Abstract
The lysine-specific demethylase (LSD1) is a flavin-dependent amine oxidase that selectively removes one or two methyl groups from histone H3 at the Lys4 position. Along with histone deacetylases 1 and 2, LSD1 is involved in epigenetically silencing gene expression. LSD1 has been implicated as a potential therapeutic target in cancer and other diseases. In this chapter, we discuss several approaches to measure LSD1 demethylase activity and their relative strengths and limitations for inhibitor discovery and mechanistic characterization. In addition, we review the principal established chemical functional groups derived from monoamine oxidase inhibitors that have been investigated in the context of LSD1 as demethylase inhibitors. Finally, we highlight a few examples of recently developed LSD1 mechanism-based inactivators and their biomedical applications.
1. INTRODUCTION AND BACKGROUND
The histone code features reversible Lys modifications as a major mechanism for the regulation of chromatin accessibility, gene expression, and cellular growth. Lys side chain acetylation and methylation are considered the dominant and best-studied PTMs in histones. Lys acetylation is regulated by histone acetyltransferases (HATs or KATs) and histone deacetylases (HDACs or KDACs), whereas Lys methylation is controlled by histone Lys methyltransferases (HMTs or KMTs) and histone demethylases (KDMs) (Cole, 2008). Whereas acetylation of the Lys side chain only occurs once per Lys residue, Lys methylation can occur as mono-, di-, and trimethylation forms. Until the report of LSD1 (lysine-specific demethylase 1) in 2004, there was some uncertainty as to whether protein Lys methylation was reversible (Shi et al., 2004). It is now generally accepted that there are at least 18 Lys demethylases, including two flavoenzymes LSD1 (KDM1A) and LSD2 (KDM1B) and the rest being nonheme iron, α-ketoglutarate-dependent JMJ oxygenases (Culhane & Cole, 2007; Thinnes et al., 2014). Common features among the histone demethylases are that they utilize molecular oxygen, catalyze oxidative demethylation, and produce formaldehyde as a by-product (Culhane & Cole, 2007).
LSD1, and its less well-studied paralog LSD2, is members of the amine oxidase enzyme family that depend on a flavin cofactor (Hou & Yu, 2010). This family includes monoamine oxidases that act to metabolize norepinephrine and related neurotransmitters and polyamine oxidases that metabolize spermidine, spermine, and other alkylamines (Edmondson, Mattevi, Binda, Li, & Hubalek, 2004). Although the precise chemical details of oxidation by amine oxidases are still being debated, functionally the reactions can be viewed as involving hydride transfer between the substrate nitrogen and the flavin cofactor (Culhane & Cole, 2007). Consequently, LSD1 and LSD2, which catalyze demethylation reactions on mono- and dimethyl Lys substrates, are incapable of demethylating trimethyl-Lys substrates because of their lack of an available electron lone pair. This contrasts the JMJ demethylase enzymes that typically process trimethyl-Lys substrates since they directly oxidize methyl groups (Hou & Yu, 2010). Upon LSD1-mediated hydride transfer, the corresponding unstable imine intermediate likely spontaneously hydrolyzes to formaldehyde and the demethylated amine (Fig. 1). In order for there to be multiple catalytic turnovers, the reduced flavin must be reoxidized, and this involves reaction with molecular oxygen, extracted out of the aerobic environment, leading to stoichiometric hydrogen peroxide as a by-product.
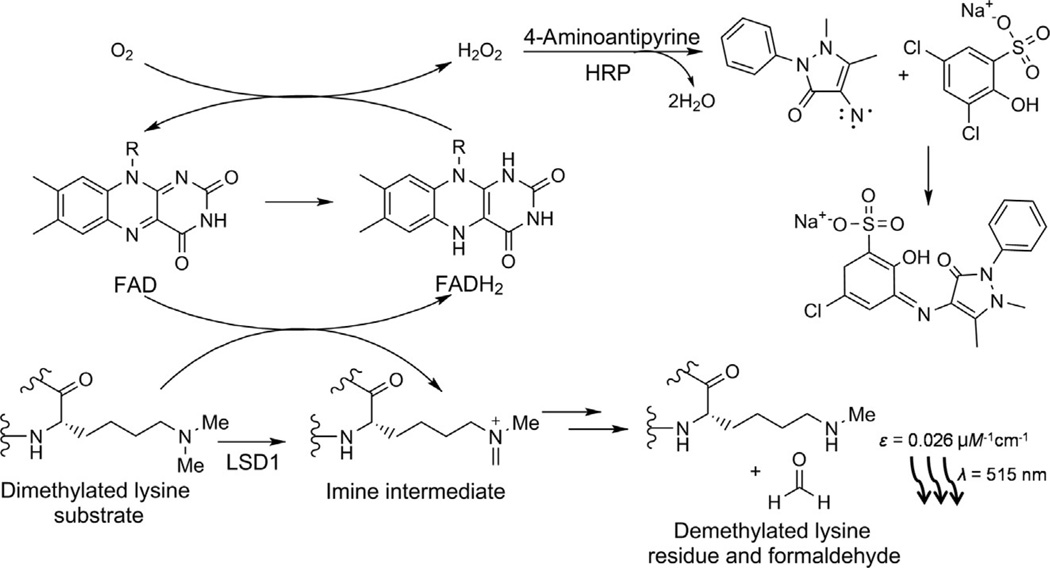
Fig. 1.
Hydrogen peroxide (HOOH) detection assay for LSD1. When a dimethylated lysine substrate (bottom left) is demethylated by LSD1, the FAD cofactor is reduced to FADH2 (center). The cofactor is reoxidized by molecular oxygen to form HOOH (top center). Horseradish peroxidase (HRP) uses this to couple 4-aminoantipyrine and dichlorohydroxybenzenesulfonate (top right) to form the chromophore (center right) that is detected at 515 nm (bottom right).
In contrast to many histone modifying enzymes that are often pleiotropic with regard to site specificity, LSD1 is highly selective for demethylation of the Lys4 site in histone H3, avoiding thousands of other methylated lysines in the cell (Forneris et al., 2006). Since H3K4 methylation is a mark of gene activation, LSD1 is most often associated with effecting gene silencing (Shi et al., 2004). In this way, LSD1 can act in concert with HDACs that are also commonly considered to serve as turn off switches for gene expression. In fact, LSD1 and HDAC1 (or its highly similar paralog HDAC2) are subunits of the well-studied CoREST repressor complex, with both enzymes interacting with the two different SANT domains of CoREST1 (Lee, Wynder, Cooch, & Shiekhattar, 2005; Yang et al., 2006). LSD1 is a 90 kDa protein that contains a SWIRM domain for targeting nucleosomes and an insert in its amine oxidase domain known as a “tower domain” that engages the SANT2 domain of CoREST (Yang et al., 2006). LSD2 lacks this tower domain, does not complex with CoREST, and has distinct biological roles (Fang et al., 2012). Recent biochemical and structural studies on the LSD1/CoREST heterodimer suggest the multilayer interactions of the demethylase with its native nucleosome substrate that include an interaction between one section of the SANT2 domain and nucleosomal DNA (Kim, Chatterjee, Jennings, Bartholomew, & Tan, 2015; Pilotto et al., 2015). Using purified LSD1, it is established that a minimum of about 20 residues of the histone H3 tail are necessary for efficient LSD1-catalyzed demethylation (Forneris, Binda, Antonietta Vanoni, Battaglioli, & Mattevi, 2005; Forneris et al., 2006). However, several different crystal structure complexes of LSD1 bound to peptides substrate analogs have offered alternative models on how such substrate recognition occurs (Forneris, Binda, Adamo, Battaglioli, & Mattevi, 2007; Yang et al., 2007).
There have been a smattering of reports that LSD1 can act on sites beyond methyl-H3K4, the most commonly discussed site being Lys9 of histone H3 (Laurent et al., 2015; Nicholson & Chen, 2009). However, these reports on targeting H3K9Me are typically based on cell-based experiments where the role of LSD1 on H3K9 demethylation may be indirect and noncatalytic. Compounding the assignment of site specificity is that antibodies for H3K9Me have been reported as lacking in reliability (Hattori et al., 2013). Although a recent report suggested that a splice variant of LSD1 may be capable of histone H3K9Me demethylation (Laurent et al., 2015), this possibility was previously rendered unlikely by an earlier enzymologic study with the purified LSD1 splice variant protein that showed similar catalytic activity and site specificity to the standard LSD1 form (Toffolo et al., 2014; Zibetti et al., 2010).
A number of lines of evidence point to LSD1 as having important functions in biology and disease. Knockout of LSD1 is embryonically lethal, and LSD1 is particularly associated with neuronal development (Zibetti et al., 2010). LSD1 may participate in mechanisms of neurologic disease and viral pathogenesis (Hill et al., 2014; Liang, Vogel, Narayanan, Peng, & Kristie, 2009; Rusconi et al., 2015; Sun et al., 2010; Wang, Telese, et al., 2015). LSD1 levels have been found to be elevated in many cancers (Wu, Hu, Du, Kong, & Pan, 2015). Consequently, there has been an intense interest in the development of LSD1 inhibitors.
2. LSD1 ASSAYS
Kinetic analysis of the LSD1 demethylase reaction is important both for understanding the mechanistic features of the reaction and for the screening and analysis of LSD1 inhibitors. Several in vitro biochemical LSD1 enzymatic assays have been developed including HOOH detection through a peroxidase-coupled assay, formaldehyde detection through a formaldehyde dehydrogenase assay, radioactive measurements to monitor changes in a radiolabeled peptide substrate, and mass spectrometry analysis to measure changes in the unlabeled substrate (Blair et al., 2011; Forneris et al., 2005; Kokura, Sun, & Fang, 2015). In addition, cellular assays have been adapted to quantify LSD1-mediated effects on histone H3K4 methylation using antibody detection (Schmitt et al., 2014).
2.1 Coupled Enzyme Assays
Mattevi and colleagues described the use of horseradish peroxidase-coupled assays for accurately measuring LSD1 activity with methylated H3 tail peptide substrates (Forneris et al., 2005). This approach uses well-established methodologies that have been applied decades ago to the study of monoamine oxidases. HOOH is an obligate side product of LSD1 catalyzed demethylation and a substrate for horseradish peroxidase. In the presence of 4-aminoantipyrine (4-AP) along with 3,5-dichloro-2-hydroxybenzenesulfonate (DHBS), horseradish peroxidase converts HOOH into a spectrophotometric absorbance change that can be monitored at 515 nm with a sizable extinction coefficient of 26,000 M−1 cm−1 (Forneris et al., 2005; Fig. 1). This is a relatively sensitive and reliable assay that can be done continuously. A more sensitive version of this assay replaces 4-AP/DHBS with Amplex red, which reacts with HOOH to generate the fluorophore resorufin that has fluorescence emission detected at 585 nm (Schmitt et al., 2014).
The other major side product of the LSD1 histone demethylase reaction is formaldehyde. Formaldehyde release can be monitored via its oxidation to formate using formaldehyde dehydrogenase where cofactor NADH formation is measured spectrophotometrically at 340 nm (Kokura et al., 2015). Both the horseradish peroxidase and formate dehydrogenase assays are relatively low cost and can be done continuously to measure LSD1 activity in real time. However, formaldehyde dehydrogenase is somewhat more complex as a coupling enzyme than horseradish peroxidase and the UV change for NAD reduction is less sensitive than HOOH detection and so the horseradish peroxidase assay is typically preferred. Because these coupled enzyme assays provide real-time kinetic information, they are particularly powerful for analyzing time-dependent inactivation, a common phenomenon of LSD1 inhibitors. Disadvantages of both the horseradish peroxidase and formaldehyde dehydrogenase coupled assays are that they are indirect and changes in reaction conditions or addition of inhibitors can be misinterpreted.
2.2 Radiolabeled Assays
Radioactive LSD1 assays typically involve the use of radiolabeled peptide substrates, usually tritiated methyl groups attached to the H3K4 side chain and scintillation counting is applied to quantify the radioactivity in the substrate and/or formaldehyde product. One example relies on lysine methyltransferases and tritiated S-adenosylmethionine (SAM) as the methyl donor to attach a radiolabeled methyl group to a synthetic histone H3 tail peptide (Kokura et al., 2015). Subsequently the radiolabeled peptide is reacted with LSD1 to afford tritiated formaldehyde, which is then converted into diacetyldihydrolutidine, extracted with pentanol and the radioactive material quantified by scintillation counting. A complementary method employs tritiated SAM and the KMT7 methyltransferase to attach a radiolabeled methyl group after LSD1 demethylation of an unlabeled methyl-substratepeptide/productmixture, to“remethylate” the demethylated site (Yu et al., 2014). These assays also allow for kinetic analysis of LSD1 and can measure both substrate and product levels concurrently. However, they are technically demanding, expensive, and discontinuous. They are not readily applicable for high-throughput assays but can be used to validate findings with the spectrophotometric coupled assays described earlier.
2.3 Antibody-Based Assays
Antibody-based assays can be used to analyze LSD1 activity on peptide and histone protein substrates. A recent version of this assay involves a heterogeneous format where biotinylated peptide substrates are incubated with LSD1 and then bound to a streptavidin microplate using primary antibodies to the site of interest and secondary antibodies conjugated to europium in an assay called DELFIA (dissociation-enhanced lanthanide fluorescent immunoassay) (Schmitt et al., 2014). An advantage of this assay is the use of the biotinylated peptide, which allows for washout of all other assay components before treatment with antibody. A related assay which uses the europium antibody is called LANCE Ultra where FRET occurs between the donor europium-labeled primary antibody to the site of interest and the acceptor ULight (Gale&Yan, 2015). Here, the peptide is N-terminally bound by primary antibody and FRET occurs between the europium and ULight at the C-terminal end. An Alpha Screen that is analogous to LANCE Ultra has also been reported (Gale&Yan, 2015). Library screening can be pursued with the DEL-FIA assay since no fluorescence or quenching of the compounds is likely to occur. However, these methods have somewhat limited quantitative robustness and depend heavily on the reliability of the Ab reagents.
2.4 Mass Spectrometry-Based Assays
Mass spectrometry has been used for many years to discover new posttranslational modifications and is frequently employed to measure demethylation in peptide and histone substrates (Huang, Lin, Garcia, & Zhao, 2015). Quantifying the four states of lysine modification: unmodified, mono-, di-, and trimethylation can be a challenge. Several methods have been developed to address the methyl-Lys population states. One of these approaches involves the reductive methylation with heavy (dideuterio) formaldehyde to convert all states to the same chemical species such that there are no intrinsic ionization differences but the mass differences are detectable (Blair et al., 2011; Blair, Avaritt, & Tackett, 2012). This method, called MassSQUIRM, which uses MALDI-TOF (matrix-assisted laser desorption ionization-time-of-flight) is a highly quantitative technique and has been used to assess LSD1 inhibition (Blair et al., 2011).
A second method for analyzing multiple methylation states uses a flow-injection-based time-of-flight mass spectrometry involving electrospray ionization. The multiply charged species of each type of modification can be separated to accurately determine abundance of each peptide form (Sakane, Ohta, & Shidoji, 2015). A third mass spectrometry method for analyzing demethylase activity applies SAMDI (self-assembled monolayer desorption/ionization) that involves biotinylated peptide absorption onto a neutravidin monolayer surface followed by release using MALDI as the ionization method and TOF–TOF as the separation method (Wigle et al., 2015). SAMDI has been used in a high-throughput screen to identify LSD1 inhibitors. Another mass spectrometry-based assay to analyze demethylation is RapidFire that uses tagged peptides, which are then pooled (Gale & Yan, 2015). Mass spectrometry-based methods are sensitive, can be made label free, and require very little amounts of enzyme and substrate. However, they are intrinsically technically demanding and expensive in requiring specialized instrumentation.
Beyond peptide substrates, some of these LSD1 approaches are applicable to more complex substrates and conditions including nucleosome demethylation and cell-based assays. For example, the formaldehyde release method using radioactive SAM incorporation can be performed on nucleosomes or histone octamers (Kokura et al., 2015). Antibody-based assays are now validated to assess LSD1 activity in cells, with the monitoring of demethylation of histone H3 being particularly diagnostic.
3. INHIBITORS OF LSD1
The first reports of LSD1 inhibitors appeared within about 18 months of the enzyme’s functional identification as a histone demethylase (Culhane et al., 2006; Lee, Wynder, Schmidt, McCafferty, & Shiekhattar, 2006; Schmidt & McCafferty, 2007; Szewczuk et al., 2007). Experiments with peptide substrate analogs and established monoamine oxidase inhibitor antidepressant drugs led to the identification of several LSD1 suicide inactivating compounds (Culhane et al., 2006; Lee et al., 2006; Paech, Salach, & Singer, 1980). Propargyl-Lys-containing peptide substrate analogs, inspired by a class of MAO inhibitors such as pargyline, are potent LSD1 inhibitors and display characteristic time-dependent mechanism-based inactivation (Culhane et al., 2006).
Mechanism-based enzyme inactivators, also known as suicide inhibitors, are latently reactive molecules that exploit the target enzymes normal chemistry to unmask a warhead that can covalently modify the active sites of these protein catalysts, leading to irreversible inhibition (Silverman, 1995; Walsh, 1984). These inhibitors are characterized using the Kitz–Wilson equation and potencies most precisely expressed as a ratio of kinact over Ki(inact) inhibitory constants (Kitz & Wilson, 1962; Walsh, 1984). The kinact is the first-order rate constant of enzyme inactivation at saturating inhibitor concentration and is analogous to the kcat for a standard enzyme reaction catalyzing conversion of substrate to product. The Ki(inact) corresponds to the apparent affinity of the inhibitor for the enzyme in the initial encounter complex, prior to enzyme processing, and is akin to the Km for a substrate in a standard enzyme reaction. The ratio kinact/Ki(inact) is a measure of overall efficiency of inactivation by a given compound (Silverman, 1995) and can be used to rank order a series of suicide inhibitors. In practice, given the complexities of these measurements, many labs report suicide inactivator potencies as IC50s, making it difficult to compare inhibitors from one study to the next (Wang, Huang, Suzuki, Liu, & Zhan, 2015). Regardless, the pharmacologic effects in the cellular environments depend on additional factors among which are cell permeability, rate of resynthesis of LSD1 protein, and partition ratio of the enzyme-generated reactive species toward modification of the target vs diffusion off of the enzyme (Silverman, 1995).
Propargyl-Lys-containing peptides are potent suicide inactivators of LSD1 that generate a covalent adduct with the active site flavin cofactor but are likely too large to enter cells, limiting their in vivo pharmacologic potential (Culhane et al., 2006; Szewczuk et al., 2007). Nevertheless, these propargyl-Lys peptides have shown utility in structural biology studies. An H3 tail peptide containing a propargyl-Lys was used to generate a crystallographic complex with LSD1 as discussed earlier (Yang et al., 2007). In more recent studies, a propargyl-Lys analog was incorporated into histone H3 and ultimately nucleosomes and was elegantly applied to capture a low-resolution structure of an LSD1/CoREST heterodimer stably bound to nucleosomes (Pilotto et al., 2015).
Although the propargyl-containing antidepressant MAO inhibitor pargyline itself is not an LSD1 inhibitor (Culhane, Wang, Yen, & Cole, 2010; Lee et al., 2006), despite cell-based pharmacologic reports that have suggested otherwise (Metzger et al., 2005), tranylcypromine and phenelzine MAO inhibitors do act as suicide inhibitors of LSD1 via their cyclopropylamine and hydrazine functional groups (Culhane et al., 2010; Lee et al., 2006; Schmidt & McCafferty, 2007). Tranylcypromine and phenelzine themselves are relatively weak LSD1 inhibitors compared to their actions vs MAOs, but in the context of elaboration into peptides or more complex small molecule analogs, these cyclopropylamine and hydrazine functionalities can confer high potency and specificity for LSD1 (Prusevich et al., 2014; Vianello et al., 2014). For tranylcypromine analogs, it has been shown that attachment of an aryl group via flexible linkers in the para position of the phenyl ring can greatly enhance the mechanism-based inactivation of LSD1 relative to other amine oxidases (Vianello et al., 2014). Such aryl-alkyl attachment also has been used to increase the potency and specificity of phenelzine compounds (Prusevich et al., 2014). N-substitution of the cyclopropylamine of tranylcypromine is another approach to improve LSD1 targeting. X-ray crystal structures have been somewhat informative in establishing the mechanistic basis for the improved LSD1 potencies of these synthetic analogs of tranylcypromine (Vianello et al., 2014).
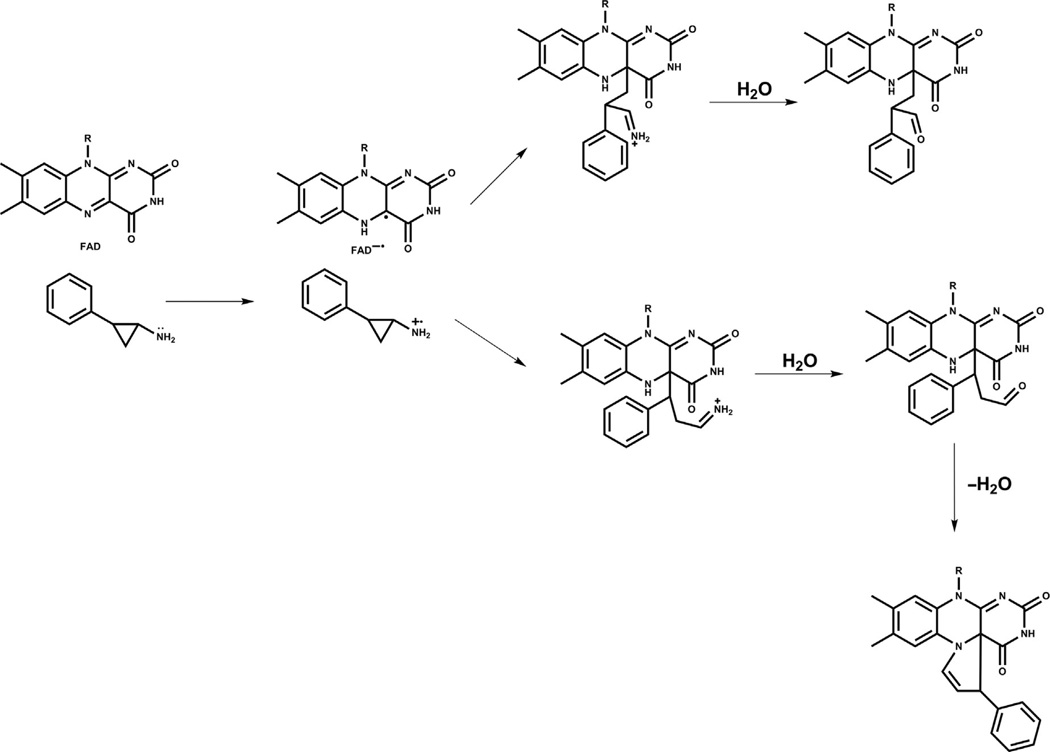
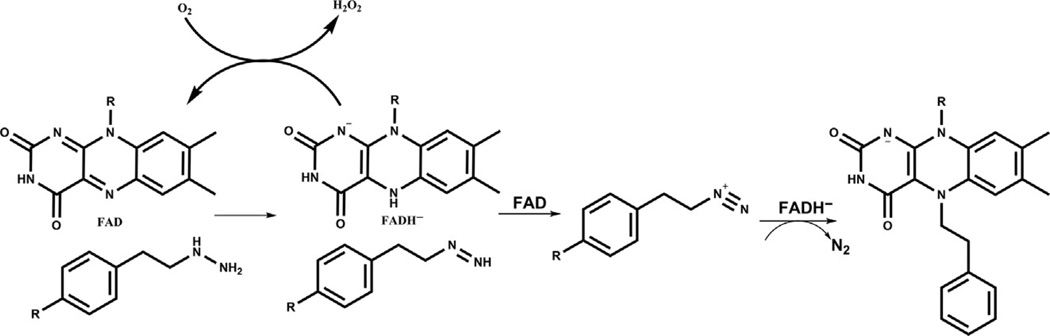
In general, cyclopropylamine analogs undergo ring opening, likely generating a benzyl radical that is stabilized by aromatic conjugation. Several different fates have been reported for the covalent bond formation of the radical intermediate and the flavin cofactor as determined by X-ray crystal structures (Binda et al., 2010; Mohammad et al., 2015; Yang et al., 2007; Fig. 2). Both the N5 and C4a atoms of the flavin have been invoked as attachment points for the benzyl scaffold, driving ring formation as well as acyclic structures (Binda et al., 2010; Yang et al., 2007). It seems plausible that more than one of these covalent adducts can be generated from a given inhibitor and X-ray crystallography of the LSD1 complexes is limited in its ability to distinguish such mixed molecular populations. For inactivation by phenelzine analogs, the precise mechanism of inhibition is also unclear. One proposed pathway for inhibition involves oxidation of the hydrazine functionality to the diazonium species, with subsequent attack by the flavin or perhaps an LSD1 enzyme residue (Culhane et al., 2010; Fig. 3). Such oxidation to the diazonium, however, requires a net four-electron transfer, which may be kinetically challenging. It is therefore possible that the two-electron oxidation, generating the aza analog or its functional equivalent, is the key species for enzyme inactivation. There have not yet been X-ray crystal structure of LSD1 inactivated by phenelzine analogs, although mass spectroscopy has suggested that a flavin adduct can be formed, at least as a minor product (Culhane et al., 2010).
Fig. 2.
Mechanism of LSD1 inactivation by cyclopropylamine analogs. Cyclopropylamine analogs form covalent bonds with the flavin cofactor of LSD1 through ring opening. The C4a and N5 atoms (top right and bottom right) serve as proposed points of attachment that occur after cyclopropyl ring opening (center).
Fig. 3.
Potential mechanism of LSD1 inactivation by hydrazine analogs. A possible mechanism of hydrazine-mediated inactivation of LSD1 involves formation of a covalent bond with the flavin cofactor. When the hydrazine moiety initially encounters the FAD cofactor (left), it may undergo a four-electron oxidation to form the diazonium species (center) which can be attacked by the cofactor or another nucleophile in the vicinity. When the flavin attacks (as shown), a covalent bond forms which inactivates the enzyme.
Other compounds beyond tranylcypromine and phenelzine analogs have been reported as LSD1 inhibitors including polyamines (Nowotarski et al., 2015) and hydrazone HCI-2509 but whose specificity and mechanisms of inhibition remain less well characterized (Wang, Huang, et al., 2015). Given that many of the in vitro LSD1 demethylase assays employ peroxidase as an indirect measure of LSD1 enzymatic activity, and the peroxidase activity can be interfered with by particular compounds, it is critical to use secondary assays such as mass spectrometry analysis that directly monitors peptide methylation status to ensure the reliability of a particular LSD1 inhibitor finding.
4. APPLICATIONS OF LSD1 INHIBITORS
Applications of LSD1 inhibitors can be considered in the context of stem cell differentiation (Eliazer et al., 2014), neurobiology (Neelamegam et al., 2012), oxidative stress (Prusevich et al., 2014), viral infectivity (Hill et al., 2014; Sakane et al., 2011), and many forms of cancer. There are now numerous reports of synthetic LSD1 inhibitors of varying mechanisms of inhibition, potencies, and selectivities being applied to biomedical discovery. Basic features including effects on histone marks and gene expression as well as functional effects on cell growth and physiologic processes have been assessed with these compounds. Here we highlight a select group of recent findings involving well-characterized LSD1 inhibitors with an emphasis on cancer (Fig. 4).
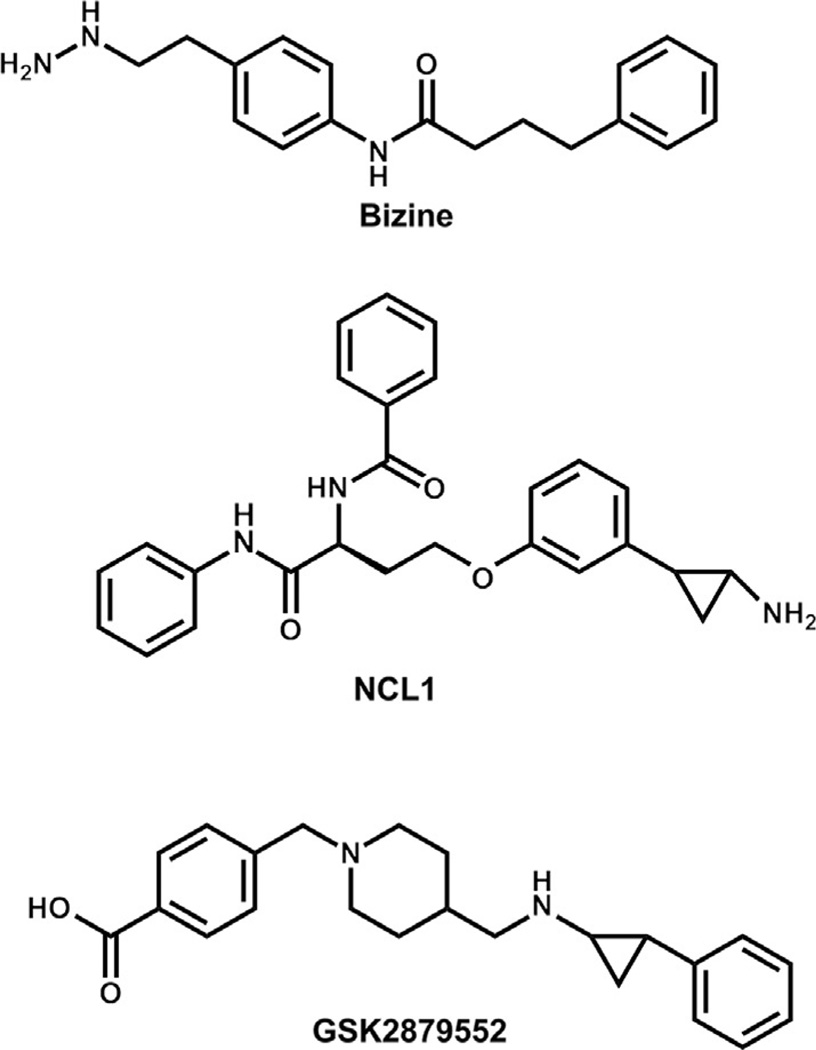
Fig. 4.
Structures of representative LSD1 inhibitors. Bizine, a selective phenelzine analog (top);NCL1, a tranylcypromine analog (middle); and GSK2879552, a recent tranylcypromine analog (bottom), have been shown to be potent and selective LSD1 inhibitors.
4.1 Bizine
Bizine is a selective and potent LSD1 inhibitor based on the monoamine oxidase inhibitor phenelzine. This inhibitor showed specific effects on H3K4Me2 in a variety of cancer cell lines including LnCAP, androgen-dependent prostate cancer, and H460 lung cancer (Prusevich et al., 2014). Interestingly, the effects on H3K4Me2 showed an oscillating kinetic behavior with increases in H3K4Me2 detected by 6 h, then absent at 12 h, and reappearing at 24 h and persisting to 72 h (Prusevich et al., 2014). The relatively rapid appearance of H3K4Me2 at 6 h suggests that basal cellular activity of LSD1 is substantial and is in equilibrium with one or more methyltransferases. The basis for cellular oscillation in H3K4Me2 after bizine treatment is not known but the rebound to baseline at 12 h suggests that compensatory effects after LSD1 inhibition may occur involving changes in methyltransferase levels or localization. Interestingly, levels of mono- and trimethylation of H3K4 were not affected by bizine treatment (Prusevich et al., 2014). These findings suggest that globally stimulating dimethylation of H3K4 does not drive H3K4 trimethylation per se. While HeK4Me3 is generally considered most critical for gene activation, perhaps a global increase in H3K4Me2 in chromatin may serve biologically key functions that are less appreciated.
Whole genome chromatin immunoprecipitation (ChIP-seq) analyzing H3K4Me2 positioning after bizine treatment suggested widespread effects on numerous gene promoters, including many tumor suppressor genes (Prusevich et al., 2014). Despite these findings, the antitumor proliferation effects of bizine were of modest potency (Prusevich et al., 2014), considerably weaker than its effects on histone marks. Combination studies of bizine with some histone deacetylase inhibitors did suggest the potential of additivity or mild synergy of antitumor effects with particular epigenetic agents (Prusevich et al., 2014). Earlier studies in glioblastoma with another LSD1 inhibitor also pointed to the potential of such combination therapies (Singh et al., 2011). Of note, bizine also resulted in neuronal protection against oxidative stress (Prusevich et al., 2014), although the mechanism for this remains unclear.
4.2 NCL1
The tranylcypromine derivative NCL1 is a potent and selective LSD1 mechanism-based inhibitor developed by Suzuki and colleagues (Ueda et al., 2009). This compound was shown to exhibit with relatively low toxicity but significant antiproliferative effects against LnCAP cells in vitro and in mouse xenograft studies (Etani et al., 2014). While the cellular potency of NCL1 on LnCAP growth was modest, the in vivo antitumor effect was relatively strong. The precise mechanisms of NCL1’s antitumor effects were unclear but appear to involve induction of apoptosis and autophagy.
4.3 GSK2879552
The GSK pharmaceutical company effort led by Kruger reported on a tranylcypromine analog—GSK2879552 (Mohammad et al., 2015) as a specific and fairly potent LSD1 inhibitor. These investigators showed GSK2879552 is a suicide inactivator and obtained a crystal structure with LSD1. GSK2879552 is most noteworthy for its very high specificity vs MAOs, likely residing from the extra substitution on the cyclopropylamine nitrogen. GSK2879552 was tested for its antitumor effects across a wide range of tumor cell lines and was shown to be potent against most acute myeloid leukemia lines investigated and a subset of small cell lung cancer lines (Mohammad et al., 2015). Like those of bizine, effects of GSK2879552 on H3K4Me2 were detected globally, and these effects as well as influence on gene expression occurred relatively rapidly compared to antiproliferative effects. It was especially noteworthy that for many cell lines, antiproliferative effects did not appear until 6 days after treatment with GSK2879552 (Mohammad et al., 2015). In some cases, cancer cell growth inhibition was incomplete and plateaued at high GSK2879552 concentration, suggesting that cells were still dividing but at a slower rate (Mohammad et al., 2015). Taken together with GSK2879552-induced gene expression changes, these data were consistent with a model of GSK2879552-induced cellular differentiation but not cell death in such cases. GSK investigators were unable to identify a gene expression signature from one cancer cell line to the next conferred by GSK2879552. After careful bioinformatics analysis, however, the GSK team was able to identify a DNA hypomethylation signature that was predictive of response to GSK2879552. The molecular mechanism for such a response was unclear, however.
5. SUMMARY AND FUTURE DIRECTIONS
Since LSD1’s discovery as a histone demethylase about a dozen years ago, our understanding of LSD1’s structure and enzymatic mechanism has advanced relatively rapidly, but elucidating the biological consequences of its catalytic activity have proved challenging. Major strides in the development of enzymatic assays and inhibitor discovery have begun to allow for the development of powerful chemical probes being brought to bear on LSD1’s apparent myriad of cellular functions. Still, how LSD1 contributes to cancer and other diseases, how LSD1 inhibitors might synergize with other epigenetic agents, and how subunits of the CoREST complex modulate LSD1 function are just some of the unresolved topics under active investigation. Other poorly understood issues relate to LSD1’s regulation by alternative splicing and posttranslational modifications. We anticipate that the vibrant research efforts devoted to LSD1 investigation will drive important discoveries in the coming years with translational and clinical impact.
Acknowledgments
We thank Dr. Jay Kalin for helpful advice and the NIH, FAMRI Foundation, and V Foundation for financial support.
REFERENCES
- Binda C, Valente S, Romanenghi M, Pilotto S, Cirilli R, Karytinos A, et al. Biochemical, structural, and biological evaluation of tranylcypromine derivatives as inhibitors of histone demethylases LSD1 and LSD2. Journal of the American Chemical Society. 2010;12:6827–6833. doi: 10.1021/ja101557k. [DOI] [PubMed] [Google Scholar]
- Blair LP, Avaritt NL, Huang R, Cole PA, Taverna SD, Tackett AJ. An assay for quantitative measurement of lysine demethylase activity. Epigenetics. 2011;6:490–499. doi: 10.4161/epi.6.4.14531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair LP, Avaritt NL, Tackett AJ. Application of MassSQUIRM for quantitative measurement of lysine demethylase activity. Journal of Visualized Experiment. 2012;61:e3604. doi: 10.3791/3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole PA. Chemical probes for histone-modifying enzymes. Nature Chemical Biology. 2008;4(10):590–597. doi: 10.1038/nchembio.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culhane JC, Cole PA. LSD1 and the chemistry of histone demethylation. Current Opinion in Chemical Biology. 2007;11:561–568. doi: 10.1016/j.cbpa.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culhane JC, Szewczuk LM, Liu X, Da G, Marmorstein R, Cole PA. A mechanism-based inactivator for histone demethylase LSD1. Journal of the American Chemical Society. 2006;128:4536–4537. doi: 10.1021/ja0602748. [DOI] [PubMed] [Google Scholar]
- Culhane JC, Wang D, Yen PM, Cole PA. Comparative analysis of small molecules and histone substrate analogues as LSD1 lysine demethylase inhibitors. Journal of the American Chemical Society. 2010;132:3164–3176. doi: 10.1021/ja909996p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson DE, Mattevi A, Binda C, Li M, Hubalek F. Structure and mechanism of monoamine oxidase. Current Medicinal Chemistry. 2004;11:1983–1993. doi: 10.2174/0929867043364784. [DOI] [PubMed] [Google Scholar]
- Eliazer S, Palacios V, Wang Z, Kollipara RK, Kittler R, Buszczak M. Lsd1 restricts the number of germline stem cells by regulating multiple targets in escort cells. PLoS Genetics. 2014;10:e1004200. doi: 10.1371/journal.pgen.1004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etani T, Suzuki T, Naiki T, Naiki-Ito A, Ando R, Iida K, et al. NCL1, a highly selective lysine-specific demethylase 1 inhibitor, suppresses prostate cancer without adverse effect. Oncotarget. 2014;6:2865–2878. doi: 10.18632/oncotarget.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang R, Chen F, Dong Z, Hu D, Barbera AJ, Clark EA, et al. LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation. Molecular Cell. 2012;49:558–570. doi: 10.1016/j.molcel.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forneris F, Binda C, Adamo A, Battaglioli E, Mattevi A. Structural basis of LSD1-CoREST selectivity in histone H3 recognition. The Journal of Biological Chemistry. 2007;282:20070–20074. doi: 10.1074/jbc.C700100200. [DOI] [PubMed] [Google Scholar]
- Forneris F, Binda C, Antonietta Vanoni M, Battaglioli E, Mattevi A. Human histone demethylase LSD1 reads the histone code. The Journal of Biological Chemistry. 2005;280:41360–41365. doi: 10.1074/jbc.M509549200. [DOI] [PubMed] [Google Scholar]
- Forneris F, Binda C, Dall’Aglio A, Fraajie MW, Battaglioli E, Mattevi A. A highly specific mechanism of histone H3-K4 recognition by histone demethylase LSD1. The Journal of Biological Chemistry. 2006;281(46):35289–35295. doi: 10.1074/jbc.M607411200. [DOI] [PubMed] [Google Scholar]
- Gale M, Yan Q. High-throughput screening to identify inhibitors of lysine demethylases. Epigenomics. 2015;7:57–65. doi: 10.2217/epi.14.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori T, Taft JM, Swist KM, Luo H, Witt H, Slattery M, et al. Recombinant antibodies to histone post-translational modifications. Nature Methods. 2013;10:992–998. doi: 10.1038/nmeth.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JM, Quenelle DC, Cardin RD, Vogel JL, Clement C, Bravo FJ, et al. Inhibition of LSD1 reduces herpesvirus infection, shedding, and recurrence by promoting epigenetic suppression of viral genomes. Science Translational Medicine. 2014;6:265ra169. doi: 10.1126/scitranslmed.3010643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H, Yu H. Structural Insights into histone lysine demethylation. Current Opinion in Structural Biology. 2010;20:739–748. doi: 10.1016/j.sbi.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Lin S, Garcia BA, Zhao Y. Quantitative proteomic analysis of histone modifications. Chemical Reviews. 2015;115:2376–2418. doi: 10.1021/cr500491u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-A, Chatterjee N, Jennings MJ, Bartholomew B, Tan S. Extranucleosomal DNA enhances the activity of the LSD1/CoREST histone demethylase complex. Nucleic Acids Research. 2015;43(10):4868–4880. doi: 10.1093/nar/gkv388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitz R, Wilson IB. Esters of methanesulfonic acid as irreversible inhibitors of acetylcholinesterase. Journal of Biological Chemistry. 1962;237:3245–3249. [PubMed] [Google Scholar]
- Kokura K, Sun L, Fang J. In vitro histone demethylase assays. In: Chellappan SP, editor. Chromatin protocols, methods in molecular biology: Vol 1288. New York: Springer; 2015. pp. 109–122. [DOI] [PubMed] [Google Scholar]
- Laurent B, Ruitu L, Mum J, Hempei K, Ferrao R, Xiang Y, et al. A specific LSD1/KDM1A isoform regulates neuronal differentiation through H3K9 demethylation. Molecular Cell. 2015;57:1–14. doi: 10.1016/j.molcel.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Schmidt DM, McCafferty DG, Shiekhattar R. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chemistry and Biology. 2006;13:565–567. doi: 10.1016/j.chembiol.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Liang Y, Vogel JL, Narayanan A, Peng H, Kristie TM. Inhibition of the histone demethylase LSD1 blocks α-herpesvirus lytic replication and reactivation from latency. Nature Medicine. 2009;15:1312–1318. doi: 10.1038/nm.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AHFM, et al. LSD1 demethylates repressive histone marks to promoter androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- Mohammad H, Smitherman KN, Kamat CD, Soong D, Federowicz KE, Van Aller GS, et al. A DNA hypomethylation signature predicts antitumor activity of LSD1 inhibitors in SCLC. Cancer Cell. 2015;28:57–69. doi: 10.1016/j.ccell.2015.06.002. [DOI] [PubMed] [Google Scholar]
- Neelamegam R, Ricq EL, Malvaez M, Patnaik D, Norton S, Carlin SM, et al. Brain-penetrant LSD1 inhibitors can block memory consolidation. ACS Chemical Neuroscience. 2012;3:120–128. doi: 10.1021/cn200104y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson TB, Chen T. LSD1 demethylates histone and non-histone proteins. Epigenetics. 2009;4(3):129–132. doi: 10.4161/epi.4.3.8443. [DOI] [PubMed] [Google Scholar]
- Nowotarski SL, Pachaiyappan B, Holshouser SL, Kutz CJ, Li Y, Huang Y, et al. Structure-activity study for (bis)ureidopropyl- and (bis)thioureidopropyldiamine LSD1 inhibitors with 3-5-3 and 3-6-3 carbon backbone architectures. Bioorganic & Medicinal Chemistry. 2015;23:1601–1612. doi: 10.1016/j.bmc.2015.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paech C, Salach JI, Singer TP. Suicide inactivation of monoamine oxidase by trans-phenylcyclopropylamine. Journal of Biological Chemistry. 1980;10:2700–2704. [PubMed] [Google Scholar]
- Pilotto S, Speranzini V, Tortorici M, Durand D, Fish A, Valente S, et al. Interplay among nucleosomal tails, histone tails, and corepressor CoREST underlies LSD1-mediated H3 demethylation. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(9):2752–2757. doi: 10.1073/pnas.1419468112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusevich P, Kalin JH, Ming SA, Basso M, Givens J, Li X, et al. A selective phenelzine analogue inhibitor of histone demethylase LSD1. ACS Chemical Biology. 2014;9:1284–1293. doi: 10.1021/cb500018s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusconi F, Paganini L, Braida D, Ponzoni L, Toffolo E, Maroli A, et al. LSD1 neurospecific alternative splicing controls neuronal excitability in mouse models of epilepsy. Cerebral Cortex. 2015;25:2729–2740. doi: 10.1093/cercor/bhu070. [DOI] [PubMed] [Google Scholar]
- Sakane N, Kwon H-S, Pagans S, Kaehlcke K, Mizusawa Y, Kamada M, et al. Activation of HIV transcription by the viral Tat protein requires a demethylation step mediated by lysine-specific demethylase 1 (LSD1/KDM1) PLoS Pathogens. 2011;7:e1002184. doi: 10.1371/journal.ppat.1002184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakane C, Ohta H, Shidoji Y. Measurement of lysine-specific demethylase-1 activity in the nuclear extracts by flow-injection based time-of-flight mass spectrometry. Journal of Clinical Biochemical Nutrition. 2015;56:123–131. doi: 10.3164/jcbn.14-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt DMZ, McCafferty DG. Trans-2-phenylcyclopropylamine is a mechanism based inactivator of demethylase LSD1. Biochemistry. 2007;46:4408–4416. doi: 10.1021/bi0618621. [DOI] [PubMed] [Google Scholar]
- Schmitt ML, Ladwein KI, Carlino L, Schulz-Fincke J, Willmann D, Metzger E, et al. Heterogeneous antibody-based activity assay for lysine specific demethylase I (LSD1) on a histone peptide substrate. Journal of Biomolecular Screening. 2014;19:973–978. doi: 10.1177/1087057114529156. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Silverman RB. Mechanism-based enzyme inactivators. In: Abelson JN, Simon MI, Purich DL, editors. Methods in enzymology: Vol 249. Academic Press: San Diego; 1995. pp. 240–283. [DOI] [PubMed] [Google Scholar]
- Singh MM, Manton CA, Bhat KP, Tsai W-W, Aldape K, Barton MC, et al. Inhibition of LSD1 sensitizes glioblastoma cells to histone deacetylase inhibitors. Neuro-oncology. 2011;13:894–903. doi: 10.1093/neuonc/nor049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Alzayady K, Stewart R, Ye P, Yang S, Wendong L, et al. Histone demethylase LSD1 regulates neural stem cell proliferation. Molecular and Cellular Biology. 2010;30:1997–2005. doi: 10.1128/MCB.01116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczuk LM, Culhane JC, Yang M, Majumdar A, Yu H, Cole PA. Mechanistic analysis of a suicide inactivator of histone demethylase LSD1. Biochemistry. 2007;46:6892–6902. doi: 10.1021/bi700414b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thinnes CC, England KS, Kawamura A, Chowdhury R, Schofield C, Hopkinson RJ. Targeting histone lysine demethylases—Progress, challenges, and the future. Biochimica et Biophysica Acta. 2014;1839:1416–1432. doi: 10.1016/j.bbagrm.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toffolo E, Rusconi F, Paganini L, Tortorici M, Pilotto S, Heisi C, et al. Phosphorylation of neuronal lysine-specific demethylase 1 LSD1/KDM1A impairs transcriptional repression by regulating interaction with CoREST and histone deacetylases HDAC1/2. Journal of Neurochemistry. 2014;128:603–616. doi: 10.1111/jnc.12457. [DOI] [PubMed] [Google Scholar]
- Ueda R, Suzuki T, Mino K, Tsumoto H, Nakagawa H, Hasegawa M, et al. Identification of cell-active lysine specific demethylase 1-selective inhibitors. Journal of the American Chemical Society. 2009;131:17536–17537. doi: 10.1021/ja907055q. [DOI] [PubMed] [Google Scholar]
- Vianello P, Botrugno OA, Cappa A, Ciossani G, Dessanti P, Mai A, et al. Synthesis, biological activity and mechanistic insights of 1-substituted cyclopropylamine derivatives: A novel class of irreversible inhibitors of histone demethylase KDM1A. European Journal of Medicinal Chemistry. 2014;86:352–363. doi: 10.1016/j.ejmech.2014.08.068. [DOI] [PubMed] [Google Scholar]
- Walsh CT. Suicide substrates, mechanism-based enzyme inactivators: Recent developments. Annual Review of Biochemistry. 1984;53:493–535. doi: 10.1146/annurev.bi.53.070184.002425. [DOI] [PubMed] [Google Scholar]
- Wang X, Huang B, Suzuki T, Liu X, Zhan P. Medicinal chemistry insights in the discovery of novel LSD1 inhibitors. Epigenomics. 2015;7:1379–1396. doi: 10.2217/epi.15.86. [DOI] [PubMed] [Google Scholar]
- Wang J, Telese F, Tan Y, Li W, Jin C, He X, et al. LSD1n is an H4K20 demethylase regulating memory formation via transcriptional elongation control. Nature Neuroscience. 2015;18:1256–1268. doi: 10.1038/nn.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigle TJ, Swinger KK, Campbell JE, Scholle MD, Sherrill J, Admirand EA, et al. A high-throughput mass spectrometry assay coupled with redox activity testing reduces artifacts and false positives in lysine demethylase screening. Journal of Biomolecular Screening. 2015;20:810–820. doi: 10.1177/1087057115575689. [DOI] [PubMed] [Google Scholar]
- Wu J, Hu L, Du Y, Kong F, Pan Y. Prognostic role of LSD1 in various cancers: Evidence from a meta-analysis. Oncotargets and Therapy. 2015;8:2565–2570. doi: 10.2147/OTT.S89597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Culhane JC, Szewczuk LM, Gocke CB, Brautigam CA, Tomchick DR, et al. Structural basis of histone demethylation by LSD1 revealed by suicide inactivation. Nature Structural and Molecular Biology. 2007;14:535–539. doi: 10.1038/nsmb1255. [DOI] [PubMed] [Google Scholar]
- Yang M, Gocke CB, Luo X, Borek D, Tomchick DR, Machius M, et al. Structural basis for CoREST-dependent demethylation of nucleosome by the human LSD1 histone demethylase. Molecular Cell. 2006;23:377–387. doi: 10.1016/j.molcel.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Yu W, Eram MS, Hajian T, Szykowska A, Burgess-Brown N, Vedadi M, et al. A scintillation proximity assay for histone demethylases. Analytical Biochemistry. 2014;463:54–60. doi: 10.1016/j.ab.2014.06.023. [DOI] [PubMed] [Google Scholar]
- Zibetti C, Adamo A, Binda C, Forneris F, Toffolo E, Verpelli C, et al. Alternative splicing of the histone demethylase LSD1/KDM1 contributes to the modulation of neurite morphogenesis in the mammalian nervous system. The Journal of Neuroscience. 2010;30(7):2521–2532. doi: 10.1523/JNEUROSCI.5500-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]