Abstract
Mutations in Artemis in both humans and mice result in severe combined immunodeficiency due to a defect in V(D)J recombination. In addition, Artemis mutants are radiosensitive and chromosomally unstable, which has been attributed to a defect in nonhomologous end joining (NHEJ). We show here, however, that Artemis-depleted cell extracts are not defective in NHEJ and that Artemis-deficient cells have normal repair kinetics of double-strand breaks after exposure to ionizing radiation (IR). Artemis is shown, however, to interact with known cell cycle checkpoint proteins and to be a phosphorylation target of the checkpoint kinase ATM or ATR after exposure of cells to IR or UV irradiation, respectively. Consistent with these findings, our results also show that Artemis is required for the maintenance of a normal DNA damage-induced G2/M cell cycle arrest. Artemis does not appear, however, to act either upstream or downstream of checkpoint kinase Chk1 or Chk2. These results define Artemis as having a checkpoint function and suggest that the radiosensitivity and chromosomal instability of Artemis-deficient cells may be due to defects in cell cycle responses after DNA damage.
Artemis is a member of the SNM1/PSO2 gene family, the archetypical member of which was identified in budding yeast (Saccharomyces cerevisiae) as a factor required for efficient DNA interstrand cross-link repair (23, 53). Members of this family, which in humans also include SNM1, SNM1B, ELAC2, and CPSF73 (15, 27, 57), share a region of homology termed the SNM1 domain, which contains a metallo-β-lactamase fold and an appended β-CASP (for metallo-β-lactamase-associated CPSF Artemis SNM1/PSO2) domain that is a predicted nucleic acid binding motif (7, 41). Outside the SNM1 domain, the sequences of the yeast and human proteins are different. The function of yeast Snm1 remains largely unresolved, although several studies have indicated that it is involved in repairing double-strand breaks (DSBs) resulting from processing of interstrand cross-links (31, 35, 40). Artemis was originally identified molecularly as deficient in a human radiosensitive severe combined immunodeficiency syndrome (RS-SCID) (41), which is characterized by a defect in V(D)J recombination resulting in premature arrest of both B- and T-cell maturation. In addition, patient cell lines exhibited greater sensitivity to ionizing radiation (IR) than normal cells (9, 42, 44). RS-SCID resembles murine SCID caused by defects in DNA-PK, a protein complex involved in both V(D)J recombination and repair of DSBs via the nonhomologous end-joining (NHEJ) pathway. These findings have also been confirmed in a mouse model in which the Artemis gene was disrupted by gene targeting (51). Biochemical studies of Artemis have shown that it possesses a 5′→3′ exonucleolytic activity on single-stranded DNA, and when complexed with DNA-PKcs, it acquires endonucleolytic activity on 5′ and 3′ overhangs and the ability to open DNA hairpins (34). This latter activity is consistent with the observed defect in coding joint formation in Artemis-deficient cells. The nuclease function of Artemis appears to reside in the conserved SNM1 domain. In addition, it was shown that Artemis is a substrate of the kinase activity of DNA-PKcs in vitro. DNA-PKcs is a member of a family of large phosphatidylinositol-3-OH kinase-like kinases (PIKKs) that includes the ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and RAD3-related (ATR) gene products (reviewed in reference 16). These findings define a role for Artemis in V(D)J recombination, and with the hypersensitivity of Artemis-deficient cells to IR irradiation, also indicate a role for this gene in the cellular response to DNA damage. It has been proposed, although not formally demonstrated, that the radiosensitivity of Artemis-deficient cells is due to a defect in NHEJ (34).
ATM and ATR are two central signaling kinases that mediate pleiotropic response to DNA damage, including activation of cell cycle checkpoints, DNA repair pathways, transcription, and apoptosis (reviewed in references 1, 16, 56, and 63). Although they have some functional redundancy, ATM and ATR have specialized roles that appear to operate in parallel in the response to DNA damage: ATM primarily responds to the induction of DSBs, while ATR is activated by many forms of DNA damage and replication inhibitors. Many downstream phosphorylation targets of ATM and/or ATR have been identified, including p53, BRCA1, Nbs1, Smc1, FANCD2, Chk1, Chk2, and Rad17. These proteins are mediators and transducers of the stress signal emanating from these two PIKKs. Some of these substrates, as well as ATM and ATR, are found together in a large multifactorial association of proteins referred to as the BRCA1-associated surveillance complex (BASC), which may function to recognize unusual or aberrant DNA structures and to activate DNA repair and checkpoint pathways (61). Thus, the BASC is thought to be a complex that both senses and transduces the DNA damage signal.
In this report we demonstrate that Artemis-deficient cells are not, in fact, significantly defective in NHEJ following exposure to IR. Rather, Artemis is shown to interact with known checkpoint proteins and to be phosphorylated by ATM and ATR in vitro. In addition, Artemis is phosphorylated in vivo after exposure of cells to genotoxic stress: this modification is dependent upon both DNA-PK and ATM after IR and upon ATR in response to UV radiation. Further, we show that Artemis-deficient cells exposed to IR are defective in a G2/M DNA damage checkpoint. These findings define a novel function for Artemis as a checkpoint protein and suggest a new basis for the radiosensitivity and chromosomal instability of Artemis-deficient cells.
MATERIALS AND METHODS
Antibodies.
A fragment encoding amino acid residues 347 to 692 of Artemis was fused to a hexahistidine tag by insertion into pET28 (Novagen). Purified recombinant protein from Escherichia coli was used to raise antiserum in rabbits using standard protocols. Antisera were affinity purified using antigen that had been blotted and immobilized on nitrocellulose paper or by affinity chromatography.
Antibodies for Nbs1 (C-19), Rad17 (H-300), and cyclin B1 were purchased from Santa Cruz Biotech. Antibodies for Mre11 (12 D7), ATM (2c-1), and DNA-PKcs were from GeneTex. A monoclonal antibody for BRCA1 (Ab-1) was from CalBiochem, and a polyclonal antibody for ATR (Ab-2) was from Oncogene. Polyclonal antibodies for phosphorylated Chk1 (phospho-Chk1) (Ser345) and phospho-Chk2 (Thr68) and a monoclonal antibody for phospho-histone H3 (P-H3) (Ser10) were purchased from Cell Signaling. A monoclonal antibody for γ-H2AX (Ser139) was from Upstate. A monoclonal antibody for DNA-PKcs (Ab-4) was from NeoMarkers. Monoclonal antibodies for α-tubulin and Cdc2 were from Molecular Probes and Transduction Laboratories, respectively. Rad50 antibodies were provided by John Petrini.
Immunoprecipitation and dephosphorylation assays.
For coimmunoprecipitation experiments, cells were grown in 100-mm-diameter plates, washed with cold phosphate-buffered saline (PBS) twice, and lysed by adding 400 μl of EBC buffer (50 mM Tris-HCl [pH 8.0], 120 mM NaCl, 0.5% Nonidet P-40 [NP-40], 1 mM phenylmethylsulfonyl fluoride [PMSF]) on ice for 20 min. In some cases, cells were transfected with a construct (pDEST27-Artemis) expressing a glutathione S-transferase (GST)-Artemis fusion protein and incubated for 2 days prior to extract preparation. The lysates were spun for 15 min at 8,200 × g, and the supernatants were mixed with 2 volumes of Net-N buffer (20 mM Tris [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5% NP-40) plus the indicated antibodies and incubated at 4°C for 60 min. For some experiments, extracts were pretreated with DNase I (0.5 mg/ml) for 10 min at room temperature. Fifteen microliters of protein A-Sepharose CL-4B beads (Amersham Pharmacia Biotech) equilibrated with Net-N buffer was then added, and the mixture was incubated for 60 min. The beads were washed five times with Net-N buffer, and bound proteins were eluted in sodium dodecyl sulfate (SDS) sample buffer and separated by SDS-polyacrylamide gel electrophoresis for immunoblotting. For some experiments, HeLa whole-cell extracts prepared by the method of Manley et al. (36) were used.
For dephosphorylation experiments, Artemis protein was immunoprecipitated from cell lysates as described above and washed with 1× alkaline phosphatase buffer (provided by the manufacturer). Protein A-Sepharose beads with bound protein were incubated with 20 U of alkaline phosphatase (Boehringer Mannheim) at 30°C for 30 min, with or without 10 mM Na3VO4, and washed twice with Net-N buffer. The proteins were eluted in SDS sample buffer and separated by SDS-polyacrylamide gel electrophoresis for immunoblotting.
In vitro kinase assay.
Kinase assays were performed essentially as described previously (8). The substrate for these experiments was recombinant Artemis prepared as described above for antibody preparation. This truncated fragment contains all the Arteims SQ motifs but is lacking the two TQ motifs in the amino terminus of the protein. The concentration of recombinant Artemis was 0.9 μM.
Cell-free nonhomologous end-joining assay.
Immunodepletion of Artemis was achieved by mixing 15 μl of equilibrated protein A-Sepharose beads and 20 μl of Artemis antiserum with 400 μl of Net-N buffer and then incubating the mixture at 4°C for 1 h. After the beads were washed three times with Net-N buffer, they were mixed with 100 μg of HeLa whole-cell extract prepared as described previously (5) and incubated at 4°C for 2 h. The beads were spun down, and the supernatant was removed for immunoblot analysis and the end-joining assay. The end-joining assays were performed as previously described (5).
In vivo assay for DSB rejoining after IR.
DNA DSB rejoining, as measured by residual lesions remaining after IR, was determined by pulsed-field gel electrophoresis as previously described (11, 29).
Preparation of ATR-deficient cells.
To prepare ATR−/− cells, ATRflox/− HCT116 cells in which one allele of ATR is disrupted and the other contains loxP sites flanking exon 2 (12) were seeded at a density of 8 × 105 per 60-mm-diameter plate and infected with an adenovirus encoding Cre recombinase at a multiplicity of infection of 150. After 8 h of incubation, fresh medium was added, and the plates were incubated for 48 h. Cells were then exposed to IR or UV, and the lysates were prepared as described above for immunoblotting.
Inhibition of expression by siRNA.
The sequence of the coding strand of the ATM small interfering RNA (siRNA) was GCACCAGUCCAGUAUUGGC. The sequence of the Artemis-1 and Artemis-2 siRNAs were CUGAAGAGAGCUAGAACAG and UUAGGAGUCCAGGUUCAUG, respectively, and the sequence of the DNA-PKcs siRNA was UGGGCCAGAAGAUCGCACC. The 53BP1 siRNA was previously described (59).
Cell cycle analysis and phospho-histone H3 staining.
Cells were exposed to IR or UV or left untreated, incubated for various times (indicated in the figures), harvested by trypsinization, and fixed with 70% ethanol. Cells were then permeabilized with 0.15% Triton 100 in PBS containing 4% bovine serum albumin for 30 min and then incubated with phospho-histone H3 antibody for 60 min. Cells were washed with PBS and incubated with fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse immunoglobulin G (IgG) for 20 min. After the cells were washed with PBS, they were stained with 50 μg of propidium iodide per ml, treated with 10 μg of DNase-free RNase per ml, and analyzed by flow cytometry.
RESULTS
Artemis is not essential for NHEJ.
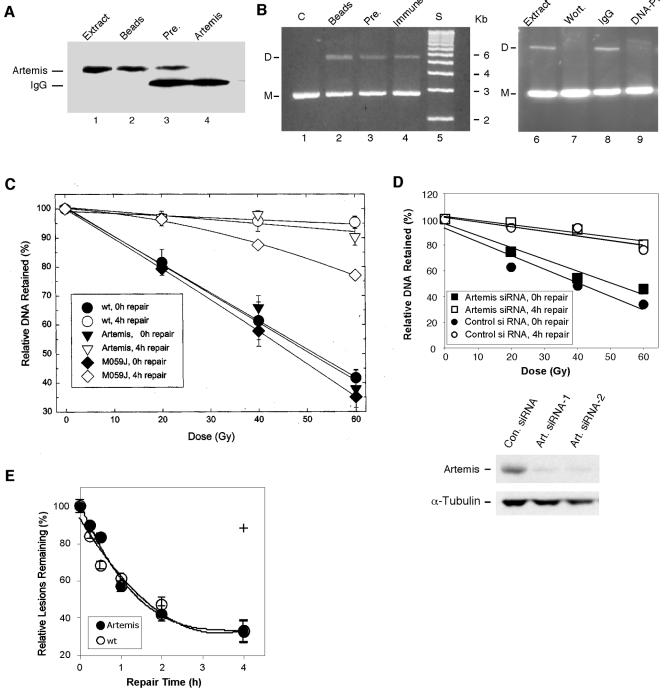
The findings that human cells defective for Artemis are deficient in V(D)J recombination and hypersensitive to IR have led to the conclusion that, as in mice with SCID, these cells have a defect in NHEJ (32, 34). Baumann and West (5) and Hanakahi et al. (21) have developed an in vitro DNA end-joining assay using human cell extracts that is dependent upon DNA-PK, Xrcc4, and DNA ligase IV, indicating that it is a measure of NHEJ. To determine whether Artemis is involved in NHEJ, we performed this end-joining assay with HeLa cell extracts, prepared as described previously (5), with or without immunodepletion of Artemis. As shown in Fig. 1A and B, removal of Artemis from HeLa extracts had little effect on the observed amount of DNA end joining of a substrate that contained complementary single-stranded tails. For controls to ensure that the assay was dependent upon DNA-PK as previously demonstrated (5), we performed the end-joining assay in the presence of wortmannin (22, 26) or a neutralizing monoclonal antibody to DNA-PKcs. In both cases, the assay was strongly inhibited, indicating that it was a measure of the DNA-PK-dependent NHEJ pathway (Fig. 1B, lanes 6 to 9).
FIG. 1.
Artemis is not essential for NHEJ. (A) Immunodepletion of Artemis from HeLa whole-cell extracts as determined by immunoblotting. HeLa whole-cell extract (lane 1); HeLa whole-cell extract precipitated with protein A-Sepharose beads only (lane 2); whole-cell extract precipitated with preimmune serum (Pre.) (lane 3); and Artemis-depleted whole-cell extract (depleted with Artemis antiserum) (lane 4). (B) Immunodepletion of Artemis from HeLa cell extracts does not affect end joining of linearized plasmid DNA. Ethidium bromide-stained agarose gels show the results of end-joining assays. Plasmid DNA was linearized by digestion with BamHI. The positions of monomer (M) and dimer (D) plasmids are indicated to the left of the gels. DNA only was used as a control (C) in lane 1. Assays shown in lanes 2 to 4 were performed with whole-cell extracts treated as described above for panel A. DNA size markers (S) are shown in lane 5. IgG, nonspecific immune serum. Controls (lanes 6 to 9) show that wortmannin (Wort.) and antibodies to DNA-PKcs inhibit the rejoining reaction. (C) DSB rejoining is normal in Artemis-deficient cells. P11 cells expressing Artemis, the simian virus 40-transformed human cell line XP2OS (considered the wild type [wt] in these experiments), and MO59J (DNA-PKcs-deficient) cells were compared as a function of IR dose for DNA rejoining by pulsed-field gel electrophoresis. (D) DSB rejoining is normal in HEK293 cells depleted of Artemis by siRNA. Depletion of Artemis by siRNA is shown by immunoblotting. Con., control; Art., Artemis. (E) DSB rejoining examined as a function of time of incubation. The + indicates the value at the 4-h time point for the MO59J cell line.
These results suggest that Artemis is not required for NHEJ; however, this is not a complete test of its possible involvement in NHEJ, since incompatible DNA termini may require nucleolytic processing that could involve Artemis in a subset of end-joining reactions. We therefore also examined the repair of DSBs in vivo after exposure to IR by the method of pulsed-field gel electrophoresis (10, 11, 29). In Fig. 1C, initial DNA lesions and DNA retained after 4 h of repair with increasing doses of radiation are shown. This technique has previously been used to detect defects in DSB repair after IR exposure in Ku80 and DNA-PKcs mutants. As shown, the Artemis-deficient cell line P11 (41) did not exhibit a significant difference in DSB repair from a similarly simian virus 40-transformed human fibroblast cell line, whereas a DNA-PKcs-deficient mutant was clearly defective. To confirm these results, Artemis was depleted from HEK293 cells by transfection with siRNA, and pulsed-field gel electrophoresis experiments were repeated. Compared with cells transfected with a nonspecific (control) siRNA, Artemis-depleted cells did not exhibit a defect in rejoining of DSBs after exposure to IR (Fig. 1D). We also examined the kinetics of DSB repair in Artemis and wild-type cells and found no significant differences (Fig. 1E). Taken together with the in vitro results, these findings indicate that Artemis is not an essential component of NHEJ, although we cannot rule out the possibility that Artemis may play a minor role in a subset of end-joining reactions. Nicolas et al. (43) had previously reached a similar conclusion; they found no defect in the kinetics of DSB repair in Artemis-deficient cells after exposure to IR.
Artemis is phosphorylated in vivo in response to both IR and UV.
Artemis contains 10 SQ/TQ motifs, nine of which are conserved in humans and mice (Fig. 2A). These motifs, particularly when clustered, have been found to be the preferred sites of phosphorylation by the ATM, ATR, and DNA-PK PIKKs (28). Artemis has been shown to be a substrate of DNA-PKcs in vitro (34). To investigate the phosphorylation of Artemis in vivo, we prepared and affinity purified antibodies against the nonconserved carboxy-terminal region of the protein. Immunoblotting indicated that these antibodies recognized a protein migrating at approximately 105 kDa in control cell lines, but not in the Artemis-deficient cell line P11, which confirmed their specificity (Fig. 2B). We then examined whether the gel migration of Artemis was altered in MCF-7 cells by treating cells with IR or UV radiation. Interestingly, both IR and UV induced a significant shift of Artemis to a slower-migrating form (Fig. 2C, lanes 1 and 4).
FIG. 2.
Artemis is phosphorylated in vivo after exposure of MCF-7 cells to IR or UV irradiation. (A) Schematic of Artemis indicating locations of S/TQ motifs. Asterisks above or below the schematic indicate motifs conserved or not conserved in humans and mice, respectively. aa, amino acids. (B) Immunoblotting shows the absence of Artemis in Artemis-deficient P11 cells. The asterisk indicates a nonspecific loading control. (C) Treatment with either IR or UV results in phosphorylation of Artemis (Artemis-P). Artemis was immunoprecipitated from extracts of cells exposed to IR (10 Gy) or UV (20 J/m2) and subsequently incubated for 2 h (lanes 1 and 4). The same immunoprecipitate was incubated with alkaline phosphatase (lanes 2 and 5) or treated with alkaline phosphatase in the presence of the phosphatase inhibitor Na3VO4 (lanes 3 and 6). The immunoprecipitate from untreated cells (UT) is shown in lane 7. (D) Phosphorylation of Artemis as a function of IR dose examined 2 h after treatment. (E) Kinetic analysis of the phosphorylation of Artemis after the cells were exposed to IR (10 Gy) for times ranging from 5 min (5m) to 17 h. (F) Phosphorylation of Artemis as a function of UV dose examined 2 h after treatment. (G) Kinetic analysis of the phosphorylation of Artemis after exposure of cells to UV (20 J/m2).
To determine whether this shift was due to phosphorylation, cellular lysates obtained from cells treated with IR or UV were incubated with calf intestinal phosphatase (CIP), with or without the addition of a phosphatase inhibitor. Incubation with CIP resulted in a shift of Artemis to the faster-migrating form (Fig. 2C, lanes 2 and 5), which did not occur in the presence of a phosphatase inhibitor (Fig. 2C, lanes 3 and 6). We have also observed modification of Artemis after exposure of MCF-7 cells to the topoisomerase I inhibitor camptothecin (24) (results not shown). Essentially similar results of DNA damage-induced modification of Artemis were obtained with HeLa, HT-1080, and HEK293 cells (not shown), and taken together, these results demonstrate that Artemis is phosphorylated in vivo in response to various types of DNA damage.
We also examined the modification of Artemis as a function of dose and observed in general that its phosphorylation increased with the level of DNA damage (Fig. 2D and F). Kinetic studies indicated that the observed phosphorylation of Artemis could be detected as early as 5 min after exposure to IR and is sustained for at least 17 h in MCF-7 cells (Fig. 2E). Phosphorylation caused by UV irradiation occurred later and was not detected until the 2-h time point (Fig. 2G), although in other experiments, phosphorylation of Artemis after UV can clearly be seen after 1 h. These findings indicate that Artemis is phosphorylated in vivo in response to DNA damage induced by both IR and UV irradiation. In particular, the induction of phosphorylation by UV was unanticipated and indicates that Artemis plays a wider role in the cellular response to genotoxic agents than suggested by previous studies.
Phosphorylation of Artemis is mediated by DNA-PK in vivo.
Artemis has been shown to be phosphorylated by DNA-PKcs in vitro (34). To determine whether the IR-induced phosphorylation of Artemis observed in vivo (Fig. 2) was DNA-PK dependent, we examined the DNA-PKcs-deficient cell line MO59J and the control line MO59K (30). Unlike what we had observed in MCF-7 or HeLa cells, a constitutive level of phosphorylation of Artemis was observed in the absence of treatment in these cell lines, suggesting a DNA-PK-independent phosphorylation of Artemis (for example, two Artemis bands can be seen in Fig. 3A, lanes 1 and 4). Treating cells with caffeine reduced the level of the top band, indicating that this modification is due to phosphorylation (lane 2). Upon treatment with IR, a decrease in the mobility of Artemis was observed in MO59K cell lysates, but not in MO59J cell lysates (Fig. 3A, compare lanes 3 and 5). However, the bottom band in the MO59J lysates disappeared, suggesting that some phosphorylation of Artemis did occur in these cells.
FIG.3.
Artemis is phosphorylated in vivo by DNA-PKcs after IR. (A) Phosphorylation of Artemis in vivo after exposure of cells to IR is deficient in MO59J cells. MO59J and MO59K cells were treated with IR (10 Gy), incubated for 1 h prior to extract preparation, and compared with untreated cells (UT). Caff., caffeine. (B) Depletion of DNA-PKcs by siRNA shows that DNA-PKcs and a caffeine-sensitive kinase(s) participate in the phosphorylation of Artemis after IR. LC, loading control. The presence (+) or absence (−) of control and DNA-PKcs siRNA, caffeine, and IR irradiation are indicated above the blots. (C) Phosphorylation of Artemis after IR treatment is inhibited by wortmannin (Wor.) in MCF-7 cells, but not by the Chk1 inhibitor UCN-01 (UCN.). (D) Artemis antibodies coimmunoprecipitate DNA-PKcs with and without exposure to IR in HCT116 cells. (E) DNA-PKcs interacts with phosphorylated and unphosphorylated forms of Artemis as determined by coimmunoprecipitation (IP) with DNA-PKcs antibodies. HEK293 cells stably transfected with pDEST27-Artemis (expressing GST-Artemis) were exposed to IR and incubated for 1 h. Protein A-Sepharose beads were used as a control.
The differences in the phosphorylation of Artemis in the MO59K and MO59J cell lines after exposure to IR suggest that Artemis is an in vivo substrate of DNA-PKcs, but the MO59J cell line contains a mutation in ATM and is reported to have about 33% of normal ATM activity (58). Therefore, to further examine the role of DNA-PKcs in the modification of Artemis, we used siRNA to deplete this kinase in HeLa cells (Fig. 3B). As expected in the absence of treatment, little change in the migration of Artemis was observed, although there may have been some increase in the level of constitutive phosphorylation of Artemis upon depletion of DNA-PKcs (compare lanes 1 and 3), perhaps due to an increase in the amount of unrepaired DNA damage. Upon treatment with IR, strong phosphorylation of Artemis is observed even with depletion of DNA-PKcs; however, this modification is reduced in the presence of caffeine (compare lanes 5 and 6), an inhibitor of ATM and ATR but not DNA-PK (54, 55). These observations clearly indicate that a caffeine-sensitive kinase(s) is at least partially responsible for the observed phosphorylation. However, caffeine did not affect the level of Artemis modification in cells treated with a control siRNA (compare lanes 7 and 8), indicating that DNA-PKcs is also involved in the phosphorylation of Artemis after IR. It should also be noted that there appears to a consistent decrease in the level of Artemis upon depletion of DNA-PKcs, suggesting that this kinase may be required for Artemis stability. Additionally, phosphorylation of Artemis after IR exposure was also inhibited in MCF-7 cells in the presence of wortmannin, which is an inhibitor of ATM, ATR, and DNA-PK (22, 26), but not in the presence of the Chk1 inhibitor UCN-01 (20) (Fig. 3C).
We also examined the interaction between Artemis and DNA-PKcs by coimmunoprecipitation from HCT116 cells (Fig. 3D). In untreated cells, Artemis antibodies coprecipitated DNA-PKcs; however, upon exposure to IR, the interaction between Artemis and DNA-PKcs increased and was maximal between 8 and 16 h after exposure to IR. For a further control for the interaction of Artemis with DNA-PKcs, we stably expressed a GST-Artemis fusion protein in HEK293 cells. Upon exposure to IR, approximately half of the overexpressed fusion protein was phosphorylated (Fig. 3E). Immunoprecipitation with DNA-PKcs antibodies showed that both phosphorylated and unphosphorylated forms of Artemis coimmunoprecipitated with DNA-PKcs. Taken together and combined with the results of previous in vitro studies (34), these results indicate that Artemis is a likely phosphorylation substrate of DNA-PK and that this modification is induced by IR in vivo. However, these findings also suggest that other kinases are involved in the phosphorylation of Artemis after DNA damage.
Artemis interacts with checkpoint proteins and is phosphorylated by ATM and ATR in vitro.
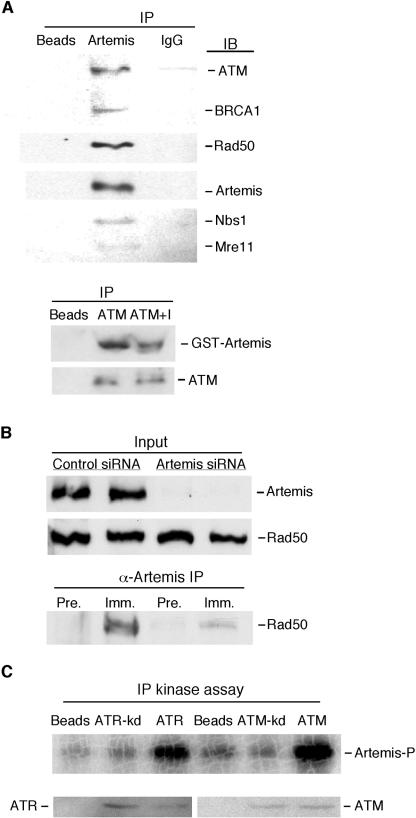
We have shown above that Artemis interacts with DNA-PK and is a likely phosphorylation substrate of this kinase in vivo. However, as indicated above, these findings also suggest that phosphorylation of Artemis may also be mediated by other kinases. ATM is a central kinase involved in the cellular response to IR-induced DNA damage (1, 16, 56). To determine whether Artemis interacts with ATM, we performed an immunoblot analysis for ATM after an immunoprecipitation from HeLa extracts with Artemis affinity-purified antibodies. As shown in Fig. 4A (top gel), this experiment indicated that ATM coimmunoprecipitated with Artemis. ATM has previously been shown to be a component of a large complex termed BASC (61). In addition to ATM and BRCA1, this complex includes Mre11, Rad50, and Nbs1 (MRN), as well as other proteins involved in DNA repair, checkpoint signaling, and chromatin remodeling (6, 19).
FIG. 4.
Artemis interacts with checkpoint proteins and is phosphorylated by ATM and ATR. (A, top blot) Artemis antibodies coimmunoprecipitate ATM, BRCA1, Rad50, Nbs1, and Mre11 from HeLa extracts. Protein A-Sepharose beads were used as a control. IP, immunoprecipitation; IgG, nonspecific immune serum. Immunoblotting (IB) was performed using antibodies to the indicated proteins. (A, lower blot) Reciprocal coimmunoprecipitation of Artemis by ATM antibodies. ATM+I, incubation of the extracts with DNase I prior to coimmunoprecipitation. (B) Depletion of Artemis in HeLa cells by transfection of siRNA eliminates the coimmunoprecipitation of Rad50 by Artemis antibodies. α-Artemis IP, immunoprecipitation with anti-Artemis antibodies; Pre., preimmune serum; Imm., Artemis antiserum. (C) Immunoprecipitation (IP) kinase assay shows that Artemis was phosphorylated by both ATM and ATR but by kinase-dead (kd) variants of the enzymes. The top shows an autoradiogram of Artemis phosphorylation by ATM or ATR. The bottom shows immunoblots of immunoprecipitated kinases used in the assays.
To determine whether Artemis interacts with other components of BASC, we immunoblotted for BRCA1 and MRN and found that all were specifically coimmunoprecipitated from HeLa cell extracts by Artemis antibodies (Fig. 4A, top blot). We were also able to demonstrate a reciprocal coimmunoprecipitation of exogenously expressed GST-Artemis by antibodies to ATM (Fig. 4A). This interaction was not mediated by DNA, since prior incubation of the extracts with DNase I did not prevent the coimmunoprecipitation. For a control to demonstrate the specificity of the Artemis antiserum, we used siRNA to deplete HeLa cells of Artemis and then repeated the coimmunoprecipitation experiment. As shown in Fig. 4B), Artemis siRNA effectively depleted HeLa cells of Artemis but did not affect Rad50 levels. In the absence of Artemis, coimmunoprecipitation of Rad50 with Artemis antibodies was greatly reduced, demonstrating that the results shown in Fig. 4A are not due to nonspecific interactions of the Artemis antibody.
Since Artemis interacts with ATM, we examined whether Artemis is a substrate of ATM or ATR by performing an immunoprecipitation kinase assay (3, 39). As shown in Fig. 4C, Artemis was phosphorylated by both ATM and ATR in vitro but not by kinase-dead variants of these enzymes. These findings, taken together with the interactions with known checkpoint proteins, suggest that Artemis has a potential role in cell signaling pathways induced by DNA damage.
Phosphorylation of Artemis in vivo in response to IR is ATM dependent.
We have shown above that Artemis interacts with ATM in vivo and is phosphorylated by this kinase in vitro. To determine whether ATM plays a role in the IR-induced phosphorylation of Artemis in vivo, we first examined ATM-deficient cells and observed altered migration of Artemis as seen in other cell lines (Fig. 5A). This result is consistent with a major role for DNA-PK in Artemis phosphorylation after exposure to IR irradiation, as discussed above, but does not rule out a role for ATM. To determine whether ATM is involved, we used transfection of siRNA oligonucleotides to eliminate expression of ATM in the DNA-PKcs-deficient cell line MO59J (Fig. 5B, top gels) and subsequently examined the phosphorylation of Artemis after IR or UV radiation (Fig. 5B, bottom gel). In cells transfected with a control siRNA, phosphorylation of Artemis due to IR or UV treatment was observed. However, in the ATM siRNA-transfected cells, phosphorylation of Artemis was reduced after IR, but not after UV (Fig. 5B, bottom gel, compare lanes 2 and 5 and lanes 3 and 6), irradiation. Taken together with our in vitro results, these findings demonstrate that ATM is involved in the modification of Artemis that occurs in response to IR and are consistent with previous studies indicating that ATM does not play a significant role in the response to UV irradiation. Thus, Artemis phosphorylation after IR exposure is dependent upon both ATM and DNA-PK.
FIG. 5.
Phosphorylation of Artemis is partially dependent upon ATM after IR irradiation and upon ATR after UV irradiation. (A) Artemis is phosphorylated in ATM-deficient cells (GM9607) after IR irradiation. UT, untreated cells. (B, top blot) Depletion of ATM expression in MO59J cells by siRNA. LC, loading control. (B, bottom blot) Phosphorylation of Artemis in MO59J cells depleted of ATM is affected after IR irradiation, but not after UV irradiation. (C, top blot) Elimination of ATR expression by infection of ATRflox/− cells with adenovirus expressing Cre recombinase. (Bottom blot) Phosphorylation of Artemis in cells depleted of ATR is affected after UV irradiation, but not after IR irradiation.
Phosphorylation of Artemis in vivo in response to UV is ATR dependent.
As shown above (Fig. 2), Artemis is phosphorylated in response to both IR and UV. The major PIKK that mediates the cellular response to UV is ATR (1, 16, 56, 63). To assess the role of ATR in UV-induced phosphorylation of Artemis, we used a recently developed HCT116 conditional null cell line (ATRflox/−) in which one allele of ATR is disrupted and the other contains loxP sites flanking exon 2 (12). Infection of the ATRflox/− cell line with an adenovirus encoding Cre recombinase reduced expression of ATR to low levels (Fig. 5C, top gels). We then examined the phosphorylation of Artemis after exposure to UV or IR in cells with or without expression of Cre recombinase. Eliminating the expression of ATR did not affect the phosphorylation of Artemis induced by IR (Fig. 5C, bottom gel, compare lanes 3 and 6) but did reduce the phosphorylation of Artemis after UV treatment (Fig. 5E, bottom gel, compare lanes 2 and 5). Also note that the phosphorylation of Artemis after UV treatment is inhibited in the presence of caffeine (Fig. 3B, lane 5). Thus, the ATR-mediated modification of Artemis induced by UV radiation is consistent with the observed in vitro phosphorylation of Artemis shown above.
Artemis is involved in a G2/M DNA damage checkpoint.
Artemis-deficient cells are hypersensitive to IR (9, 42, 44), but as shown above (Fig. 1), this sensitivity does not appear to be due to a significant defect in NHEJ. Another possible explanation for this phenotype is loss of checkpoint function, particularly since both IR and UV irradiation induce phosphorylation of Artemis (Fig. 2). IR induces an ATM-dependent G2/M checkpoint arrest in mammalian cells. To assess this checkpoint, we used siRNA to deplete HEK293 cells of Artemis. Cells depleted of Artemis by transfection with siRNA and control transfected cells were subsequently irradiated with IR (6 Gy), and DNA content and level of phospho-histone H3 (P-H3) were measured as a function of time (Fig. 6A). Transfection with Artemis siRNA resulted in a slight accumulation in the G1 population in untreated cells compared to transfection with the control siRNA (see the 0-h time points in Fig. 6A). This accumulation, which was consistently observed in all cell lines examined, could be due to a fraction of arrested cells caused by increased spontaneous chromosomal damage in the absence of Artemis. Rooney et al. (51) observed significantly higher levels of chromosomal aberrations in Artemis-deficient mouse embryo fibroblasts compared with wild-type cells. One hour after IR irradiation, both Artemis-depleted and control cells exhibited a dramatic reduction in P-H3 staining; however, at later time points, Artemis-depleted cells exhibited a higher level of P-H3 staining than did the control cells, suggesting a failure of the former cells to fully arrest or to maintain an arrest in G2/M (Fig. 6A and B). This trend was also reflected in the cell cycle data, where 16 h after IR irradiation, 51.6% of the Artemis-depleted cells were in G2/M compared with 72.5% of the control cells.
FIG.6.
Artemis is required for a normal DNA damage-induced G2/M cell cycle checkpoint. (A) Analysis of the G2/M checkpoint after IR. Fluorescence-activated cell sorting profile of Artemis-depleted and control HEK293 cells stained with propidium iodide and anti-phospho-histone H3 (P-H3) at the indicated times after IR irradiation (6 Gy). The percentages of cells in the G1/S and G2/M phases are shown. (B) Graphical analysis of P-H3 staining indicates a defect in the G2/M checkpoint in Artemis-depleted cells. The fraction of P-H3-positive cells is expressed as a percentage of that measured at the 0-h time point. (C) P-H3 accumulation is not due to differential exit from mitosis. Values of P-H3 levels for the untreated sample (UT) were set at 1.0, and results for the other samples are shown as a percentage of this value. Nocodazole was added 30 min after IR treatment, and data were collected after 16 h. (D) Comparison of the cell cycle fractions of Artemis- and DNA-PKcs-depleted cells 16 h after IR (graphs at the top). The levels of cyclin B in the same cells are shown (middle blot). Immunoblotting showing depletion of DNA-PKcs by siRNA in HEK293 cells is shown (bottom blot). LC, loading control.
To ensure that the increased levels of P-H3 staining observed in Artemis-depleted cells was not due to a failure to exit mitosis with normal kinetics, we performed the experiment in the presence of nocodazole 16 h after IR irradiation. As shown in Fig. 6C, the increase in P-H3 staining in Artemis-depleted cells also occurred in the presence of nocodazole, indicating that it is due to premature transition from G2 to M and not to a differential exit from mitosis. Finally, these results have been confirmed by depletion of Artemis in HEK293 cells with a second siRNA and by similar experiments conducted with HCT116 and HeLa cells (not shown) and indicate that Artemis is required for a normal G2/M cell cycle delay that occurs after IR irradiation.
To further distinguish the roles of Artemis and DNA-PKcs, we depleted expression of each of these proteins separately by siRNA transfection and compared the fraction of G2/M cells 16 h after IR irradiation. As expected, Artemis-depleted cells exhibited an incomplete arrest, whereas cells transfected with a control or DNA-PKcs siRNA showed normal and slightly enhanced enforcement of the checkpoint, respectively (Fig. 6D). This conclusion was also confirmed by examination of cyclin B levels 16 h after IR irradiation. With control siRNA, cyclin B levels were higher in IR-treated cells than in untreated cells. This effect was even more pronounced in cells depleted of DNA-PKcs, whereas Artemis-depleted cells showed little increase in cyclin B levels after IR irradiation (Fig. 6D), indicating a failure to delay in G2/M for the latter cells. Taken together, these findings indicate that Artemis is involved in a G2/M checkpoint and are consistent with a requirement for ATM and ATR in the DNA damage-induced phosphorylation of Artemis.
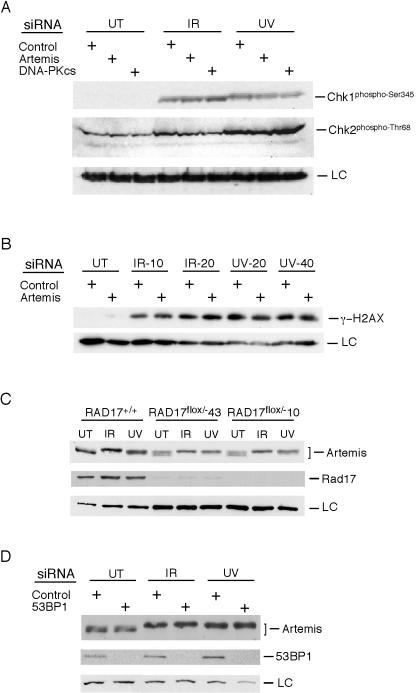
A host of factors have been identified in the mediation of DNA damage-induced G2/M checkpoint responses in mammalian cells (13, 52, 63). Two kinases that act downstream of ATM and ATR in checkpoint responses are Chk1 and Chk2. Since Artemis is rapidly phosphorylated after IR irradiation (Fig. 2), we examined whether the phosphorylation of either Chk1 or Chk2 was affected in Artemis-depleted cells. As shown in Fig. 7A, no differences in the phosphorylation of these proteins were observed after exposure to IR or UV compared with control cells or cells depleted of DNA-PKcs, indicating that Artemis does not act upstream of these checkpoint kinases. H2AX is a variant histone that is rapidly phosphorylated after DNA damage (γ-H2AX) and is required for the formation of foci by other checkpoints proteins, such as Nbs1, 53BP1, and BRCA1 (48, 49, 50, 59). Examination of the phosphorylation of H2AX showed that it occurred normally in both Artemis-depleted and control cells (Fig. 7B).
FIG. 7.
Artemis is not involved in established G2/M checkpoint pathways. Where not indicated, the IR dose was 20 Gy and the UV dose was 50 J/m2. All experiments were performed 2 h after irradiation. (A) Phosphorylation of Chk1 (phospho-Ser345) and Chk2 (phospho-Thr68) are unaffected in Artemis-depleted cells after IR or UV irradiation. UT, untreated cells; LC, loading control. (B) Phosphorylation of H2AX is unaffected in Artemis-depleted cells. Numbers shown after the indicated treatment specify the dosage in grays (IR) or joules per square meter (UV). (C) Phosphorylation of Artemis is unaffected in Rad17-depleted cells. Depletion of Rad17 in RAD17flox/− cells is shown after infection with adenovirus expressing Cre recombinase. (D) Phosphorylation of Artemis is unaffected in 53BP1-depleted cells.
Both Rad17, a component of the checkpoint clamp-loading complex, and 53BP1, a BRCT repeat-containing protein, are rapidly phosphorylated after DNA damage and have been shown to be required for G2/M checkpoint responses (4, 14, 18, 59). To determine whether the phosphorylation of Artemis depends upon either of these proteins, Rad17 was depleted from HCT116 RAD17flox/− cells by introduction of Cre recombinase (60), and 53BP1 was depleted from HEK293 cells by siRNA transfection (59). As shown in Fig. 7C and D, the phosphorylation of Artemis after IR or UV exposure was not affected by the depletion of these checkpoint proteins. Thus, while phosphorylation of Artemis after DNA damage is dependent upon ATM and ATR, it does not appear that it is a component of the well-established G2/M checkpoint pathway that functions downstream of these PIKKs (13, 52, 63).
DISCUSSION
Previous studies of Artemis have shown that it plays a crucial role in V(D)J recombination because in its functional absence, T and B lymphocytes fail to mature, leading to SCID syndrome (9, 41, 42, 44). The role of Artemis in V(D)J recombination has been defined as an endonuclease that cleaves the hairpins at coding joints, producing unmistakable DSBs that are subsequently repaired by the NHEJ pathway (34). Additionally, Artemis-expressing cells are radiosensitive, suggesting the attractive concept that Artemis also plays a role in rejoining IR-induced DSBs via NHEJ (34). This hypothesis, although seemingly widely accepted in the literature, has, to our knowledge, never been directly demonstrated; in fact, Artemis-deficient cells have been reported to have normal repair kinetics of DSBs in vivo (43). Consistent with these earlier findings, we have shown here that Artemis is not an essential element of DSB rejoining in vitro and that Artemis-deficient cells are not significantly defective in vivo in restoration of DSBs after IR treatment, although we cannot completely rule out the possibility that Artemis may be involved in a minor subset of reactions in this pathway. Nevertheless, these findings indicate that the significant radiosensitivity of Artemis-deficient cells is unlikely to be due to defects in NHEJ. In addition to our findings, several other lines of evidence also support the conclusion that Artemis does not play a major role in NHEJ. First, Artemis-deficient cells are not defective in signal joint rejoining, although DNA-PKcs-deficient cells are defective in the fidelity of this process (41, 51). Second, inactivating mutations in other components of NHEJ, including Ku70, Ku80, DNA-PKcs, Xrcc4, and DNA ligase 4, have not been observed in human immunodeficiencies, presumably due to the essential nature of NHEJ in human cells, whereas Artemis is clearly involved in human SCID (41). Third, a recent report on the transposition of the Sleeping Beauty element showed that NHEJ, but not Artemis, is required for this recombination pathway (25).
Phosphorylation of Artemis in response to genotoxic stress is mediated by ATM, ATR, and DNA-PK.
Artemis has been shown to be a phosphorylation substrate of DNA-PKcs in vitro (34). We have extended these studies to show that phosphorylation of Artemis in vivo after IR treatment is mediated by DNA-PK. However, these experiments also indicated that an additional kinase or kinases were involved in the phosphorylation of Artemis after IR irradiation and that Artemis was also phosphorylated after UV irradiation. We also showed that Artemis interacts with some components of BASC, a multifactorial complex that plays a central role in the mediation of both cell cycle checkpoints and DNA repair pathways (61) and that Artemis is an in vitro substrate of ATM and ATR. Consistent with the defined roles of these two PIKKs in the cellular response to DNA damage, in vivo phosphorylation of Artemis mediated by ATM and ATR was shown to be induced by IR and UV treatment, respectively. These findings suggest a wider role for Artemis in the cellular response to DNA damage than has been previously appreciated. It is unlikely that this role involves a direct participation in DNA repair pathways, because DNA damage induced by IR or UV radiation results in lesions of distinctly different types, and their repair is mediated by disparate pathways that have little overlap in mechanism or components. Thus, a direct role for Artemis in multiple pathways of DNA repair appears less plausible than a role in the mediation of cell cycle checkpoints in response to various types of DNA damage. Consistent with this interpretation, we showed that Artemis-deficient cells are defective in a G2/M cell cycle checkpoint induced by IR.
The observation of ATM/ATR-mediated phosphorylation of Artemis induced by genotoxic stress is consistent with a defective checkpoint response that we have described here; however, the phosphorylation of Artemis after IR irradiation by DNA-PK remains unexplained. Our findings and those reported previously by others (2) indicate that DNA-PKcs is not required for a G2/M DNA damage checkpoint induced by IR. In fact, in the absence of DNA-PKcs, the checkpoint appears to be enhanced, presumably due to a decreased level of repair of DSBs. DNA-PKcs interacts with both the unphosphorylated and phosphorylated forms of Artemis, and the association between the two proteins appears to increase until it reaches a maximum 8 to 16 h after IR irradiation. This result suggests the possibility that DNA-PK is required to maintain the phosphorylation of Artemis after DNA damage.
Artemis is involved in cell cycle checkpoint pathways.
The conclusion that Artemis functions in cell cycle checkpoint pathways is based on the following observations. (i) Both IR and UV induce phosphorylation of Artemis. (ii) Artemis interacts with checkpoint proteins and is phosphorylated by the checkpoint kinases ATM and ATR in vitro. (iii) Phosphorylation of Artemis in vivo after genotoxic stress requires ATM and ATR as well as DNA-PK. (iv) Artemis is required for a normal G2/M arrest after IR irradiation. The mechanism of the Artemis-mediated checkpoint arrest does not appear to involve the well-established pathways of G2/M delay that function downstream of ATM and ATR (13, 52, 63). Although Artemis is rapidly phosphorylated after IR irradiation, it is not required for the damage-induced phosphorylation of Chk1, Chk2, or H2AX. In addition, the phosphorylation of Artemis is not dependent upon Rad17 or 53BP1 and was not inhibited by the Chk1 inhibitor UCN-01. Rad17 has recently been shown to be important for ATR-dependent but not ATM-dependent checkpoint signaling through Chk1 (60) and may facilitate the phosphorylation of other targets downstream of ATR (64). Thus, our findings suggest that Artemis does not act downstream of Chk1. There is, however, precedence for Rad17-independent checkpoint signaling mediated by ATR homologues in both budding and fission yeasts (37, 38). In addition, the checkpoint phenotype of ATR-deficient cells is more severe than that of Rad17-deficient cells, indicating that a Rad17-independent pathway(s) exists in mammalian cells (60). 53BP1 is a mediator of DNA damage-induced checkpoints that acts downstream of ATM and ATR and is required for phosphorylation of BRCA1 and Chk2 (14, 18, 59). Our results indicate that Artemis does not function downstream of 53BP1 and thereby suggest that it does not act downstream of Chk2.
Artemis has been shown to possess both endonuclease and exonuclease functions that act on DNA hairpins and broken ends (34). It is possible that the checkpoint function of Artemis could be mediated through its nucleolytic activity. Recently, it has been shown that single-stranded DNA is the activating signal for the ATR- and possibly ATM-mediated checkpoint responses (45, 65). Also, there is precedence for a factor which contains a nuclease function and which is also involved in checkpoint mechanisms. The MRN complex possesses exonuclease and hairpin-opening activities (46, 47) and has been shown to function in a DNA damage-induced checkpoint in S phase (17, 33, 62). Alternatively, since Artemis does not appear to act through the canonical pathways of G2/M checkpoint arrest that are highly conserved between yeast and mammals, it is possible that Artemis may be in a pathway involved in the maintenance of checkpoint arrest, rather than in its initiation. Studies of yeast have shown that initiation and maintenance of checkpoint signaling are functionally separable (13). This hypothesis is supported by our finding that Artemis-deficient cells are not defective in the initial imposition of cell cycle arrest that occurs within 1 h after IR irradiation but rather appear unable to maintain the arrest at later time points. Such a pathway would likely monitor DNA repair to ensure its completion before releasing the checkpoint. Maintenance pathways are not well understood, but conceivably the interaction between Artemis and DNA-PKcs, which is maximal at these later time points, could be part of a mechanism to monitor completion of DSB repair and provide a signal to relieve the arrest and allow cells to proceed through the cell cycle. Obviously, an important element of defining the role of Artemis in cell cycle responses to genotoxic agents will be to correlate the phosphorylation of Artemis mediated by ATM and ATR to the observed failure to properly arrest after DNA damage.
Acknowledgments
We thank the following investigators for gifts of reagents. The ATR and ATM expression constructs were provided by Karlene Cimprich and Michael Kastan, respectively. P11 and P14 cell lines were provided by J. P. de Villartay. The ATRflox/− cells were provided by Stephen Elledge, and the RAD17flox/− cells were provided by Lei Li. We thank Lei Li for comments on the manuscript.
This work was supported in part by grants from the NIH (CA52461, CA96574, CA90270, and ES07784).
REFERENCES
- 1.Abraham, R. T. 2001. Cell cycle checkpoints signaling through the ATM and ATR kinases. Genes Dev. 15:2177-2196. [DOI] [PubMed] [Google Scholar]
- 2.Allalunis-Turner, J., G. M. Barron, and R. S. Day III. 1997. Intact G2-phase checkpoint in cells of a human cell line lacking DNA-dependent protein kinase activity. Radiat. Res. 147:284-287. [PubMed] [Google Scholar]
- 3.Banin, S., L. Moyal, S. Shieh, Y. Taya, C. W. Anderson, L. Chessa, N. I. Smorodinsky, C. Prives, Y. Reiss, Y. Shiloh, and Y. Ziv. 1998. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281:1674-1677. [DOI] [PubMed] [Google Scholar]
- 4.Bao, S., R. S. Tibbetts, K. M. Brumbaugh, Y. Fang, D. A. Richardson, A. Ali, S. M. Chen, R. T. Abraham, and X. F. Wang. 2001. ATR/ATM-mediated phosphorylation of human Rad17 is required for genotoxic stress responses. Nature 411:969-974. [DOI] [PubMed] [Google Scholar]
- 5.Baumann, P., and S. C. West. 1998. DNA end-joining catalyzed by human cell-free extracts. Proc. Natl. Acad. Sci. USA 95:14066-14070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bochar, D. A., L. Wang, H. Beniya, A. Kinev, Y. Sue, W. S. Lane, W. Wang, F. Kashanchi, and R. Shiekhattar. 2000. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell 102:257-265. [DOI] [PubMed] [Google Scholar]
- 7.Callebaut, I., D. Moshous, J. P. Mornon, and J. P. de Villartay. 2002. Metallo-β-lactamase fold within nucleic acids processing enzymes: the β-CASP family. Nucleic Acids Res. 30:3592-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canman, C. E., D. S. Lim, K. A. Cimprich, Y. Taya, K. Tamai, K. Sakaguchi, E. Appella, M. B. Kastan, and J. D. Siliciano. 1998. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281:1677-1679. [DOI] [PubMed] [Google Scholar]
- 9.Cavazzana-Calva, M., F. Le Deist, G. De Saint Basile, D. Papadopoulo, J. P. De Villartay, and A. Fischer. 1993. Increased radiosensitivity of granulocyte macrophage colony-forming units and skin fibroblasts in human autosomal recessive severe combined immunodeficiency. J. Clin. Investig. 91:1214-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan, D. W., B. P. Chen, S. Prithivirajsingh, A. Kurimasa, M. D. Story, J. Qin, and D. J. Chen. 2002. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 16:2333-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, F., S. R. Peterson, M. D. Story, and D. J. Chen. 1996. Disruption of DNA-PK in Ku80 mutant xrs-6 and the implications in DNA double-strand break repair. Mutat. Res. 362:9-19. [DOI] [PubMed] [Google Scholar]
- 12.Cortez, D., S. Guntuku, J. Qin, and S. J. Elledge. 2001. ATR and ATRIP: partners in checkpoint signaling. Science 294:1713-1716. [DOI] [PubMed] [Google Scholar]
- 13.Cuddihy, A. R., and M. J. O'Connell. 2003. Cell-cycle responses to DNA damage in G2. Int. Rev. Cytol. 222:99-140. [DOI] [PubMed] [Google Scholar]
- 14.DiTullio, R. A., Jr., T. A. Mochan, M. Venere, J. Bartkova, M. Sehested, J. Bartek, and T. D. Halazonetis. 2002. 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nat. Cell Biol. 4:998-1002. [DOI] [PubMed] [Google Scholar]
- 15.Dronkert, M. L., J. de Wit, M. Boeve, M. L. Vasconcelos, H. van Steeg, T. L. Tan, J. H. Hoeijmakers, and R. Kanaar. 2000. Disruption of mouse SNM1 causes increased sensitivity to the DNA interstrand cross-linking agent mitomycin C. Mol. Cell. Biol. 20:4553-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durocher, D., and S. P. Jackson. 2001. DNA-PK, ATM and ATR as sensors of DNA damage: variation on a theme. Curr. Opin. Cell Biol. 13:225-231. [DOI] [PubMed] [Google Scholar]
- 17.Falck, J., J. H. Petrini, B. R. Williams, J. Lukas, and J. Bartek. 2002. The DNA damage-dependent intra-S phase checkpoint is regulated by parallel pathways. Nat. Genet. 30:290-294. [DOI] [PubMed] [Google Scholar]
- 18.Fernandez-Capetillo, O., H. T. Chen, A. Celeste, I. Ward, P. J. Romanienko, J. C. Morales, K. Naka, Z. Xia, R. D. Camerini-Otero, N. Motoyama, P. B. Carpenter, W. M. Bonner, J. Chen, and A. Nussenzweig. 2002. DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nat. Cell Biol. 4:993-997. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Higuera, I., T. Taniguchi, S. Ganesan, M. S. Meyn, C. Timmers, J. Hejna, M. Grompe, and A. D. D'Andrea. 2001. Interaction of the fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell 7:249-262. [DOI] [PubMed] [Google Scholar]
- 20.Graves, P. R., L. Yu, J. K. Schwarz, J. Gales, E. A. Sausville, P. M. O'Connor, and H. Piwnica-Worms. 2000. The Chk1 protein kinase and the Cdc25C regulatory pathways are targets of the anticancer agent UCN-01. J. Biol. Chem. 275:5600-5605. [DOI] [PubMed] [Google Scholar]
- 21.Hanakahi, L. A., M. Bartlet-Jones, C. Chappell, D. Pappin, and S. C. West. 2000. Binding of inositol phosphate to DNA-PK and stimulation of double-strand break repair. Cell 102:721-729. [DOI] [PubMed] [Google Scholar]
- 22.Hartley, K. D., D. Gell, G. C. M. Smith, H. Zhang, N. Devecha, M. A. Connelly, A. Admon, S. P. Lees-Miller, C. W. Anderson, and S. P. Jackson. 1995. DNA-dependent protein kinase catalytic subunit a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell 82:849-856. [DOI] [PubMed] [Google Scholar]
- 23.Henriques, J. A., and E. Moustacchi. 1980. Isolation and characterization of pso mutants sensitive to photo-addition of psoralen derivatives in Saccharomyces cerevisiae. Genetics 95:273-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsiang, Y. H., R. Hertzberg, S. Hecht, and L. F. Lieu. 1985. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J. Biol. Chem. 260:14873-14878. [PubMed] [Google Scholar]
- 25.Izsvak, Z., E. E. Stuwe, D. Fiedler, A. Katzer, P. A. Jeggo, and Z. Ivics. 2004. Healing the wounds inflicted by Sleeping Beauty transposition by double-strand break repair in mammalian somatic cells. Mol. Cell 13:279-290. [DOI] [PubMed] [Google Scholar]
- 26.Izzard, R. A., S. P. Jackson, and G. C. M. Smith. 1999. Competitive and noncompetitive inhibition of the DNA-dependent protein kinase. Cancer Res. 59:2581-2586. [PubMed] [Google Scholar]
- 27.Jenny, A., L. Minvielle-Sebastia, P. J. Preker, and W. Keller. 1996. Sequence similarity between the 73 kilodalton protein of mammalian CPSF and a subunit of yeast polyadenylation factor I. Science 274:1514-1517. [DOI] [PubMed] [Google Scholar]
- 28.Kim, S. T., D. S. Lim, C. E. Canman, and M. B. Kastan. 1999. Substrate specificities and identification of putative substrates of ATM kinase family members. J. Biol. Chem. 274:37538-37543. [DOI] [PubMed] [Google Scholar]
- 29.Kurimasa, A., S. Kumano, N. V. Boubnov, M. D. Story, C.-S. Tung, S. R. Peterson, and D. J. Chen. 1999. Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol. Cell. Biol. 19:3877-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lees-Miller, S. P., R. Godbout, D. W. Chan, M. Weinfeld, R. S. Day, G. M. Barron, and J. Allalinis-Turner. 1995. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science 267:1183-1185. [DOI] [PubMed] [Google Scholar]
- 31.Li, X., and R. E. Moses. 2003. The β-lactamase motif in Snm1 is required for repair of DNA double strand breaks caused by interstrand cross-links in S. cerevisiae. DNA Repair 2:121-129. [DOI] [PubMed] [Google Scholar]
- 32.Lieber, M. R., Y. Ma, U. Pannicke, and K. Schwarz. 2003. Mechanism and regulation of human nonhomologous DNA end-joining. Nat. Rev. Mol. Cell Biol. 4:712-720. [DOI] [PubMed] [Google Scholar]
- 33.Lim, D. S., S. T. Kim, B. Xu, R. S. Maser, J. Lin, J. H. Petrini, and M. B. Kastan. 2000. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature 404:613-617. [DOI] [PubMed] [Google Scholar]
- 34.Ma, Y., U. Pannicke, K. Schwarz, and M. R. Lieber. 2002. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 108:781-794. [DOI] [PubMed] [Google Scholar]
- 35.Magana-Schwencke, N., J. A. Henriques, R. Chanet, and E. Moustacchi. 1982. The fate of 8-methoxypsoralen photoinduced crosslinks in nuclear and mitochondrial yeast DNA: comparison of wild-type and repair-deficient strains. Proc. Natl. Acad. Sci. USA 79:1722-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manley, J. L., A. Fire, A. Cano, P. A. Sharp, and M. L. Gefter. 1980. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc. Natl. Acad. Sci. USA 77:3855-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maringele, L., and D. Lydall. 2002. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yKU70Δ mutants. Genes Dev. 16:1919-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinho, R. G., H. D. Lindsay, G. Flaggs, A. J. DeMaggio, M. F. Hoekstra, A. M. Carr, and N. J. Bentley. 1998. Analysis of Rad3 and Chk1 protein kinases defines different checkpoint responses. EMBO J. 17:7239-7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuoka, S., G. Rotman, A. Ogawa, Y. Shilogh, K. Tamai, and S. J. Elledge. 2000. Ataxia telangiectasis-mutated phosphorylates Chk2 in vivo and in vitro. Proc. Natl. Acad. Sci. USA 97:10389-10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meniel, V., N. Magana-Schwencke, and D. Averbeck. 1995. Preferential repair in Saccharomyces cerevisiae rad mutants after induction of interstrand cross-links by 8-methoxypsoralen plus UVA. Mutagenesis 10:543-548. [DOI] [PubMed] [Google Scholar]
- 41.Moshous, D., I. Callebaut, R. de Chasseval, B. Corneo, M. Cavazzana-Calvo, F. Le Deist, I. Tezcan, O. Sanal, Y. Bertrand, N. Philippe, A. Fischer, and J. P. de Villartay. 2001. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 105:177-186. [DOI] [PubMed] [Google Scholar]
- 42.Moshous, D., L. Li, R. de Chasseval, N. Philippe, N. Jabado, M. J. Cowan, A. Fischer, and J. P. de Villartay. 2000. A new gene involved in DNA double-strand break repair and V(D)J recombination is located on human chromosome 10p. Hum. Mol. Genet. 9:583-588. [DOI] [PubMed] [Google Scholar]
- 43.Nicolas, N., N. J. Finnie, M. Cavazzana-Calvo, D. Papadopoulo, F. Le Deist, A. Fischer, S. P. Jackson, and J. P. de Villartay. 1996. Lack of detectable defect in DNA double-strand break repair and DNA-dependent protein kinase activity in radiosensitive human severe combined immunodeficiency fibroblasts. Eur. J. Immunol. 26:1118-1122. [DOI] [PubMed] [Google Scholar]
- 44.Nicolas, N., D. Moshous, M. Cavazzana-Calvo, D. Papadopoulo, R. de Chasseval, F. Le Deist, A. Fischer, and J. P. de Villartay. 1998. A human severe combined immunodeficiency (SCID) condition with increased sensitivity to ionizing radiations and impaired V(D)J rearrangements defines a new DNA recombination/repair deficiency. J. Exp. Med. 188:627-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nur, E., A. Kamal, T. K. Li, A. Zhang, H. Qi, E. S. Hars, and L. F. Liu. 2003. Single-stranded DNA induces ataxia telangiectasia mutant (ATM)/p53-dependent DNA damage and apoptotic signals. J. Biol. Chem. 278:12475-12481. [DOI] [PubMed] [Google Scholar]
- 46.Paull, T. T., and M. Gellert. 1998. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell 1:969-979. [DOI] [PubMed] [Google Scholar]
- 47.Paull, T. T., and M. Gellert. 1999. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 13:1276-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paull, T. T., E. P. Rogakou, E. P. Yamazaki, C. U. Kirchgessner, M. Gellert, and W. M. Bonner. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10:886-895. [DOI] [PubMed] [Google Scholar]
- 49.Rogakou, E. P., C. Boon, C. Redon, and W. M. Bonner. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146:905-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rogakou, E. P., D. R. Pilch, A. H. Orr, V. S. Ivanova, and W. M. Bonner. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858-5868. [DOI] [PubMed] [Google Scholar]
- 51.Rooney, S., J. Sekiguchi, C. Zhu, H. L. Cheng, J. Manis, S. Whitlow, J. DeVido, D. Foy, J. Chaudhuri, D. Lombard, and F. W. Alt. 2002. Leaky scid phenotype associated with defective V(D)J coding end processing in Artemis-deficient mice. Mol. Cell 10:1379-1390. [DOI] [PubMed] [Google Scholar]
- 52.Rouse, J., and S. P. Jackson. 2002. Interfaces between the detection, signaling, and repair of DNA damage. Science 297:547-551. [DOI] [PubMed] [Google Scholar]
- 53.Ruhland, A., M. Kircher, F. Wilborn, and M. Brendel. 1981. A yeast mutant specifically sensitive to bifunctional alkylation. Mutat. Res. 91:457-462. [DOI] [PubMed] [Google Scholar]
- 54.Sarkaria, J. N., E. C. Busby, R. S. Tibbetts, P. Roos, Y. Yaya, M. Karnitz, and R. T. Abraham. 1999. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 59:4375-4382. [PubMed] [Google Scholar]
- 55.Schild-Poulter, C., L. Pope, W. Giffin, J. C. Kochan, J. K. Ngsee, J. K. Traykova-Andonova, and R. J. Hache. 2001. The binding of Ku antigen to homeodomain proteins promotes their phosphorylation by DNA-dependent protein kinase. J. Biol. Chem. 276:16848-16856. [DOI] [PubMed] [Google Scholar]
- 56.Shiloh, Y. 2003. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer 3:155-168. [DOI] [PubMed] [Google Scholar]
- 57.Tavtigian, S. V., J. Simard, D. H. Teng, V. Abtin, M. Baumgard, A. Beck, N. J. Camp, A. R. Carillo, Y. Chen, P. Dayananth, et al. 2001. A candidate prostate cancer susceptibility gene at chromosome 17p. Nat. Genet. 27:172-180. [DOI] [PubMed] [Google Scholar]
- 58.Tsuchida, R., Y. Yamada, M. Takagi, A. Shimada, C. Ishioka, Y. Katsuki, T. Igarashi, L. Chessa, D. Delia, H. Teraoka, and S. Mizutani. 2002. Detection of ATM mutation in human glioma cell line MO59J by a rapid frameshift/stop codon assay in yeast. Radiat. Res. 158:195-201. [DOI] [PubMed] [Google Scholar]
- 59.Wang, B., S. Matsuoka, P. B. Carpenter, and S. J. Elledge. 2002. 53BP1, a mediator of the DNA damage checkpoint. Science 298:1435-1438. [DOI] [PubMed] [Google Scholar]
- 60.Wang, X., L. Zou, H. Zheng, Q. Wei, S. J. Elledge, and L. Li. 2003. Genomic instability and endoreduplication triggered by RAD17 deletion. Genes Dev. 17:965-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, Y., D. Cortez, P. Yazdi, N. Neff, S. J. Elledge, and J. Qin. 2000. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 14:927-939. [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao, S., Y. C. Weng, S. S. Yuan, Y. T. Lin, H. C. Hsu, S. C. Lin, E. Gerbino, M. H. Song, M. Z. Zdzienicka, R. A. Gatti, J. W. Shay, Y. Ziv, Y. Shiloh, and E. Y. Lee. 2000. Functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products. Nature 405:473-477. [DOI] [PubMed] [Google Scholar]
- 63.Zhou, B.-B.S., and S. J. Elledge. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433-439. [DOI] [PubMed] [Google Scholar]
- 64.Zou, L., D. Cortez, and S. J. Elledge. 2002. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 16:198-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zou, L., and S. J. Elledge. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542-1548. [DOI] [PubMed] [Google Scholar]