgt1 Δ1::HIS31 pRS315-SGT1(D220V) | This study |
| KSC674 | matα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 sgt1 Δ1::HIS31 pRS315-SGT1(E364K) | This study |
| KSC967 | matα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 GAL1-3XHA-HIS3::CTF13 pRS315-SGT1 | This study |
| KSC968 | matα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 GAL1-3XHA-HIS3::CTF13 pRS315-sgt1-3 | This study |
| KSC969 | matα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 GAL1-3XHA-HIS3::CTF13 pRS315-sgt1-5 | This study |
| KSC1199 | matα ura3-52 lys2-801 ade2-101 trp1-631 his3-Δ200 leu2-Δ1 pRS315-GALI-SGT1 | This study |
| KSC1200 | matα ura3-52 lys2-801 ade2-101 trp1-631 his3-Δ200 leu2-Δ1 pRS315-GALI-SGT1 (1-268) | This study |
| KSC1202 | matα ura3-52 lys2-801 ade2-101 trp1-631 his3-Δ200 leu2-Δ1 pRS315 | This study |
| KSC1631 | matα ura3-52 lys2-801 ade2-101 trp1-631 his3-Δ200 leu2-Δ1 pRS315-GALI-sgt1-5 | This study |
| KSC1632 | matα ura3-52 lys2-801 ade2-101 trp1-631 his3-Δ200 leu2-Δ1 pRS315-GALI-sgt1-5(L31P) | This study |
| KSC1035 | mataura3-1 his3-1115 leu2-3,12 trp1-1 can1-100 ade1-1 Δ1 sc82::LEU2 Δhsp82::LEU pTGPD/P82 (HSP82) GAL1-3XHA-HIS3::CTF13 | This study |
| KSC1037 | mataura3-1 his3-11,15 leu2-3,12 trp1-1 can1-100 ade1-1 Δhsc82::LEU2 Δhsp82::LEU pTGPD/T101I (hsp82-T101I) GAL1-3XHA-HIS3::CTF13 | This study |
| KSC1039 | mataura3-1 his3-11,15 leu2-3,12 trp1-1 can1-100 ade1-1 Δhsc82::LEU2 Δhsp82::LEU pTGPD/G170D (hsp82-G170D) GAL1-3XHA-HIS3::CTF13 | This study |
Yeast growth and extract preparation.
Yeast strains were grown in standard YEP medium supplemented with the indicated carbon source at 2% (15). Temperature-sensitive strains were grown to mid-log phase at 25°C and then shifted to 37°C for 3 h, unless otherwise indicated. To generate extracts, cells were prepared as previously described and broken in extract buffer (50 mM bis-Tris-propane [pH 7.2], 200 mM KCl, 5 mM EGTA, 5 mM EDTA, 100 mM β-glycerophosphate, 10% glycerol, 14 mM 2-mercaptoethanol, 10 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM tosylsulfonyl phenylalanyl chloromethyl ketone [TPCK], 10 μg each of leupeptin, pepstatin, and chymostatin per ml) (19).
In vitro binding assay.
Sgt1p was translated at 30°C in rabbit reticulocyte lysates in the presence of [35S]methionine (Amersham Biosciences) with the TNT Quick Coupled Transcription/Translation System as recommended by the manufacturer (Promega). Templates for transcription were generated by PCR with 5′ oligonucleotides containing the T7 promoter and 3′ oligonucleotides that insert a stop codon at the indicated position.
Glutathione S-transferase (GST)-Ctf13p and GST-Skp1p were expressed in High Five insect cells (Invitrogen Corp.) by infecting cells with baculoviruses and harvesting as previously described (18, 30). Recombinant GST-Ctf13p and GST-Skp1p were purified by incubating cell extracts with glutathione Sepharose (Amersham Biosciences) under the manufacturer's recommended conditions. Maltose binding protein (MBP)-Hsp82p was produced in Escherichia coli with the pMAL-C2X plasmid and purified with amylose resin as recommended by the manufacturer (New England Biolabs). Binding reaction mixtures were prepared with 15 μl of beads containing equal molar amounts of GST fusion (2.5 μg), GST, MBP-Hsp82p, or MBP alone in 500 μl of binding buffer (50 mM HEPES [pH 7.4], 150 mM NaCl, 50 mM NaF, 50 mM β-glycerophosphate, 0.2 mM EDTA, 1% Triton X-100, 10% glycerol, 1 mM dithiothreitol, 1 mM PMSF, 1 mM TPCK, 10 μg each of leupeptin, pepstatin, and chymostatin per ml) and incubated at 30°C for 1 h and then at 4οC for 3 h with end-over-end tube rotation. Beads were recovered by centrifugation and washed three times with binding buffer. Bead-associated proteins were resolved by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-PAGE), and levels of translated protein were quantified with a Storm imaging system (Molecular Dynamics).
Trypsin analysis of Sgt1p.
Recombinant GST-Sgt1p was produced in E. coli with the pGEX-6p2 plasmid, purified with glutathione Sepharose, and eluted with PreScission protease in accordance with the manufacturer's (Amersham Biosciences) instructions. Purified Sgt1p was treated with 2.5 μg of trypsin (Boehringer Mannheim) per ml in 50 mM HEPES (pH 8.0)-300 mM KCl-1 mM EDTA-30 mM NaF-30 mM β-glycerophosphate-1% Triton X-100 at room temperature. Aliquots were removed at different time points, and proteolysis was stopped by boiling the sample in Laemmli buffer. Accumulation of distinct proteolytic fragments was visualized following SDS-PAGE by Coomassie staining. Microsequencing of proteolytic fragments was performed at the Molecular Structure Laboratory at the University of California Davis after transfer of fragments to PVDF membranes.
Sgt1p IP and purification from yeast extracts.
To isolate Sgt1p complexes, yeast extracts (2 mg of total protein) were diluted into immunoprecipitation (IP) buffer (50 mM Tris [pH 8.0], 50 mM KCl, 0.2% Triton X-100, 14 mM 2-mercaptoethanol, 1 mM PMSF, 1 mM TPCK, 10 μg each of leupeptin, pepstatin, and chymostatin per ml) and incubated with 2 μl of antibody for 1 h at 4°C and then with 20 μl of protein A-Sepharose (Amersham Biosciences) for 30 min at 4°C. Beads were washed three times in IP buffer, and bound proteins were eluted and analyzed by immunoblotting as previously reported (30). Chemiluminescence signals were detected with film or captured by a charge-coupled device camera with FluorChem 8900 software (Alpha Innotech). Band intensities were quantified with ImageQuant 5.0 software, and scanned images of film were derived from multiple exposures or multiple captured images to ensure linearity. The Sgt1p-TAP fusion protein was purified from 5 mg of yeast extract with immunoglobulin G (IgG) Sepharose (Amersham Biosciences) as previously described (27, 28), eluted in Laemmli buffer, and analyzed by immunoblotting as described above.
CBF3 band shift assays.
CBF3 band shift assays were performed by incubating 40 to 120 μg of protein from the indicated yeast extracts with an 88-bp radiolabeled CEN DNA as previously described (8, 18, 19, 30). Active coexpressed Ctf13p and Skp1p (Ctf13p/Skp1p) and coexpressed Cep3p and Ndc10p (Cep3p/Ndc10p) were produced in insect cells following coinfection with the indicated recombinant baculoviruses as previously described (18).
Nucleotide dependence of Sgt1p-Skp1p interactions.
An MBP fusion to the amino terminus of Sgt1p was expressed in E. coli with the pMAL-C2X vector and purified on amylose resin in accordance with the manufacturer's (New England Biolabs) recommendations. Equal molar amounts of MBP-Sgt1p (1.0 μg) or MBP alone immobilized on 15 μl of resin were suspended in binding buffer (50 mM HEPES [pH 8.0], 150 mM NaCl, 50 mM NaF, 50 mM β-glycerophosphate, 0.2 mM EDTA, 1% Triton X-100, 10% glycerol, 1 mM dithiothreitol, 1 mM PMSF, 1 mM TPCK, 10 μg each of leupeptin, pepstatin, and chymostatin per ml) supplemented with 1.0 mg of the indicated yeast extract and nucleotide (5 mM) and then incubated for 10 to 60 min at 30°C. Beads were washed three times in binding buffer and incubated in elution buffer (20 mM Tris [pH 8.0], 200 mM KCl, 1 mM EDTA, 10 mM maltose) for 20 min at 4°C and then analyzed by immunoblotting.
RESULTS
The amino terminus of Sgt1p interacts with Skp1p and Ctf13p.
The reported association between Sgt1p and a number of proteins implicated in CBF3 function led us to characterize the domains of Sgt1p that mediate these interactions (21). To this end, we established an in vitro binding assay with Sgt1p translated in rabbit reticulocyte lysates supplemented with [35S]methionine. The ability of translated Sgt1p to bind to GST-Skp1p, GST-Ctf13p, and GST alone was evaluated following an in vitro binding assay (see Materials and Methods). Previous results have suggested that Sgt1p binds to Skp1p and to Ctft13p, although biochemical studies suggest that the Sgt1p-Skp1p complex includes only a minor proportion of the total Skp1p in the cell (21, 29). Interestingly, translated Sgt1p also showed a weak affinity for GST-Skp1p and GST-Ctf13p (Fig. 1A, ∼5 to 7% of translated Sgt1p bound). This interaction was specific, as Sgt1p did not significantly bind to GST or to GST-Skp1-4p, a mutation previously shown to block Skp1p-Sgt1p interaction (Fig. 1A and data not shown). The integrity of both the GST fusion proteins was confirmed on the basis of the ability of GST-Skp1p to interact with reticulocyte lysate-translated Ctf13p and that of GST-Ctf13p to interact with translated Skp1p (>25% of load bound; data not shown). Thus, we conclude that Sgt1p has relatively low affinity for both Skp1p and Ctf13p, consistent with observations of endogenous protein complexes in yeast (21).
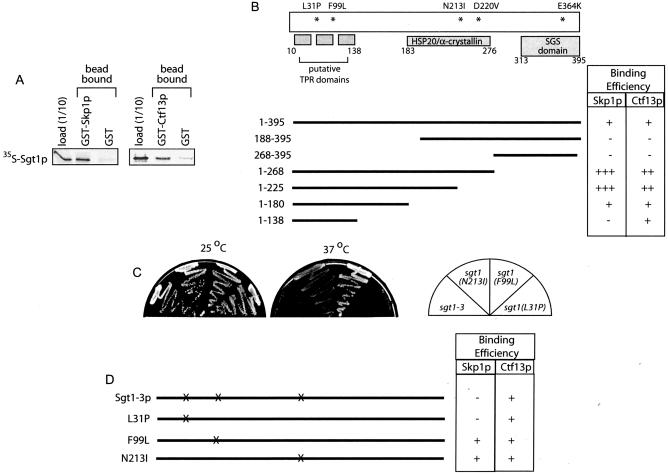
FIG. 1.
Domain mapping of Sgt1p sequences required for Skp1p and Ctf13p interaction. (A) In vitro-translated 35S-Sgt1p was incubated with either GST-Skp1p, GST-Ctf13p, or GST immobilized on glutathione Sepharose beads. Bound 35S-Sgt1p was quantified after SDS-PAGE by Phosphorimager analysis. (B) A schematic showing the relative positions of the putative TPR motifs, the HSP20/α-crystallin fold, and the unique, well-conserved region of SGT1 (SGS) is shown. Fragments of Sgt1p were translated in reticulocyte lysates and tested for the ability to bind GST-Skp1p, GST-Ctf13p, or GST as in panel A. The levels of Sgt1p specifically bound to the GST fusion proteins are expressed as a percentage of the input and coded as follows: +++, >25% bound; ++, 11 to 25% bound; +, 5 to 10% bound; −, <5% bound. (C) Temperature-sensitive growth of SGT1 alleles was assessed following 3 days of growth at the indicated temperatures. (D) SGT1 alleles encoding the indicated point mutations were translated and analyzed as in panel A.
We used this assay to map the regions in Sgt1p important for binding Skp1p and Ctf13p. Deletion of the first 188 amino acids of Sgt1p (Sgt1p188-395) eliminates the putative TPR domain (proposed to include three TPR motifs [21]) and fails to bind to either Skp1p or Ctf13p (Fig. 1B and Fig. S1A in the supplemental material), arguing that the amino terminus and potentially the TPR domain of Sgt1p is important for these interactions. Truncations of carboxy-terminal sequences suggest that Skp1p and Ctf13p interact with the first 225 amino acids of Sgt1p, a region notably larger than that proposed to include the three TPR motifs. A carboxy-terminal deletion (amino acids 1 to 138) that only includes the three putative TPR motifs bound to Ctf13p but failed to bind to Skp1p. These results suggest that the TPR domain is required but not sufficient for Skp1p binding and further suggest that Skp1p and Ctf13p interact with distinct regions of Sgt1p. Because deletions can cause unpredictable changes in the folding of protein domains, we analyzed how the point mutations found in the temperature-sensitive allele sgt1-3 affect binding. The sgt1-3 allele consists of three amino acid changes (L31P, F99L, and N213I), fails to bind Skp1p, and causes cells to arrest in G2/M at 37°C (21). SGT1 alleles containing each single mutation were created and placed over a deletion of SGT1 to assess temperature-sensitive growth. Only the L31P substitution conferred temperature-sensitive growth on yeast (Fig. 1C). Similarly, only L31P inhibited Sgt1p binding to Skp1p, suggesting that amino acids in this region are important for interaction with Skp1p (Fig. 1D and Fig. S1B in the supplemental material). Significantly, the interaction with Ctf13p was not affected by L31P, further arguing that Skp1p and Ctf13p bind to separate domains in Sgt1p. It may be that these distinct binding regions represent separately folded domains or multiple binding sites in the same folded domain. In summary, we conclude that both Sgt1p binding partners, Skp1p and Ctf13p, interact with regions in the amino terminus of Sgt1p that include but are not limited to the putative TPR domain.
The carboxy terminus of Sgt1p regulates Skp1p and Ctf13p association.
Although the carboxy-terminal fragment of Sgt1p (Sgt1p268-395) has no measurable affinity for either Skp1p or Ctf13p, we noticed that deleting this region had a dramatic effect on Skp1p and Ctf13p binding (Fig. 2A and Fig. S1A in the supplemental material). Quantification shows that eliminating the carboxy terminus of Sgt1p results in a five- to eightfold increase in binding to GST-Skp1p and GST-Ctf13p compared to that of the wild type (Fig. 2A, Sgt1p1-268, and data not shown). This observation confirms that Skp1p and Ctf13p binding domains lie in the amino terminus of Sgt1p and suggests that the carboxy terminus of Sgt1p acts to inhibit binding of Skp1p and Ctf13p. If the carboxy terminus of Sgt1p is directly responsible for inhibiting protein binding to amino-terminal domains, we reasoned that more subtle mutations in the carboxy terminus might also affect Skp1p binding. The sgt1-5 allele contains two amino acid substitutions (D220V and E364K) and is temperature sensitive for growth, causing cells to arrest in G1 at 37°C (21). While the E364K mutation was sufficient to confer temperature-sensitive growth, both mutations were necessary to alter the affinity of Sgt1p for Skp1p (Fig. 2A). Remarkably, the double mutation allowed Sgt1p to bind to Skp1p with the same affinity as the Sgt1p1-268 truncation, suggesting that mutations in the HSP20/α-crystallin domain and in conserved carboxy-terminal amino acids cooperate to perturb the inhibitory activity of the carboxy terminus (Fig. 2A, Sgt1-5p). One possibility is that the carboxy terminus physically occludes the binding domains in the amino terminus of Sgt1p. To test this idea, we used limited proteolysis to probe the accessibility of trypsin sites located in Sgt1p and analyzed fragments by protein sequencing. When the full-length Sgt1p protein was exposed to trypsin (Fig. 2C; arrow 1), it was rapidly cleaved in the region of amino acid 270, eliminating the carboxy terminus and producing an amino-terminal Sgt1p fragment (40.2 kDa; Fig. 2C, arrow 2). Kinetic analysis revealed that only after release of the carboxy terminus did amino-terminal trypsin cleavage sites become accessible (Fig. 2C, arrow 3, and D). While this result could indicate that the amino terminus of Sgt1p is highly structured and therefore more resistant to proteolysis, taken together with the mapping data, we propose that the carboxy terminus of Sgt1p occludes amino-terminal Skp1p and Ctf13p binding sites (see model in Fig. 6). A more complete understanding of how the carboxy terminus of Sgt1p inhibits Skp1p binding requires detailed structural analyses of the Sgt1p-Skp1p complex.
FIG.2.
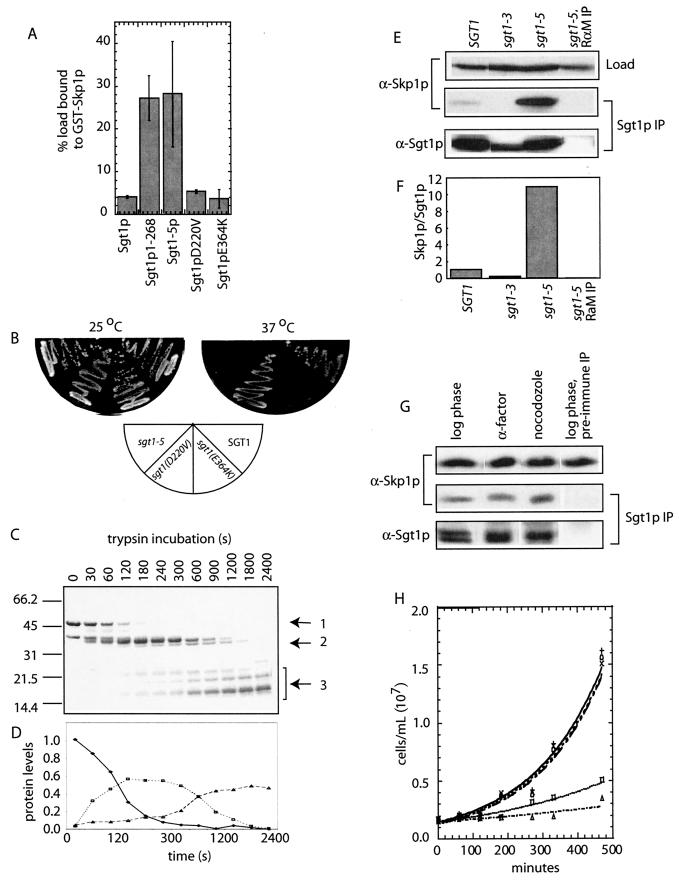
The carboxy terminus of Sgt1p occludes amino-terminal binding domains. (A) The indicated SGT1 alleles were translated, and their affinity for GST-Skp1p was determined after SDS-PAGE by Phosphorimager analysis. (B) Temperature-sensitive growth of SGT1 alleles was assessed following 3 days of growth on YPD plates at the indicated temperatures. (C) Sgt1p proteolytic fragments were visualized by SDS-PAGE and Coomassie staining following treatment with trypsin for the indicated times (seconds). (D) The levels of full-length Sgt1p (arrow 1 in panel C; ⧫), a fragment representing amino acids 1 to 270 (arrow 2 in panel C; □), and small amino-terminal fragments (arrow 3 in panel C; Δ) were quantified with ImageQuant 5.0 and plotted as a function of time. Protein levels are expressed as a fraction of the input (zero time point). (E) Extracts from SGT1, sgt1-3, and sgt1-5 mutant yeast strains grown at 37°C for 3 h were equalized for Sgt1p and immunoprecipitated with antibodies against Sgt1p. Immune complexes were eluted and analyzed by immunoblotting with Skp1p polyclonal antibodies. The specificity of immunoprecipitates was confirmed with rabbit anti-mouse antibodies (RαM) in place of the Sgt1p antibody. Load represents 2% of the protein used for each IP. (F) Levels of Skp1p that were copurified in panel E were quantified with ImageQuant 5.0, and the data were normalized to the level of Sgt1p immunoprecipitated. (G) Sgt1p was immunoprecipitated from extracts of cells grown to log phase or grown in the presence of α factor (5 μg/ml) or nocodazole (15 μg/ml) for 2 h. Immune complexes were eluted and analyzed by immunoblotting with Skp1p polyclonal antibodies. The specificity of immunoprecipitates was confirmed with preimmune antibodies instead of Sgt1p antibody. Load represents 2% of the protein used for each IP. (H) Yeast cells transformed with a plasmid containing a GAL1 promoter and SGT1 (X), SGT1(1-268) (□), sgt1-5 (Δ), sgt1-5, L31P (○), or the vector alone (+) were grown at 30°C in the presence of galactose. Cell growth was monitored for 9 h, and the number of cells per milliliter was plotted. The lines graphed for the vector alone, SGT1, and sgt1-5, L31P overlap.
FIG. 6.
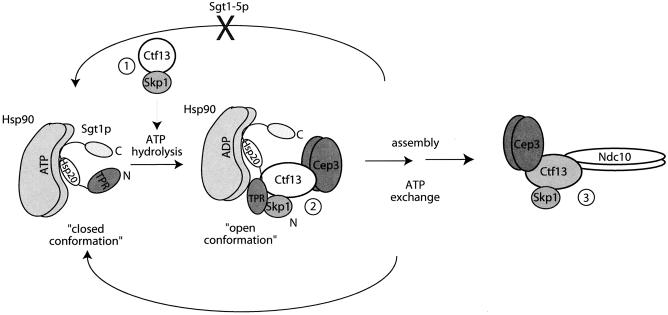
HSP90 activity mediates the transient assembly of intermediate CBF3 complexes. HSP90 bound to ATP interacts with Sgt1p and keeps it in a closed conformation, unable to bind to Skp1p and Ctf13p via its amino terminus. Following ATP hydrolysis, HSP90 contributes to the open conformation of Sgt1p, allowing Skp1p and Ctf13p to interact. The nucleotide exchange returns HSP90 to its ATP-bound state and Sgt1p to its closed conformation. The release of Sgt1p-HSP90 allows the final step in CBF3 assembly to occur. The Sgt1-5p mutant is unable to achieve the closed conformation and therefore cannot release Sgt1p-HSP90 from the Ctf13p complex.
To address whether the carboxy terminus of Sgt1p modulates Skp1p binding in cell extracts, we immunoprecipitated Sgt1p from extracts derived from wild-type or sgt1-5 yeast grown at 37°C. When equal amounts of Sgt1p and Sgt1-5p were isolated, we observed a dramatic enrichment (10-fold) in the levels of Skp1p that were copurified with Sgt1-5p compared to Sgt1p (Fig. 2E and F). To address whether the increase in Sgt1p-Skp1p affinity was a consequence of the G1 cell cycle arrest caused by sgt1-5, we examined the association between Sgt1p and Skp1p during different stages of the cell cycle. Sgt1p was isolated from extracts of wild-type yeast grown to log-phase, arrested in G1 with α factor, or arrested in G2/M with the microtubule poison nocodazole. Similar levels of Skp1p were copurified from each of these extracts, suggesting that the level of Sgt1p in complex with Skp1p does not vary during the cell cycle (Fig. 2G).
The increase in affinity between Sgt1-5p and Skp1p is consistent with an inhibitory role for the carboxy terminus of Sgt1p and raises the possibility that maintaining low levels of Sgt1p-Skp1p complexes in cells is important. Consistent with this possibility, growth of wild-type cells was inhibited by the GAL1-induced overexpression of sgt1-5 or sgt1(1-268) but not wild-type SGT1 (Fig. 2H). If inhibition of cell growth results from the high-affinity interaction between Sgt1-5p and Skp1p, we predicted that preventing Skp1p binding would alleviate the growth defect caused by overexpression of Sgt1-5p. To test this prediction, we introduced the L31P mutation into sgt1-5 to eliminate Skp1p binding (Fig. S1B in the supplemental material). Overexpression of sgt1-5, L31P did not inhibit cell growth, arguing that the high-affinity Sgt1p-Skp1p interaction is detrimental to cell growth (Fig. 2H; sgt1-5, L31P).
The stabilized interaction between Sgt1-5p and Skp1p decreases CBF3 levels.
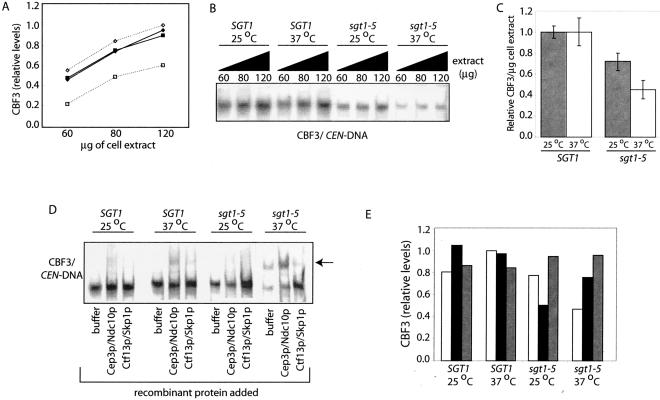
Previous work has demonstrated that CBF3 is maintained at remarkably constant levels by maintaining a balance of Ctf13p activation and ubiquitin-mediated degradation (18, 29, 30). Our recent observation that CBF3 is continually assembled during the cell cycle argues that it may be important to recycle complexes involved in Ctf13p activation (29). In this light, we reasoned that increasing the affinity between Sgt1p and Skp1p may cause them to become limiting for Ctf13p activation. To test this possibility, we carefully compared the levels of CBF3 complexes in extracts from wild-type and sgt1-5 cells grown at permissive and nonpermissive temperatures. To ensure that we could accurately measure changes in the steady-state levels of CBF3 complexes, we confirmed that the CBF3 band shift assay could detect a linear relationship between the amount of extract added and the level of the CBF3-CEN DNA complex measured (Fig. 3A). With this range of extract concentrations, we measured the relative levels of CBF3 in extracts derived from SGT1 and sgt1-5 mutant strains (Fig. 3B). Immunoblot analyses of Skp1p and tubulin were used to ensure that equal amounts of cell extract were analyzed under each condition (data not shown). Quantification showed that sgt1-5 extracts had 25 and 50% decreases in the levels of CBF3 at permissive and nonpermissive temperatures, respectively (Fig. 3B and C). To identify the limiting CBF3 subunit in sgt1-5 extracts, we performed biochemical complementation in extracts with recombinant CBF3 subunits produced in insect cells; specifically, we used insect cell extracts that contain Ctf13p/Skp1p or Cep3p/Ndc10p as previously described (29). Mixing of recombinant Ctf13/Skp1p with Cep3p/Ndc10p reconstituted CBF3 complexes in the absence of yeast extract, confirming that these subunits are active and sufficient to form CBF3 complexes, as previously reported (data not shown and reference 18). Addition of recombinant Ctf13p/Skp1p restored CBF3 to wild-type levels in sgt1-5 extracts, indicating that the levels of active Ctf13p are decreased in sgt1-5 cells (Fig. 3D and E). In contrast, addition of recombinant Cep3p/Ndc10p had no effect on the total levels of CBF3 in sgt1-5 extracts. It is interesting that the level of extended CBF3 complexes that include an additional dimer of Ndc10p, was increased when sgt1-5 is grown at a nonpermissive temperature (Fig. 3D, arrow). Formation of the extended CBF3 complex is favored specifically under conditions under which Ctf13p is limiting, thus offering additional evidence that Ctf13p activity is compromised in sgt1-5 cells (8). Although the decrease in CBF3 levels observed in sgt1-5 is not sufficient to compromise chromosome segregation (21), the observation that sgt1-5 fails to complement the sgt1-3 defect in a heteroallelic diploid strain argues that sgt1-5 also has a G2/M role and raises the intriguing possibility that subtle changes in CBF3 levels compromise other mitotic processes (21; see Discussion).
FIG. 3.
CBF3 levels are decreased in sgt1-5 mutant cells. (A) The linear range of the CBF3 band shift assay was confirmed by calculating the relative levels of CBF3-CEN DNA complexes detected with different amounts of extracts added to the reaction mixture from wild-type (⧫) and sgt1-5 mutant (▪) cells grown at room temperature (closed symbol) or 37°C (open symbol) for 3 h. (B) CBF3 levels were measured in the indicated amounts of extracts from wild-type or sgt1-5 mutant cells grown at room temperature or 37°C for 3 h by band shift assay. (C) The average amounts of CBF3-CEN DNA complexes were calculated per microgram of extracts made from SGT1 and sgt1-5 cells grown at 25°C (░⃞) or 37°C (□). Average values were statistically significantly different (paired Student t test, P < 0.005) between SGT1 and sgt1-5 at both temperatures. (D) Equal amounts of yeast extracts from the indicated strains were supplemented with recombinant Cep3p/Ndc10p or recombinant Ctf13p/Skp1p and analyzed for the CBF3 complex by band shift assay. (E) Relative levels of CBF3 measured when extracts were supplemented with buffer alone (□), recombinant Cep3p/Ndc10 (▪), or recombinant Ctf13p/Skp1p (░⃞) were quantified as for panel C.
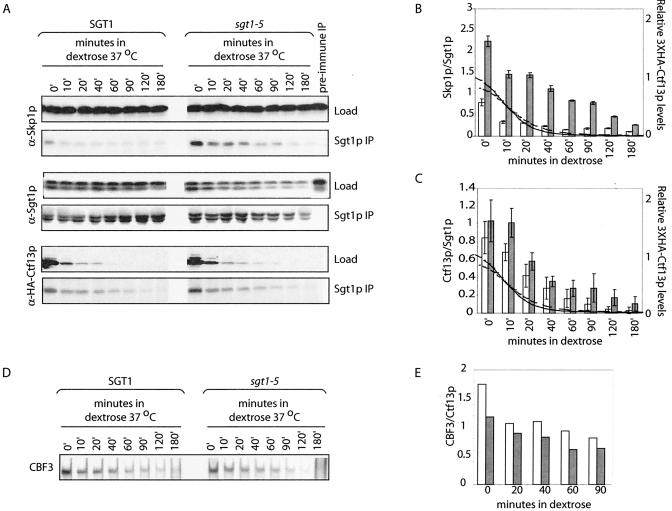
We speculate that the decrease in CBF3 levels observed in sgt1-5 is caused by the increased affinity between Sgt1-5p and Skp1p, effectively trapping complexes critical for Ctf13p activation. To test this possibility, we took advantage of a strain in which the assembly and turnover of CBF3 complexes can be easily monitored. As previously demonstrated, fusing a GAL1 promoter upstream of 3XHA-CTF13 allows depletion of 3XHA-Ctf13p when cells are grown in medium containing dextrose and its rapid induction when cells are grown in medium containing galactose (29). The very short half-life of 3XHA-Ctf13p and the contemporaneous decrease in CBF3 levels imply that as 3XHA-Ctf13p is degraded, CBF3 complexes turn over. For example, when cells are grown in the presence of galactose, significant levels of Sgt1p-Skp1p complexes can be detected; however, growing cells in dextrose inhibits de novo CBF3 assembly and rapidly depletes Sgt1p-Skp1p complexes (29). Therefore, with this system it is possible to monitor the half-life of complexes in the absence of de novo CBF3 assembly. To monitor the stability of Sgt1p and Sgt1-5p complexes, extracts from SGT1 and sgt1-5 cells were harvested at different time points following the transfer of cells to medium containing dextrose. As expected, 3XHA-Ctf13p levels rapidly decreased in SGT1 cells, exhibiting a half-life of 12 min (Fig. 4A, bottom). Interestingly, 3XHA-Ctf13p showed a similar half-life in sgt1-5 cells, even though this mutation has been reported to compromise the SCF-ubiquitin ligase complex required for Ctf13p degradation (18, 21). The levels of all other CBF3 subunits were unaltered by the growth of cells in dextrose (Fig. 4A, top, and data not shown). To monitor the levels of Sgt1p complexes in the absence of de novo CBF3 assembly, Sgt1p was purified from extracts with a polyclonal antibody and the purified proteins were immunoblotted to detect CBF3 subunits. At each time point, fewer Sgt1p-Skp1p than Sgt1-5-Skp1p complexes were observed (Fig. 4A, top, and B). While the levels of Sgt1p-Skp1p quickly decrease in the absence of CBF3 assembly, Sgt1-5p-Skp1p complexes persist throughout the time course. This stabilization appears to be independent of Ctf13p, as the rate of Ctf13p dissociation from Sgt1p mirrors its degradation in both wild-type and sgt1-5 mutant cells (Fig. 4B). Together, these data are consistent with the possibility that Sgt1-5p traps complexes involved in the assembly of CBF3.
FIG. 4.
Sgt1-5p increases the stability of an Sgt1p-Skp1p-Ctf13p complex. (A) SGT1 and sgt1-5 mutant cells containing the GAL1 promoter upstream of 3XHA-CTF13 were cultured at 37°C in medium containing galactose for 1.5 h, transferred to medium containing dextrose, and maintained at 37°C for 0, 10, 20, 40, 60, 90, 120, and 180 min. Extracts were immunoprecipitated with Sgt1p antibody, and immune complexes were eluted and immunoblotted with antibodies against Skp1p, Sgt1p, and HA to detect 3XHA-Ctf13p. The specificity of immunoprecipitates was confirmed with rabbit preimmune sera in place of the Sgt1p antibody. Load represents 2% of the protein used for each IP. (B) Levels of Skp1p immunoprecipitated with Sgt1p (□) or Sgt1-5p (▪) were quantified as described in the legend to Fig. 2F. To ensure accuracy, IPs were repeated in duplicate. Data presented are the mean and standard deviation calculated from duplicate experiments. Levels of 3XHA-Ctf13p in extract from SGT1 (solid line) or sgt1-5 (dashed line) mutant cells were quantified and normalized to the level in SGT1 mutant cell extract harvested at 0 min. (C) Levels of 3XHA-Ctf13p immunoprecipitated with Sgt1p (□) or Sgt1-5p (▪) depicted in panel A were quantified as described above. Lines represent relative levels of 3XHA-Ctf13p, as described for panel B. (D) CBF3 levels were measured in extracts from SGT1 and sgt1-5 mutant cells by band shift assay and quantified (E) relative to the amount of 3XHA-Ctf13p in SGT1 (□) or sgt1-5 (▪) mutant cell extracts.
We propose that the decreased levels of CBF3 observed in sgt1-5 extracts result from compromised Ctf13p activation, due to trapped Sgt1-5p-Skp1p complexes. Alternatively, it is possible that CBF3 complexes are less stable in sgt1-5 cells, contributing to the decrease in steady-state levels. To distinguish between these possibilities, we examined the turnover of CBF3 complexes following the inhibition of de novo CBF3 assembly. Following the switch of cells to medium containing dextrose, CBF3 levels were observed to decrease with similar kinetics in both SGT1 and sgt1-5 cells (Fig. 4D and E). We conclude that the disassociation of Sgt1p-Skp1p complexes is required to continually activate Ctf13p and assemble CBF3. Inhibition of assembly while maintaining a constant rate of CBF3 turnover therefore results in the lower steady-state levels of CBF3 observed in sgt1-5.
HSP90 activity regulates the interaction between Sgt1p and Skp1p.
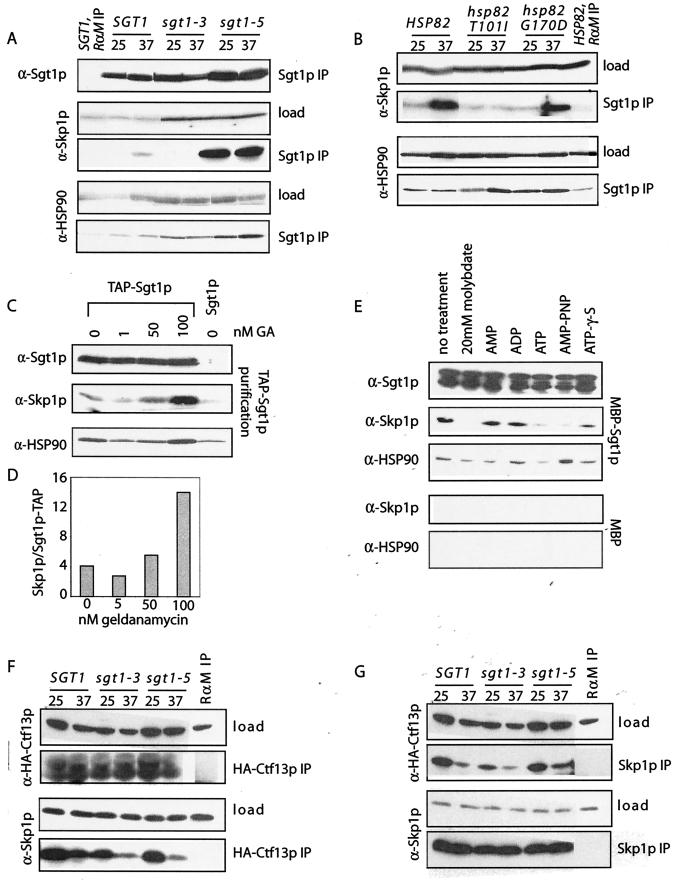
The carboxy terminus of Sgt1p inhibits its interaction with Skp1p and thus may serve as a switch to maintain the proper balance of these complexes. The recent finding that Sgt1p interacts with HSP90 chaperones raises the intriguing possibility that chaperones might regulate the interaction between Sgt1p and Skp1p (17, 22, 35). To address this possibility, we examined the relationship between Sgt1p and the HSP90 chaperones in yeast, encoded by HSP82 and HSC82. Following IP of equal amounts of Sgt1p from SGT1, sgt1-3, and sgt1-5 cells grown at permissive or nonpermissive temperatures, immunoblot analysis revealed that a small but significant percentage of HSP90 chaperones specifically were copurified with Sgt1p (Fig. 5A). This interaction is independent of Skp1p, as similar levels of HSP90 were copurified with Sgt1-3p, a mutant protein that fails to bind Skp1p (Fig. 5A). Similarly, the Sgt1-5p mutation does not affect its ability to interact with HSP90, arguing that the carboxy terminus of Sgt1p does not regulate HSP90 interaction as it does for Skp1p and Ctf13p. These findings were confirmed with an in vitro Hsp82p binding assay (Fig. S2 in the supplemental material) and are consistent with the recent demonstration that HSP90 interacts with the HSP20/α-crystallin fold (22). We conclude from these experiments that the interaction between Sgt1p and HSP90 chaperones is distinct from its interaction with either Skp1p or Ctf13p.
FIG.5.
HSP90 regulates the interaction between Skp1p and Sgt1p. (A) Extracts from SGT1, sgt1-3, and sgt1-5 mutant yeast strains grown at 37°C for 3 h were equalized for Sgt1p and immunoprecipitated with antibodies against Sgt1p. Immune complexes were eluted and analyzed by immunoblotting with antibodies against Skp1p or HSP90. The specificity of immunoprecipitates was confirmed with rabbit anti-mouse antibodies (RαM) in place of the Sgt1p antibody. Load represents 2% of the protein used for each IP. (B) Extracts from HSP82, hsp82-T101I, and hsp82-G170D mutant yeast strains grown at 37°C for 3 h were equalized for Sgt1p and analyzed as for panel A. (C) Sgt1-TAP was purified from extracts with IgG Sepharose in the presence of increasing amounts of geldanamycin (GA) and analyzed by immunoblotting with antibodies against Skp1p, Sgt1p, or HSP90. As a control, extract containing untagged Sgt1p was incubated with IgG Sepharose in the presence of dimethyl sulfoxide. (D) Levels of Skp1p and Sgt1p immunoprecipitated in panel C were quantified as described in the legend to Fig. 2F. (E) Purified recombinant MBP-Sgt1p or MBP was prebound to amylose resin and added to extracts from wild-type yeast cells supplemented with the indicated ATP analogue at 5 mM. Following the elution of MBP-Sgt1p, the levels of Skp1p, HSP90, and Sgt1p were determined by immunoblotting with the appropriate antibody. (F) Extracts from SGT1, sgt1-3, and sgt1-5 mutant yeast cells containing the GAL1 promoter upstream of 3XHA-CTF13 and grown at 25 or 37°C in galactose medium for 3 h were immunoprecipitated with antibodies against HA, and immune complexes were eluted and immunoblotted with antibodies against Skp1p and HA to detect 3XHA-Ctf13p. The specificity of immunoprecipitates was confirmed with rabbit anti-mouse antibodies (labeled RαM) in place of the HA antibody. Load represents 2% of the protein used for each IP. (G) Yeast extracts described in panel F were immunoprecipitated with Skp1p antibody, and immune complexes were eluted and immunoblotted against Skp1p and HA to detect 3XHA-Ctf13p.
It is possible that Sgt1p is an HSP90 client, requiring the activity of the chaperone to properly fold and interact with Skp1p. To address this possibility, we examined the effect HSP82 mutants have on the ability of Sgt1p to associate with Skp1p. Strains carrying deletions of HSC82 and HSP82 were kept alive by plasmids containing wild-type HSP82 or one of two previously characterized mutant alleles, hsp82-T101I or hsp82-G170D (24, 25). When grown at nonpermissive temperatures, Sgt1p immunoprecipitates from hsp82-T101I contained little Skp1p compared to the wild type (Fig. 5B). In contrast, Sgt1p immunoprecipitates from hsp82-G170D contained levels of Skp1p comparable to those of the wild type. The differential effect of the HSP82 mutations on the Sgt1p-Skp1p interaction led us to consider how these HSP82 mutations affect the activity of the chaperone. The T101I change lies in the catalytic core of Hsp82p and has been previously shown to slow the rate of ATP hydrolysis (37). In contrast, the G170D change lies in the ATP binding domain and inhibits the ability of ATP to bind to Hsp82p (G. Hutchins and B. Errede, personal communication). Thus, it is possible that the interaction between Sgt1p and Skp1p is sensitive to the nucleotide-bound state of Hsp82p. To test this possibility, we isolated affinity-tagged Sgt1p (Sgt1p-TAP) in the absence or the presence of increasing concentrations of the drug geldanamycin. Geldanamycin competes with ATP for binding, resulting in HSP90 chaperones with no nucleotide bound (33), a state that we presume to be analogous to that of the hsp82-G170D allele. In the absence of geldanamycin, low levels of Skp1p were copurified with Sgt1p-TAP, consistent with the relatively weak interaction between these two proteins (Fig. 5C; compare to Fig. 1). With increasing concentrations of geldanamycin, we observed a nearly fourfold increase in the levels of Skp1p that were copurified with Sgt1p-TAP (Fig. 5C and D). Similar levels of Sgt1p and associated HSP90 chaperones were purified under all of the conditions tested, suggesting that only the association of Sgt1p and Skp1p is affected by the nucleotide-bound state of HSP90.
To further examine the relationship between nucleotide hydrolysis and the Sgt1p-Skp1p interaction, we established an in vitro binding assay with recombinant Sgt1p fused to MBP (MBP-Sgt1p) and yeast cell extracts. Equal amounts of recombinant MBP-Sgt1p immobilized on amylose resin were incubated with extracts alone or in the presence of nucleotide analogues and then subject to immunoblot analysis. With no treatment, low but significant levels of Skp1p associated with MBP-Sgt1p in extracts (Fig. 5E). Importantly, HSP90 was also observed to associate with recombinant MBP-Sgt1p, implying that the in vitro binding assay reassembles a complex containing at least Sgt1p, Skp1p, and HSP90. Control incubations with recombinant MBP demonstrated the specificity of the binding assay (Fig. 5E; bottom). To test the sensitivity of these interactions to the nucleotide-bound state of HSP90 chaperones, various nucleotide analogues were added to the purifications. Treating extracts with 20 mM molybdate, which inhibits ATP hydrolysis by HSP90, completely inhibited the copurification of Skp1p with MBP-Sgt1p, suggesting that the ATP-bound form of HSP90s inhibits Sgt1p-Skp1p interactions. Consistent with this possibility, the addition of ATP or nonhydrolyzable analogues of ATP (AMP-PNP and ATP-γ-S) eliminated or reduced the interaction between MBP-Sgt1p and Skp1p. In contrast, addition of AMP or ADP enhanced the interaction between MBP-Sgt1p and Skp1p, arguing that hydrolysis of ATP by HSP90 is required for the stabilization of Sgt1p and Skp1p complexes. Consistent with our observations with HSP82 mutants, the nucleotide-bound state of HSP90 did not affect its affinity for MBP-Sgt1p, suggesting a non-client-like interaction between Sgt1p and HSP90 chaperones. Together, these data support a role for HSP90 in regulating the interaction between Sgt1p and Skp1p, a role that we suggest is critical for maintaining the proper balance of assembled Sgt1p-Skp1p complexes.
Through its association with HSP90, Sgt1p has been proposed to function as a cochaperone that facilitates the formation of a Ctf13p-Skp1p heterodimeric complex required for Ctf13p activation (34). This hypothesis predicts that mutations that disrupt the interaction between Sgt1p and Skp1p will fail to form the Ctf13p-Skp1p heterodimer. To test this possibility, we examined the formation of Ctf13p-Skp1p heterodimers in sgt1-3; as shown above, Sgt1-3p interacts with HSP90 but not with Skp1p and prevents Ctf13p activation (21). SGT1, sgt1-3, and sgt1-5 mutant strains expressing 3XHA-Ctf13p were grown at a permissive or nonpermissive temperature. The interaction between 3XHA-Ctf13p and Skp1p was monitored by immunoblotting following the isolation of Skp1p or 3XHA-Ctf13p complexes. In all backgrounds, we observed a decrease in the levels of 3XHA-Ctf13p-Skp1p complexes at 37°C, with slightly fewer complexes observed in the SGT1 mutants. However, there were no significant differences in the levels of 3XHA-Ctf13p-Skp1p complexes isolated from sgt1-3 and sgt1-5 mutant strains, arguing that changes in the affinity between Sgt1p and Skp1p do not alter the ability of Ctf13p and Skp1p to interact (Fig. 5F and G). These results are consistent with previous observations that Ctf13p interacts normally with Skp1-4p, an allele that fails to bind Sgt1p (18, 29). Together these results argue that interaction between Sgt1p and Skp1p is not required to form Ctf13p-Skp1p heterodimers.
DISCUSSION
The need to maintain transient interactions between proteins has been associated with systems that require plasticity, either to ensure that activated signaling complexes do not persist or to reassemble protein complexes to carry out cyclical tasks. Here we show that the HSP90 class of chaperones is intimately linked to the formation of a transient Sgt1p-Skp1p complex. Our data suggest that the transient nature of this interaction is maintained through the carboxy terminus of Sgt1p. Compromising this regulatory region traps assembled Sgt1p complexes and impairs the ability of cells to maintain normal levels of CBF3. We speculate that Sgt1p acts to transiently link HSP90 chaperones to protein complexes, ensuring that the assembly of complexes is coupled to their turnover.
Although mutations that trap Sgt1p complexes (i.e., sgt1-5) do not compromise chromosome segregation, our observations suggest that even minor changes in the levels of CBF3 impair cytokinesis and cell wall integrity (A. Gillis, S. Thomas, and K. Kaplan, unpublished observations). These observations are consistent with the finding that in Schizosaccharomyces pombe, mutations in the SGT1 homologue git7 produce defects in cell wall integrity and cytokinesis (31). It is tempting to speculate that changes in the levels of CBF3 or other kinetochore complexes may have important regulatory consequences, possibly affecting cell cycle advance and the completion of mitosis. In this light, it will be interesting to compare SGT1 alleles with other mutants that affect CBF3 assembly to determine if CBF3 is directly linked to the regulation of other mitotic events.
While we have focused on the role of Sgt1p in CBF3 assembly, genetic studies suggest that SGT1 is involved in regulating the activity of protein kinase A and the SCF ubiquitin ligase. Although it remains possible that the Sgt1p-Skp1p interaction is important for these other pathways, mutations that disrupt the Sgt1p-Skp1p interaction appear to specifically affect CBF3 and not SCF or protein kinase A (7, 21, 29). Nonetheless, it remains possible that other pathways require proper regulation of the Sgt1p-Skp1p interaction. In this light, it will be interesting to determine whether HSP90 regulates the association of other Sgt1p-interacting proteins important for the function of protein kinase A or SCF.
Sgt1p is a multidomain, HSP90-associated protein.
Sgt1p is a highly conserved protein that contains several recognizable protein motifs. The amino terminus of Sgt1p has homologies to a TPR domain and an HSP20/α-crystallin fold, domains that are known to mediate protein-protein interactions and are found in a class of HSP90-interacting proteins called cochaperones. Our data obtained with Sgt1p deletions and point mutations suggest that the putative TPR domain in Sgt1p is involved in both Skp1p and Ctf13p binding. The binding of HSP90 to Sgt1p is independent of Skp1p binding, as the sgt1-3 mutation eliminates Skp1p but not HSP90 interactions. These data are consistent with the recent report that human Sgt1p interacts with HSP90 via the middle region containing the HSP20/α-crystallin fold (22). Identifying the precise protein fold required for binding Skp1p and Ctf13p requires further mutational and structural characterizations. In total, our data strongly argue that Sgt1p interacts with target proteins (e.g., Skp1p and Ctf13p) via its amino-terminal TPR domain.
Our data implicate the carboxy terminus of Sgt1p in ensuring its transient and low-affinity interaction with Skp1p. We propose that the carboxy terminus inhibits protein interactions by occluding amino-terminal binding domains (Fig. 6). A number of possible models can explain the inhibitory function of the carboxy terminus of Sgt1p. The presence of an exposed loop in the middle of Sgt1p argues that the carboxy terminus is flexible and thus may fold to mask amino-terminal binding sites through intramolecular interactions. The flexibility of Sgt1p domains has also recently been suggested on the basis of nuclear magnetic resonance spectroscopy (22). Alternatively, it is possible that Sgt1p forms oligomers and thus occludes amino-terminal binding sites through an intermolecular interaction. Finally, it is possible that the carboxy terminus specifically associates with another cellular protein(s) that inhibits binding of Skp1p and Ctf13p. However, purified recombinant Sgt1p produced in bacteria also has a low affinity for Skp1p, arguing that any associated regulator is not specific to eukaryotes and would have to act at submolar ratios.
One intriguing implication of our work is that the conformation of Sgt1p is regulated by HSP90 chaperones. In the model outlined in Fig. 6, we propose that HSP90 binds to the HSP20/α-crystallin domain of Sgt1p; ATP hydrolysis stabilizes an intermediate folding state that allows the association of Skp1p. Following ATP exchange, the carboxy terminus of Sgt1p refolds to occlude Skp1 binding sites, thus ensuring the transient nature of this interaction. Mutations in Sgt1p that perturb the conformation of the carboxy terminus may bypass the requirement for HSP90-assisted folding to bind Skp1p. In this light, it will be interesting to test if HSP82 mutants still inhibit CBF3 in the sgt1-5 background.
While this model suggests that Sgt1p is a client of HSP90, the presence of the conserved HSP20/α-crystallin domain raises the possibility that Sgt1p functions as a cochaperone. Consistent with this possibility, recent studies have suggested that HSP90 interacts with Sgt1p through its p23-like HSP20/α-crystallin domain (22). However, unlike the p23 cochaperone, which preferentially interacts with the ATP-bound form of HSP90 (9), Sgt1p-HSP90 interaction is nucleotide independent. For similar reasons, the nucleotide-independent association between Sgt1p and HSP90 suggests that Sgt1p is not a typical chaperone client. Rather, our data are most consistent with Sgt1p belonging to a novel class of HSP90-interacting proteins. On the basis of our observations, we propose that Sgt1p acts as a scaffold that may link together a number of proteins critical for CBF3 assembly. In this model, we propose that the role of HSP90 is to maintain the transient interaction of these proteins, ensuring that a constant pool of these proteins is available for continual protein complex assembly.
A role for Sgt1p in maintaining the balance of protein complex assembly and turnover.
Once recruited to a complex by Sgt1p, HSP90 may act to ensure that Sgt1p is only transiently associated. Alternatively, together Sgt1p and HSP90 may stabilize folding intermediates critical in the stepwise assembly of protein complexes. Recent work suggests that HSP90 stabilizes the Ctf13p-Skp1p interaction and speculates that Sgt1p may link HSP90 to Ctf13p (34). However, our data suggest that Sgt1p binding to Skp1p is not required to stabilize the Ctf13p-Skp1p interaction. It is possible that Sgt1p recruits HSP90 to the preformed Ctf13p-Skp1p heterodimer or perhaps recruits other cellular proteins to this complex. One possibility is that HSP90 activity is important to transport partially assembled Ctf13p complexes into the nucleus for final assembly and binding to CEN DNA. Such a role would be analogous to the role of HSP90 in nuclear hormone receptor regulation. In this model, HSP90 promotes the release of Sgt1p from the complex (step 2 in Fig. 6) and a transition in Ctf13p conformation that supports nuclear import and the binding to the Ndc10p subunit (step 3 in Fig. 6). This model is consistent with the recently characterized steps involved in CBF3 assembly; Ctf13p initially binds Cep3p, Skp1p, and Sgt1p, which recruits HSP90; release of Sgt1p and therefore HSP90 allows Ctf13p to assume a new conformation that promotes Ndc10p binding, the final step in CBF3 assembly (29).
Previous data clearly demonstrate the importance of the Sgt1p-Skp1p interaction for Ctf13p function and CBF3 assembly (18, 21). However, our data suggest that the dissociation of Sgt1p and Skp1 is also important; stabilization of the Sgt1p-Skp1p interaction traps complexes that we propose are then unavailable for de novo assembly of CBF3 complexes (Fig. 4). Although the initial analysis of sgt1-5 did not remark on the decrease in CBF3 levels, we note that there is a clear decrease in the CBF3 level in sgt1-5 cells grown at 37°C compared to 25°C (21). In addition, our previous observation that Skp1-3p also forms a stable complex with Sgt1p and stabilizes CBF3 suggests that the process of Sgt1p-Skp1p release is coupled to the turnover of CBF3 complexes by mechanisms that are not completely understood (29). Perhaps the turnover of CBF3 complexes is analogous to the previously observed turnover of F box subunits in the ubiquitin ligase complex SCF (38). In this complex, the turnover of F box subunits is critical for forming de novo SCF complexes required to target different substrates throughout the cell cycle. The association of Sgt1p with SCF raises the possibility that Sgt1p may function generally in linking complex assembly with turnover. Further characterization of protein complex turnover is required to fully appreciate the role of Sgt1p in maintaining the proper balance of protein complexes involved in cell signaling and chromosome segregation.
Supplementary Material
Acknowledgments
We acknowledge the American Cancer Society (RSG-02-035-01-CCG) and the Sidney Kimmel Cancer Foundation for funding of K.B.K. L.B.L. was supported in part by an NIGMS MORE Institutional Research and Academic Career Development Award to UC Davis and San Francisco State University (grant K12GM00679).
We thank members of the Kaplan laboratory for helpful discussion and editorial comments on the manuscript.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Austin, M. J., P. Muskett, K. Kahn, B. J. Feys, J. D. Jones, and J. E. Parker. 2002. Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295:2077-2080. [DOI] [PubMed] [Google Scholar]
- 2.Azevedo, C., A. Sadanandom, K. Kitagawa, A. Freialdenhoven, K. Shirasu, and P. Schulze-Lefert. 2002. The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295:2073-2076. [DOI] [PubMed] [Google Scholar]
- 3.Boeke, J. D., J. Trueheart, G. Natsoulis, and G. R. Fink. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154:164-175. [DOI] [PubMed] [Google Scholar]
- 4.Buchner, J. 1999. Hsp90 & Co.—a holding for folding. Trends Biochem. Sci. 24:136-141. [DOI] [PubMed] [Google Scholar]
- 5.Caplan, A. J. 1999. Hsp90's secrets unfold: new insights from structural and functional studies. Trends Cell Biol. 9:262-268. [DOI] [PubMed] [Google Scholar]
- 6.Cope, G. A., and R. J. Deshaies. 2003. COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell 114:663-671. [DOI] [PubMed] [Google Scholar]
- 7.Dubacq, C., R. Guerois, R. Courbeyrette, K. Kitagawa, and C. Mann. 2002. Sgt1p contributes to cyclic AMP pathway activity and physically interacts with the adenylyl cyclase Cyr1p/Cdc35p in budding yeast. Eukaryot. Cell 1:568-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Espelin, C. W., K. B. Kaplan, and P. K. Sorger. 1997. Probing the architecture of a simple kinetochore using DNA-protein crosslinking. J. Cell Biol. 139:1383-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felts, S. J., and D. O. Toft. 2003. p23, a simple protein with complex activities. Cell Stress Chaperones 8:108-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freeman, B. C., and K. R. Yamamoto. 2001. Continuous recycling: a mechanism for modulatory signal transduction. Trends Biochem. Sci. 26:285-290. [DOI] [PubMed] [Google Scholar]
- 11.Freeman, B. C., and K. R. Yamamoto. 2002. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science 296:2232-2235. [DOI] [PubMed] [Google Scholar]
- 12.Frydman, J., and J. Hohfeld. 1997. Chaperones get in touch: the Hip-Hop connection. Trends Biochem. Sci. 22:87-92. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Ranea, J. A., G. Mirey, J. Camonis, and A. Valencia. 2002. p23 and HSP20/alpha-crystallin proteins define a conserved sequence domain present in other eukaryotic protein families. FEBS Lett. 529:162-167. [DOI] [PubMed] [Google Scholar]
- 14.Gietz, R. D., and R. A. Woods. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350:87-96. [DOI] [PubMed] [Google Scholar]
- 15.Guthrie, C., and G. R. Fink. 1991. Guide to yeast genetics and molecular biology. Academic Press, San Diego, Calif.
- 16.Hohfeld, J., D. M. Cyr, and C. Patterson. 2001. From the cradle to the grave: molecular chaperones that may choose between folding and degradation. EMBO Rep. 2:885-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hubert, D. A., P. Tornero, Y. Belkhadir, P. Krishna, A. Takahashi, K. Shirasu, and J. L. Dangl. 2003. Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J. 22:5679-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplan, K. B., A. A. Hyman, and P. K. Sorger. 1997. Regulating the yeast kinetochore by ubiquitin-dependent degradation and Skp1p-mediated phosphorylation. Cell 91:491-500. [DOI] [PubMed] [Google Scholar]
- 19.Kaplan, K. B., and P. K. Sorger. 1997. Purification of sequence-specific DNA-binding proteins, p. 245-278. In T. E. Creighton (ed.), Protein function: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 20.Kirschner, M. 1999. Intracellular proteolysis. Trends Cell Biol. 9:M42-M45. [PubMed] [Google Scholar]
- 21.Kitagawa, K., D. Skowyra, S. J. Elledge, J. W. Harper, and P. Hieter. 1999. SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol. Cell 4:21-33. [DOI] [PubMed] [Google Scholar]
- 22.Lee, Y. T., J. Jacob, W. Michowski, M. Nowotny, J. Kuznicki, and W. J. Chazin. 2004. Human Sgt1 binds HSP90 through the CS domain and not the TPR domain. J. Biol. Chem. 279:16511-16517. [DOI] [PubMed] [Google Scholar]
- 23.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 24.Nathan, D. F., and S. Lindquist. 1995. Mutational analysis of Hsp90 function: interactions with a steroid receptor and a protein kinase. Mol. Cell. Biol. 15:3917-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nathan, D. F., M. H. Vos, and S. Lindquist. 1997. In vivo functions of the Saccharomyces cerevisiae Hsp90 chaperone. Proc. Natl. Acad. Sci. USA 94:12949-12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prodromou, C., G. Siligardi, R. O'Brien, D. N. Woolfson, L. Regan, B. Panaretou, J. E. Ladbury, P. W. Piper, and L. H. Pearl. 1999. Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. EMBO J. 18:754-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riedel, N., J. A. Wise, H. Swerdlow, A. Mak, and C. Guthrie. 1986. Small nuclear RNAs from Saccharomyces cerevisiae: unexpected diversity in abundance, size, and molecular complexity. Proc. Natl. Acad. Sci. USA 83:8097-8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 29.Rodrigo-Brenni, M. C., S. Thomas, D. C. Bouck, and K. B. Kaplan. 2004. Sgt1p and Skp1p modulate the assembly and turnover of CBF3 complexes required for proper kinetochore function. Mol. Biol. Cell 15:3366-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russell, I. D., A. S. Grancell, and P. K. Sorger. 1999. The unstable F-box protein p58-Ctf13 forms the structural core of the CBF3 kinetochore complex. J. Cell Biol. 145:933-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schadick, K., H. M. Fourcade, P. Boumenot, J. J. Seitz, J. L. Morrell, L. Chang, K. L. Gould, J. F. Partridge, R. C. Allshire, K. Kitagawa, P. Hieter, and C. S. Hoffman. 2002. Schizosaccharomyces pombe Git7p, a member of the Saccharomyces cerevisiae Sgtlp family, is required for glucose and cyclic AMP signaling, cell wall integrity, and septation. Eukaryot. Cell 1:558-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheufler, C., A. Brinker, G. Bourenkov, S. Pegoraro, L. Moroder, H. Bartunik, F. U. Hartl, and I. Moarefi. 2000. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 101:199-210. [DOI] [PubMed] [Google Scholar]
- 32a.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle rectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stebbins, C. E., A. A. Russo, C. Schneider, N. Rosen, F. U. Hartl, and N. P. Pavletich. 1997. Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell 89:239-250. [DOI] [PubMed] [Google Scholar]
- 34.Stemmann, O., A. Neidig, T. Kocher, M. Wilm, and J. Lechner. 2002. Hsp90 enables Ctf13p/Skp1p to nucleate the budding yeast kinetochore. Proc. Natl. Acad. Sci. USA 99:8585-8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi, A., C. Casais, K. Ichimura, and K. Shirasu. 2003. HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. USA 100:11777-11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tor, M., P. Gordon, A. Cuzick, T. Eulgem, E. Sinapidou, F. Mert-Turk, C. Can, J. L. Dangl, and E. B. Holub. 2002. Arabidopsis SGT1b is required for defense signaling conferred by several downy mildew resistance genes. Plant Cell 14:993-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young, J. C., and F. U. Hartl. 2000. Polypeptide release by Hsp90 involves ATP hydrolysis and is enhanced by the co-chaperone p23. EMBO J. 19:5930-5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou, P., and P. M. Howley. 1998. Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol. Cell 2:571-580. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.