Summary
We identified a polyclonal CD8+ T-cell response against mutant KRAS G12D in tumor-infiltrating lymphocytes obtained from a patient with metastatic colorectal cancer. We observed objective regression of all seven lung metastases after the infusion of approximately 1.11×1011 HLA-C*08:02–restricted tumor-infiltrating lymphocytes that were composed of four different T-cell clonotypes that specifically targeted KRAS G12D. However, one of these lesions had progressed on evaluation 9 months after therapy. The lesion was resected and found to have lost the chromosome 6 haplotype encoding the HLA-C*08:02 class I major histocompatibility complex (MHC) molecule. The loss of expression of this molecule provided a direct mechanism of tumor immune evasion. Thus, the infusion of CD8+ cells targeting mutant KRAS mediated effective antitumor immunotherapy against a cancer that expressed mutant KRAS G12D and HLA-C*08:02.
Adoptive cell therapy using ex vivo expanded tumor-infiltrating lymphocytes has led to durable complete regression of tumors in 20 to 25% of patients with metastatic melanoma.1,2 This effect is probably mediated by T cells that specifically target mutant peptides encoded by de novo somatic mutations, which are known as neoepitopes.3–8 Correlative evidence suggests that clinical responses in patients with cancer after the administration of immune checkpoint inhibitors may also be mediated by neoepitope-reactive T cells.9–14 Direct evidence of the therapeutic utility of the targeting of neoepitopes was observed in a patient with metastatic cholangiocarcinoma who had tumor regression that lasted for 35 months after the infusion of a 95% pure population of CD4+ T cells that targeted a mutated ERBB2IP epitope expressed by her tumors.15 Thus, strategies that harness a T-cell response against mutated tumor antigens may be of clinical benefit in patients with cancer.
The targeting of driver mutations is conceptually attractive, since they are tumor-specific, biologically important for tumor progression, and likely to be expressed by all tumor cells.16 Mutations in the KRAS oncogene are frequent and contribute to the formation and progression of many human cancers. The vast majority of KRAS mutations are recurrent “hot-spot” driver mutations that occur at codon 12, 13, or 61, with codon 12 being the most frequent site of mutation. A conversion of the amino acid glycine (G) to aspartic acid (D) at this site, hereafter referred to as KRAS G12D, is the most frequent KRAS mutant in human gastrointestinal cancers and has been identified in approximately 45% of pancreatic cancers and 13% of colorectal cancers.17,18 Despite decades of study, there is currently no drug or vaccine that can effectively target the KRAS G12D protein in humans. Here, we describe the clinical and biologic findings in a patient with metastatic colorectal cancer who underwent tumor regression after the administration of cytotoxic T cells targeting mutant KRAS G12D.
CASE REPORT
A 50-year-old woman with metastatic colorectal cancer (Patient 4095) was enrolled in our ongoing phase 2 trial (ClinicalTrials.gov number, NCT01174121), which was designed to test whether the adoptive transfer of ex vivo expanded tumor-infiltrating lymphocytes containing T cells targeting personalized cancer neoepitopes (cell therapy) can mediate regression of metastatic solid cancers. (Details about the trial are provided in the protocol, available with the full text of this article at NEJM.org.) In this trial, we screen cultures of tumor-infiltrating lymphocytes obtained from each patient for reactivity against all identified mutant neoepitopes expressed by their autologous tumor. If we identify neoepitope-reactive cultures, these cultures are selected and used in autologous cell therapy, regardless of the identity of the targeted neoepitope.
In Patient 4095, baseline computed tomography (CT) revealed lung disease as the sole source of cancer progression. Three of 10 lung lesions (with maximum diameters of 0.6 cm, 0.8 cm, and 1.0 cm) were resected with the use of video-assisted thoracoscopic surgery (VATS), and 24 individual cultures of tumor-infiltrating lymphocytes were generated from multiple tumor fragments. Samples of the 3 lesions also underwent whole-exomic sequencing (median sequencing depth of lesions: 128, 131, and 163) and transcriptome sequencing to identify mutations expressed by the tumors. (See the Methods section and Table S1 in the Supplementary Appendix, available at NEJM.org. The whole-exome and transcriptome sequence data are available through the National Center for Biotechnology Information BioProject database at identification number PRJNA342632.)
We evaluated each culture for reactivity against these mutant neoepitopes and found that the tumor-infiltrating lymphocytes contained CD8+ T cells that specifically recognized mutant KRAS G12D (Fig. S1A and S1B in the Supplementary Appendix). We selected the culture that showed the highest frequency of CD8+ T cells that were reactive to the G12D mutant and expanded it for treatment (Fig. S1B and S1C in the Supplementary Appendix). Before cell infusion, the patient received a nonmyeloablative, lymphodepleting chemotherapy regimen consisting of cyclophosphamide (at a dose of 60 mg per kilogram of body weight) for 2 days, followed by fludarabine (25 mg per square meter of body-surface area) for 5 days.19
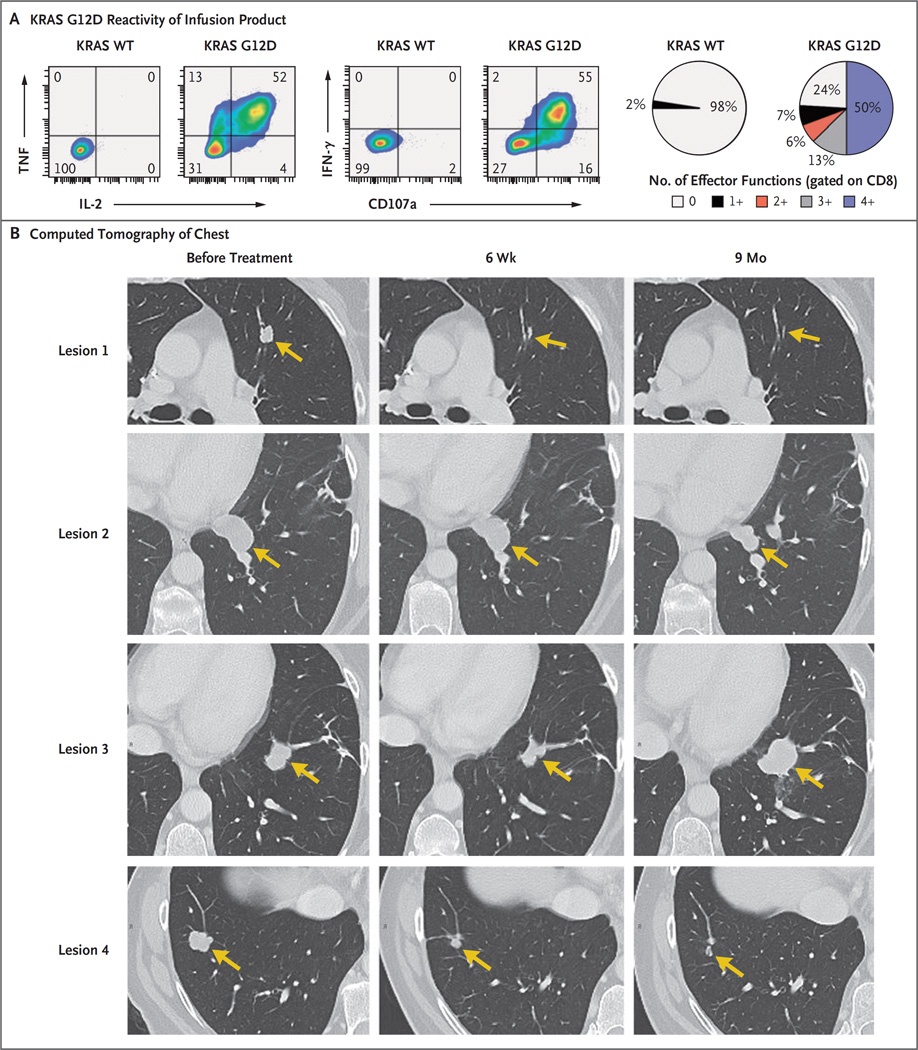
The patient received a single infusion of 1.48×1011 tumor-infiltrating lymphocytes, which consisted of approximately 75% CD8+ T cells (1.11×1011 cells) that were reactive to mutant KRAS G12D and that had no detectable reactivity against the patient’s other mutations (Fig. S1C in the Supplementary Appendix). The majority of these T cells produced multiple effector cytokines (interferon-γ, tumor necrosis factor, and interleukin-2) and showed cytolytic potential (Fig. 1A). This therapy was followed by the administration of five doses of interleukin-2 (720,000 IU per kilogram), which was limited because of fatigue reported by the patient.
Figure 1. Adoptive Transfer of KRAS G12D–Specific T Cells.
Panel A shows the flow cytometric analysis of the effector function of tumor-infiltrating lymphocytes in the infusion product with the use of intracellular cytokine staining (including interferon-γ [IFN-γ], tumor necrosis factor [TNF], and interleukin-2 [IL-2]), and cell-surface mobilization of the degranulation marker CD107a after 6-hour coculture with autologous dendritic cells incubated overnight with wild-type (WT) KRAS or KRAS G12D peptides consisting of 24 amino acids. Data are gated on CD8+ T cells. Pie charts represent the percentages of CD8+ cells that expressed the indicated number of effector functions. Panel B shows contrast-enhanced computed tomographic scans of the chest of Patient 4095 before and approximately 6 weeks and 9 months after the infusion of 1.48×1011 tumor-infiltrating lymphocytes; at least 75% of these cells were specific for mutant KRAS G12D. Arrows highlight lung lesions before and after therapy. Shown are four of seven lesions; the remaining three lesions (not shown) had completely regressed at 9 months.
All seven metastatic lung lesions had regressed at the first follow-up visit conducted 40 days after cell therapy, and the patient had a 9-month partial response (according to the Response Evaluation Criteria in Solid Tumors) until one lesion (lesion 3) had progression (Fig. 1B). This lesion was resected, and the patient remained clinically disease-free 4 months after the lung resection.
METHODS
IDENTIFICATION OF NEOEPITOPE-REACTIVE T CELLS AND GENERATION OF INFUSION PRODUCT
To test whether tumor-infiltrating lymphocytes that were obtained from the patient recognized somatic neoepitopes expressed by the metastatic lung tumors, we used a method that has been described previously.3,15,20 (Details are provided in the Methods section in the Supplementary Appendix.) Culture number 6 of the tumor-infiltrating lymphocytes contained the highest frequency of CD8+ T cells that were reactive to the KRAS G12D mutant; these cells underwent a 2-week rapid-expansion procedure before cell infusion, as described previously.15
IN VIVO TRACKING OF T-CELL CLONES
We performed deep sequencing on the variable region of the T-cell receptor beta chain on genomic DNA isolated from the patient’s infusion product, on three separate lung nodules before treatment, on the progressing lesion (lesion 3), and on peripheral blood before and at various times after cell infusion (Adaptive Biotechnologies) to interrogate the frequency of T-cell receptors that were reactive to the G12D mutant.
ASSESSING REACTIVITY OF T-CELL RECEPTORS
We identified four T-cell receptors that were reactive to mutant KRAS G12D. (See the Methods section and Figs. S2 and S3 in the Supplementary Appendix.) We synthesized the alpha- and beta-chain sequences of the T-cell receptors, which were then cloned into the MSGV1 retroviral vector (GenScript). Retroviral supernatants encoding the T-cell receptors were generated and used to transduce autologous peripheralblood T cells, as described previously.20 T cells that were transduced with the T-cell receptors were then cocultured with autologous peripheral-blood mononuclear cells (PBMCs) loaded with titrated doses of various KRAS peptides or KRAS G12D–positive pancreatic-cancer cell lines that either stably expressed or did not express the restricting HLA-C*08:02 allele.20 T-cell reactivity was determined the next day by means of interferon-γ enzyme-linked immunosorbent spot (ELISPOT) assay and flow cytometric analysis of the T-cell activation markers 4-1BB and OX40.20
CHROMOSOMAL COPY-NUMBER ANALYSIS
We used Sequenza software21 to determine aberrations in tumor chromosome copy numbers, with normal samples as reference and hg19 coordinates. We determined tumor HLA-C allele frequencies as described in the Methods section in the Supplementary Appendix.
RESULTS
CLINICAL EVALUATION
At the first follow-up 40 days after cell therapy, we found that all seven lung metastases had regressed on CT. Of these lesions, six had either completely regressed or showed continuing regression 9 months after therapy (Fig. 1B). The therapy had no adverse effects after the patient had recovered from the effects of interleukin-2 treatment. The patient was discharged from the hospital 2 weeks after cell therapy. A VATS resection of the left lower lung was performed at approximately 9 months after cell therapy to remove the sole progressing lesion (lesion 3) as well as a responding lesion (lesion 2), which was negative on positron-emission tomography. Pathological analysis of tissue sections from lesion 2 (performed with hematoxylin and eosin staining) showed extensive necrosis and no live tumor cells.
IN VIVO FREQUENCY OF KRAS G12D–REACTIVE CD8+ T CELLS
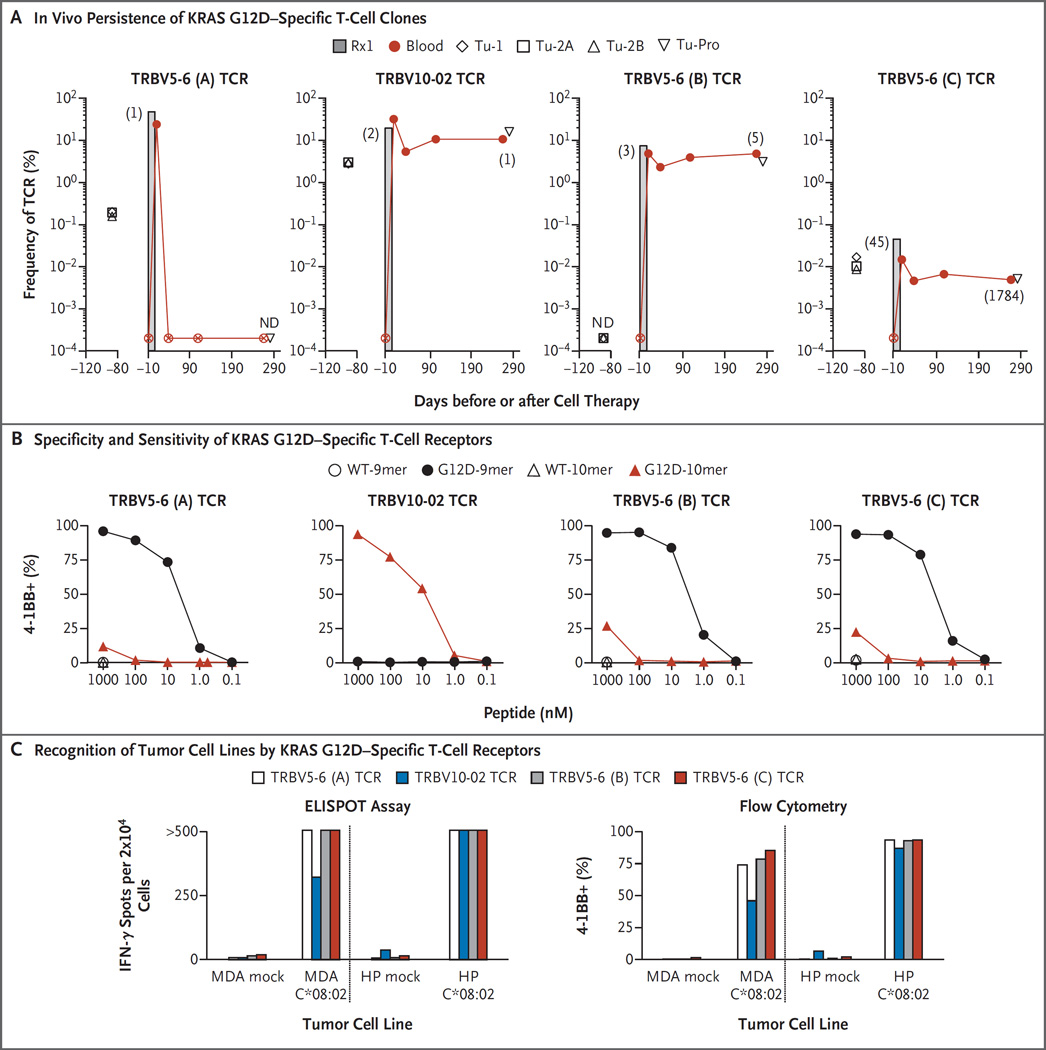
The infusion product contained at least four T-cell clonotypes that were reactive against the G12D mutant. The three highest-frequency T-cell receptors in the infusion product were reactive to KRAS G12D and made up 49.5%, 19.1%, and 6.9% of the infusion bag, respectively. A fourth KRAS G12D–reactive T-cell receptor was the 45th most frequent and was present at only 0.04% of the infusion bag (Fig. 2A, and Table S2 in the Supplementary Appendix). None of these KRAS G12D–reactive T-cell receptors were detected in the patient’s peripheral blood 1 week before infusion (frequency, <0.0002%). After cell therapy, dramatic differences in the engraftment of the KRAS G12D–reactive T-cell receptors were observed. The most dominant infused T-cell clonotype (approximately 7.3×1010 cells) was not detected in the blood 40 days after cell therapy, whereas the remaining KRAS G12D–reactive T-cell clones were detected at this time point. The T-cell clones that persisted in the peripheral blood represented 10.4%, 4.5%, and 0.005% of all peripheral-blood T cells at approximately 9 months after cell therapy, and the most dominant T-cell clone in the peripheral blood at that time was the TRBV10-02 mutant KRAS G12D–reactive T-cell receptor (Fig. 2A, and Table S2 in the Supplementary Appendix). There did not appear to be an enrichment of the T-cell clones that were reactive against KRAS G12D in the progressing tumor relative to the peripheral blood.
Figure 2. In Vivo Persistence and Reactivity Profile of KRAS G12D–Specific T-Cell Clones in the Infusion Product.
Panel A shows the results of deep sequencing of the variable region of the T-cell receptor (TCR) beta chain to measure the frequency of each of the four identified KRAS G12D–reactive T-cell clones in the infusion product (Rx1), in three metastatic lung samples before cell transfer (designated Tu-1, Tu-2A, and Tu-2B), in the one progressing lesion after cell therapy (designated Tu-Pro), and in the peripheral blood of Patient 4095 before and at various times after cell therapy. T-cell receptors are identified according to their gene names (TRBV5-6 and TRBV10-02, with A, B, and C denoting different T-cell receptor clonotypes that share the same T-cell receptor beta variable gene family). The numbers in parentheses indicate the rank of the T-cell receptor sequence in the given sample. A circle with an X and ND denote not detected (<0.0002%). Autologous peripheral-blood T cells were genetically engineered to express each of the indicated KRAS G12D–reactive T-cell receptors that were present in the infusion product. Panel B shows the expression of the T-cell activation marker 4-1BB on T cells engineered with the indicated T-cell receptor after overnight coculture with autologous peripheral-blood mononuclear cells that were incubated with titrated amounts of wild-type KRAS or G12D mutant peptides consisting of either 9 or 10 amino acids (9mer or 10mer). Panel C shows interferon-γ (IFN-γ) production (left) and 4-1BB expression (right) of T cells that were genetically engineered with the indicated T-cell receptor after overnight coculture with two KRAS G12D–positive pancreatic-cancer cell lines not expressing the HLA-C*08:02 allele (mock) or expressing the HLA-C*08:02 allele with the use of enzyme-linked immunosorbent spot (ELISPOT) assay and flow cytometry, respectively. MDA denotes MDA Panc48 cell line, and HP HPAC cell line. Flow cytometry data are gated on CD8+ KRAS G12D–reactive cells. Values of more than 500 on the ELISPOT assay were not considered to be accurate.
SPECIFICITY AND SENSITIVITY OF KRAS G12D–REACTIVE T-CELL RECEPTORS
We cloned the four KRAS G12D–reactive T-cell receptors and genetically modified autologous peripheral-blood T cells to express each of these receptors (Fig. S4 in the Supplementary Appendix). Three of the four T-cell receptors were preferentially reactive against the KRAS G12D peptide GADGVGKSA (consisting of 9 amino acids [9mer]), whereas one T-cell receptor was reactive only against the KRAS G12D peptide GADGVGKSAL (consisting of 10 amino acids [10mer]) (Fig. 2B). All T-cell receptors were specific for the mutant and did not recognize the wild-type KRAS peptides. In peptide titration experiments, we found that the T-cell receptors recognized peptides at concentrations ranging from 1 nM to 10 nM when pulsed onto autologous PBMCs (Fig. 2B). Although we did not have any autologous tumor cell lines to use as targets, we found that the T-cell receptors specifically recognized allogeneic pancreatic-cancer cell lines only when they expressed both mutant KRAS G12D and the HLA-C*08:02 allele (Fig. 2C).
GENETIC ANALYSIS OF TUMORS
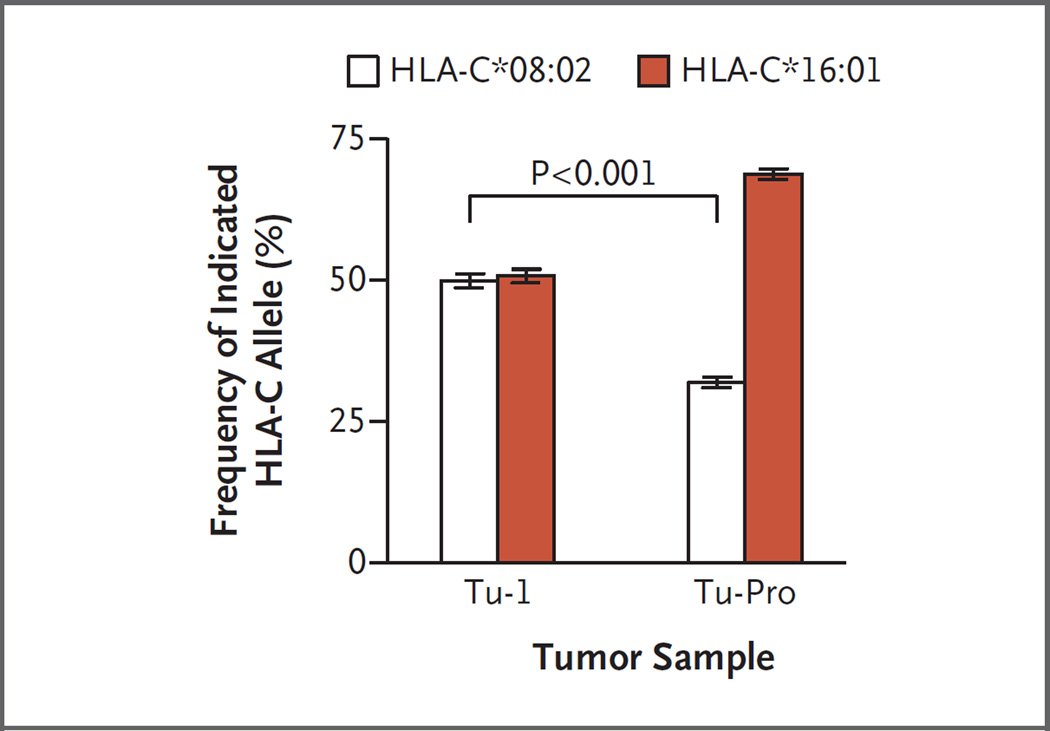
Whole-exome and transcriptome sequencing revealed that the sole progressing lesion still expressed the KRAS G12D mutation (Table S3 in the Supplementary Appendix). This finding showed that the loss of the targeted antigen was not responsible for the progressive nature of the lesion. However, chromosomal copy-number analysis showed that the progressing lesion harbored several genetic abnormalities distinct from the other tumors that had been resected before cell therapy, including a copy-neutral loss of heterozygosity at chromosome 6, which encodes the HLA locus (Fig. S5 in the Supplementary Appendix). The copy of chromosome 6 that was lost encoded the HLA-C*08:02 allele (Fig. 3). Thus, the genetic loss of the HLA-restriction element that was required for recognition by the KRAS G12D–reactive T cells provided a direct mechanism of immune evasion by the progressing lesion.
Figure 3. Analysis of the Frequency of HLA-C Alleles in Tumor Samples.
Shown is the frequency of the two HLA-C alleles in a representative metastatic lesion resected from Patient 4095 before cell therapy (Tu-1) and in the progressing lesion (Tu-Pro). The Tu-1 and Tu-Pro samples were estimated to contain 53% and 34% tumor, respectively, and thus contained some normal cells that also contributed to the calculation of the HLA-C allele frequency. As shown, the progressing lesion harbored cells with a genetic loss of the HLA-C*08:02 allele, which provided a direct mechanism of immune evasion by the tumor, since the infused KRAS G12D–reactive T cells require this molecule for direct tumor recognition. The I bars represent standard errors. The P value for the comparison of the allele frequency of a metastatic lesion before therapy and the progressing lesion after therapy was calculated with the use of the Wilcoxon matched-pairs signed-rank test.
DISCUSSION
We report the regression of metastatic colon cancer followed by immune evasion of one lesion after the adoptive transfer of T cells specifically targeting mutant KRAS G12D. The four identified KRAS G12D–specific T-cell clonotypes showed highly variable levels of engraftment in the peripheral blood after treatment. Flow cytometric analysis of the infused tumor-infiltrating lymphocytes with the use of KRAS G12D–specific tetramers showed that the highest persisting reactive T-cell clone, which recognized the KRAS G12D 10mer epitope, contained an increased frequency of T cells expressing CD28 and CD57 as well as T cells that had a central memory phenotype (CD45RO+CD62L+) that was less differentiated than those of other T cells in the infusion product (Fig. S6 in the Supplementary Appendix).
Currently, 12 patients with metastatic gastrointestinal cancers have been treated with various frequencies of prospectively identified T cells targeting personalized mutated neoantigens. However, the patient who is described in this report was the only one who was treated with an enriched population of T cells targeting mutant KRAS. We have identified endogenous CD8+ T-cell reactivity against mutant KRAS in only one other patient with metastatic colon cancer (Patient 3995),20 and this patient did not have a response to cell therapy. However, in Patient 3995, only 0.002% of the infused cells targeted KRAS G12D, a possible reason for the absence of an antitumor response. The KRAS G12D–specific T-cell receptor that was isolated from Patient 3995 was also restricted by HLA-C*08:02 and, intriguingly, shared the identical T-cell receptor alpha-chain complementarity determining region 3 (CDR3) sequence with two of the KRAS G12D–reactive T-cell receptors that were identified from Patient 4095 (Table S2 and Fig. S7 in the Supplementary Appendix). The sharing of identical T-cell–receptor chains between individuals is thought to be an extremely rare event but can occur in so-called public T-cell responses, which can dominate a response to a given shared antigen, such as those derived from common viruses.22
The identification of HLA-C*08:02–restricted T-cell receptors that target KRAS G12D neoepitopes provides an opportunity to develop T-cell–receptor gene therapy directed against a common driver mutation found in multiple cancer types. Such therapy has the advantage of targeting a molecule that is biologically important for tumor progression and that is likely to be clonally expressed in the tumor while not being expressed in normal tissues. In the United States, the HLA-C*08:02 allele is expressed in approximately 8% of whites and 11% of blacks, whereas the KRAS G12D mutation is most frequently found in pancreatic cancers (approximately 45%) and colorectal cancers (approximately 13%). Thus, the high incidence of KRAS G12D expression in these and other cancers means that in the United States alone, thousands of patients per year would potentially be eligible for T-cell–based immunotherapy targeting KRAS G12D. HLA-A*11:01–restricted KRAS G12D and KRAS G12V–reactive T-cell receptors have been isolated from immunized HLA-A*11:01 transgenic mice,23 although these receptors have not yet been evaluated in clinical trials.
In Patient 4095, we found direct evidence of tumor immune evasion in one lesion through loss of heterozygosity of the copy of chromosome 6 that encoded HLA-C*08:02 (Fig. 3, and Fig. S5 in the Supplementary Appendix). The presence of this molecule is required for direct tumor recognition by the transferred KRAS G12D–specific T cells. Loss of heterozygosity at the HLA locus has been observed in human cancers and may represent a hurdle that needs to be overcome to enhance immunotherapeutic efficacy in some patients.24 In such cases, targeting antigens that are presented by the remaining HLA class I alleles or exploiting the CD4+ T-cell response or the innate immune system against cancer may be necessary to induce a durable clinical response.
Supplementary Material
Acknowledgments
Supported by the Center for Cancer Research Intramural Research Program of the National Cancer Institute.
We thank Mona El-Gamil, Jessica Crystal, and Yong F. Li for their technical assistance; Robert P. Somerville, John R. Wunderlich, and the TIL (Tumor-Infiltrating Lymphocytes) Laboratory for generation of the infusion product; Arnold Mixon and Shawn Farid for flow-cytometry support; James C. Yang for CT scan images; Alena Gros and other members of the Surgery Branch for helpful discussions; and the Tetramer Core Facility of the National Institutes of Health for provision of MHC class I tetramers.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
REFERENCES
- 1.Rosenberg SA, Yang JC, Sherry RM, et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin Cancer Res. 2011;17:4550–4557. doi: 10.1158/1078-0432.CCR-11-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goff SL, Dudley ME, Citrin DE, et al. Randomized, prospective evaluation comparing intensity of lymphodepletion before adoptive transfer of tumor-infiltrating lymphocytes for patients with metastatic melanoma. J Clin Oncol. 2016;34:2389–2397. doi: 10.1200/JCO.2016.66.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu YC, Yao X, Crystal JS, et al. Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. Clin Cancer Res. 2014;20:3401–3410. doi: 10.1158/1078-0432.CCR-14-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu YC, Yao X, Li YF, et al. Mutated PPP1R3B is recognized by T cells used to treat a melanoma patient who experienced a durable complete tumor regression. J Immunol. 2013;190:6034–6042. doi: 10.4049/jimmunol.1202830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robbins PF, Lu YC, El-Gamil M, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19:747–752. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gros A, Parkhurst MR, Tran E, et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat Med. 2016;22:433–438. doi: 10.1038/nm.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gros A, Robbins PF, Yao X, et al. PD-1 identifies the patient-specific CD8+ tumor-reactive repertoire infiltrating human tumors. J Clin Invest. 2014;124:2246–2259. doi: 10.1172/JCI73639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen CJ, Gartner JJ, Horovitz-Fried M, et al. Isolation of neoantigen-specific T cells from tumor and peripheral lymphocytes. J Clin Invest. 2015;125:3981–3991. doi: 10.1172/JCI82416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology: mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Rooij N, van Buuren MM, Philips D, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol. 2013;31(32):e439–e442. doi: 10.1200/JCO.2012.47.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGranahan N, Furness AJ, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tran E, Turcotte S, Gros A, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344:641–645. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGranahan N, Favero F, de Bruin EC, Birkbak NJ, Szallasi Z, Swanton C. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci Transl Med. 2015;7:283ra54. doi: 10.1126/scitranslmed.aaa1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bryant KL, Mancias JD, Kimmelman AC, Der CJ. KRAS: feeding pancreatic cancer proliferation. Trends Biochem Sci. 2014;39:91–100. doi: 10.1016/j.tibs.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaughn CP, Zobell SD, Furtado LV, Baker CL, Samowitz WS. Frequency of KRAS, BRAF, and NRAS mutations in colorectal cancer. Genes Chromosomes Cancer. 2011;50:307–312. doi: 10.1002/gcc.20854. [DOI] [PubMed] [Google Scholar]
- 19.Dudley ME, Wunderlich JR, Yang JC, et al. Adoptive cell transfer therapy following non-myeloablative but lymphodepleting chemotherapy for the treatment of patients with refractory metastatic melanoma. J Clin Oncol. 2005;23:2346–2357. doi: 10.1200/JCO.2005.00.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tran E, Ahmadzadeh M, Lu YC, et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science. 2015;350:1387–1390. doi: 10.1126/science.aad1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Favero F, Joshi T, Marquard AM, et al. Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data. Ann Oncol. 2015;26:64–70. doi: 10.1093/annonc/mdu479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venturi V, Price DA, Douek DC, Davenport MP. The molecular basis for public T-cell responses? Nat Rev Immunol. 2008;8:231–238. doi: 10.1038/nri2260. [DOI] [PubMed] [Google Scholar]
- 23.Wang QJ, Yu Z, Griffith K, Hanada K, Restifo NP, Yang JC. Identification of T-cell receptors targeting KRAS-mutated human tumors. Cancer Immunol Res. 2016;4:204–214. doi: 10.1158/2326-6066.CIR-15-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garrido F, Aptsiauri N, Doorduijn EM, Garcia Lora AM, van Hall T. The urgent need to recover MHC class I in cancers for effective immunotherapy. Curr Opin Immunol. 2016;39:44–51. doi: 10.1016/j.coi.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.