Abstract
We investigated the role of RNA polymerase II (pol II) carboxy-terminal domain (CTD) phosphorylation in pre-mRNA processing coupled and uncoupled from transcription in Xenopus oocytes. Inhibition of CTD phosphorylation by the kinase inhibitors 5,6-dichloro-1β-d-ribofuranosyl-benzimidazole and H8 blocked transcription-coupled splicing and poly(A) site cleavage. These experiments suggest that pol II CTD phosphorylation is required for efficient pre-mRNA splicing and 3′-end formation in vivo. In contrast, processing of injected pre-mRNA was unaffected by either kinase inhibitors or α-amanitin-induced depletion of pol II. pol II therefore does not appear to participate directly in posttranscriptional processing, at least in frog oocytes. Together these experiments show that the influence of the phosphorylated CTD on pre-mRNA splicing and 3′-end processing is mediated by transcriptional coupling.
In eukaryotic cells, pol II synthesizes pre-mRNA that is processed in the nucleus to become mature mRNA and is then exported to the cytoplasm. Capping, splicing, and 3′-end processing are interdependent and often occur cotranscriptionally on the nascent transcript at the DNA template (2, 5, 32). Processing can also occur posttranscriptionally after release from the site of transcription (3, 37). The carboxy-terminal domain (CTD) of the largest subunit of pol II (Rpb1) provides an important link between transcription and processing by acting as a landing pad that binds directly to processing factors and localizes them to the site of transcription (4, 7, 13, 14, 23, 31). In mammalian cells, pol II lacking the CTD produces transcripts that are not efficiently capped, spliced, or cleaved at poly(A) sites (24, 25). Furthermore, in vitro the CTD can enhance capping, splicing, and poly(A) site cleavage uncoupled from transcription (15-17, 33, 40, 42, 43). These results suggest that the CTD of pol II that is not transcriptionally engaged can act as an allosteric activator of pre-mRNA processing reactions. Although the CTD is important for pre-mRNA processing, pol II transcription is by no means essential. RNA precursors can be processed in vitro and, in some cases, in vivo in the absence of transcription. Introns appearing early in the pre-mRNA of Chironomus BR1 and BR3 genes are predominantly spliced at the site of transcription, whereas introns close to the 3′ end are spliced after the transcript has been released (3, 37). It is also possible that cleavage and polyadenylation occurs posttranscriptionally, because cleavage frequently does not precede termination (29). It is not known if pol II that is not transcriptionally engaged can facilitate pre-mRNA processing in vivo after release from the site of transcription.
During transcription, the CTD undergoes extensive phosphorylation and dephosphorylation on Ser2 and Ser5 residues of the heptad repeats (YSPTSPS). CTD hyperphosphorylation by CDK7 and CDK9 is associated with the transition from initiation to elongation (19, 21). Protein kinase inhibitors, including 5,6-dichloro-1β-d-ribofuranosyl-benzimidazole (DRB) and H8, reduce CTD phosphorylation by inhibiting CDK7 and CDK9 and prevent efficient transcriptional elongation (9, 30, 39, 44). In vitro, the hyperphosphorylated CTD can stimulate splicing more than the hypophosphorylated form (16). The phosphorylated CTD is specifically bound by the capping enzyme guanylyltransferase and the putative splicing factor CA150 (6, 35). Although DRB reduced pol II phosphorylation in mammalian cells, it did not strongly inhibit capping (26), consistent with the fact that low-level phosphorylation is sufficient for binding of capping enzymes (24). CTD phosphorylation is required for 3′-end processing of U2 snRNA (18, 26). Little is known about the importance of CTD phosphorylation for splicing and 3′-end processing of mRNAs in vivo; however, inhibition of kinases that phosphorylate Ser2 residues causes a modest inhibition of poly(A) site cleavage in Drosophila melanogaster and budding yeast (1, 28).
Cotranscriptional processing has not been directly compared with posttranscriptional processing of the same transcript in vivo. Xenopus oocytes have the unique advantage that processing can be assessed coupled and uncoupled from transcription by injecting either a DNA template (38) or an in vitro-synthesized capped pre-mRNA (12). In vitro, coupling with pol II transcription accelerates the splicing reaction (11). We show that splicing and poly(A) site cleavage of human β-globin pre-mRNA requires CTD phosphorylation when coupled to transcription but not when processing occurs uncoupled from transcription.
MATERIALS AND METHODS
Oocyte injections.
Oocyte nuclei were injected with 1 ng of plasmid or 2.3 ng of capped pre-mRNA in 23 nl of water, except where noted. α-Amanitin was injected at 25 μg/ml. The pol III-transcribed pSPVA plasmid used as a control for nuclear injection efficiency and RNA recovery was injected at 1 pg/oocyte. RNA was isolated using RNA-Bee (Tel-Test Inc.) or as previously described (39) followed by DNase I treatment. Oocytes were incubated in modified Barth's solution containing increasing amounts of DRB or H8 for 3 h prior to injection.
RNA analysis.
Capped pre-mRNA was synthesized and DNase treated using the Ambion MEGAscript kit with diguanosine triphosphate cap analogue. The template was produced by PCR incorporating a T7 promoter using forward oligonucleotide 5′ CCT AAT ACG ACT CAC TAT AGG ACA TTT GCT TCT GAC ACA AC 3′, complementary to human β-globin starting at position −50 relative to the ATG, and reverse oligonucleotide 5′ CCG GAT CCA CTG ACC TCC CAC ATT CC 3′, complementary to positions +65 to +84 relative to the AATAAA motif at the poly(A) site. RNase protection and fractionation of capped and uncapped RNA by binding to glutathione S-transferase (GST)-eIF4E was as described previously (24). Globin and VA RNase protection probes have already been described (10, 24). In all cases undigested probe (data not shown) was clearly resolved from RNase protection products. Results were quantified by a Molecular Dynamics PhosphorImager. The percent processed was calculated as the signal for spliced or cleaved product divided by the sum of processed plus unprocessed signals after compensating for 32P-U content.
Western blotting.
Oocytes were homogenized in a solution of 0.1 M NaCl, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride (PMSF), 20 mM Tris-HCl (pH 7.6), 1 mM dithiothreitol (DTT), 10 mM β-glycerophosphate, 2 mM EGTA, and 2 mM benzamidine. Proteins were blotted to Immobilon filters, and signals were visualized by chemiluminescence (Perkin Elmer Western Lighting).
Antibodies.
8WG16, anti-CstF p77, and anti-CTD Ser5-PO4 antibodies have already been described (10, 34, 36). Anti-B10 was purchased from Chemicon.
Plasmids.
pAT7Rpb1wtAmS, pAT7Rpb1wtAmR, and pAT7Rpb1ΔCTDAmR were described previously (10, 27).
pCMV β-globin was constructed by cloning a human β-globin fragment from the transcriptional start (+1) to the PstI site (+2164) into pCI-neo (Promega) between SacI and BamHI sites. The pCMV β-globin synthetic poly(A) site (SPA) was made from pCMV β-globin by inserting the oligonucleotide 5′ TCG AGA ATA AAA GAT CTT TAT TTT CAT TAG ATC TGT GTG TTG GTT TTT TGT GTG G 3′ into an XhoI site engineered 10 bases 3′ of the termination codon. pBS β-globin SPA, which was used to make the RNase protection probe, contains a PCR fragment extending 116 bases 5′ of the SPA site and 64 bases 3′ of the β-globin poly(A) cleavage site in Bluescript KS+.
pBS CMV β-globin used to make a 5′-end mapping probe contained a PCR fragment derived from pCMV β-globin corresponding to the region from +138 relative to the β-globin start site to −102 relative to the CMV start site.
RESULTS
pol II ΔCTD inhibits transcription-coupled processing but not processing uncoupled from transcription.
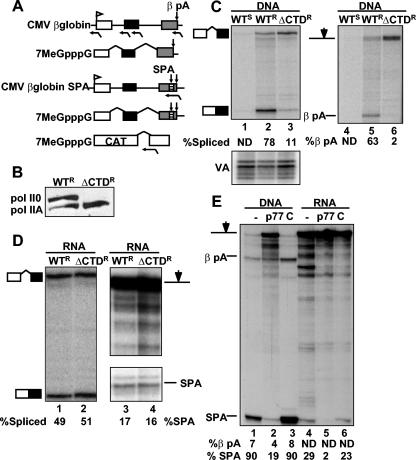
We compared the effects of expressing the pol II large subunit, Rpb1, lacking the CTD on pre-mRNA processing of human β-globin transcripts coupled to and uncoupled from transcription in Xenopus oocytes. Epitope-tagged α-amanitin-resistant full-length Rpb1 (Rpb1WTR) and a deletion that removed all 52 heptads in the CTD (Rpb1ΔCTDR) was expressed in oocytes by microinjection of CMV-driven expression vectors. As a control, an expression vector for α-amanitin-sensitive full-length Rpb1 (Rpb1WTS) was also injected. Expression of these pol II large subunits was confirmed by Western blotting (Fig. 1B). Fifteen hours after injection of the Rpb1 expression vectors, CMV β-globin plasmid (Fig. 1A) was injected along with α-amanitin to inhibit endogenous pol II. VA plasmid was also injected as a control for injection efficiency. After a second 15-h incubation, RNA was isolated and assayed for splicing and poly(A) site cleavage by RNase protection assay (RPA). β-Globin pre-mRNA made in the oocyte by wild-type pol II was accurately spliced at introns 1 and 2 (Fig. 1C, lane 2, and data not shown) and was cleaved at the poly(A) site (Fig. 1C, lane 5). Consistent with previous results with mammalian cells (25), RNA precursors transcribed by pol II ΔCTD were not efficiently spliced or cleaved (Fig. 1C, lanes 3 and 6) relative to the control with intact CTD (Fig. 1C, lanes 2 and 5). When fractionated by binding to GST-eIF4E, transcripts made by pol II ΔCTD partitioned mostly to the uncapped fraction (data not shown). We conclude that, as in mammalian cells, the pol II CTD is essential for cotranscriptional capping, splicing, and poly(A) site cleavage in Xenopus oocytes.
FIG. 1.
pol II ΔCTD does not support cotranscriptional pre-mRNA processing and does not inhibit processing uncoupled from transcription in Xenopus oocytes. (A) Maps of the CMV β-globin gene and capped (7MeGpppG) pre-mRNAs with and without synthetic poly(A) site (SPA, striped box) in exon 3. Poly(A) cleavage sites βpA and SPA are marked by downward arrows. Capped CAT pre-mRNA containing the SV40 t-intron is shown at the bottom. Antisense RNase protection probes are marked below the maps. (B) Western blot of Rpb1WTR (WTR) and Rpb1ΔCTDR (ΔCTDR) expressed in oocytes detected with antibody against the B10 epitope tag. Hyper- and hypophosphorylated wild-type pol II0 and pol IIA are indicated. (C) RPA of CMV β-globin transcripts in α-amanitin-injected oocytes expressing α-amanitin-sensitive (WTS) or -resistant Rpb1 with full-length (WTR) or deleted CTD (ΔCTDR) with intron 1 and poly(A) site probes. VA RNA transcribed by pol III is a control for injection efficiency. The CMV β-globin (0.2 ng in 4.6 nl) and VA plasmids were coinjected with α-amanitin 15 h after injection of the Rpb1 expression plasmids (0.2 ng in 4.6 nl). Percent spliced or cleaved relative to total RNA (see Materials and Methods) is noted below lane numbers. Values too small to be accurately determined are labeled ND. (D) RPA of β-globin SPA pre-mRNA coinjected with α-amanitin into oocytes expressing Rpb1WTR or Rpb1ΔCTDR with intron 1 and SPA probes. Note that expression of Rpb1ΔCTDR has no effect on processing uncoupled from transcription. (E) RPA of CMV β-globin SPA (lanes 1 to 3) or injected β-globin SPA transcripts with SPA probe (lanes 4 to 6). Oocytes were coinjected with either no antibody (−), anti-CstF p77 (p77) (5.3 ng/oocyte), or irrelevant control (C) rabbit immunoglobulin G (5.3 ng/oocyte).
To investigate whether pol II ΔCTD affected processing uncoupled from transcription, we injected expression vectors for Rpb1WTR and Rpb1ΔCTDR followed by a second injection of α-amanitin plus capped T7 transcripts of β-globin pre-mRNA containing a strong synthetic poly(A) site (SPA) (22) inserted into exon 3 upstream of the natural β-globin poly(A) site, βpA (Fig. 1A, CMV β-globin SPA). Side-by-side 5′-end mapping showed that the T7 transcripts initiate less than 5 bases upstream of the CMV globin transcripts synthesized in oocytes (data not shown). Expression of pol II ΔCTD had no effect on processing of injected pre-mRNA (Fig. 1D, lanes 1 to 4). Overall we observed a lower processing efficiency for injected pre-mRNA, probably because of the lack of association with the transcription machinery. Note that processing efficiencies for injected pre-mRNA are minimum values, because capping of pre-mRNA synthesized in vitro is not complete. We conclude from these results that the CTD of pol II is necessary for cotranscriptional pre-mRNA processing in Xenopus oocytes and that expression of Rpb1ΔCTD has no dominant-negative effect on processing uncoupled from transcription.
To demonstrate that processing in oocyte nuclei is accessible to inhibitors, we injected antibody against the C-terminal 17 residues of human CstF p77. This antibody cross-reacts strongly with Xenopus CstF p77 (GenBank accession no. BJ077499) in Western blots (data not shown) as expected, because the proteins are identical at 16 of the 17 C-terminal residues. Cleavage at the SPA both in injected pre-mRNA and transcript from the CMV β-globin SPA plasmid was reduced 5- to 10-fold by anti-CstF p77 relative to a control antibody (Fig. 1E, lanes 2, 3, 5, and 6). The inhibition of poly(A) site cleavage in injected pre-mRNA by anti-CstF p77 shows that injected RNA is not sequestered in an inaccessible compartment within the nucleus.
CTD phosphorylation is required for processing coupled to transcription but not for processing uncoupled from transcription.
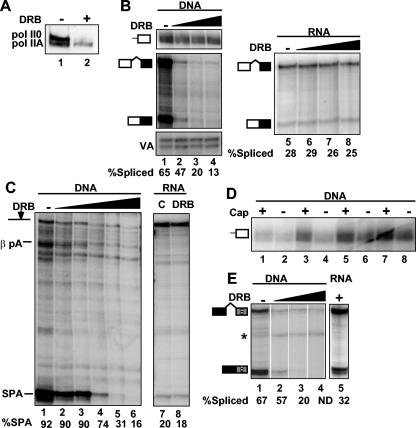
We investigated the role of CTD phosphorylation in pre-mRNA processing by using inhibitors of CTD kinases. Oocytes treated with 75 μM DRB for 3 h were analyzed by Western blot using antibody against the pol II CTD Ser 5 phosphoisoform. DRB inhibited CTD phosphorylation as demonstrated by reduced reactivity to this antibody and the shift toward the faster migrating pol IIA isoform (Fig. 2A). The effect of DRB on cotranscriptional processing was assayed in oocytes injected with CMV β-globin SPA plasmid (Fig. 1A). DRB inhibited elongation as shown by the ratio of signal from the 5′ end to that at exon 2 (Fig. 2B, lanes 1 to 4; compare the top and middle panels). Those β-globin SPA transcripts that extended to the 3′ end in the presence of DRB were not efficiently spliced or cleaved at the poly(A) sites relative to RNA made in the absence of DRB (Fig. 2B, compare lanes 2 to 4 with lane 1, and C, compare lanes 2 to 6 with lane 1). We assessed pre-mRNA processing uncoupled from transcription under conditions of increasing amounts of DRB by injecting β-globin pre-mRNA. DRB inhibition of CTD phosphorylation had no significant effect on splicing of intron 1, intron 2, or cleavage at the SPA when processing was uncoupled from transcription (Fig. 2B, lanes 5 to 8, and C, lanes 7 and 8, and data not shown). The failure of DRB to inhibit processing of injected pre-mRNA is not because the RNA is inaccessible, because poly(A) site cleavage was inhibited by injected anti-CstF p77 antibody (Fig. 1E, lanes 4 to 6). It is possible that inhibition of cotranscriptional splicing and 3′-end processing by DRB is a secondary effect of reduced capping. We therefore analyzed CMV globin SPA transcripts that had been fractionated by binding to the cap binding protein GST-eIF4E (Fig. 2D). DRB did not markedly reduce the ratio of capped:uncapped RNAs (Fig. 2D) (26). The cap-selected fraction from each sample was analyzed for splicing of intron 2. As expected, spliced transcripts were enriched in the capped fraction relative to the uncapped fraction (data not shown), and, most importantly, capped transcripts from oocytes treated with DRB were spliced far less efficiently than capped transcripts from untreated controls (Fig. 2E, lanes 1 to 4). Inhibition of splicing and 3′-end processing by DRB is therefore not an indirect consequence of poor capping. Rather, we propose that inhibition of CTD phosphorylation by DRB directly inhibited cotranscriptional splicing and poly(A) site cleavage but had no effect on processing uncoupled from transcription. Note that capped transcripts made by hypophosphorylated pol II in the presence of DRB were processed significantly less effectively than synthetic pre-mRNAs injected into DRB-treated oocytes (Fig. 2E, compare lanes 3 and 4 with lane 5).
FIG. 2.
Inhibition of CTD phosphorylation by DRB inhibits cotranscriptional processing but not processing uncoupled from transcription. (A) Western blot of Rpb1 from oocytes preincubated without (−) or with (+) DRB (75 μM) using anti-pol II CTD Ser 5-PO4. (B) RPA of β-globin RNAs from oocytes incubated in 0, 15, 30, and 60 μM DRB and injected with pCMV β-globin SPA or capped β-globin SPA pre-mRNA. 5′ end (lanes 1 to 4, top panel) and intron 1 probes (lanes 1 to 4, middle panel, and lanes 5 to 8) were used for RPA. VA is a control for injection efficiency. (C) RPA of CMV β-globin SPA transcripts from oocytes treated with DRB at 0, 3.75, 7, 15, 30, and 60 μM with SPA probe (lanes 1 to 5). β-globin RNAs from oocytes incubated in 0 and 75 μM DRB and injected capped β-globin SPA pre-mRNA are also shown (lanes 6 and 7). (D) RPA of capped (+) and uncapped (−) transcripts from panel B selected by GST-eIF4E binding with 5′ CMV β-globin probe (amount of DRB in lanes 1 and 2, 0 μM; lanes 3 and 4, 15 μM; lanes 5 and 6, 30 μM; lanes 7 and 8, 60 μM). (E) Cap-selected transcripts from the RPA shown in panel B were assessed for splicing of β-globin intron 2. Note that capped transcripts made in 30 and 60 μM DRB are predominantly unspliced (lanes 3 and 4), whereas those made with in the absence of DRB (lane 1) are predominantly spliced. Capped β-globin pre-mRNA was injected into oocytes treated with 75 μM DRB (lane 6). The asterisk indicates an irrelevant undigested probe.
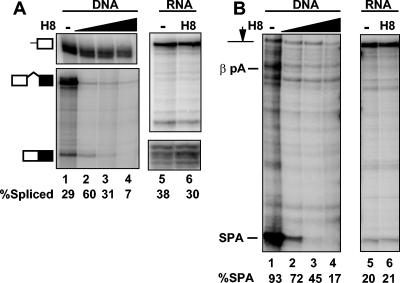
We also tested how the kinase inhibitor H8 affected pre-mRNA processing. Consistent with previous results (39), synthesis of long transcripts was reduced by H8 as determined by the ratio of 5′:3′ RPA signals (Fig. 3A, lanes 1 to 4; compare top and bottom panels). CMV β-globin SPA plasmid was injected into oocytes pretreated with increasing amounts of H8. H8 inhibited splicing of intron 1 (Fig. 3A, lanes 3 to 4) and cleavage at the β-globin and SPA poly(A) sites (Fig. 3B, lanes 2 to 4). H8 did not alter the transcription start site as determined by 5′-end mapping (Fig. 3A, lanes 1 to 4, top panel). We did observe variation in the absolute level of pre-mRNA processing between batches of oocytes, but inhibition of cotranscriptional processing was consistently observed in multiple batches of oocytes from different frogs. In contrast to transcription-coupled processing, splicing and 3′-end processing of injected β-globin SPA pre-mRNA was not significantly affected by H8 (Fig. 3A and B lanes 5 and 6). Together the results in Fig. 2 and 3 show that processing of pre-mRNAs transcribed from a DNA template clearly differs from processing of injected pre-mRNAs uncoupled from transcription in its sensitivity to kinase inhibitors DRB and H8. CTD phosphorylation is apparently vital for cotranscriptional splicing and 3′-end processing but not for processing uncoupled from transcription.
FIG. 3.
The protein kinase inhibitor H8 specifically inhibits pre-mRNA processing coupled to transcription. (A) RPA of β-globin intron 1 splicing from oocytes injected with CMV β-globin SPA and treated with 0, 25, 50, or 100 μg of H8/ml (lanes 1 to 4). RPA of capped β-globin pre-mRNA injected into oocyte treated with 0 or 200 μg of H8/ml (lanes 5 and 6). (B) RPA of SPA site cleavage with the same RNA samples as those used for the experiment depicted in panel A.
pol II depletion does not inhibit processing uncoupled from transcription.
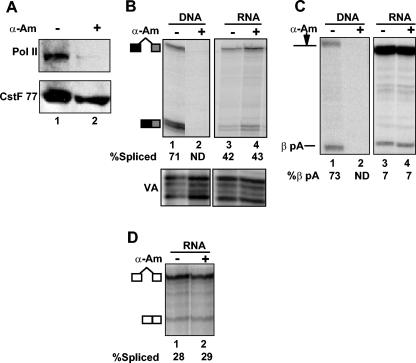
Does pol II that is not engaged in transcription serve to facilitate pre-mRNA processing uncoupled from transcription? To address this question we induced degradation of pol II by prolonged treatment of oocytes with α-amanitin (27). Western blotting showed that α-amanitin induced specific loss of pol II relative to CstF p77 after a 15-h incubation (Fig. 4A). We cannot eliminate the possibility that the level of CstF p77 is slightly reduced by prolonged α-amanitin treatment; however, it is reduced to an extent far less than that of pol II Rpb1, consistent with the results with somatic cells (27). Oocytes were injected with α-amanitin and incubated for 15 h and then were reinjected with β-globin pre-mRNA and incubated for an additional 12 h. α-Amanitin-induced degradation of pol II had no significant effect on splicing of intron 2 or cleavage at the β-globin poly(A) site of injected pre-mRNA (Fig. 4B and C, lanes 3 and 4). Transcription of the CMV β-globin plasmid was completely inhibited relative to that of the VA control in oocytes pretreated with α-amanitin (Fig. 4B and C, lanes 2). To assess processing uncoupled from transcription of a pre-mRNA other than β-globin, we injected in vitro-synthesized chloramphenicol acetyltransferase (CAT) pre-mRNA (Fig. 1A) containing the simian virus 40 (SV40) t intron into oocytes with and without α-amanitin treatment. Consistent with our observations for β-globin pre-mRNA, α-amanitin-induced pol II degradation did not effect splicing of the SV40 t intron (Fig. 4D). We cannot exclude the possibility that the small amount of residual pol II remaining after α-amanitin-induced depletion contributes to uncoupled pre-mRNA processing. However, the results depicted in Fig. 4 show that extensive degradation of pol II did not inhibit in vivo pre-mRNA processing uncoupled from transcription.
FIG. 4.
Degradation of RNA pol II by α-amanitin does not inhibit pre-mRNA processing uncoupled from transcription. (A) Western blot of total protein from oocytes with (+) and without (−) α-amanitin (α-Am) injection. The blot was probed with anti-pol II 8WG16 and anti-CstF p77 as a loading control. (B) RPA of β-globin intron 2 splicing from oocytes injected with either CMV β-globin DNA or β-globin pre-mRNA with (+) or without (−) preinjection of α-amanitin 15 h beforehand. VA is a control for injection efficiency. (C) RPA of β-globin poly(A) site cleavage with the same RNA samples as those used for the experiment shown in panel B. (D) RPA of SV40 t intron splicing in oocytes injected with capped CAT pre-mRNA (Fig. 1A) with (+) or without (−) preinjection of α-amanitin. ND, not determined.
DISCUSSION
We have shown that cotranscriptional pre-mRNA processing by splicing and poly(A) site cleavage requires CTD phosphorylation and that this requirement does not apply to processing uncoupled from transcription in the same cell. Processing of human β-globin pre-mRNA transcribed from an injected plasmid in Xenopus oocytes required the phosphorylated pol II CTD by the criterion that it was inhibited by treatment with two CTD kinase inhibitors, DRB and H8 (Fig. 2 and 3). In contrast, processing of β-globin pre-mRNA injected into the oocyte nucleus was unaffected by either expression of pol II ΔCTD (Fig. 1), treatment with CTD kinase inhibitors (Fig. 2 and 3), or α-amanitin-induced degradation of the pol II large subunit (Fig. 4). Processing of injected pre-mRNA was, however, inhibited by injection of anti-CstF p77 antibody (Fig. 1E). These differences imply that the mechanisms, which operate in vivo for cotranscriptional and posttranscriptional processing, differ from one another. The results strongly suggest that phosphorylated pol II CTD is essential for processing coupled to transcription but is dispensable for processing uncoupled from transcription. This distinction indicates that processing at sites of transcription operates differently from processing at other locations in the nucleus. Our results do not eliminate the possibility that CTD phosphorylation is important for cotranscriptional marking of transcripts for posttranscriptional processing. DRB and H8 can inhibit kinases other than CDK7 and CDK9, including casein kinase II (41), and they may alter the phosphorylation of processing factors such as SRp55 (20). Our experiments, therefore, do not exclude the possibility that phosphorylation of proteins other than pol II is also required for transcription-coupled processing. In vitro protein interaction studies have not revealed an effect of CTD phosphorylation on binding to the 3′-end processing factor CstF (10, 25). However, these studies using in vitro-phosphorylated recombinant CTD do not eliminate the possibility that interaction of 3′ processing factors with intact pol II in vivo is influenced by physiological patterns of CTD phosphorylation.
Unexpectedly, capped transcripts made by hypophosphorylated pol II in the presence of kinase inhibitors were processed less effectively than capped transcripts made in vitro and then injected into the nucleus (Fig. 2 and 3). Nascent pre-mRNAs made by pol II with an abnormally modified CTD may be packaged with hnRNPs into a form that is not available to the processing machinery. It is also possible that pre-mRNAs made by pol II with an incorrectly programmed pattern of CTD phosphorylation are sequestered in a nuclear compartment that is inaccessible to some processing factors. Mutations of human β-globin that prevent splicing or 3′-end formation cause the transcripts to be retained at the site of transcription in murine erythroleukemia cells (8). A functional link between processing and release from the site of transcription is also suggested by the fact that poly(A) site cleavage and splicing of the terminal exon of Chironomus BR1 transcripts closely coincides with release from the DNA (2). Nascent transcripts made by pol II with an improperly phosphorylated CTD may be retained at the site of transcription, and this local environment may exclude some essential splicing and 3′ processing factors unless they are specifically recruited there by interaction with the CTD.
Our results are consistent with experiments showing that CTD phosphorylation enhances an early step in splicing in HeLa extracts (16, 42). They are also consistent with the impairment of 3′ processing caused by inhibition of CDK9 in Drosophila and CTK1 in yeast (1, 28). The experiments in microinjected oocytes reveal a difference between cotranscriptional and posttranscriptional processing that is not evident in vitro. The phosphorylated recombinant CTD can stimulate processing in extracts that is uncoupled from transcription (15, 16, 42), whereas CTD phosphorylation in oocytes had no effect on processing uncoupled from transcription. Although we think it unlikely, it is possible that processing of transcripts from the full-length β-globin gene is inherently different from that of the partial gene transcripts previously assayed in vitro. It is also possible that this discrepancy reflects a unique property of Xenopus oocyte nuclei that contain high levels of transcription and processing factors relative to their genomic DNA content. The difference between in vitro and in vivo results could be due to compartmentalization in the nucleus that makes pol II unavailable for posttranscriptional processing. The concentration of pol II CTD in the in vitro reactions may have been high enough to mimic a situation which is only achieved in vivo in the context of cotranscriptional processing. We conclude that phosphorylated pol II CTD is essential for splicing and poly(A) site cleavage reactions in vivo when processing occurs at the site of transcription but not when it is uncoupled from transcription. CTD phosphorylation could specifically influence cotranscriptional processing directly by affecting localization of splicing and 3′-end formation factors close to the nascent transcript. It is also possible that CTD phosphorylation could influence cotranscriptional processing indirectly, for example, by affecting the rate of transcriptional elongation.
Acknowledgments
We thank T. Blumenthal and J. Jaehning for helpful discussions. We are grateful to the UCHSC Cancer Center sequencing facility, the UCHSC Center for Laboratory Animal Care, and to N. Fong for anti-CstF p77.
This work was supported by NIH grant GM58163 to D.B., a predoctoral training grant in molecular biology (NIH T32-GM0870) to G.B., and a Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation fellowship (DRG-1626) to D.A.R.Z.
REFERENCES
- 1.Ahn, S. H., M. Kim, and S. Buratowski. 2004. Phosphorylation of serine 2 within the rna polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell. 13:67-76. [DOI] [PubMed] [Google Scholar]
- 2.Bauren, G., S. Belikov, and L. Wieslander. 1998. Transcriptional termination in the Balbiani ring 1 gene is closely coupled to 3′-end formation and excision of the 3′-terminal intron. Genes Dev. 12:2759-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauren, G., and L. Wieslander. 1994. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell 76:183-192. [DOI] [PubMed] [Google Scholar]
- 4.Bentley, D. 2002. The mRNA assembly line: transcription and processing machines in the same factory. Curr. Opin. Cell Biol. 14:336-342. [DOI] [PubMed] [Google Scholar]
- 5.Beyer, A. L., and Y. N. Osheim. 1988. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 2:754-765. [DOI] [PubMed] [Google Scholar]
- 6.Carty, S. M., A. C. Goldstrohm, C. Sune, M. A. Garcia-Blanco, and A. L. Greenleaf. 2000. Protein-interaction modules that organize nuclear function: FF domains of CA150 bind the phosphoCTD of RNA polymerase II. Proc. Natl. Acad. Sci. USA 97:9015-9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corden, J., and C. Ingles. 1992. Carboxy-terminal domain of the largest subunit of eukaryotic RNA polymerase II, p. 81-108. In S. McKnight and K. Yamamoto (ed.), Transcriptional regulation, vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 8.Custodio, N., M. Carmo-Fonseca, F. Geraghty, H. S. Pereira, F. Grosveld, and M. Antoniou. 1999. Inefficient processing impairs release of RNA from the site of transcription. EMBO J. 18:2855-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egyhazi, E., A. Ossoinak, A. Pigon, C. Holmgren, J. M. Lee, and A. L. Greenleaf. 1996. Phosphorylation dependence of the initiation of productive transcription of balbiani ring-2 genes in living cells. Chromosoma 104:422-433. [DOI] [PubMed] [Google Scholar]
- 10.Fong, N., and D. Bentley. 2001. Capping, splicing and 3′ processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD. Genes Dev. 15:1783-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghosh, S., and M. A. Garcia-Blanco. 2000. Coupled in vitro synthesis and splicing of RNA polymerase II transcripts. RNA 6:1325-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green, M. R., T. Maniatis, and D. A. Melton. 1983. Human beta-globin pre-mRNA synthesized in vitro is accurately spliced in Xenopus oocyte nuclei. Cell 32:681-694. [DOI] [PubMed] [Google Scholar]
- 13.Greenleaf, A. L. 1993. Positive patches and negative noodles: linking RNA processing to transcription? Trends Biochem. Sci. 18:117-119. [DOI] [PubMed] [Google Scholar]
- 14.Hirose, Y., and J. L. Manley. 2000. RNA polymerase II and the integration of nuclear events. Genes Dev. 14:1415-1429. [PubMed] [Google Scholar]
- 15.Hirose, Y., and J. L. Manley. 1998. RNA polymerase II is an essential mRNA polyadenylation factor. Nature 395:93-96. [DOI] [PubMed] [Google Scholar]
- 16.Hirose, Y., R. Tacke, and J. L. Manley. 1999. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev. 13:1234-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho, C., and S. Shuman. 1999. Distinct Effector roles for Ser2 and Ser5 phosphorylation of the RNA polymerase II CTD in the recruitment and allosteric activation of mammalian capping enzyme. Mol. Cell. 3:405-411. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs, E. Y., I. Ogiwara, and A. M. Weiner. 2004. Role of the C-terminal domain of RNA polymerase II in U2 snRNA transcription and 3′ processing. Mol. Cell. Biol. 24:846-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobor, M., and J. Greenblatt. 2002. Regulation of transcription elongation by phosphorylation. Biochim. Biophys. Acta 1577:261. [DOI] [PubMed] [Google Scholar]
- 20.Lai, M. C., R. I. Lin, and W. Y. Tarn. 2003. Differential effects of hyperphosphorylation on splicing factor SRp55. Biochem. J. 371:937-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laybourn, P. J., and M. E. Dahmus. 1990. Phosphorylation of RNA polymerase IIA occurs subsequent to interaction with the promoter and before the initiation of transcription. J. Biol. Chem. 265:13165-13173. [PubMed] [Google Scholar]
- 22.Levitt, N., D. Briggs, A. Gil, and N. J. Proudfoot. 1989. Definition of an efficient synthetic poly(A) site. Genes Dev. 3:1019-1025. [DOI] [PubMed] [Google Scholar]
- 23.Maniatis, T., and R. Reed. 2002. An extensive network of coupling among gene expression machines. Nature 416:499-506. [DOI] [PubMed] [Google Scholar]
- 24.McCracken, S., N. Fong, E. Rosonina, K. Yankulov, G. Brothers, D. Siderovski, A. Hessel, S. Foster, Amgen EST Program, S. Shuman, and D. Bentley. 1997. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 11:3306-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCracken, S., N. Fong, K. Yankulov, S. Ballantyne, G. H. Pan, J. Greenblatt, S. D. Patterson, M. Wickens, and D. L. Bentley. 1997. The C-terminal domain of RNA polymerase II couples messenger RNA processing to transcription. Nature 385:357-361. [DOI] [PubMed] [Google Scholar]
- 26.Medlin, J. E., P. Uguen, A. Taylor, D. L. Bentley, and S. Murphy. 2003. The C-terminal domain of pol II and a DRB-sensitive kinase are required for 3′ processing of U2 snRNA. EMBO J. 22:925-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen, V. T., F. Giannoni, M. F. Dubois, S. J. Seo, M. Vigneron, C. Kedinger, and O. Bensaude. 1996. In vivo degradation of RNA polymerase II largest subunit triggered by alpha-amanitin. Nucleic Acids Res. 24:2924-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ni, Z., B. E. Schwartz, J. Werner, J. R. Suarez, and J. T. Lis. 2004. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol. Cell. 13:55-65. [DOI] [PubMed] [Google Scholar]
- 29.Osheim, Y. N., N. J. Proudfoot, and A. L. Beyer. 1999. EM visualization of transcription by RNA polymerase II: downstream termination requires a poly(A) signal but not transcript cleavage. Mol. Cell. 3:379-387. [DOI] [PubMed] [Google Scholar]
- 30.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 32.Rasmussen, E. B., and J. T. Lis. 1993. In-vivo transcriptional pausing and cap formation on 3 drosophila heat-shock genes. Proc. Natl. Acad. Sci. USA 90:7923-7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryan, K., K. G. Murthy, S. Kaneko, and J. L. Manley. 2002. Requirements of the RNA polymerase II C-terminal domain for reconstituting pre-mRNA 3′ cleavage. Mol. Cell. Biol. 22:1684-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schroeder, S., B. Schwer, S. Shuman, and D. Bentley. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14:2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shuman, S. 1997. Origins of mRNA identity: capping enzymes bind to the phosphorylated C-terminal domain of RNA polymerase II. Proc. Natl. Acad. Sci. USA 94:12758-12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson, N., D. Aronson, and R. Burgess. 1990. Purification of eukaryortic RNA polymerase II by immunoaffinity chromatography. J. Biol. Chem. 265:7069-7077. [PubMed] [Google Scholar]
- 37.Wetterberg, I., G. Bauren, and L. Wieslander. 1996. The intranuclear site of excision of each intron in Balbiani ring 3 pre-mRNA is influenced by the time remaining to transcription termination and different excision efficiencies for the various introns. RNA 2:641-651. [PMC free article] [PubMed] [Google Scholar]
- 38.Wickens, M. P., and J. B. Gurdon. 1983. Post-transcriptional processing of simian virus 40 late transcripts in injected frog oocytes. J. Mol. Biol. 163:1-26. [DOI] [PubMed] [Google Scholar]
- 39.Yankulov, K. Y., M. Pandes, S. Mccracken, D. Bouchard, and D. L. Bentley. 1996. TFIIH functions in regulating transcriptional elongation by RNA-polymerase II in Xenopus oocytes. Mol. Cell. Biol. 16:3291-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuryev, A., M. Patturajan, Y. Litingtung, R. Joshi, C. Gentile, M. Gebara, and J. Corden. 1996. The CTD of RNA polymerase II interacts with a novel set of SR-like proteins. Proc. Natl. Acad. Sci. USA 93:6975-6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zandomeni, R., M. C. Zandomeni, D. Shugar, and R. Weinmann. 1986. Casein kinase type II is involved in the inhibition by 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole of specific RNA polymerase II transcription. J. Biol. Chem. 261:3414-3419. [PubMed] [Google Scholar]
- 42.Zeng, C., and S. M. Berget. 2000. Participation of the C-terminal domain of RNA polymerase II in exon definition during pre-mRNA splicing. Mol. Cell. Biol. 20:8290-8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou, M., L. Deng, F. Kashanchi, J. N. Brady, A. J. Shatkin, and A. Kumar. 2003. The Tat/TAR-dependent phosphorylation of RNA polymerase II C-terminal domain stimulates cotranscriptional capping of HIV-1 mRNA. Proc. Natl. Acad. Sci. USA 100:12666-12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu, Y. R., T. Peery, T. M. Peng, Y. Ramanathan, N. Marshall, T. Marshall, B. Amendt, M. B. Mathews, and D. H. Price. 1997. Transcription elongation factor P-TEFb is required for HIV-1 Tat transactivation in vitro. Genes Dev. 11:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]