Abstract
Estrogen-related receptors (ERRs) are orphan nuclear receptors activated by the transcriptional coactivator peroxisome proliferator-activated receptor γ (PPARγ) coactivator 1α (PGC-1α), a critical regulator of cellular energy metabolism. However, metabolic target genes downstream of ERRα have not been well defined. To identify ERRα-regulated pathways in tissues with high energy demand such as the heart, gene expression profiling was performed with primary neonatal cardiac myocytes overexpressing ERRα. ERRα upregulated a subset of PGC-1α target genes involved in multiple energy production pathways, including cellular fatty acid transport, mitochondrial and peroxisomal fatty acid oxidation, and mitochondrial respiration. These results were validated by independent analyses in cardiac myocytes, C2C12 myotubes, and cardiac and skeletal muscle of ERRα−/− mice. Consistent with the gene expression results, ERRα increased myocyte lipid accumulation and fatty acid oxidation rates. Many of the genes regulated by ERRα are known targets for the nuclear receptor PPARα, and therefore, the interaction between these regulatory pathways was explored. ERRα activated PPARα gene expression via direct binding of ERRα to the PPARα gene promoter. Furthermore, in fibroblasts null for PPARα and ERRα, the ability of ERRα to activate several PPARα targets and to increase cellular fatty acid oxidation rates was abolished. PGC-1α was also shown to activate ERRα gene expression. We conclude that ERRα serves as a critical nodal point in the regulatory circuitry downstream of PGC-1α to direct the transcription of genes involved in mitochondrial energy-producing pathways in cardiac and skeletal muscle.
The essential role of nuclear receptors in regulating various cellular metabolic pathways is becoming increasingly evident. In recent years, various nuclear receptors that do not respond to classical endocrine ligands, including peroxisome proliferator-activated receptors (PPARs), liver X receptors, farnesoid X receptors, and retinoid X receptors, have been shown to be activated by low-affinity diet-derived ligands (6, 11, 26, 44). Activation of these receptors by metabolite ligands such as fatty acids, oxysterols, and bile acids elicits downstream transcriptional regulation of pathways involved in synthesis and catabolism of these ligands. The remaining receptors, designated orphan receptors because endogenous ligands have not been identified, comprise the largest subcategory of nuclear receptors. It is likely that orphan receptors serve additional roles in regulating intermediary metabolism. Linking orphan receptors to target genes is an important goal in the field of nuclear receptor biology. Target gene profiling will also provide insights for determining what metabolites serve as endogenous ligands for these receptors and, in turn, for developing pharmacologic interventions designed to regulate cellular metabolism.
One group of orphan receptors recently identified as candidate regulators of cellular metabolism are the estrogen-related receptor (ERR) family. There are three members of the ERR family, ERRα, ERRβ, and ERRγ (13, 16, 18). Early descriptions of the tissue and developmental expression patterns of ERR isoforms in mammalian organisms suggests that they are involved in the regulation of cellular energy metabolism. For example, in adults, ERRα and ERRγ expression is enriched in tissues that rely primarily on mitochondrial oxidative metabolism for energy generation, such as heart, brown adipose, and slow-twitch skeletal muscle (16, 43, 47). During embryonic development, both isoforms are expressed in heart and skeletal muscle, suggesting that they serve functional roles during development as well as for maintaining function in differentiated muscle. We demonstrated a dramatic increase in cardiac ERRα expression after birth, coincident with the postnatal switch to fatty acids as an energy substrate and the global upregulation of enzymes involved in cellular fatty acid uptake and mitochondrial oxidation (19). Moreover, ERRα binds the 5′ regulatory region of the gene encoding medium-chain acyl coenzyme A dehydrogenase (MCAD), which catalyzes the first step of the mitochondrial β-oxidation pathway and therefore was implicated in the direct regulation of fatty acid oxidation pathways (43, 47).
Early attempts failed to demonstrate ERRα-mediated activation of MCAD gene transcription in various cell culture models, suggesting that the cells lacked a key functional component of ERRα signaling. Recently, we and others identified members of the PPARγ coactivator 1 (PGC-1) family of transcriptional coactivators as potent coactivators for ERRα and ERRγ (17, 19, 21, 42). Three PGC-1 isoforms have been characterized, PGC-1α, PGC-1β, and PRC. PGC-1α is a key regulator of an array of cellular energy metabolic pathways, but its primary effect in target tissues is to enhance mitochondrial oxidative metabolism (24, 37). PGC-1α increases cellular mitochondrial number, fatty acid oxidation, and respiration via coactivation of a number of nuclear receptor and non-nuclear receptor transcription factor partners (29, 38, 49). PGC-1β is also thought to activate oxidative metabolism in tissues, though it does so through a relatively restricted set of transcriptional partners compared to PGC-1α (31, 45).
Shared PGC-1α and PGC-1β partners include ERRα and ERRγ, nuclear respiratory factor 1 (NRF-1), hepatocyte nuclear factor 4, estrogen receptor α, and peroxisome proliferator-activated receptor α (PPARα) (17, 19, 21, 30, 42, 46, 49). Hence, ERR isoforms likely confer PGC-1-mediated regulation on ERR target genes in tissues where ERRα, ERRγ, and PGC-1 coactivators are coexpressed, such as heart and skeletal muscle. Indeed, we demonstrated that the ERRα/PGC-1α complex directly activated the MCAD gene promoter through the ERRα binding site identified in earlier studies and that ERRα overexpression activated endogenous MCAD gene expression in NIH 3T3 cells (19). Collectively, the published results to date suggest that ERRs serve as a component of the regulatory circuitry downstream of PGC-1 and have stimulated interest in defining the metabolic roles of ERRα and related isoforms. However, the specific target genes and related metabolic pathways regulated by ERR isoforms have not been defined.
In order to identify potential target genes of ERRα, transcriptional profiling studies were performed in rat neonatal cardiac myocytes overexpressing ERRα. Validation studies were performed in cell culture and in vivo in heart and skeletal muscle of ERRα null mice. These studies unveiled several key regulatory functions for ERRα. First, we found that ERRα activates genes involved in multiple key energy production pathways, including cellular fatty acid uptake, fatty acid oxidation, and mitochondrial electron transport/oxidative phosphorylation. Second, ERRα-mediated regulation of fatty acid utilization genes occurs, at least in part, through direct activation of PPARα gene transcription, a mechanism that is coactivated by PGC-1α. Collectively, these results identify ERRα as a critical regulator of energy metabolism in heart and skeletal muscle.
MATERIALS AND METHODS
Plasmid constructs.
The wild-type and mutated forms of the human PPARα promoter-reporter plasmids have been described (35, 36). The mammalian expression vectors expressing human ERRα and mouse PGC-1α, pcDNA3.1-ERRα and pcDNA3.1-myc/his.PGC-1α, have also been described (19, 46). The pSG5-HA-ERRγ expression vector was a kind gift from M. Stallcup (University of Southern California).
Mammalian cell culture and transient transfections.
CV1 cells were maintained at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium-10% fetal calf serum. Transient transfections were performed by the calcium phosphate coprecipitation method (9). Reporter plasmids (4 μg/ml) were cotransfected with RSV-βGal (0.5 μg/ml), expressing the β-galactosidase gene driven by the Rous sarcoma virus promoter, to control for transfection efficiency. For cotransfection experiments, mammalian expression vectors expressing ERR isoforms, PGC-1α, or the corresponding empty vectors were used. Cells were collected and assayed 48 h posttransfection, and luciferase and β-galactosidase activities were measured as described previously (5).
Ventricular cardiac myocytes were prepared from 1-day-old Harlan-Sprague-Dawley rats as described previously (9). After 24 h, cells were infected with adenovirus expressing green fluorescent protein (GFP), ERRα, PGC-1α, or PPARα driven by a cytomegalovirus promoter. The experimental constructs also express GFP from an independent promoter. Infection rates of 90 to 95% were achieved by 18 h as assessed by quantitation of GFP-expressing cells with fluorescence microscopy. RNA and whole-cell protein extracts were prepared from cells 48 and 72 h postinfection, respectively. The construction and propagation of adenovirus expressing GFP, human ERRα, and mouse PGC-1α or mouse PPARα have been described (2, 15, 19, 20, 28).
Primary mouse fibroblasts were prepared from tail tissue of wild-type, ERRα−/− (ERR knockout), or ERRα−/− PPARα−/− (double-knockout) mice. Isolation and culturing of fibroblasts were performed in complete medium (Dulbecco's modified Eagle's medium, 20% fetal calf serum, 2 mM l-glutamine, 1 mM nonessential amino acids, and 100 μg of penicillin/streptomycin per ml). In brief, tail clips (1 cm) were soaked 10 min in complete medium with 500 μg/ml penicillin/streptomycin per ml, rinsed, and minced in medium with no penicillin/streptomycin. Tissue pieces were digested overnight at 37°C in complete medium plus 500 U collagenase. Cells were liberated by triturating with a 5-ml pipette, collected by centrifugation, and cultured in T25 flasks in complete medium. After 48 h cells were subcultured and expanded to determine appropriate viral titration and used at passage two or three for overexpression experiments.
DNA microarray.
Total RNA was isolated from rat neonatal cardiac myocytes with RNAzol (Tel-Test Inc.) followed by an RNeasy kit (Qiagen Inc.) clean-up. Double-stranded cDNA was synthesized from 12 μg of total RNA that was first reverse transcribed with Superscript II (Invitrogen Corp.) with a T7 promoter-poly(A) primer (T7T24) followed by second-strand synthesis according to the manufacturer's protocol. Biotin-labeled cRNA was synthesized with T7-coupled ENZO BioArray High-Yield RNA transcript labeling kit (ENZO Diagnostics Inc.) following the manufacturer's protocol.
The Alvin Siteman Cancer Center's Multiplexed Gene Analysis Core at Washington University School of Medicine performed hybridization to the Affymetrix rat U34A chip. Affymetrix MAS 5.0 software was used for the initial data analysis and background normalization. Subsequent data manipulations were performed in Excel. Probe sets that were called absent by Affymetrix software in both GFP and ERRα overexpression conditions were excluded from subsequent data analysis. Signal intensities were normalized to the average intensity for all probe sets. Signal intensity ratios were calculated as the ratio of ERRα-expressing to GFP-expressing cells in order to detect changes due to ERRα overexpression. Three independent trials were performed. Signal intensity ratios that increased ≥2-fold (induced) or decreased ≤0.5-fold (repressed) in at least two trials were considered potentially regulated in ERRα-expressing cells.
Northern and immunoblot analyses.
Total cellular RNA isolation and blotting were performed as described previously (5). Blots were hybridized with radiolabeled probes derived from cDNA mouse clones for MCAD, ERRγ, PPARα, and PGC-1α. In addition, human ERRα, rat M-CPT I, and universal actin probes were used. Protein extracts were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (7.5%) and transferred to nitrocellulose membranes. Immunodetection of MCAD and acyl-coenzyme A oxidase was performed with the previously described anti-MCAD and anti-acyl-coenzyme A oxidase antibodies (7, 22). The fatty acyl coenzyme A synthetase 1 antibody was generously provided by J. E. Schaffer (Washington University School of Medicine).
Real-time quantitative reverse transcription-PCR.
Real-time PCR was performed as described previously (40). Total RNA isolated from the soleus muscle of 8-week-old wild-type or ERRα−/− mice was reverse transcribed with Taqman reverse transcription reagents (Applied Biosystems) with oligo(dT) and random hexamers (1:1 ratio). Reactions were performed in triplicate in the 96-well format with Taqman core reagents and a Prism 7700 sequence detector (Applied Biosystems). The PGC-1α primer-probe set has been described (3). The following mouse-specific primer-probe sets were used to detect specific gene expression: MCAD forward, 5′-GGAAATGATCAACAAAAAAAGAAGTATTT-3′; MCAD reverse, 5′-ATGGCCGCCACATCAGA-3′; MCAD probe, 5′-TGTCACACAGTAAGGACACATCATTGGCTG-3′; M-CPT I forward, 5′-TCTAGGCAATGCCGTTCAC-3′; M-CPT I reverse, 5′-GAGCACATGGGCACCATAC-3′; M-CPT I probe, 5′-TCAAGCCGGTCATGGCACTGG-3′; CD36 forward, 5′-CGGACATTGAGATTCTTTTCC-3′; CD36 reverse, 5′-TCCTTTAAGGTCGATTTCAGATC-3′; CD36 probe, 5′-ACAGCGTAGATAGACCTGCAAATG-3′; ERRγ forward, 5′-TGACTTGGCTGACCGAG-3′; ERRγ reverse, 5′-CCGAGGATCAGAATCTCC-3′; ERRγ probe, 5′-CATATTCCAGGCTTCTCCACACTG-3′; PPARα forward, 5′-ACTACGGAGTTCACGCATGTG-3′; PPARα reverse, 5′-TTGTCGTACACCAGCTTCAGC-3′; PPARα probe, 5′-AGGCTGTAAGGGCTTCTTTCGGCG-3; PPARβ forward, 5′-TCACCGGCAAGTCCAGCCA-3′; PPARβ reverse, 5′-ACACCAGGCCCTTCTCTGCCT-3′; and PPARβ probe, 5′-AACGCACCCTTTGTCATCCACGA-3′. The rRNA (VIC) probe set was included in all reactions as an internal correction control, and corrected data were normalized to β-actin expression (Applied Biosystems).
Electrophoretic mobility shift assays and chromatin immunoprecipitation.
Double-stranded complementary oligonucleotides corresponding to HNF-4-responsive element (5′GATCCTGGAGGGTGGGGCAAAGTTCACCATAGGTA-3′) or HNF-4REmut (5′GATCCTGGAGGGTGCAGCAAAGTTCACCATAGGTA-3′) of the human PPARα promoter were used to generate probes to assay ERR isoform binding in vitro. Probes were 32P labeled by a Klenow fill-in reaction. Recombinant proteins for human ERRα and mouse ERRγ were synthesized with the TNT Quick T7-coupled translation kit (Promega). Synthesis of appropriately sized proteins was verified in reactions incorporating [35S]methionine followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis and autoradiographic detection. Electrophoretic mobility shift assay reactions were performed as described previously (9).
Chromatin immunoprecipitation assays were performed following previously published methods in which specific buffer recipes used in the protocol can be found (4, 48). In brief, L6 rat myoblasts (1 × 107 to 2 × 107) were infected with the adenovirus construct Ad-ERRα. Cross-linking was performed 48 h postinfection by incubating cells with 1% formaldehyde (10 min) followed by addition of 0.125 M glycine to halt cross-linking. Cells were washed and collected in cold phosphate-buffered saline and pelleted by centrifugation, followed by lysis and disruption via Dounce homogenization. Nuclei were pelleted and resuspended in a 1% sodium dodecyl sulfate nuclear lysis buffer. Chromatin was sheared by four 30-s sonication cycles with a Branson 250 sonifier. Aliquots were analyzed by agarose gel electrophoresis to verify enrichment of the 400- to 1,000-bp subpopulation. Sheared chromatin was precleared with bovine serum albumin-blocked Pansorbin (Calbiochem), followed by dilution of an aliquot in immunoprecipitation dilution buffer and incubation overnight at 4°C with immunoglobulin G or polyclonal antibody against ERRα (43).
Antibody-chromatin complexes were bound with Pansorbin and pelleted by centrifugation. An aliquot from the immunoglobulin G sample was reserved as “input” control. The pellets were washed twice in dialysis buffer followed by four washes in a 500 mM LiCl immunoprecipitation buffer. Immunoprecipitated complexes were eluted by two 15-min incubations with elution buffer. Cross-links were reversed by incubating samples at 65οC for 5 h, during which time samples were also RNase A treated. Samples were phenol and CHCl3 extracted, precipitated overnight, and then proteinase K treated. A final phenol-CHCl3 purification and precipitation was performed to yield template for PCR (30 cycles with standard temperature conditions). Primers were designed to amplify a 310-bp amplicon corresponding to the region of the rat PPARα promoter flanking the HNF-4-responsive element (nucleotides −1304 to −1613). Primers amplifying 250 bp of the TFIID gene were used to control for nonspecific enrichment of genomic DNA in the immunoprecipitation. PCR-amplified bands were analyzed on a 1.3% agarose gel, and relative band intensities were quantified by densitometry.
Animal studies.
All animal protocols were approved by the Animal Studies Committee at Washington University School of Medicine. The ERRα−/− mice have recently been described (32). The original background strain of the ERRα−/− mice was a hybrid strain (C57BL/6/SvJ129). For baseline comparisons, littermate wild-type and ERRα−/− mice were generated from heterozygous breeders to control for strain background. Heart and skeletal muscle (gastrocnemius and soleus) were isolated from fed wild-type and ERRα−/− mice during the daytime (1000 to 1200 h). ERRα−/− backcrossed to C57BL/6 were bred with a C57BL/6 strain of PPARα−/− mice (3, 27) to generate doubly heterozygous mice that were then intercrossed to generate the ERRα−/− PPARα−/− (double-knockout) mouse lines that were used to isolate primary fibroblasts.
Palmitate oxidation assays.
Measurement of palmitate oxidation rates was performed with [9,10-3H]palmitic acid as described previously (10). Cells (≈5 × 103) were cultured in 24-well plates and infected with adenovirus expressing GFP (Ad-GFP), Ad-ERRα, Ad-PGC-1α, or Ad-PPARα 24 h later. At 84 h postinfection, palmitate oxidation assays were performed. To demonstrate the specificity of the assay for measuring fatty acid oxidation, 50 μM sodium etomoxir, a CPT I inhibitor, was added to half of the labeling reactions. Incubations were performed for 2 h at 37°C. Data were normalized for background and for total cellular protein quantitated by the bicinchoninic acid method. Rates were calculated as nanomoles of [3H]palmitate oxidized per hour per milligram of protein.
Neutral lipid detection.
Rat neonatal cardiac myocytes were infected with Ad-GFP or Ad-ERRα for 24 h before addition of 35 μM oleic acid conjugated to bovine serum albumin. Oil red O staining was performed by the Digestive Disease Histology Core at Washington University School of Medicine as described previously (1).
RESULTS
ERRα induces expression of genes involved in cellular fatty acid uptake, oxidation, and mitochondrial respiration.
To characterize the regulatory effects of ERRα on cellular metabolism, ERRα was overexpressed in primary rat neonatal cardiac myocytes and gene expression changes were profiled with the Affymetrix rat U34A array in three independent trials. The criteria for designating a gene regulated by ERRα was a present call in ERRα-expressing myocytes and a signal intensity twofold or greater above that of GFP-expressing myocytes in at least two of three independent trials. Overall, ERRα induced 90 distinct genes, a significant number of which encode enzymes involved in cellular energy metabolic pathways (Table 1). A complete list of ERRα-upregulated genes is available from the authors (unpublished data).
TABLE 1.
Genes upregulated by ERRα in rat neonatal cardiac myocytes
| Category | Accession no. | Identification or gene symbola | Change (fold) |
|---|---|---|---|
| Cellular fatty acid import/oxidation | AA799326/AB005743 | CD36 antigen (Cd36) | 3.2 |
| D90109/AA893242 | Fatty acyl-CoA ligase, long chain 2 (Facl2) | 3.0 | |
| J02791 | Acyl-CoA dehydrogenase, medium chain (Acadm) | 2.7 | |
| D30647 | Acyl-CoA dehydrogenase, very long chain (Acadvl) | 2.7 | |
| X98225 | Enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase (Hadha) | 2.1 | |
| AI237731 | Lipoprotein lipase (Lpl) | 2.4 | |
| J02773 | Fatty acid binding protein, heart (Fabp3) | 2.1 | |
| J02752 | Acyl-CoA oxidase (Acox1) | 2.3 | |
| AI013834 | Peroxisomal multifunctional enzyme II (Hsd17b4) | 2.0 | |
| AF036761 | Stearoyl-CoA desaturase 2 (Scd2) | 3.0 | |
| AF007107 | Cytochrome b5, soluble (Cyb5) | 2.2 | |
| M23995 | Aldehyde dehydrogenase, phenobarbital inducible (Aldh1a4) | 4.6 | |
| Mitochondrial respiratory chain | K00750 | Cytochrome c (Cycs) | 2.6 |
| X64827 | Cytochrome c oxidase VIIIh (Cox8h) | 2.3 | |
| AA799336 | NADH dehydrogenase (ubiquinone) | ||
| Acyl carrier chain (ACP/CI-SDAP) | 2.5 | ||
| AI176422 | Flavoprotein-ubiquinone oxidoreductase (Etfdh) | 2.3 | |
| AA892863 | Mitochondrial carrier homolog 2 (Mtch2) | 2.8 | |
| Glucose/glycogen metabolism | J04526 | Hexokinase (Hk1) | 3.1 |
| AI103238 | Protein phophatase 2A, β regulatory subunit (Pppr2b2) | 3.1 | |
| S90449 | Protein phosphatase 2C (Pp2c2) | 2.3 | |
| Cholesterol metabolism | AI177004 | Hydroxymethylglutaryl-CoA synthase I (Hmgcs1) | 2.3 |
| X55286 | Hydroxymethylglutaryl-CoA reductase (Hmgcr) | 2.7 | |
| AF003835 | Isopentenyl diphosphate-dimethylallyl diphosphate isomerase (Idi1) | 2.1 | |
| S81497 | Cholesterol esterase, lysosomal acid lipase (Lip1) | 2.9 | |
| Other metabolic pathways | AF062740 | Pyruvate dehydrogenase phosphatase I (Pdp1) | 2.0 |
| M57664 | Creatine kinase B (Ckb) | 2.0 | |
| J03190 | Aminolevulinic acid synthase (Alas1) | 2.0 | |
| D10262 | Choline kinase (Chk) | 3.0 | |
| AA997614 | Cytochrome P450 subfamily 51 (Cyp51) | 2.1 | |
| AA800062 | N-Acylsphingosine amidohydrolase (acid ceramidase) (Asah) | 2.2 | |
| AF048687 | UDP-Gal:glucosylceramide β-1,4-galactosyltransferase (B4galt6) | 3.7 | |
| J04171 | Aspartate aminotransferase, soluble (Got1) | 3.3 | |
| Gene regulation | AA799412 | ERRα (Esrra) | 7.3 |
| U44948 | Smooth muscle cell LIM protein, cysteine-rich protein (Csrp2) | 2.3 | |
| D84418 | High-mobility-group box 2 (Hmgb2) | 2.3 | |
| X99723 | Brahma homolog, SWI/SNF-related actin-dependent regulator of chromatin, subfamily a, member 4 (Smarca4) | 2.7 | |
| AI231164 | Transformer-2-related, splicing factor arginine/serine-rich 10 (Sfrs10) | 2.2 | |
| Contractile associated | AA799276 | Ca2+ transporting ATPase, cardiac, slow twitch 2 (Atp2a2) | 2.5 |
| L03382 | Phospholamban (Pln) | 2.2 | |
| AI104924 | Myosin heavy-chain α (Myh6) | 3.7 | |
| X15939 | Myosin heavy-chain β (Myh7) | 5.1 |
CoA, coenzyme A.
Notably, we found a number of genes involved in the cellular uptake and mitochondrial oxidation of fatty acids. In addition, several genes involved in mitochondrial respiratory function were also activated. These results were of interest because mitochondrial fatty acid oxidation serves as the chief source of energy in adult heart. Specifically, genes encoding lipoprotein lipase, the fatty acid transporter, CD36/fatty acid transporter, and heart-specific fatty acid binding protein FABP3 were upregulated in rat cardiac myocytes. In addition, fatty acyl coenzyme A synthetase, which facilitates fatty acid uptake by coupling transport to esterification at the plasma membrane, was induced in response to ERRα overexpression.
As shown previously, the gene encoding MCAD, a key enzyme in the fatty acid oxidation cycle, was increased by ERRα. Additional genes encoding enzymes of mitochondrial (very long chain acyl-coenzyme A dehydrogenase, long-chain hydroxyacyl coenzyme A dehydrogenase) and peroxisomal (acyl coenzyme A oxidase) β-oxidation were also activated in parallel with MCAD. A subset of the genes encoding enzymes and proteins involved in mitochondrial electron transport and oxidative phosphorylation were also induced, including cytochrome c, the muscle-specific cytochrome oxidase VIIIh subunit, NADH (ubiquinone) dehydrogenase, and flavoprotein-ubiquinone oxidoreductase. Additional mitochondrion-associated enzymes, such as aminolevulinic acid synthase, involved in heme synthesis, hydroxymethylglutaryl coenzyme A synthase, creatine kinase, and aldehyde dehydrogenase, were also induced in response to ERRα overexpression. Notably, components of other metabolic pathways, such as glycolysis and cholesterol metabolism, were also activated by ERRα. However, no metabolic pathways appeared to be more uniformly regulated by ERRα in cardiac myocytes than those involved in fatty acid uptake and oxidation. Finally, a number of muscle-specific genes associated with the contractile apparatus were upregulated in ERRα-expressing cardiac myocytes, including Ca2+-transporting ATPase, phospholamban, and myosin heavy-chain isoforms. Collectively, the results suggest that ERRα upregulates genes involved in cellular fatty acid utilization and mitochondrial oxidation.
The putative ERRα targets were validated by independent analytical methods (Fig. 1). Upregulation of lipoprotein lipase, CD36/fatty acid transporter, and FABP3 in ERRα-expressing myocytes was demonstrated in RNA prepared from independent overexpression trials performed in cardiac myocytes (Fig. 1A). ERRα-mediated regulation of genes involved in mitochondrial energy production was compared with that of PGC-1α, which has been shown to increase the expression of a number of nucleus- and mitochondrion-encoded genes involved in mitochondrial fatty acid oxidation and electron transport/oxphos (Fig. 1B) (28). Expression of genes encoding mitochondrial fatty acid oxidation enzymes (muscle carnitine palmitoyltransferase I [M-CPT I, CPT Iβ] and MCAD) and the peroxisomal enzyme acyl coenzyme A oxidase was increased in response to ERRα or PGC-1α expression. In addition, ERRα modestly induced the expression of cytochrome oxidase IV, cytochrome c, and ATP synthase β. Although less robust, these results paralleled that of PGC-1α-mediated regulation. Analysis of whole-cell protein extracts from ERRα-overexpressing cells verified induction of MCAD, acyl coenzyme A oxidase, and fatty acyl coenzyme A synthetase 1 protein by ERRα (Fig. 1C). Collectively, the results indicate that ERRα regulates a subset of PGC-1α targets, mainly genes involved in mitochondrial oxidative fatty acid catabolism or respiratory function.
FIG. 1.
Validation of putative ERRα target genes involved in cellular fatty acid utilization and mitochondrial respiratory pathways. (A) Expression analysis of fatty acid uptake enzyme genes. Northern blotting was performed with 15 μg of total RNA isolated from Ad-GFP (GFP) or Ad-ERRα (ERRα)-infected cardiac myocytes. Blots were sequentially hybridized with probes specific for ERRα, fatty acid transporter (FAT)/CD36, FABP3, and lipoprotein lipase (LPL). (B) Induced expression of mitochondrial enzymes by ERRα or PGC-1α. Northern analysis was performed as above with probes against PGC-1α, MCAD, acyl coenzyme A oxidase (ACO), M-CPT I, cytochrome oxidase IV (COXIV), cytochrome c (Cyt. c), and ATP synthase β, with RNA from cardiac myocytes overexpressing either GFP, ERRα, or PGC-1α, as indicated. (C) Western analysis of 30 μg of whole-cell extract (WCE) prepared from rat neonatal cardiac myocytes infected with adenovirus vectors expressing GFP or ERRα. FACS-1, fatty acyl coenzyme A synthetase 1.
ERRα increases cardiac myocyte fatty acid uptake and oxidation.
We next investigated the physiologic relevance of the observed induction of lipid transport and uptake and mitochondrial oxidative enzyme genes by ERRα. Cardiac myocytes overexpressing GFP (control) or ERRα were incubated with oleic acid complexed to bovine serum albumin and stained with Oil Red O to detect intracellular neutral lipid (Fig. 2). . ERRα-overexpressing myocytes showed increased accumulation of small lipid droplets. Increased lipid accumulation was also observed at the same concentration of bovine serum albumin-oleate in cardiac myocytes overexpressing either PGC-1α or PPARα, which were included as positive controls (data not shown).
FIG. 2.
ERRα expression causes lipid accumulation in primary cardiac myocytes. Oil Red O staining of primary rat neonatal cardiac myocytes expressing either GFP (Ad-GFP) or ERRα (Ad-ERRα) and cultured in the presence of 35 μM bovine serum albumin-complexed oleic acid. The red droplets (inset) represent accumulated neutral lipid. Magnification, ×400.
To determine if fatty acid utilization was also increased, oxidation of [3H-9,10]palmitic acid was measured (Table 2). A significant increase in mean palmitate oxidation rates (49.3 ± 7.8%) was observed in the ERRα-overexpressing cells compared to GFP. Oxidation rates were inhibited by 85% (± 0.4%) by the CPT I inhibitor etomoxir, verifying that the assay was specifically measuring mitochondrial fatty acid oxidation (data not shown). With the same assay, overexpression of PPARα or PGC-1α increased palmitate oxidation 41% and 92%, respectively, as expected (Table 2). These data are consistent with the observed effects of PPARα/PGC-1α on fatty acid oxidation in NIH 3T3 cells (46). These physiologic effects reflect the observed gene expression changes and demonstrate that ERRα increases cardiac myocyte fatty acid uptake and oxidative capacity.
TABLE 2.
Palmitate oxidation in rat neonatal cardiac myocytes overexpressing ERRα
| Infection | Mean ± SEM [3H]palmitate oxidation rate (nmol [mg of protein]−1 h−1) | Mean % changea (relative to GFP) ± SEM |
|---|---|---|
| Ad-GFP | 11.25 (± 0.83) | 100 |
| Ad-ERRα | 16.80 (± 0.88) | 149.3 (± 7.8)* |
| Ad-PGC-1α | 21.58 (± 1.66) | 191.7 (± 14.8)* |
| Ad-PPARα | 15.90 (± 0.70) | 141.3 (± 6.2)* |
*, P < 0.05 compared to the control (Ad-GFP).
Effects of ERRα gene deletion on in vivo expression of fatty acid utilization genes in cardiac and skeletal muscle.
The ERRα knockout mouse was recently described (32). The phenotype of ERRα knockout mice includes a derangement in white adipocyte lipid metabolism that manifests as resistance to diet-induced obesity. The results shown above suggest that ERRα plays a role in the regulation of genes involved in skeletal muscle and cardiac energy metabolism in vivo. To explore this possibility, the expression of several putative ERRα target genes involved in cellular fatty acid uptake and oxidation was characterized in cardiac and skeletal muscle of wild-type and littermate ERRα knockout animals (Fig. 3). The levels of transcripts encoding MCAD or PPARα were not different in the hearts of ERRα knockout mice compared to wild-type controls (Fig. 3A). Interestingly, the levels of PGC-1α and ERRγ mRNA were significantly increased in ERRα knockout hearts compared to wild-type hearts, suggesting a compensatory response mediated by the ERRγ/PGC-1α complex.
FIG. 3.
Deletion of the ERRα gene has differential effects on expression of fatty acid utilization enzyme genes mouse heart and skeletal muscle. (A) Northern blot studies performed with 15 μg of total RNA isolated from the hearts of wild-type (WT) or ERRα−/− mice. Blots were hybridized sequentially with probes corresponding to MCAD, PGC-1α, PPARα, and ERRγ. Phosphorimage quantification of Northern signal intensities is shown on the right. Data represent mean intensity values (± standard error) normalized to wild-type values (= 1.0). (B) Northern analysis of total RNA comparing expression of ERR isoforms, PGC-1α, PPARα, and MCAD in the vastus lateralis muscle, comprised predominantly of fast-twitch glycolytic fibers, versus the soleus muscle, comprised of slow-twitch oxidative fibers. (C) (Left) Northern analysis of total RNA isolated from the soleus of wild-type and ERRα−/− mice. Representative pairs of samples from each genotype are shown. (Right) Real-time PCR (Taqman) analysis of soleus gene expression in wild-type (n = 6) and ERRα−/− (n = 6) mice. In addition to the transcripts detected in the Northern panel, quantitative analysis of mRNA encoding the cellular fatty acid utilization enzyme M-CPT I and the PPARβ isoform is shown. Data represent mean arbitrary units (± standard error) corrected to the β-actin transcript and normalized to values in the wild type (= 1.0). Asterisks indicate significant differences (P < 0.05) compared to the control.
The effect of ERRα gene deletion on the expression of the putative ERRα target genes was different in skeletal muscle compared to heart. As a preliminary step, the relative expression levels of ERR isoforms were assessed in the vastus lateralis, which contains mostly glycolytic fast-twitch fibers, and soleus, which is predominantly a slow-twitch oxidative muscle, in wild-type mice (Fig. 3B). Expression of ERR isoforms, PPARα, PGC-1α, and MCAD (used as a marker of mitochondrial fatty acid oxidative capacity) was significantly higher in the soleus compared to the vastus in the wild-type mice. These results suggested that any effects of ERRα deletion on expression of putative ERRα targets would most likely be observed in the soleus.
In contrast to the heart, transcript levels for MCAD, a known PPARα gene target, were lower in ERRα−/− soleus compared to wild-type control soleus. However, no change in several other known PPARα targets (CD36/fatty acid transporter or M-CPT I) was observed in the soleus of ERRα null compared to wild-type mice (Fig. 3C). Regarding possible compensatory changes, PGC-1α expression was significantly increased, similar to the effect of ERRα deletion in the heart. However, ERRγ and PPARα were unchanged in the ERRα−/− mouse soleus. This distinct pattern of compensatory gene regulation, particularly the lack of induction of the ERRγ gene, may contribute to the differential effects of ERRα deletion on PPARα targets in the soleus compared to the heart. Taken together, the results of the ERRα deletion and overexpression studies support a role for ERRα in the regulation of muscle mitochondrial oxidative metabolism.
ERRα activates the PPARα gene regulatory pathway.
The ERRα overexpression studies described above revealed upregulation of a number of genes involved in fatty acid utilization pathways, most of which are known target genes for the fatty acid-responsive nuclear receptor PPARα (8). These results suggested that ERRα drives a metabolic regulatory program that overlaps PPARα or that ERRα might regulate the PPARα signaling pathway. To explore this potential mechanism, the effect of ERRα on the expression of PPARα and related transcriptional activators was investigated.
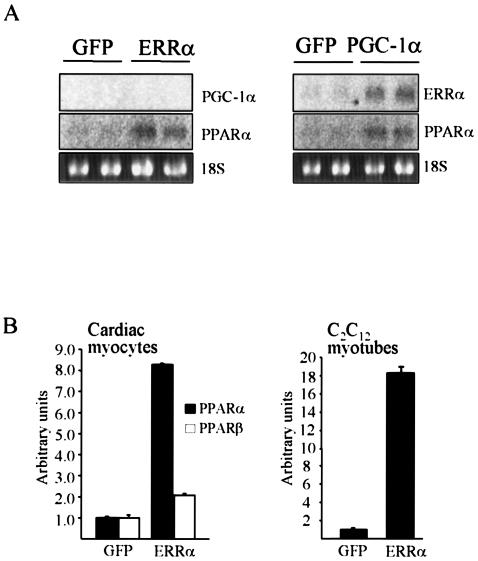
The PPARα transcript was called absent in the array analyses, suggesting that this assay was not sensitive enough to detect endogenous levels of PPARα in cardiac myocytes. Therefore, we used more sensitive Northern and real-time PCR analyses to look at changes in PPARα expression (Fig. 4). Forced expression of ERRα increased endogenous PPARα transcript levels 8.3-fold in cardiac myocytes. The ERRα-mediated regulation was most robust for the PPARα isoform, although we also observed an approximately twofold increase in PPARβ expression in the ERRα-overexpressing cardiocytes (Fig. 4B). PPARα expression was also upregulated by ERRα in C2C12 myotubes. The expression of endogenous PGC-1α was unaffected by ERRα, indicating that the metabolic effects of ERRα were not mediated through the known effects of PPARα coactivation by PGC-1α (Fig. 4A) (46). In contrast, forced expression of PGC-1α led to upregulated of expression of both ERRα and PPARα in cardiac myocytes, suggesting a potential feedforward regulation involving PGC-1α, ERRα, and PPARα.
FIG. 4.
ERRα induces endogenous PPARα expression. (A) Northern analyses to characterize the expression of regulators of mitochondrial fatty acid oxidation, PPARα, PGC-1α, and ERRα in cardiac myocytes expressing GFP, ERRα, or PGC-1α. (B) Quantification of PPARα and PPARβ mRNA levels in response to ERRα overexpression in cardiac myocytes and C2C12 myotubes by real-time PCR. Data represent mean arbitrary units (± standard error) corrected to the glyceraldehhyde-3-phosphate dehydrogenase (GAPDH) transcript and normalized to the values in GFP-expressing myocytes (= 1.0).
To determine whether ERRα directly regulates the transcriptional activity of the PPARα gene, a human PPARα promoter-reporter construct, pα(H-H)pGL3, containing 1,664 bp of the 5′-flanking region, was cotransfected with ERRα in the absence or presence of PGC-1α into CV1 cells. Cotransfection of ERRα alone activated the PPARα promoter almost fivefold (Fig. 5A). As expected, addition of the coactivator, PGC-1α, significantly enhanced ERRα-mediated activation of the PPARα promoter. We also tested the related isoform, ERRγ, which shares many target genes with ERRα (Fig. 5A). ERRγ activated the PPARα promoter to a similar magnitude as ERRα. Αlthough we have shown that PGC-1α and ERRγ form a functional complex (19), the activity of ERRγ was only modestly enhanced by addition of PGC-1α on the PPARα promoter.
FIG. 5.
ERR isoforms activate the PPARα gene promoter through a conserved nuclear receptor binding site. (A) Transient cotransfection studies in CV1 cells to analyze ERR regulation of the PPARα promoter. (Left) The reporter construct pα(H-H)-pGL3, containing the −1664 to +83 region of the human PPARα gene promoter, was cotransfected with empty expression vector (−ERRα) or pcDNA3.1-hERRα (+ERRα) in the presence or absence of the coactivator PGC-1α. (Right) Activation by ERRγ was analyzed with the same conditions as for ERRα. A β-galactosidase (β-gal) expression construct was cotransfected to control for transfection efficiency. (B) Transient transfections were performed with either the wild-type PPARα promoter-reporter construct or with constructs mutated at the HNF-4-responsive element (HNF4-REmut) or farnesoid X-responsive element (FXREmut) sites. Experiments were performed as described for panel A. All bars represent mean (± standard error) corrected relative light units (RLU) for β-galactosidase normalized to the activity of the reporter cotransfected with pcDNA3.1(−) (= 1.0). Data represent at least three independent trials performed in triplicate. (C) Electrophoretic mobility shift assays were performed with 32P-labeled probes corresponding to the wild-type HNF-4-responsive element contained in the human PPARα promoter or a mutated HNF-4-responsive element (Mut). The Mut probe contains the same nucleotide substitutions as the HNF4-REmut promoter-reporter analyzed in B. Probes were incubated with 1 or 3 μl of recombinant human ERRα (left) or mouse ERRγ (right) synthesized in a rabbit reticulocyte lysate. Control reactions included probe alone (−) and probe incubated with unprogrammed reticulocyte lysate (rl). (D) (Top) L6 myoblasts were infected with Ad-ERRα for 48 h. Cross-linked chromatin was immunoprecipitated with nonspecific antibody (immunoglobulin G) or anti-human ERRα (ERRα) antibody. Amplicons corresponding to the 310-bp region of the PPARα promoter containing the HNF-4-responsive element (PPARα) or a 210-bp nonspecific region of the TFIID promoter (control, Cont) were amplified by PCR. Input represents 0.2% of the total chromatin used in the immunoprecipitation reactions. A representative trial from multiple experiments is shown. (Bottom) Band intensities from chromatin immunoprecipitation experiments were quantified by densitometry. Data represent mean band intensity in arbitrary units (± standard error) from three independent trials with PCR performed in triplicate normalized to immunoglobulin G (= 1.0). The asterisk indicates a significant difference compared to the control (immunoglobulin G).
The human PPARα gene promoter contains two independent nuclear receptor response elements within the 1,664-bp region shown to be responsive to ERRα. The distal site comprises direct repeat 1 (DR1), through which several receptors, including hepatocyte nuclear factor 4 (HNF-4) and chicken ovalbumin upstream promoter-transcription factor I, as well as PPARα itself, activate or repress the activity of the promoter (36). The proximal site binds the farnesoid X receptor and confers bile acid responsiveness on the PPARα promoter (35). To determine whether either of the previously characterized nuclear receptor binding sites was involved in ERRα/PGC-1α-mediated activation of the PPARα gene, cotransfection experiments were repeated with full-length mutant PPARα promoter constructs in which either the HNF-4 or farnesoid X receptor had been disrupted (Fig. 5B). FXREmut was activated by ERRα/PGC-1α to the same degree as the wild-type promoter. However, disruption of the HNF-4-responsive element (HNF4-REmut) abolished induction of the human PPARα promoter by ERRα/PGC-1α. Interestingly, the basal activity of the HNF4-REmut construct was 20-fold higher than that of the wild type, suggesting that the HNF-4-responsive element is occupied by a repressor in CV1 cells that is displaced by activators, like ERRα or HNF-4. Despite the enhanced baseline activity of HNF4-REmut, induction by the farnesoid X receptor ligands chenodeoxycholic acid and taurocholic acid, which occurs through the downstream farnesoid X receptor, was still observed, demonstrating that the altered baseline activity does not prevent further activation through other elements (data not shown).
We then sought to determine whether ERR isoforms bound directly to the PPARα promoter. Electrophoretic mobility shift assays were performed with radiolabeled probes corresponding to the wild-type and mutated HNF-4-responsive element and recombinant ERRα and ERRγ proteins synthesized in a reticulocyte lysate. ERRα and ERRγ bound to the HNF-4-responsive element in a concentration-dependent manner (Fig. 5C). In contrast, the mutated HNF-4-responsive element, which contains the same mutation that abolished ERRα responsiveness, did not form a complex with either ERR isoform. Parallel binding assays were performed with wild-type and mutated farnesoid X-responsive element probes, but no binding to ERRα or ERRγ was observed (data not shown).
To determine whether ERRα bound the PPARα promoter in cells, chromatin immunoprecipitation assays were performed on cross-linked chromatin isolated from L6 myoblasts expressing ERRα (Fig. 5D). Precipitation with ERRα antibody enriched for chromatin containing the HNF-4-responsive element (approximately threefold) compared to precipitations performed in parallel with nonspecific antibody (immunoglobulin G). Collectively, these studies show that ERRα activates PPARα gene expression via transcriptional regulation through a nuclear receptor response element that is conserved in the human and rodent PPARα genes.
ERRα-mediated activation of fatty acid oxidation enzyme genes is dependent on the presence of PPARα.
Our data indicated that ERRα overexpression induces PPARα targets involved in cellular fatty acid utilization and is capable of directly activating PPARα gene transcription. Therefore, the observed metabolic regulation in response to ERRα may occur through direct regulation of metabolic target genes or through modulation of the PPARα pathway. To determine whether certain ERRα-mediated regulatory events are dependent on PPARα, we evaluated the effects of ERRα overexpression on PPARα target gene expression in the presence and absence of PPARα.
To this end, ERRα was overexpressed with adenovirus in primary fibroblasts isolated from ERRα−/− (ERRα knockout) mice or PPARα−/− ERRα−/− (double-knockout) mice (Fig. 6). ERRα knockout cells were used as the PPARα-expressing control in order to maximize detection of ERRα-mediated activation of downstream targets. As expected, the expression of known PPARα targets involved in fatty acid oxidation, MCAD and acyl coenzyme A oxidase, was induced by ERRα in the ERRα knockout cells (Fig. 6A). In striking contrast, expression of these fatty acid oxidation target genes was not induced by ERRα in double-knockout cells. Similar results were observed with an additional PPARα target, M-CPT I, although induction by ERRα in ERRα knockout cells required coexpression of the ERRα coactivator PGC-1α (Fig. 6B). As expected, endogenous PPARα was induced in ERRα-expressing ERRα knockout fibroblasts relative to the GFP control (data not shown). In addition, PPARα target genes (MCAD and acyl coenzyme A oxidase) were induced in response to PPARα overexpression in double-knockout cells (data not shown). Thus, ERRα-mediated induction of at least a subset of genes involved in cellular fatty acid utilization requires PPARα.
FIG. 6.
ERRα induction of β-oxidation enzymes genes is dependent on the presence of PPARα. Primary fibroblasts isolated from ERRα−/− PPARα−/− (double-knockout, DKO) or ERRα−/− (ERR knockout, ERRKO) mice were infected with an adenoviral construct expressing GFP (−) or ERRα (+) as indicated. PGC-1α was included in some conditions to enhance ERRα activity. Real-time PCR was used to quantify the expression of endogenous MCAD and acyl coenzyme A oxidase (ACO) (A) or M-CPT I (B) in total RNA isolated from these cells. Data are reported as mean (± standard error) arbitrary units normalized to the GFP condition (= 1.0) for three independent trials performed in triplicate. (C) Palmitate oxidation rates were measured in the same primary fibroblasts as above expressingGFP or ERRα. Data were normalized to GFP (= 1.0), and values represent means (± standard error) for three overexpression trials performed with cells from two independent isolations. Asterisks indicate a significant difference compared to the controls (minus ERRα).
The effects of ERRα overexpression on palmitate oxidation rates were also measured in the ERRα knockout and double-knockout fibroblasts (Fig. 6C). In the ERRα knockout cells, ERRα overexpression increased oxidation rates by 63%. In contrast, ERRα expression elicited only a 15% increase in the palmitate oxidation rate in double-knockout fibroblasts. Interestingly, PGC-1α overexpression, used as a positive control, was competent to induce palmitate oxidation in both ERRα knockout and double-knockout cells (data not shown), suggesting that another PGC-1α partner besides ERRα and PPARα is able to mediate activation of fatty acid oxidation. These results, which are consistent with the gene expression studies, demonstrate that ΕRRα-mediated induction of fatty acid oxidation in fibroblasts requires PPARα.
DISCUSSION
Despite the fact that the ERRs were the first family of orphan nuclear receptors cloned, their biological function has remained uncertain (12, 13). Recent evidence has implicated ERRα and ERRγ in the transcriptional regulation of cellular energy metabolism. ERRα is enriched in adult mammalian tissues with high oxidative metabolic capacity, such as the heart, slow-twitch skeletal muscle, and brown adipose. ERRα and ERRγ have also recently been shown to serve as functional partners for the PGC-1 family of coactivators (17, 19, 21, 42), which have emerged as key regulators of mitochondrial metabolism and biogenesis (23, 37). We hypothesized that ERR isoforms serve as key regulators of heart and skeletal muscle energy metabolism downstream of PGC-1α. To this end, gene expression profiling experiments were conducted in cardiac myocytes. ERRα overexpression was shown to increase the expression of genes involved in multiple pathways involved in cellular fatty acid utilization, including fatty acid uptake and intracellular binding and mitochondrial oxidation. In addition, a subset of genes involved in mitochondrial electron transport and oxidative phosphorylation were upregulated by ERRα.
Heart and slow-twitch skeletal muscle meet high ATP demands predominantly through oxidation of fatty acids in mitochondria. Our results demonstrating ERRα as an activator of oxidative metabolism in myocytes are consistent with the enriched expression of this nuclear receptor in heart and slow-twitch skeletal muscle. Indeed, ERRα null mice exhibit a compensatory increase in PGC-1α and ERRγ expression in heart and a reduction in expression of MCAD, a key fatty acid metabolic target gene, in the soleus, where no change in ERRγ expression was observed. These findings strongly suggest that ERR isoforms contribute to the high-level basal expression of fatty acid utilization genes in oxidative tissues.
Consistent with this conclusion, ERRα was recently shown, in a combined proteomic and gene expression profiling study, to be coregulated with proteins and enzymes physically associated with the mitochondria, further supporting its role as a regulator of energy metabolism (33). Interestingly, metabolic function in white adipose, a predominantly glycolytic tissue, is impaired in ERRα null animals and is associated with increased MCAD expression (32). These apparently disparate results support a complex tissue-selective metabolic regulatory function for ERRα, with the activity of ERRα being dependent on the complement of cofactors coexpressed in a given tissue. However, our in vivo gene expression data do not exclude indirect effects, such as metabolic derangements related to the ERRα-deficient state, influencing the expression of some putative gene targets.
We found that the metabolic regulatory effects of ERRα overexpression in cardiac myocytes displayed considerable overlap with those of the nuclear receptor PPARα. The following lines of evidence indicate that this overlap is due, at least in part, to direct activation of PPARα gene expression by ERRα: (i) overexpression of ERRα induced PPARα gene expression in cardiac myocytes, C2C12 skeletal myotubes, and primary mouse fibroblasts; (ii) ERRα directly activated the PPARα gene promoter in transient cotransfection assays through a conserved nuclear receptor response element to which it bound in vitro and in cells; and (iii) ERRα-mediated regulation of a subset of its fatty acid oxidation targets in primary fibroblasts absolutely required the presence of PPARα. Specifically, ERRα overexpression in PPARα null fibroblasts had no effect on the expression of PPARα targets, including M-CPT I, MCAD, or acyl-coenzyme A oxidase, yet ERRα activated these targets in PPARα-expressing cells. These results strongly suggest that in tissues where ERRα and PPARα are coexpressed, like skeletal muscle and heart, activation of PPARα by ERRα is an important mechanism to control the expression of certain genes involved in cellular fatty acid metabolism.
Collectively, our data and the results of recently published studies suggest that ERRα regulates mitochondrial metabolism through the activation of several downstream transcriptional regulatory cascades. While this manuscript was in review, two studies presented additional evidence for ERRα as a key component of the PGC-1α-mediated regulation of mitochondrial metabolism. Schreiber et al. demonstrated that induction of mitochondrial proliferation and respiratory chain enzyme gene expression by PGC-1α is impaired by RNA interference inhibition of ERRα expression (41), indicating that ERRα is downstream of PGC-1α in regulating certain mitochondrial biogenic programs. Studies by Mootha et al. found a similar role for ERRα downstream of PGC-1α in C2C12 myotubes (34). The latter study suggested that ERRα activates the NRF cascade through direct activation of the Gabpa gene promoter, which encodes a component of the NRF-2 complex. These data are consistent with our findings, which demonstrate that ERRα activates the mitochondrial fatty acid oxidation in cardiac myocytes by converging on the PPARα regulatory pathway.
Our data do not exclude the possibility that, in addition to activating PPARα, ERRα plays a direct role in the regulation of certain target genes. The results of our gene expression profiling studies demonstrated that, in addition to the fatty acid oxidation enzyme genes, a number of genes involved in cellular fatty acid uptake and mitochondrial respiration were also activated by ERRα. It is likely that a number of these genes are directly regulated by ERRα. Indeed, our previous work demonstrated that ERRα with PGC-1α directly activates the MCAD gene promoter in transient transfection assays through NRRE-1, a pleiotropic nuclear receptor response element that has been shown to bind both PPARα and ERRα (14, 19, 43, 47). We also observed modest activation of the lipoprotein lipase promoter by ERRα (J. Huss, unpublished observation). Furthermore, recent studies have demonstrated that ERRα with PGC-1α directly activates the ATP synthase β and cytochrome c gene promoters through consensus ERRα response elements (41). It is therefore likely that ERRα regulates cellular metabolism through multiple pathways, including indirect regulation via other transcription factors and direct regulation of metabolic target genes.
Our results and studies by others have shown that ERRα expression is upregulated by PGC-1α in cultured cells (Fig. 4A) (42) and in vivo in the hearts of mice overexpressing PGC-1α (39; L. Russell and J. Huss, unpublished observation). These results place ERRα in a central position within the PGC-1α regulatory network (Fig. 7). We propose that ERRα transduces PGC-1α-derived signals to transcription factors such as PPARα and NRFs as well as directly to target genes involved in energy metabolism (34, 41). The extent of the role of ERR isoforms in mediating the actions of PGC-1α on cellular metabolism and physiology is unknown. However, the inducibility of PGC-1α by fasting, exercise, and cold exposure suggests that this regulatory circuit serves a critical role in the physiologic regulation of cardiac and skeletal muscle energy metabolism. Given that derangements in mitochondrial oxidative metabolism occur in pathophysiologic states such as skeletal muscle insulin resistance and cardiac hypertrophy, ERRs may prove to be an attractive therapeutic target for common human diseases such as diabetes and heart failure.
FIG. 7.
Role of ERRα in regulating cellular oxidative capacity. PGC-1α coactivates and regulates the expression of a number of transcription factors, including ERRα, PPARα, and NRFs, involved in mediating PGC-1α effects on cellular metabolism. PGC-1α regulation of ERRα and NRF-2 expression involves both cross- and auto-regulatory mechanisms (25, 34). Data presented in the current study demonstrated that ERRα likely directs PGC-1α upregulation of PPARα. In response to PGC-1α, activation of the NRF cascade regulates genes involved in mitochondrial respiration and biogenesis, whereas activation of the PPARα pathway regulates fatty acid uptake and mitochondrial oxidation enzymes. ERRα may also directly regulate metabolic target genes in both pathways. FAO, fatty acid oxidation; ACO, acyl coenzyme A oxidase; mtTFA, mitochondrial transcription factor A; oxid. phos., oxidative phosphorylation.
Acknowledgments
This work is supported by NIH grants DK45416 and HL58493, Digestive Diseases Core Center grant P30DK52574 (D.P.K), and an operating grant from the Canadian Institutes for Health Research (V.G.). J.M.H. is supported by NIH 1K01 DK063051-01 and the Washington University School of Medicine Diabetes Research Training Center P60 DK20579.
We thank the Alvin Siteman Cancer Center Multiplexed Gene Analysis Core at Washington University School of Medicine for performing DNA microarray analyses and the Digestive Disease Histology Core for histologic studies. We thank Mary Wingate for expert assistance in preparing the manuscript.
REFERENCES
- 1.Barger, P. M., J. M. Brandt, T. C. Leone, C. J. Weinheimer, and D. P. Kelly. 2000. Deactivation of peroxisome proliferator-activated receptor-α during cardiac hypertrophic growth. J. Clin. Investig. 105:1723-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barger, P. M., A. C. Browning, A. N. Garner, and D. P. Kelly. 2001. p38 MAP kinase activates PPARα: a potential role in the cardiac metabolic stress response. J. Biol. Chem. 276:44495-44501. [DOI] [PubMed] [Google Scholar]
- 3.Bernal-Mizrachi, C., S. Weng, C. Feng, B. N. Finck, R. H. Knutsen, T. C. Leone, T. Coleman, R. P. Mecham, D. P. Kelly, and C. F. Semenkovich. 2003. Glucocorticoid induction of hypertension and diabetes is PPAR-α dependent in mice. Nat. Med. 9:1069-1075. [DOI] [PubMed] [Google Scholar]
- 4.Boyd, K. E., J. Wells, J. Gutman, S. M. Bartley, and P. J. Farnham. 1998. c-Myc target gene specificity is determined by a post-DNA-binding mechanism. Proc. Natl. Acad. Sci. USA 95:13887-13892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandt, J., F. Djouadi, and D. P. Kelly. 1998. Fatty acids activate transcription of the muscle carnitine palmitoyltransferase I gene in cardiac myocytes via the peroxisome proliferator-activated receptor α. J. Biol. Chem. 273:23786-23792. [DOI] [PubMed] [Google Scholar]
- 6.Chawla, A., J. J. Repa, R. M. Evans, and D. J. Mangelsdorf. 2001. Nuclear receptors and lipid physiology: opening the X-files. Science 294:1866-1870. [DOI] [PubMed] [Google Scholar]
- 7.Chu, R., N. Usuda, M. K. Reddy, C. Liu, T. Hashimoto, K. Alvares, M. S. Rao, and J. K. Reddy. 1994. Functional expression of rat peroxisomal acyl-CoA oxidase in Spodoptera frugiperda cells. Biochem. Biophys. Res. Commun. 200:178-186. [DOI] [PubMed] [Google Scholar]
- 8.Desvergne, B., and W. Wahli. 1999. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocrine Rev. 20:649-688. [DOI] [PubMed] [Google Scholar]
- 9.Disch, D. L., T. A. Rader, S. Cresci, T. C. Leone, P. M. Barger, R. Vega, P. A. Wood, and D. P. Kelly. 1996. Transcriptional control of a nuclear gene encoding a mitochondrial fatty acid oxidation enzyme in transgenic mice: role for nuclear receptors in cardiac and brown adipose expression. Mol. Cell. Biol. 16:4043-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Djouadi, F., J.-P. Bonnefont, A. Munnich, and J. Bastin. 2003. Characterization of fatty acid oxidation in human muscle mitochondria and myoblasts. Mol. Genet. Metab. 78:112-118. [DOI] [PubMed] [Google Scholar]
- 11.Francis, G. A., E. Fayard, F. Picard, and J. Auwerx. 2003. Nuclear receptors and the control of metabolism. Annu. Rev. Physiol. 65:261-311. [DOI] [PubMed] [Google Scholar]
- 12.Giguère, V. 2002. To ERR in the estrogen pathway. Trends Endocrinol. Metab. 13:220-225. [DOI] [PubMed] [Google Scholar]
- 13.Giguère, V., N. Yang, P. Segui, and R. M. Evans. 1988. Identification of a new class of steroid hormone receptors. Nature 331:91-94. [DOI] [PubMed] [Google Scholar]
- 14.Gulick, T., S. Cresci, T. Caira, D. D. Moore, and D. P. Kelly. 1994. The peroxisome proliferator activated receptor regulates mitochondrial fatty acid oxidative enzyme gene expression. Proc. Natl. Acad. Sci. USA 91:11012-11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heard, D. J., P. L. Norbu, J. Holloway, and H. Vissing. 2000. Human ERRγ, a third member of the estrogen receptor-related receptor (ERR) subfamily of orphan nuclear receptors: Tissue-specific isoforms are expressed during development and in the adult. Mol. Endocrinol. 14:383-392. [DOI] [PubMed] [Google Scholar]
- 17.Hentschke, M., U. Susens, and U. Borgmeyer. 2002. PGC-1 and PERC, coactivators of the estrogen receptor-related receptor gamma. Biochem. Biophys. Res. Commun. 299:872-879. [DOI] [PubMed] [Google Scholar]
- 18.Hong, H., L. Yang, and M. R. Stallcup. 1999. Hormone-independent transcriptional activation and coactivator binding by novel orphan nuclear receptor ERR3. J. Biol. Chem. 274:22618-22626. [DOI] [PubMed] [Google Scholar]
- 19.Huss, J. M., R. P. Kopp, and D. P. Kelly. 2002. PGC-1α coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-α and -γ. J. Biol. Chem. 277:40265-40274. [DOI] [PubMed] [Google Scholar]
- 20.Huss, J. M., F. H. Levy, and D. P. Kelly. 2001. Hypoxia inhibits the PPARα/RXR gene regulatory pathway in cardiac myocytes. J. Biol. Chem. 276:27605-27612. [DOI] [PubMed] [Google Scholar]
- 21.Kamei, Y., H. Ohizumi, Y. Fujitani, T. Nemoto, T. Tanaka, N. Takahashi, T. Kawada, M. Miyoshi, O. Ezaki, and A. Kakizuka. 2003. PPARγ coactivator 1β/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc. Natl. Acad. Sci. USA 100:12378-12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly, D. P., J. J. Kim, J. J. Billadello, B. E. Hainline, T. W. Chu, and A. W. Strauss. 1987. Nucleotide sequence of medium-chain acyl-CoA dehydrogenase mRNA and its expression in enzyme-deficient human tissue. Proc. Natl. Acad. Sci. USA 84:4068-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly, D. P., and R. C. Scarpulla. 2004. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 18:357-368. [DOI] [PubMed] [Google Scholar]
- 24.Knutti, D., and A. Kralli. 2001. PGC-1, a versatile coactivator. Trends Endocrinol. Metab. 12:360-365. [DOI] [PubMed] [Google Scholar]
- 25.Laganiere, J., G. B. Tremblay, C. R. Dufour, S. Giroux, F. Rousseau, and V. Giguere. 2004. A polymorphic autoregulatory hormone response element in the human estrogen-related receptor α (ERRα) promoter dictates peroxisome proliferator-activated receptor γ coactivator-1α control of ERRα expression. J. Biol. Chem. 279:18504-18510. [DOI] [PubMed] [Google Scholar]
- 26.Lee, C.-H., P. Olson, and R. M. Evans. 2003. Lipid metabolism, metabolic diseases, and peroxisome proliferator-activated receptors. Endocrinology 144:2201-2207. [DOI] [PubMed] [Google Scholar]
- 27.Lee, S. S. T., T. Pineau, J. Drago, E. J. Lee, J. W. Owens, D. L. Kroetz, P. M. Fernandez-Salguero, H. Westphal, and F. J. Gonzalez. 1995. Targeted disruption of the α isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol. Cell. Biol. 15:3012-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lehman, J. J., P. M. Barger, A. Kovacs, J. E. Saffitz, D. Medeiros, and D. P. Kelly. 2000. PPARγ coactivator-1 (PGC-1) promotes cardiac mitochondrial biogenesis. J. Clin. Investig. 106:847-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lehman, J. J., and D. P. Kelly. 2002. Gene regulatory mechanisms governing energy metabolism during cardiac hypertrophic growth. Heart Failure Rev. 7:175-185. [DOI] [PubMed] [Google Scholar]
- 30.Lin, J., P. Puigserver, J. Donovan, P. Tarr, and B. M. Spiegelman. 2002. Peroxisome proliferator-activated receptor γ coactivator 1β (PGC-1β), a novel PGC-1-related transcription coactivator associated with host cell factor. J. Biol. Chem. 277:1645-1648. [DOI] [PubMed] [Google Scholar]
- 31.Lin, J., P. T. Tarr, R. Yang, J. Rhee, P. Puigserver, C. B. Newgard, and B. M. Spiegelman. 2003. PGC-1beta in the regulation of hepatic glucose and energy metabolism. J. Biol. Chem. 278:30843-30848. [DOI] [PubMed] [Google Scholar]
- 32.Luo, J., R. Sladek, J. Carrier, J. A. Bader, D. Richard, and V. Giguere. 2003. Reduced fat mass in mice lacking orphan nuclear receptor estrogen-related receptor alpha. Mol. Cell. Biol. 23:7947-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mootha, V. K., J. Bunkenborg, J. V. Olsen, M. Hjerrid, J. R. Wisniewski, E. Stahl, M. S. Bolouri, H. N. Ray, S. Sihag, M. Kamal, N. Patterson, E. S. Lander, and M. Mann. 2003. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell 115:629-640. [DOI] [PubMed] [Google Scholar]
- 34.Mootha, V. K., C. Handschin, D. Arlow, X. Xie, J. St. Pierre, S. Sihag, W. Yang, D. Altshuler, P. Puigserver, N. Patterson, P. J. Willy, I. G. Schulman, R. A. Heyman, E. S. Lander, and B. M. Spiegelman. 2004. ERRα and Gabpa/b specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. USA 101:6472-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pineda Torra, I., T. Claudel, C. Duval, V. Kosykh, J. C. Fruchart, and B. Staels. 2003. Bile acids induce the expression of the human peroxisome proliferator-activated receptor alpha gene via activation of the farnesoid X receptor. Mol. Endocrinol. 17:259-272. [DOI] [PubMed] [Google Scholar]
- 36.Pineda Torra, I., Y. Jamshidi, D. M. Flavell, J.-C. Fruchart, and B. Staels. 2002. Characterization of the human PPARalpha promoter: identification of a functional nuclear receptor response element. Mol. Endocrinol. 16:1013-1028. [DOI] [PubMed] [Google Scholar]
- 37.Puigserver, P., and B. M. Spiegelman. 2003. Peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α): transcriptional coactivator and metabolic regulator. Endocrine Rev. 24:78-90. [DOI] [PubMed] [Google Scholar]
- 38.Puigserver, P., Z. Wu, C. W. Park, R. Graves, M. Wright, and B. M. Spiegelman. 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92:829-839. [DOI] [PubMed] [Google Scholar]
- 39.Russell, L. K., C. M. Mansfield, J. J. Lehman, A. Kovacs, M. Courtois, J. E. Saffitz, D. M. Medeiros, M. L. Valencik, J. A. McDonald, and D. P. Kelly. 2004. Cardiac-specific induction of the transcriptional coactivator peroxisome proliferator-activated receptor γ coactivator-1α promotes mitochondrial biogenesis and reversible cardiomyopathy in a developmental stage-dependent manner. Circ. Res. 94:525-533. [DOI] [PubMed] [Google Scholar]
- 40.Schoenfeld, J. R., M. Vasser, P. Jhurani, P. Ng, J. J. Hunter, J. Ross Jr., K. R. Chien, and D. G. Lowe. 1998. Distinct molecular phenotypes in muring cardiac muscle development, growth, and hypertrophy. J. Mol. Cell. Cardiol. 30:2269-2280. [DOI] [PubMed] [Google Scholar]
- 41.Schreiber, S. N., R. Emter, M. B. Hock, D. Knutti, J. Cardenas, M. Podvinec, E. J. Oakeley, and A. Kralli. 2004. The estrogen-related receptor alpha (ERRα) functions in PPARγ coactivator 1α (PGC-1α)- induced mitochondrial biogenesis. Proc. Natl. Acad. Sci. USA 101:6472-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schreiber, S. N., D. Knutti, K. Brogli, T. Uhlmann, and A. Kralli. 2003. The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor α (ERRα). J. Biol. Chem. 278:9013-9018. [DOI] [PubMed] [Google Scholar]
- 43.Sladek, R., J.-A. Bader, and V. Giguère. 1997. The orphan nuclear receptor estrogen-related receptor α is a transcriptional regulator of the human medium-chain acyl coenzyme a dehydrogenase gene. Mol. Cell. Biol. 17:5400-5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sladek, R., and V. Giguère. 2000. Orphan nuclear receptors: An emerging family of metabolic regulators. Adv. Pharmacol. 47:23-87. [DOI] [PubMed] [Google Scholar]
- 45.St-Pierre, J., J. Lin, S. Krauss, P. T. Tarr, R. Yang, C. B. Newgard, and B. M. Spiegelman. 2003. Bioenergetic analysis of peroxisome proliferator-activated receptor γ coactivators 1α and 1β (PGC-1α and PGC-1β) in muscle cells. J. Biol. Chem. 278:26597-26603. [DOI] [PubMed] [Google Scholar]
- 46.Vega, R. B., J. M. Huss, and D. P. Kelly. 2000. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor α in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol. Cell. Biol. 20:1868-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vega, R. B., and D. P. Kelly. 1997. A role for estrogen-related receptor alpha in the control of mitochondrial fatty acid beta-oxidation during brown adipocyte differentiation. J. Biol. Chem. 272:31693-31699. [DOI] [PubMed] [Google Scholar]
- 48.Weinmann, A. S., S. M. Bartley, T. Zhang, M. Q. Zhang, and P. Farnham. 2001. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell. Biol. 21:6820-6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu, Z., P. Puigserver, U. Andersson, C. Zhang, G. Adelmant, V. Mootha, A. Troy, S. Cinti, B. Lowell, R. C. Scarpulla, and B. M. Spiegelman. 1999. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98:115-124. [DOI] [PubMed] [Google Scholar]