Abstract
The understanding of melanoma malignancy mechanisms is essential for patient survival, because melanoma is responsible for ca. 75% of deaths related to skin cancers. Enhanced formation of invadopodia and extracellular matrix (ECM) degradation are two important drivers of cell invasion, and actin dynamics facilitate protrusive activity by providing a driving force to push through the ECM. We focused on the influence of epidermal growth factor (EGF), hepatocyte growth factor (HGF) and transforming growth factor β (TGFβ) on melanoma cell invasiveness, since they are observed in the melanoma microenvironment. All three factors stimulated invasion of A375 and WM1341D cells derived from primary tumor sites. In contrast, only EGF and HGF stimulated invasion of WM9 and Hs294T cells isolated from lymph node metastasis. Enhanced formation of invadopodia and ECM degradation underlie the increased amount of invasive cells after stimulation with the tested agents. Generally, a rise in invasive potential was accompanied by a decrease in actin polymerization state (F:G ratio). The F:G ratio remained unchanged or was even increased in cell lines from a metastasis treated with TGFβ. Our findings indicate that the effects of stimulation with EGF, HGF and TGFβ on melanoma cell invasiveness could depend on melanoma cell progression stage.
Key words: EGF, HGF, TGFβ, actin cytoskeleton, melanoma, invasion, metastasis
Introduction
The widespread metastases are the main cause of death in melanoma patients. The critical moment is when the lesion progresses to a vertical-growth phase in which some of the cells develop the ability to invade the dermis. Malignant lesions are composed of neoplastic transformed cells and stroma consisting of cells and extracellular matrix (ECM).1 Some protein components of ECM contain binding sites for signaling molecules (SMs), which results in a local increase of SM concentration. In the case of TGFβ, its sequestration in ECM is considered crucial for its proper mobilization and activation. Matrix metalloproteinases (MMPs) MMP-2 and -9 are responsible for active TGFβ release during cell invasion.2 The crosstalk via SMs between tumor cells, cells of neoplastic stroma and ECM modulates the cancer microenvironment, supporting the tumor growth and invasion potential.3 The model of this crosstalk in melanoma tumor, simplified to three tested signaling proteins, is presented in Figure 1. In cancer cells receptor activation by the autocrine stimulation is frequently observed. Melanoma cells of different progression stages are characterized by varying production of signaling molecules including EGF, HGF and TGFβ.4,5 Secretion of cytokines and growth factors by melanoma cells affects both cancer and stromal cells, especially fibroblasts.6 The paracrine stimulation mainly by TGFβ induces recruitment of fibroblasts to the tumor microenvironment and their activation. On the other hand, it has been reported that HGF is responsible for fibroblast activation in gastric cancer.7 Such fibroblasts create a subpopulation of cells in the tumor environment known as cancer associated fibroblasts (CAFs).8 Activated fibroblasts are characterized by increased SMs secretion and in melanoma HGF is among the most important ones.6 However, in advanced melanoma TGFβ is more important.9
Figure 1.
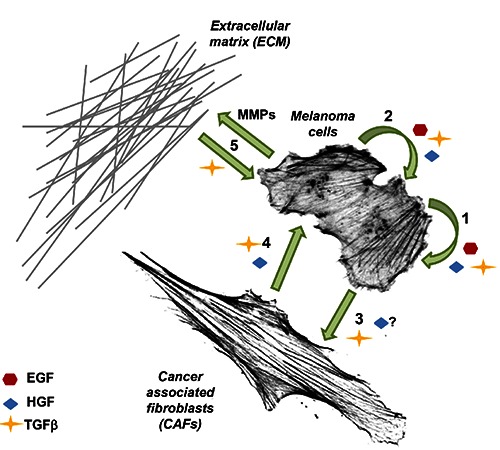
Intercellular communication involving signalling molecules (SMs) in melanoma tumour microenvironment simplified to three tested in this publication SMs. 1, The autocrine stimulation of a melanoma cell by EGF, HGF andTGFβ.4,5 2, The paracrine stimulation of neighbouring melanoma cells by all three growth factors.4,5 3, Activation of fibroblasts by TGFβ, which is secreted by melanoma cells.6 Stimulation of CAFs by HGF secreted by melanoma cells7 remains to be verified. 4, Secretion of HGF6 and TGFβ9 by CAFs, which influence melanoma cells. 5, Sequestration of TGFβ by ECM proteins and its release is induced by e.g. matrix degradation caused by matrix metalloproteases.2
In our study we focused on the influence of epidermal growth factor (EGF), hepatocyte growth factor (HGF) and transforming growth factor β (TGFβ) on melanoma cells’ invasiveness. Although altered EGF expression was observed in melanoma cells of different progression stages, its role in forming melanoma metastases has not been elucidated yet.5 HGF has been described as a melanocyte mitogen10 and is correlated with pigment cells’ escape from keratinocyte-mediated control during early melanoma progression stages.4 TGFβ is the main player in cancer-stroma interaction and epithelial-mesenchymal transition and in advanced melanoma TGFβ seems to be a key SM.9 We decided to evaluate the influence of these signaling molecules on melanoma cell invasion abilities because different expression patterns of EGF, HGF and TGFβ receptors have been reported in primary tumors and in metastases. Analysis of Kaplan-Meier plot survival curves of melanoma patients,11 deposited in the Prognoscan database, for high and low HGF receptor (MET) expression groups indicated a drastic decrease in overall survival of patients with high MET expression levels. Patients with a high TGFβ receptor 1 (TGFβR1) expression level had poorer prognosis than a TGFβR1 low expression group, although this group was characterized by longer overall survival than patients with high MET expression. Analysis of Kaplan-Meier plots in the case of EGF receptor (EGFR) expression level does not give a clear result. The data indicate either poorer or better prognosis for patients with high EGFR expression.
Some tumor cells develop an invasive phenotype through acquisition of the ability to actively protrude to overcome the basement membrane barrier. Cytoskeletal dynamics, especially actin cytoskeleton remodeling, is indispensable for cell shape changes and formation of membrane protrusions. These structures are driven by localized polymerization of actin filaments under the cell membrane.12 Among them there are invadopodia, located typically in the close vicinity of the cell nucleus and responsible for ECM degradation due to MMP secretion.13
In our study we intended to evaluate the influence of EGF, HGF and TGFβ on melanoma cells’ invasiveness. Thus, we analyzed the relative invasion ratio, actin polymerization state, invadopodia formation and ECM degradation upon stimulation with SMs. We tested four melanoma cell lines, two isolated from a primary tumor site and the other two from a lymph node metastasis.
Materials and Methods
Antibodies and dyes
Rabbit anti-EGFR (1005), rabbit anti-MET (C-12), rabbit anti-cortactin (H-191) and rabbit anti-TGFβRI (V-22) antibodies were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). Mouse anti-γ actin IgG1 (2-2.1.14.17) antibodies were purchased from Sigma-Aldrich (Warsaw, Poland). Donkey anti-rabbit-Alexa Fluor 488 and anti-rabbit-Alexa Fluor 633 antibodies, Alexa Fluor 488- and 568-labeled phalloidin, Alexa Fluor 594 DNase I conjugate and Hoechst 33342 were from Invitrogen (Carlsbad, CA, USA). Anti-rabbit and anti-mouse HRP-linked antibodies were from Cell Signaling Technology (Danvers, MA, USA).
Cell lines and culture conditions
Cell lines A375 and Hs294T were from ATCC. WM9 and WM1341D cells were a kind gift of Prof. Andrzej Mackiewicz from Greater Poland Cancer Center in Poznan, Poland. These cell lines are available from Rockland Immunochemicals, Inc. All cell lines were cultured according to resellers’ recommendations and were authenticated within last 6 months.
EGF, HGF and TGFβ stimulation
The cells after reaching the desired confluence were serum-starved for 24 h. Then the medium was replaced with medium containing only 20% fetal bovine serum (FBS) (control) or medium containing 20% FBS and one of the tested SMs - EGF (BD Biosciences, Warsaw, Poland), HGF (Sigma-Aldrich) and TGFβ (BD Biosciences) - at a final concentration of 31 ng/mL, 31 ng/mL and 0.1 ng/mL, respectively. Twenty-four (24) h later the cells were subjected to further analyses.
Isolation of cellular extracts and Western blot analysis
We used the procedure previously described.14 Cell lysates were prepared on ice by scraping the cells into cytoskeletal-bound protein extraction buffer (10 mM Tris-HCl pH 7.4, 100 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM NaF, 20 mM Na4P2O7, 2 mM Na3VO4, 1% Triton X-100, 10% glycerol, 0.1% SDS, 0.5% sodium deoxycholate) with addition of protease inhibitor cocktail (AEBSF, Aprotinin, Bestatin, E-64, Leupeptin, Pepstatin A) (Sigma-Aldrich). Next the samples were three times frozen and thawed and centrifuged at 12,000 × g for 5 min at 4°C. The supernatants were collected and stored at -80°C. Protein concentration was determined by the standard Bradford procedure.15 Samples containing 30 μg of protein were separated in 10% polyacrylamide gel by SDS-PAGE16 then transferred to nitrocellulose.17 Transfer efficiency and control analysis of protein loading were performed by Ponceau S staining (BioShop, Burlington, Canada). Immunoblots were blocked for 1 h in 5% skimmed milk in TBS-T (50 mM Tris-HCl pH 7.6, 150 mM NaCl, 0.1% Tween 20) and then incubated with primary antibodies overnight at 4°C. The next day, immunoblots were incubated for 1 h at room temperature (RT) with secondary antibodies conjugated to horseradish peroxidase (HRP). The immunoblots were developed using the Clarity Western ECL Substrate (Bio-Rad, Warsaw, Poland), visualized by the ChemiDoc MP System (Bio-Rad) and then analyzed using Image Lab 4.0 software (Bio-Rad).
Immunocytochemistry
The cells were seeded onto 12 mm coverslips in full medium. After 24 h the cells were fixed with 4% formaldehyde (FA) for 20 min. Cells were permeabilized with 0.1% Triton X- 100 in PBS for 6 min. Next all coverslips were blocked with 1% bovine serum albumin (BSA) in PBS for 45 min. For EGFR, MET and TGFβR1/F-actin staining the coverslips were first incubated with primary antibodies diluted in 1% BSA in PBS overnight at 4°C and next incubated for 1 h at RT with secondary antibodies conjugated to Alexa Fluor 488 and Alexa Fluor 568-labeled phalloidin diluted in 1% BSA in PBS. For simultaneous G-actin and F-actin visualization the coverslips were incubated with Alexa Fluor 594 DNase I conjugates and Alexa Fluor 488-labeled phalloidin diluted in 1% BSA in PBS for 1 h at RT. All coverslips were incubated with Hoechst 33342 (Invitrogen) diluted in 1% BSA in PBS for 10 min at RT to visualize cell nuclei. Finally, the coverslips were mounted with Dako mounting medium on glass slides. Slides were analyzed using a Zeiss LSM 510 confocal laser scanning microscope (Zeiss, Jena, Germany). Images were processed with the Zen 2008 software (Zeiss). For every fluorophore an independent track was used. Images were captured sequentially track by track and every track had assigned its own parameters such as laser, filter or beam splitters. The possibility of cross-talk between fluorescent dyes was reduced by: choice of fluorophore significantly different in excitation and emission wavelength; analysis of all fluorophore fluorescence spectra by Fluorescence SpectraViewer (tool available on Thermo Fisher Scientific website); and the use of specific lasers, filters and beam splitters assigned to every track.
Invasion assay
The cell invasion assay was performed using Transwell filters (BD Bioscience) coated with Matrigel (BD Bioscience) at a final concentration of 1 mg/mL diluted in a serum-free medium.18 Twenty-four (24) h later, after serum starvation, 3.5 × 104 cells were seeded onto a Transwell filter with a Matrigel layer in 500 μL of serum-free medium. As a chemoattractant, placed in a lower compartment, medium containing only 20% FBS or containing 20% FBS with addition of EGF, HGF or TGFβ was used. Upon 24 h incubation at 37°C, cells remaining on the upper side of the filter were removed together with Matrigel. The cells on the lower side of the filter were fixed with 4% FA for 20 min at RT, stained with Hoechst 33342 and counted under a fluorescent microscope. The cells with invasion ability are presented as a relative invasion factor (fold), with 1 taken as the number of cells invading under control conditions. The experiments were performed three times, each as an independent experiment.
MTT assay
The MTT assay was used to compare the viability of the cells seeded into a 96-well plate in medium containing only 20% FBS (control) or containing 20% FBS with addition of EGF, HGF or TGFβ at the same concentrations as used in the invasion assay. Cells were grown under these conditions for 24 h. Next the medium was replaced with MTT (Sigma-Aldrich) working solution (0.5 mg/mL in serum-free medium). The cells were incubated for 3 h at 37°C. Subsequently, MTT solution was removed and DMSO was added for 10 min with gentle agitation to dissolve purple formazan crystals. Absorbance was measured with an ELISA plate reader (BIO-TEK Instruments, Inc.) using a wavelength of 570 nm, with a reference wavelength of 630 nm. Viability of control cells is presented as 1, and the results obtained for stimulated cells are compared to control values. The assay was performed three times, each time as an independent experiment.
Isolation of cytosolic fraction and evaluation of actin polymerization state
The cells were homogenized and the cytosolic fraction was prepared as described previously.19 The cells were seeded onto ϕ 60 mm Petri dishes. After reaching 80-90% confluency the cells were starved for 24 h in serum-free medium. Then, the medium was replaced with medium containing only 20% FBS (control) or containing 20% FBS with addition of EGF, HGF or TGFβ at the same concentrations as used in the other experiments here. After 24 h the cells were collected with a scraper from Petri dishes into 2.5 mL ice-cold PBS and centrifuged at 1000 × g for 5 minutes at 4°C. Pellets were resuspended in G-actin stabilizing buffer A [10 mM Tris-HCl pH 7.4, 0.1 mM CaCl2, 0.25 M sucrose, 1 mM DTT (added prior to use) and 0.1 mM ATP (added prior to use)] in a ratio of 1:2 and mechanically homogenized in the Dounce tissue grinder. Then the homogenates were centrifuged at 105,000 × g for 1 h at 4°C. Supernatants (cytosolic fractions) were stored at -80°C. The actin content was determined by the inhibition of DNase I from bovine pancreas under standard assay conditions.19 The concentration of monomeric (G) actin was estimated by DNase I inhibition, directly in the cytosolic fraction of the cells. Total (T) actin content was measured after dilution of the samples with G-actin stabilizing buffer (buffer A) [10 mM Tris-HCl pH 7.4, 0.1 mM CaCl2, 0.25 M sucrose, 1 mM DTT (added prior to use) and 0.1 mM ATP (added prior to use)]. For the measurement of maximal inhibition a specific dilution below the critical actin concentration had to be applied to completely depolymerize the filamentous (F) actin. The amount of Factin was calculated by subtracting the amount of G-actin from the total actin (F = T - G). The state of actin polymerization was defined by the F-actin to G-actin ratio (F:G). One unit of DNase I inhibitor (actin) is the amount that reduces the activity of 20 ng of DNase I by 10% under standard assay conditions.19,20 Actin concentration was expressed in units of DNase I inhibitor per mg of sample protein. All experiments were done in triplicate. Each independent experiment consisted of three measurements/probes. The F:G ratio for control cells is presented as 1, and the results obtained for stimulated cells are compared to control values and presented as the relative F:G actin ratio (fold).
Fluorescent-gelatin degradation assay
The experiment was conducted according to the procedure described elsewhere21 to observe formation of active invadopodia by the cells. Poly-L-lysine coated coverslips (BD Biosciences) were washed with PBS and fixed with 0.5% glutaraldehyde for 15 min at RT, followed by extensive washing. Then, the coverslips were inverted on a 40 μl drop of gelatin conjugated with FITC (Invitrogen) and incubated for 10 min at RT. After washing with PBS, the residual reactive groups were quenched with 5 mg/mL sodium borohydride for 1 min at RT followed by extensive washing with PBS. The cells were first seeded onto ϕ 35 mm Petri dishes. After reaching 60% confluency the cells were starved for 24 h in serum-free medium. Next the cells were seeded onto coverslips coated with fluorescently labeled gelatin in 24-well plates in medium containing only 20% FBS (control) or 20% FBS with addition of EGF, HGF or TGFβ at the same concentrations as used in other experiments here and incubated at 37°C. In the case of matrix metalloproteases inhibition the cells were treated similarly, however instead of a SM GM6001 at a final concentration of 25 μM was added to the medium supplemented with 20% FBS. After 16 h the cells were fixed with 4% FA in PBS for 20 min and permeabilized for 5 min with 0.1% Triton X-100 in PBS. Alexa Fluor 568-labeled phalloidin was used for F-actin visualization. Confocal images were collected using an Olympus FV500 confocal laser scanning microscope. Enzymatic activity was indicated by gelatin degradation areas, which were characterized by the lack of a fluorescence signal. Gelatin degradation areas were analyzed using ImageJ software. Thirty cells were analyzed for each condition. The coverslips prepared for the gelatin degradation assay were also used in evaluation of the impact of tested SMs on invadopodia formation. The number of invadopodia per cell was calculated using ImageJ software. Thirty cells were analyzed for each condition.
Statistical analysis
For data presented as standard bar charts there are provided error bars representing standard deviations (+/- SD). The data are partially presented as box plots to indicate the degree of dispersion within obtained results. Statistical significance was determined in the two-tailed, unpaired Student’s t test. The significance levels were set at P≤0.05 (one asterisk), P≤0.01 (two asterisks) and P≤0.001 (three asterisks). Bar charts were prepared using Excel 2013 (Microsoft) and box plots were plotted with ggplot2 in R for iOS, ver. 3.2.4.22
Results
Tested melanoma cells express EGFR, MET and TGFβR1
As a cell model we used two cell lines isolated from primary tumor sites (A375 and WM1341D) and another two derived from a lymph node metastasis (WM9 and Hs294T). We evaluated the expression level of EGFR, MET and TGFβR1 by Western blot analysis. We noted that all these receptors were expressed in tested cells (Figure 2A). Immunocytochemical analysis of cells (Figure 2B) corroborated these results.
Figure 2.
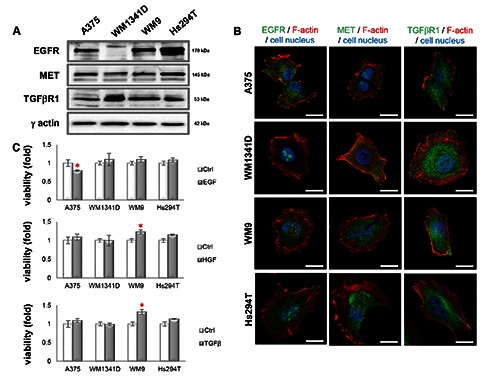
Expression level and subcellular localization of EGFR, MET and TGFβR1 in melanoma cell lines and effects of EGF, HGF or TGFβ on melanoma cell lines’ viability. A) Western blot analysis of EGFR, MET and TGFβR1 levels in cell lysates. The blots were probed with rabbit anti-EGFR, anti-MET or anti-TGFβR1 antibodies. Visualization of γ-actin served as a loading control; 30 μg of protein was loaded on every lane. B) Immunostaining was performed on cells with antibodies directed against EGFR, MET or TGFβR1. The cells were counterstained with Alexa Fluor 568-labeled phalloidin and Hoechst 33342. Scale bar: 20 μm. C) Viability of cells treated with SMs was compared to viability of non-treated control cells. Results expressed as the mean ±SD are representative for at least three independent experiments. Statistical significance was determined in two-tailed, unpaired Student’s t-test. Significance levels were set at P≤0.05 (one asterisk).
Effect of tested cytokine and growth factors on melanoma cells’ viability
TGFβ is capable to either stimulate or inhibit cells’ proliferation23,24 and both EGF25 and HGF26 are known as mitogens for mammalian cells. An MTT assay was performed to exclude the possibility that the results obtained in an invasion assay could be influenced by changed viability due to SM stimulation. As control cells we used the cells cultured in medium containing only 20% FBS. On charts (Figure 2C) control cells’ viability is presented as 100% and viability of cells after SM stimulation is compared to that value. Generally, we did not observe any statistically significant differences in viability after EGF, HGF and TGFβ treatment. However, we noted slightly lowered viability after EGF incubation in the case of A375 cells and higher WM9 cells number upon HGF and TGFβ addition.
Influence of tested cytokine and growth factors on melanoma invasion abilities
According to the literature, growth factors are usually used at the following concentrations: 4-100 ng/mL of EGF;27 6.25-50 ng/mL of HGF28 or 50 ng/mL of EGF or HGF.29 To our knowledge, lower concentrations are not used. Our previous experience21,30 indicates that the optimal EGF concentration to stimulate cell migration/invasion is 31 ng/mL (5 nM). We decided to use HGF at the same concentration as EGF. In the case of TGFβ in the preliminary experiments we tested different concentrations of TGFβ and we noted a prominent effect on invasiveness already at the concentration of 0.1 ng/mL. The obtained results from invasion assays are presented in Figure 3. All three SMs positively stimulated invasiveness of A375 and WM1341D cells. The strongest effects were observed for TGFβ stimulation. In the case of Hs294T and WM9 cell lines EGF and HGF caused a significant increase in the number of invasive cells. However, there were over fifteen times more WM9 cells able to invade upon HGF stimulation, whereas Hs294T cells were only moderately stimulated by EGF and HGF to invade under assay conditions. Interestingly, TGFβ, which was the most invasion-enhancing agent for A375 and WM1341D cells, acted as an inhibitor in the case of WM9 and Hs294T cells. The number of WM9 and Hs294T cells with invasive potential was significantly lower after addition of TGFβ to the medium.
Figure 3.
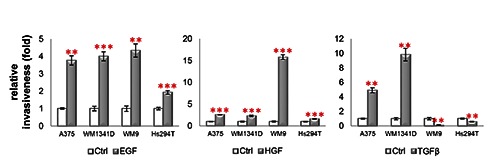
Effects of EGF, HGF or TGFβ on melanoma cell lines' invasiveness. Invasive potential of cells treated with SM was compared to invasive potential of control cells not treated with SM. Results expressed as the mean ±SD are representative for at least three independent experiments. Statistical significance was determined in two-tailed, unpaired Student’s t-test. Significance levels were set at P≤0.05 (one asterisk), P≤0.01 (two asterisks) and P≤0.001 (three asterisks).
Influence of EGF, HGF and TGFβ on ECM degradation
To evaluate the impact of SM stimulation on invadopodia formation and metalloproteinases activity, the cells were seeded onto FITC-conjugated gelatin and incubated with SMs for 16 h. Microscopic evaluation of stained coverslips revealed the presence of active, degrading ECM invadopodia in all cell lines both under control conditions and upon stimulation with all three SMs (Figure 4). We observed very clearly colocalizing F-actin with a degraded area of FITC-gelatin, and cortactin, an invadopodium marker, was observed within these structures (Supplementary Figure 1). We could not detect any gelatin-FITC degradation only in the case of WM1341D cells, either in control cells or after incubation with EGF. As a negative control we used GM6001, a paninhibitor of matrix metalloproteases (MMPs). Cells incubated with this agent showed no gelatin-FITC degradation activity (Figure 4). Next we performed quantitative analysis of invadopodia formation (Figure 5A). In A375 cells we observed a significant increase in the number of formed invadopodia in cells treated with all tested SMs. In the case of WM1341D only TGFβ stimulation had an influence on invadopodia number, but TGFβ did not affect the number of invadopodia in WM9 cells. In this cell line a significantly increased number of invadopodia was noted after EGF and HGF stimulation. In the case of Hs294T cells tested growth factors and TGFβ significantly stimulated invadopodia formation. The usage of GM6001 resulted in either no changes in the amount of invadopodia (A375 and WM1341D cells) or lowered number of invadopodia in cell lines obtained from a lymph node metastasis (WM9 and Hs294T). Simultaneously, we estimated the impact of SM treatment on the gelatin digestion area (Figure 5B). It showed that in the case of A375 cells the average digested area was significantly larger upon stimulation with all SMs, but for WM1341D it was found only for incubation of the cells with TGFβ. The opposite situation was noted in cell lines from a lymph node metastasis (WM9 and Hs294T). In both cell lines after TGFβ treatment no significant differences were observed in gelatin degradation in comparison to control cells. On the other hand, in WM9 and Hs294T cells EGF and HGF stimulation correlated with an increase in metalloproteinases activity. Incubation of the cells with GM6001 abrogated the MMPs activity in all tested cell lines.
Figure 4.
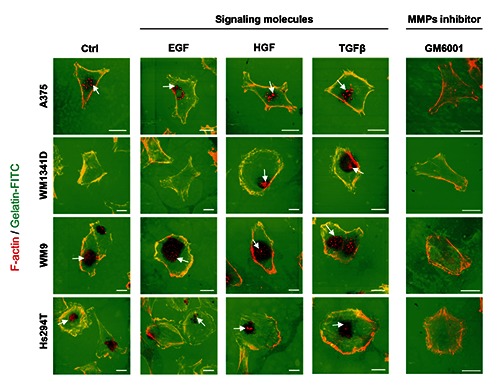
Microscopical analysis of effects of EGF, HGF or TGFβ on melanoma cell lines’ invadopodia formation and extracellular matrix degradation potential. Melanoma cells were seeded in medium containing only 20% FBS (control) or containing 20% FBS with addition of EGF, HGF or TGFβ onto coverslips coated with fluorescently labeled gelatin. As a negative control the cells were treated with medium containing 20% FBS and 25 μM GM6001, a pan-inhibitor of matrix metalloproteases. 16 h later the cells were fixed and stained with Alexa Fluor 568-labeled phalloidin. Invadopodia are indicated with white arrows. Gelatin degradation is visualized by lack of a green fluorescence signal. Scale bar: 20 μm.
Figure 5.
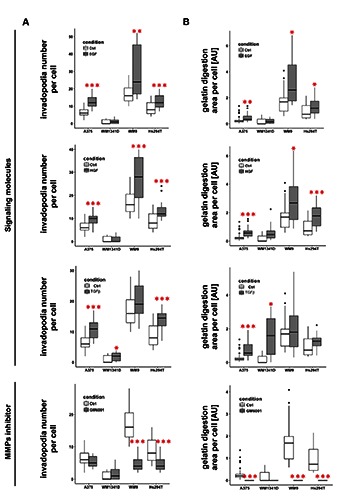
Quantitative analysis of effects of EGF, HGF or TGFβ on melanoma cell lines’ invadopodia formation and extracellular matrix degradation potential. A-B: Upon capture of the images (Figure 4) number of invadopodia (A) and the size of digested area (B) were calculated using ImageJ software. Thirty cells were analyzed for each condition. Boxes show the median values of RQ±interquartile range. Statistical significance was determined in two-tailed, unpaired Student’s t-test. Significance levels were set at P≤0.05 (one asterisk), P≤0.01 (two asterisks) and P≤0.001 (three asterisks).
The actin polymerization state after treatment of the cells with signaling molecules
Our final concern referred to the influence of tested signaling molecules on actin polymerization state, since actin dynamics is very important for cell movement. In A375 cells we observed a decrease in the filamentous (F) to monomeric (G) actin ratio upon stimulation with all tested SMs (Figure 6A). In the case of WM1341D cells only after HGF and TGFβ stimulation we noted the same statistically significant decreasing effect. In the case of cell lines derived from a lymph node metastasis only in WM9 cells we observed a drop in the F:G actin ratio after incubation with EGF and HG. Interestingly, the TGFβ treatment in WM9 cells had no influence on the F:G actin ratio, whereas in Hs294T cells it caused a significant rise in the amount of polymerized actin. Simultaneous visualization of G- and F-actin pools (Figure 6B) showed that F-actin was mainly concentrated under the cell membrane and in stress fibers. In contrast, G-actin was mainly dispersed in the cytoplasm, in cell nuclei and in motile protrusions, lamellipodia. The cells probably accumulate a G-actin pool in the lamellipodia to provide proper conditions for fast actin polymerization, and thus quick actin rearrangements crucial for cell movement.
Figure 6.
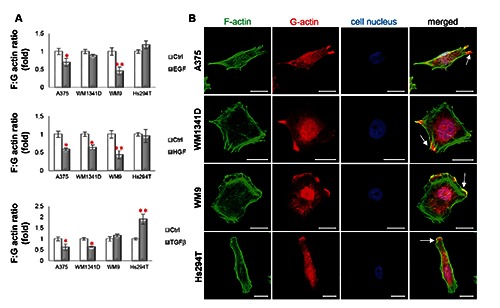
Actin polymerization state in melanoma cells is affected by SMs. A) F:G ratio of control cells is presented as 1. B) Confocal microscopy analysis was performed on cells, which were stained with Alexa Fluor 488-labeled phalloidin, Alexa Fluor 594-labeled DNase I and Hoechst 33342 to visualize F-actin, G-actin and nuclei. White arrows indicate the lamellipodia rich in F- and G-actin. Scale bar: 20 μm.
Discussion
The role of various signaling molecules and their receptors in tumor progression and metastasis formation has been evaluated for decades in different tumor types. Those studies contributed to the development of new strategies in cancer treatment such as targeted therapy against overexpressed or mutated growth factor receptors. Unfortunately, in the case of melanoma clinicians are not dealing with a single receptor being overexpressed or mutated. On the contrary, there takes place cooperation of many signaling pathways triggered by several compounds including cytokines and growth factors.31 Despite numerous studies focusing on involvement of SMs in melanoma progression, there is no study regarding the influence of a panel of SMs on the invasive potential of melanoma cells at different progression stages. Our aim was to investigate this unexplored area.
For our studies we chose cytokine TGFβ and two growth factors, EGF and HGF, because all three compounds have been demonstrated to be involved in melanoma. We observed expression of EGFR, MET and TGFβR1 receptors in melanoma cell lines originating from primary tumor and a lymph node metastasis. We found that the melanoma cells at different tumorigenesis stages respond to EGF, HGF and TGFβ differently in terms of their invasiveness. EGF and HGF positively stimulated invasiveness of all cell lines. However, TGFβ treatment resulted in the highest number of A375 and WM1341D cells with invasive potential. But in the case of cell lines originating from lymph node metastasis, TGFβ acted as an inhibitor of invasion. These differences were not reflected in EGFR, MET and TGFβR1 expression level or their cellular localization. To evaluate differences in the influence of EGF, HGF and TGFβ on melanoma cell invasion potential, we decided to investigate their impact on invadopodia formation, ECM degradation and actin polymerization state. A schematic representation of obtained results is presented in Figure 7.
Figure 7.
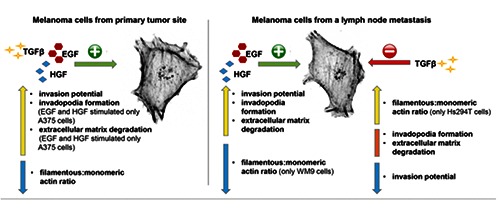
Schematic representation of effects caused by EGF, HGF and TGFβ on melanoma cells isolated from primary tumor site (A375 and WM1341D) and from a lymph node metastasis (WM9 and Hs294T). A yellow arrow represents a rise, a orange bar reflects unchanged values and a blue arrow represents a decrease.
The role of EGF in invadopodia formation has been thoroughly described, especially for breast cancer.32 Both HGF33 and TGFβ34 were also shown to be able to trigger invadopodia formation. For this process actin polymerization is essential. In our study we observed a correlation between increasing invasive potential and a greater amount of invadopodia per cell after EGF, HGF and TGFβ treatment. In cell lines originating from primary tumor sites, all three SMs increased the invasiveness and invadopodia formation, although in the case of WM1341D cells only TGFβ stimulation led to a significant rise in invadopodia number. In both cell lines originating from a lymph node metastasis after EGF and HGF treatment we observed an increase in both invasive potential and the number of invadopodia per cell. On the other hand, TGFβ affecting the invasiveness of WM9 and Hs294T cells did not affect the number of invadopodia in WM9 cells, although in the case of Hs294T cells it significantly stimulated invadopodia formation.
EGF positively stimulated gelatin degradation in three melanoma cell lines - A375, WM9 and Hs294T - although the invasion potential was significantly increased in all four tested cell lines. It has been reported previously that EGF induces MMP-935 and MMP-136 expression in breast cancer and glioma cells, respectively. HGF treatment, significantly raising the invasive potential in four melanoma cell lines, caused growth of the gelatin degradation area only in three cell types: A375, WM9 and Hs294T. HGF in human gliomas contributes to tumor progression by triggering MMP-2 and MT-MMP-1 production.37 In the case of TGFβ treatment we observed different cellular responses depending on the cell line origin. In cell lines derived from primary tumors we observed enhanced ECM degradation after TGFβ stimulation, which was positively correlated with increased invasive potential. But in cell lines derived from metastases, whose invasive potential TGFβ was affecting, we did not note any statistically significant impact on gelatin degradation. This is interesting, since in Hs294T cells TGFβ significantly stimulated invadopodia formation in comparison to control conditions. It is possible that in Hs294T cells upon TGFβ treatment formation of fully active invadopodia is impaired. It has been shown that TGFβ in highly invasive breast cancer cells increased the mRNA level of metalloproteinases (MMP-2, MMP-9 and MT-MMP-1) as well as of MMP inhibitors (TIMP-2 and RECK).38 But at the protein level enhanced expression of MMP-2, MMP-9 and TIMP-2 was observed, although the amount of RECK was lowered. These results suggest that TGFβ is able to either enhance or inhibit ECM degradation. The exact mechanism leading to TGFβ’s diverse action has not been elucidated yet.
Analysis of the results obtained from invasion assay, quantification of invadopodia number and ECM degradation area gives an interesting observation. EGF, HGF and TGFβ having a pro-invasive effect on A375, WM9 and Hs294T cells, caused an increase in invadopodia number and enhanced ECM degradation in these cell lines. However, in the case of WM1341D cells, although EGF and HGF were significantly increasing the number of invading cells, formation of invadopodia and ability to degrade ECM remained unchained upon EGF and HGF stimulation. It was shown for several cell types including melanoma cells that populations of tumor cells are heterogenous in regard to migration mode and phenotype, thus being partly mesenchymal and ameboid-like.39 Mesenchymal mode of motility is characterized by slower migratory speed, spindle-like shape and MMPs secretion, whereas for the ameboid-like mode characteristic are round morphology, higher speed of migration and independence of MMPs activity.39,40 In the case of WM1341D cells TGFβ could stimulate the cells to move in a mesenchymal mode, whereas EGF and HGF might be chemoattractive for cells moving rather in an ameboid-like mode. Moreover, a signaling molecule could cause a shift in a mode of cell’s movement due to plasticity of tumor cells’ motility.40 Bergert and colleagues41 proved that alterations in actin protrusivity and actomysoin contractility can cause conversion of the motility mode. Vast of signaling pathways have varying effects on actin cytoskeleton42 and different external stimuli trigger diverse intracellular signaling pathways having impact on cells’ motility.43 On the other hand EGF was shown to trigger dismantle of focal adhesions, what leaded to lowered adhesion of the cells and thus increased cells’ motility44,45 and decreased adhesion favors ameboid-like mode of motility.39,46 Triggering of divergent motility modes in melanoma cells by tested here SMs should be further studied.
We found that tested SMs, elevating the number of cells able to invade, caused a decrease in F:G actin ratio. On contrary, in cell lines originating from a lymph node metastasis TGFβ treatment resulted not only in inhibition of their invasiveness but also in an unchanged or even increased F:G actin ratio. Cell migration is based on the actin treadmill, which depends on G-actin recycling from the cell rear to the front.47 Moreover, it has been shown that G-actin concentration in the lamellipodium of a moving cell is about 1500 times higher than the actin monomer concentration required for filament elongation.48 This indicates that in cell movement both the monomeric and filamentous actin plays an important role. In this context, our data seem plausible. The EGF, HGF and TGFβ tested by us stimulated melanoma cell invasiveness, which was correlated with increased invadopodia formation involving localized actin polymerization. Probably, the lowered F:G actin ratio in the cytosolic fraction of the cells of higher invasive potential reflects the shift of F-actin towards the plasma membrane, where it is incorporated into F-actin structures forming e.g. invadopodia. On the other hand, in the case of stimulation of WM9 and Hs294T cells with TGFβ, which acts as an invasion inhibitor on these cells isolated from metastases, the F:G ratio remains unaltered or is even higher in the cytosolic fraction.
To our knowledge, we show here for the first time that the cells at different progression stages react in a varying manner to the same extracellular signal. At this moment using the TGFβR1 and growth factor receptor inhibitors in melanoma treatment could bring divergent effects depending on the melanoma progression stage in a given patient. Hence it seems to be crucial to design further experiments to study the effects of extracellular signals on melanoma cell invasion and actin dynamics and not only on proliferation rate and cell survival - especially if we bear in mind that already a single melanoma tumor initiating cell is capable to form a tumor.49
Acknowledgments
This work was supported by the Foundation for Polish Science within the HOMING Plus Programme (HOMING Plus/2010-2/8) and by the National Science Centre, Poland (project no. 2014/15/B/NZ5/01467).
References
- 1.Egeblad M, Nakasone ES, Werb Z. Tumors as organs: complex tissues that interface with the entire organism. Dev Cell 2010;18:884-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horiguchi M, Ota M, Rifkin DB. Matrix control of transforming growth factor-β function. J Biochem 2012;152:321-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg R. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [DOI] [PubMed] [Google Scholar]
- 4.Li G, Schaider H, Satyamoorthy K, Hanakawa Y, Hashimoto K, Herlyn M. Downregulation of E-cadherin and Desmoglein 1 by autocrine hepatocyte growth factor during melanoma development. Oncogene 2001;20:8125-35. [DOI] [PubMed] [Google Scholar]
- 5.Elias EG, Hasskamp JH, Sharma BK. Cytokines and growth factors expressed by human cutaneous melanoma. Cancers (Basel) 2010;2:794-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li G, Satyamoorthy K, Meier F, Berking C, Bogenrieder T, Herlyn M. Function and regulation of melanoma-stromal fibroblast interactions: when seeds meet soil. Oncogene 2003;22):3162-71. [DOI] [PubMed] [Google Scholar]
- 7.Wu X, Chen X, Zhou Q, Li P, Yu B, Li J, et al. Hepatocyte growth factor activates tumor stromal fibroblasts to promote tumorigenesis in gastric cancer. Cancer Lett 2013;335:128-35. [DOI] [PubMed] [Google Scholar]
- 8.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer 2006;6:392-401. [DOI] [PubMed] [Google Scholar]
- 9.Zhou L, Yang K, Andl T, Wickett RR, Zhang Y. Perspective of targeting cancer-associated fibroblasts in melanoma. J Cancer 2015;6:717-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsumoto K, Tajima H, Nakamura T. Hepatocyte growth factor is a potent stimulator of human melanocyte DNA synthesis and growth. Biochem Biophys Res Commun 1991;176:45-51. [DOI] [PubMed] [Google Scholar]
- 11.Bogunovic D, O’Neill DW, Belitskaya-Levy I, Vacic V, Yu Y-L, Adams S, et al. Immune profile and mitotic index of metastatic melanoma lesions enhance clinical staging in predicting patient survival. Proc Natl Acad Sci USA 2009; 106:20429-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamazaki D, Kurisu S, Takenawa T. Regulation of cancer cell motility through actin reorganization. Cancer Sci 2005;96:379-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Linder S, Wiesner C, Himmel M. Degrading devices: invadosomes in proteolytic cell invasion. Annu Rev Cell Dev Biol 2011;27:185-211. [DOI] [PubMed] [Google Scholar]
- 14.Mazur AJ, Radaszkiewicz T, Makowiecka A, Malicka-Blaszkiewicz M, Mannherz HG, Nowak D. Gelsolin interacts with LamR, hnRNP U, nestin, Arp3 and β-tubulin in human melanoma cells as revealed by immunoprecipitation and mass spectrometry. Eur J Cell Biol 2015;95:26-41. [DOI] [PubMed] [Google Scholar]
- 15.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976;72:248-54. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli UK. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970;227:680-5. [DOI] [PubMed] [Google Scholar]
- 17.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 1979;76:4350-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simiczyjew A, Mazur AJ, Popow-Wozniak A, Malicka-Blaszkiewicz M, Nowak D. Effect of overexpression of β and γ-actin isoforms on actin cytoskeleton organization and migration of human colon cancer cells. Histochem Cell Biol 2014;142:307-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malicka-Blaszkiewicz M, Roth JS. Some factors affecting the interaction between actin in leukemic L1210 cells and DNASE I. Biochem Biophys Res Commun 1981; 102:594-601. [DOI] [PubMed] [Google Scholar]
- 20.Malicka-Blaszkiewicz M. DNase 1-like activity and actin content in the liver of some vertebrates. Comp Biochem Physiol B 1986;84:207-9. [DOI] [PubMed] [Google Scholar]
- 21.Simiczyjew A, Mazur AJ, Ampe C, Malickablaszkiewicz M, van Troys M, Nowak D. Active invadopodia of mesenchymally migrating cancer cells contain both β and γ cytoplasmic actin isoforms. Exp Cell Res 2015; 339:206-219. [DOI] [PubMed] [Google Scholar]
- 22.Hadley W. Ggplot2: elegrant graphics for data analysis. Springer-Verlag, New York; 2009. [Google Scholar]
- 23.Moses HL, Yang EY, Pietenpol JA. TGF-| stimulation and inhibition of cell proliferation: New mechanistic insights. Cell 1990;63:245-7. [DOI] [PubMed] [Google Scholar]
- 24.Seoane J. Escaping from the TGFbeta antiproliferative control. Carcinogenesis 2006;27:2148-56. [DOI] [PubMed] [Google Scholar]
- 25.Yewale C, Baradia D, Vhora I, Patil S, Misra A. Epidermal growth factor receptor targeting in cancer: a review of trends and strategies. Biomaterials 2013;34:8690-707. [DOI] [PubMed] [Google Scholar]
- 26.Nakamura T, Mizuno S. The discovery of hepatocyte growth factor (HGF) and its significance for cell biology, life sciences and clinical medicine. Proc Jpn Acad Ser B Phys Biol Sci 2010;86:588-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paulsen CE, Truong TH, Garcia FJ, Homann A, Gupta V, Leonard SE, et al. Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nat Chem Biol 2012;8:57-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 2012;487:500-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature 2012;487:505-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Popow-Wozniak A, Mazur AJ, Mannherz HG, Malicka-Blaszkiewicz M, Nowak D. Cofilin overexpression affects actin cytoskeleton organization and migration of human colon adenocarcinoma cells. Histochem Cell Biol 2012;138:725-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zbytek B, Carlson JA, Granese J, Ross J, Mihm MC, Slominski A. Current concepts of metastasis in melanoma. Expert Rev Dermatol 2008;3:569-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamaguchi H, Lorenz M, Kempiak S, Sarmiento C, Coniglio S, Symons M, et al. Molecular mechanisms of invadopodium formation: The role of the N-WASP-Arp2/3 complex pathway and cofilin. J Cell Biol 2005;168:441-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rajadurai C V, Havrylov S, Zaoui K, Vaillancourt R, Stuible M, Naujokas M, et al. Met receptor tyrosine kinase signals through a cortactin-Gab1 scaffold complex, to mediate invadopodia. J Cell Sci 2012;125:2940-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pignatelli J, Tumbarello DA, Schmidt RP, Turner CE. Hic-5 promotes invadopodia formation and invasion during TGF-β-induced epithelial-mesenchymal transition. J Cell Biol 2012;197:421-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim S, Choi JH, Lim HI, Lee S-K, Kim WW, Cho S, et al. EGF-induced MMP-9 expression is mediated by the JAK3/ERK pathway, but not by the JAK3/STAT-3 pathway in a SKBR3 breast cancer cell line. Cell Signal 2009;21:892-8. [DOI] [PubMed] [Google Scholar]
- 36.Anand M, Van Meter TE, Fillmore HL. Epidermal growth factor induces matrix metalloproteinase-1 (MMP-1) expression and invasion in glioma cell lines via the MAPK pathway. J Neurooncol 2011;104: 679-87. [DOI] [PubMed] [Google Scholar]
- 37.Hamasuna R, Kataoka H, Moriyama T, Itoh H, Seiki M, Koono M. Regulation of matrix metalloproteinase-2 (MMP-2) by hepatocyte growth factor/scatter factor (HGF/SF) in human glioma cells: HGF/SF enhances MMP-2 expression and activation accompanying up-regulation of membrane type-1 MMP. Int J Cancer 1999;82(274-81. [DOI] [PubMed] [Google Scholar]
- 38.Gomes LR, Terra LF, Wailemann RA, Labriola L, Sogayar MC. TGF-β1 modulates the homeostasis between MMPs and MMP inhibitors through p38 MAPK and ERK1/2 in highly invasive breast cancer cells. BMC Cancer 2012;12:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clark AG, Vignjevic DM. Modes of cancer. cell invasion and the role of the microenvironment. Curr Opin Cell Biol 2015;36:13-22. [DOI] [PubMed] [Google Scholar]
- 40.Friedl P, Wolf K. Plasticity of cell migration: A multiscale tuning model. J Cell Biol 2010;188:11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bergert M, Chandradoss SD, Desai R a, Paluch E. Cell mechanics control rapid transitions between blebs and lamellipodia during migration. Proc Natl Acad Sci USA 2012;109:14434-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J. Actin dynamics, architecture, and mechanics in cell motility. Physiol Rev 2014;94:235-63. [DOI] [PubMed] [Google Scholar]
- 43.Harrison SM, Knifley T, Chen M, O’Connor KL. LPA, HGF, and EGF utilize distinct combinations of signaling pathways to promote migration and invasion of MDA-MB- 231 breast carcinoma cells. BMC Cancer 2013;13:501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie H, Pallero M a, Gupta K, Chang P, Ware MF, Witke W, et al. EGF receptor regulation of cell motility: EGF induces disassembly of focal adhesions independently of the motility-associated PLCgamma signaling pathway. J Cell Sci 1998;111:615-24. [DOI] [PubMed] [Google Scholar]
- 45.Lu Z, Jiang G, Blume-jensen P, Hunter T. Epidermal growth factor-induced tumor cell invasion and metastasis initiated by dephosphorylation and downregulation of focal adhesion kinase. Society 2001;21:4016-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sroka J, Krecioch I, Zimolag E, Lasota S, Rak M, Kedracka-Krok S, et al. Lamellipodia and membrane blebs drive efficient electrotactic migration of rat walker carcinosarcoma cells WC 256. PLoS One 2016;11:1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mogilner A, Oster G. Polymer motors: pushing out the front and pulling up the back. Curr Biol 2003;13:R721-33. [DOI] [PubMed] [Google Scholar]
- 48.Koestler SA, Rottner K, Lai F, Block J, Vinzenz M, Small JV. F- and G-actin concentrations in lamellipodia of moving cells. PLoS One 2009;4:e4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature 2008;456:593-8. [DOI] [PMC free article] [PubMed] [Google Scholar]


