Abstract
Bacterial pathogens encode a wide variety of effectors and toxins that hijack host cell structure and function. Of particular importance are virulence factors that target actin cytoskeleton dynamics critical for cell shape, stability, motility, phagocytosis, and division. In addition, many bacteria target organelles of the general secretory pathway (e.g., the endoplasmic reticulum and the Golgi complex) and recycling pathways (e.g., the endolysosomal system) to establish and maintain an intracellular replicative niche. Recent research on the biochemistry and structural biology of bacterial effector proteins and toxins has begun to shed light on the molecular underpinnings of these host-pathogen interactions. This exciting work is revealing how pathogens gain control of the complex and dynamic host cellular environments, which impacts our understanding of microbial infectious disease, immunology, and human cell biology.
Keywords: bacterial effectors, bacterial toxins, actin cytoskeleton, Golgi complex, endosomal/lysosomal trafficking
OVERVIEW
Bacteria interact and take control of their environment by delivering macromolecules across the inner membranes and outer membranes of gram-positive and gram-negative bacteria, respectively. At least seven secretory systems (SS) have evolved for this purpose. However, the type 3 SS (T3SS), type 4 SS (T4SS), and type 6 SS (T6SS) have the unique ability to translocate virulent effector proteins from the bacterial cytoplasm directly to the cytoplasm of host cells (Costa et al. 2015, Hayes et al. 2010). These three secretion systems are found primarily in pathogenic bacteria, and the loss of their protein delivery capacities results in avirulent phenotypes. By extension, the function of secreted effector proteins is essential to both animal and plant infectious disease.
The term effector protein is a generic description of any bacterial protein delivered into the host cytoplasm by the T3SS, T4SS, and T6SS apparatus. This term also distinguishes the mode of host cell delivery from that of classic toxins that are first secreted into the extracellular space prior to host cell uptake by endocytic-type mechanisms. However, there are several critical differences between effectors and toxins. Unlike large, multidomain bacterial toxins, effectors are typically small proteins composed of a short secretory signal, which is required for host translocation and is linked to a minimal enzymatic or functional domain (Dean 2011). Once delivered into the host cell, effector proteins recognize specific substrates and typically catalyze chemical reactions that disrupt substrate functions. Although the inhibitory activities of bacterial effector proteins have deleterious effects on host cell architecture and signal transduction, many effector proteins have evolved to directly activate substrates for the purpose of reprogramming cell behavior. These reprogramming events allow pathogens to access new cell sites, such as the cytoplasm or intracellular vacuole, that can promote dissemination across tissue barriers and/or evasion of the host immune defense systems. Thus, the action of bacterial effector proteins allows pathogens to exploit host environments that are typically inaccessible to normal flora.
In this review, our goal is to distill the wealth of information on bacterial effector proteins and toxins, with a specific emphasis on detailing the molecular mechanisms of substrate recognition and the chemical modifications that alter host cell structure and function. Although some historical context is discussed, we primarily emphasize new studies that reveal how pathogens gain control of two major structural elements of the host cell: the actin cytoskeleton and intracellular organelles.
THE ACTIN CYTOSKELETON: A TARGET OF BACTERIAL PATHOGENS THAT CONTROLS HOST STRUCTURE AND FUNCTION
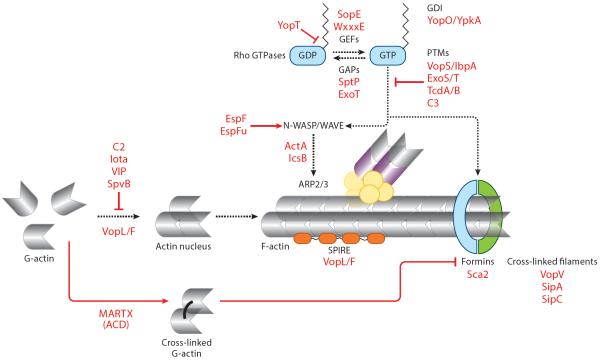
Actin is one of the most abundant proteins in eukaryotic cells and is involved in multiple essential cell processes, including motility, division, organelle function, and the maintenance of cell architecture. As such, the regulatory components of actin filaments are attractive targets of bacterial pathogens. Microbial perturbation of actin dynamics often results in profound changes in host cell structure that can result in opposing phenotypic outcomes, such as the induction versus inhibition of phagocytosis. Actin is a monomeric ATPase (G-actin) that can undergo cycles of spontaneous self-assembly into filaments (F-actin), ATP hydrolysis, and depolymerization. In the cell, the transition from G-actin to F-actin is tightly controlled by numerous actin-binding proteins (ABPs) and their upstream activators, which together produce all actin architectures required for cell function. Actin dynamics are controlled by the activity of the Rho family of small GTPases (Spiering & Hodgson 2011). Once activated by upstream stimuli, such as growth factor receptor engagement, Rho-family GTPases signal to downstream effectors, including nucleation-promoting factors (NPFs). NPFs in turn activate the Arp2/3 complex, the actin nucleator responsible for the generation of branched actin filaments. Interestingly, pathogens have evolved toxins and effector proteins to directly mimic the function of many ABPs and their regulators. These processes can be broadly categorized into three classes of pathogenic regulators: (a) toxins/effectors that directly regulate actin filament assembly, bundling, or depolymerization; (b) toxins/effector proteins that indirectly regulate F-actin by mimicking actin nucleators or their upstream NPFs; and (c) toxins/effectors that target the Rho-family GTPases, master regulators of signal transduction to the actin cytoskeleton (Figure 1).
Figure 1.
Regulation of actin dynamics by bacterial toxins and effector proteins. Bacterial pathogens have evolved multiple strategies to both positively and negatively regulate actin dynamics. Actin polymerization is inhibited through ADP-ribosylation of G-actin into nonpolymerizable monomers by the C2 toxin, the iota toxin, the VIP (vegetative insecticidal protein) toxin, and the Salmonella effector SpvB. ACD (actin cross-linking domain) toxin–mediated actin cross-linking poisons the formin family of actin nucleators. Actin polymerization is inhibited by bacterial effectors through the removal of membrane-bound Rho GTPases via either sequestration of the lipid group by YopO/YpkA or proteolytic cleavage of the lipid group from Rho GTPases by YopT. Additionally, inactivation of Rho GTPases via effector GAP (GTPase-activating protein) activity of SptP and ExoT shuts off actin polymerization. Rho GTPases undergo posttranslational modification (PTM) by ADP-ribosylation via C3 toxins and effectors ExoS and ExoT and by AMPylation by VopS and IbpA, preventing Rho GTPase activation of actin polymerization. Bacterial pathogens have also evolved mechanisms to induce actin dynamics through various mechanisms. VopL/F, Sca2, and SipC directly nucleate actin. Additionally, VopV, SipC, and SipA stabilize actin filaments by cross-linking. Bacterial effectors directly activate or function as nucleation-promoting factors to promote Arp2/3-mediated actin nucleation. Activation of Rho GTPase signaling by SopE- and WxxxE-family members of bacterial GEFs (guanine nucleotide exchange factors) also induces actin polymerization.
Bacterial Effectors Directly Promote Actin Polymerization by Mimicking Host Factors
To precisely control the actin dynamics required for sophisticated cell processes, the nucleation of actin is tightly regulated within the cell. Actin polymerization into F-actin filaments begins with the formation of an actin nucleus, a stable complex of actin monomers that structurally resemble the arrangement of subunits in a filament. Once the actin nucleus forms, additional actin monomers readily bind to the barbed end of the filament, and elongation occurs. Because nucleus formation is the rate-limiting step in the polymerization reaction, pathogens have evolved effector proteins that promote actin nucleation. Perhaps the greatest insights into this process have occurred through the study of bacterial pathogens that hijack actin polymerization for intracellular motility and cell-to-cell spread. One of the best-studied pathogens in this regard, Listeria monocytogenes, activates host NPF Arp2/3 (discussed in more detail below). In contrast, several pathogens, including Rickettsia and some Vibrio species, secrete effector proteins that directly polymerize actin.
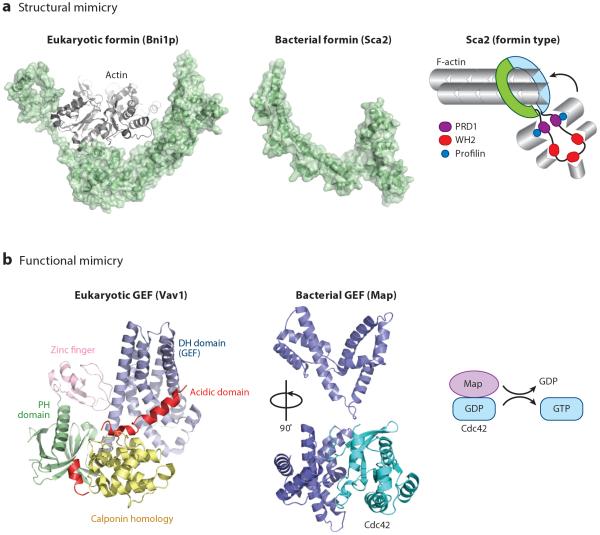
Rickettsia are gram-negative, obligate intracellular pathogens that infect humans via an arthropod vector; these pathogens are responsible for multiple diseases, including typhus and spotted fever. Like Listeria, Rickettsia depends upon actin-based motility for its intracellular propulsion. Rickettsia was the first pathogen found to bypass Arp2/3 activation for motility and instead encodes the autotransporter Sca2, a formin-like actin nucleator (Haglund et al. 2010, Madasu et al. 2013). Sca2 mimics formins in its ability to function as an elongation factor by remaining processively associated with the barbed end of the growing actin filament, by requiring profilin for efficient elongation, and by inhibiting actin filament–capping proteins. Recently, the N-terminal domain of Sca2 was crystallized and shown to adopt a previously uncharacterized fold that has a series of helix-loop-helix repeats and exhibits a crescent moon structure (Figure 2a) (Madasu et al. 2013). It is predicted that the C-terminal domain also contains helix-loop-helix repeats and adopts a crescent shape similar to that of host formins (Otomo et al. 2005). The interaction of the N- and C-terminal domains may result in a ringlike structure reminiscent of the ringlike structure of the formin FH2 homodimer. By extension, these domains are predicted to be responsible for organizing actin monomers into an actin nucleus. Sca2 also contains three WASP homology 2 (WH2) domains for actin monomer recruitment and proline-rich motifs (PRMs) for profilin-dependent elongation. Interestingly, Rickettsia motility requires Sca2 as well as RickA, a Rickettsia NPF, but these molecules act independently at different stages throughout the infection process (Reed et al. 2014).
Figure 2.
Bacterial effectors mimic eukaryotic actin-binding proteins and regulators. Bacterial effector proteins can structurally or functionally mimic host proteins. (a) The Rickettsia actin nucleator Sca2 (PDB: 4J7O) is a structural mimic of eukaryotic formin Bni1p (PDB: 1Y64) (Otomo et al. 2005). Formins adopt a ringlike structure that aids in barbed-end polymerization. (b) Several classes of bacterial guanine nucleotide exchange factors (GEFs), including SopE- and WxxxE-family members (Escherichia coli Map shown here; PDB: 3GCG), functionally mimic eukaryotic GEFs (Vav1 shown here; PDB: 3KY9). Both bacterial and eukaryotic GEFs of the Dbl family induce similar structural rearrangements in Rho GTPase proteins, but they do so using completely distinct structural architectures and catalytic residues. In addition, unlike eukaryotic GEFs, which are highly decorated with various accessory domains, bacterial GEFs are compact and encode minimal accessory domains. Abbreviations: DH, Dbl homology domain; PH, pleckstrin homology domain.
The T3SS effectors VopL from Vibrio parahaemolyticus and VopF (a homolog of VopL) from Vibrio cholerae nucleate actin in vitro and, during infection, generate actin stress fibers and filopodia, respectively (Liverman et al. 2007, Yu et al. 2011). Recent structural insights into VopL have provided the strongest evidence for the mechanism by which actin nucleation occurs. VopL is a multidomain protein encoding a VopL C-terminal domain (VCD) and an array of three WH2 domains interspaced with PRMs. Although the VCD domain is sufficient for nucleation, its activity is greatly enhanced by the WH2 domains (Namgoong et al. 2011, Yu et al. 2011). The structure of the VCD dimer was solved in complex with three actin monomers, and this structure revealed that actin monomers bound by the VCD dimer are arranged in a manner similar to that of actin subunits found within a short-pitch actin filament (Zahm et al. 2013). However, as Zahm et al. (2013) point out, the actin monomers are arranged in a confirmation that is not conducive for polymerization, suggesting that additional structural rearrangements required for filament elongation may be responsible for the rapid release of VopL from the growing filament. Competing models of VopL/F-actin structures have been proposed (Avvaru et al. 2015, Namgoong et al. 2011). Nevertheless, VopL-mediated actin nucleation occurs through the cooperation between its VCD actin organization domain and actin-binding WH2 domains. These distinct actin-binding and actin organization properties are shared among eukaryotic nucleation factors, including Arp2/3 and formins, indicating that VopL functionally mimics eukaryotic proteins.
Bundling and Cross-Linking Actin by Bacterial Toxins and Effectors
Actin filaments can be bundled into thick fibers or cross-linked into a weblike architecture that supports cell structures. Pathogens have evolved effectors and toxins to produce similar actin phenotypes for virulence. For example, the V. parahaemolyticus effector VopV is secreted by the T3SS2, and its ability to bind and bundle F-actin filaments is required for its enterotoxicity (Hiyoshi et al. 2011). VopV consists of a central long repetitive (LR) domain that confers these interaction capabilities, and recently, the structure of one of the repeat units found within the LR domain of VopV (VopVrep1) was solved in complex with F-actin (Nishimura et al. 2015). The 9.6-Å cryoEM structure revealed that VopVrep1 undergoes a disorder-to-order transition upon binding across the interstrand region of F-actin. Interestingly, the interstrand region is also the binding site for the Salmonella effector SipA, which is discussed in more detail below (Galkin et al. 2002).
Salmonella encodes two T3SS: the SPI-1 system for bacterial invasion and the SPI-2 system for intracellular survival and replication. The SPI-1 effectors SipA and SipC act cooperatively to mediate actin polymerization and actin filament bundling (McGhie et al. 2001) and, along with additional effectors such as SopE and SptP (discussed below), are responsible for Salmonella invasion. SipA promotes actin polymerization in several ways. First, SipA stabilizes F-actin and lowers the critical concentration for actin polymerization tenfold (Zhou et al. 1999). Electron microscopy revealed that the actin-binding fragment of SipA stabilizes filaments by binding along the interstrand region (Galkin et al. 2002). Second, SipA inhibits ADF/cofilin depolymerization through competitive binding. Finally, SipA inhibits and reverses gelsolin-mediated actin severing (McGhie et al. 2004). This multitude of activities are also coordinated with the action of SipC, a component of the Salmonella T3SS machinery (Kaniga et al. 1995a) that nucleates actin polymerization (Hayward & Koronakis 1999) and bundles actin filaments (Myeni & Zhou 2010). Although SipC oligomerization has been shown to be important for its nucleation properties (Chang et al. 2007), the lack of structural studies currently precludes the determination of its mechanism of nucleation. However, homologs of SipA and SipC are found in the intracellular pathogens Shigella flexneri and Burkholderia pseudomallei (Druar et al. 2008; Kaniga et al. 1995a,b), suggesting that these effectors may also be key factors in host cell invasion.
Bacterial Toxins and Effectors Catalyzing Actin Depolymerization
Historically, many of the most important bacterial toxins were found to directly depolymerize actin filaments by ADP-ribosylation. The C2 toxin from Clostridium botulinum (Aktories et al. 1986), the iota toxin from Clostridium perfringens (Stiles & Wilkins 1986), the Clostridium difficile transferase toxin (Popoff et al. 1988), the Clostridium spiroforme toxin (Simpson et al. 1989), the vegetative insecticidal proteins from Bacillus cereus (Han et al. 1999), and the Salmonella SPI-2 T3SS effector SpvB (Otto et al. 2000) have been shown to ADP-ribosylate G-actin. These toxins reversibly modify G-actin at Arg177 through the addition of an ADP-ribose moiety from NAD (Aktories & Wegner 1989). The functional result of this modification is the conversion of G-actin into a polymerization-deficient subunit. Incorporation of this modified G-actin into the growing filament effectively caps the growing filament, leading to subsequent filament depolymerization.
Bacterial Effectors Mimicking Actin Nucleation-Promoting Factors
It has long been known that intracellular pathogens such as L. monocytogenes propel themselves throughout eukaryotic cells on so-called actin comet tails (and can penetrate the plasma membrane and thereby move from cell to cell). This observation has been the subject of intense investigation over the past two decades and has revolutionized our understanding of actin filament dynamics (Pollard et al. 2000). Importantly, mechanistic studies on Listeria motility have been essential for defining the minimal components required for sustained actin polymerization. Such studies led to the first identification and activation mechanism of the Arp2/3 nucleation complex (Machesky et al. 1994, Welch et al. 1997) as well as the conceptual and experimental framework for the complete reconstitution of cell motility in vitro (Loisel et al. 1999). In addition to these pioneering studies, intracellular actin motility has been described for unrelated bacterial pathogens that activate actin nucleators (e.g., Arp2/3) or their upstream NPFs (e.g., N-WASP), including S. flexneri (Goldberg & Theriot 1995), Burkholderia (Benanti et al. 2015), and Mycobacterium marinum (Stamm et al. 2003). Extracellular pathogens also secrete T3SS effector proteins; notably, enterohemorrhagic Escherichia coli (EHEC) O157:H7 effectors EspF and EspFu (also known as TccP) activate N-WASP (Alto et al. 2007, Sallee et al. 2008).
Bacteria Targeting Rho-Family GTPases
Rho-family GTPases are small-molecular guanine nucleotide–binding proteins that function as molecular switches, existing in an inactive GDP-bound state and an active GTP-bound state. Controlling the interconversion between these two states are guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs). GEFs activate Rho-family GTPases by mediating the exchange of GDP for GTP and GAPs, which inactivate GTPases by promoting the hydrolysis of the bound GTP to GDP (recently reviewed in Cherfils & Zeghouf 2013). In their GTP-bound state, Rho-family GTPases activate their downstream effectors, including the WASP family of NPFs (Rohatgi et al. 1999). Thus, the cycles of guanine nucleotide exchange and hydrolysis are directly coupled to the induction of actin polymerization. In addition, the location of Rho GTPase function is controlled by long-chain fatty acid modifications of the C-terminal CaaX motif; such modifications target the GTPases to the plasma membrane and various intracellular organelles. Remarkably, bacterial pathogens have evolved toxins and effector proteins to target nearly every step of the Rho GTPase signaling cascade through molecular mimicry of GTPase regulatory proteins [GEFs, GAPs, and guanine nucleotide dissociation inhibitors (GDIs)] and through posttranslational modifications (PTMs) (discussed in detail below).
Bacterial GEFs
The first bacterial GEF identified was SopE, a T3SS effector from Salmonella species that mediates bacterial invasion (Hardt et al. 1998). SopE activates Rac1 and, along with its later identified homolog SopE2, can also activate Cdc42 to induce actin polymerization and ultimately internalization of the invading bacterium (Friebel et al. 2001, Rudolph et al. 1999, Stender et al. 2000). Homologs of SopE include CopE, which is found in Chromobacteria violaceum (Miki et al. 2011), and BopE, which is encoded by B. pseudomallei (Stevens et al. 2003). Structural studies on SopE showed that, unlike the eukaryotic Rho-family GTPase GEFs that contain a Dbl-homology (DH) domain or Dock Homology region 2 (DHR-2) domains (Yang et al. 2009, Yu et al. 2010), SopE adopts a V-shaped fold that consists of two intersecting bundles of α-helices connected by an extended catalytic loop (Figure 2b) (Buchwald et al. 2002). Although structurally distinct, both eukaryotic and bacterial GEFs induce similar nucleotide-free transition states that are required for efficient nucleotide exchange. More than a decade later, a second family of bacterial GEFs was identified. This family shares an invariant Trp-X-X-X-Glu (WxxxE) signature motif, and members include IpgB1 and IpgB2 from Shigella, SifA and SifB from Salmonella, and Map and EspM from E. coli. Although the SopE-type GEFs and WxxxE GEFs share no sequence homology, they adopt nearly identical V-shaped structures (Figure 2b). Clearly, these families have diverged from a common evolutionary ancestor, yet surprisingly they are structurally distinct from eukaryotic GEFs (Figure 2b). Recent studies suggest that bacterial GEFs are further endowed with accessory motifs that drive their subcellular localization and their ability to locally amplify actin polymerization (Orchard et al. 2012). This is an open area of investigation that could help explain how effector proteins secreted at very low concentration can induce robust changes in cell architecture and signal transduction.
Bacterial GAPs
Pathogenic bacteria have also evolved effector proteins that mimic GAPs to dampen Rho-family GTPase signaling. One of the most intriguing functions of bacterial GAPs is their ability to suppress bacterial GEFs required for pathogenic invasion. As Salmonella is an intracellular organism, profound and long-lasting Rho-family GTPase activation by SopE and SopE2 would be counterproductive to Salmonella's pathogenicity. Therefore, Salmonella has evolved the GAP SptP to reverse the extensive effector-mediated actin polymerization following internalization (Fu & Galan 1999, Stebbins & Galan 2000). The translocation of SopE prior to SptP coupled with the relatively rapid degradation of SopE after translocation confers a sophisticated degree of temporal regulation of Rho-family GTPase signaling during infection (Kubori & Galan 2003, Van Engelenburg & Palmer 2008, Winnen et al. 2008). Much like eukaryotic GAPs, bacterial GAPs commonly use a conserved arginine finger to induce efficient GTP hydrolysis, switching the Rho-family GTPases into their inactive state. Bacterial GAPs are found encoded in Salmonella spp., Yersinia spp., and Pseudomonas aeruginosa (Aili et al. 2002, Andor et al. 2001, Sundin et al. 2001).
Regulation of GTPase Localization by Bacterial GDIs and Prenyl-Hydrolases
In addition to directly controlling the nucleotide cycling of the Rho-family GTPases, bacterial pathogens have evolved the ability to remove Rho-family GTPases from cell membranes. Rho-family GTPases are acylated at their C terminus, and proper membrane localization is critical for GTPase function (Cherfils & Zeghouf 2013). When the acyl group is embedded into the membrane, Rho-family GTPases are able to interact with their GEFs and, once activated, can engage downstream signaling molecules. GDIs carry out the removal of a GTPase from the membrane through the sequestration of the prenylated CaaX motif (Hoffman et al. 2000, Scheffzek et al. 2000). The Yersinia effector YopO (also known as YpkA) acts as a GDI mimic for Rac1 and RhoA (Juris et al. 2000, Prehna et al. 2006). Furthermore, another Yersinia effector, YopT, functions as a cysteine protease and disrupts the actin cytoskeleton by cleaving lipid-modified CaaX sequence from the GTPase (Shao et al. 2002). Liberation of Rho GTPases from the membrane irreversibly inhibits signaling to the actin cytoskeleton.
Posttranslational Modification of GTPases
Protein PTM through the covalent addition or removal of functional groups is a key regulator of protein activity and localization and controls the activation of downstream substrates. It is therefore not surprising that bacterial pathogens have evolved effectors and toxins to chemically modify Rho GTPases. As discussed in more detail below, most PTMs are found in the switch I or switch II loops that regulate interactions with guanine nucleotides (GDP/GTP), GEFs, GAPs, and GTPase effectors.
The large family of clostridial toxins—including the botulinum (C3) toxin (Mohr et al. 1992), as well as the P. aeruginosa effectors ExoS and ExoT (Fleiszig et al. 1997)—ADP-ribosylate Rho GTPases. C3 toxins were the first Rho GTPase–modifying enzymes identified and as such were instrumental for distinguishing the roles of different Rho-family isoforms. These toxins and effector proteins ADP-ribosylate RhoA, RhoB, and RhoC on the invariant Asn41 residue found within the nucleotide-binding switch I region (Aktories 2011). The resulting modification enhances the interaction between the Rho GTPase and its GDI, resulting in the sequestration of the GTPase into the cytosol and preventing activation by GEFs (Sehr et al. 1998). Several C3-related toxins, including the Edin toxin of Staphylococcus aureus (Wilde et al. 2001) and C3cer in B. cereus (Just et al. 1995), have been identified. Interestingly, the insect pathogen Photorhabdus luminescens uses a C3-like toxin complex (PTC5) to ADP-ribosylate the switch II region of Rac and Cdc42, preventing GTP hydrolysis and therefore generating a constitutively active GTPase (Lang et al. 2010). Rho GTPases are also activated by the cytotoxic necrotizing factors CNF from Yersinia pseudo-tuberculosis and CNF1, CNF2, and CNF3 from E. coli and by the Vibrio T3SS2 effector VopC (Flatau et al. 1997, Okada et al. 2014, Schmidt et al. 1997). These toxins use their C-terminal domain to catalyze deamination of Gln61 or Gln63 found in the switch II region of Rho GTPases; conversion to a glutamic acid residue prevents GTP hydrolysis, thereby locking the GTPase in an activated site.
Originally described more than 40 years ago (Kingdon et al. 1967), protein modification by addition of adenosine monophosphate (AMP) was recently rediscovered in the V. parahaemolyticus effector VopS (Yarbrough et al. 2009) and in Histophilus somni IbpA (Zekarias et al. 2010). Both VopS and IpbA proteins contain the FIC (filamentation induced by cAMP) domain, which hydrolyzes the α-β phosphate bond of ATP and transfers AMP to the hydroxyl group of serine, threonine, or tyrosine residues of Rho-family GTPases. AMPylation (or adenylylation, as the reaction was originally termed) of Rho GTPases on the switch I region prevents interactions with their downstream effectors, leading to a disorganization of the actin cytoskeleton. It is now clear that FIC-domain proteins can utilize a range of chemical cofactors as donors for PTMs (Roy & Cherfils 2015). The common feature of these cofactors is a diphosphate moiety that can include nucleotides as well as CDP-choline, which is cleaved during the linkage of the adduct to the target protein.
Toxins A and B (also termed TcdA and TcdB, respectively) from C. difficile, the lethal and hemorrhagic toxins from Clostridium sordellii, the α-toxin from Clostridium novyi, and the recently discovered TpeL from C. perfringens glycosylate Rho GTPases, leading to their inactivation and their inability to signal to the actin cytoskeleton (Nagahama et al. 2011, von Eichel–Streiber et al. 1996). All these bacterial enzymes target the switch I region, Thr35 in Rac and Cdc42 or Thr37 of Rho isoforms, with either mono-O-glycosylation or N-acetylglucosamination. Modification at this residue results in a GEF- and GAP-insensitive GTPase (Sehr et al. 1998). Therefore, glycosylated Rho GTPases are unable to participate in nucleotide cycling, resulting in actin cytoskeleton disruption.
ORGANELLE ARCHITECTURE: CONTROLLING MEMBRANE TRANSPORT SYSTEMS BY BACTERIAL PATHOGENS
Membrane-bound organelles are the major feature distinguishing eukaryotic from prokaryotic cells. Such organelles not only sequester chemical reactions but also organize protein and lipid transport systems that support complex cell structures. Like the actin cytoskeleton discussed above, membrane transport systems are heavily targeted by bacterial pathogens. Examples of such systems are the secretory system made up of distinct organelles [including the endoplasmic reticulum (ER), the Golgi apparatus, and the plasma membrane], endolysosomal compartments, and autophagosomes. By specifically manipulating critical aspects of host organelle biogenesis and function, bacteria have the ability to inhibit immune cytokine secretion and immune detection, to establish a replicative vacuole in the host cytoplasm, and/or to avoid destruction through regulation of autophagy, all of which are discussed in more detail below.
Pathogen Targeting the ER-to-Golgi Trafficking Network
The general secretory pathway (GSP), composed of the ER, Golgi apparatus, and cell vesicles, is responsible for processing, sorting, and distributing proteins and lipids throughout the cell. During a host's battle with pathogens, a functional GSP mediates the secretion of cytokines, chemokines, and antimicrobial peptides that are crucial for host defense. Microbial pathogens have therefore developed several strategies to antagonize or exploit the GSP to assist in their infection. It has been known for many years that certain classes of bacterial toxins traffic along a retrograde pathway, eventually being excreted from the ER compartment (Mukhopadhyay & Linstedt 2013). Although the GSP pathway is clearly hijacked for toxin delivery, other pathogens directly regulate ER and Golgi trafficking networks to avoid immune detection or establish a replicative niche within the host cytoplasm. Here we highlight some recent findings detailing how bacteria regulate the intracellular secretory pathway, especially focusing on the modulation of ARF- and Rab-family GTPases, which are master regulators of organelle structure and ER-to-Golgi vesicle transport.
Golgi Destruction by Coordinated Regulation of ARF and Rab GTPases
GTPases of the ARF and Rab families are master regulators of membrane traffic, organelle structure, and cargo transport (Donaldson & Jackson 2011, Hutagalung & Novick 2011). Like the Rho-family GTPases discussed above, ARFs and Rabs cycle between an inactive GDP-bound form and an active GTP-bound form on the basis of their interactions with GEFs and GAPs. In their active form, ARFs and Rabs associate with ER and Golgi membranes, to which these proteins recruit downstream effectors that promote vesicle fission or membrane docking and fusion events (Cherfils & Zeghouf 2013).
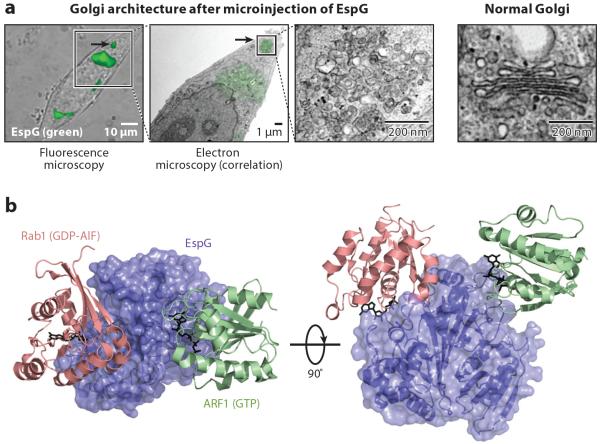
Recent studies have identified bacterial virulence factors that have profound consequences on Golgi structure and function. EspG/VirA-family T3SS effector proteins encoded by EPEC, EHEC, and Shigella (Elliott et al. 2001) cause massive Golgi vesiculation when ectopically expressed in host cells (Selyunin et al. 2011) or when secreted by the pathogen (Figure 3a) (Dong et al. 2012). Structural studies indicated that EspG specifically binds to the switch I loop and nucleotide-binding pocket of GTP-bound ARFs (Figure 3b) (Selyunin et al. 2011). The unique architecture of this interaction prevents GAP-catalyzed GTP hydrolysis, allowing EspG to stabilize active ARF on host membranes (Selyunin et al. 2011). In addition, EspG is a Rab1-specific GAP, thereby targeting two critical components of the host secretory pathway (Dong et al. 2012). Selyunin et al. (2014) described a concerted model for EspG function, highlighting both ARF and Rab interactions (Figure 3b). By stabilizing ARF-GTP on Golgi membranes, EspG promotes the recruitment of vesicle tethering factors such as GMAP210 that link incoming vesicles to recipient membranes; simultaneously, EspG prevents vesicle fusion by its Rab1-GAP activity. This concurrent activity of EspG toward ARF and Rab1 is required for EspG to fully inhibit intracellular trafficking and induce the clustered vesicle phenotype observed in cells (Selyunin et al. 2014). Interestingly, Shigella VirA exhibits similar Rab1-GAP activity and induces Golgi fragmentation (Dong et al. 2012), yet it does not have the structural elements to support ARF1 binding (Dong et al. 2012, Selyunin et al. 2011). Thus, VirA may coordinate Rab1 inactivation with other signaling components (Selyunin et al. 2014). The Golgi matrix protein GM130 has been identified as a binding partner for EspG in a yeast two-hybrid screen, but whether the EspG-GM130 interaction is linked to EspG-mediated disruption of vesicular transport is unclear (Clements et al. 2011). The molecular study of EspG/VirA function in arresting Golgi trafficking opens new avenues of investigation into the potential roles of these proteins in bacteria pathogenesis. EspG inhibits the secretion of the proinflammatory cytokine IL-8 from HeLa cells during EHEC infection, thus enhancing bacterial intracellular survival (Dong et al. 2012). EspG1 and -G2 mediate tight junction (TJ) disruption during EPEC infection (Glotfelty & Hecht 2012); the potential of EspG1/G2 to block secretion of TJ proteins may be one undetermined mechanism. For Shigella, inactivation of Rab1 by VirA inhibits autophagy-mediated host defense (Dong et al. 2012).
Figure 3.
Bacterial effector regulation of organelle structure and function. (a) As shown by correlative light electron microscopy, the Golgi apparatus vesiculates shortly after microinjection of the Escherichia coli effector EspG. A normal Golgi apparatus is shown at the right. (b) The structure of EspG in complex with Rab1 and ARF1 has provided important insights into the mechanism of EspG-mediated Golgi vesiculation (PDB: 4FME).
Elimination of N-Myristoyl Modification of ARF GTPases
Besides VirA, another Shigella T3SS effector protein named IpaJ has recently been implicated in the inhibition of cargo transport through the GSP. According to structure-based bioinformatics analysis, IpaJ exhibits similarities with the C39-like cysteine peptidase family and harbors a classic Cys/His/Asp catalytic triad required for peptide bond cleavage by this family. Substitutions of the three residues abolished IpaJ's ability to induce Golgi fragmentation during Shigella infection of host cells, indicating that this function depends on the cysteine protease activity of IpaJ (Burnaevskiy et al. 2013). Further study indicated that IpaJ specifically cleaves the myristoylated N-terminal glycine of host proteins, and one potential target is ARF1, a small GTPase critical for Golgi apparatus structure and function (Burnaevskiy et al. 2013). ARF1 regulates cargo transport through the Golgi apparatus by binding to the Golgi membranes (through N-myristoylation) and recruiting the COPI coat complex for vesicle fission (Kahn 2009). IpaJ cleaves the peptide bond between N-myristoylated Gly2 and Asn3 of human ARF1, which releases ARF1 from the Golgi membranes and causes fragmentation of the Golgi and disruption of host cargo trafficking (Burnaevskiy et al. 2013).
Profiling the host cell “myristoylome” along with detailed functional studies revealed that IpaJ recognizes and cleaves most N-myristoylated proteins in vitro yet exhibits high specificity in vivo, such as targeting ARF and ARL GTPase families in the context of Shigella infection (Burnaevskiy et al. 2015). IpaJ recognizes two diverse structural elements on ARF proteins: One element (myristoylated glycine) is common to all myristoylated proteins, whereas the other (the GTPase domain) is unique to GTPase-family proteins (Burnaevskiy et al. 2015). Further work uncovered the ability of IpaJ to inhibit ER exit of STING to antagonize the cGAS/STING cytosolic DNA-sensing pathway involved in the type I interferon response (Dobbs et al. 2015). IpaJ blocks the translocation of STING from ER to ERGIC (ER-Golgi intermediate compartment), which is critical for STING activation. IpaJ also suppresses STING activation of disease-associated STING mutants that causes constitutive ER exit. These functions of IpaJ are dependent on its activity to demyristoylate ARF GTPase (Dobbs et al. 2015).
Hijacking Host ER-to-Golgi Vesicular Transport by Manipulating Rab1 Activity
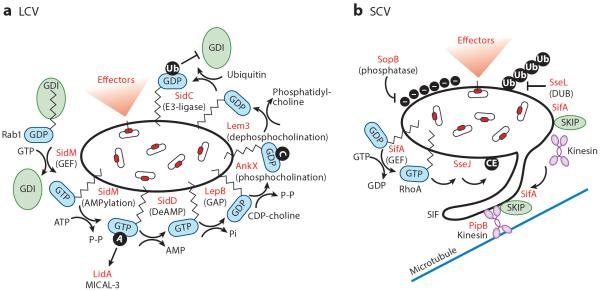
In addition to blocking the secretory pathway, some pathogens exploit the trafficking system to establish intracellular persistence. Legionella is an intracellular pathogen that hijacks ER-derived vesicles to create a Legionella-containing vacuole (LCV) for replication. The biogenesis and maturation of an LCV involve rewiring the destination of secretory vesicles derived from the ER; such rewiring is promoted by acquisition of Rab1 GTPase. The molecular mechanisms underlying exploitation of host cell Rab1 by Legionella have been well studied and involve both regulation of the Rab1 guanine nucleotide activation cycle and chemical modifications by bacterial enzymes (Figure 4a). The SidM/DrrA effector, which possesses both GDI displacement factor activity and GEF activity, promotes the recruitment of Rab1 to the LCV (Brombacher et al. 2009, Machner & Isberg 2007). Because Rab1 can rapidly undergo GAP-mediated hydrolysis, SidM/DrrA further stabilizes GTP-active Rab1 on the LCV by AMPylating at Tyr77 of the switch loop. This PTM blocks access of GAPs to the nucleotide-binding pocket, thereby stabilizing membrane-bound GTP-Rab1 (Muller et al. 2010). Importantly, this chemical modification can be reversed by the deAMPylating enzyme SidD (Neunuebel et al. 2011, Tan & Luo 2011), and subsequently Rab1 can be inactivated by the Legionella GAP LepB (Schuerch et al. 2005). Ultimately, the regulation of Rab1 by both GEFs/GAPs and AMPylation controls the timing and location of Rab1 signal transduction from the LCV. Additional Legionella enzymes targeting Rab1 include AnkX and LegA8, which catalyze the covalent attachment of CDP-choline to Rab1 (phosphocholination); Lem3 can reverse this process (Goody et al. 2012, Tan et al. 2011). In addition, the effector SidC possesses ubiquitin ligase activity that is correlated with Rab1 ubiquitination, yet direct ubiquitination of Rab1 by SidC has not been shown (Horenkamp et al. 2014, Hsu et al. 2014). Thus, the ubiquitination of Rab1 is likely mediated through an indirect unknown mechanism. Crystal structure analysis suggests that C-terminal Rab1 ubiquitination may interfere with Rab1-GDI interaction, but the exact function of this modification in Legionella infection requires further investigation (Horenkamp et al. 2014).
Figure 4.
Effector-mediated remodeling of pathogen-containing vacuoles: Legionella-containing vacuoles (LCV) and Salmonella-containing vacuoles (SCV). (a) Legionella encodes multiple effectors that regulate Rab1 activity. SidM is a multidomain effector with both Rab1 GEF (guanine nucleotide exchange factor) activity and AMPylation activity (black circle labeled A). SidD can deAMPylate Rab1. Rab1 can be inactivated by the GAP (GTPase-activating protein) LepB. Furthermore, Rab1 can be modified by the addition of phosphocholine by AnkX through its FIC (filamentation induced by cAMP) domain (black circle labeled C) or by the removal of the phosphocholine by Lem3. Lastly, the ubiquitin ligase SidC is thought to ubiquitinate (black circle labeled Ub) Rab1, preventing its interaction with its GDI (guanine nucleotide dissociation inhibitor). (b) Salmonella modifies its SCV through the action of multiple effectors. The SPI-1-secreted effector SopB, a phosphoinositide phosphatase, is involved in invasion and alters the surface charge (black circles labeled with minus signs) of the SCV to prevent lysosomal fusion. SifA is a key effector regulating SCV maintenance and Salmonella-induced filament (SIF) formation. SifA, a SPI-2 effector, interacts with the host protein SKIP (SifA kinesin-interacting protein) to retain SCV perinuclear positioning through kinesin and PipB2 interactions. In addition, the GEF domain of SifA is thought to activate RhoA GTPase at the SCV, which subsequently induces the acyltransferase activity of the SPI-2 effector SseJ, generating cholesterol esters on the SCV (black circle labeled CE). These events are thought to promote SIF formation. SseL, through its deubiquitinase (DUB) activity, reverses SCV ubiquitination and host cell recognition by autophagosomes.
Pathogen Remodeling of the Endolysosomal System
Much like Legionella's interaction with the host secretory pathway, many bacteria have evolved strategies to remodel the host endolysosomal system to create a replication-competent vacuole. Although the individual virulence factors may be different among bacterial species, these pathogens have evolved common strategies to modify the vacuole to form a degradation-resistant and replication-permissive microenvironment. Many pathogens deliver effector proteins to target host cell proteins or lipids to modify the vacuolar membrane composition or promote selective interactions with the endolysosomal system, which prevents the pathogens from degradation by lysosomes, yet allows interactions with host organelles that are required for both vacuole expansion and nutrient uptake.
Salmonella is perhaps the best-studied pathogen that exploits the host endolysosomal system. Immediately following internalization, these bacteria establish a Salmonella-containing vacuole (SCV) that provides three major advantages. First, the manipulation of the vacuole components restricts the fusion of the vacuole with the host lysosome. Second, the vacuole slows replication with the host, thereby providing a stable niche to persist in the host cell environment while avoiding cytosolic immune receptors. Finally, the SCV is trafficked to a perinuclear region of the cell, from where it acquires nutrients from its environment. This process is mediated predominantly by effector proteins secreted by the SPI-2 T3SS, although several SPI-1 effectors link invasion to SCV biogenesis (Figure 4b). For example, the phosphoinositide phosphatase SopB, which is required for actin-mediated Salmonella internalization, decreases the negative surface charge of the vacuolar membrane shortly after invasion (Bakowski et al. 2010). This event leads to the dissociation of several trafficking proteins from the vacuole, helping to prevent lysosome fusion. The SCV then migrates to the perinuclear region in close proximity to the Golgi network. A main feature of SCV maturation is the formation of Salmonella-induced filaments (SIFs) that extend from the vacuole at late stages of infection.
A key effector protein for SCV maintenance and SIF formation is SifA (Beuzon et al. 2000). Shortly after secretion, SifA is targeted via C-terminal acylation to the SCV, from where SifA interacts with two host proteins: SKIP (SifA kinesin-interacting protein) and RhoA GTPase. The interaction with SKIP has two seemingly opposing roles in SCV biogenesis. First, SKIP was originally shown to inhibit plus end–directed microtubule kinesin-1 activity at the SCV, thereby restricting microtubule motility of the SCV toward the cell periphery and maintaining its position in a perinuclear region permissive for Salmonella replication (Boucrot et al. 2005). Second, SifA recruitment of SKIP to the SCV is required for SIF formation, which maintains SCV integrity and is a kinesin-1-dependent process. Although the order of events required for SIF formation remains obscure, several additional effector proteins, including PipB2, coordinate kinesin-1 recruitment to SIFs (Henry et al. 2006). It has been suggested that SifA recruits inactive kinesin-1 to membranes and that PipB2 activates the motor protein to generate SIF protrusions from the vacuole. In addition, the SifA/SKIP complex sequesters Rab9 and subverts Rab9-dependent retrograde trafficking of mannose-6-phosphate receptors, thereby attenuating lysosome function (McGourty et al. 2012). Two recent reports also identified PLEKHM1 as a new host target for SifA. PLEKHM1 localizes to distinct regions of the SCV, where it interacts directly with both Rab7 and SifA to regulate membrane biogenesis of the SCV and maintain the normal morphology of this compartment (McEwan et al. 2015a,b).
Another SPI-2 T3SS effector, SseJ, acts complementarily with SifA to regulate dynamics of the SCV membrane. Unlike sifA− Salmonella strains that escape the SCV, sifA−/sseJ− double-mutant strains do not lose vacuole integrity during infection (Ruiz-Albert et al. 2002). In addition, ectopic expression of SifA and SseJ in HeLa cells induces endosomal tubulation, similar to the case for SIFs that form during Salmonella infection. This endosomal tubulation is also observed when SseJ is coexpressed with GTP-bound RhoA, RhoB, and RhoC, and SifA likely activates RhoA-family GTPases that coordinate SseJ activity (Christen et al. 2009). Structural studies indicated that, in addition to binding SKIP, SifA belongs to the WxxxE family of Rho GEFs. It has therefore been proposed that SseJ, SifA, SKIP, and RhoA constitute a complex that promotes vacuole tabulation as a precursor to SIF formation (Ohlson et al. 2008). SseJ may also modulate the composition of cholesterol on SCV membranes by acting as a RhoA-dependent glycerophospholipid-cholesterol acyltransferase enzyme (Christen et al. 2009, Lossi et al. 2008, Nawabi et al. 2008). Besides SseJ, additional SPI-2 effectors, including SopD2, SpvB, SteA, SseF, and SseG, are involved in SIF formation (Birmingham et al. 2005; Domingues et al. 2014; Knodler & Steele-Mortimer 2005; Krieger et al. 2014; Schroeder et al. 2010, 2011). SseF and SseG are also implicated in the perinuclear positioning of SCVs by recruiting dynein (Birmingham et al. 2005, Deiwick et al. 2006, Kuhle et al. 2006, Salcedo & Holden 2003). As work over the past decade has indicated, SCV positioning and maintenance are highly complex processes requiring the concerted action of both bacterial effector proteins and host substrates.
Like Salmonella, Brucella has developed several strategies to restrict fusion of the Brucella-containing vacuole (BCV) with lysosome compartments through the function of the T4SS effector VirB (Comerci et al. 2001, de Figueiredo et al. 2015). Legionella delivers multiple effectors to escape phagolysosomal killing, as well as effector-independent mechanisms (Prashar & Terebiznik 2015). Strikingly different from other vacuolar pathogens that avoid fusion of the vacuole with the lysosome, the Coxiella burnetii–containing vacuole is loaded with lysosome enzymes. C. burnetii is unique in its ability to survive in this harsh environment; it may exploit the acidic pH of the lysosome-like vacuole for metabolic activation (Ghigo et al. 2012, van Schaik et al. 2013).
Interference of Autophagy Signaling
Autophagy can be a host defense mechanism to target intracellular pathogens for lysosomal degradation. Although some bacteria are targeted and eventually destroyed by autophagy, several pathogens have developed strategies to avoid autophagy or even hijack the host's autophagic machinery for intracellular growth and survival.
Inhibition and evasion of autophagy
Several virulence factors have evolved to disrupt autophagosome formation. For example, the Eis (enhanced intracellular survival) protein secreted by Mycobacterium tuberculosis inhibits JNK-dependent autophagy activation (Kim et al. 2012, Shin et al. 2010). Rab1 inactivation by VirA suppresses host autophagy signaling (Dong et al. 2012) because Rab1 also plays a role in autophagosome formation (Huang et al. 2011). Listeria phospholipases PlcA and -B inhibit autophagic flux and autophagosome formation, preventing efficient targeting of cytosolic bacteria by autophagy machinery (Tattoli et al. 2013). Legionella delivers an effector protein, RavZ, which is a cysteine protease to deconjugate Atg8/LC3 from phosphatidylethanolamine and inhibit autophagosome formation (Choy et al. 2012). Additionally, Salmonella can escape from autophagy-mediated degradation by promoting mTOR reactivation on the surface of SCV, although the bacterial factors and mechanisms involved in this process remain unknown (Kim et al. 2012). Finally, some pathogens like Bacillus anthracis and V. cholerae express toxins that inhibit autophagy induction by increasing cyclic AMP levels (Shahnazari et al. 2011).
Pathogens can also avoid autophagic degradation by blocking autophagosome maturation. M. marinum recruits LC3 to phagosomes and can regulate the fate of these phagosomes. These LC3-decorated phagosomes do not acquire lysosomal enzymes, indicating that the infecting bacteria block autophagosomal maturation (Lerena & Colombo 2011). Similarly, Helicobacter pylori (Raju et al. 2012), Yersinia pestis (Pujol et al. 2009), Chlamydia trachomatis (Al-Younes et al. 2004, Yasir et al. 2011), and Porphyromonas gingivalis (Dorn et al. 2001) disrupt autophagosomal maturation by blocking autophagosome acidification or lysosomal degradation, although the molecular mechanisms have not been elucidated.
Certain pathogens have evolved the ability to mask themselves from autophagy recognition. For example, when Listeria enters the cytosol, its surface protein ActA recruits host proteins of the actin polymerization machinery (such as actin, Arp2/3, and VASP) to polymerize actin and form actin tails at the bacterial surface. A higher proportion of ΔactA mutant bacteria are sequestered in LC3-positive vacuoles than are wild-type bacteria (Birmingham et al. 2007, Yoshikawa et al. 2009). Listeria also uses the virulence factor InlK to recruit a host protein named major vault protein (MVP) to decorate its surface for autophagy evasion, and this new strategy to avoid autophagy is independent of ActA (Dortet et al. 2011, Neves et al. 2013). Shigella also counters autophagic attack through effector protein IcsB. IcsB competes with host ATG5 for binding to the bacterial surface protein IcsA, thereby masking the bacteria from autophagy recognition (Ogawa et al. 2005). IcsB may also prevent Shigella from autophagic targeting by masking the bacteria from septin binding (Mostowy et al. 2010). A recent study has shown that Salmonella utilizes a deubiquitinase, SseL, to mask the presence of ubiquitinated aggregates formed during the infection of epithelial cells or macrophages and to reduce the recruitment of autophagic components (Mesquita et al. 2012, Thomas et al. 2012). Francisella may avoid autophagy recognition by using the surface polysaccharidic O-antigen through an as-yet-unidentified mechanism (Case et al. 2014).
Exploitation of autophagy
In some cases, bacteria even exploit the host's autophagic machinery to benefit their own infection. For example, Coxiella replicates in a vacuole decorated by autophagic markers LC3, Beclin 1, and RAB24 (Gutierrez et al. 2005, Vazquez & Colombo 2010). Induction of autophagy promotes the infectivity of Coxiella, and blocking of autophagy impairs bacterial replication. Autophagic proteins play important roles in the maturation of the vacuole that supports Coxiella replication (Beron et al. 2002, Gutierrez et al. 2005). Infection of Coxiella actively modulates the autophagic pathway, including steps such as LC3 processing and Beclin 1 recruitment to the vacuole, although the bacterial factors involved in these processes remain to be further elucidated (Romano et al. 2007, Vazquez & Colombo 2010). Coxiella actively recruits and fuses autophagosomes to the vacuole to deliver nutrients for pathogen replication; the bacterial T4SS and effector Cig2 is required for this process (Newton et al. 2014, Winchell et al. 2014).
Different from pathogens that exploit autophagy to acquire host cell nutrients and support their own replication, Brucella subverts autophagy machinery to promote cell-to-cell spreading. At a late stage of infection, the replicative Brucella-containing vacuole is converted into a compartment with autophagic features, the autophagic Brucella-containing vacuole (aBCV). This conversion requires the activity of proteins that initiate autophagy, including ULK1, Beclin 1, ATG14L, and PI3K, but is independent of autophagy elongation factors ATG5, ATG16L1, ATG4B, ATG7, and LC3B. BCV formation does not contribute to bacterial proliferation but promotes bacterial cell-to-cell spread. Further studies are needed to clarify the mechanisms by which aBCV mediates bacterial release and reinfection (Starr et al. 2012).
V. parahaemolyticus is the first extracellular pathogen characterized to exploit autophagy for its own benefit. Two V. parahaemolyticus T3SS1 effectors, Vp1659 (Zhou et al. 2010) and VopQ, and one T6SS effector, VgrG2 (Yu et al. 2015), have been found to be involved in the induction of autophagy. VopQ induces acute autophagy, causes the infected cell to digest itself, and prevents phagocytosis of the bacteria (Burdette et al. 2008, 2009). Molecularly, VopQ alters autophagic flux by binding to the V-ATPase and disrupting host lysosome ion homeostasis (Sreelatha et al. 2013). VopQ-mediated autophagy also interferes with the NLRC4 inflammasome, thus contributing to bacteria evasion from inflammasome-mediated host immune responses (Higa et al. 2013).
OUTLOOK
This article details individual effector proteins and toxins that target animal cell structure and function. The virulence factors described often exhibit novel enzymatic mechanisms that alter actin cytoskeleton dynamics and membrane architecture. Importantly, progress in elucidating effector/toxin activities has required reductionist approaches and isolated biochemical or cell systems. As pathogens rely on the synchronized activities of multiple effectors, studying the mechanisms by which each effector functions individually paints an incomplete picture of the infection processes. New experimental approaches are needed to clarify the location and timing of effector activity, which will begin to elucidate how bacteria hijack signaling systems in a coordinated manner. The future understanding of these complex host-pathogen interactions will require more holistic approaches that bridge the gap between reconstitution studies in vitro, cell biology models of infection, and animal models of pathogenesis. Prior to achieving this goal, however, it remains important to elucidate the structure and enzymatic mechanisms of poorly characterized effectors and toxins, as bacterial infection is the sum of all parts of the system. Finally, the study of toxins and effectors that alter cell architecture provides significant insight into fundamental biological processes and holds great promise to reveal new leads for drug discovery and biotechnology.
BACTERIA CONVERTING ACTIN INTO PATHOGENIC SECOND MESSENGERS.
Because actin is one of the most abundant proteins in cells, it has been unclear how low concentrations of toxins or effector proteins delivered into host cytoplasm may have such a robust effect on cell architecture and function. Exciting new studies suggest that actin can be converted into a second messenger that amplifies low abundant toxin function (Heisler et al. 2015). The actin cross-linking domain (ACD) found in toxins such as the multifunctional autoprocessing repeats-in-toxin (MARTX)-containing toxins covalently cross-links actin into oligomers that are structurally incompatible for polymerization, resulting in rapid cell rounding with loss of all polymerized actin (Fullner & Mekalanos 2000, Lin et al. 1999). It was previously thought that ACD toxicity was a consequence of cross-linking the G-actin pool into polymerization-deficient oligomers (Cordero et al. 2006, Fullner & Mekalanos 2000, Lin et al. 1999). However, due to the inconsistencies between in vitro ACD activity (which is low) and cell G-actin concentrations (which are high), Heisler et al. (2015) proposed that toxicity is caused by ACD cross-linked actin oligomers targeting upstream actin regulator proteins. Indeed, they found that actin oligomers poison the formin family of actin nucleators. Therefore, bacterial toxins not only can target existing signaling networks but also can amplify pathogenic signaling cascades by generating novel toxic second messengers.
PATHOGENS TRIGGERING HOST IMMUNITY BY REGULATION OF THE ACTIN CYTOSKELETON.
Receptors that recognize conserved microbial-associated molecular patterns (MAMPs) carry out pathogen detection. As many of the classical MAMPS, such as peptidoglycan and LPS, are shared among commensals and pathogens, the mere presence of the bacterial product is not indicative of a pathogenic infection. To distinguish friend from foe, immune receptors monitor the cytosol for evidence of bacterial pathogenic activity. Recent studies suggest that the innate immune system monitors bacterial regulation of Rho GTPase activity. For example, the Salmonella GEF SopE induces NF-κB activation in a NOD1-dependent manner (Keestra et al. 2013). Normally, NOD1 and NOD2 recognize the presence of peptidoglycan in the cytosol (Keestra & Baumler 2014, Tattoli et al. 2007). However, excessive activation of Rho GTPases during Salmonella invasion may be the signal for NOD1 activation during infection. Furthermore, the Pyrin inflammasome is activated by chemical modifications of the switch I loop in Rho GTPase isoforms (Xu et al. 2014). These modifications include glycosylation by the large clostridial TcdA/B toxins, AMPylation by FIC domain–encoding effectors, ADP-ribosylation by the C2 or C3 toxin, and deamidation by an unidentified Burkholderia cenocepacia T6SS effector (Xu et al. 2014). Future work will likely reveal the mechanism behind NOD1 and Pyrin activation via Rho GTPase regulation.
ACKNOWLEDGMENTS
We would like to apologize to our colleagues whose work we failed to mention due to space limitations. A.J. is supported by the National Science Foundation Graduate Research Fellowships Program (#20133167214). Research in the Alto lab is further supported by grants from the National Institutes of Health (AI083359 and GM100486), the Welch Foundation (#I-1704), the Burroughs Wellcome Fund, and the Hartwell Foundation.
Glossary
- Globular actin (G-actin)
monomeric actin
- Filamentous actin (F-actin)
composed of a double-stranded helical polymer of actin subunits
- Nucleation-promoting factors (NPFs)
proteins (e.g., N-WASP/WAVE) that activate actin nucleators such as the Arp2/3 complex or formins
- Arp2/3 complex
a seven-subunit protein complex that creates branched actin networks by nucleating actin polymerization from the sides of existing actin filaments
- Actin-based motility
intracellular pathogens that hijack actin polymerization machinery for movement in and between cells
- Formins
actin nucleators composed of formin homology (FH2) domains that polymerize actin at the plus end of a filament
- Wasp homology 2 (WH2) domains
18-amino-acid sequences that bind to actin monomers and profilin and aid polymerization
- SPI-1 and SPI-2
Salmonella encodes two T3SSs; SPI-1 secretes effectors required for invasion, and SPI-2 secretes effectors required for SCV biogenesis
- ADF/cofilin
an actin-binding protein that disassembles actin filaments
- Gelsolin
an actin filament-severing protein
- ADP-ribosylation
the addition of ADP-ribose to target proteins
- Nicotinamide adenine dinucleotide (NAD)
the NAD+ cofactor is required for ADP-ribosylation
- CaaX motif
an amino acid sequence found at the C terminus of proteins that is prenylated and is required for membrane anchoring of Ras-family GTPases
- Tethering factors
structural proteins such as GMAP210 and GM130 that tether vesicles to organelles or maintain organelle integrity
- Myristoylation
the addition of a 14-carbon saturated fatty acid to the N-terminal glycine of proteins
- STING
an ER resident protein that is activated by cyclic dinucleotides and signals to activate the interferon response
- Legionella- containing vacuole (LCV)
exists in a cell compartment between the ER and Golgi and exhibits properties of ER membranes
- Phospholipase
an enzyme that hydrolyzes phospholipids to produce fatty acids and molecules often used in signal transduction
Footnotes
DISCLOSURE STATEMENT The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Aili M, Hallberg B, Wolf-Watz H, Rosqvist R. GAP activity of Yersinia YopE. Methods Enzymol. 2002;358:359–70. doi: 10.1016/s0076-6879(02)58102-7. [DOI] [PubMed] [Google Scholar]
- Aktories K. Bacterial protein toxins that modify host regulatory GTPases. Nat. Rev. Microbiol. 2011;9:487–98. doi: 10.1038/nrmicro2592. [DOI] [PubMed] [Google Scholar]
- Aktories K, Barmann M, Ohishi I, Tsuyama S, Jakobs KH, Habermann E. Botulinum C2 toxin ADP-ribosylates actin. Nature. 1986;322:390–92. doi: 10.1038/322390a0. [DOI] [PubMed] [Google Scholar]
- Aktories K, Wegner A. ADP-ribosylation of actin by clostridial toxins. J. Cell Biol. 1989;109:1385–87. doi: 10.1083/jcb.109.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Younes HM, Brinkmann V, Meyer TF. Interaction of Chlamydia trachomatis serovar L2 with the host autophagic pathway. Infect. Immun. 2004;72:4751–62. doi: 10.1128/IAI.72.8.4751-4762.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alto NM, Weflen AW, Rardin MJ, Yarar D, Lazar CS, et al. The type III effector EspF coordinates membrane trafficking by the spatiotemporal activation of two eukaryotic signaling pathways. J. Cell Biol. 2007;178:1265–78. doi: 10.1083/jcb.200705021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andor A, Trulzsch K, Essler M, Roggenkamp A, Wiedemann A, et al. YopE of Yersinia, a GAP for Rho GTPases, selectively modulates Rac-dependent actin structures in endothelial cells. Cell. Microbiol. 2001;3:301–10. doi: 10.1046/j.1462-5822.2001.00114.x. [DOI] [PubMed] [Google Scholar]
- Avvaru BS, Pernier J, Carlier MF. Dimeric WH2 repeats of VopF sequester actin monomers into non-nucleating linear string conformations: an X-ray scattering study. J. Struct. Biol. 2015;190:192–99. doi: 10.1016/j.jsb.2015.03.008. [DOI] [PubMed] [Google Scholar]
- Bakowski MA, Braun V, Lam GY, Yeung T, Heo WD, et al. The phosphoinositide phosphatase SopB manipulates membrane surface charge and trafficking of the Salmonella-containing vacuole. Cell Host Microbe. 2010;7:453–62. doi: 10.1016/j.chom.2010.05.011. [DOI] [PubMed] [Google Scholar]
- Benanti EL, Nguyen CM, Welch MD. Virulent Burkholderia species mimic host actin polymerases to drive actin-based motility. Cell. 2015;161:348–60. doi: 10.1016/j.cell.2015.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beron W, Gutierrez MG, Rabinovitch M, Colombo MI. Coxiella burnetii localizes in a Rab7-labeled compartment with autophagic characteristics. Infect. Immun. 2002;70:5816–21. doi: 10.1128/IAI.70.10.5816-5821.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuzon CR, Meresse S, Unsworth KE, Ruiz-Albert J, Garvis S, et al. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 2000;19:3235–49. doi: 10.1093/emboj/19.13.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmingham CL, Canadien V, Gouin E, Troy EB, Yoshimori T, et al. Listeria monocytogenes evades killing by autophagy during colonization of host cells. Autophagy. 2007;3:442–51. doi: 10.4161/auto.4450. [DOI] [PubMed] [Google Scholar]
- Birmingham CL, Jiang X, Ohlson MB, Miller SI, Brumell JH. Salmonella-induced filament formation is a dynamic phenotype induced by rapidly replicating Salmonella enterica serovar Typhimurium in epithelial cells. Infect. Immun. 2005;73:1204–8. doi: 10.1128/IAI.73.2.1204-1208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucrot E, Henry T, Borg JP, Gorvel JP, Meresse S. The intracellular fate of Salmonella depends on the recruitment of kinesin. Science. 2005;308:1174–78. doi: 10.1126/science.1110225. [DOI] [PubMed] [Google Scholar]
- Brombacher E, Urwyler S, Ragaz C, Weber SS, Kami K, et al. Rab1 guanine nucleotide exchange factor SidM is a major phosphatidylinositol 4-phosphate–binding effector protein of Legionella pneumophila. J. Biol. Chem. 2009;284:4846–56. doi: 10.1074/jbc.M807505200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwald G, Friebel A, Galan JE, Hardt WD, Wittinghofer A, Scheffzek K. Structural basis for the reversible activation of a Rho protein by the bacterial toxin SopE. EMBO J. 2002;21:3286–95. doi: 10.1093/emboj/cdf329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette DL, Seemann J, Orth K. Vibrio VopQ induces PI3-kinase-independent autophagy and antagonizes phagocytosis. Mol. Microbiol. 2009;73:639–49. doi: 10.1111/j.1365-2958.2009.06798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette DL, Yarbrough ML, Orvedahl A, Gilpin CJ, Orth K. Vibrio parahaemolyticus orchestrates a multifaceted host cell infection by induction of autophagy, cell rounding, and then cell lysis. PNAS. 2008;105:12497–502. doi: 10.1073/pnas.0802773105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnaevskiy N, Fox TG, Plymire DA, Ertelt JM, Weigele BA, et al. Proteolytic elimination of N-myristoyl modifications by the Shigella virulence factor IpaJ. Nature. 2013;496:106–9. doi: 10.1038/nature12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnaevskiy N, Peng T, Reddick LE, Hang HC, Alto NM. Myristoylome profiling reveals a concerted mechanism of ARF GTPase deacylation by the bacterial protease IpaJ. Mol. Cell. 2015;58:110–22. doi: 10.1016/j.molcel.2015.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case ED, Chong A, Wehrly TD, Hansen B, Child R, et al. The Francisella O-antigen mediates survival in the macrophage cytosol via autophagy avoidance. Cell. Microbiol. 2014;16:862–77. doi: 10.1111/cmi.12246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Myeni SK, Lin TL, Wu CC, Staiger CJ, Zhou D. SipC multimerization promotes actin nucleation and contributes to Salmonella-induced inflammation. Mol. Microbiol. 2007;66:1548–56. doi: 10.1111/j.1365-2958.2007.06024.x. [DOI] [PubMed] [Google Scholar]
- Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 2013;93:269–309. doi: 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
- Choy A, Dancourt J, Mugo B, O'Connor TJ, Isberg RR, et al. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science. 2012;338:1072–76. doi: 10.1126/science.1227026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen M, Coye LH, Hontz JS, LaRock DL, Pfuetzner RA, et al. Activation of a bacterial virulence protein by the GTPase RhoA. Sci. Signal. 2009;2:ra71. doi: 10.1126/scisignal.2000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements A, Smollett K, Lee SF, Hartland EL, Lowe M, Frankel G. EspG of enteropathogenic and enterohemorrhagic E. coli binds the Golgi matrix protein GM130 and disrupts the Golgi structure and function. Cell. Microbiol. 2011;13:1429–39. doi: 10.1111/j.1462-5822.2011.01631.x. [DOI] [PubMed] [Google Scholar]
- Comerci DJ, Martinez-Lorenzo MJ, Sieira R, Gorvel JP, Ugalde RA. Essential role of the VirB machinery in the maturation of the Brucella abortus–containing vacuole. Cell. Microbiol. 2001;3:159–68. doi: 10.1046/j.1462-5822.2001.00102.x. [DOI] [PubMed] [Google Scholar]
- Cordero CL, Kudryashov DS, Reisler E, Satchell KJ. The actin cross-linking domain of the Vibrio cholerae RTX toxin directly catalyzes the covalent cross-linking of actin. J. Biol. Chem. 2006;281:32366–74. doi: 10.1074/jbc.M605275200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa TR, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, et al. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat. Rev. Microbiol. 2015;13:343–59. doi: 10.1038/nrmicro3456. [DOI] [PubMed] [Google Scholar]
- de Figueiredo P, Ficht TA, Rice-Ficht A, Rossetti CA, Adams LG. Pathogenesis and immunobiology of brucellosis: review of Brucella-host interactions. Am. J. Pathol. 2015;185:1505–17. doi: 10.1016/j.ajpath.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean P. Functional domains and motifs of bacterial type III effector proteins and their roles in infection. FEMS Microbiol. Rev. 2011;35:1100–25. doi: 10.1111/j.1574-6976.2011.00271.x. [DOI] [PubMed] [Google Scholar]
- Deiwick J, Salcedo SP, Boucrot E, Gilliland SM, Henry T, et al. The translocated Salmonella effector proteins SseF and SseG interact and are required to establish an intracellular replication niche. Infect. Immun. 2006;74:6965–72. doi: 10.1128/IAI.00648-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbs N, Burnaevskiy N, Chen D, Gonugunta VK, Alto NM, Yan N. STING activation by translocation from the ER is associated with infection and autoinflammatory disease. Cell Host Microbe. 2015;18:157–68. doi: 10.1016/j.chom.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues L, Holden DW, Mota LJ. The Salmonella effector SteA contributes to the control of membrane dynamics of Salmonella-containing vacuoles. Infect. Immun. 2014;82:2923–34. doi: 10.1128/IAI.01385-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat. Rev. Mol. Cell Biol. 2011;12:362–75. doi: 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N, Zhu Y, Lu Q, Hu L, Zheng Y, Shao F. Structurally distinct bacterial TBC-like GAPs link Arf GTPase to Rab1 inactivation to counteract host defenses. Cell. 2012;150:1029–41. doi: 10.1016/j.cell.2012.06.050. [DOI] [PubMed] [Google Scholar]
- Dorn BR, Dunn WA, Jr, Progulske-Fox A. Porphyromonas gingivalis traffics to autophagosomes in human coronary artery endothelial cells. Infect. Immun. 2001;69:5698–708. doi: 10.1128/IAI.69.9.5698-5708.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dortet L, Mostowy S, Samba-Louaka A, Gouin E, Nahori MA, et al. Recruitment of the major vault protein by InlK: a Listeria monocytogenes strategy to avoid autophagy. PLOS Pathog. 2011;7:e1002168. doi: 10.1371/journal.ppat.1002168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druar C, Yu F, Barnes JL, Okinaka RT, Chantratita N, et al. Evaluating Burkholderia pseudomallei Bip proteins as vaccines and Bip antibodies as detection agents. FEMS Immunol. Med. Microbiol. 2008;52:78–87. doi: 10.1111/j.1574-695X.2007.00345.x. [DOI] [PubMed] [Google Scholar]
- Elliott SJ, Krejany EO, Mellies JL, Robins-Browne RM, Sasakawa C, Kaper JB. EspG, a novel type III system–secreted protein from enteropathogenic Escherichia coli with similarities to VirA of Shigella flexneri. Infect. Immun. 2001;69:4027–33. doi: 10.1128/IAI.69.6.4027-4033.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatau G, Lemichez E, Gauthier M, Chardin P, Paris S, et al. Toxin-induced activation of the G protein p21 Rho by deamidation of glutamine. Nature. 1997;387:729–33. doi: 10.1038/42743. [DOI] [PubMed] [Google Scholar]
- Fleiszig SM, Wiener-Kronish JP, Miyazaki H, Vallas V, Mostov KE, et al. Pseudomonas aeruginosa–mediated cytotoxicity and invasion correlate with distinct genotypes at the loci encoding exoenzyme S. Infect. Immun. 1997;65:579–86. doi: 10.1128/iai.65.2.579-586.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebel A, Ilchmann H, Aepfelbacher M, Ehrbar K, Machleidt W, Hardt WD. SopE and SopE2 from Salmonella typhimurium activate different sets of RhoGTPases of the host cell. J. Biol. Chem. 2001;276:34035–40. doi: 10.1074/jbc.M100609200. [DOI] [PubMed] [Google Scholar]
- Fu Y, Galan JE. A Salmonella protein antagonizes Rac-1 and Cdc42 to mediate host-cell recovery after bacterial invasion. Nature. 1999;401:293–97. doi: 10.1038/45829. [DOI] [PubMed] [Google Scholar]
- Fullner KJ, Mekalanos JJ. In vivo covalent cross-linking of cellular actin by the Vibrio cholerae RTX toxin. EMBO J. 2000;19:5315–23. doi: 10.1093/emboj/19.20.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, VanLoock MS, Zhou D, Galan JE, Egelman EH. The bacterial protein SipA polymerizes G-actin and mimics muscle nebulin. Nat. Struct. Biol. 2002;9:518–21. doi: 10.1038/nsb811. [DOI] [PubMed] [Google Scholar]
- Ghigo E, Colombo MI, Heinzen RA. The Coxiella burnetii parasitophorous vacuole. Adv. Exp. Med. Biol. 2012;984:141–69. doi: 10.1007/978-94-007-4315-1_8. [DOI] [PubMed] [Google Scholar]
- Glotfelty LG, Hecht GA. Enteropathogenic E. coli effectors EspG1/G2 disrupt tight junctions: new roles and mechanisms. Ann. N. Y. Acad. Sci. 2012;1258:149–58. doi: 10.1111/j.1749-6632.2012.06563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MB, Theriot JA. Shigella flexneri surface protein IcsA is sufficient to direct actin-based motility. PNAS. 1995;92:6572–76. doi: 10.1073/pnas.92.14.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goody PR, Heller K, Oesterlin LK, Muller MP, Itzen A, Goody RS. Reversible phosphocholination of Rab proteins by Legionella pneumophila effector proteins. EMBO J. 2012;31:1774–84. doi: 10.1038/emboj.2012.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez MG, Vazquez CL, Munafo DB, Zoppino FC, Beron W, et al. Autophagy induction favours the generation and maturation of the Coxiella-replicative vacuoles. Cell. Microbiol. 2005;7:981–93. doi: 10.1111/j.1462-5822.2005.00527.x. [DOI] [PubMed] [Google Scholar]
- Haglund CM, Choe JE, Skau CT, Kovar DR, Welch MD. Rickettsia Sca2 is a bacterial formin-like mediator of actin-based motility. Nat. Cell Biol. 2010;12:1057–63. doi: 10.1038/ncb2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Craig JA, Putnam CD, Carozzi NB, Tainer JA. Evolution and mechanism from structures of an ADP-ribosylating toxin and NAD complex. Nat. Struct. Biol. 1999;6:932–36. doi: 10.1038/13300. [DOI] [PubMed] [Google Scholar]
- Hardt WD, Chen LM, Schuebel KE, Bustelo XR, Galan JE. S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell. 1998;93:815–26. doi: 10.1016/s0092-8674(00)81442-7. [DOI] [PubMed] [Google Scholar]
- Hayes CS, Aoki SK, Low DA. Bacterial contact–dependent delivery systems. Annu. Rev. Genet. 2010;44:71–90. doi: 10.1146/annurev.genet.42.110807.091449. [DOI] [PubMed] [Google Scholar]
- Hayward RD, Koronakis V. Direct nucleation and bundling of actin by the SipC protein of invasive Salmonella. EMBO J. 1999;18:4926–34. doi: 10.1093/emboj/18.18.4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler DB, Kudryashova E, Grinevich DO, Suarez C, Winkelman JD, et al. ACD toxin–produced actin oligomers poison formin-controlled actin polymerization. Science. 2015;349:535–39. doi: 10.1126/science.aab4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry T, Couillault C, Rockenfeller P, Boucrot E, Dumont A, et al. The Salmonella effector protein PipB2 is a linker for kinesin-1. PNAS. 2006;103:13497–502. doi: 10.1073/pnas.0605443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa N, Toma C, Koizumi Y, Nakasone N, Nohara T, et al. Vibrio parahaemolyticus effector proteins suppress inflammasome activation by interfering with host autophagy signaling. PLOS Pathog. 2013;9:e1003142. doi: 10.1371/journal.ppat.1003142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyoshi H, Kodama T, Saito K, Gotoh K, Matsuda S, et al. VopV, an F-actin-binding type III secretion effector, is required for Vibrio parahaemolyticus–induced enterotoxicity. Cell Host Microbe. 2011;10:401–9. doi: 10.1016/j.chom.2011.08.014. [DOI] [PubMed] [Google Scholar]
- Hoffman GR, Nassar N, Cerione RA. Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell. 2000;100:345–56. doi: 10.1016/s0092-8674(00)80670-4. [DOI] [PubMed] [Google Scholar]
- Horenkamp FA, Mukherjee S, Alix E, Schauder CM, Hubber AM, et al. Legionella pneumophila subversion of host vesicular transport by SidC effector proteins. Traffic. 2014;15:488–99. doi: 10.1111/tra.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F, Luo X, Qiu J, Teng YB, Jin J, et al. The Legionella effector SidC defines a unique family of ubiquitin ligases important for bacterial phagosomal remodeling. PNAS. 2014;111:10538–43. doi: 10.1073/pnas.1402605111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Birmingham CL, Shahnazari S, Shiu J, Zheng YT, et al. Antibacterial autophagy occurs at PI3P-enriched domains of the endoplasmic reticulum and requires Rab1 GTPase. Autophagy. 2011;7:17–26. doi: 10.4161/auto.7.1.13840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 2011;91:119–49. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juris SJ, Rudolph AE, Huddler D, Orth K, Dixon JE. A distinctive role for the Yersinia protein kinase: actin binding, kinase activation, and cytoskeleton disruption. PNAS. 2000;97:9431–36. doi: 10.1073/pnas.170281997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Just I, Selzer J, Jung M, van Damme J, Vandekerckhove J, Aktories K. Rho-ADP-ribosylating exoenzyme from Bacillus cereus. Purification, characterization, and identification of the NAD-binding site. Biochemistry. 1995;34:334–40. doi: 10.1021/bi00001a041. [DOI] [PubMed] [Google Scholar]
- Kahn RA. Toward a model for Arf GTPases as regulators of traffic at the Golgi. FEBS Lett. 2009;583:3872–79. doi: 10.1016/j.febslet.2009.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniga K, Trollinger D, Galan JE. Identification of two targets of the type III protein secretion system encoded by the inv and spa loci of Salmonella typhimurium that have homology to the Shigella IpaD and IpaA proteins. J. Bacteriol. 1995a;177:7078–85. doi: 10.1128/jb.177.24.7078-7085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniga K, Tucker S, Trollinger D, Galan JE. Homologs of the Shigella IpaB and IpaC invasins are required for Salmonella typhimurium entry into cultured epithelial cells. J. Bacteriol. 1995b;177:3965–71. doi: 10.1128/jb.177.14.3965-3971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keestra AM, Baumler AJ. Detection of enteric pathogens by the nodosome. Trends Immunol. 2014;35:123–30. doi: 10.1016/j.it.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keestra AM, Winter MG, Auburger JJ, Frassle SP, Xavier MN, et al. Manipulation of small Rho GTPases is a pathogen-induced process detected by NOD1. Nature. 2013;496:233–37. doi: 10.1038/nature12025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, An DR, Song J, Yoon JY, Kim HS, et al. Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7. PNAS. 2012;109:7729–34. doi: 10.1073/pnas.1120251109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingdon HS, Shapiro BM, Stadtman ER. Regulation of glutamine synthetase. 8. ATP: glutamine synthetase adenylyltransferase, an enzyme that catalyzes alterations in the regulatory properties of glutamine synthetase. PNAS. 1967;58:1703–10. doi: 10.1073/pnas.58.4.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler LA, Steele-Mortimer O. The Salmonella effector PipB2 affects late endosome/lysosome distribution to mediate Sif extension. Mol. Biol. Cell. 2005;16:4108–23. doi: 10.1091/mbc.E05-04-0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger V, Liebl D, Zhang Y, Rajashekar R, Chlanda P, et al. Reorganization of the endosomal system in Salmonella-infected cells: the ultrastructure of Salmonella-induced tubular compartments. PLOS Pathog. 2014;10:e1004374. doi: 10.1371/journal.ppat.1004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubori T, Galan JE. Temporal regulation of Salmonella virulence effector function by proteasome-dependent protein degradation. Cell. 2003;115:333–42. doi: 10.1016/s0092-8674(03)00849-3. [DOI] [PubMed] [Google Scholar]
- Kuhle V, Abrahams GL, Hensel M. Intracellular Salmonella enterica redirect exocytic transport processes in a Salmonella pathogenicity island 2–dependent manner. Traffic. 2006;7:716–30. doi: 10.1111/j.1600-0854.2006.00422.x. [DOI] [PubMed] [Google Scholar]
- Lang AE, Schmidt G, Schlosser A, Hey TD, Larrinua IM, et al. Photorhabdus luminescens toxins ADP-ribosylate actin and RhoA to force actin clustering. Science. 2010;327:1139–42. doi: 10.1126/science.1184557. [DOI] [PubMed] [Google Scholar]
- Lerena MC, Colombo MI. Mycobacterium marinum induces a marked LC3 recruitment to its containing phagosome that depends on a functional ESX-1 secretion system. Cell. Microbiol. 2011;13:814–35. doi: 10.1111/j.1462-5822.2011.01581.x. [DOI] [PubMed] [Google Scholar]
- Lin W, Fullner KJ, Clayton R, Sexton JA, Rogers MB, et al. Identification of a Vibrio cholerae RTX toxin gene cluster that is tightly linked to the cholera toxin prophage. PNAS. 1999;96:1071–76. doi: 10.1073/pnas.96.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liverman AD, Cheng HC, Trosky JE, Leung DW, Yarbrough ML, et al. Arp2/3-independent assembly of actin by Vibrio type III effector VopL. PNAS. 2007;104:17117–22. doi: 10.1073/pnas.0703196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401:613–16. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- Lossi NS, Rolhion N, Magee AI, Boyle C, Holden DW. The Salmonella SPI-2 effector SseJ exhibits eukaryotic activator-dependent phospholipase A and glycerophospholipid: cholesterol acyltransferase activity. Microbiology. 2008;154:2680–88. doi: 10.1099/mic.0.2008/019075-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Atkinson SJ, Ampe C, Vandekerckhove J, Pollard TD. Purification of a cortical complex containing two unconventional actins from Acanthamoeba by affinity chromatography on profilin-agarose. J. Cell Biol. 1994;127:107–15. doi: 10.1083/jcb.127.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machner MP, Isberg RR. A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science. 2007;318:974–77. doi: 10.1126/science.1149121. [DOI] [PubMed] [Google Scholar]
- Madasu Y, Suarez C, Kast DJ, Kovar DR, Dominguez R. Rickettsia Sca2 has evolved formin-like activity through a different molecular mechanism. PNAS. 2013;110:E2677–86. doi: 10.1073/pnas.1307235110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan DG, Popovic D, Gubas A, Terawaki S, Suzuki H, et al. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol. Cell. 2015a;57:39–54. doi: 10.1016/j.molcel.2014.11.006. [DOI] [PubMed] [Google Scholar]
- McEwan DG, Richter B, Claudi B, Wigge C, Wild P, et al. PLEKHM1 regulates Salmonella-containing vacuole biogenesis and infection. Cell Host Microbe. 2015b;17:58–71. doi: 10.1016/j.chom.2014.11.011. [DOI] [PubMed] [Google Scholar]
- McGhie EJ, Hayward RD, Koronakis V. Cooperation between actin-binding proteins of invasive Salmonella: SipA potentiates SipC nucleation and bundling of actin. EMBO J. 2001;20:2131–39. doi: 10.1093/emboj/20.9.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhie EJ, Hayward RD, Koronakis V. Control of actin turnover by a Salmonella invasion protein. Mol. Cell. 2004;13:497–510. doi: 10.1016/s1097-2765(04)00053-x. [DOI] [PubMed] [Google Scholar]
- McGourty K, Thurston TL, Matthews SA, Pinaud L, Mota LJ, Holden DW. Salmonella inhibits retrograde trafficking of mannose-6-phosphate receptors and lysosome function. Science. 2012;338:963–67. doi: 10.1126/science.1227037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesquita FS, Thomas M, Sachse M, Santos AJ, Figueira R, Holden DW. The Salmonella deubiquitinase SseL inhibits selective autophagy of cytosolic aggregates. PLOS Pathog. 2012;8:e1002743. doi: 10.1371/journal.ppat.1002743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T, Akiba K, Iguchi M, Danbara H, Okada N. The Chromobacterium violaceum type III effector CopE, a guanine nucleotide exchange factor for Rac1 and Cdc42, is involved in bacterial invasion of epithelial cells and pathogenesis. Mol. Microbiol. 2011;80:1186–203. doi: 10.1111/j.1365-2958.2011.07637.x. [DOI] [PubMed] [Google Scholar]
- Mohr C, Koch G, Just I, Aktories K. ADP-ribosylation by Clostridium botulinum C3 exoenzyme increases steady-state GTPase activities of recombinant rhoA and rhoB proteins. FEBS Lett. 1992;297:95–99. doi: 10.1016/0014-5793(92)80335-e. [DOI] [PubMed] [Google Scholar]
- Mostowy S, Bonazzi M, Hamon MA, Tham TN, Mallet A, et al. Entrapment of intracytosolic bacteria by septin cage-like structures. Cell Host Microbe. 2010;8:433–44. doi: 10.1016/j.chom.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Linstedt AD. Retrograde trafficking of AB5 toxins: mechanisms to therapeutics. J. Mol. Med. 2013;91:1131–41. doi: 10.1007/s00109-013-1048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller MP, Peters H, Blumer J, Blankenfeldt W, Goody RS, Itzen A. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science. 2010;329:946–49. doi: 10.1126/science.1192276. [DOI] [PubMed] [Google Scholar]
- Myeni SK, Zhou D. The C terminus of SipC binds and bundles F-actin to promote Salmonella invasion. J. Biol. Chem. 2010;285:13357–63. doi: 10.1074/jbc.M109.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahama M, Ohkubo A, Oda M, Kobayashi K, Amimoto K, et al. Clostridium perfringens TpeL glycosylates the Rac and Ras subfamily proteins. Infect. Immun. 2011;79:905–10. doi: 10.1128/IAI.01019-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namgoong S, Boczkowska M, Glista MJ, Winkelman JD, Rebowski G, et al. Mechanism of actin filament nucleation by Vibrio VopL and implications for tandem W domain nucleation. Nat. Struct. Mol. Biol. 2011;18:1060–67. doi: 10.1038/nsmb.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawabi P, Catron DM, Haldar K. Esterification of cholesterol by a type III secretion effector during intracellular Salmonella infection. Mol. Microbiol. 2008;68:173–85. doi: 10.1111/j.1365-2958.2008.06142.x. [DOI] [PubMed] [Google Scholar]
- Neunuebel MR, Chen Y, Gaspar AH, Backlund PS, Jr., Yergey A, Machner MP. De-AMPylation of the small GTPase Rab1 by the pathogen Legionella pneumophila. Science. 2011;333:453–56. doi: 10.1126/science.1207193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves D, Job V, Dortet L, Cossart P, Dessen A. Structure of internalin InlK from the human pathogen Listeria monocytogenes. J. Mol. Biol. 2013;425:4520–29. doi: 10.1016/j.jmb.2013.08.010. [DOI] [PubMed] [Google Scholar]
- Newton HJ, Kohler LJ, McDonough JA, Temoche-Diaz M, Crabill E, et al. A screen of Coxiella burnetii mutants reveals important roles for Dot/Icm effectors and host autophagy in vacuole biogenesis. PLOS Pathog. 2014;10:e1004286. doi: 10.1371/journal.ppat.1004286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura M, Fujii T, Hiyoshi H, Makino F, Inoue H, et al. A repeat unit of Vibrio diarrheal T3S effector subverts cytoskeletal actin homeostasis via binding to interstrand region of actin filaments. Sci. Rep. 2015;5:10870. doi: 10.1038/srep10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–31. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- Ohlson MB, Huang Z, Alto NM, Blanc MP, Dixon JE, et al. Structure and function of Salmonella SifA indicate that its interactions with SKIP, SseJ, and RhoA family GTPases induce endosomal tubulation. Cell Host Microbe. 2008;4:434–46. doi: 10.1016/j.chom.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada R, Zhou X, Hiyoshi H, Matsuda S, Chen X, et al. The Vibrio parahaemolyticus effector VopC mediates Cdc42-dependent invasion of cultured cells but is not required for pathogenicity in an animal model of infection. Cell. Microbiol. 2014;16:938–47. doi: 10.1111/cmi.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orchard RC, Kittisopikul M, Altschuler SJ, Wu LF, Suel GM, Alto NM. Identification of F-actin as the dynamic hub in a microbial-induced GTPase polarity circuit. Cell. 2012;148:803–15. doi: 10.1016/j.cell.2011.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MK. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433:488–94. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- Otto H, Tezcan-Merdol D, Girisch R, Haag F, Rhen M, Koch-Nolte F. The spvB gene-product of the Salmonella enterica virulence plasmid is a mono(ADP-ribosyl)transferase. Mol. Microbiol. 2000;37:1106–15. doi: 10.1046/j.1365-2958.2000.02064.x. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 2000;29:545–76. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- Popoff MR, Rubin EJ, Gill DM, Boquet P. Actin-specific ADP-ribosyltransferase produced by a Clostridium difficile strain. Infect. Immun. 1988;56:2299–306. doi: 10.1128/iai.56.9.2299-2306.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prashar A, Terebiznik MR. Legionella pneumophila: homeward bound away from the phagosome. Curr. Opin. Microbiol. 2015;23:86–93. doi: 10.1016/j.mib.2014.11.008. [DOI] [PubMed] [Google Scholar]
- Prehna G, Ivanov MI, Bliska JB, Stebbins CE. Yersinia virulence depends on mimicry of host Rho-family nucleotide dissociation inhibitors. Cell. 2006;126:869–80. doi: 10.1016/j.cell.2006.06.056. [DOI] [PubMed] [Google Scholar]
- Pujol C, Klein KA, Romanov GA, Palmer LE, Cirota C, et al. Yersinia pestis can reside in autophagosomes and avoid xenophagy in murine macrophages by preventing vacuole acidification. Infect. Immun. 2009;77:2251–61. doi: 10.1128/IAI.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju D, Hussey S, Ang M, Terebiznik MR, Sibony M, et al. Vacuolating cytotoxin and variants in Atg16L1 that disrupt autophagy promote Helicobacter pylori infection in humans. Gastroenterology. 2012;142:1160–71. doi: 10.1053/j.gastro.2012.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed SC, Lamason RL, Risca VI, Abernathy E, Welch MD. Rickettsia actin-based motility occurs in distinct phases mediated by different actin nucleators. Curr. Biol. 2014;24:98–103. doi: 10.1016/j.cub.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, et al. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–31. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- Romano PS, Gutierrez MG, Beron W, Rabinovitch M, Colombo MI. The autophagic pathway is actively modulated by phase II Coxiella burnetii to efficiently replicate in the host cell. Cell. Microbiol. 2007;9:891–909. doi: 10.1111/j.1462-5822.2006.00838.x. [DOI] [PubMed] [Google Scholar]
- Roy CR, Cherfils J. Structure and function of Fic proteins. Nat. Rev. Microbiol. 2015;13:631–40. doi: 10.1038/nrmicro3520. [DOI] [PubMed] [Google Scholar]
- Rudolph MG, Weise C, Mirold S, Hillenbrand B, Bader B, et al. Biochemical analysis of SopE from Salmonella typhimurium, a highly efficient guanosine nucleotide exchange factor for RhoGTPases. J. Biol. Chem. 1999;274:30501–9. doi: 10.1074/jbc.274.43.30501. [DOI] [PubMed] [Google Scholar]
- Ruiz-Albert J, Yu XJ, Beuzon CR, Blakey AN, Galyov EE, Holden DW. Complementary activities of SseJ and SifA regulate dynamics of the Salmonella typhimurium vacuolar membrane. Mol. Microbiol. 2002;44:645–61. doi: 10.1046/j.1365-2958.2002.02912.x. [DOI] [PubMed] [Google Scholar]
- Salcedo SP, Holden DW. SseG, a virulence protein that targets Salmonella to the Golgi network. EMBO J. 2003;22:5003–14. doi: 10.1093/emboj/cdg517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallee NA, Rivera GM, Dueber JE, Vasilescu D, Mullins RD, et al. The pathogen protein EspFU hijacks actin polymerization using mimicry and multivalency. Nature. 2008;454:1005–8. doi: 10.1038/nature07170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffzek K, Stephan I, Jensen ON, Illenberger D, Gierschik P. The Rac-RhoGDI complex and the structural basis for the regulation of Rho proteins by RhoGDI. Nat. Struct. Biol. 2000;7:122–26. doi: 10.1038/72392. [DOI] [PubMed] [Google Scholar]
- Schmidt G, Sehr P, Wilm M, Selzer J, Mann M, Aktories K. Gln 63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor-1. Nature. 1997;387:725–29. doi: 10.1038/42735. [DOI] [PubMed] [Google Scholar]
- Schroeder N, Henry T, de Chastellier C, Zhao W, Guilhon AA, et al. The virulence protein SopD2 regulates membrane dynamics of Salmonella-containing vacuoles. PLOS Pathog. 2010;6:e1001002. doi: 10.1371/journal.ppat.1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder N, Mota LJ, Meresse S. Salmonella-induced tubular networks. Trends Microbiol. 2011;19:268–77. doi: 10.1016/j.tim.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Schuerch DW, Wilson-Kubalek EM, Tweten RK. Molecular basis of listeriolysin O pH dependence. PNAS. 2005;102:12537–42. doi: 10.1073/pnas.0500558102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehr P, Joseph G, Genth H, Just I, Pick E, Aktories K. Glucosylation and ADP ribosylation of Rho proteins: effects on nucleotide binding, GTPase activity, and effector coupling. Biochemistry. 1998;37:5296–304. doi: 10.1021/bi972592c. [DOI] [PubMed] [Google Scholar]
- Selyunin AS, Reddick LE, Weigele BA, Alto NM. Selective protection of an ARF1-GTP signaling axis by a bacterial scaffold induces bidirectional trafficking arrest. Cell Rep. 2014;6:878–91. doi: 10.1016/j.celrep.2014.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selyunin AS, Sutton SE, Weigele BA, Reddick LE, Orchard RC, et al. The assembly of a GTPase-kinase signalling complex by a bacterial catalytic scaffold. Nature. 2011;469:107–11. doi: 10.1038/nature09593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahnazari S, Namolovan A, Mogridge J, Kim PK, Brumell JH. Bacterial toxins can inhibit host cell autophagy through cAMP generation. Autophagy. 2011;7:957–65. doi: 10.4161/auto.7.9.16435. [DOI] [PubMed] [Google Scholar]
- Shao F, Merritt PM, Bao Z, Innes RW, Dixon JE. A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell. 2002;109:575–88. doi: 10.1016/s0092-8674(02)00766-3. [DOI] [PubMed] [Google Scholar]
- Shin DM, Jeon BY, Lee HM, Jin HS, Yuk JM, et al. Mycobacterium tuberculosis Eis regulates autophagy, inflammation, and cell death through redox-dependent signaling. PLOS Pathog. 2010;6:e1001230. doi: 10.1371/journal.ppat.1001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson LL, Stiles BG, Zepeda H, Wilkins TD. Production by Clostridium spiroforme of an iota-like toxin that possesses mono(ADP-ribosyl)transferase activity: identification of a novel class of ADP-ribosyltransferases. Infect. Immun. 1989;57:255–61. doi: 10.1128/iai.57.1.255-261.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiering D, Hodgson L. Dynamics of the Rho-family small GTPases in actin regulation and motility. Cell Adh. Migr. 2011;5:170–80. doi: 10.4161/cam.5.2.14403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreelatha A, Bennett TL, Zheng H, Jiang QX, Orth K, Starai VJ. Vibrio effector protein, VopQ, forms a lysosomal gated channel that disrupts host ion homeostasis and autophagic flux. PNAS. 2013;110:11559–64. doi: 10.1073/pnas.1307032110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm LM, Morisaki JH, Gao LY, Jeng RL, McDonald KL, et al. Mycobacterium marinum escapes from phagosomes and is propelled by actin-based motility. J. Exp. Med. 2003;198:1361–68. doi: 10.1084/jem.20031072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T, Child R, Wehrly TD, Hansen B, Hwang S, et al. Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe. 2012;11:33–45. doi: 10.1016/j.chom.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins CE, Galan JE. Modulation of host signaling by a bacterial mimic: structure of the Salmonella effector SptP bound to Rac1. Mol. Cell. 2000;6:1449–60. doi: 10.1016/s1097-2765(00)00141-6. [DOI] [PubMed] [Google Scholar]
- Stender S, Friebel A, Linder S, Rohde M, Mirold S, Hardt WD. Identification of SopE2 from Salmonella typhimurium, a conserved guanine nucleotide exchange factor for Cdc42 of the host cell. Mol. Microbiol. 2000;36:1206–21. doi: 10.1046/j.1365-2958.2000.01933.x. [DOI] [PubMed] [Google Scholar]
- Stevens MP, Friebel A, Taylor LA, Wood MW, Brown PJ, et al. A Burkholderia pseudomallei type III secreted protein, BopE, facilitates bacterial invasion of epithelial cells and exhibits guanine nucleotide exchange factor activity. J. Bacteriol. 2003;185:4992–96. doi: 10.1128/JB.185.16.4992-4996.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles BG, Wilkins TD. Clostridium perfringens iota toxin: synergism between two proteins. Toxicon. 1986;24:767–73. doi: 10.1016/0041-0101(86)90101-7. [DOI] [PubMed] [Google Scholar]
- Sundin C, Henriksson ML, Hallberg B, Forsberg A, Frithz-Lindsten E. Exoenzyme T of Pseudomonas aeruginosa elicits cytotoxicity without interfering with Ras signal transduction. Cell. Microbiol. 2001;3:237–46. doi: 10.1046/j.1462-5822.2001.00108.x. [DOI] [PubMed] [Google Scholar]
- Tan Y, Arnold RJ, Luo ZQ. Legionella pneumophila regulates the small GTPase Rab1 activity by reversible phosphorylcholination. PNAS. 2011;108:21212–17. doi: 10.1073/pnas.1114023109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, Luo ZQ. Legionella pneumophila SidD is a deAMPylase that modifies Rab1. Nature. 2011;475:506–9. doi: 10.1038/nature10307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattoli I, Sorbara MT, Yang C, Tooze SA, Philpott DJ, Girardin SE. Listeria phospholipases subvert host autophagic defenses by stalling pre-autophagosomal structures. EMBO J. 2013;32:3066–78. doi: 10.1038/emboj.2013.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattoli I, Travassos LH, Carneiro LA, Magalhaes JG, Girardin SE. The Nodosome: Nod1 and Nod2 control bacterial infections and inflammation. Semin. Immunopathol. 2007;29:289–301. doi: 10.1007/s00281-007-0083-2. [DOI] [PubMed] [Google Scholar]
- Thomas M, Mesquita FS, Holden DW. The DUB-ious lack of ALIS in Salmonella infection: A Salmonella deubiquitinase regulates the autophagy of protein aggregates. Autophagy. 2012;8:1824–26. doi: 10.4161/auto.21742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Engelenburg SB, Palmer AE. Quantification of real-time Salmonella effector type III secretion kinetics reveals differential secretion rates for SopE2 and SptP. Chem. Biol. 2008;15:619–28. doi: 10.1016/j.chembiol.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schaik EJ, Chen C, Mertens K, Weber MM, Samuel JE. Molecular pathogenesis of the obligate intracellular bacterium Coxiella burnetii. Nat. Rev. Microbiol. 2013;11:561–73. doi: 10.1038/nrmicro3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez CL, Colombo MI. Coxiella burnetii modulates Beclin 1 and Bcl-2, preventing host cell apoptosis to generate a persistent bacterial infection. Cell Death Differ. 2010;17:421–38. doi: 10.1038/cdd.2009.129. [DOI] [PubMed] [Google Scholar]
- von Eichel–Streiber C, Boquet P, Sauerborn M, Thelestam M. Large clostridial cytotoxins—a family of glycosyltransferases modifying small GTP-binding proteins. Trends Microbiol. 1996;4:375–82. doi: 10.1016/0966-842X(96)10061-5. [DOI] [PubMed] [Google Scholar]
- Welch MD, Iwamatsu A, Mitchison TJ. Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature. 1997;385:265–69. doi: 10.1038/385265a0. [DOI] [PubMed] [Google Scholar]
- Wilde C, Chhatwal GS, Schmalzing G, Aktories K, Just I. A novel C3-like ADP-ribosyltransferase from Staphylococcus aureus modifying RhoE and Rnd3. J. Biol. Chem. 2001;276:9537–42. doi: 10.1074/jbc.M011035200. [DOI] [PubMed] [Google Scholar]
- Winchell CG, Graham JG, Kurten RC, Voth DE. Coxiella burnetii type IV secretion-dependent recruitment of macrophage autophagosomes. Infect. Immun. 2014;82:2229–38. doi: 10.1128/IAI.01236-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnen B, Schlumberger MC, Sturm A, Schupbach K, Siebenmann S, et al. Hierarchical effector protein transport by the Salmonella Typhimurium SPI-1 type III secretion system. PLOS ONE. 2008;3:e2178. doi: 10.1371/journal.pone.0002178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Yang J, Gao W, Li L, Li P, et al. Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature. 2014;513:237–41. doi: 10.1038/nature13449. [DOI] [PubMed] [Google Scholar]
- Yang J, Zhang Z, Roe SM, Marshall CJ, Barford D. Activation of Rho GTPases by DOCK exchange factors is mediated by a nucleotide sensor. Science. 2009;325:1398–402. doi: 10.1126/science.1174468. [DOI] [PubMed] [Google Scholar]
- Yarbrough ML, Li Y, Kinch LN, Grishin NV, Ball HL, Orth K. AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science. 2009;323:269–72. doi: 10.1126/science.1166382. [DOI] [PubMed] [Google Scholar]
- Yasir M, Pachikara ND, Bao X, Pan Z, Fan H. Regulation of chlamydial infection by host autophagy and vacuolar ATPase-bearing organelles. Infect. Immun. 2011;79:4019–28. doi: 10.1128/IAI.05308-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat. Cell Biol. 2009;11:1233–40. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- Yu B, Cheng HC, Brautigam CA, Tomchick DR, Rosen MK. Mechanism of actin filament nucleation by the bacterial effector VopL. Nat. Struct. Mol. Biol. 2011;18:1068–74. doi: 10.1038/nsmb.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Martins IR, Li P, Amarasinghe GK, Umetani J, et al. Structural and energetic mechanisms of cooperative autoinhibition and activation of Vav1. Cell. 2010;140:246–56. doi: 10.1016/j.cell.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Fang L, Zhang Y, Sheng H, Fang W. VgrG2 of type VI secretion system 2 of Vibrio parahaemolyticus induces autophagy in macrophages. Front. Microbiol. 2015;6:168. doi: 10.3389/fmicb.2015.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahm JA, Padrick SB, Chen Z, Pak CW, Yunus AA, et al. The bacterial effector VopL organizes actin into filament-like structures. Cell. 2013;155:423–34. doi: 10.1016/j.cell.2013.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zekarias B, Mattoo S, Worby C, Lehmann J, Rosenbusch RF, Corbeil LB. Histophilus somni IbpA DR2/Fic in virulence and immunoprotection at the natural host alveolar epithelial barrier. Infect. Immun. 2010;78:1850–58. doi: 10.1128/IAI.01277-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Mooseker MS, Galan JE. Role of the S. typhimurium actin-binding protein SipA in bacterial internalization. Science. 1999;283:2092–95. doi: 10.1126/science.283.5410.2092. [DOI] [PubMed] [Google Scholar]
- Zhou X, Konkel ME, Call DR. Vp1659 is a Vibrio parahaemolyticus type III secretion system 1 protein that contributes to translocation of effector proteins needed to induce cytolysis, autophagy, and disruption of actin structure in HeLa cells. J. Bacteriol. 2010;192:3491–502. doi: 10.1128/JB.01493-09. [DOI] [PMC free article] [PubMed] [Google Scholar]